Abstract
Genome-wide association studies of inflammatory bowel disease have identified several risk loci in genes that regulate autophagy, and studies have provided insight into the functional effects of these polymorphisms. We review the mechanisms by which autophagy contributes to intestinal homeostasis, focusing on its cell type-specific roles in regulating gut ecology, restricting pathogenic bacteria, and controlling inflammation. Based on this information, we are beginning to understand how alterations in autophagy can contribute to intestinal inflammation.
Keywords: IBD, pathogenesis, cell biology, genetics
Introduction
Autophagy is an essential cellular process for the maintenance of cellular and tissue homeostasis. Under basal conditions, damaged cellular components destined for removal are targeted by a variety of protein adaptors, which engage autophagy machinery to engulf and deliver cargo to the lysosome for degradation. Autophagy is induced under conditions of nutrient starvation, liberating energy stores and promoting cellular survival. Defects in autophagy have been associated with multiple human diseases including inflammatory bowel diseases (IBD), nonalcoholic steatohepatitis, pancreatitis, and several types of gastrointestinal cancers, reflecting the fundamental role of autophagy in supporting normal physiology.1
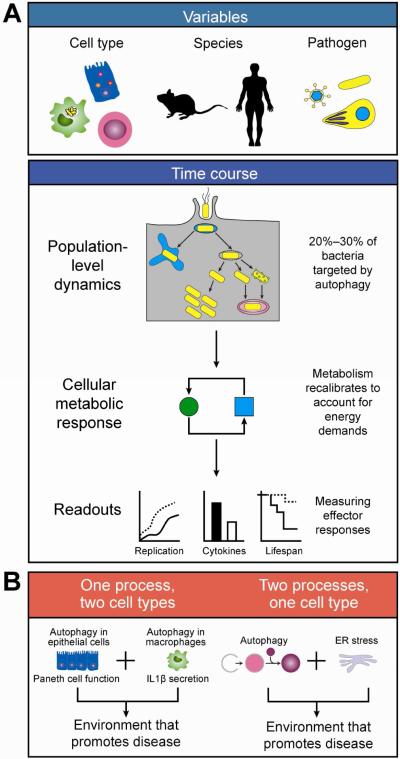
In the context of IBD, genome-wide association studies have identified variants in autophagy genes ATG16L1 and IRGM, as risk alleles for Crohn's disease (CD),2-4 with subsequent studies suggesting that these risk variants function to reduce selective autophagy but not basal autophagy.2, 3, 5 Both genes have been shown to play roles as restriction factors for pathogens through autophagy. These studies have employed the common approach of studying pathogenic bacteria to gain insight to the role of autophagy in microbial defense, although dysbiosis of commensal microbes more likely contributes to IBD pathogenesis. In fact, host genetics, including variants in ATG16L1 and NOD2, can influence the composition of the gut microbiome in some but not all studies.6, 7,8 Nonetheless, studies of pathogens in the context of gut immunity have provided an important framework for understanding the interactions between autophagy and microbes. It is important to recognize that interpretation of results related to pathogen-induced autophagy must take into consideration the cell type, host species, pathogen, and time point being studied, as these factors can have profound impacts on phenotypic readouts (Figure 1).
Figure 1. Challenges to understanding pathogen-induced autophagy.
(A) When studying autophagy triggered by pathogen infection, a number of variables must be considered, with cell type, host species, and pathogen biology of primary importance (top). Differences in cell type will determine which E3 ligases function in ubiquitination of autophagy substrates (e.g., LRSAM1 in epithelial cells, SMURF1 in macrophages). Species context is important to consider, as several autophagy proteins are not conserved between mice and humans (e.g., IRGM, NDP52). Lastly, pathogen biology is relevant as different pathogens (e.g., virus, bacteria, parasites) have different intracellular lifestyles that impact different cellular pathways. In assessing the impact of pathogen-related autophagy factors (bottom), one must take into account both population-level dynamics well as cellular metabolic responses. As only a small percentage of pathogens are targeted for autophagy, a strong phenotype may translate to a subtle effect at the population level. Hallmarks of infection will induce cellular metabolic recalibration, so the amount of time an infection is allowed to proceed must also be considered. In the context of these complex dynamics, phenotypic readouts of both host and pathogen effects may be subtle. (B) To understand how perturbations to a single factor may lead to disease, one may consider “multiple hits” models. In the first model, a single factor has a function that impacts multiple cellular pathways (e.g,. autophagy and ER stress). In the second model, a single factor plays a role in multiple cell types (e.g., Paneth cells and macrophages).
