Abstract
The pathogenesis of pulmonary fibrosis is a complicated and complex process that involves phenotypic abnormalities of a variety of cell types and dysregulations of multiple signaling pathways. There are numerous genetic, epigenetic and post-transcriptional mechanisms that have been identified to participate in the pathogenesis of this disease. However, efficacious therapeutics developed from these studies have been disappointingly limited. In the past several years, a group of new molecules, i.e., non-coding RNAs (ncRNAs), has been increasingly appreciated to have critical roles in the pathological progression of lung fibrosis. In this review, we summarize the recent findings on the roles of ncRNAs in the pathogenesis of this disorder. We analyze the translational potential of this group of molecules in treating lung fibrosis. We also discuss challenges and future opportunities of studying and utilizing ncRNAs in lung fibrosis.
Keywords: Idiopathic pulmonary fibrosis (IPF), microRNA, Long non-coding RNA (lncRNA)
Introduction
Tissue fibrosis represents one of the major threats to human health. It is estimated that 45 % of all mortality in the United States can be attributed, directly or indirectly, to fibrotic organ disorders [1]. Pulmonary fibrosis includes a heterogeneous group of lung diseases characterized by an excess accumulation of extracellular matrix (ECM) that leads to scarring and stiffening of lung tissue, loss of alveolar architecture, and ultimate disruption of gas exchange and lethal respiratory failure [1–4].
Idiopathic pulmonary fibrosis (IPF) is the most common form of lung fibrosis that has unknown etiology and a life expectancy of 2–6 years after diagnosis [3, 5]. IPF is also considered to be an age-related disease because it strikes mostly individuals older than 60 years [6]. To date, despite decades of extensive studies, effective treatments for IPF are still disappointingly limited.
IPF pathogenesis is a complicated and complex process that involves phenotypic abnormalities of a variety of cell types and dysregulations of multiple signaling pathways [2–5]. It has been shown that alveolar epithelial cells (AECs), lung fibroblasts (myofibroblsts), bone marrow-derived fibrocytes, and pulmonary macrophages, smooth muscle cells, and endothelial cells are all closely involved in the onset and progression of IPF [2, 7]. There are numerous genetic, epigenetic and post-transcriptional mechanisms that have been identified to participate in the pathogenesis of this disease. Specifically, aberrations at the molecular level of the signaling pathways of a number of growth factors, transforming growth factor (TGF)-β in particular, AKT, Wnt/β-catenin, Notch, etc., are well recognized to be critical aspects of the disease mechanisms. Additionally, dysregulations at the cellular level of senescence, endoplasmic reticulum stress, autophagy, and metabolism are emerging as major contributors to the pathogenesis of IPF [2, 7–10].
In the past several years, a group of new molecules, i.e., non-coding RNAs (ncRNAs), has been increasingly appreciated to have critical roles in IPF pathogenesis [11–14]. Instead of acting alone, ncRNAs are probably entangled with every aspect of the pathological mechanisms of IPF through their various targets and binding partners. In this review, we summarized the recent findings on the roles of ncRNAs in IPF pathogenesis. We analyzed the translational potential of this group of molecules in IPF therapeutics. We also discussed challenges and future opportunities of studying and utilizing ncRNAs in lung fibrosis.
Non-coding RNAs: biogenesis and mode of action
The recently developed high throughput transcriptome analysis revealed that mammalian genomes are pervasively transcribed to produce many types of RNAs. However, only less than 2 % of these transcripts are messenger RNAs (mRNAs) that encode proteins [15, 16]. >98 % of all RNA transcripts do not possess protein encoding capabilities [15, 16]. A large fraction of ncRNAs is the “housekeeping” ncRNAs, such as transfer RNAs (tRNAs), ribosome RNAs (rRNAs) and small nuclear RNAs, all of which are involved in protein synthesis and ribosome modification. The rest of the ncRNAs species, including microRNAs (miRNAs), piwi-interacting RNAs (piRNAs) and long ncRNAs (lncRNAs), have been shown to play regulatory roles in various biological processes, and thus being recognized as regulatory ncRNAs [17–19].
miRNAs
Based on their length, ncRNAs can be divided into two sub-classes: long ncRNAs (lncRNAs) (>200 nucleotides-n.t.) and small/short ncRNAs (<200 n.t.). Small ncRNAs include miRNAs and piRNAs [20–22]. Unlike piRNAs that mainly mediate gene silencing in germlines, miRNAs are expressed in all types of cells and are shown to participate in numerous molecular and cellular processes [17–19].
microRNAs (miRNAs) are a class of 20–25 n.t. small ncRNAs that can regulate gene expression at the post-transcriptional level [23, 24]. The first miRNA lin-4 was discovered in 1993 in Caenorhabditis elegans and found to control larval development by regulating the translation of lin-14 through its interaction with the 3′ untranslated region (UTR) of the mRNA transcript [25]. Ever since, thousands of miRNAs have been discovered and characterized in various biological species [23, 26].
miRNAs are primarily transcribed by RNA polymerase II from respective genomic loci [23, 27–29]. The primary transcripts (pri-miRNA) range from ~200 to several thousand n.t. in length [17, 23, 27, 30]. Pri-miRNAs are cleaved by RNase III endoribonuclease Drosha/DGCR8, generating ~60–120 n.t. long hairpin RNA molecules called precursor miRNAs (pre-miRNA) [17, 23, 27, 30]. Pre-miRNAs are then exported to the cytoplasm by Ran-GTP and Exportin 5 and are further excised by the endoribonuclease Dicer to generate ~22 bp miRNA duplexes [17, 23, 27, 30]. The miRNAs duplexes are assembled into the miRNA-induced silencing complexes (miRISC) that consist of Argonaute proteins, human immunodeficiency virus (HIV) transactivating response RNA (TAR) binding protein (TRBP), protein activator of the interferon induced protein kinase (PACT), where the two strands are separated. The leading strands are preserved to form mature miRNAs, while the passenger strands (miRNA*) are unstable and normally degraded. To function, the “seed” sequence (n.t. 2-8 from the 5′ end) of miRNAs acts as a guide for miRISC to specifically recognize the 3′UTR of target mRNAs through Watson–Crick base-pairing. Upon binding, miRNA can promote mRNA degradation and/or repress mRNA translation, and thus downregulating the expression of target proteins [17, 23, 27, 30].
miRNAs have been recognized to be key players in numerous biological processes, such as cell proliferation, differentiation, apoptosis, oncogenesis and organ development [23–25, 31, 32]. Their roles in the initiation and progression of respiratory diseases, including pulmonary fibrosis, were also under intensive investigation in the past several years [11–14, 33].
LncRNAs
LncRNAs do not possess protein-coding capability due to lack of significant open reading frame (ORF). LncRNAs have been known for 30 years, but not until very lately were their functional significance recognized and their roles in various biological processes appreciated [34, 35]. Recent studies have identified far more than 10,000 lncRNAs in the human transcriptome, suggesting that our current understanding has just scratched the surface in the complex world of lncRNAs [34, 35]. Majority of lncRNAs are transcribed by RNA Polymerase II and normally contain 5′ m7G-cap and 3′ poly(A) tail [36]. Therefore, the expression of lncRNAs is subject to similar regulations to that of protein-coding genes and miRNAs.
