Abstract
Purpose
We report the experience with 2,000 consecutive patients with advanced cancer who underwent testing on a genomic testing protocol, including the frequency of actionable alterations across tumor types, subsequent enrollment onto clinical trials, and the challenges for trial enrollment.
Patients and Methods
Standardized hotspot mutation analysis was performed in 2,000 patients, using either an 11-gene (251 patients) or a 46- or 50-gene (1,749 patients) multiplex platform. Thirty-five genes were considered potentially actionable based on their potential to be targeted with approved or investigational therapies.
Results
Seven hundred eighty-nine patients (39%) had at least one mutation in potentially actionable genes. Eighty-three patients (11%) with potentially actionable mutations went on genotype-matched trials targeting these alterations. Of 230 patients with PIK3CA/AKT1/PTEN/BRAF mutations that returned for therapy, 116 (50%) received a genotype-matched drug. Forty patients (17%) were treated on a genotype-selected trial requiring a mutation for eligibility, 16 (7%) were treated on a genotype-relevant trial targeting a genomic alteration without biomarker selection, and 40 (17%) received a genotype-relevant drug off trial. Challenges to trial accrual included patient preference of noninvestigational treatment or local treatment, poor performance status or other reasons for trial ineligibility, lack of trials/slots, and insurance denial.
Conclusion
Broad implementation of multiplex hotspot testing is feasible; however, only a small portion of patients with actionable alterations were actually enrolled onto genotype-matched trials. Increased awareness of therapeutic implications and access to novel therapeutics are needed to optimally leverage results from broad-based genomic testing.
INTRODUCTION
The increasing availability of next-generation sequencing combined with the availability of molecular therapeutics targeting genomically defined populations has created a growing interest in using multiplexed genomic profiling for routine cancer care and, in particular, for directing patients to relevant clinical trials. However, implementation of genomically informed therapy requires not only access to genomic profiling, but also the availability of molecularly targeted therapies matched to the genomic testing results. Availability of clinical trials may not only differ from institution to institution, but may also differ between tumor types. Enrollment onto clinical trials is also limited by trial eligibility criteria, as well as availability of slots.
As a result of growing physician and patient demand for genomic profiling at The University of Texas MD Anderson Cancer Center, we initiated a prospective clinical study where physicians were able to enroll patients who they felt would benefit from multiplex genomic testing and where patients were likely to consider enrollment onto therapeutic clinical trials. Patients with any malignancy were eligible for the study and underwent genomic testing on a Clinical Laboratory Improvement Amendments (CLIA) –compliant platform after informed consent. Here, we report the experience with the first 2,000 patients who underwent testing on the genomic testing protocol, including the frequency of actionable alterations across tumor types, subsequent enrollment onto clinical trials, and the challenges for subsequent trial enrollment.
PATIENTS AND METHODS
Patient Selection and Enrollment
Patients were enrolled onto an institutional review board–approved prospective protocol for genomic profiling, Molecular Testing for the MD Anderson Cancer Center Personalized Cancer Therapy Program (NCT01772771), after informed consent. The study was piloted in the Nellie B. Connally Breast Center and the GI Center through research nurse/coordinator identification of patients with metastatic breast and colorectal cancer. After 5 months, patient identification was transitioned to the treating oncologists, and patients perceived as likely to benefit from genomic characterization were enrolled. The majority of patients had metastatic, inoperable locally advanced or locally recurrent disease or were otherwise considered high risk. Patients were mainly accrued in disease centers with genomically relevant trials; also enrolled were patients with diseases for which there were no disease-specific trials but the treating physicians expressed interest in referring patients for phase I trial enrollment. Notably, patients with diseases for which multiplex genomic testing is accepted as standard of care (eg, lung cancer) were often tested without protocol enrollment and are under-represented.
We reviewed the first 2,000 patients who underwent genomic profiling; clinical information was collected from electronic medical records and prospective databases. Data acquisition was locked on August 26, 2014.
Genomic Analysis
Samples were evaluated using hematoxylin and eosin staining for tumor cellularity. DNA was extracted, purified, and quantified. Genomic analysis was performed by mass spectroscopy–based multiplex assay to assess the mutational status of hotspot regions in 11 genes (first 251 patients) or with next-generation sequencing using the Ion Ampliseq Cancer Panel (Life Technologies, Grand Island, NY) to assess hotspot mutations in 46 genes (Appendix Table A1, online only). In the last few months, testing expanded to 50 genes by adding EZH2, IDH2, GNA11, and GNAQ (33 patients, Appendix Table A1).
Sequence alignment and base calling were performed by Torrent Suite software V2.0.1 (Life Technologies) with Human Genome Build 19 (Hg19) as reference. Torrent Variant Caller software V1.0 (Life Technologies) was used for variant detection, and the Integrative Genomics Viewer (http://www.broadinstitute.org/igv/) was used to visualize variants. OncoSeek software was used to integrate the data.1 Routine germline testing was not performed. Germline variants were defined based on relative prevalence within the MD Anderson Cancer Center patient population and by comparing the data with dbSNP v.138 (http://www.ncbi.nlm.nih.gov/projects/SNP/) global minor allele frequency numbers where available, as previously described.2 Variants classified as likely to be germline are listed in Appendix Table A2 (online only).