Autophagy consists of four steps: initiation, nucleation, elongation, and fusion. These complex steps require the coordinated action of core autophagy-related (ATG) proteins. During initiation, the cell recognizes the need for removal of particular cellular constituents. This process is mediated by a series of phosphorylation and dephosphorylation events by a complex containing ULK1, Atg13, and FIP200.9 Nucleation involves recruitment of autophagy proteins to the pre-autophagosomal structure, an event that is coordinated by beclin 1 in a complex with class III phosphatidylinositol-3-OH kinase (PI(3)K), Vps34, and several other proteins, followed by formation of an isolation membrane.10 During elongation, two ubiquitin-like conjugation systems facilitate elongation of the isolation membrane to encompass the items targeted for degradation. The first pathway culminates in the conjugation of Atg5 to Atg12, while the second pathway acts to lipidate LC3-I via conjugation to phosphatidylethanolamine, thereby generating LC3-II; Atg7 is required for both pathways.10 The Atg5-Atg12 complex binds to Atg16l1, facilitating recruitment of autophagy machinery to sites of autophagosome formation.1, 10 Fusion refers to the docking and fusion of a fully formed autophagosome to a lysosome, resulting in delivery of lytic enzymes to the newly formed autolysosome. This fusion occurs through the action of a group of proteins including the HOPS complex, syntaxin 17, SNAP-29, and R SNARE protein VAMP8 or VAMP7, all of which are required for this process.11-13
Multiple adaptor proteins mediate cargo selection and targeting to autophagosomes. Several cargo adaptors simultaneously associate with intracellular bacteria or their surrounding membranes along with lipidated LC3, thereby mediating recruitment of autophagosomes. Antibacterial autophagy can be triggered directly by recognition of bacterial pathogens, but also by recognition of hallmarks of pathogen invasion including membrane damage, membrane remnants, amino acid starvation, cytosolic protein aggregate formation, and the presence of bacterial DNA. This recognition process can be ubiquitin-dependent or -independent. In ubiquitin-dependent pathways, NDP52, SQSTM1/p62, and OPTN act as adaptors to target ubiquitinated bacteria for autophagic destruction, with E3 ligases mediating ubiquitination of cargo in cell type-specific contexts. Ubiquitin-independent recognition of damaged membranes by galectin 8 can also target bacteria for destruction through autophagy.14 Several adaptor proteins have been recently identified that mediate cargo selection for autophagy induced by a number of cellular stresses including DNA damage, hypoxia, redox stress, endoplasmic reticulum (ER) stress, and mitochondrial damage.15 Future studies will determine how core autophagy proteins interact with effectors of selective autophagy in a time-dependent and stimulus-dependent manner. The timeline of autophagy induction likely defines the mechanism by which autophagy is regulated: although post-transcriptional mechanisms control acute activation of autophagy, sustained autophagy is regulated by a network of transcription factors including transcription factor EB (TFEB), farnesoid X receptor (FXR), peroxisome proliferator-activated receptor-a (PPARα), and cAMP response element-binding protein (CREB).16-18 TFEB has been shown to play a pivotal role in pathogen defense through induction of autophagy at the transcriptional level highlighting the importance of autophagy in innate defense pathways.16
Cell type-specific roles of autophagy
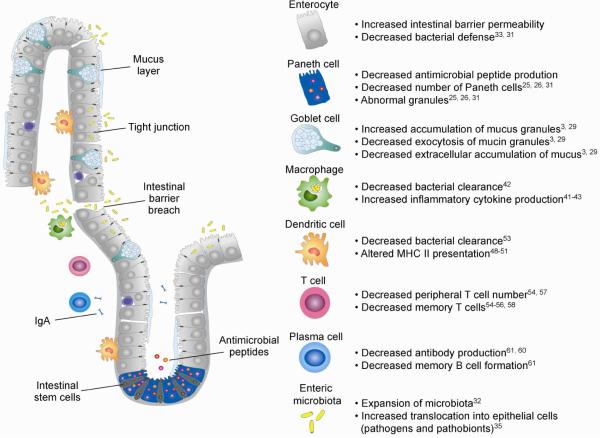
The gut can be viewed as an organ system with the physiological challenge of absorbing nutrients while maintaining a delicate balance between defense and tolerance against high microbial loads. This balance is disturbed in IBD, a chronic inflammation of the intestine. Evidence suggests that pathogenesis involves altered intestinal homeostasis that results in inappropriate host immune responses to commensal flora at the mucosal barrier. This barrier has several layers of defense, including specialized functions of multiple epithelial cell types, tight junctions that maintain paracellular permeability, and rapid cellular turnover mediated by the intestinal stem cell niche.19 Phagocytic cells reside below the epithelial layer to perform surveillance and elimination of any microbiota that pass through breaches in the epithelium. In IBD, altered barrier permeability leads to downstream events that culminate in an imbalance between defense and tolerance in interactions with the intestinal microbiota (Figure 2).
Figure 2. Defective intestinal homeostasis associated with perturbations in the autophagy system.
Perturbations to the autophagy system result in breakdown of multiple aspects of innate and adaptive immunity that support the maintenance of intestinal homeostasis. This schematic illustrates the state of the intestinal epithelium in the context of perturbations to the autophagy system. Changes include a defective mucus layer, decreased AMP production, increased paracellular permeability resulting in increased bacterial translocation, expansion of the microbiota, defective antigen presentation and adaptive immune cell priming, and decreased IgA production. Crosstalk between autophagy and immune cells supports intestinal homeostasis, and breakdowns in this communication are associated with defective intestinal barrier function. References included in the right panel list primary literature further detailing specific changes in crosstalk associated with changes in autophagy.
Intestinal Epithelial Cells
Paneth cells are a well-established example of the intersection between mucosal defense, autophagy, and IBD. These cells reside at the base of the crypts of Lieberkuhn in the small intestine, where they secrete antimicrobial peptides (AMPs) to control the intestinal microbiota.20 Demonstrating the importance of these factors in maintaining intestinal homeostasis, decreased expression of human defensins alpha 5 and 6 (DEFA5 and DEFA6, also known as HD-5 and HD-6) have been associated with ileal CD.21 Some AMPs, including HD-6 and Reg3γ (and its human orthologue REG3α), also provide physical protective barriers to the epithelium.22, 23 Reg3γ is a soluble C-type lectin that specifically targets gram-positive bacteria to maintain an approximately 50-μm zone between the intestinal microbiota and the intestinal epithelial cell surface, and loss of this spatial segregation leads to increased bacterial colonization of the intestinal epithelial surface.24
Evidence for a role of autophagy in Paneth cell function originated from studies of ATG16L1 and the CD-associated allele ATG16L1 T300A. This protein plays a key role in Paneth cell function, and studies of hypomorphic mice and CD-associated alleles of ATG16L1 exhibit significant abnormalities in Paneth cells, including fewer and profoundly disorganized granules and diffuse lysozyme staining, and transcriptional profiling of these cells revealed an increase in inflammatory mediators.25 More recent studies of knock-in mice and patients expressing ATG16L1 T300A have demonstrated similar defects in Paneth cell number, granule structure, secretion, and organoid assembly.3, 5 These Paneth cell phenotypes are not unique to studies of ATG16L1, as deletion of Atg5, Atg7 or Atg4B leads to similar Paneth cell abnormalities.26, 27 Taken together, these data demonstrate a critical role for autophagy in the generation, packaging, and secretion of Paneth cell granules into the crypt lumen. Given the critical importance of AMPs in the maintenance of proper microbial ecology in the intestine,28 a link may be drawn between perturbations in autophagy and the microbial dysbiosis associated with IBD.
