Abstract
Background & Aims
The ratio of liver size to body weight (hepatostat) is tightly controlled, but little is known about how the physiologic functions of the liver help determine its size. Livers of mice repopulated with human hepatocytes (humanized livers) grow to larger than normal; the human hepatocytes do not recognize fibroblast growth factor-15 (FGF15) produced by mouse intestine. This results in upregulation of bile acid synthesis in the human hepatocytes and enlargement of the bile acid pool. We investigated whether abnormal bile acid signaling affects the hepatostat in mice.
Methods
We crossed Fah−/−, Rag2−/−, Il2r−/− mice with NOD mice to create FRGN mice, whose livers can be fully repopulated with human hepatocytes. We inserted the gene for human FGF19 (ortholog to mouse Fgf15), including regulatory sequences, into the FRGN mice to create FRGN19+ mice. Livers of FRGN19+ mice and their FRGN littermates were fully repopulated with human hepatocytes. Liver tissues were collected and bile acid pool sizes and RNA sequences were analyzed and compared with those of mice without humanized livers (controls).
Results
Livers were larger in FRGN mice with humanized livers (13% of body weight), compared to control FRGN mice; they also had much larger bile acid pools and aberrant bile acid signaling. Livers from FRGN19+ normalized to 7.8% of body weight, and their bile acid pool and signaling more closely resembled that of control FRGN19+ mice. RNA sequence analysis showed activation of the Hippo pathway, and immunohistochemical and transcription analyses revealed increased hepatocyte proliferation, but not apoptosis, in the enlarged humanized livers of FRGN mice. Cell sorting experiments showed that although healthy human liver does not produce FGF19, non-parenchymal cells from cholestatic livers produce FGF19.
Conclusions
In mice with humanized livers, expression of an FGF19 transgene corrects bile acid signaling defects, resulting in normalization of bile acid synthesis, the bile acid pool, and liver size. These findings indicate that liver size is, in part, regulated by the size of the bile acid pool that the liver must circulate.
Keywords: mouse model, regeneration, signal transduction, CYP7A
Introduction
The mammalian liver is capable of marked growth after liver injury, typified by partial hepatectomy, in which the remnant liver grows back to within 5% of the original size in 7–10 days in rodents1 and 4–8 weeks in humans. Many studies have demonstrated that liver mass is proportional to body size,2 a phenomenon termed the “hepatostat.”3 Some molecular signals for control of organ size have been elucidated, including the YAP/Hippo signaling pathway.4–6 It is presumed, however, that liver-to-body weight ratio depends on some physiologic function(s) the liver performs.7
Mice with humanized livers have aberrant bile acid signaling, wherein the rate-controlling enzyme for bile acid synthesis, CYP7A, is highly upregulated in the human hepatocytes.8 Human CYP7A expression could be downregulated with administration of FGF19, human ortholog to mouse Fgf15, thus demonstrating that human hepatocytes do not recognize Fgf15 and explaining the aberrant signaling in this model. Unregulated bile acid synthesis led to an enlargement of the bile acid pool, and we hypothesized that the increased liver size we and others see in humanized chimeric livers might be due to this short-circuit in bile acid signaling.
Bile acids and attendant signaling have recently gained traction as potential drivers of liver regeneration9–13 and the related governance of liver size. Mice fed cholic acid had modest increases in liver size,9 while rats undergoing partial hepatectomy after drainage of the bile acid pool had significantly less regrowth of the liver which could be reversed with administration of bile acids.10
In order to determine whether abnormal bile acid signaling was related to the hepatostat we introduced a long segment of human genomic DNA into the immune-deficient Fah−/− (FRGN) mouse model for liver repopulation. The human DNA fragment contained the entire genomic FGF19 sequence and its regulatory sequences, thus conferring physiological control of the FGF19 gene. When human hepatocytes were transplanted into these FRGN19+ mice, we found near-normalization of CYP7A in the human hepatocytes, a significant decrease in the total bile acid pool, and a marked reduction in liver size to near-expected size for body weight. We thus propose that one of the main determinants of the hepatostat is the amount of bile acids the liver must circulate.
Materials and Methods
Additional methods are available in Supplemental Materials and Methods.
Animals
Fah−/−, Rag2−/−, Il2r−/− mice (“FRG”)14 were crossed onto the NOD mouse strain to create “FRGN” mice (Yecuris)15, whose livers can be fully repopulated with human hepatocytes.8 Using the FRGN background, we generated mice with a bacterial artificial chromosome (BAC, RP11-266K14, Invitrogen) containing ~164 kb of human genomic DNA in the middle of which sits the genomic sequence for FGF19 (see Supp. Material & Methods for full details). Presence of the entire BAC was confirmed with a mapping strategy using specific PCR primer sets designed to detect relevant sequences in the human genome (Figure 1). FRGN FGF19+ (“FGF19+”) and their FRGN littermates without the transgene (“FGF19-“) were used in these experiments.
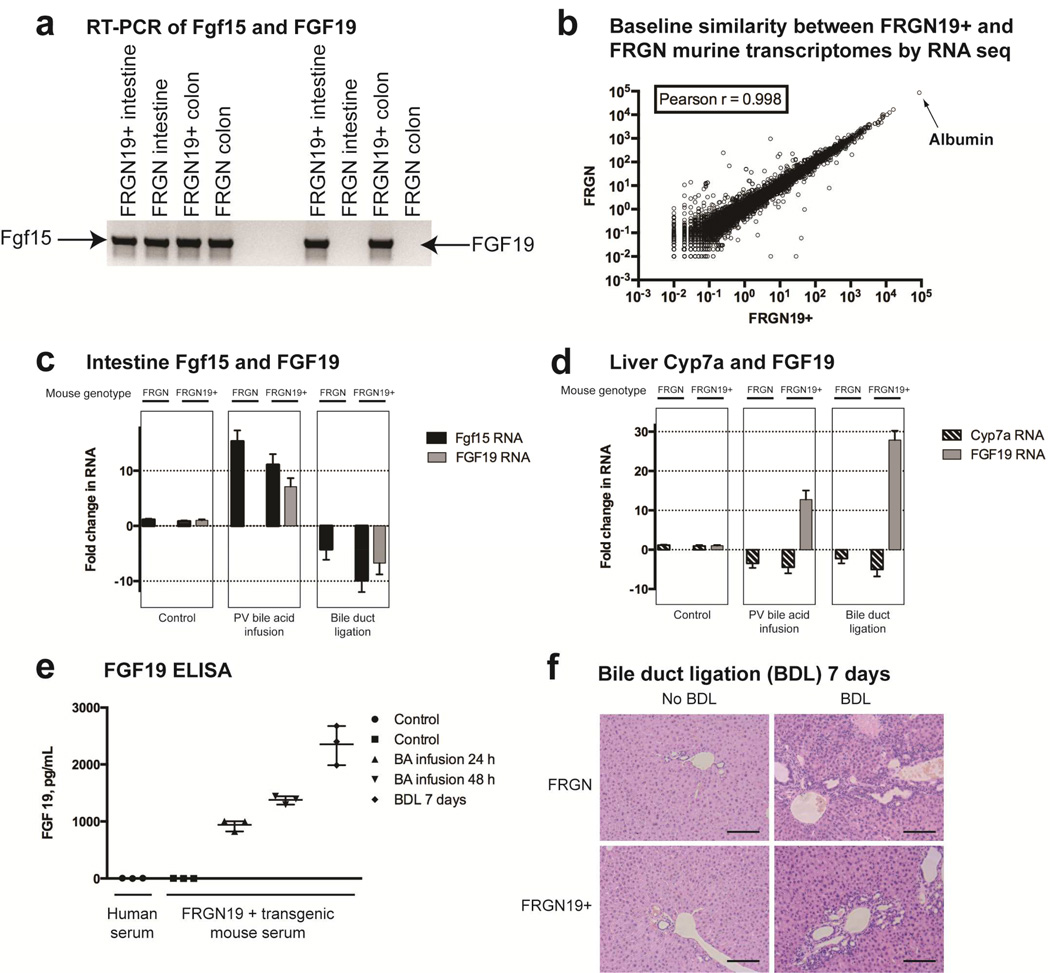
Figure 1. FRGN19+ transgenic mice demonstrate physiological regulation of FGF19 and bile acid signaling.
