Abstract
Liver cancer is one of the most common cancers worldwide with high morbidity and mortality. Its molecular mechanism hasn’t been fully understood though many studies have been conducted and thus further researches are still needed to improve the prognosis of liver cancer. Firstly, differentially expressed genes (DEGs) between six Mdr2-knockout (Mdr2-KO) mutant mice samples (3-month-old and 12-month-old) and six control mice samples were identified. Then, the enriched GO terms and KEGG pathways of those DEGs were obtained using the Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/). Finally, protein-protein interactions (PPI) network of those DEGs were constructed using STRING database (http://www.string-db.org/) and visualized by Cytoscape software, at the same time, genes with high degree were selected out. Several novel biomarkers that might play important roles in liver cancer were identified through the analysis of gene microarray in GEO. Also, some genes such as Tyrobp, Ctss and pathways such as Pathways in cancer, ECM-receptor interaction that had been researched previously were further confirmed in this study. Through the bioinformatics analysis of the gene microarray in GEO, we found some novel biomarkers of liver cancer and further confirmed some known biomarkers.
Keywords: Liver cancer, Bioinformatics, Biomarker, Pathway
Introduction
Liver cancer is one of the most common malignancies. It has a high morbidity and mortality, especially in sub-Saharan Africa and eastern Asia. The incidence of liver cancer has doubled or even more in the past 15 years [1]. However, the molecular mechanism of liver cancer is still largely unknown. For above reasons, an increasing number of researches on liver cancer have been conducted in recent years. Different molecular mechanism and various biomarkers related to liver cancer have been identified. Through qRT-PCR and Western blotting, JianXin et al. [2] have inferred that GOLPH3, which has higher expression level in gene and protein level of liver cancer patients compared with that of the normal population, is a new biomarker for liver cancer. Mah et al. [3] have found that the inflammation-related pathway NFkB plays an important role in liver cancer by analyzing the methylation profile of 59 liver cancer patients. Despite a great number of previous researches, molecular mechanism of liver cancer has not been fully grasped. Hence, further researches of molecular level, such as researches of gene or protein, are still needed to find out new molecular mechanism or biomarkers in an effort to improve the prognosis, diagnosis and treatment of liver cancer.
Mdr2-knockout (Mdr2-KO) mice lack the liver-specific P-glycoprotein responsible for phosphatidylcholine transport across the canalicular membrane, which may result in dysfunctional phospholipid secretion [4]. Signs of inflammation are accompanied by an increase in plasma transaminase levels and followed by enhanced connective tissue storage and fibrosis progression. As a consequence of chronic inflammation and progressing fibrosis, Mdr2-knockout mice may develop liver cancer between the ages of 12 and 15 months [5].
With the rapid growth of microarry and its implication in cancer research, a lot of genes that are related to cancers (including liver cancer) have been verified. For example, Yang et al. [6] have found that Gα12 is an important therapeutic target for liver cancer through cDNA microarray analysis. Xu et al. [7] have verified, through microarray and RT-PCR technology, the role of CXCL5 in liver cancer migration and invasion.
In this research, by analyzing the gene expression microarray of liver cancer in GEO database, we further confirmed the molecular mechanism and some biomarkers of liver cancer that had been investigated previously. Moreover, genes, which had not been researched but also had a great importance to liver cancer, were also included in this research. Also, most of the enriched GO terms and KEGG pathways of those genes were related to liver cancer, especially cell cycle, immune response, inflammatory response, pathways in cancer, MAPK signaling pathway, Cell adhesion molecules and etc. In conclusion, our finding can improve our understanding of liver cancer and provide potential therapeutic targets for further studies.
Materials and Methods
Gene Expression Microarray Data
In this study, the gene expression microarray data set GSE4612 was downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). GSE4612 [8] is a gene expression profile data including six Mdr2 knockout (Mdr2-KO) mutant mice samples(3-month-old and 12-month-old) and six control mice samples(3-month-old and 12-month-old). The platform of this microarray data is GPL339 [MOE430A] Affymetrix Mouse Expression 430A Array.