Another cell type that illustrates connections between autophagy and IBD, goblet cells are primarily responsible for generation and maintenance of the mucus layer as the first line of defense for the epithelial barrier. These cells secrete mucins, Fc-gamma binding protein (Fcgbp), and trefoil factors. MUC2 is the principal structural unit of the mucus layer, Fcgbp functions as a crosslinker to stabilize the MUC2 structure, and trefoil factors are involved in mucosal defense. Knockout mouse studies of Atg5, Atg7, and Lc3b have demonstrated a common role for these genes in regulating goblet cell mucin secretion, likely through regulation of reactive oxygen species production.29 In mice expressing ATG16L1 T300A, goblet cells exhibit blunted versions of the phenotypes associated with complete knockout of core autophagy genes. Notably, goblet cells in ATG16L1 T300A mice are enlarged with accumulated mucin granules in the epithelial cuff, although goblet cells in the intestinal crypts do not show these defects.3
Absorptive enterocytes, which make up the majority of intestinal epithelial cells, are responsible for forming a barrier that prevents translocation of bacteria while absorbing nutrients from ingested food. In these cells, autophagy operates as part of the cell-intrinsic innate immunity program to restrict bacterial replication and dissemination. Defects in the restriction of bacterial replication have been linked to IBD, as proteins involved in autophagic bacterial clearance have decreased expression in epithelial cells from patients with active IBD.30 Studies to date have used Salmonella as a model of infectious bacteria that are susceptible to autophagy. Mice with epithelial cell-specific deletion of Atg16l1 (Atg16l1ΔIEC) or Atg5 (Atg5ΔIEC) showed significant defects in their ability to target Salmonella for autophagy, resulting in greater sensitivity to Salmonella infection and increased bacterial dissemination to other tissues in these mice, highlighting the importance of bacterial restriction in the epithelium.31, 32 Infection of germ-free mice with Enterococcus faecalis, an opportunistically invasive pathobiont, also resulted in bacterial targeting to autophagy, suggesting a role for autophagy in restriction of invasive intestinal microbiota under certain conditions. Studies of another gram-negative pathogen, adherent-invasive Escherichia coli (AIEC), an opportunistic pathogen in IBD, have demonstrated the importance of autophagy for proper handling of AIEC in intestinal epithelial cells.33 Notably, AIEC survival in multiple cell types has been linked to increased intestinal inflammation: ATG5, SQSTM1/p62, and HIF-1α, a hypoxic transcription factor and inducer of autophagy, are all required for autophagy of AIEC in vitro. Further studies will be required to determine whether commensal microbes induce autophagy and to assess the impact of autophagy on restriction of intestinal microbiota in IBD patients. IgA-SEQ (a flow-cytometry-based bacterial cell sorting combined with 16S rRNA sequencing to characterize taxa-specific IgA) has resulted in identification of inflammatory commensal bacteria that preferentially exacerbate intestinal disease.34 Further studies utilizing IgA-SEQ combined with microbial reconstitution of T300A knock-in mice will facilitate evaluation of the impact of disease-associated polymorphisms on restriction of inflammatory microbiota.34 Epithelial barrier permeability and prevention of bacterial translocation is maintained by tight junctions. One recent report showed that starvation-induced autophagy reduced permeability of the intestinal epithelium, suggesting a role for autophagy in maintenance of barrier function under stress conditions.35
The absorptive function of enterocytes is mediated by microvilli, microscopic membrane protrusions that increase surface area of the apical side of these cells. A key role for autophagy in microvillus function is suggested by microvillus inclusion disease, a disease involving the shortening of microvilli that is thought to be caused by increased autophagy of the apical membrane of enterocytes. Although the role of autophagy in the development and maintenance of microvilli remains unclear, parallels may be drawn with recent studies uncovering a role for autophagy in ciliogenesis.36-38
Antigen-Presenting Cells
The gut hosts the largest pool of resident macrophages, which play important roles in immune surveillance and bacterial clearance. Intestinal macrophages constitutively clear microorganisms and cellular debris, and promote epithelial renewal and regulatory T cell expansion by secreting PGE2 and IL-10, respectively.39, 40 Several studies have highlighted the role of autophagy in intestinal macrophages for clearance of fungi, bacteria, and parasites. For example, autophagic degradation of A20, an NF-κB inhibitor, is required for macrophage expression of neutrophil-recruiting cytokines needed for the clearance of Candida albicans.41 Moreover, studies of both Pseudomonas aeruginosa and the parasite Toxoplasma gondii have highlighted respective roles for Atg7 and Atg5 in pathogen clearance by macrophages.42, 43 These studies illustrate important roles of autophagy in the proper surveillance and destruction of microbial pathogens.
