Abstract
Background and purpose
Remote ischemic conditioning (RIC) is a phenomenon in which short periods of non-fatal ischemia in one tissue confers protection to distant tissues. Here we performed a longitudinal human pilot study in patients with aneurysmal subarachnoid hemorrhage (aSAH) undergoing RIC by limb ischemia to compare changes in DNA methylation and transcriptome profiles before and after RIC.
Methods
Thirteen patients underwent 4 RIC sessions over 2–12 days after rupture of an intracranial aneurysm. We analyzed whole blood transcriptomes using RNA sequencing and genome-wide DNA methylomes using reduced representation bisulfite sequencing, both before and after RIC. We tested differential expression (DE) and differential methylation (DM) using an intra-individual paired study design, and then overlapped the DE and DM results for analyses of functional categories and protein-protein interactions.
Results
We observed 164 DE genes and 3,493 DM CpG sites after RIC, of which 204 CpG sites overlapped with 103 genes, enriched for pathways of cell cycle (P<3.8×10−4) and inflammatory responses (P<1.4×10−4). The cell cycle pathway genes form a significant protein-protein interaction network of tightly co-expressed genes (P<0.00001).
Conclusions
Gene expression and DNA methylation changes in aSAH patients undergoing RIC are involved in coordinated cell cycle and inflammatory responses.
Keywords: aneurysm, subarachnoid hemorrhage, preconditioning, genomics, transcriptome, remote ischemic conditioning, RIC, DNA methylation
Introduction
Remote ischemic conditioning (RIC) is a phenomenon where non-lethal ischemic exposure in a peripheral tissue induces a systemic protection of subsequent injuries in distant organs and tissues.1 RIC has shown encouraging results in animal models by providing cardio- and neuroprotective effects against an ischemic injury, and thus RIC is emerging as an attractive novel therapeutic for clinical trials.2–4 Recent human studies have confirmed the safety and feasibility of lower limb RIC in patients with aneurysmal subarachnoid hemorrhage (aSAH).5,6 Based on our separate study (Laiwalla et al., unpublished data), the OR of a good outcome for patients with RIC is 5.17 (95% CI: 1.21–25.02) when compared with matched controls with SAH.
The effectiveness of RIC is likely to be caused by its multifactorial effects, and rodent studies suggest that these are mediated in part by a cascade of transcriptional and translational changes.7 Activation of basic cell survival responses to transient ischemia causes a shift towards a protective genetic profile, leading to a differential regulation of genes involved in inflammation, neurotransmitter excitotoxicity, apoptosis, and cerebrovascular perfusion.8–13 Nevertheless, the mechanisms by which RIC provides neuroprotective effects in human are not well understood. The involvement of humoral factors has been demonstrated in animals as the protection can be transferred from a RIC animal to a non-conditioned animal by whole blood transfusion.14 Thus, genetic and epigenetic studies in human blood could elucidate the humeral processes catalyzed by RIC, and furthermore provide potential diagnostic and therapeutic targets for the treatment and prevention of ischemic injury.
In this human pilot study, we performed a prospective longitudinal evaluation in a group of patients with aneurysmal subarachnoid hemorrhage (aSAH) undergoing RIC to study the induced genomic responses by identifying and comparing blood DNA methylation and gene expression profiles before RIC and one week after RIC. Identification of factors altered by a transient limb RIC can provide insights into the mechanisms of neuroprotective action, and ultimately, may yield biomarkers for SAH prognosis and treatment.
Methods
Study Samples
Patients with aSAH were enrolled from the “Remote Ischemic Preconditioning in Subarachnoid Hemorrhage (RIPC-SAH) Trial” [Clinicaltrials.gov # NCT01158508]. The study was approved by the local institutional review board, and all participants gave a written informed consent. Patients 18–80 years old with SAH confirmed by computed tomography (CT) or lumbar puncture, and presence of a ruptured intracranial aneurysm confirmed by CT, magnetic resonance (MR), or catheter angiography were considered for enrollment in this study. Patients that were pregnant or with a history or physical exam findings of peripheral vascular disease, deep venous thrombosis, peripheral neuropathy, or lower extremity bypass were excluded. Clinical characteristics are provided in table 1.
Table 1.
Clinical characteristics of the aSAH patients.
| Subject ID | Age | Sex | Smoking | Alcohol | Hypertension | T2D | Vasospasm | Clinical functional outcome |
|---|---|---|---|---|---|---|---|---|
| SAH 551 | 61 | F | No | No | No | No | N | Improved or no change |
| SAH 553 | 77 | F | No | No | No | No | Y | Improved or no change |
| SAH 554 | 56 | M | Former | No | No | No | Y | Improved or no change |
| SAH 555 | 53 | F | No | No | No | No | Y | Deteriorated |
| SAH 556 | 23 | F | No | No | No | No | N | Improved or no change |
| SAH 557 | 47 | F | No | No | Yes | No | Y | Deteriorated |
| SAH 558 | 65 | F | No | No | Yes | No | Minimal | Improved or no change |
| SAH 559 | 43 | F | Yes | Yes | Yes | No | Y | Improved or no change |
| SAH 5510 | 36 | M | Yes | Yes | No | No | Y | Improved or no change |
| SAH 5511 | 51 | M | Yes | Yes | Yes | Yes | N | Improved or no change |
| SAH 5512 | 43 | M | Yes | No | Yes | Yes | Y | Improved or no change |
| SAH 5513 | 60 | F | No | No | Yes | Yes | Y | Deteriorated |
| SAH 5514 | 51 | M | Yes | Yes | Yes | No | Suspected | Improved or no change |
T2D indicates type 2 diabetes.