(a) RT-PCR shows presence of RNA for Fgf15 in the small intestine and colon of both FRGN19+ transgenic mice and their FRGN littermates, while only showing FGF 19 RNA in the transgenic mice. (b) Scatterplot of RNA sequencing of mouse transcriptomes from FRGN19+ transgenic mice and their FRGN littermates are near indistinguishable (Pearson r = 1 signifies identical datasets). (c) Intestinal regulation of Fgf 15 and FGF 19 under 3 conditions: control, portal vein (PV) bile acid infusion and bile duct ligation (BDL), n = 3 mice of each genotype for each condition. (d) Liver regulation of Cyp 7a and FGF 19 under 3 conditions: control, portal vein (PV) bile acid infusion and bile duct ligation (BDL), n = 3 mice of each genotype for each condition. (e) ELISA for serum FGF19 on human and mouse serum. (f) H&E histology of liver (10×) with BDL for 7 days in FRGN19+ and FRGN mice, showing typical biliary ductal proliferation seen in BDL. Scale bar = 100 µm.
Human hepatocytes (Celsis) were transplanted via intrasplenic technique and allowed to repopulate the mouse liver by withdrawing NTBC, a medication that prevents hepatocyte death when FAH deficiency is present.14 In this model, donor (in this case human) hepatocytes with intact FAH have a selective advantage compared to the recipient FAH deficient hepatocytes, leading to repopulation of the liver with donor cells. The success and degree of human hepatocyte repopulation is noted by absence of liver failure and monitoring production of human albumin via specific ELISA.8 Mice with human hepatocyte repopulated livers (“humanized”) were sacrificed after serum human albumin levels indicated a high degree of repopulation (4 months post-transplant).
Bile duct ligation, bile acid infusion, intestinal bile acid pool and tissue analysis methods are described in Supplemental Materials and Methods.
All studies were approved by the Oregon Health & Science University Institutional Animal Care and Use Committee as set forth in the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health.
Analysis and visualization of RNA sequencing data
After transformation of RNA sequencing data in to standardized Reads Per Kilobase Million (RPKM), data was analyzed using Gene Set Enrichment Analysis (GSEA) as described.16 Hierarchical cluster analysis was performed with GENE E.17
Statistical Analysis
Prism 6 (GraphPad Prism) was used to graph all noted data except for RNA sequence data (see Supplementary Materials and Methods). Results are expressed as mean values ± SD. P values were calculated by two-sided independent t-test, and a p value < 0.05 was considered significant.
Results
Generation of transgenic mice with human FGF 19
Bacterial artificial chromosome (BAC) RP11-266K14 contains a 164 kb fragment of human DNA containing the genomic sequence for the FGF19 gene, along with its promoter and regulatory elements. This BAC was linearized and introduced into Fah−/−, Rag2−/−, Il2r−/−, NOD mice (“FRGN”), whose livers can be fully repopulated with transplanted human hepatocytes.8 The presence of the full human sequence in the transgenic mice was confirmed by specific PCR amplicons (Fig. S1a and S1b) present in transgenic (FRGN19+) mice but not their non-transgenic littermates (FRGN).
FRGN19+ transgenic mice demonstrate physiological regulation of FGF19 and bile acid signaling
RT-PCR of intestines from FRGN19+ and FRGN mice showed presence of Fgf15 (mouse ortholog of human FGF19) in both strains, while FGF19 RNA transcripts were found only in transgenic mice (Fig. 1a). Scatterplot comparison of complete mouse liver transcriptomes of FRGN19+ and FRGN mice are nearly identical, showing that introduction of the human genomic DNA did not significantly alter homeostatic liver gene expression (Fig. 1b).
Intestinal Fgf15 and FGF19 expression were assayed in two different physiologic experiments. Sodium taurocholate (NaTCA) was infused into the portal vein (PV) for 24 hours at three times the normal PV bile acid concentration, exceeding the liver’s capacity for bile acid uptake and secretion, and resulting in liver cholestasis and markedly increased intestinal bile acid concentrations. As expected, both Fgf15 and FGF19 mRNA were significantly upregulated in this example of intestinal bile acid excess. Seven days after bile duct ligation (BDL), both Fgf15 and FGF19 in the intestine were nearly undetectable consistent with absence of bile acids in the intestine (Fig. 1c).
Humans express FGF19 in the liver as well as the intestine,18 but mice express Fgf15 only in the intestine.19 The cellular source of FGF19 (human) and Fgf15 (mouse) in the intestine is the ileal enterocyte;20 the cellular source of FGF19 in human liver under cholestatic conditions, however, is unclear.18 Human liver under normal conditions does not produce FGF19.21 Consistent with this expression pattern, FGF19 RNA was expressed in the transgenic mouse liver in both cholestatic conditions, PV bile acid infusion and BDL, and no Fgf15 was detected in the livers of any mice (Fig. 1d). An ELISA for FGF19 protein did not detect the hormone in normal human blood nor in the blood of the FRGN19+ transgenic mice. Increasing levels of serum FGF19 were detectable, however, with increasing states of cholestasis (Fig. 1e). Histology (H&E) of FRGN19+ and FRGN mice 7 days after BDL showed proliferation of bile ducts and a decrease in bile infarcts in FRGN19+ mice (Fig. 1f), consistent with a prior report where recombinant FGF19 protected mice from injury associated with BDL.22 RNA sequence data showed that the bile acid machinery in the liver (Fig. S5a) and intestine (Fig. S5b) responded appropriately to cholestasis (liver) and bile acid excess (intestine) in FRGN19+ transgenic mice.
Taken together, the above data show that the FRGN19+ mice generated here responded in a physiologically relevant human-specific manner in regards to bile acid signaling, including with tissue specificity known to be different between humans and mice.
Liver size is corrected in humanized FRGN19+ mice
FRGN19+ transgenic mice and their FRGN littermates were transplanted with human hepatocytes (two cohorts transplanted at different times with different single donor human hepatocytes), and the mice killed when liver repopulation was complete. Ex vivo appearance of the repopulated livers revealed larger livers in FRGN mice (Fig. 2a). In this model, repopulating hepatocytes are FAH positive and stain brown, as shown in repopulated livers from both mouse genotypes (Fig. 2b). Microscopically the repopulated livers appeared similar in FRGN and FRGN19+ mice, with most parenchymal cells replaced by brown FAH+ human hepatocytes. When mouse Fah + hepatocytes are transplanted, repopulated livers had a similar appearance microscopically regardless of recipient mouse FGF19 status (Fig. 2c).