Preprocessing of the Microarray Data
Unwanted noise of the raw microarray data was filtered out in the preprocessing stage. The normalization of raw data and background correction was conducted via affy[9] package in R. Moreover, multiple probes that corresponded to one gene symbol were summarized–taking the average expression values of those probes as the expression value of this gene. There were a total of 22,690 probes in the microarray and 13,687 gene symbols that had no duplicate before and after preprocessing.
Get the Differentially Expressed Genes
After the preprocessing, the critical step was to get the differentially expressed genes (DEGs) between the case samples and the control samples. The tool used in this study was the limma [10] package in R. t-test was conducted on the gene expression values between case samples and control samples and the genes with P value < 0.05 and |log2(fold change)|>1 were selected out. According to those criteria, in the first step, the DEGs between the case samples and the control samples were selected out from 3-month-old mice and 12-month-old mice respectively, then the overlapped genes between those two list DEGs were selected out. The heatmap of the overlapped DEGs was obtained through gplots package in R to visualize their expression value in different samples.
GO Enrichment and KEGG Pathway Analysis of the DEGs
After getting the DEGs, GO enrichment and KEGG pathway analysis of the DEGs were conducted. Here, the tool used in this study was DAVID (http://david.abcc.ncifcrf.gov/) (Database for Annotation, Visualization and Integrated Discovery). It could be used to do functional annotation for a list of genes, gene functional classfication or gene ID conversion. In this study, the module used in this study was the functional annotation. First, we submitted the DEGs list into the database and selected Mus musculus in species column. Finally, the GO terms and the KEGG pathways with P value smaller than 0.05 and at least five genes were selected out as the enriched function of DEGs.
Construct the PPI Network of DEGs
To further investigate the molecular mechanism of liver cancer, PPI network of the DEGs was constructed through STRING database (http://www.string-db.org/). STRING is a database that infers the interaction between genes through analyzing the genomic data that comes from different sources, such as high-throughput experiments, co-expression data and the previous data and etc. Also, it has a unique scoring framework which assigns the interaction an integrated score to represent its confidence through combining the score of the different sources. Here, we selected the gene-gene interactions, whose integrated scores were bigger than 0.4 (the default threshold in the STRING database), to construct the PPI network and Cytoscape [11] was used for visualization.
Select the Core Gene in the Network