RIC Protocol
Patients underwent 4 RIC sessions over 2–12 days after aneurysm rupture. RIC sessions were performed on the lower limb with a large adult-sized blood pressure cuff. Each session consisted of four inflation cycles lasting 5 minutes, followed by 5-minute deflations. Cuff pressure was originally inflated at 20 mmHg over the patient’s baseline systolic blood pressure, then increased until the dorsalis pedis pulse was abolished, as confirmed by a Doppler ultrasonography. This pressure was maintained for 5-minutes throughout the duration of the inflation cycle.
Peripheral blood samples were drawn from aSAH patients at two different time points: before RIC (baseline) and after 4 sessions of the RIC treatment. DNA and RNA were isolated according to standard protocols.
Aneurysm controls
We included 24 control individuals with a history of intracranial aneurysms who never received RIC treatment. The blood collection and sample processing were performed in the same way as described for the aSAH cases above.
RNA sequencing
We included 13 aSAH sample pairs and 24 aneurysm controls in the study after the initial quality control of the blood RNA (RIN value>7, RNA concentration>10ng/ul). The blood RNA sequencing libraries were prepared using Illumina TruSeq RNA library kit, and sequencing of the paired-end, 100-bp reads was performed using the Illumina Hiseq2000 platform, resulting in on average 46.1 M reads per sample. We used STAR15 to align the fastq files to the human GRCh37/hg19 reference genome with the following settings: the maximum intron size was set at 500 kb; the minimum intron size was set at 20; and we allowed for four mismatches. We used HTSeq (version HTSeq-0.6.1)16 to produce raw counts.
Differential Expression (DE) using EdgeR and DESeq2
We employed both EdgeR17 and DESeq218 R-packages to identify differentially expressed (DE) genes using the paired sample design, and focused on their overlap to obtain a set of highly confident DE genes. First, using EdgeR we excluded the genes that did not have one count per million reads (CPM) in at least 50% of the samples. We normalized the read count values using trimmed means of M value and estimated common, trended, and tagwise dispersions using R software (version RX64 3.0.2). Together these quality control steps removed genes with low expression and normalized the libraries for library size and biological variability, resulting in 14,816 genes for our subsequent analyses. We determined DE using the generalized linear model (GLM) likelihood ratio test employing a significance threshold of FDR<0.05.
Second, similarly as in EdgeR, we used a multi-factor design with DESeq2. We estimated the size factors, dispersions, and performed negative binomial GLM fitting for the “sample” as a factor and Wald statistics for DE. We used Benjamini-Hochberg adjusted P<0.05 as a threshold for significance.
To compare the aSAH patients with aneurysm controls, we considered only the genes DE between the aSAH baseline and after the treatment. We performed two separate DE analyses using negative binomial and determined DE using Wald test for 1) the aSAH baseline group versus the controls; and 2) the aSAH RIC treatment group versus the controls (Supplemental figure I). The genes changing the DE status between the two analyses (i.e., the genes that were not DE between the aSAH baseline group and controls, but became DE when comparing the aSAH treatment group with the controls) were carried forward to subsequent analyses.
Methylation
We analyzed blood DNA methylation profiles by reduced representation bisulfite sequencing (RRBS). RRBS libraries from human genomic DNA were prepared as previously described.19 Briefly, we treated blood DNA with sodium bisulfite (Epitecht Illumina), digested it with the MspI enzyme, and selected fragments averaging 100–250 bp. We multiplexed four samples per lane and sequenced the libraries using single-end 100-bp reads with the Illumina Hiseq2000 platform, resulting in on average 25.1 M reads per sample.
We performed initial QC for fastq files using FastaQC. We utilized BS-seeker220 with Bowtie221 for RRBS alignment using hg19 as a reference genome. For alignment, we considered in silico MspI fragments between 40–500 bp to cover all possible MspI fragments from the RRBS libraries. We aligned the reads using the Bowtie2 end-to-end alignment mode by allowing four mismatches. We called the methylation status of the individual CpG sites (percentage of methylated cells) by requiring at least 10 reads per a CpG site. Pearson correlation coefficient was used to estimate pair-wise correlations in methylation sites between the individuals. Paired Students t-tests were conducted to compare between-group and within-group differences. The CpG sites passing a 2-tailed nominal P<0.01 were considered significant, and carried forward for subsequent analyses. Finally, we used BEDTOOLS22 to overlap the DE genes with methylated regions.
Functional annotation and co-expression of the pathway genes
We utilized DAVID23,24 to search for functional categories of the DE genes. To highlight the most relevant gene ontology terms associated with the overlapped DE and DM gene lists, we performed a batch annotation and gene-GO term enrichment analysis. We searched for protein-protein interaction (PPI) networks using STRING v9.1.25 We used Pearson correlation coefficient to estimate correlations between the pathway genes, and ggplot2 and reshape2 to visualize these results. Reactome26,27 was used to explore specific pathways.