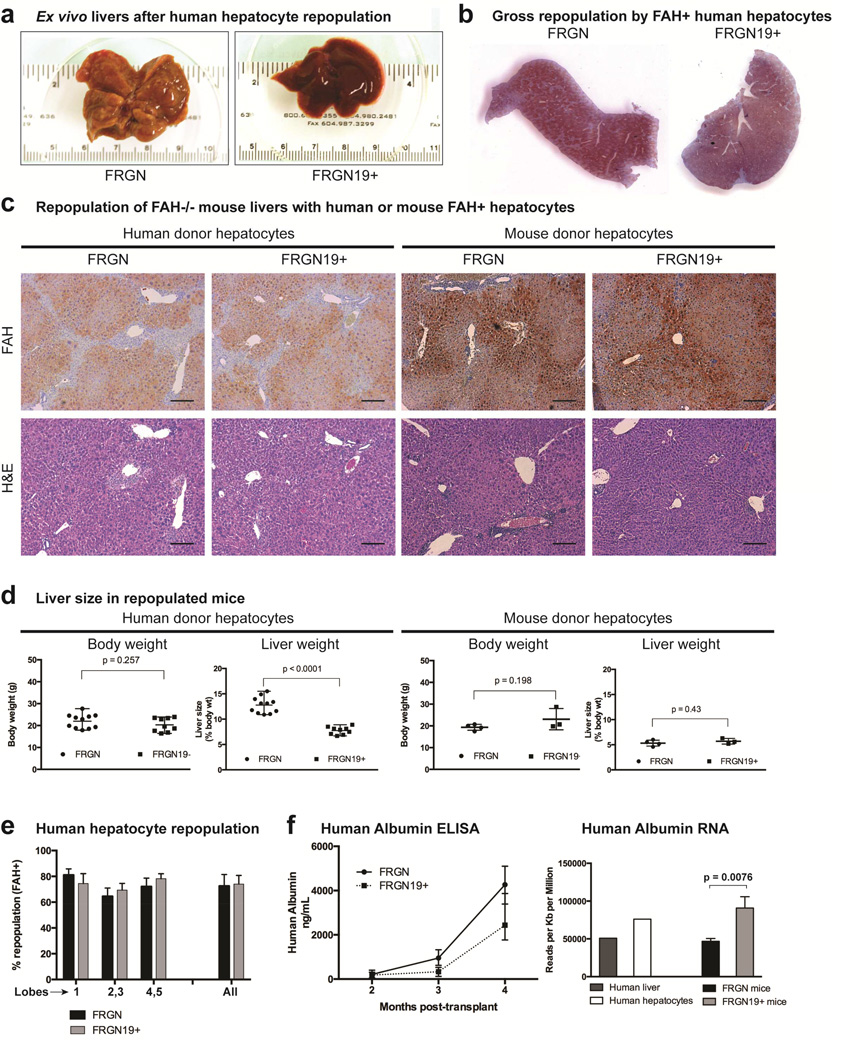
Figure 2. Human hepatocyte repopulated mouse livers are larger in FGF 19 – mice compared to FRGN19+ mice.
(a) Representative gross pictures of human hepatocyte repopulated mouse livers 4 months after hepatocyte transplantation. (b) Low powered view of representative lobes of human hepatocyte repopulated livers stained for FAH (brown) in FRGN and FRGN19+ mice. (c) FAH staining (brown) identifies transplanted human or mouse hepatocytes in FRGN and FRGN19+ transgenic mice. H&E staining of repopulated livers shows normal parenchymal architecture. Scale bar = 100 µm. (d) Body and liver weights of mice with livers fully repopulated with either human or mouse hepatocytes in FRGN and FRGN19+ mice. (e) Percent of liver lobes positive for FAH (indicating level of repopulation) by morphometric analysis. (f) ELISA for human albumin in serum of FRGN and FRGN19+ mice transplanted with human hepatocytes, n = 10 FRGN mice and n = 9 FRGN19+ mice. Human albumin RNA quantified by RNA sequencing from human hepatocytes 4 months after transplant in FRGN and FRGN19+, n = 6 FRGN and n = 6 FRGN19+ mice.
Overall bodyweight was similar in FRGN19+ and FRGN littermates when livers were repopulated with human hepatocytes. Surprisingly, the FRGN19+ mice had livers that were about half the size of their FRGN littermates (Fig. 2d). We have previously noted that FRG and FRGN mice transplanted with human hepatocytes have large livers, roughly three times the expected size, similar to human chimeric livers in uPA/SCID mice.23 When Fah + mouse donor hepatocytes were used to repopulate the liver, there was no difference in liver size between FRGN and FRGN19+ mice, and the repopulated livers were the expected size (~5% of body weight) (Fig. 2d). Morphometric analysis of human hepatocyte repopulated livers showed that there was no difference in the percentage of repopulation between FRGN and FRGN19+ mice (Fig. 2e). Together, these data indicate human hepatocyte repopulated mouse livers are significantly larger (~14% body weight) than normal mouse livers (~5% body weight), and that this phenomenon could be largely corrected with the introduction of the human FGF19 gene under physiological control. Similar high degrees of repopulation argue against a defect in regeneration or repopulating ability for human hepatocytes transplanted in FRGN19+ mice. Serum human albumin production was lower in FRGN19+ mice, but human albumin mRNA expression per cell was much higher (Fig. 2f) suggesting that transplanted human hepatocytes in FRGN mice made less albumin mRNA on a per cell basis, but more albumin overall due to the increase in hepatocyte mass.
FRGN19+ mice with humanized livers show normalized bile acid signaling
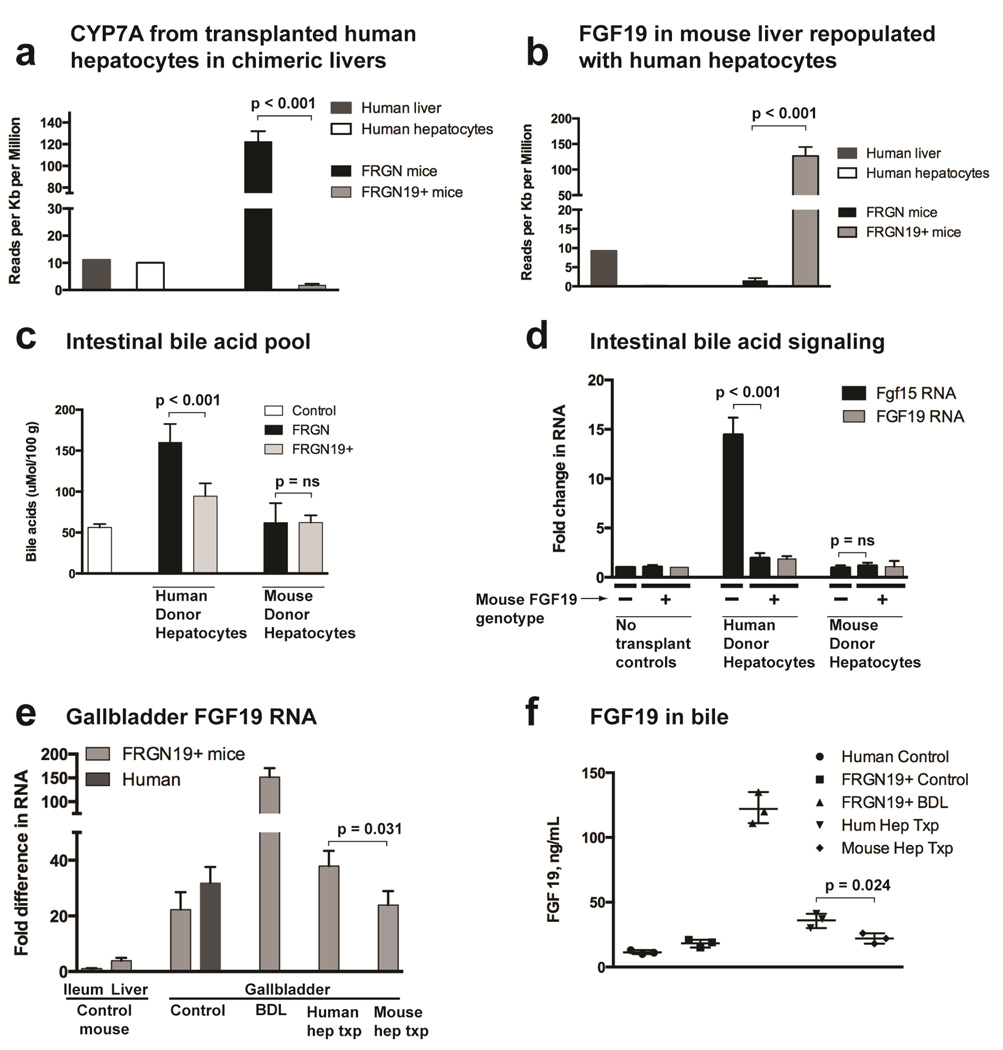
CYP7A in livers of human hepatocyte transplanted mice is highly up-regulated, and was correctable by exogenous FGF19 administration.8 This finding was confirmedhere as shown by a marked increase in human CYP7A in humanized livers of FRGN mice. Importantly, however, CYP7A in repopulated livers of FRGN19+ mice, however, was 70 times lower, near levels of normal control human liver (Fig. 3a). FGF19 RNA was detected at robust levels in FRGN19+ human hepatocyte repopulated livers, but not in livers of FRGN littermates (Fig. 3b). These findings clearly indicate that FGF19 production in FRGN19+ mice down-regulates CYP7a in transplanted human hepatocytes, correcting a bile acid signaling short-circuit in mice expressing only Fgf15, which is not recognized by human hepatocytes. Correction of aberrant bile acid signaling was confirmed by measuring the bile acid pool, markedly enlarged in FRGN mice (a phenomenon not seen when livers are repopulated with mouse donor hepatocytes) and restored near normal levels in FRGN19+ mice (Fig. 3c). Measurement of other contributors to the bile acid pool (portal venous, systemic venous, and hepatic compartments, Fig. S6a–c) showed findings similar to those seen in the intestinal compartment. Importantly, the elevated portal venous bile acid concentration in non-transgenic humanized livers indicates a significant increase in the circulating bile acid pool, suggesting that the increased bile acid production in the liver is not simply being lost in the stool. As expected from the enlarged bile acid pool in FRGN mice, expression of intestinal Fgf15 was upregulated compared to controls, and normalized when the bile acid pool returns toward normal in FRGN19+ mice (Fig. 3d).