Recent studies demonstrated that lncRNAs play essential roles in a variety of pathobiological processes, such as oncogenesis [37, 38], reprogramming of induced pluripotent stem cells [39], cell apoptosis and differentiation [40, 41], X-inactivation and parental imprinting [42–44], and adipogenesis [45]. LncRNAs regulate gene expression at the epigenetic, post-transcriptional, and translational levels. In addition, some lncRNAs can serve as molecular scaffolds to maintain subnuclear structure [46]. Given the universal presence and regulatory versatility of lncRNAs, it is predictable that lncRNAs are also involved in pulmonary disorders. Indeed, investigation of the role of lncRNAs in pulmonary diseases, including pulmonary fibrosis, has been already started [11, 47–49].
miRNAs and lung fibrosis
The systemic study of miRNAs in tissue fibrosis, including that in the lungs, is made possible only after the technical development of miRNA profiling. There have been a number of studies that tried to identify differentially expressed miRNAs in fibrotic human lungs, in lungs of animals with experimental pulmonary fibrosis, and in in vitro models using relevant cells [50–55]. Many of the miRNAs with altered expression discovered by these efforts have been further studied. The roles of these miRNAs participating in the disease mechanisms and, in some cases, their potentials as therapeutic targets have been determined [51–53]. These studies significantly advanced our understanding of the pathogenesis of this pernicious disease.
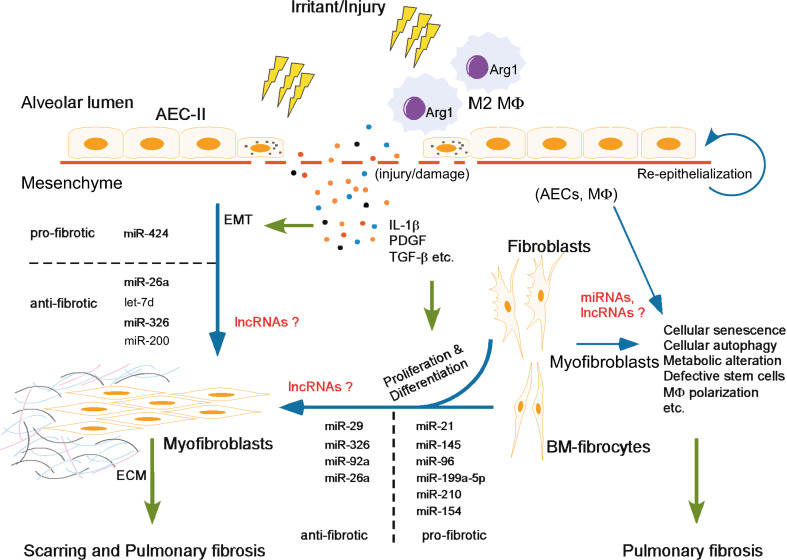
Given that miRNAs function almost solely by modulating the expression of their target proteins, it is conceivable that they are involved in the pathogenesis of pulmonary fibrosis in a way related to those pro- or antifibrotic mediators in this disease. Contingently, miRNAs can participate in the fibrotic process as either positive or negative players (Fig. 1; Table 1). As also well recognized, the pathological process of pulmonary fibrosis involves many types of cells in the lungs and also from circulation [2, 7]. Dysregulations of miRNAs in these cells are certain to contribute to the phenotypic and functional aberrations that are necessary for the onset and progression of pulmonary fibrosis.
Fig. 1.
Overview of the participation of ncRNAs in the pathogenesis of lung fibrosis: ncRNAs participate in the pathogenesis of lung fibrosis through regulating various pro- or antifibrotic mechanisms
Table 1.
miRNAs that participate in pulmonary fibrosis
| Fibro-miRs | miRNAs | Targets/mediators | Upstream regulators and functional effects |
|---|---|---|---|
| Profibrotic miRNAs | miR-21 | Smad7 | Upregulated by TGF-β1 in lung fibroblasts; promotes myofibroblast differentiation [52] |
| miR-199a-5p | Caveolin 1 | Upregulated by TGF-β1 in lung fibroblasts; promotes myofibroblast differentiation [73] | |
| miR-145 | KLF4 | Upregulated by TGF-β1 in lung fibroblasts; promotes α-SMA expression, myofibroblast differentiation and latent TGF-β1 activation [76] | |
| miR-424 | Smurf2 | Upregulated by TGF-β1 in lung epithelial cells; promotes TGF-β1-induced EMT in lung epithelial cells [77] | |
| miR-210 | MNT | Upregulated by HIF-2α in lung fibroblasts exposed to hypoxia; enhances hypoxia-induced lung fibroblasts proliferation [82] | |
| miR-96 | FoxO3a | Enhances the proliferative and antiapoptotic phenotype in IPF fibroblasts [91] | |
| miR-154 | Wnt/β-catenin pathway | Upregulated by TGF-β in Smad3-dependent manner; promotes proliferation and migration of lung fibroblasts [94] | |
| Antifibrotic miRNAs | miR-29 | Collagens, laminins and integrins | Downregulated by TGF-β in Smad3-dependent manner; inhibits the expression of ECM components [56–59] |
| miR-26a | TGF-β2, TGF-βR1, Smad4, HMGA2, CTGF, collagen 1, cyclin D2 | Downregulated by TGF-β in Smad3-dependent manner; negatively regulates TGF-β and CTGF signaling; Inhibits EMT, myofibroblast differentiation, collagen 1 expression and lung fibroblast proliferation [51, 60, 63] | |
| miR-326 | TGF-β1, Smad3, Ets1, MMP9 | Inhibits TGF-β1 expression; negatively regulates TGF-β1 signaling and other fibrosis-related pathways [65] | |
| Let-7d | HMGA2 | Downregulated by TGF-β in Smad3-dependent manner; inhibits alveolar EMT and the profibrotic activities of myofibroblasts [53, 69] | |
| miR-200 | GATA3, ZEB1/2 | Downregulated by TGF-β in AECs; inhibits alveolar EMT and the fibrogenic activity of lung fibroblasts [70] | |
| miR-92a | WISP1 | Negatively regulates lung fibrosis by targeting WISP1-mediated alveolar EMT [71] | |
| miR-27b | Gremlin1 | Downregulated by TGF-β1 in lung fibroblasts; inhibits TGF-β1 signaling and promotes BMP signaling [72] |
miRNAs that play a negative regulatory role in lung fibrosis
miR-29
The miR-29 family, consisting of miR-29a, miR-29b and miR-29c, was initially identified as miRNAs downregulated in the lungs of mice with bleomycin-induced pulmonary fibrosis [56]. In 2011, Cushing and colleagues found that miR-29 is exclusively expressed in a subset of interstitial cells located in the fibrotic areas of lungs and miR-29 levels were inversely correlated to the severity of lung fibrosis. miR-29 expression was suppressed by TGF-β1 treatment in human lung fibroblasts. More importantly, downregulation of miR-29 de-repressed multiple targets that are profibrotic, such as several types of collagens. miR-29 also targeted other fibrosis-associated genes, including laminins and integrins [56]. A later study further confirmed that miR-29 is a direct transcriptional target of Smad3 and negatively regulated by TGF-β/Smad signaling during pulmonary fibrosis [57]. Ectopic expression of miR-29 by Sleeping Beauty (SB) gene transfer technology was found to prevent and reverse bleomycin-induced pulmonary fibrosis in mice [57]. The therapeutic benefits of miR-29 were again tested by a different approach. In that study, it was first confirmed that a single intravenous delivery of synthetic miR-29 mimics can substantially increase miR-29 levels in various organs, including kidney, spleen, heart, liver and lung. The elevated expression of miR-29 in these organs could last 2–4 days. More importantly, therapeutic delivery of the miR-29 mimics effectively blunted bleomycin-induced pulmonary fibrosis in mice [58]. miR-29 has also been shown to mediate reciprocal regulations between lung fibroblasts and ECM in IPF lungs [59]. Lung fibroblasts grown on ECM derived from IPF lungs have expression of genes enriched for those encoding ECM proteins detected in IPF tissue, which is likely mediated by downregulated expression of miR-29 in this culture [59]. All these findings suggest a great potential for miR-29 as a novel therapeutic target for treating pulmonary fibrosis.