Results
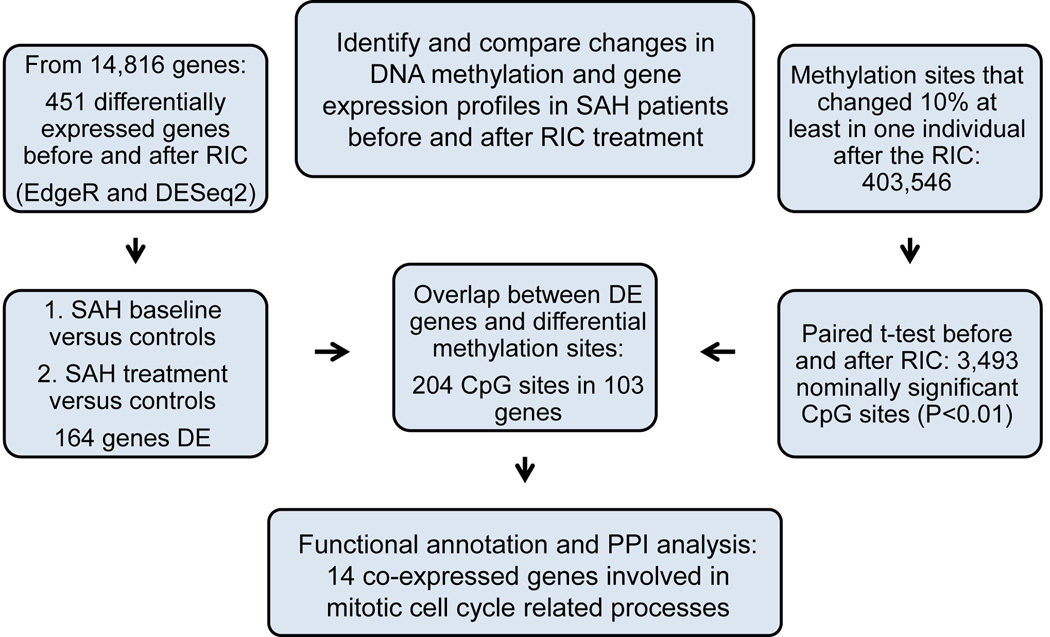
The overall study design is shown in figure 1. To identify genomic mechanisms for the effects of RIC in aSAH patients, we employed a paired sample design where each patient gave blood samples before and after 4 RIC sessions. Using this longitudinal study design, each individual functions as a control for him-/herself in the differential expression (DE) and differential methylation (DM) analyses. Accordingly, we were able to adjust for potential confounding factors, such as age, smoking, medication, and ethnicity using this intraindividual paired design. We analyzed the blood RNA expression and DNA methylation profiles of each patient before RIC and one week after the RIC treatment started. We compared these profiles to the ones of the controls who did not receive any RIC treatments. Lastly, we overlapped the DE genes with DM sites and performed functional annotations and protein-protein interaction analysis (PPIs) of the overlapping genes (figure 1).
Figure 1.
A schematic overview of study design and results.
Differential expression (DE)
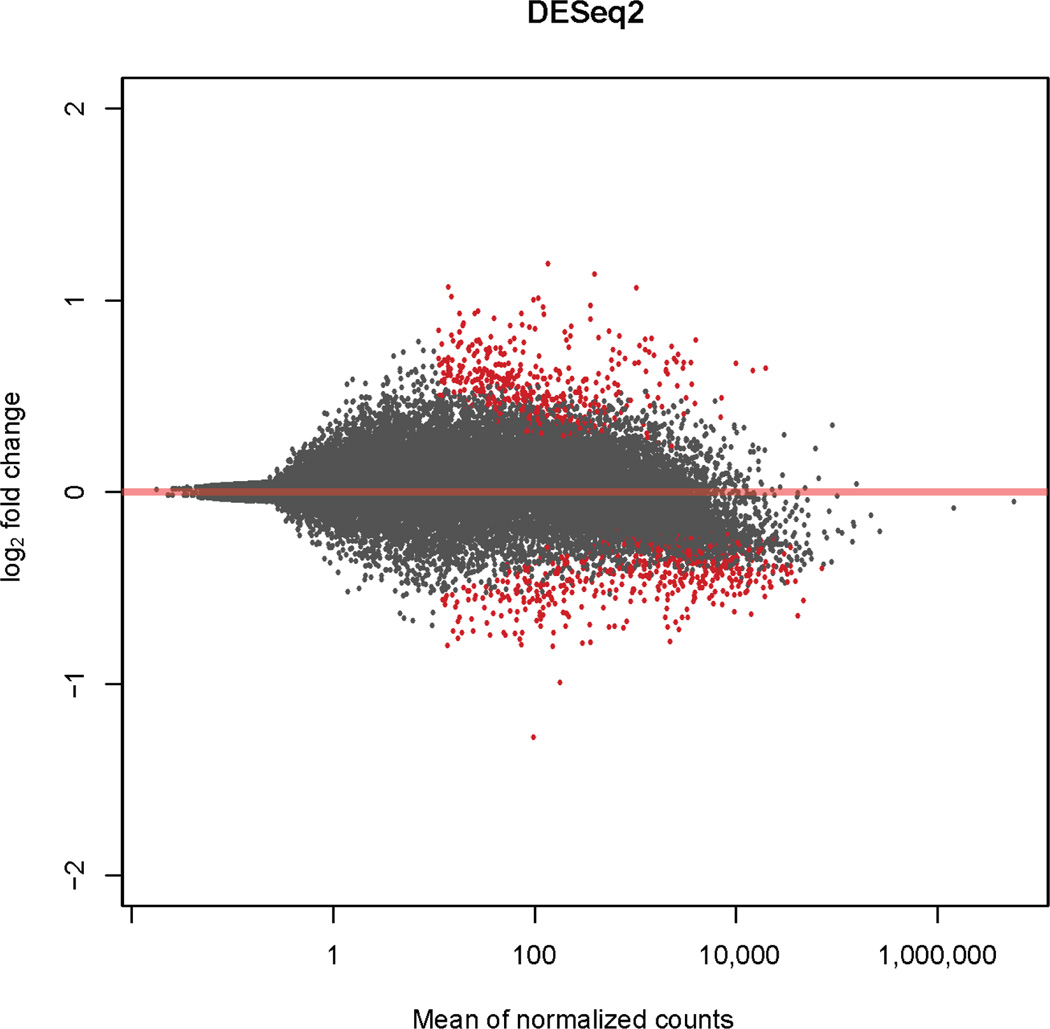
We found 451 DE genes after RIC (FDR<0.05) consistently using both EdgeR and DESeq2, of which 205 were upregulated following the RIC treatment and 246 were down-regulated, respectively (Figure 2 and Supplemental table I). Next, to identify genes responding to the RIC treatment, we tested the expression of the 451 genes in the controls for DE against their expression at both the aSAH baseline and after the RIC treatment, considering a Bonferroni corrected P<1.1×10−4 (P<0.05/451 DE genes) significant. We found 164 DE genes (see Supplemental table II for the list of DE genes) between the controls and aSAH patients before and after RIC treatment, suggesting that these genes may contribute to the response to the RIC treatment.
Figure 2.