To select core genes (the genes that might be more likely involved in liver cancer) from PPI network, we analyzed the topological structure of the network and obtained the degree (the number of genes that directly interact with the gene) of each gene. Here, we selected the genes whose degree is beyond 10 as the core genes in the network.
Results
Differentially Expressed Genes (DEGs)
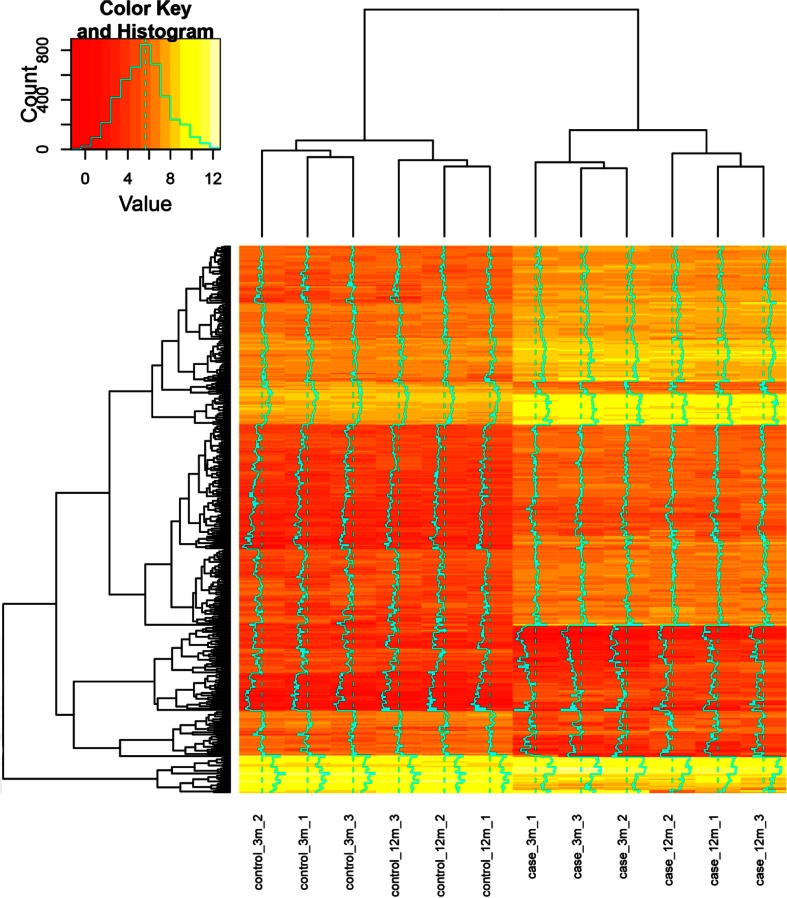
There were 1898 DEGs in the 3-month-old mice and 864 DEGs in the 12-month-old mice between the case samples and control samples. A total of 380 overlapped DEGs between those two DEG lists were identified. From the heatmap (Fig. 1), we could get that the gene expression of Mdr2 knockout samples were distinguished from the control samples, meanwhile, the gene expression of 3-month-old samples were distinguished from the 12-month-old samples, indicating that obvious differences existed in these groups.
Fig. 1.
The heatmap of the DEGs. In the figure ‘control_3m_n (n = 1, 2, 3)’ is the 3-month-old mice in control samples; ‘control_12m_n (n = 1, 2, 3)’ is the 12-month-old mice in control samples; ‘case_3m_n (n = 1,2,3)’ is the 3-month-old mice in case samples; ‘case_12m_n (n = 1,2,3)’ is the 12-month-old mice in case samples
Enriched GO Terms and KEGG Pathways of DEGs
In this study, a total of 128 enriched GO terms and 23 KEGG pathways were obtained. The top 10 enriched GO terms of the DEGs according to P value were shown (Table 1). Table 1 indicated that the main enriched GO terms was the biological process of cell, such as cell adhesion, regulation of cell growth, regulation of cell cycle. Besides the cell biological process, there were also some enriched GO terms related to immune response, inflammatory response and etc.
Table 1.
The top 10 enriched GO terms of DEGs, which were sorted by P value in ascending
| Category | GOID | GO name | P value | Gene number |
|---|---|---|---|---|
| CC | GO:0005578 | proteinaceous extracellular matrix | 2.74E-08 | 25 |
| CC | GO:0005576 | extracellular region | 5.41E-08 | 70 |
| CC | GO:0031012 | extracellular matrix | 5.82E-08 | 25 |
| CC | GO:0044421 | extracellular region part | 7.71E-08 | 42 |
| BP | GO:0006955 | immune response | 1.98E-05 | 26 |
| BP | GO:0006952 | defense response | 2.54E-05 | 25 |
| BP | GO:0007155 | cell adhesion | 5.24E-05 | 28 |
| BP | GO:0022610 | biological adhesion | 5.37E-05 | 28 |
| MF | GO:0004197 | cysteine-type endopeptidase activity | 2.65E-04 | 8 |
| BP | GO:0006954 | inflammatory response | 2.88E-04 | 15 |
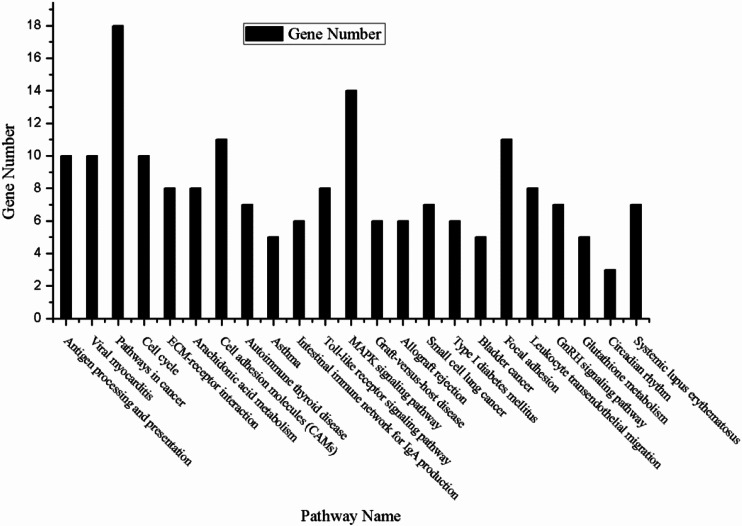
The enriched KEGG pathways of the DEGs were shown in Table 2. A few enriched KEGG pathways were directly related to cancer, such as Pathways in cancer, Small cell lung cancer, Bladder cancer. What’s more, it was possible that other pathways had an important influence on the progression of cancer via some biological process, such as Toll-like receptor signaling, EMC-receptor interaction, MAPK signaling pathway and etc. The KEGG pathways and their corresponding gene number were shown in Fig. 2.