miR-26a
miR-26a is another miRNA downregulated in the lungs of IPF patients and mice with experimental pulmonary fibrosis [51, 60]. A study by Liang et al. found that downregulation of miR-26a in fibrotic lungs is inversely correlated with the expression of its target genes, such as CTGF and collagen 1 [60]. Inhibition of miR-26a promoted, while overexpression of miR-26a attenuated bleomycin-induced pulmonary fibrosis in mice. In human lung fibroblasts, miR-26a was downregulated by TGF-β1 in Smad3-dependent manner. Moreover, miR-26a overexpression inhibited TGF-β1-induced differentiation of fibroblasts into myofibroblasts. Mechanistically, miR-26a inhibited TGF-β1 signaling through directly targeting Smad4, and thus decreasing the nuclear translocation of Smad3 and myofibroblast activation [60]. Meanwhile, they also showed that miR-26a regulates epithelial to mesenchymal transition (EMT) in pulmonary fibrosis. Inhibition of miR-26a induced, whereas overexpression of miR-26a diminished EMT in human lung epithelial cells [51]. The inhibitory role of miR-26a in EMT is likely through targeting HMGA2, a transcriptional factor that has been previously shown to play a role in this process [51, 61, 62]. Alternatively, miR-26a could inhibit the proliferative activity of human lung fibroblasts by directly targeting cyclin D2, which leads to G1 arrest [63]. miR-26a is also involved in the regulation of TGF-β2, TGF-β receptor 1, as well as other key profibrotic mediators including collagen and CTGF [63]. These findings suggest that downregulation of miR-26a in fibrotic lungs could serve as a positive feedback mechanism to enhance myofibroblast differentiation and proliferation and EMT. Therefore, replenishment of the decreased miR-26a in the lungs may represent an effective therapeutic strategy to treat pulmonary fibrosis.
miR-326
Although the role of TGF-β1 in promoting pulmonary fibrosis has been well established, application of neutralizing antibodies to TGF-β1 in treating lung fibrosis has been disappointing [64]. These findings suggest the complexity of TGF-β1 activity in lung fibrosis and the need for better strategies to target TGF-β1 in the lungs. A recent study found that miR-326 has diminished expression in human IPF specimens [65]. miR-326 also showed an inversed correlation with TGF-β1 expression in the lungs of mice with bleomycin-induced pulmonary fibrosis [65]. In vitro studies found that miR-326 regulates TGF-β1 expression in multiple human cell lines. Furthermore, intranasal delivery of miR-326 mimics was effective to inhibit TGF-β1 expression and attenuate the fibrotic response in the lungs of mice that are treated with bleomycin [65]. A number of profibrotic mediators that are involved in TGF-β1 signaling and fibrosis-related pathways, such as Ets1, Smad3, and matrix metalloproteinase 9, were identified as miR-326 targets [65]. These data suggest that miR-326 regulates pulmonary fibrosis by de-repressing multiple profibrotic genes. These data also indicate the antifibrotic therapeutic potential of miR-326 replacement/replenishment in the lungs.
Let-7d
The EMT of AECs has long been implicated in the pathogenesis of pulmonary fibrosis [66–68]. Let-7d is the first miRNA identified to participate in this process. Using miRNA array, Kaminski’s laboratory performed miRNA profiling on 10 normal control lungs and 10 IPF lungs and found that the expression of let-7d, along with 17 other miRNAs, was decreased in IPF lung tissues [53]. Let-7d was primarily expressed in the AECs in normal lungs and its expression was almost lost in these cells in the fibrotic area of IPF lungs. Let-7d expression was inhibited by TGF-β1, which was mediated by direct binding of Smad3 to the let-7d promoter. Inhibition of let-7d caused mesenchymal phenotypic changes in multiple lung epithelial cell lines. Furthermore, blocking let-7d in mouse lungs promoted lung fibrosis probably through enhancing EMT, indicated by elevated expression of collagens, α-SMA, and thickening of alveolar septa and decreased expression of epithelial markers E-cadherin and TJP1. Mechanistically, the inhibitory effect of let-7d on EMT was likely mediated by HMGA2, a positive regulator of this process [53, 61, 62]. In addition to its role in EMT, introduction of let-7d into lung fibroblasts was able to diminish the profibrotic activities of myofibroblasts [69], suggesting that promotion of lung fibrosis by let-7d blockade is a complex result of enhanced alveolar EMT and augmented myofibroblast differentiation.
miR-200
Recently, our group found that miR-200 family members, including miR-200a, miR-200b, and miR-200c are significantly downregulated in the lungs of mice with bleomycin-induced fibrosis [70]. The expression of miR-200a and miR-200c was also diminished in human IPF lungs. Similar to let-7d, miR-200 was primarily expressed in AECs, but not in lung fibroblasts in normal lungs. miR-200 expression was decreased in AECs from mice with bleomycin-induced pulmonary fibrosis [53]. Overexpression of miR-200 family members inhibited TGF-β1-induced EMT of AECs and was able to reverse the fibrogenic activity of myofibroblasts from fibrotic mouse lungs and human IPF lungs. We found that miR-200 directly targets multiple key positive regulators of EMT, such as GATA3, ZEB1, and ZEB2. Moreover, intratracheal delivery of miR-200c mimics inhibited experimental pulmonary fibrosis in mice [70]. The findings with let-7d and miR-200 suggest that inhibition of EMT by manipulating miRNAs is a novel therapeutic approach to treat pulmonary fibrotic disorders [70].
There are several additional miRNAs that have also been shown to play inhibitory roles in pulmonary fibrosis by targeting various signaling pathways. miR-92a was identified to be downregulated in lungs from patient with IPF. miR-92a directly targeted WNT1-inducible signaling pathway protein 1 (WISP1), a profibrotic mediator in IPF, and thereby inhibiting pulmonary fibrosis [71]. miR-27b, another miRNA downregulated in TGF-β1 treated lung fibroblasts, was found to diminish the expression of several profibrotic genes likely by targeting Gremlin1. Gremlin1 has been shown to promote pulmonary fibrosis through decreasing BMP signaling, but elevating TGF-β1 signaling [72].
In general, most of the negative regulatory miRNAs in pulmonary fibrosis are downregulated in fibrotic lungs, which leads to derepression of profibrotic mediators, and thereby promoting pulmonary fibrosis. A replacement or enhancement of these miRNAs may be novel therapeutic strategies in treating this disease.
miRNAs that promote lung fibrosis
Besides acting as a negative regulator in pulmonary fibrosis, some miRNAs can also play profibrotic roles by targeting the negative mediators in pulmonary fibrosis.
miR-199a-5p
A recent study by Lino et al. found that miR-199a-5p is significantly increased in the lungs of mice with bleomycin-induced fibrosis and in IPF lungs [73]. In situ hybridization analysis demonstrated that miR-199a-5p is selectively expressed in myofibroblasts in fibrotic lung tissues, suggesting its participation in myofibroblast differentiation. miR-199a-5p was induced by TGF-β1 in vitro in lung fibroblasts. Overexpression of miR-199a-5p markedly enhanced myofibroblast differentiation. This was likely achieved by downregulating caveolin 1, an antifibrotic mediator that was previously shown to ameliorate bleomycin-induced pulmonary fibrosis through modulating JNK kinase activation [73, 74]. Caveolin 1 expression had an inverse correlation with that of miR-199a-5p in fibrotic lungs and caveolin 1 was demonstrated to be a bona fide miR-199a-5p target. Aside from directly targeting caveolin 1, miR-199a-5p also regulates pulmonary fibrosis through caveolin 1-independent mechanisms, as suggested by additional profibrotic effects caused by miR-199a-5p overexpression that was not seen with caveolin 1 knockdown. This is a reflection of multifaceted activities of miRNAs in that they regulate a biological process generally by targeting multiple targets [73]. Of note, miR-199a-5p was also upregulated in experimental models of kidney fibrosis and liver fibrosis, suggesting that miR-199a-5p is a common therapeutic target in treating organ fibrotic disorders [73].