The differentially expressed (DE) genes between the aSAH baseline and a week after RIC treatment. Red dots indicate DE genes with an FDR<0.05.
Differential methylation (DM)
We were able to map on average 66% of reads/sample to the human genome, which is in accordance with previous RRBS studies.20,28 The resulting methylation profiles per sample covered on average 1,764,402 CpG sites, of which 676,543 were assayed in all individuals. The overall methylation status changed very little within an individual (~98%) and between individuals (~97.5%) (Supplemental figure II), suggesting that methylation is a stable phenomenon and only a small number of sites are actively responding to environmental factors. We focused on the 403,546 CpG sites that altered by more than 10% in at least one individual following RIC. To test DM cytosines between the baseline and after treatment, we used two-tailed paired student’s t-test. A total of 3,493 CpG sites were DM (P<0.01).
Overlapping the DE and DM regions
When we overlapped (defined +/−250kb from the each DE gene) the DM CpGs with the DE genes, we found 204 CpG sites corresponding to 103 DE and DM genes, suggesting methylation as a potential mechanism for DE (Supplemental table III). Furthermore, 52 of the genes had more than one nearby DM site.
Functional annotation and co-expression of the pathway genes
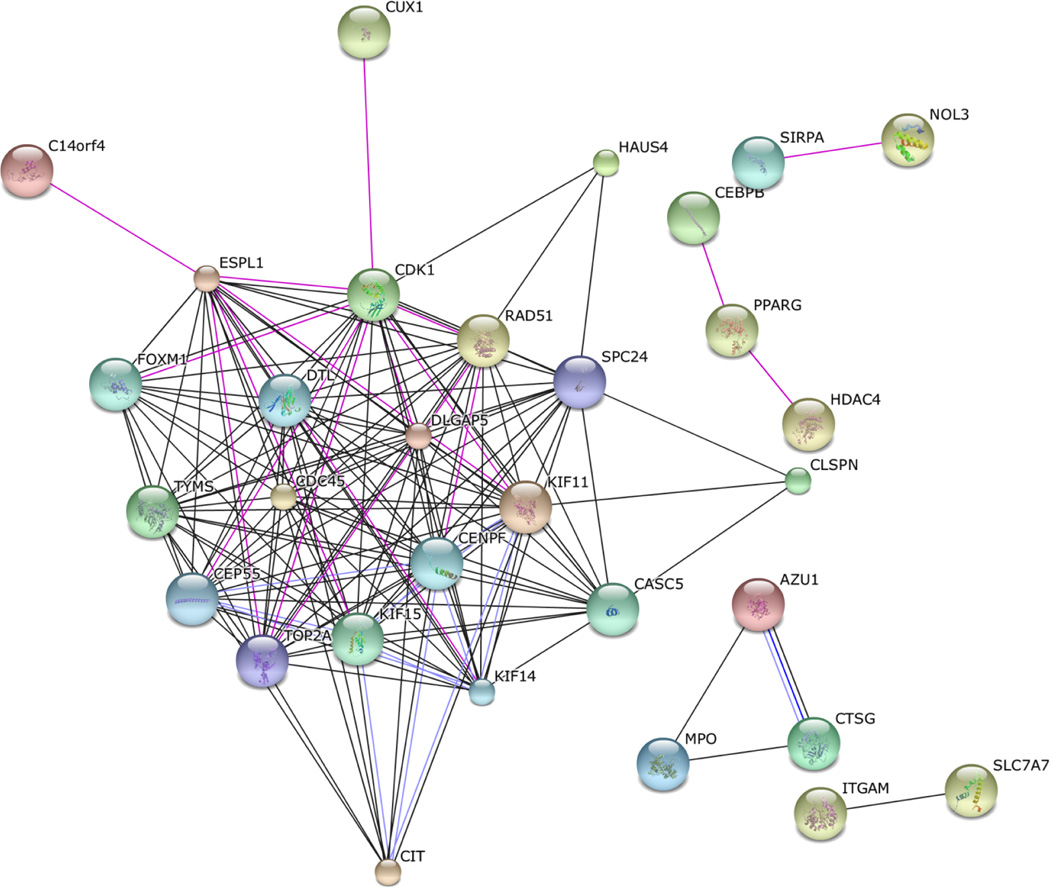
Functional annotation with DAVID software showed that the overlapping 103 DE and DM genes are enriched for defense and inflammatory responses (Benjamini-Hochberg (B-H) corrected P<1.4×10−4) and for cell cycle and mitosis (B-H corrected P<3.8×10−4) (table 2). In addition, we examined the protein-protein interactions (PPIs) of the 103 DE genes using String (figure 3). We found a significant enrichment for PPIs and one large network consisting of 21 DE and DM genes (figure 3) of which 14 are part of the cell cycle pathway from the functional enrichment analysis (table 2). We also found two smaller PPIs consisting of 3 proteins each; CEBPB, HDAC4, PPARG and AZU1, CTSG, MPO (figure 3), all present in the significant pathways of defense and inflammatory response mechanisms (table 2).