Table 2.
The KEGG pathways enriched in DEGs
| Category | Pathway name | Gene number | P value |
|---|---|---|---|
| KEGG_PATHWAY | mmu04612:Antigen processing and presentation | 10 | 3.22E-04 |
| KEGG_PATHWAY | mmu05416:Viral myocarditis | 10 | 4.11E-04 |
| KEGG_PATHWAY | mmu05200:Pathways in cancer | 18 | 0.001908471 |
| KEGG_PATHWAY | mmu04110:Cell cycle | 10 | 0.003677215 |
| KEGG_PATHWAY | mmu04512:ECM-receptor interaction | 8 | 0.003769465 |
| KEGG_PATHWAY | mmu00590:Arachidonic acid metabolism | 8 | 0.003769465 |
| KEGG_PATHWAY | mmu04514:Cell adhesion molecules (CAMs) | 11 | 0.003969349 |
| KEGG_PATHWAY | mmu05320:Autoimmune thyroid disease | 7 | 0.007639543 |
| KEGG_PATHWAY | mmu05310:Asthma | 5 | 0.007822698 |
| KEGG_PATHWAY | mmu04672:Intestinal immune network for IgA production | 6 | 0.00947486 |
| KEGG_PATHWAY | mmu04620:Toll-like receptor signaling pathway | 8 | 0.009821668 |
| KEGG_PATHWAY | mmu04010:MAPK signaling pathway | 14 | 0.011527368 |
| KEGG_PATHWAY | mmu05332:Graft-versus-host disease | 6 | 0.012714159 |
| KEGG_PATHWAY | mmu05330:Allograft rejection | 6 | 0.012714159 |
| KEGG_PATHWAY | mmu05222:Small cell lung cancer | 7 | 0.016578151 |
| KEGG_PATHWAY | mmu04940:Type I diabetes mellitus | 6 | 0.017724229 |
| KEGG_PATHWAY | mmu05219:Bladder cancer | 5 | 0.018106115 |
| KEGG_PATHWAY | mmu04510:Focal adhesion | 11 | 0.021349818 |
| KEGG_PATHWAY | mmu04670:Leukocyte transendothelial migration | 8 | 0.024856275 |
| KEGG_PATHWAY | mmu04912:GnRH signaling pathway | 7 | 0.029651851 |
| KEGG_PATHWAY | mmu00480:Glutathione metabolism | 5 | 0.036428615 |
| KEGG_PATHWAY | mmu04710:Circadian rhythm | 3 | 0.038159626 |
| KEGG_PATHWAY | mmu05322:Systemic lupus erythematosus | 7 | 0.038193344 |
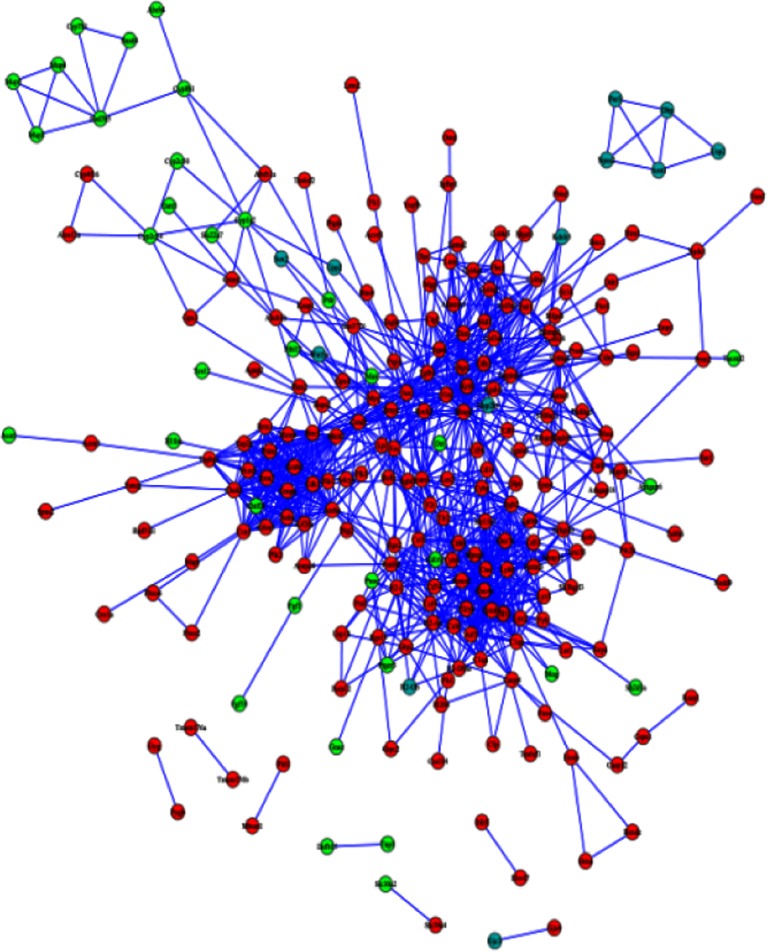
Fig. 2.
The PPI network of the DEGs. The network contains 244 nodes and 1053 edges. The 198 crimson nodes are the genes that have higher expression values in the case samples compared with the control samples in both 3-month-old mice and 12-month-old mice; the 34 bright green nodes are the genes that have lower expression values in the case samples compared with the control samples in both 3-month-old mice and 12-month-old mice; the 12 blue nodes represent the genes that have contradictory expression trend between case samples and control samples in 3-month-old mice and 12-month-old mice
PPI Network of the DEGs and Core Genes in the PPI Network