miR-21
Our group first demonstrated that miR-21 is upregulated in the lungs of mice with bleomycin-induced fibrosis and in IPF lungs and miR-21 is primarily expressed in myofibroblasts in human and mouse fibrotic lungs [52]. We found that miR-21 is induced by TGF-β1 in primary lung fibroblasts. Furthermore, overexpression of miR-21 promoted, whereas inhibition of miR-21 diminished TGF-β1-induced differentiation of lung fibroblasts into myofibroblasts. Consistent with its profibrotic activity in vitro, miR-21 antisense probes significantly inhibited the severity of bleomycin-induced lung fibrosis in mice. Mechanistically, we identified Smad7, an inhibitory Smad in the TGF-β signaling pathway, as a direct target of miR-21. Taken together, these data suggest that miR-21 functions in a positive feed-back loop to promote pulmonary fibrosis [52]. The upregulation of miR-21 was validated independently in several lung fibrosis models as well as in human subjects, such as in the serum of patient with IPF [54, 75]. All these findings suggest the critical role of miR-21 in the pathogenesis of pulmonary fibrosis and also indicate miR-21 as a putative therapeutic target for developing miRNA-based strategies for IPF treatment.
miR-145
The role of miR-145 in pulmonary fibrosis was also initially reported by our group [76]. We found that miR-145 is induced by TGF-β1 in human lung fibroblasts. miR-145 expression was similarly increased in the lungs of patients with IPF. Overexpression of miR-145 in lung fibroblasts promoted TGF-β1-induced myofibroblast differentiation, as demonstrated by elevated α-SMA expression, enhanced contractility, and increased formation of focal and fibrillar adhesions. In contrast, miR-145 downregulation diminished the characteristic phenotype of TGF-β1-induced myofibroblasts [76]. We found that miR-145 directly targets KLF4, a known negative regulator of α-SMA expression, and thereby promoting myofibroblast differentiation. Likely as the result of the increased contractility, miR-145 enhanced the activation of latent TGF-β1. More significantly, we found that miR-145(-/-) mice develop less lung fibrosis than miR-145(+/+) controls after intratracheal administration of bleomycin [76].
miR-424
miR-424 was first reported to participate in TGF-β1-induced EMT of AECs. Xiao et al. recently identified 6 upregulated and 3 downregulated miRNAs in lung epithelial cell line A549 that undergoes EMT [77]. One of the 6 upregulated miRNAs was miR-424. Overexpression of miR-424 enhanced TGF-β1 activity and promoted TGF-β1-induced mesenchymal phenotype in A549 cells, as reflected by increased expression of α-SMA, CTGF and fibronectin. miR-424 was shown to target multiple key negative regulators of the TGF-β signaling, such as Smurf2. Smurf2 is a HECT (homologous to the E6-accessory protein C-terminus) domain containing E3 ubiquitin ligase that negatively regulates TGF-β1 and BMP signaling by degrading receptor-regulated Smads [77, 78]. These data suggest that miR-424 functions via a positive feedback mechanism to promote TGF-β1-induced EMT of lung epithelial cells [77]. However, the role of miR-424 in myofibroblast differentiation and the pathogenesis of lung fibrosis, and the potential of miR-424 as a therapeutic target in treating lung fibrosis should be further examined.
miR-210
There is increasing evidence indicating the participation of hypoxia in the pathogenesis of tissue fibrosis [79]. However, the mechanism by which hypoxia contributes to the progression of pulmonary fibrosis has not been well elucidated. In the past 5 years, a number of miRNAs were identified to be upregulated by hypoxia and were thus designated as “hypoxamirs”. It was found that the induction of most hypoxamirs is not only modest, but also largely cellular-context dependent. However, hypoxia-induced miR-210 is generally robust in many types of cells. miR-210 has been well recognized to regulate cellular responses to hypoxia, such as altered proliferation, differentiation, apoptosis, metabolism and antitumor immune response [80, 81]. In the study to characterize the role of hypoxamirs in pulmonary fibrosis, Henke’s group found that miR-210 expression is elevated in IPF fibroblasts exposed to hypoxia. They showed that knockdown of miR-210 diminishes hypoxia-induced proliferation of IPF fibroblasts. They also showed that HIF-2α, but not HIF-1α, regulates miR-210 expression by directly binding to the hypoxic response element (HRE) in the miR-210 promoter [82]. Mechanistically, miR-210 promoted IPF fibroblast proliferation by targeting MNT, a c-myc suppressor that can inhibit cell cycle entry and proliferation [82, 83]. Moreover, they found that miR-210 expression is located to cells that are also positive with hypoxic marker carbonic anhydrase-IX and HIF-2α within the IPF fibrotic reticulum, indicating an interactive relationship among miR-210, hypoxia and IPF pathogenesis [82]. Taken together, these findings suggest that miR-210 is one of the hypoxamirs that mediates the profibrotic roles of hypoxia by promoting profibrotic phenotype of IPF fibroblasts in the lungs.
miR-96
The PTEN/PI3K/AKT axis has been well known to play a role in the pathogenesis of pulmonary fibrosis [84–86]. A number of studies found that IPF lung fibroblasts have increased AKT activity, which leads to elevated phosphorylation and functional inhibition of FoxO3a, a transcriptional factor regulating many cellular activities, such as cell proliferation, differentiation, apoptosis, and cellular metabolism [87, 88]. The suppressed FoxO3a activity has also been shown to be closely associated with the maintenance of the myofibroblastic phenotype of IPF fibroblasts in collagen-rich matrix [89, 90]. As a FoxO3a targeting miRNA, the expression of miR-96 was significantly induced in IPF fibroblasts cultured on collagen matrix, compared to that in normal lung fibroblasts. Furthermore, miR-96 expression is inversely correlated with that of FoxO3a in the fibroblastic foci in IPF lungs. Indeed, miR-96 suppressed the expression of FoxO3a and FoxO3a targeted p21, p27 and Bim in IPF lung fibroblasts. The decreased expression of these negative regulators of cell cycle and apoptosis promoted the proliferative and antiapoptotic phenotype in IPF fibroblasts [91]. These studies suggest that miR-96 promotes fibrotic progression by downregulating FoxO3a in IPF lungs [91].
Chromosome 14q32 microRNA cluster
miRNAs that reside in close proximity in the genome are called a miRNA cluster. The expression of clustered miRNAs is often under same regulations [23]. Furthermore, although individual miRNA in a cluster shares no seed targeting sequence, there was evidence that miRNA clusters can function synergistically to regulate biological events [92, 93]. There is currently very little information regarding the role of a cluster of miRNAs in pulmonary disorders. It was found that a number of miRNAs located at the human chromosome 14q32 microRNA cluster is upregulated in the lungs of IPF patients [94]. These miRNAs were also induced by TGF-β1 in normal human lung fibroblasts, likely in a Smad3-binding dependent manner. Among these, miR-154 was shown to enhance the proliferation and migration of lung fibroblasts by activating the Wnt/β-Catenin pathway [94]. These data suggest that the 14q32 microRNA cluster may be able to act in a synergistic mechanism to promote pulmonary fibrosis. Therefore, it is worthwhile to further investigate the role of all miRNAs at this locus and determine their functional interactions in the pathogenesis of lung fibrosis.
Extracellular miRNAs and lung fibrosis
In addition to acting in the cytoplasm, miRNAs can be secreted/released into the extracellular environment by various types of cells. These extracellular miRNAs are normally included in exosomes or microvesicles and can travel long distance in circulation [95]. miRNAs in exosomes can be transferred from cell to cell, and regulate gene expression in the recipient cells through canonical actions. miRNA transfer between cells is thought to be an important component of intercellular communications [95].