Table 2.
Functional annotations of the 103 identified DE genes using the David pathway tool.
| Enrichment Score | 4.35 | Gene count | B-H corrected p-value | |
| Cluster 1 | Defense response | 18 | 1.4×10−4 | |
| Inflammatory response | 10 | 1.7×10−2 | ||
| Enrichment Score | 3.95 | Gene count | B-H corrected p-value | |
| Cluster 2 | Cell cycle | 19 | 3.8×10−4 | |
| M phase | 12 | 1.7×10−3 | ||
| Cell cycle phase | 13 | 1.9×10−3 | ||
| Nuclear division | 10 | 1.7×10−3 | ||
| Mitosis | 10 | 1.7×10−3 | ||
B-H indicates P-value after Benjamini-Hochberg correction for false discovery rate.
Figure 3.
Protein-protein interactions (PPIs) of the 103 differentially expressed (DE) genes using String. We observed a statistically significant (P<0.00001) enrichment of PPIs of the DE genes residing in the cell cycle, defense and inflammatory response pathways, all passing the Benjamini correction as shown in Table 2.
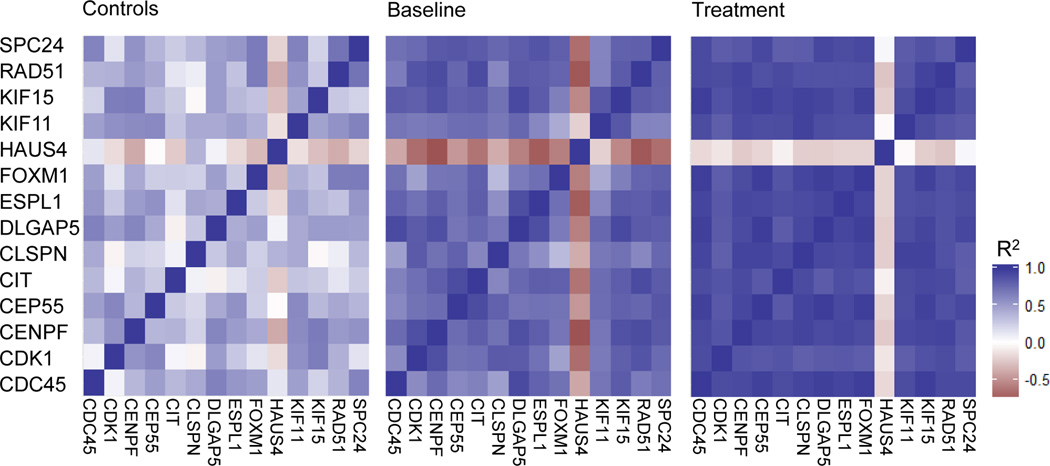
Next, we further examined the 14 cell cycle pathway genes for correlations between their gene expressions. These genes exhibited highly dynamic correlation shifts, with substantially tighter correlations after the RIC treatment (figure 4), suggesting that different phases of cell cycle pathway are turned on due to RIC. Interestingly, when we visualized the co-expression of these genes in the control group, we observed a clear difference in their correlations when compared to the aSAH patients at baseline and even more after the treatment (figure 4), indicating the involvement of these genes in aSAH, and the potential influence of the RIC treatment.