Figure 3. FRGN19+ mice with humanized livers show normalized bile acid signaling compared to FRGN mice.
(a) Complete RNA sequencing of chimeric human hepatocyte repopulated mouse livers showed a marked decrease in human CYP7A in FRGN19+ compared to FRGN mice, indicating restoration of intestine-liver signaling of bile acid homeostasis (n = 3 each genotype; n = 1 each for human liver and human hepatocytes). (b) RNA sequencing of chimeric livers revealed FGF 19 in liver tissue from FRGN19+ mice, but no FGF 19 in chimeric livers from FRGN mice, indicating that human hepatoctyes do not produce FGF 19 in this model (n = 3 each genotype). (c) Intestinal contents were collected from control C57/B6 (n = 3), human hepatocyte-transplanted FRGN (n = 5) and FRGN19+ mice (n = 5), and mouse hepatocyte-transplanted FRGN (n = 4) and FRGN19+ (n = 3) mice, and the bile acid pool quantitated after ethanol extraction of bile acids. (d) Fgf 15 and FGF 19 RNA was quantitated by qPCR in small intestine tissue, fold change normalized to un-transplanted controls; human hepatocyte-transplanted FRGN (n = 10), FRGN19+ (n = 9), and mouse hepatocyte-transplanted FRGN (n = 4) and FRGN19+ (n = 3) mice. (e) FGF 19 RNA was quantitated by qPCR in gallbladder tissue, normalized to FGF 19 RNA in the ileum of an FRGN19+ mouse; FRGN19+ controls (n = 4), human GB controls (n = 3), FRGN19+ mice after 7 days of BDL (n = 3), FRGN19+ mice repopulated with human hepatocytes (n = 4) and FRGN19+ mice repopulated with mouse hepatocytes (n = 3). (f) FGF 19 ELISA of bile from human controls (n = 3, obtained during resection of hepatic hemangioma n = 1 or adenoma n = 2), and FRGN19+ mice (controls, 7 days after BDL, liver repopulated with human hepatocytes or liver repopulated with mouse hepatocytes).
Gallbladders of FRGN19+ mice expressed more FGF19 RNA than either ileum or liver, with highest expression occurring in the BDL model. FGF19 expression was modestly but significantly higher in FRGN19+ mice and human hepatocyte repopoulated livers compared to mouse repopulated livers (Fig. 3e). This finding is consistent with the still modestly increased bile acid pool size in human hepatocyte repopulated livers. FGF19 has been found at high levels in human bile,24 a finding also observed in FRGN19+ mice (Fig. 3f). We assessed Fgf15 (RT-PCR and RNA sequencing) in the gallbladders of mice after BDL and humanization of the liver, and no measureable Fgf15 was detected.
In sum, these results indicate an uncoupling of bile acid signaling between mouse intestine and transplanted human hepatocyte resulting in upregulation of bile acid synthesis in transplanted human hepatocytes. The aberrant signaling was corrected with the FGF19 transgene.
Transcriptional and proliferative differences in human hepatocytes when transplanted into FRGN or FRGN19+ mice
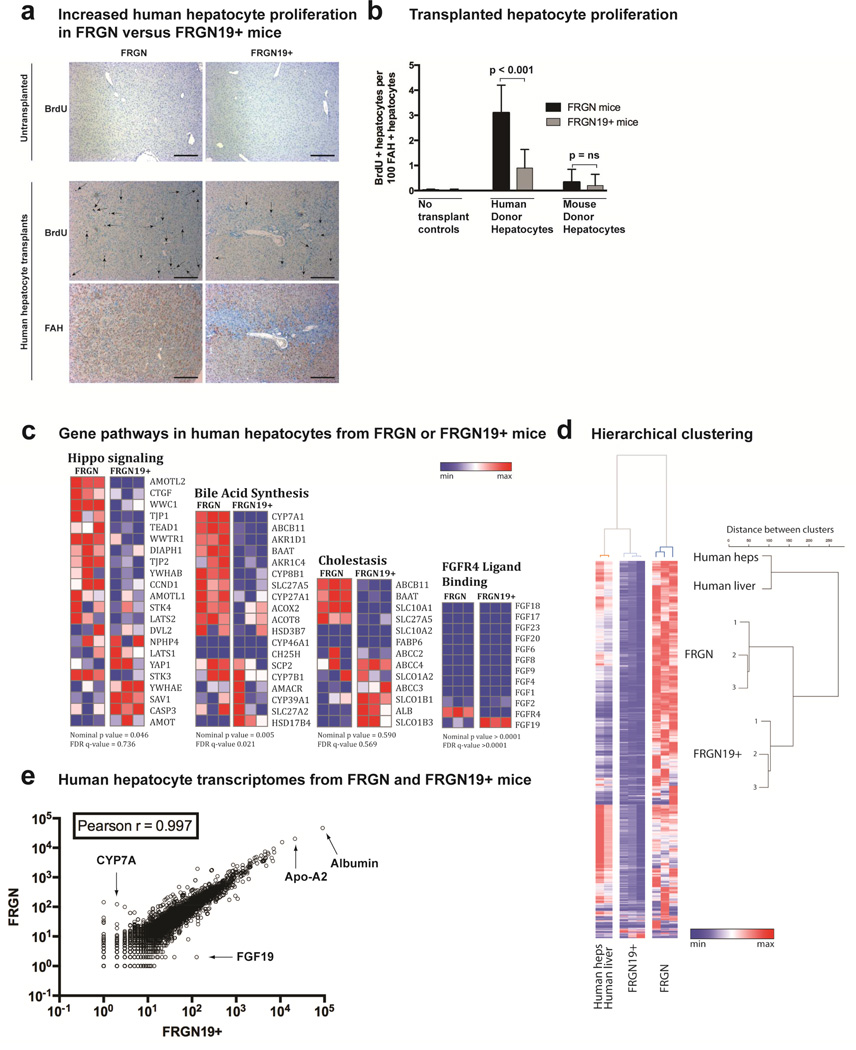
Livers of FRGN or FRGN19+ mice have nearly no hepatocyte proliferation in the quiescent state (Fig. 4a). Human hepatocyte repopulated livers, however, showed significant proliferation as shown by BrdU incorporation; the proliferation was significantly decreased in FRGN19+ mice versus their non-transgenic littermates (Fig. 4a, 4b). Activation of gene pathways for DNA replication and cell cycle are significantly increased in FRGN mice compared to FRGN19+ mice, but there was no difference in apoptosis pathways (Fig. S2). These results suggest that the larger liver size in FRGN mice with human hepatocyte repopulated livers was the result of increased hepatocyte proliferation absent increased apoptosis.