Unlike in other tissues, macrophages in the intestine show little inflammatory response to bacterial ingestion or Toll-like receptor engagement, thereby limiting excessive inflammation despite chronic exposure to microbial stimuli.44 Aberrant macrophage responses to commensal bacteria are implicated in the pathogenesis of IBD, suggesting that this hypo-responsiveness is critical to maintaining intestinal homeostasis. During colitis, macrophage populations can become skewed toward pro-inflammatory subsets. Monocytes expressing the CD-associated risk variant ATG16L1 T300A exhibit increased IL-1β production, suggesting that autophagy plays a role in preventing skewing towards pro-inflammatory macrophages in the intestine.3
As with macrophages, the ability of dendritic cells to initiate appropriate responses to microbes has important implications for maintenance of intestinal health. This function is achieved by antigen uptake and transport to mesenteric lymph nodes for presentation. In this context, autophagy is important for efficient MHC-I and MHC-II presentation by dendritic cells as well as antigen-specific CD4+ T cell priming.45-48 In a study of peptide citrullination, a post-translational modification implicated in several autoimmune diseases,49 chemical or genetic inhibition of autophagy blocked citrullination of peptides presented by dendritic cells on MHC-II, suggesting that presentation of citrullinated peptides is a result of an autophagy response.50 In a mouse model of graft versus host disease, deletion of Atg16l1 resulted in global dendritic cell activation, leading to increased T cell stimulation.51 On the other hand, dendritic cells lacking Atg5 were unable to induce the differentiation of cytokine-producing CD4+ T cells during T. gondii infection.52 Furthermore, dendritic cells from CD patients carrying risk variants in NOD2 and ATG16L1 showed defects in autophagy induction and MHC-II antigen presentation.53 Future studies are needed to determine how autophagy in dendritic cells expressing IBD risk variants alters the T cell repertoire and what implications this has for chronic inflammation and disease.
Lymphocytes
Effector T cells have a profound impact on the maintenance of a healthy intestinal environment. Alterations in T cell repertoire have been demonstrated in IBD, including increased Th1 and Th17 cells in CD. Autophagy plays an important role in several T cell-specific functions, including development, polarization, and memory.51, 54-58
Autophagy functions at each stage of T cell development, including hematopoetic stem cells, common lymphoid precursors, thymocytes, and effector and memory T cells.55 Conditional knockout of Atg5 and Atg7 in T cells resulted in lower numbers of thymic and peripheral T cells due to defects in mitophagy.57 Deletion of Atg5 or Atg7 during T cell development resulted in fewer peripheral T cells and less proliferative activity after T cell receptor engagement.58 Lastly, deletion of Atg5 or Atg7 also compromised the formation of memory CD8+ T cells after viral challenge.55, 58 Interestingly, defects in autophagy observed in T cells from aged mice could be reversed through addition of the compound spermidine, suggesting a potential avenue for immunomodulation to enhance T cell responses in the elderly.55 These reports suggest a model in which autophagy can influence the inflammatory state of the intestinal environment by inducing differential proor anti-inflammatory T cell effector populations as well as by modulating the functionality and memory of effector T cells. Further studies will be required to determine the specific T cell populations associated with IBD risk variants in autophagy genes.
The predominant B cell population in the intestinal epithelium is composed of IgA-secreting plasma cells. IgA levels have been linked to IBD, as IBD patients show a decrease in secreted IgA in intestinal tissues. IgA is a critical component of the first-line defense in the intestine by blocking pathogen adherence, coating bacteria to facilitate clearance, directly inhibiting pathogen virulence, and controlling the intestinal microbiota.59 Thus, a decrease in the presence of IgA can lead to aberrant immune responses by other cell types. Studies of B cell-specific knockout of Atg5 have shown that Atg5 is required for maintenance of plasma cell homeostasis and viability,60 and for generating antibody responses during antigen-specific stimulation, parasite infection, and mucosal inflammation.61
Many of the studies described above that examine the role of autophagy in specialized intestinal cells have utilized deletion of core autophagy genes rather than disease-associated polymorphisms as models. Nonetheless, cell-specific knockout studies have uncovered unanticipated patterns in autophagy gene function, including phenotypic similarities in function of core autophagy genes Atg5 and Atg7 in epithelial cells but differential effects in immune cells.52
Autophagy in the maintenance of intestinal homeostasis
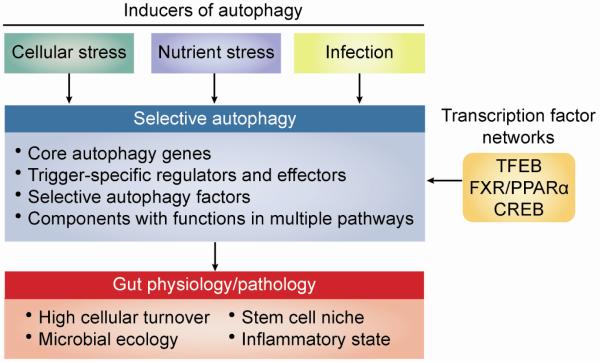
Autophagy is induced by internal and external environmental cues to orchestrate cell-intrinsic functions and survival strategies under conditions of cellular stress (Figure 3). Core autophagy proteins interact positively and negatively in pathways regulating cellular output related to metabolic sensors, quality control, and danger signals. Together these functions of autophagy promote nutrient utilization in the intestinal epithelium while controlling cellular responses to block overactive inflammation.
Figure 3. Framework for building a gut-autophagy interactome.
To understand the multiple pathways involved in maintenance of gut homeostasis, one must consider a number of pathways that converge to determine healthy or disease states in this organ system. Multiple cellular, nutrient, and pathogenic stressors induce autophagy. At the same time, transcriptional regulation affects expression of multiple factors involved in these pathways. Stressors and transcription converge on the selective autophagy pathway. This pathway is modulated by the actions of well-studied core autophagy genes. Additional factors, many of which have yet to be identified, include trigger-specific regulators and effectors of autophagy, selective autophagy factors, and components that function in multiple pathways that can be co-opted by invading pathogens. Together the actions of these proteins have profound effects on the cellular turnover, microbial ecology, stem cell niche, and inflammatory state in physiology and disease states that determine the homeostatic properties of the gut.