Figure 4.
Co-expression analysis of the 14 mitotic cell cycle genes identified in the Protein-protein interactions (PPIs) and functional enrichment analyses.
Based on a more detailed Reactome pathway analysis (Supplemental table IV), eight of the 14 genes (FDR<1.0×10−5) are involved in the cell cycle pathway (SPC24, ESPL1, CLSPN, CDC45, CENPF, FOXM1, CDK1, RAD51). In the Reactome analysis, CDK1 acts as a key regulator of specific mitotic cell cycle pathways. For instance, we observed that CDK1 is involved in G2/M transition and mitotic G2-G2/M phases with CENPF and FOXM1, regulating the G2/M checkpoints with CLSPN and CDC45. In addition, CDK1 is involved in processes such as kinetochore assembly in mitotic prometaphase and M Phase with SPC24, CENPF, and ESPL1 (Supplemental table IV). CDK1 is also present in numerous activation and signaling pathways within mitotic cell cycle pathway (Supplemental table IV).
Discussion
We performed the first longitudinal and systematic genome-wide pilot study in humans, comparing gene expression and methylation changes following RIC in aSAH. We found 164 DE genes and 3,493 DM CpG sites that are modified, potentially due to RIC. When we overlapped these regions, we observed 204 DM CpG sites corresponding to 103 DE genes, suggesting methylation as a potential mechanism regulating gene expression. These genes were enriched for cell cycle related processes, as well as for defense and inflammatory responses. Furthermore, the identified 14 cell cycle genes exhibited highly correlated expression signals after RIC (figure 4). Overall, these findings provide first insights into the neuroprotective molecular mechanisms underlying RIC in humans.
Our prior work has demonstrated RIC-induced metabolic changes in the preconditioned limb as well as cerebral tissue.29,30 Muscle microdialysis during RIC showed an increase in lactate/pyruvate ratio and lactate, without change in glycerol.29 Cerebral microdialysis during RIC showed a decrease in lactate/pyruvate ratio and glycerol, which persisted after the last RIC session.30 Identification of markers of the RIC effects beyond local factors is imperative for determining appropriate endpoints in future RIC clinical studies.
Whole-genome transcriptional analysis has been applied to uncover genetic changes underlying ischemia-induced neuroprotective effects in animal models.31,32 DNA methylation changes of gene promoter regions have also been investigated to uncover preconditioning induced epigenetic changes, contributing to neuroprotection in mice.33 While proof of concept animal studies have given great insight into the potential mechanisms of RIC, they do not necessarily translate directly to humans, and thus human studies are essential to evaluate effects in clinical settings.
Cell cycle machinery and related molecules have been previously implicated in ischemic neuronal death,34,35 and irregular cell cycle activation has been implicated in stroke.36–38 We show evidence for involvement of genes in cell cycle processes regulated by CDK1 in the acute stage of aSAH, possibly modified by RIC (figure 4). We postulate that this pathway may be important in various forms of ischemia. Furthermore, we hypothesize that RIC may induce a release of substances from the ischemic limb muscles to blood. These in turn stimulate white blood cells, such as macrophages, to increase the expression of genes involved in cell cycle and cell proliferation. Subsequently, these white blood cells may stream to the ischemic location in brain and release substances, including growth factors and other cytokines, to protect brain from further apoptosis. This mechanism could lead to the neuroprotective effects of RIC, although additional functional studies are warranted to verify the underlying mechanisms.
One of the mechanisms proposed for RIC is inflammatory responses.39,40 In accordance with this, our DAVID pathway analysis implicated a set of 18 both DE and DM genes in defense response pathways (CEBPB, AZU1, BPI, CTSG, CRISP3, CYSLTR1, HDAC4, INHBA, IL1R1, IL10RB, LTF, MPO, OLR1, PPARG, PROK2, STAT5B, STAB1, and TLR5). Six of these genes were also involved in two separate PPIs (figure 3).
A recent study exploring human plasma proteome in RIC found that cysteine-rich secretory protein 3 (CRISP-3) was increased in serum after RIC in six adults.41 This is consistent with our finding of over a two-fold increase of CRISP3 gene expression in blood followed by RIC (Supplemental table I), suggesting its role as a humoral RIC mediator and surrogate marker. CRISP-3 is a glycoprotein present in exocrine secretions, bone marrow, secretory granules of neutrophils, and in plasma bound to a1B-glycoprotein.42,43 Although its complete function is unknown, it is thought to act in innate immune response and as a prostate cancer marker.42,43
In summary, in this first pilot study, employing a longitudinal design to investigate genome-wide expression and methylation changes in aSAH patients after RIC, we found evidence for coordinated expression and methylation changes of a small set of key genes in mitotic cell cycle, defense, and inflammatory responses. We have limitations in this study and therefore, the results presented here should be further investigated and verified in future considerably larger genomic studies. In addition to the small sample size, we recognize that some of the observed changes in genes expression and methylation are potentially due to other medical treatments these patients received in the hospital, and hence future studies should comprise a randomization that includes patients not receiving any RIC treatment as controls. We also recognize that differences in blood cell types may contribute to the changes in DNA methylation and gene expression, and thus future RIC studies should include analysis of separate FACS sorted cells. Nevertheless, longitudinal genome-wide studies of stroke and especially SAH, integrating expression and methylation changes at the genome-wide level are still very sparse, and thus our study provides valuable initial data, starting to elucidate the largely unknown mechanisms underlying RIC in humans.