Figure 4. Transcriptional and proliferative differences in human hepatocytes when transplanted into FRGN and FRGN19+ mice.
(a) Immunohistochemistry showing BrdU or FAH staining in untransplanted or human-hepatocyte repopulated livers 4 months after transplant. Serial sections used for BrdU and FAH staining in repopulated livers, and arrows indicate BrdU positive hepatocyte nuclei in FAH + hepatocytes. Scale bar = 100 µm. (b) Quantitation of BrdU positive nuclei in FAH + hepatocytes in FRGN and FRGN19+ mice, controls (n = 3 each), human hepatocyte-transplanted FRGN (n = 4) and FRGN19+ (n = 4) mice, and mouse hepatocyte-transplanted FRGN (n = 3) and FRGN19+ (n = 3) mice. (c) Gene pathways indicated from RNA sequencing data (human transcriptome only) from liver tissue of human hepatocyte repopulated mice, either FRGN (n = 3) or FRGN19+ (n = 3) mice; statistics generated through GSEA, heatmap generated with GENEE. (d) Clustering heatmap of human transcriptomes from liver tissue of human hepatocyte repopulated FGF 19 – or + mice (n = 3 each); Ward’s unsupervised hierarchical clustering shows two distinct transcriptome clusters for single-donor human hepatocytes when transplanted into FRGN versus FRGN19+ mice. (e) Scatterplot of RNA sequencing of human hepatoctye transcriptomes (single donor) from FRGN19+ transgenic mice and their FRGN littermates are similar; arrows indicate specific genes.
The Hippo pathway is important for regulation of liver size,4, 25, 26 so it was not surprising to find activation of this pathway in the enlarged FRGN human hepatocyte repopulated mice (Fig. 4c). A recent study linked bile acids to YAP activation through a scaffolding intermediate, IQGAP1.27 RNA sequence data showed IQGAP1 to be significantly elevated in livers exposed to bile acid excess (portal vein bile acid infusion, Fig. S7), though there was no difference between the two genotypes. In human hepatocyte repopulated livers, however, IQGAP1 was elevated in FRGN compared to FRGN19+ mice, paralleling the activation of YAP genes. As expected, the transcriptiona signature for bile acid synthesis also was significantly different between the two genotypes. The FGF19 receptor FGFR4 was significantly upregulated in FRGN mice, as expected in absence of FGF19 (Fig. 4c).
Examination of other gene pathways involved in liver regeneration showed that only the FOXM1 pathway was significantly upregulated in human hepatocytes in the larger FRGN livers (Fig. S3). This is not surprising given that FOXM1 is a transcriptional target of FXR,28 and increase in FOXM1 expression increases hepatocyte proliferation,29 and suggests a potential mechanism by which increased bile acids ultimately lead to hepatocyte proliferation in vivo. Western blot of FOXM1 from fully repopulated livers showed only modest levels of expression for FRGN mice, and no significant expression in FRGN19+ mice, while normal human liver also had no significant expression (Fig. S3). Interestingly, in mice undergoing active repopulation (two months prior to full repopulation), FoxM1 protein was significantly increased in both genotypes, paralleling the increase in hepatocyte proliferation in this phase.
A recent study wherein siRNA was used to knock down Fgfr4 in the liver prior to partial hepatectomy also found Foxm1 to increase with hepatocyte proliferation, which was initially reduced in Fgfr4 knockdown mice after hepatectomy. These mice had massive hepatocyte necrosis thought due to the marked bile acid excess seen with Fgfr4 knockdown, with hepatocyte proliferation markedly attenuated at 48 hours, but significantly increased hepatocyte proliferation at 72 hours, along with elevated Foxm1 expression.30
Notably, gene sets specific for inflammatory pathways were not different between the two genotypes. The IL6 pathway (Fig. S3) was slightly (though not significantly) less activated in the FRGN19+ humanized mice, but gene sets reflecting AKT, NFκB, AP1, p38, and ERK pathways were not activated (data not shown).
Hierarchical clustering of human hepatocyte RNA transcriptomes from FRGN and FRGN19+ mice showed differential clustering, highlighting the influence of external signaling on human hepatocytes originating from a single donor (Fig. 4d). The overall transcriptomes were remarkably similar in a scatterplot of the two data sets, though CYP7A and FGF19 could clearly be distinguished as outliers (Fig. 4e). Multiple genes involved with various hepatic functions were compared (Fig. S4) in human hepatocytes from FRGN and FRGN19+ mice, mostly showing similar regulation of functions, with the exception of bile acid synthesis and transport. RNA for the membrane bile acid receptor TGR5 was not present in any of the liver samples (but was noted at high levels in the gallbladders of mice that underwent BDL).
Location of FGF19 expression in human liver
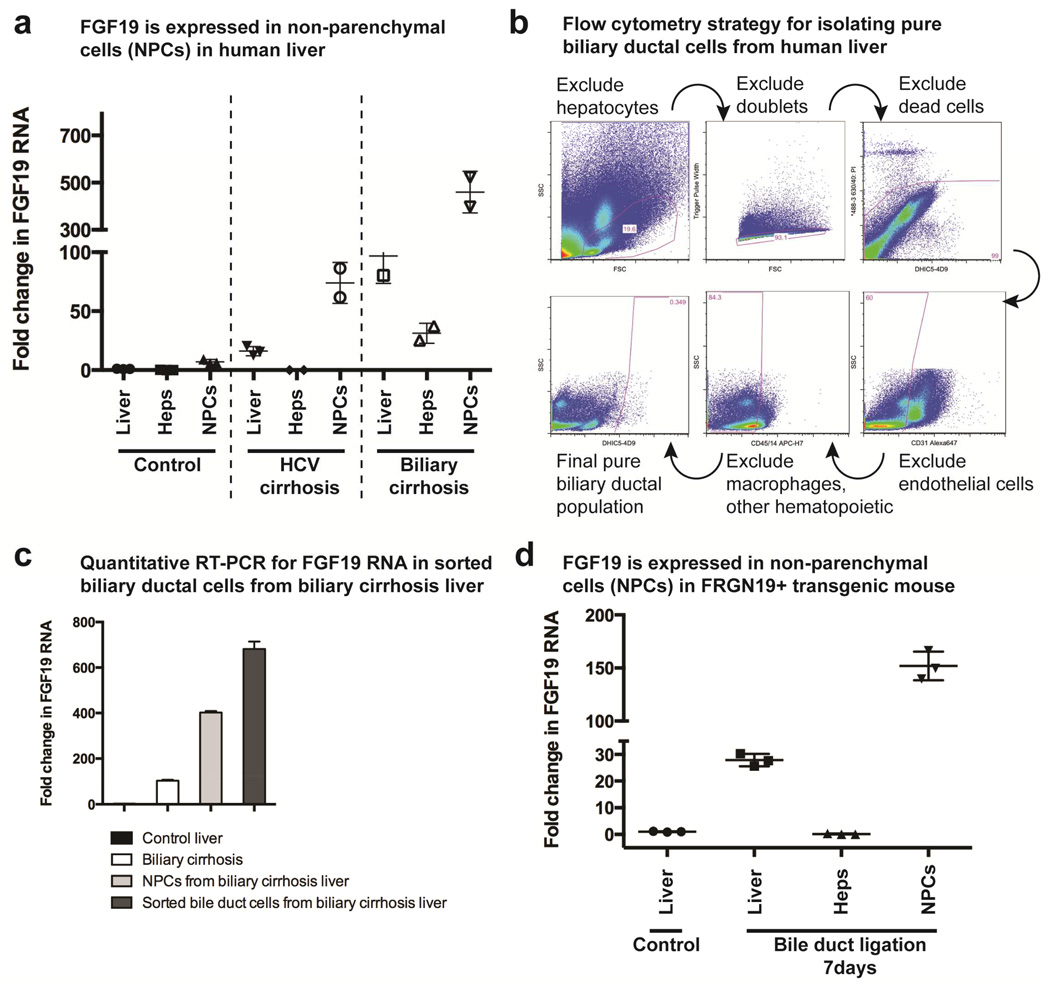
We obtained several human liver samples, and separated the hepatocytes from the non-parenchymal cell (NPC) fraction. In control liver, nearly no FGF19 RNA was detected. In 2 patients with HCV cirrhosis and 2 patients with biliary cirrhosis (explant livers), FGF19 RNA was increased, but this was found to originate in the NPC rather than hepatocyte fraction (Fig. 5a). Sorted bile duct cells from a human liver with biliary cirrhosis had the highest levels of FGF19 RNA (Fig. 5b,c). FGF19 produced in transgenic FGF19 mice (BDL, Fig. 5d) also comes from the non-parenchymal cells (NPCs), not hepatocytes. We thus conclude that FGF19 produced in the human liver in vivo likely originates not from hepatocytes but from the NPC fraction, mostly from biliary ductal cells. The FGF19 transgenic mouse described will be useful for studying human cholestatic diseases compared to mice absent this signaling mechanism.