There has been systemic effort to identify circulating miRNAs in IPF patients. A recent study found 47 differentially expressed miRNAs in the sera of patients with rapid or slowly progressive IPF compared to those in healthy controls [96]. Further bioinformatic analysis showed that 53 KEGG biological processes were enriched among the differentially expressed circulating miRNAs, such as signaling pathways associated with TGF-β, MAPK, PI3K-AKT, Wnt, and Notch, all of which are well recognized to have important roles in the pathological fibrogenesis in the lungs [96]. They also validated the profiling in a second cohort of samples and demonstrated that miR-21, miR-199a-5p, and miR-200c were significantly increased while miR-31, let-7a, and let-7d were significantly decreased in the sera of IPF patients compared to healthy controls [96]. The elevated level of miR-21 in the sera of IPF patients was confirmed by another study [33]. All of these findings suggest that circulating miRNAs may be used as biomarkers for diagnosis as well as for evaluation of treatment efficacy and prognosis.
However, except for identification of the association of circulating miRNAs with IPF, there is currently no knowledge about the roles of these miRNAs in the disease mechanisms. It is unknown where the miRNAs originally come from, lung or circulation? If from lung, what cell populations secrete them? Whether these miRNAs can transfer from one cell type to the other in the lung? Given the critical role of intercellular interactions, such as those between AECs and myofibroblasts, in IPF pathogenesis, investigation into how one cell population affects the other’s phenotype through these secreted miRNAs will definitely shed new light on the disease mechanism.
miRNA-based therapeutics and lung fibrosis
The frequent aberrations of miRNAs in various pathogeneses, including lung fibrosis, and their small size and sequence conservation make them ideal and easy targets for therapeutics [97]. There have been numerous instances of pre-clinical targeting of miRNAs in animals, such as those in models of pulmonary fibrosis as described above. There are also several trials underway at different clinical phases [97]. The one that could become the first miRNA-based therapy is treating HCV by inhibition of miR-122. Two independent clinical trials using miR-122 antisense oilgos with different modifications so far have shown very promising efficacies and drug tolerance [98] (http://www.regulusrx.com). Therefore, there is every reason to believe that we will soon experience a new era of miRNA-based therapeutics.
The basic principle of miRNA-based therapeutics is restoring to the homeostasis of dysregulated miRNA expression, i.e., reconstitution of downregulated or inhibition of upregulated miRNAs. There are currently two strategies to therapeutically restore the activities of a lost or downregulated miRNA [97]. One approach is to use synthetic modified double-stranded miRNA mimics and the other is to use lenti-, adeno, or adeno-associated virus to express specific miRNAs [97]. The passenger strand of miRNA mimics is often conjugated with cholesterol and chemically modified to facilitate intracellular entry and to prevent incorporation into miRISC. Overall, this type of miRNA duplexes has been proven to be bio-safe, but there is a report that they can potentially activate interferon response through Toll-like receptors [99]. There are also two approaches that have been successfully employed to inhibit miRNAs in vivo [97, 100]. One is to express antisense RNAs that bind to the “seed” region of miRNAs using the various viral vehicles described above. These molecules are called miRNA sponges [101]. However, the easier and more efficient approach is to use synthetic single stranded antisense oligos called antimiRs [97]. Ideal antimiRs have high binding affinity, biostability and pharmacokinetic properties. To achieve these objectives, chemical modifications of these oligos are required. General approaches include substitution of the phosphodiester backbone linkages with phosphorothiorate linkages and sugar modifications, such as 2′-O-methyl, 2′-O-Methoxyethyl, 2′-fluoro, and locked nucleic acid [97, 100].
A particularly important point that needs careful consideration when therapeutically targeting miRNAs is how to restore optimal expression of miRNAs that renders maximal efficacy and minimal side effects. Because it has been indicated that effective targeting of mRNAs by specific miRNAs depends on threshold points set by the relative levels of miRNA and the target mRNAs [102], therapeutic restoration of miRNAs that is at sub- or supraphysiological level could lead to targeting a quite different set of genes from those under normal conditions and thus causing less favorable effects and/or more unwanted outcomes. Similarly, blocking in excess of pathologically upregulated miRNAs may also de-repress genes that otherwise have no or low expression in cells and thus bringing deleterious side effects. Another common challenge for therapeutic reconstitution and inhibition of miRNAs in vivo lies in tissue- and/or cell-specific delivery of agents, although it also exists with most other treatment modalities. Several approaches have been tested to achieve targeted delivery of miRNA mimics/antimiR molecules. For example, conjugation of these molecules with antibodies that recognize specific cell surface proteins or with peptides that bind to cell surface receptors improved the access to targeted tissues or cell types [97, 100]. Expression of miRNAs driven by cell-specific promoters is another way to achieve targeted delivery [97]. In addition, local delivery, such as that directly to lungs via inhalation, can improve tissue-specific introduction of miRNA molecules [97].
Although there is currently no miRNA-based clinical trials underway for treating lung fibrosis, one should be optimistic that this type of new approaches will be soon tested in the field. Of note, as an industrial pioneer, the biotech company miRagen recently initiated Investigational New Drug (IND) application for miR-29 to treat pulmonary fibrosis (http://miragentherapeutics.com).
LncRNAs in pulmonary fibrosis
To date, studies on lncRNAs in pulmonary diseases almost exclusively focused on lung cancers [11]. The knowledge about the role of lncRNAs in pulmonary fibrotic disorders is nearly absent. However, the lack of understanding is about to change soon. A recent study by Lv et al. identified a large number of lncRNAs that have altered expression in the lungs of rat with experimental fibrosis. This was the first piece of evidence suggesting the participation of lncRNAs in the pathogenesis of pulmonary fibrosis [48]. Among these lncRNAs, several have been previously studied in different biological settings, such as H19, RNA component of mitochondrial RNA processing endoribonuclease (RMRP), and telomerase RNA component (TERC) [103–106]. H19 is an imprinted maternally expressed lncRNA, which is located at the H19/IGF2 locus. Further studies found that H19 regulates the expression of IGF-2, modulates IGF signaling and tumorigenesis, including those in the lungs, and even produces two miRNAs (miR-675-5p and miR-675-3p) [103, 104]. RMRP and TERC were previously reported to be associated with human telomerase reverse transcriptase (TERT) and thereby regulating small interfering RNA production and telomerase activity, respectively [105, 106]. Moreover, it was found that a mutation in the CCAAT box of the TERC lncRNA promoter contributes to disorders associated with abnormal telomere, such as pulmonary fibrosis [107]. Thus, it is predictable that these lncRNAs may participate in the pathogenesis of pulmonary fibrosis through their associated protein partners, such as TERT. Additionally, a later study from the same group examined the potential functional interactions between the previously identified lncRNAs and their adjacent or homologous protein-coding genes [47]. They found that those lncRNAs that have altered expression in fibrotic lungs have the potential to regulate gene expression by acting as competing endogenous RNAs (ceRNA), a mechanism that RNAs serve as a “sponge” for miRNAs and thereby preventing miRNAs from binding to their mRNA targets [47].
Although our understanding of the role of lncRNAs in pulmonary fibrosis is very limited, there have been already a number of elegant and thorough studies on their activities and functional mechanisms in many other pathophysiological processes [108]. Therefore, we have every reason to be optimistic that we will gain significant insight into how lncRNAs regulate the pathogenesis of lung fibrosis in the coming years. These studies will certainly unravel the complexity of lncRNAs and suggest novel therapeutic targets for treating pulmonary disorders, including fibrosis.
Conclusions and perspectives
In just a couple years, ncRNA studies in the lungs have generated a wealth of knowledge supporting the critical role of ncRNAs in modulating lung disorders, including pulmonary fibrosis. With the demonstration that specific miRNAs play key roles via negative or positive regulatory mechanisms in this disease, the field has opened new avenues for identifying novel therapeutic targets in treating pulmonary fibrosis.