Supplementary Material
Acknowledgments
Sources of Funding
This work was supported by the Ruth and Raymond Stotter Endowement, the National Institutes of Health NINDS grant K23NS079477, NIBIB grant R01EB000362, and the NHLBI grants HL-28481 and HL-095056.
Footnotes
Disclosure
The authors have no personal, financial, or institutional interests in any of the drugs, materials, or devices described in this article.
References
- 1.Przyklenk K, Bauer B, Ovize M, Kloner RA, Whittaker P. Regional ischemic 'preconditioning' protects remote virgin myocardium from subsequent sustained coronary occlusion. Circulation. 1993;87:893–899. doi: 10.1161/01.cir.87.3.893. [DOI] [PubMed] [Google Scholar]
- 2.Ren C, Gao X, Steinberg GK, Zhao H. Limb remote-preconditioning protects against focal ischemia in rats and contradicts the dogma of therapeutic time windows for preconditioning. Neuroscience. 2008;151:1099–1103. doi: 10.1016/j.neuroscience.2007.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li SJ, Wu YN, Kang Y, Yin YQ, Gao WZ, Liu YX, et al. Noninvasive limb ischemic preconditioning protects against myocardial I/R injury in rats. J Surg Res. 2010;164:162–168. doi: 10.1016/j.jss.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 4.Wei D, Ren C, Chen X, Zhao H. The chronic protective effects of limb remote preconditioning and the underlying mechanisms involved in inflammatory factors in rat stroke. PLoS One. 2012;7:e30892. doi: 10.1371/journal.pone.0030892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koch S, Katsnelson M, Dong C, Perez-Pinzon M. Remote ischemic limb preconditioning after subarachnoid hemorrhage: a phase Ib study of safety and feasibility. Stroke. 2011;42:1387–1391. doi: 10.1161/STROKEAHA.110.605840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez NR, Connolly M, Dusick JR, Bhakta H, Vespa P. Phase I clinical trial for the feasibility and safety of remote ischemic conditioning for aneurysmal subarachnoid hemorrhage. Neurosurgery. 2014;75:590–598. doi: 10.1227/NEU.0000000000000514. discussion 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarabi AS, Shen H, Wang Y, Hoffer BJ, Bäckman CM. Gene expression patterns in mouse cortical penumbra after focal ischemic brain injury and reperfusion. J Neurosci Res. 2008;86:2912–2924. doi: 10.1002/jnr.21734. [DOI] [PubMed] [Google Scholar]
- 8.Konstantinov IE, Arab S, Kharbanda RK, Li J, Cheung MM, Cherepanov V, et al. The remote ischemic preconditioning stimulus modifies inflammatory gene expression in humans. Physiol Genomics. 2004;19:143–150. doi: 10.1152/physiolgenomics.00046.2004. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Qian H, Zhao P, Hong SS, Xia Y. Rapid hypoxia preconditioning protects cortical neurons from glutamate toxicity through delta-opioid receptor. Stroke. 2006;37:1094–1099. doi: 10.1161/01.STR.0000206444.29930.18. [DOI] [PubMed] [Google Scholar]
- 10.Liu YX, Zhang M, Liu LZ, Cui X, Hu YY, Li WB. The role of glutamate transporter-1a in the induction of brain ischemic tolerance in rats. Glia. 2012;60:112–124. doi: 10.1002/glia.21252. [DOI] [PubMed] [Google Scholar]
- 11.Ding ZM, Wu B, Zhang WQ, Lu XJ, Lin YC, Geng YJ, et al. Neuroprotective Effects of Ischemic Preconditioning and Postconditioning on Global Brain Ischemia in Rats through the Same Effect on Inhibition of Apoptosis. Int J Mol Sci. 2012;13:6089–6101. doi: 10.3390/ijms13056089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang N, Yin Y, Han S, Jiang J, Yang W, Bu X, et al. Hypoxic preconditioning induced neuroprotection against cerebral ischemic injuries and its cPKCγ-mediated molecular mechanism. Neurochem Int. 2011;58:684–692. doi: 10.1016/j.neuint.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura H, Katsumata T, Nishiyama Y, Otori T, Katsura K, Katayama Y. Effect of ischemic preconditioning on cerebral blood flow after subsequent lethal ischemia in gerbils. Life Sci. 2006;78:1713–1719. doi: 10.1016/j.lfs.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Dickson EW, Reinhardt CP, Renzi FP, Becker RC, Porcaro WA, Heard SO. Ischemic preconditioning may be transferable via whole blood transfusion: preliminary evidence. J Thromb Thrombolysis. 1999;8:123–129. doi: 10.1023/a:1008911101951. [DOI] [PubMed] [Google Scholar]