Figure 5. Location of FGF 19 expression in human liver.
(a) Quantitative RT-PCR (qPCR) for FGF 19 was performed on whole liver tissue or specific populations of cells (hepatocytes or non-parenchymal cells, NPCs) from the same liver tissue; control humam liver, n = 3, obtained during resection of hepatic hemangioma n = 1 or adenoma n = 2, HCV cirrhotic liver and biliary cirrhosis liver (n = 2 each) obtained from explants prior to liver transplantation. (b) Flow cytometry isolated pure biliary ductal cells from an explanted human liver with biliary cirrhosis from biliary atresia. (c) Quantitative RT-PCR for FGF19 RNA done on human biliary cirrhosis liver, NPC cell fraction, and sorted bile duct cells (all from 1 liver, 3 qPCR replicates). (d) Similar FGF 19 qPCR was done in FRGN19+ transgenic mice 7 days after bile duct ligation (BDL) had been performed (n = 3 mice).
Discussion
The liver-to-body weight ratio is tightly regulated (hepatostat), but the mechanism for this homeostatic regulation of liver size remains uncertain. Signaling pathways implicated in hepatostat governance include cytokines responsible for termination of liver regeneration (TGFβ31 and others3), involvement of the extracellular matrix (e.g. integrin receptors32) and general mechanisms controlling organ size (YAP/hippo4, 25, 33). While these signaling pathways are undoubtedly involved in the complex regulation of liver size, it seems likely that performance of some metabolic function ultimately drives the necessity for a particular hepatic mass. Given the hundreds of metabolic functions the liver performs, identification of the function(s) responsible for regulating the hepatostat has been challenging.
Herein we used a mouse model where the liver was repopulated with human hepatocytes.14 We confirmed our previous findings that human hepatocyte repopulated livers are significantly larger than livers repopulated with mouse hepatocytes or control livers. We further found a marked increase in the size of the bile acid pool, driven by unregulated bile acid synthesis from transplanted human hepatocytes. In a prior study8 we hypothesized that the unregulated bile acid synthesis in this model was due to failure of human hepatocytes to recognize Fgf15, the mouse inhibitory hormone produced in the intestine, and showed that the transplanted human hepatocytes were responsive to exogenous FGF19, the human ortholog to Fgf15.34 Here we produced a transgenic mouse with genomic human DNA containing the FGF19 gene and regulatory sequences. At baseline, FRGN19+ mice behaved phenotypically the same as control mice with regard to bile acid signaling, and had similar baseline liver gene expression compared to control mice. When donor mouse hepatoctyes repopulated FRGN19+ mice livers, liver size remained the same as in controls, and there were no perturbations in bile acid pool size or signaling. When repopulating hepatocytes were human, however, FRGN19+ mice exhibited a marked normalization of bile acid synthesis and pool size compared to FRGN littermates. Importantly, this also led to reduction of liver size to near-normal expected for the mouse.
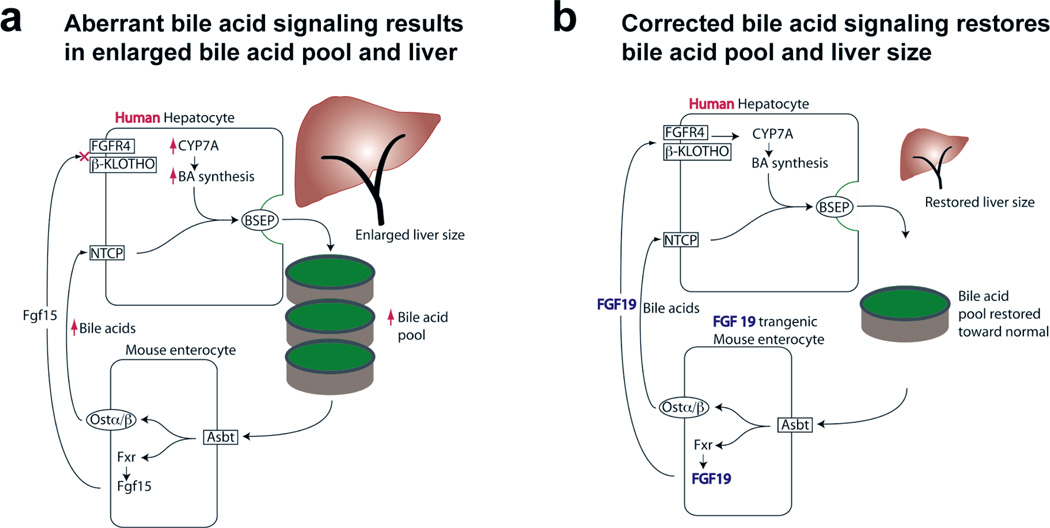
Though continually circulating, the bile acid pool is relatively fixed in size and proportional to the size of the organism (mice and rats 13 µmol/gram liver,35, 36 humans 50–60 mmol/kg body weight37). The size of the bile acid pool is controlled by concentrations in the ileum, where enterocytes reabsorb bile acids and provide negative feedback for bile acid synthesis in the liver by elaborating Fxr-dependent Fgf15 (rodent)20 or FGF19 (human). In addition, the liver has a finite capacity to secrete bile acids (e.g. maximum secretory rate for rodent liver is 120 nmol/min/gram liver35). Our data provide novel support for a model (Fig. 6) where liver growth occurs when the bile acid pool exceeds the liver’s maximum secretory capacity, and thus provides insight into a metabolic function of the liver that regulates the hepatostat. We have previously shown that mice with humanized livers have a relatively hydrophobic composition of the bile acid pool (Ellis et al), a finding reversed with administration of recombinant FGF19. Further studies are underway to understand if similar findings occur in FGF19+ transgenic mice, and if such bile composition changes may affect the hepatostat.
Figure 6. Relationship between bile acid pool size and liver size.
(a) Aberrant signaling perturbs bile acid homeostasis when transplanted human hepatocytes fail to recognize Fgf15, the mouse intestinal signal for inhibiting bile acid synthesis in the liver. This leads to an enlarged bile acid pool and consequent increase in hepatocyte mass needed to circulate the pool. (b) Introduction of the human inhibitory signal, FGF19, allows transplanted human hepatocytes to properly regulate bile acid synthesis, and the bile acid pool is normalized. Hepatocyte mass is decreased, proportional to the bile acid pool which is circulated.