The PPI (Fig. 3) network contained 244 nodes and 1053 edges. The nodes represented the DEGs and the edges represented the interactions between the DEGs. A great number of genes of higher degree, which were the core genes in the PPI network, might relate to liver cancer more closely. The core genes and their corresponding degree were shown in Table 3. Among those core genes, Ctss and Tyrobp had the highest degree and there were 28 genes whose degree was beyond 20.
Fig. 3.
The KEGG pathway and their corresponding gene number
Table 3.
The core genes and their corresponding degree
| Gene | Degreea | Gene | Degree | Gene | Degree | Gene | Degree |
|---|---|---|---|---|---|---|---|
| Lum | 10 | Fbn1 | 15 | Mcm5 | 19 | Cdh1 | 24 |
| Tpm4 | 10 | Itga6 | 15 | Cdca5 | 19 | Ccnd1 | 25 |
| Lyz1 | 10 | Lgals1 | 15 | Tlr2 | 20 | Jun | 25 |
| Gbp2 | 10 | S100a6 | 15 | Serpinh1 | 20 | Fos | 25 |
| Max | 11 | Fermt3 | 16 | Lyz2 | 20 | Cd86 | 25 |
| Gm3776 | 11 | P2ry6 | 16 | Cd53 | 20 | Rmcs2 | 25 |
| Col4a2 | 11 | Ccnf | 17 | Clec4n | 20 | Birc5 | 25 |
| Ctgf | 12 | Chtf18 | 17 | H2-Aa | 20 | Ccnb2 | 25 |
| Arhgdib | 12 | C1qa | 17 | Prim1 | 20 | Ccl5 | 26 |
| Cd14 | 12 | Anln | 17 | Mcm6 | 20 | Ly86 | 26 |
| H2-L | 12 | Col5a2 | 17 | Rrm2 | 20 | Icam1 | 27 |
| Cd74 | 12 | Cybb | 17 | Kif20a | 20 | Mki67 | 27 |
| F2r | 12 | Col3a1 | 18 | Slc15a3 | 21 | Aurka | 27 |
| Efemp2 | 13 | Dcn | 18 | Clec7a | 22 | Itgb2 | 27 |
| Cyba | 13 | C1qc | 18 | Plk1 | 22 | Mmp2 | 28 |
| Rock2 | 13 | Myc | 18 | Chek1 | 22 | Cdk1 | 30 |
| Ccl9 | 13 | Psmb8 | 18 | Cd52 | 22 | Vim | 31 |
| Topbp1 | 14 | Fstl1 | 19 | Bgn | 23 | Tyrobp | 37 |
| Col4a1 | 14 | Ccl6 | 19 | Aif1 | 24 | Ctss | 37 |
| Nis1 | 14 | Rrm1 | 19 | Mpeg1 | 24 | ||
| Cenpq | 15 | Top2a | 19 | Tgfb1 | 24 |
aThe number of genes that directly interact with the genes in the PPI network
Discussion
Although researchers have made considerable efforts in disclosing the mechanisms of liver cancer,current understanding of the genetic alterations associated with the progression of liver cancer has not yet to be elucidated. In this study, we conducted genome-wide gene expression analysis by a high throughput method to identify the DEGs from liver cancer compared with normal liver tissues. Here, a total number of 380 overlapped DEGs from original dataset of two groups (3-month-old group and 12-month-old group) were identified, including 289 overexpressed genes, 66 down-regulated genes and 25 genes that had contradictory expression trend.