The role of miRNAs in regulating the phenotypic dysregulations of pulmonary fibroblasts and epithelial cells in lung fibrosis seems to be well established. However, it is completely unknown if miRNAs or lncRNAs participate in emerging, but important disease mechanisms, such as the contributions from aberrant cellular senescence, defective lung stem cells, viral infection, metabolic dysregulation and alternative macrophage activation [109–113]. There has been plenty of information about the roles of ncRNAs in all of the above mentioned processes [114–118]. Therefore, it shall be extremely rewarding to investigate how ncRNAs regulate lung fibrosis via these mechanisms.
As discussed above, it is certain that more miRNAs will be identified to participate in the pathogenesis of lung fibrosis. However, it is equally important to understand how these many miRNAs can collaborate to regulate the pathological process. Another lingering challenge is to identify the specific targets of miRNA. Most past studies usually chose one or two targets that are known to participate in tissue fibrogenesis. This approach could potentially exaggerate the role of certain mediators, but likely miss other presently unknown but important regulators.
So far, almost all miRNA profiling analyses have been performed on healthy or diseased total lung tissues. One should be aware that findings from these assays sometimes could be misleading. It is apparent that fibrotic lung tissues have a cellular composition vastly different from healthy controls. In addition, different cell types may have distinct miRNA profiles. Therefore, the conventional approaches may only identify miRNAs that reflects the cell population alterations in the fibrotic tissues and run a significant risk of missing important miRNAs that have real changes in specific cell types during pulmonary fibrogenesis. To overcome these limitations, improved methodologies, such as laser capture microdissection and microarray expression analysis, and single cell sequencing, should be considered in future studies.
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitations. Sources of funding: NIH grants HL114470 (VJT), HL105473 (GL) and HL076206 (GL).
Conflict of interest
None.
References
- 1.Thannickal VJ, Zhou Y, Gaggar A, Duncan SR. Fibrosis: ultimate and proximate causes. J Clin Investig. 2014;124(11):4673–4677. doi: 10.1172/JCI74368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkauskas CE, Noble PW. Cellular mechanisms of tissue fibrosis. 7. New insights into the cellular mechanisms of pulmonary fibrosis. Am J Physiol Cell Physiol. 2014;306(11):C987–C996. doi: 10.1152/ajpcell.00321.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noble PW, Barkauskas CE, Jiang D. Pulmonary fibrosis: patterns and perpetrators. J Clin Investig. 2012;122(8):2756–2762. doi: 10.1172/JCI60323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strieter RM, Mehrad B. New mechanisms of pulmonary fibrosis. Chest. 2009;136(5):1364–1370. doi: 10.1378/chest.09-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolters PJ, Collard HR, Jones KD. Pathogenesis of idiopathic pulmonary fibrosis. Annual review of pathology. 2014;9:157–179. doi: 10.1146/annurev-pathol-012513-104706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thannickal VJ. Mechanistic links between aging and lung fibrosis. Biogerontology. 2013;14(6):609–615. doi: 10.1007/s10522-013-9451-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lekkerkerker AN, Aarbiou J, van Es T, Janssen RA. Cellular players in lung fibrosis. Curr Pharm Des. 2012;18(27):4093–4102. doi: 10.2174/138161212802430396. [DOI] [PubMed] [Google Scholar]
- 8.Chilosi M, Poletti V, Zamo A, Lestani M, Montagna L, Piccoli P, Pedron S, Bertaso M, Scarpa A, Murer B, Cancellieri A, Maestro R, Semenzato G, Doglioni C. Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol. 2003;162(5):1495–1502. doi: 10.1016/s0002-9440(10)64282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu K, Moghal N, Egan SE. Notch signaling in lung development and disease. Adv Exp Med Biol. 2012;727:89–98. doi: 10.1007/978-1-4614-0899-4_7. [DOI] [PubMed] [Google Scholar]
- 10.Yan Z, Kui Z, Ping Z. Reviews and prospectives of signaling pathway analysis in idiopathic pulmonary fibrosis. Autoimmun Rev. 2014;13(10):1020–1025. doi: 10.1016/j.autrev.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 11.Booton R, Lindsay MA. Emerging role of MicroRNAs and long noncoding RNAs in respiratory disease. Chest. 2014;146(1):193–204. doi: 10.1378/chest.13-2736. [DOI] [PubMed] [Google Scholar]
- 12.Pagdin T, Lavender P. MicroRNAs in lung diseases. Thorax. 2012;67(2):183–184. doi: 10.1136/thoraxjnl-2011-200532. [DOI] [PubMed] [Google Scholar]
- 13.Pandit KV, Milosevic J (2015) MicroRNA regulatory networks in idiopathic pulmonary fibrosis. Biochem Cell Biol Biochimie et biologie cellulaire. doi:10.1139/bcb-2014-0101 [DOI] [PubMed]
- 14.Pandit KV, Milosevic J, Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Transl Res J Lab Clin Med. 2011;157(4):191–199. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220(2):126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 16.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5(4):e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 19.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carninci P, Yasuda J, Hayashizaki Y. Multifaceted mammalian transcriptome. Curr Opin Cell Biol. 2008;20(3):274–280. doi: 10.1016/j.ceb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10(12):833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 22.Carninci P. Molecular biology: the long and short of RNAs. Nature. 2009;457(7232):974–975. doi: 10.1038/457974b. [DOI] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 24.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 25.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 26.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38(Suppl):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 27.Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25(46):6156–6162. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- 28.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13(12):1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 30.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 31.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9(3):219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 32.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 33.Li P, Li J, Chen T, Wang H, Chu H, Chang J, Zang W, Wang Y, Ma Y, Du Y, Zhao G, Zhang G. Expression analysis of serum microRNAs in idiopathic pulmonary fibrosis. Int J Mol Med. 2014;33(6):1554–1562. doi: 10.3892/ijmm.2014.1712. [DOI] [PubMed] [Google Scholar]
- 34.Brosius J. Waste not, want not—transcript excess in multicellular eukaryotes. Trends Genet. 2005;21(5):287–288. doi: 10.1016/j.tig.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 35.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14(2):103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 36.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451(7175):202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42(12):1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13(11):971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23(16):1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415(6873):810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 44.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding rnas in health and disease. Cell. 2013;152(6):1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 45.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG, Yuan B, Kellis M, Lodish HF, Rinn JL. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci USA. 2013;110(9):3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106(8):2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song X, Cao G, Jing L, Lin S, Wang X, Zhang J, Wang M, Liu W, Lv C. Analysing the relationship between lncRNA and protein-coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. J Cell Mol Med. 2014;18(6):991–1003. doi: 10.1111/jcmm.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao G, Zhang J, Wang M, Song X, Liu W, Mao C, Lv C. Differential expression of long non-coding RNAs in bleomycin-induced lung fibrosis. Int J Mol Med. 2013;32(2):355–364. doi: 10.3892/ijmm.2013.1404. [DOI] [PubMed] [Google Scholar]