- 15.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen PY, Ganguly A, Rubbi L, Orozco LD, Morselli M, Ashraf D, et al. Intrauterine calorie restriction affects placental DNA methylation and gene expression. Physiol Genomics. 2013;45:565–576. doi: 10.1152/physiolgenomics.00034.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, et al. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 2013;14:774. doi: 10.1186/1471-2164-14-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milacic M, Haw R, Rothfels K, Wu G, Croft D, Hermjakob H, et al. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers (Basel) 2012;4:1180–1211. doi: 10.3390/cancers4041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doherty R, Couldrey C. Exploring genome wide bisulfite sequencing for DNA methylation analysis in livestock: a technical assessment. Front Genet. 2014;5:126. doi: 10.3389/fgene.2014.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bilgin-Freiert A, Dusick JR, Stein NR, Etchepare M, Vespa P, Gonzalez NR. Muscle microdialysis to confirm sublethal ischemia in the induction of remote ischemic preconditioning. Transl Stroke Res. 2012;3:266–272. doi: 10.1007/s12975-012-0153-1. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez NR, Hamilton R, Bilgin-Freiert A, Dusick J, Vespa P, Hu X, et al. Cerebral hemodynamic and metabolic effects of remote ischemic preconditioning in patients with subarachnoid hemorrhage. Acta Neurochir Suppl. 2013;115:193–198. doi: 10.1007/978-3-7091-1192-5_36. [DOI] [PubMed] [Google Scholar]
- 31.Stenzel-Poore MP, Stevens SL, Xiong Z, Lessov NS, Harrington CA, Mori M, et al. Effect of ischaemic preconditioning on genomic response to cerebral ischaemia: similarity to neuroprotective strategies in hibernation and hypoxia-tolerant states. Lancet. 2003;362:1028–1037. doi: 10.1016/S0140-6736(03)14412-1. [DOI] [PubMed] [Google Scholar]
- 32.Stenzel-Poore MP, Stevens SL, Simon RP. Genomics of preconditioning. Stroke. 2004;35:2683–2686. doi: 10.1161/01.STR.0000143735.89281.bb. [DOI] [PubMed] [Google Scholar]
- 33.Zhang S, Zhang Y, Jiang S, Liu Y, Huang L, Zhang T, et al. The effect of hypoxia preconditioning on DNA methyltransferase and PP1γ in hippocampus of hypoxia preconditioned mice. High Alt Med Biol. 2014;15:483–490. doi: 10.1089/ham.2014.1042. [DOI] [PubMed] [Google Scholar]
- 34.Rashidian J, Iyirhiaro GO, Park DS. Cell cycle machinery and stroke. Biochim Biophys Acta. 2007;1772:484–493. doi: 10.1016/j.bbadis.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Wang W, Bu B, Xie M, Zhang M, Yu Z, Tao D. Neural cell cycle dysregulation and central nervous system diseases. Prog Neurobiol. 2009;891:1–17. doi: 10.1016/j.pneurobio.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Osuga H, Osuga S, Wang F, Fetni R, Hogan MJ, Slack RS, et al. Cyclin-dependent kinases as a therapeutic target for stroke. Proc Natl Acad Sci U S A. 2000;97:10254–10259. doi: 10.1073/pnas.170144197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F, Corbett D, Osuga H, Osuga S, Ikeda JE, Slack RS, et al. Inhibition of cyclin-dependent kinases improves CA1 neuronal survival and behavioral performance after global ischemia in the rat. J Cereb Blood Flow Metab. 2002;22:171–182. doi: 10.1097/00004647-200202000-00005. [DOI] [PubMed] [Google Scholar]
- 38.Rashidian J, Iyirhiaro G, Aleyasin H, Rios M, Vincent I, Callaghan S, et al. Multiple cyclin-dependent kinases signals are critical mediators of ischemia/hypoxic neuronal death in vitro and in vivo. Proc Natl Acad Sci U S A. 2005;102:14080–14085. doi: 10.1073/pnas.0500099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowen KK, Naylor M, Vemuganti R. Prevention of inflammation is a mechanism of preconditioning-induced neuroprotection against focal cerebral ischemia. Neurochem Int. 2006;49:127–135. doi: 10.1016/j.neuint.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 40.Carr-White G, Koh T, DeSouza A, Haxby E, Kemp M, Hooper J, et al. Chronic stable ischaemia protects against myocyte damage during beating heart coronary surgery. Eur J Cardiothorac Surg. 2004;25:772–778. doi: 10.1016/j.ejcts.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 41.Helgeland E, Breivik LE, Vaudel M, Svendsen ØS, Garberg H, Nordrehaug JE, et al. Exploring the human plasma proteome for humoral mediators of remote ischemic preconditioning--a word of caution. PLoS One. 2014;9:e109279. doi: 10.1371/journal.pone.0109279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellias MF, Zainal Ariffin SH, Karsani SA, Abdul Rahman M, Senafi S, Megat Abdul Wahab R. Proteomic analysis of saliva identifies potential biomarkers for orthodontic tooth movement. Scientific World Journal. 2012;2012:647240. doi: 10.1100/2012/647240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Udby L, Sørensen OE, Pass J, Johnsen AH, Behrendt N, Borregaard N, et al. Cysteine-rich secretory protein 3 is a ligand of alpha1B-glycoprotein in human plasma. Biochemistry. 2004;43:12877–12886. doi: 10.1021/bi048823e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.