GO analyses revealed that the significant ontology categories included immune response, cell adhesion, inflammatory response and so on. Immune effector process, nuclear division, cell division, mitotic cell cycle and positive regulation of cellular component organization were obviously overrepresented in the up-regulated genes according to the functional enrichment analysis. In the immune response, for example, TLR2 could enhance ovarian cancer stem cell self-renewal and eventually promote tumor repair and recurrence [12]. ICAM-1 is a transmembrane glycoprotein in the immunoglobulin superfamily, which participates in oral cancer progression and induces macrophage/SCC-cell adhesion [13]. Ciftci et al. [14] indicated that serum TGFB1 level might be elevated in breast cancer patients and had a favorable prognostic value. CDH1, involved in cell adhesion, can code the adhesion protein E-cadherin that plays a central part in the process of epithelial morphogenesis [15]. CCL5 belongs to the CC-chemokine family and plays a pivotal role in the invasion and metastasis of human cancer cells. Huang et al. reported that CCL5 stimulation could increase lung cancer migration via induced phosphorylation of the p85α subunit of PI3K and serine 473 of Akt [16].
DEGs were then used in KEGG pathway analyses and 23 pathways were screened out, such as Cell adhesion molecules, Toll-like receptor signaling, EMC-receptor interaction, MAPK signaling pathway and etc. Previous researches reported that most of these pathways were involved in cancer progression. The immune system played a critical role in body defense system, and the dysfunction of immune system might result in cancer. Stimulation of various Toll-like receptors induced specific patterns of gene expression, which resulted in the activation of innate immunity and the development of antigen-specific acquired immunity [17]. Moreover, MAPK signal molecules participated in the amplification and specificity of the transmitted signals that finally activated a number of regulatory molecules in the cytoplasm and the nucleus to initiate cellular processes such as proliferation, differentiation, and development [18].
Furthermore, the topological structure analysis of PPI network suggested that Ctss, Tyrobp, Vim, Cdk1 were the top 4 core genes, which might be potential therapeutic targets for future research. Cathepsin S (Ctss), a key enzyme in major histocompatibility complex class II (MHC-II) mediating antigen presentation, might be involved in malignant progression of lung cancer [19]. CD47 positive liver cancer cells preferentially secreted cathepsin S (CTSS), which regulated liver tumor-initiating cells through the CTSS/protease-activated receptor 2 (PAR2) loop [20]. Shabo indicated that Tyrobp (DAP12) in breast cancer was associated with an advanced tumor grade and higher rates of skeletal and liver metastases [21, 22]. Costa reported that Vim could associate with GDF15 and TMEFF2 to predict bladder cancer [23].