- 49.Han L, Zhang EB, Yin DD, Kong R, Xu TP, Chen WM, Xia R, Shu YQ, De W. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:e1665. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Honeyman L, Bazett M, Tomko TG, Haston CK. MicroRNA profiling implicates the insulin-like growth factor pathway in bleomycin-induced pulmonary fibrosis in mice. Fibrogenesis Tissue Repair. 2013;6(1):16. doi: 10.1186/1755-1536-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang H, Gu Y, Li T, Zhang Y, Huangfu L, Hu M, Zhao D, Chen Y, Liu S, Dong Y, Li X, Lu Y, Yang B, Shan H. Integrated analyses identify the involvement of microRNA-26a in epithelial-mesenchymal transition during idiopathic pulmonary fibrosis. Cell Death Dis. 2014;5:e1238. doi: 10.1038/cddis.2014.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207(8):1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, Richards T, Selman M, Watkins SC, Pardo A, Ben-Yehudah A, Bouros D, Eickelberg O, Ray P, Benos PV, Kaminski N. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182(2):220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie T, Liang J, Guo R, Liu N, Noble PW, Jiang D. Comprehensive microRNA analysis in bleomycin-induced pulmonary fibrosis identifies multiple sites of molecular regulation. Physiol Genomics. 2011;43(9):479–487. doi: 10.1152/physiolgenomics.00222.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuchuan H, Ya D, Jie Z, Jingqiu C, Yanrong L, Dongliang L, Changguo W, Kuoyan M, Guangneng L, Fang X, Lanlan T, Bo Q. Circulating miRNAs might be promising biomarkers to reflect the dynamic pathological changes in smoking-related interstitial fibrosis. Toxicol Ind Health. 2014;30(2):182–191. doi: 10.1177/0748233712452606. [DOI] [PubMed] [Google Scholar]
- 56.Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lu J. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45(2):287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao J, Meng XM, Huang XR, Chung AC, Feng YL, Hui DS, Yu CM, Sung JJ, Lan HY. miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther J Am Soc Gene Ther. 2012;20(6):1251–1260. doi: 10.1038/mt.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montgomery RL, Yu G, Latimer PA, Stack C, Robinson K, Dalby CM, Kaminski N, van Rooij E. MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol Med. 2014;6(10):1347–1356. doi: 10.15252/emmm.201303604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parker MW, Rossi D, Peterson M, Smith K, Sikstrom K, White ES, Connett JE, Henke CA, Larsson O, Bitterman PB. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J Clin Investig. 2014;124(4):1622–1635. doi: 10.1172/JCI71386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang H, Xu C, Pan Z, Zhang Y, Xu Z, Chen Y, Li T, Li X, Liu Y, Huangfu L, Lu Y, Zhang Z, Yang B, Gitau S, Lu Y, Shan H, Du Z. The antifibrotic effects and mechanisms of microRNA-26a action in idiopathic pulmonary fibrosis. Mol Ther J Am Soc Gene Ther. 2014;22(6):1122–1133. doi: 10.1038/mt.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J Cell Biol. 2006;174(2):175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008;283(48):33437–33446. doi: 10.1074/jbc.M802016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X, Liu L, Shen Y, Wang T, Chen L, Xu D, Wen F. MicroRNA-26a modulates transforming growth factor beta-1-induced proliferation in human fetal lung fibroblasts. Biochem Biophys Res Commun. 2014;454(4):512–517. doi: 10.1016/j.bbrc.2014.10.106. [DOI] [PubMed] [Google Scholar]
- 64.Varga J, Pasche B. Antitransforming growth factor-beta therapy in fibrosis: recent progress and implications for systemic sclerosis. Curr Opin Rheumatol. 2008;20(6):720–728. doi: 10.1097/BOR.0b013e32830e48e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Das S, Kumar M, Negi V, Pattnaik B, Prakash YS, Agrawal A, Ghosh B. MicroRNA-326 regulates profibrotic functions of transforming growth factor-beta in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2014;50(5):882–892. doi: 10.1165/rcmb.2013-0195OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coward WR, Saini G, Jenkins G. The pathogenesis of idiopathic pulmonary fibrosis. Ther Adv Respir Dis. 2010;4(6):367–388. doi: 10.1177/1753465810379801. [DOI] [PubMed] [Google Scholar]
- 67.Bartis D, Mise N, Mahida RY, Eickelberg O, Thickett DR. Epithelial-mesenchymal transition in lung development and disease: does it exist and is it important? Thorax. 2014;69(8):760–765. doi: 10.1136/thoraxjnl-2013-204608. [DOI] [PubMed] [Google Scholar]
- 68.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214(2):199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huleihel L, Ben-Yehudah A, Milosevic J, Yu G, Pandit K, Sakamoto K, Yousef H, LeJeune M, Coon TA, Redinger CJ, Chensny L, Manor E, Schatten G, Kaminski N. Let-7d microRNA affects mesenchymal phenotypic properties of lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2014;306(6):L534–L542. doi: 10.1152/ajplung.00149.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang S, Banerjee S, de Freitas A, Sanders YY, Ding Q, Matalon S, Thannickal VJ, Abraham E, Liu G. Participation of miR-200 in pulmonary fibrosis. Am J Pathol. 2012;180(2):484–493. doi: 10.1016/j.ajpath.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berschneider B, Ellwanger DC, Baarsma HA, Thiel C, Shimbori C, White ES, Kolb M, Neth P, Konigshoff M. miR-92a regulates TGF-beta1-induced WISP1 expression in pulmonary fibrosis. Int J Biochem Cell Biol. 2014;53:432–441. doi: 10.1016/j.biocel.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 72.Graham JR, Williams CM, Yang Z. MicroRNA-27b targets gremlin 1 to modulate fibrotic responses in pulmonary cells. J Cell Biochem. 2014;115(9):1539–1548. doi: 10.1002/jcb.24809. [DOI] [PubMed] [Google Scholar]
- 73.Lino Cardenas CL, Henaoui IS, Courcot E, Roderburg C, Cauffiez C, Aubert S, Copin MC, Wallaert B, Glowacki F, Dewaeles E, Milosevic J, Maurizio J, Tedrow J, Marcet B, Lo-Guidice JM, Kaminski N, Barbry P, Luedde T, Perrais M, Mari B, Pottier N. miR-199a-5p Is upregulated during fibrogenic response to tissue injury and mediates TGFbeta-induced lung fibroblast activation by targeting caveolin-1. PLoS Genet. 2013;9(2):e1003291. doi: 10.1371/journal.pgen.1003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang XM, Zhang Y, Kim HP, Zhou Z, Feghali-Bostwick CA, Liu F, Ifedigbo E, Xu X, Oury TD, Kaminski N, Choi AM. Caveolin-1: a critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. J Exp Med. 2006;203(13):2895–2906. doi: 10.1084/jem.20061536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li P, Zhao GQ, Chen TF, Chang JX, Wang HQ, Chen SS, Zhang GJ. Serum miR-21 and miR-155 expression in idiopathic pulmonary fibrosis. J Asthma. 2013;50(9):960–964. doi: 10.3109/02770903.2013.822080. [DOI] [PubMed] [Google Scholar]
- 76.Yang S, Cui H, Xie N, Icyuz M, Banerjee S, Antony VB, Abraham E, Thannickal VJ, Liu G. miR-145 regulates myofibroblast differentiation and lung fibrosis. FASEB J. 2013;27(6):2382–2391. doi: 10.1096/fj.12-219493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiao X, Huang C, Zhao C, Gou X, Senavirathna LK, Hinsdale M, Lloyd P, Liu L. Regulation of myofibroblast differentiation by miR-424 during epithelial-to-mesenchymal transition. Arch Biochem Biophys. 2015;566:49–57. doi: 10.1016/j.abb.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Izzi L, Attisano L. Regulation of the TGFbeta signalling pathway by ubiquitin-mediated degradation. Oncogene. 2004;23(11):2071–2078. doi: 10.1038/sj.onc.1207412. [DOI] [PubMed] [Google Scholar]