Metabolic sensors allow cells to monitor the presence of environmental macro-and micronutrients and adjust cellular metabolic pathways accordingly. Autophagy has been implicated in both nutrient sensing and turnover. In one recent paper, Efeyan et al. suggested a role for Rag GTPases as “multi-input nutrient sensors” that signal levels of glucose and amino acids upstream of mTORC1, the central cellular regulator of signaling in response to nutrient levels. In this model, decreased levels of glucose and amino acids lead to inhibition of mTORC1, resulting in the induction of autophagy to generate amino acids that are then utilized to maintain adequate glucose concentrations.62 Two recent studies have suggested that other micronutrients, such as iron, can be recycled by autophagy.63, 64 In these studies, NCOA4 was identified as an autophagy adaptor that colocalized with autolysosomes and bound ferritin, targeting ferritin to the autophagy pathway during starvation or iron depletion in a process called ferritinophagy.63, 64 The role of autophagy in iron homeostasis also has implications for pathogen defense, as bacteria scavenge iron from infected host cells and a “molecular arms race” has been posited in the evolution of host and bacterial iron-binding proteins.65 Autophagy has also been demonstrated to mediate recycling of lipids in the form of lipid droplets, as will be discussed below.
Nutrient processing and availability are influenced by the intestinal microbiota, which process dietary proteins to produce metabolites including short-chain fatty acids and succinate that influence intestinal health.66 IBD is characterized by a microbial dysbiosis with decreased populations of obligate anaerobes and increased populations of facultative anaerobes. The cause of this dysbiosis may be linked to changes in the oxygen levels in the intestine, a hypothesis supported by recent findings showing that intestinal oxygenation directly influences intestinal microbial ecology.67 The link between autophagy and modulation of the intestinal microbiota therefore suggests a connection between autophagy and nutrient processing by intestinal microbes.
In addition to the role of autophagy in response to bacterial infection, autophagy contributes to the modulation of cellular output to cope with a number of other cellular stressors including buildup of protein aggregates, organelle damage, hypoxic stress, and ER stress. In the context of IBD, patients show increased hypoxic stress and inflammatory cytokines in mucosal tissues.68 One study found that hypoxic stress in macrophages from damaged mucosa of IBD patients exhibited increased expression of HIF-1α and Wnt1. Expression of Wnt1 in macrophages initiated a Wnt signaling cascade in epithelial cells, resulting in activation of mTOR and inhibition of autophagy.69 These results demonstrate how an environmental stress characteristic of IBD can result in intercellular signaling cascades that inhibit autophagy in epithelial cells, resulting in loss of function of the epithelial barrier. ER stress is high at baseline in a number of secretory cell types in the intestinal epithelium, including Paneth cells and goblet cells,70 and has been implicated as a cause of intestinal inflammation in mouse studies of Xbp1, 71 In a recent study examining the regulation of the ER stress response, loss of the unfolded protein response and loss of autophagy were shown to reciprocally engage one another. Interestingly, loss of both pathways (Atg7/Xbp1ΔIEC or Atg16l1/Xbp1ΔIEC) resulted in intestinal inflammation in mice, highlighting the potential role for autophagy as a mechanism to cope with ER stress in the intestinal environment.72 Other studies have also suggested a mechanism by which autophagy limits inflammation-associated cellular injury by preventing the induction of apoptosis through intracellular HMGB1.73
Thus it is clear that autophagy plays a central role in the ability of cells to respond to and cope with their environment, highlighting how a breakdown in this process can lead to the pathophysiology associated with IBD.
Function of IBD risk variants in disease
Among complex genetic diseases, IBD has been a success story for genome-wide association studies. One of the first identified IBD risk alleles was ATG16L1 T300A.74 As discussed above, studies of the CD-associated ATG16L1 risk allele have yielded insight into the importance of Paneth cells in disease. Two recent reports have shed light on the mechanism by which ATG16L1 T300A results in defects in antibacterial autophagy by identifying a caspase cleavage site immediately preceding the T300A polymorphism.3, 5 ATG16L1 T300A is more susceptible to caspase cleavage than ATG16L1 T300T, resulting in an approximately 50% decrease in ATG16L1 protein level and consequent defects in bacterial targeting to autophagy and increased expression of IL-1β,3, 5 although efforts to target IL-1β to treat IBD have not been successful. More recent studies have uncovered a link between endogenous microbiota and the ATG16L1 T300A risk allele: patients homozygous for the risk allele had increased numbers of Fusobacteriaceae, while those homozygous for the non-risk allele had fewer Bacteroidaceae and Enterobacteriaceae, but increased Lachnospiraceae.6 Investigations into the intersection of ATG16L1 and the microbiome in IBD have also demonstrated a link to vitamin D. Conditional deletion of the vitamin D receptor resulted in increased E. coli and Bacteroides and decreased n-butyrate-producing microbes in the intestines of VDRΔIEC mice as well as increased susceptibility to colitis.75 VDRΔIEC mice exhibited an increase in abnormal Paneth cells and decreased expression of ATG16L1 and lysozyme.75 Addition of the bacterial product butyrate in the context of this mouse model ameliorated the development of colitis, suggesting that the alterations in microbiota were directly linked to the development of colitis.