Overall, with a microarray data set from the GEO database, a range of DEGs were obtained in liver cancer and normal tissues. These genes might be functionally relevant to pathogenesis of liver cancer. Functional analysis revealed mitotic cell cycle, proteinaceous extracellular matrix and MAPK signaling pathway participated in biological processes as the significant items for liver cancer. These results could provide a valuable data base for further investigation of liver cancer research. Of course, further experiments are still needed to further confirm the potential function of these genes.
Acknowledgements
This work was supported by the Health Bureau Science and Technology Foundation of Tianjin (No. 2012KZ063) National Natural Science Foundation of China (Grant No. 81302082, 81272685, 31301151, 81172355).
Footnotes
Pengfei Liu and Wenhua Jiang are joint first authors.
Contributor Information
Pengfei Liu, Email: 9957600@163.com.
Jihui Hao, Phone: 86-022-23340123-3072, Email: haojihui@tjmuch.com.
References
- 1.Bertino G, Demma S, Ardiri A, Proiti M, Gruttadauria S, Toro A, Malaguarnera G, Bertino N, Malaguarnera M, Di Carlo I. Hepatocellular carcinoma: novel molecular targets in carcinogenesis for future therapies. Biomed Res Int. 2014;2014:203693. doi: 10.1155/2014/203693. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.JianXin J, Cha Y, ZhiPeng L, Jie X, Hao Z, Meiyuan C, ChengYi S. GOLP3 is a predictor of survival in patients with hepatocellular carcinoma. Clin Invest Med. 2014;37(4):E233. [PubMed] [Google Scholar]
- 3.Mah WC, Thurnherr T, Chow PK, Chung AY, Ooi LL, Toh HC, Teh BT, Saunthararajah Y, Lee CG. Methylation profiles reveal distinct subgroup of hepatocellular carcinoma patients with poor prognosis. PLoS One. 2014;9(8):e104158. doi: 10.1371/journal.pone.0104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smit JJ, Schinkel AH, Oude Elferink RP, Groen AK, Wagenaar E, van Deemter L, Mol CA, Ottenhoff R, van der Lugt NM, van Roon MA, et al. Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell. 1993;75(3):451–462. doi: 10.1016/0092-8674(93)90380-9. [DOI] [PubMed] [Google Scholar]
- 5.Katzenellenbogen M, Mizrahi L, Pappo O, Klopstock N, Olam D, Barash H, Domany E, Galun E, Goldenberg D. Molecular mechanisms of the chemopreventive effect on hepatocellular carcinoma development in Mdr2 knockout mice. Mol Cancer Ther. 2007;6(4):1283–1291. doi: 10.1158/1535-7163.MCT-06-0420. [DOI] [PubMed] [Google Scholar]
- 6.Yang YM, Lee WH, Lee CG, An J, Kim ES, Kim SH, Lee SK, Lee CH, Dhanasekaran DN, Moon A, Hwang S, Lee SJ, Park JW, Kim KM, Kim SG (2014) Galpha gep oncogene deregulation of p53-responsive microRNAs promotes epithelial-mesenchymal transition of hepatocellular carcinoma. Oncogene 0 [DOI] [PubMed]
- 7.Xu X, Huang P, Yang B, Wang X, Xia J. Roles of CXCL5 on migration and invasion of liver cancer cells. J Transl Med. 2014;12:193. doi: 10.1186/1479-5876-12-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katzenellenbogen M, Pappo O, Barash H, Klopstock N, Mizrahi L, Olam D, Jacob-Hirsch J, Amariglio N, Rechavi G, Mitchell LA, Kohen R, Domany E, Galun E, Goldenberg D. Multiple adaptive mechanisms to chronic liver disease revealed at early stages of liver carcinogenesis in the Mdr2-knockout mice. Cancer Res. 2006;66(8):4001–4010. doi: 10.1158/0008-5472.CAN-05-2937. [DOI] [PubMed] [Google Scholar]