Data from prior studies from mice with genetically altered bile acid signaling have produced mixed results in regard to liver size. Mice lacking Fgfr4, the primary receptor for Fgf15, have enlarged bile acid pools but normal sized livers.38 These mice, however, have an enormous daily fecal bile acid loss, and it is thus unclear if there is a significant increase in bile acids returning to the liver for circulation.39 Similarly, transgenic mice overexpressing Cyp7a1 had an enlarged bile acid pool and normal liver size,40 but hepatic bile acid concentrations were not elevated,41 suggesting the measured bile acid pool may be largely excreted in feces rather than circulated. Conversely, mice lacking Asbt, responsible for intestinal reabsorption of bile acids have smaller bile acid pools as well as significantly smaller livers than controls.42 Rats with depletion of the bile acid pool do not grow to normal size after partial hepatectomy,10, 43 and mice fed cholic acid have a modest increase in liver size.9, 18 A recent study examining mice with FXR deficiency and repletion in the intestine strongly support our conclusions.44 In this study, mice with deletion of intestinal FXR had little Fgf15, abrogating the negative regulation of bile acid synthesis in the liver, similar to our model though by a different mechanism. Consistent with our findings, these mice had enlarged bile acid pools as well as markedly enlarged livers, similar to findings presented here. Interestingly, restoration of bile acid regulation prevented the development of liver tumors otherwise seen when the bile acid pool was deranged.
Models of robust hepatocyte proliferation pose a potential for hepatocarcinogenesis. Our laboratory has been transplanting hepatocytes for repopulation in this model for many years, and has yet to uncover a liver cancer originating from transplanted hepatocytes (rat, mouse, or human). Occasionally, however, a cancer will arise from recipient Fah−/− hepatocytes in aged (> 1 year) transplanted mice.
This study has also expanded our understanding of human FGF19 signaling. While mice express Fgf15 only in the intestine,19 humans clearly produce FGF19 in the gallbladder24 and liver,18 especially under cholestatic conditions. The cell of origin for FGF19 production outside the intestine has however been unclear. Primary human hepatocytes exposed in vitro to chenodeoxycholic acid or an FXR agonist reportedly induced FGF19 mRNA,45, 46 but in vivo confirmation is lacking. Our data shows that human hepatocytes after liver repopulation in the mouse produce no FGF19 despite the condition of marked CYP7A upregulation and bile acid excess (Fig. 3b). We found that FGF19 was expressed mainly in the non-parenchymal cells in human viral hepatitis and biliary cirrhosis, and further specification by FACS sorting revealed the cell of origin to be biliary ductal cells. Some FGF19 did seem to arise from hepatocytes in the biliary cirrhosis samples, though the amount was much smaller than seen arising from biliary ductal cells in the same donor.
Mice with humanized livers have been developed over the last 20 years by taking advantage of methods to selectively injure recipient hepatocytes thus producing a regenerative milieu for transplanted hepatocytes.47 In the context of genetic immune suppression, hepatocytes from different species, including human, can be transplanted into these mouse models, and high levels of human hepatocyte repopulation attained. At least 4 mouse models have been constructed in which the recipient mouse liver can be significantly repopulated with human hepatocytes.14, 48–50 Such human hepatocyte repopulated liver mice have a growing portfolio of applications, including the study of infectious diseases, gene therapy, stem cell biology, drug metabolism, and disease modeling.47 We have shown that humanized liver mice have aberrant liver-intestine bile acid signaling, a phenomenon which is likely present in all such models. For example, human hepatocyte repopulated uPA transgenic mice had livers 3 times the size of livers repopulated by rat hepatocytes,23 but bile acids/signaling were not measured in this study. We anticipate that genetic correction (transgenic human FGF19) of bile acid signaling in this model will further extend its applicability to human diseases, especially relevant given the recent connection of Non-Alcoholic Fatty Liver Disease (NAFDL) to bile acid homeostasis and FGF19 signaling.51–53
Supplementary Material
Acknowledgments
We are grateful to Yingming Wang for skillful technical assistance in the generation of the transgenic mice.
Funding
The authors and study were funded by the NIDDK, DK083507 (WN) and DK051592 (MG).
Abbreviations used in this paper
- FAH
fumarylacetoacetate hydrolase
- FGF19
Fibroblast growth factor 19
- Fgf15
Fibroblast growth factor 15
- FGFR4
Fibroblast growth factor receptor 4
- FXR
farnesoid X receptor
- BAC
Bacterial artificial chromosome
- PV
Portal vein
- BDL
Bile duct ligation
- NPC
Non-parenchymal cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare no conflicts of interest related to this manuscript.
Author contributions
W.E.N. and M.G. are responsible for study concept and design; W.E.N. conducted the acquisition, interpretation and analysis of data, W.E.N. and M.G. are responsible for the drafting and revision of the manuscript; B.D.T. assisted in acquisition of RNA sequence data as well as critical analysis; L.M.F. was responsible for the generation of transgenic mice; C.P. handled RNA sequence data and clustering analysis; M.T. provided hepatocyte and NPC separation from human liver tissue; B.L. sorted human biliary ductal cells; and J.D. assisted with the PV bile acid infusion and BDL experiments as well as data acquisition.
References
- 1.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 2.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 3.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camargo FD, Gokhale S, Johnnidis JB, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 5.Anakk S, Watanabe M, Ochsner SA, et al. Combined deletion of Fxr and Shp in mice induces Cyp17a1 and results in juvenile onset cholestasis. J Clin Invest. 2011;121:86–95. doi: 10.1172/JCI42846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baek JY, Hur W, Wang JS, et al. Selective COX-2 inhibitor, NS-398, suppresses cellular proliferation in human hepatocellular carcinoma cell lines via cell cycle arrest. World J Gastroenterol. 2007;13:1175–1181. doi: 10.3748/wjg.v13.i8.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, Rudnick DA. Elucidating the metabolic regulation of liver regeneration. Am J Pathol. 2014;184:309–321. doi: 10.1016/j.ajpath.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis EC, Naugler WE, Parini P, et al. Mice with chimeric livers are an improved model for human lipoprotein metabolism. PLoS One. 2013;8:e78550. doi: 10.1371/journal.pone.0078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang W, Ma K, Zhang J, et al. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 10.Naugler WE. Bile Acid flux is necessary for normal liver regeneration. PLoS One. 2014;9:e97426. doi: 10.1371/journal.pone.0097426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otao R, Beppu T, Isiko T, et al. External biliary drainage and liver regeneration after major hepatectomy. Br J Surg. 2012;99:1569–1574. doi: 10.1002/bjs.8906. [DOI] [PubMed] [Google Scholar]
- 12.Uriarte I, Fernandez-Barrena MG, Monte MJ, et al. Identification of fibroblast growth factor 15 as a novel mediator of liver regeneration and its application in the prevention of post-resection liver failure in mice. Gut. 2013;62:899–910. doi: 10.1136/gutjnl-2012-302945. [DOI] [PubMed] [Google Scholar]
- 13.Pean N, Doignon I, Garcin I, et al. The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology. 2013;58:1451–1460. doi: 10.1002/hep.26463. [DOI] [PubMed] [Google Scholar]