IRGM, another gene with important roles in autophagy, was also identified in a genome-wide association study for CD,76 although no such association has been identified for ulcerative colitis. A linkage disequilibrium analysis of SNP data revealed a large deletion upstream of the IRGM risk allele;77 this deletion leads to decreased IRGM expression and decreased antibacterial autophagy. Additional insight on this genetic variant was generated by a report that revealed that a synonymous mutation within IRGM changes a binding site for the microRNA miR-196.2 IRGM has also been implicated in governing the core autophagy machinery to promote bacterial defense.78 The CD-associated risk genes PRDM and NDP52 have also been associated with decreased plasma cells and overactive NFκB signaling, respectively.79 Additional studies of Irgm in mice have revealed defects in Paneth cell function;80 however, due to the differences in gene structure between human IRGM and murine Irgm, the functional implications of these studies for human physiology remain unclear.
The first gene to be associated with IBD,81 NOD2 encodes a cytoplasmic pattern recognition receptor that recognizes bacterial peptidoglycan in the cell cytoplasm. Although not an autophagy gene, NOD2 directs autophagic proteins through recruitment of ATG16L1 to the plasma membrane at bacterial entry sites.82 Several CD-associated risk polymorphisms have been identified in NOD2, including a truncated version of the protein that cannot recruit ATG16L1 to the plasma membrane.4, 81 The functional significance of these risk alleles has been examined through evaluation of Paneth cell phenotypes in a cohort of patient tissue samples, which revealed a correlation between the presence of NOD2 risk alleles and proportion of abnormal Paneth cells, while the ATG16L1 T300A polymorphism showed an additive effect on the number of abnormal Paneth cells.83 The Paneth cell defects associated with ATG16L1 T300A and NOD2 polymorphisms are phenotypically distinct. Importantly, increased Paneth cell abnormalities were also associated with shorter times to disease relapse after surgery. Studies demonstrating associations between specific patient genotypes and disease phenotypes are particularly promising in their potential use for generating a comprehensive analysis of IBD to determine disease treatment.
Conclusions
Autophagy has evolved as a system for cellular defense, microbial tolerance, and metabolic control that directly influences cellular and organ-level homeostasis. Studies of autophagy have highlighted conserved functions across similar cell types within different organ systems; for example, secretory cells of the pancreas and intestine share a conserved function of autophagy in granule secretion. In addition to its cell type-specific functions, autophagy is also implicated in cell-cell communication, as in the example of changes in antigen presentation by dendritic cells skewing the effector T cell repertoire and altering cytokine secretion. Cell-type specific functions of autophagy interact with one another to build an organ level “circuit” with inputs and outputs related to environmental sensing and metabolic responses.
Several studies have suggested that restoration of autophagy through chemical or genetic intervention could provide therapeutic benefit for some diseases by bringing the cellular environment back to a basal/homeostatic state. For example, a novel autophagy-inducing peptide, Tat-beclin1,84 can induce autophagy in vitro and inhibits both bacterial and viral replication.
To properly develop autophagy modulators as therapeutics, a comprehensive evaluation of their effect on multiple pathways must be assembled. One recent screen evaluated small molecules for induction of basal autophagy as well as IL-1β secretion and effects on T cell populations including Th1, Th17, and Treg subtypes,85 providing a framework for comprehensive evaluation of the effect of these small molecules on multiple pathways implicated in human disease. Such studies allow for selection of highly specific pathways for therapeutic intervention.
Acknowledgments
Grant support: This work was supported by National Institutes of Health grants DK102557, DK097485, and DK092405 (R.J.X.). We thank A. Nicole Desch and Kara G. Lassen for providing guidance, useful critiques, and supporting text. Editorial assistance was provided by Natalia Nedelsky (DK043351 to R.J.X.).
Abbreviations used in this paper
- PI(3)K
class III phosphoinositol 3 OH kinase
- AIEC
adherent invasive Escherichia coli
- ALCAT1 (LCAT1)
lysocardiolipin acyltransferase 1
- AMP
antimicrobial peptide
- AMPK
protein kinase, AMP activated, alpha 1 catalytic subunit
- Atg5
autophagy related 5
- Atg12
autophagy related 12
- Atg13
autophagy related 13
- ATG16L1
autophagy related 16 like 1
- Atg2
autophagy related 2
- Atg4
autophagy related 4
- Atg7
autophagy related 7
- Atg9
autophagy related 9
- CD
Crohn's disease
- CREB
cAMP response element-binding protein
- CRIP
cysteine-rich intestinal polypeptide
- DC
dendritic cell
- DNA
deoxyribonucleic acid
- ER
endoplasmic reticulum
- Fcgbp
Fc gamma binding protein
- FIP200 (RB1CC1, RB1)
inducible coiled coil 1
- FXR
farnesoid X receptor
- Galectin 8 (LGALS8)
galactoside binding lectin, soluble, 8
- GCN2
general control nonderepressible 2 kinase
- HD5
human defensin 5
- HD6
human defensin 6
- HIF1
hypoxia inducible factor 1
- HMGB1
high mobility group box 1
- IBD
inflammatory bowel disease
- IL1β
interleukin 1 beta
- IRGM
immunity related GTPase family M
- LC3 (MAP1LC3)
microtubule associated protein 1 light chain 3
- LPS
lipopolysaccharide
- LY75
lymphocyte antigen 75
- MAGI-1
membrane associated guanylate kinase, WW and PDZ domain containing
- MHC II
major histocompatibility complex two
- MiR196
micro RNA 196
- MMP7
matrix metalloproteinase 7
- mRNA
messenger ribonucleic acid
- MTOR
mechanistic target of rapamycin
- mTORC1
mammalian target of rapamycin complex 1
- MUC2
mucin 2
- NAFLD
non-alcoholic fatty liver disease
- NCOA4
nuclear receptor coactivator 4
- NDP52 (CALCOCO2)
calcium coiled coil domain 2
- NFκB
nuclear factor of kappa light polypeptide gene enhancer in B cells
- NOD2
nucleotide binding oligomerization domain containing 2
- OFD1
oral facial digital syndrome 1
- OPTN
optineurin
- P62
SQSTM1, sequestosome 1
- PCM1
pericentriolar material 1
- PPARα
peroxisome proliferator-activated receptor-alpha
- PRDM
PR domain containing
- RegIIIγ
regenerating islet derived protein 3 gamma
- ROS
reactive oxygen species
- siRNA
small interfering ribonucleic acid
- Smurf1
SMAD specific E3 ubiquitin protein ligase 1
- SNAP29
synaptosomal associated protein, 29kDa
- SNP
single nucleotide polymorphism
- sPLA2
secretory phospholipase A2
- TFEB
Transcription factor EB
- UC
ulcerative colitis
- ULK1
unc 51 like autophagy activating kinase 1
- VAMP
vesicle associated membrane protein
- VMP1
vacuole membrane protein 1
- Vps34
phosphatidylinositol 3 kinase, catalytic subunit type 3
- WIPI2b
WD repeat domain, phosphoinositide interacting 2
- Wnt
wingless type MMTV integration site family
- Xbp1
X box binding protein 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: The authors report no conflict of interest.