- 79.Tzouvelekis A, Harokopos V, Paparountas T, Oikonomou N, Chatziioannou A, Vilaras G, Tsiambas E, Karameris A, Bouros D, Aidinis V. Comparative expression profiling in pulmonary fibrosis suggests a role of hypoxia-inducible factor-1alpha in disease pathogenesis. Am J Respir Crit Care Med. 2007;176(11):1108–1119. doi: 10.1164/rccm.200705-683OC. [DOI] [PubMed] [Google Scholar]
- 80.Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle. 2010;9(6):1072–1083. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grosso S, Doyen J, Parks SK, Bertero T, Paye A, Cardinaud B, Gounon P, Lacas-Gervais S, Noel A, Pouyssegur J, Barbry P, Mazure NM, Mari B. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 2013;4:e544. doi: 10.1038/cddis.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bodempudi V, Hergert P, Smith K, Xia H, Herrera J, Peterson M, Khalil W, Kahm J, Bitterman PB, Henke CA. miR-210 promotes IPF fibroblast proliferation in response to hypoxia. Am J Physiol Lung Cell Mol Physiol. 2014;307(4):L283–L294. doi: 10.1152/ajplung.00069.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hurlin PJ, Queva C, Eisenman RN. Mnt: a novel Max-interacting protein and Myc antagonist. Curr Top Microbiol Immunol. 1997;224:115–121. doi: 10.1007/978-3-642-60801-8_11. [DOI] [PubMed] [Google Scholar]
- 84.Xia H, Khalil W, Kahm J, Jessurun J, Kleidon J, Henke CA. Pathologic caveolin-1 regulation of PTEN in idiopathic pulmonary fibrosis. Am J Pathol. 2010;176(6):2626–2637. doi: 10.2353/ajpath.2010.091117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Conte E, Gili E, Fruciano M, Korfei M, Fagone E, Iemmolo M, Lo Furno D, Giuffrida R, Crimi N, Guenther A, Vancheri C (2013) PI3K p110gamma overexpression in idiopathic pulmonary fibrosis lung tissue and fibroblast cells: in vitro effects of its inhibition. Lab Invest J Tech Method Pathol 93(5):566–576. doi:10.1038/labinvest.2013.6 [DOI] [PubMed]
- 86.Lu Y, Azad N, Wang L, Iyer AK, Castranova V, Jiang BH, Rojanasakul Y. Phosphatidylinositol-3-kinase/akt regulates bleomycin-induced fibroblast proliferation and collagen production. Am J Respir Cell Mol Biol. 2010;42(4):432–441. doi: 10.1165/rcmb.2009-0002OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120(Pt 15):2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 88.Nho RS, Hergert P. FoxO3a and disease progression. World J Biol Chem. 2014;5(3):346–354. doi: 10.4331/wjbc.v5.i3.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nho RS, Hergert P, Kahm J, Jessurun J, Henke C. Pathological alteration of FoxO3a activity promotes idiopathic pulmonary fibrosis fibroblast proliferation on type i collagen matrix. Am J Pathol. 2011;179(5):2420–2430. doi: 10.1016/j.ajpath.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nho RS, Peterson M, Hergert P, Henke CA. FoxO3a (Forkhead Box O3a) deficiency protects idiopathic pulmonary fibrosis (IPF) fibroblasts from type I polymerized collagen matrix-induced apoptosis via caveolin-1 (cav-1) and Fas. PLoS One. 2013;8(4):e61017. doi: 10.1371/journal.pone.0061017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nho RS, Im J, Ho YY, Hergert P. MicroRNA-96 inhibits FoxO3a function in IPF fibroblasts on type I collagen matrix. Am J Physiol Lung Cell Mol Physiol. 2014;307(8):L632–L642. doi: 10.1152/ajplung.00127.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yan X, Chen X, Liang H, Deng T, Chen W, Zhang S, Liu M, Gao X, Liu Y, Zhao C, Wang X, Wang N, Li J, Liu R, Zen K, Zhang CY, Liu B, Ba Y. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer. 2014;13:220. doi: 10.1186/1476-4598-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Milosevic J, Pandit K, Magister M, Rabinovich E, Ellwanger DC, Yu G, Vuga LJ, Weksler B, Benos PV, Gibson KF, McMillan M, Kahn M, Kaminski N. Profibrotic role of miR-154 in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2012;47(6):879–887. doi: 10.1165/rcmb.2011-0377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol. 2012;23(2):91–97. doi: 10.1097/MOL.0b013e328350a425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang G, Yang L, Wang W, Wang J, Wang J, Xu Z. Discovery and validation of extracellular/circulating microRNAs during idiopathic pulmonary fibrosis disease progression. Gene. 2015;562(1):138–144. doi: 10.1016/j.gene.2015.02.065. [DOI] [PubMed] [Google Scholar]
- 97.van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med. 2014;6(7):851–864. doi: 10.15252/emmm.201100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 99.Peacock H, Fucini RV, Jayalath P, Ibarra-Soza JM, Haringsma HJ, Flanagan WM, Willingham A, Beal PA. Nucleobase and ribose modifications control immunostimulation by a microRNA-122-mimetic RNA. J Am Chem Soc. 2011;133(24):9200–9203. doi: 10.1021/ja202492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13(8):622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 101.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43(9):854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Molecular cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, Penn LZ. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66(10):5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 105.Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, Kasim V, Hayashizaki Y, Hahn WC, Masutomi K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461(7261):230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17(4):498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 107.Aalbers AM, Kajigaya S, van den Heuvel-Eibrink MM, van der Velden VH, Calado RT, Young NS. Human telomere disease due to disruption of the CCAAT box of the TERC promoter. Blood. 2012;119(13):3060–3063. doi: 10.1182/blood-2011-10-383182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Beers MF, Morrisey EE. The three R’s of lung health and disease: repair, remodeling, and regeneration. J Clin Investig. 2011;121(6):2065–2073. doi: 10.1172/JCI45961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hecker L, Logsdon NJ, Kurundkar D, Kurundkar A, Bernard K, Hock T, Meldrum E, Sanders YY, Thannickal VJ (2014) Reversal of persistent fibrosis in aging by targeting Nox4-Nrf2 redox imbalance. Sci Transl Med 6(231):231ra247. doi:10.1126/scitranslmed.3008182 [DOI] [PMC free article] [PubMed]
- 111.Kottmann RM, Kulkarni AA, Smolnycki KA, Lyda E, Dahanayake T, Salibi R, Honnons S, Jones C, Isern NG, Hu JZ, Nathan SD, Grant G, Phipps RP, Sime PJ. Lactic acid is elevated in idiopathic pulmonary fibrosis and induces myofibroblast differentiation via pH-dependent activation of transforming growth factor-beta. Am J Respir Crit Care Med. 2012;186(8):740–751. doi: 10.1164/rccm.201201-0084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tanjore H, Blackwell TS, Lawson WE. Emerging evidence for endoplasmic reticulum stress in the pathogenesis of idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012;302(8):L721–L729. doi: 10.1152/ajplung.00410.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tao B, Jin W, Xu J, Liang Z, Yao J, Zhang Y, Wang K, Cheng H, Zhang X, Ke Y. Myeloid-specific disruption of tyrosine phosphatase Shp2 promotes alternative activation of macrophages and predisposes mice to pulmonary fibrosis. J Immunol. 2014;193(6):2801–2811. doi: 10.4049/jimmunol.1303463. [DOI] [PubMed] [Google Scholar]
- 114.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Muller OJ, Potente M, Zeiher AM, Hermeking H, Dimmeler S. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495(7439):107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 115.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kalamvoki M, Du T, Roizman B. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc Natl Acad Sci USA. 2014;111(46):E4991–E4996. doi: 10.1073/pnas.1419338111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu G, Abraham E. MicroRNAs in immune response and macrophage polarization. Arterioscler Thromb Vasc Biol. 2013;33(2):170–177. doi: 10.1161/ATVBAHA.112.300068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tian Y, Zhang Y, Hurd L, Hannenhalli S, Liu F, Lu MM, Morrisey EE. Regulation of lung endoderm progenitor cell behavior by miR302/367. Development. 2011;138(7):1235–1245. doi: 10.1242/dev.061762. [DOI] [PMC free article] [PubMed] [Google Scholar]