- 14.Azuma H, Paulk N, Ranade A, et al. Robust expansion of human hepatocytes in Fah−/− /Rag2−/−/Il2rg−/− mice. Nat Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson EM, Bial J, Tarlow B, et al. Extensive double humanization of both liver and hematopoiesis in FRGN mice. Stem Cell Res. 2014;13:404–412. doi: 10.1016/j.scr.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression2 profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gould J. Volume 2014: Broad Institute; GENE-E. https:// http://www.broadinstitute.org/cancer/software/GENE-E/index.html. [Google Scholar]
- 18.Schaap FG, van der Gaag NA, Gouma DJ, et al. High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology. 2009;49:1228–1235. doi: 10.1002/hep.22771. [DOI] [PubMed] [Google Scholar]
- 19.Fon Tacer K, Bookout AL, Ding X, et al. Research resource: Comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol Endocrinol. 2010;24:2050–2064. doi: 10.1210/me.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inagaki T, Choi M, Moschetta A, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Nishimura T, Utsunomiya Y, Hoshikawa M, et al. Structure and expression of a novel human FGF, FGF-19, expressed in the fetal brain. Biochim Biophys Acta. 1999;1444:148–151. doi: 10.1016/s0167-4781(98)00255-3. [DOI] [PubMed] [Google Scholar]
- 22.Modica S, Petruzzelli M, Bellafante E, et al. Selective activation of nuclear bile acid receptor FXR in the intestine protects mice against cholestasis. Gastroenterology. 2012;142:355–365. e1–e4. doi: 10.1053/j.gastro.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 23.Utoh R, Tateno C, Kataoka M, et al. Hepatic hyperplasia associated with discordant xenogeneic parenchymal-nonparenchymal interactions in human hepatocyte-repopulated mice. Am J Pathol. 2010;177:654–665. doi: 10.2353/ajpath.2010.090430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zweers SJ, Booij KA, Komuta M, et al. The human gallbladder secretes fibroblast growth factor 19 into bile: towards defining the role of fibroblast growth factor 19 in the enterobiliary tract. Hepatology. 2012;55:575–583. doi: 10.1002/hep.24702. [DOI] [PubMed] [Google Scholar]
- 25.Dong J, Feldmann G, Huang J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yimlamai D, Christodoulou C, Galli GG, et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anakk S, Bhosale M, Schmidt VA, et al. Bile acids activate YAP to promote liver carcinogenesis. Cell Rep. 2013;5:1060–1069. doi: 10.1016/j.celrep.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen WD, Wang YD, Zhang L, et al. Farnesoid X receptor alleviates age-related proliferation defects in regenerating mouse livers by activating forkhead box m1b transcription. Hepatology. 2010;51:953–962. doi: 10.1002/hep.23390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiang D, Liu CC, Wang MJ, et al. Non-viral FoxM1 gene delivery to hepatocytes enhances liver repopulation. Cell Death Dis. 2014;5:e1252. doi: 10.1038/cddis.2014.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padrissa-Altes S, Bachofner M, Bogorad RL, et al. Control of hepatocyte proliferation and survival by Fgf receptors is essential for liver regeneration in mice. Gut. 2014 doi: 10.1136/gutjnl-2014-307874. [DOI] [PubMed] [Google Scholar]
- 31.Russell WE, Coffey RJ, Jr, Ouellette AJ, et al. Type beta transforming growth factor reversibly inhibits the early proliferative response to partial hepatectomy in the rat. Proc Natl Acad Sci U S A. 1988;85:5126–5130. doi: 10.1073/pnas.85.14.5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Apte U, Gkretsi V, Bowen WC, et al. Enhanced liver regeneration following changes induced by hepatocyte-specific genetic ablation of integrin-linked kinase. Hepatology. 2009;50:844–851. doi: 10.1002/hep.23059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu L, Li Y, Kim SM, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright TJ, Ladher R, McWhirter J, et al. Mouse FGF15 is the ortholog of human and chick FGF19, but is not uniquely required for otic induction. Dev Biol. 2004;269:264–275. doi: 10.1016/j.ydbio.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 35.Sainz GR, Monte MJ, Barbero ER, et al. Bile secretion by the rat liver during synchronized regeneration. Int J Exp Pathol. 1997;78:109–116. doi: 10.1046/j.1365-2613.1997.d01-246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarz M, Russell DW, Dietschy JM, et al. Marked reduction in bile acid synthesis in cholesterol 7alpha-hydroxylase-deficient mice does not lead to diminished tissue cholesterol turnover or to hypercholesterolemia. J Lipid Res. 1998;39:1833–1843. [PubMed] [Google Scholar]
- 37.Jansen PL, Sturm E, Muller M. Disorders of Bile Acid Transport. In: Trauner M, Jansen PL, editors. Molecular Pathogenesis of Cholestasis. New York: Kluwer Academic/Plenum; 2004. pp. 170–185. [Google Scholar]
- 38.Yu C, Wang F, Kan M, et al. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem. 2000;275:15482–15489. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]
- 39.Xue W, Chen S, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyake JH, Doung XD, Strauss W, et al. Increased production of apolipoprotein B-containing lipoproteins in the absence of hyperlipidemia in transgenic mice expressing cholesterol 7alpha-hydroxylase. J Biol Chem. 2001;276:23304–23311. doi: 10.1074/jbc.M101853200. [DOI] [PubMed] [Google Scholar]
- 41.Li T, Owsley E, Matozel M, et al. Transgenic expression of cholesterol 7alpha-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology. 2010;52:678–690. doi: 10.1002/hep.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung D, Inagaki T, Gerard RD, et al. FXR agonists and FGF15 reduce fecal bile acid excretion in a mouse model of bile acid malabsorption. J Lipid Res. 2007;48:2693–2700. doi: 10.1194/jlr.M700351-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Dong X, Zhao H, Ma X, et al. Reduction in bile acid pool causes delayed liver regeneration accompanied by down-regulated expression of FXR and c-Jun mRNA in rats. J Huazhong Univ Sci Technolog Med Sci. 2010;30:55–60. doi: 10.1007/s11596-010-0110-8. [DOI] [PubMed] [Google Scholar]
- 44.Degirolamo C, Modica S, Vacca M, et al. Prevention of spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice by intestinal-specific farnesoid X receptor reactivation. Hepatology. 2015;61:161–170. doi: 10.1002/hep.27274. [DOI] [PubMed] [Google Scholar]
- 45.Holt JA, Luo G, Billin AN, et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song KH, Li T, Owsley E, et al. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49:297–305. doi: 10.1002/hep.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grompe M, Strom S. Mice with human livers. Gastroenterology. 2013;145:1209–1214. doi: 10.1053/j.gastro.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Rhim JA, Sandgren EP, Degen JL, et al. Replacement of diseased mouse liver by hepatic cell transplantation. Science. 1994;263:1149–1152. doi: 10.1126/science.8108734. [DOI] [PubMed] [Google Scholar]
- 49.Hasegawa M, Kawai K, Mitsui T, et al. The reconstituted 'humanized liver' in TK-NOG mice is mature and functional. Biochem Biophys Res Commun. 2011;405:405–410. doi: 10.1016/j.bbrc.2011.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Washburn ML, Bility MT, Zhang L, et al. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology. 2011;140:1334–1344. doi: 10.1053/j.gastro.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wojcik M, Janus D, Dolezal-Oltarzewska K, et al. A decrease in fasting FGF19 levels is associated with the development of non-alcoholic fatty liver disease in obese adolescents. J Pediatr Endocrinol Metab. 2012;25:1089–1093. doi: 10.1515/jpem-2012-0253. [DOI] [PubMed] [Google Scholar]
- 52.Schreuder TC, Marsman HA, Lenicek M, et al. The hepatic response to FGF19 is impaired in patients with nonalcoholic fatty liver disease and insulin resistance. Am J Physiol Gastrointest Liver Physiol. 2010;298:G440–G445. doi: 10.1152/ajpgi.00322.2009. [DOI] [PubMed] [Google Scholar]
- 53.Mudaliar S, Henry RR, Sanyal AJ, et al. Efficacy and safety of the farnesoid X receptor agonist obeticholic acid in patients with type 2 diabetes and nonalcoholic fatty liver disease. Gastroenterology. 2013;145:574–582. e1. doi: 10.1053/j.gastro.2013.05.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.