- 9.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 10.Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7:252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chefetz I, Alvero AB, Holmberg JC, Lebowitz N, Craveiro V, Yang-Hartwich Y, Yin G, Squillace L, Gurrea Soteras M, Aldo P, Mor G. TLR2 enhances ovarian cancer stem cell self-renewal and promotes tumor repair and recurrence. Cell Cycle. 2013;12(3):511–521. doi: 10.4161/cc.23406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Usami Y, Ishida K, Sato S, Kishino M, Kiryu M, Ogawa Y, Okura M, Fukuda Y, Toyosawa S. Intercellular adhesion molecule-1 (ICAM-1) expression correlates with oral cancer progression and induces macrophage/cancer cell adhesion. Int J Cancer. 2013;133(3):568–578. doi: 10.1002/ijc.28066. [DOI] [PubMed] [Google Scholar]
- 14.Ciftci R, Tas F, Yasasever CT, Aksit E, Karabulut S, Sen F, Keskin S, Kilic L, Yildiz I, Bozbey HU, Duranyildiz D, Vatansever S. High serum transforming growth factor beta 1 (TGFB1) level predicts better survival in breast cancer. Tumour Biol. 2014;35(7):6941–6948. doi: 10.1007/s13277-014-1932-y. [DOI] [PubMed] [Google Scholar]
- 15.Liu YY, Han JY, Lin SC, Liu ZY, Jiang WT (2014) Effect of CDH1 gene methylation on transforming growth factor (TGF-beta)-induced epithelial-mesenchymal transition in alveolar epithelial cell line A549. Genet Mol Res 13(4):8568–8576 [DOI] [PubMed]
- 16.Huang CY, Fong YC, Lee CY, Chen MY, Tsai HC, Hsu HC, Tang CH. CCL5 increases lung cancer migration via PI3K, Akt and NF-kappaB pathways. Biochem Pharmacol. 2009;77(5):794–803. doi: 10.1016/j.bcp.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Leavy O. Innate immune signalling: TIRAP diversifies the sites of TLR signalling. Nat Rev Immunol. 2014;14(4):211. doi: 10.1038/nri3645. [DOI] [PubMed] [Google Scholar]
- 18.Medeiros MC, Frasnelli SC, Bastos Ade S, Orrico SR, Rossa C. Modulation of cell proliferation, survival and gene expression by RAGE and TLR signaling in cells of the innate and adaptive immune response: role of p38 MAPK and NF-KB. J Appl Oral Sci. 2014;22(3):185–193. doi: 10.1590/1678-775720130593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kos J, Sekirnik A, Kopitar G, Cimerman N, Kayser K, Stremmer A, Fiehn W, Werle B. Cathepsin S in tumours, regional lymph nodes and sera of patients with lung cancer: relation to prognosis. Br J Cancer. 2001;85(8):1193–1200. doi: 10.1054/bjoc.2001.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee TK, Cheung VC, Lu P, Lau EY, Ma S, Tang KH, Tong M, Lo J, Ng IO. Blockade of CD47-mediated cathepsin S/protease-activated receptor 2 signaling provides a therapeutic target for hepatocellular carcinoma. Hepatology. 2014;60(1):179–191. doi: 10.1002/hep.27070. [DOI] [PubMed] [Google Scholar]
- 21.Shabo I, Svanvik J. Expression of macrophage antigens by tumor cells. Adv Exp Med Biol. 2011;714:141–150. doi: 10.1007/978-94-007-0782-5_7. [DOI] [PubMed] [Google Scholar]
- 22.Shabo I, Olsson H, Stal O, Svanvik J. Breast cancer expression of DAP12 is associated with skeletal and liver metastases and poor survival. Clin Breast Cancer. 2013;13(5):371–377. doi: 10.1016/j.clbc.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Costa VL, Henrique R, Danielsen SA, Duarte-Pereira S, Eknaes M, Skotheim RI, Rodrigues A, Magalhaes JS, Oliveira J, Lothe RA, Teixeira MR, Jeronimo C, Lind GE. Three epigenetic biomarkers, GDF15, TMEFF2, and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin Cancer Res. 2010;16(23):5842–5851. doi: 10.1158/1078-0432.CCR-10-1312. [DOI] [PubMed] [Google Scholar]