References
- 1.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brest P, Lapaquette P, Souidi M, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet. 2011;43:242–5. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 3.Lassen KG, Kuballa P, Conway KL, et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A. 2014;111:7741–6. doi: 10.1073/pnas.1407001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 5.Murthy A, Li Y, Peng I, et al. A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506:456–62. doi: 10.1038/nature13044. [DOI] [PubMed] [Google Scholar]
- 6.Sadaghian Sadabad M, Regeling A, de Goffau MC, et al. The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn's disease patients. Gut. 2014 doi: 10.1136/gutjnl-2014-307289. [DOI] [PubMed] [Google Scholar]
- 7.Knights D, Silverberg MS, Weersma RK, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6:107. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson SJ, Zhou JY, Geddes K, et al. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes. 2013;4:222–31. doi: 10.4161/gmic.24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–70. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–69. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Jiang P, Nishimura T, Sakamaki Y, et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25:1327–37. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima N. Sugar modification inhibits autophagosome-lysosome fusion. Nat Cell Biol. 2014;16:1132–3. doi: 10.1038/ncb3078. [DOI] [PubMed] [Google Scholar]
- 14.Thurston TL, Wandel MP, von Muhlinen N, et al. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–8. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogov V, Dotsch V, Johansen T, et al. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–78. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Visvikis O, Ihuegbu N, Labed SA, et al. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity. 2014;40:896–909. doi: 10.1016/j.immuni.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seok S, Fu T, Choi SE, et al. Transcriptional regulation of autophagy by an FXR CREB axis. Nature. 2014;516:108–11. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JM, Wagner M, Xiao R, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–5. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 20.Salzman NH, Hung K, Haribhai D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wehkamp J, Salzman NH, Porter E, et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci U S A. 2005;102:18129–34. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu H, Pazgier M, Jung G, et al. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. 2012;337:477–81. doi: 10.1126/science.1218831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukherjee S, Hooper LV. Antimicrobial defense of the intestine. Immunity. 2015;42:28–39. doi: 10.1016/j.immuni.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 24.Vaishnava S, Yamamoto M, Severson KM, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–8. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabrera S, Fernandez AF, Marino G, et al. ATG4B/autophagin-1 regulates intestinal homeostasis and protects mice from experimental colitis. Autophagy. 2013;9:1188–200. doi: 10.4161/auto.24797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cadwell K, Patel KK, Komatsu M, et al. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250–2. doi: 10.4161/auto.5.2.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cullen TW, Schofield WB, Barry NA, et al. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–5. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel KK, Miyoshi H, Beatty WL, et al. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J. 2013;32:3130–44. doi: 10.1038/emboj.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuster AT, Homer CR, Kemp JR, et al. CAP-D3 Promotes Bacterial Clearance in Human Intestinal Epithelial Cells by Repressing Expression of Amino Acid Transporters. Gastroenterology. 2015 doi: 10.1053/j.gastro.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conway K, Kuballa P, Song J. Atg16L1 is Required for Autophagy in Intestinal Epithelial Cells and Protection of Mice from Salmonella Infection. Gastroenterology. 2013;145:1–20. doi: 10.1053/j.gastro.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamin JL, Sumpter R, Jr., Levine B, et al. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe. 2013;13:723–34. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mimouna S, Bazin M, Mograbi B, et al. HIF1A regulates xenophagic degradation of adherent and invasive Escherichia coli (AIEC). Autophagy. 2014;10:2333–45. doi: 10.4161/15548627.2014.984275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–10. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nighot PK, Hu CA, Ma TY. Autophagy enhancement of intestinal epithelial tight junction barrier function by targeting claudin-2 degradation. J Biol Chem. 2015 doi: 10.1074/jbc.M114.597492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pampliega O, Orhon I, Patel B, et al. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Z, Lin MG, Stowe TR, et al. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature. 2013;502:254–7. doi: 10.1038/nature12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam HC, Cloonan SM, Bhashyam AR, et al. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J Clin Invest. 2013;123:5212–30. doi: 10.1172/JCI69636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kayama H, Ueda Y, Sawa Y, et al. Intestinal CX3C chemokine receptor 1(high) (CX3CR1(high)) myeloid cells prevent T-cell-dependent colitis. Proc Natl Acad Sci U S A. 2012;109:5010–5. doi: 10.1073/pnas.1114931109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hadis U, Wahl B, Schulz O, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–46. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 41.Kanayama M, Inoue M, Danzaki K, et al. Autophagy enhances NFkappaB activity in specific tissue macrophages by sequestering A20 to boost antifungal immunity. Nat Commun. 2015;6:5779. doi: 10.1038/ncomms6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Ye Y, Zhou X, et al. Atg7 Enhances Host Defense against Infection via Downregulation of Superoxide but Upregulation of Nitric Oxide. J Immunol. 2015;194:1112–21. doi: 10.4049/jimmunol.1401958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Z, Fux B, Goodwin M, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–69. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smythies LE, Sellers M, Clements RH, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jagannath C, Lindsey DR, Dhandayuthapani S, et al. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15:267–76. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- 47.Lee HK, Mattei LM, Steinberg BE, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–39. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ravindran R, Khan N, Nakaya HI, et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 2014;343:313–7. doi: 10.1126/science.1246829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klareskog L, Ronnelid J, Lundberg K, et al. Immunity to citrullinated proteins in rheumatoid arthritis. Annu Rev Immunol. 2008;26:651–75. doi: 10.1146/annurev.immunol.26.021607.090244. [DOI] [PubMed] [Google Scholar]
- 50.Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Exp Med. 2011;208:2625–32. doi: 10.1084/jem.20110640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hubbard-Lucey VM, Shono Y, Maurer K, et al. Autophagy gene Atg16L1 prevents lethal T cell alloreactivity mediated by dendritic cells. Immunity. 2014;41:579–91. doi: 10.1016/j.immuni.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu E, Van Grol J, Subauste CS. Atg5 but not Atg7 in dendritic cells enhances IL-2 and IFN-gamma production by Toxoplasma gondii-reactive CD4 T cells. Microbes Infect. 2015 doi: 10.1016/j.micinf.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooney R, Baker J, Brain O, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 54.Nedjic J, Aichinger M, Emmerich J, et al. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 55.Puleston DJ, Zhang H, Powell TJ, et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. Elife. 2014:3. doi: 10.7554/eLife.03706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salio M, Puleston DJ, Mathan TS, et al. Essential role for autophagy during invariant NKT cell development. Proc Natl Acad Sci U S A. 2014;111:E5678–87. doi: 10.1073/pnas.1413935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stephenson LM, Miller BC, Ng A, et al. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy. 2009;5:625–35. doi: 10.4161/auto.5.5.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu X, Araki K, Li S, et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat Immunol. 2014;15:1152–61. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mantis NJ, Rol N, Corthesy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–11. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pengo N, Scolari M, Oliva L, et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013;14:298–305. doi: 10.1038/ni.2524. [DOI] [PubMed] [Google Scholar]
- 61.Conway KL, Kuballa P, Khor B, et al. ATG5 regulates plasma cell differentiation. Autophagy. 2013;9:528–37. doi: 10.4161/auto.23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Efeyan A, Zoncu R, Chang S, et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–83. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dowdle WE, Nyfeler B, Nagel J, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16:1069–79. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 64.Mancias JD, Wang X, Gygi SP, et al. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–9. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barber MF, Elde NC. Nutritional immunity. Escape from bacterial iron piracy through rapid evolution of transferrin. Science. 2014;346:1362–6. doi: 10.1126/science.1259329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferreyra JA, Wu KJ, Hryckowian AJ, et al. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16:770–7. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Albenberg L, Esipova TV, Judge CP, et al. Correlation Between Intraluminal Oxygen Gradient and Radial Partitioning of Intestinal Microbiota in Humans and Mice. Gastroenterology. 2014;147:1055–1063. e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Glover LE, Colgan SP. Hypoxia and metabolic factors that influence inflammatory bowel disease pathogenesis. Gastroenterology. 2011;140:1748–55. doi: 10.1053/j.gastro.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ortiz-Masia D, Cosin-Roger J, Calatayud S, et al. Hypoxic macrophages impair autophagy in epithelial cells through Wnt1: relevance in IBD. Mucosal Immunol. 2014;7:929–38. doi: 10.1038/mi.2013.108. [DOI] [PubMed] [Google Scholar]
- 70.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaser A, Lee AH, Franke A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–56. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adolph TE, Tomczak MF, Niederreiter L, Hyun-Jeong Ko, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–6. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu X, Messer JS, Wang Y, et al. Cytosolic HMGB1 controls the cellular autophagy/apoptosis checkpoint during inflammation. J Clin Invest. 2015 doi: 10.1172/JCI76344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 75.Wu S, Zhang YG, Lu R, et al. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut. 2014 doi: 10.1136/gutjnl-2014-307436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCarroll SA, Huett A, Kuballa P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107–12. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chauhan S, Mandell MA, Deretic V. IRGM Governs the Core Autophagy Machinery to Conduct Antimicrobial Defense. Mol Cell. 2015;58:507–21. doi: 10.1016/j.molcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ellinghaus D, Zhang H, Zeissig S, Lipinski S, Till A, et al. Association between variants of PRDM1 and NDP52 and Crohn's disease, based on exome sequencing and functional studies. Gastroenterology. 2013;145:339–47. doi: 10.1053/j.gastro.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu B, Gulati AS, Cantillana V, et al. Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2013;305:G573–84. doi: 10.1152/ajpgi.00071.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 82.Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 83.VanDussen KL, Liu TC, Li D, et al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn's disease. Gastroenterology. 2014;146:200–9. doi: 10.1053/j.gastro.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shoji-Kawata S, Sumpter R, Leveno M, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–6. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shaw SY, Tran K, Castoreno AB, et al. Selective modulation of autophagy, innate immunity, and adaptive immunity by small molecules. ACS Chem Biol. 2013;8:2724–33. doi: 10.1021/cb400352d. [DOI] [PMC free article] [PubMed] [Google Scholar]