Analysis
Alterations potentially targetable with established or investigational therapeutics directly or indirectly (eg, inhibiting downstream signaling) were considered actionable. The actionable genes are designated by asterisks in Appendix Table A1. The therapeutic implications of potentially actionable genes are listed in Appendix Table A3 (online only).
Categorical variables were summarized in frequency tables. Mutation rates were calculated based on the tested samples. The association between the presence or absence of actionable mutations and enrollment onto a clinical trial after the genomic testing was assessed using a Pearson χ2 test.
RESULTS
From March 2012 to July 2013, 2,601 patients were enrolled; 601 patients (23%) did not undergo testing as a result of inadequate tissue or DNA quantity or quality. Two thousand patients underwent testing, 251 by an 11-gene Sequenom panel (Sequenom, San Diego, CA) and 1,716 by the 46-gene and 33 by the 50-gene Ampliseq panel. Of these 2,000 patients, 84 had already undergone sequencing of one or more genes for standard of care (eg, BRAF V600 melanoma) or for clinical trial eligibility testing for specific trials.
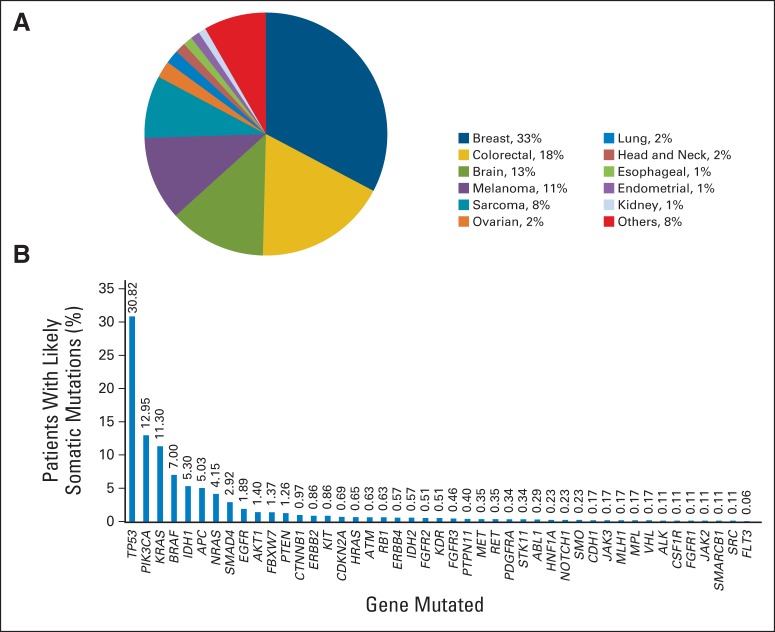
Median age was 55 years (range, 3 to 95 years). The most common cancer types tested are shown in Figure 1A. The most frequently mutated genes (with likely somatic mutations) are shown in Figure 1B.
Fig 1.
(A) The most common cancer diagnosis for patients who underwent genomic testing. Each diagnosis consisted of a variety of different histologic subtypes. (B) The most common (likely somatic) mutations observed in the overall cohort.
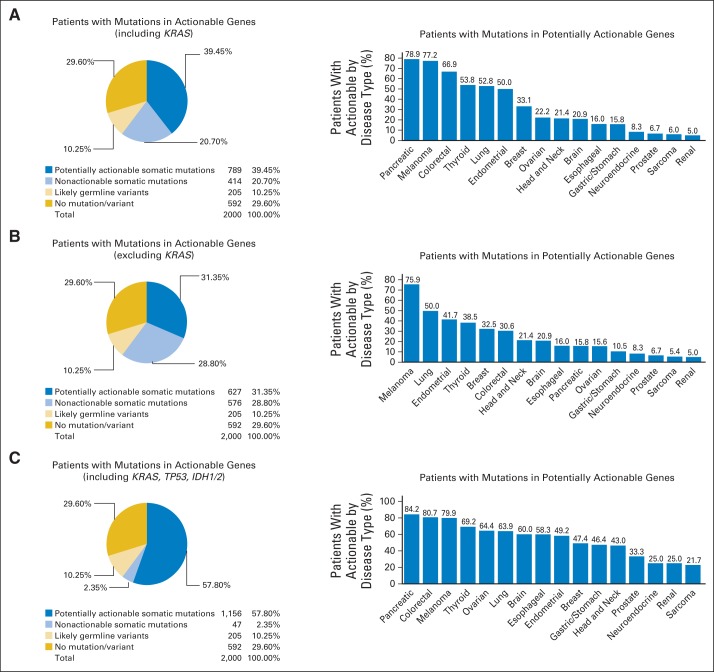
Of the 2,000 patients, 789 (39%) had at least one mutation in a potentially actionable gene, 414 (21%) had a presumed somatic mutation in a gene that was not actionable, 205 (10%) had a likely germline variant, and 592 (30%) had no mutations/variants identified (Fig 2A). Notably, most alterations considered as likely germline noted on this panel were not known functional polymorphisms or pathogenic germline mutations and were not considered actionable. One hundred forty-five patients (7.3%) had two or more potentially actionable alterations. It should be noted that for this analysis, we considered TP53, IDH1, and IDH2 mutations not actionable because of lack of genotype-matched trials for solid tumors during that time period, and we considered KRAS actionable because of availability of trials targeting MEK1, with or without KRAS selection. If KRAS was also considered not actionable, 627 patients (31%) had a mutation in a potentially actionable gene (Fig 2B). If TP53, IDH1, and IDH2 were considered potentially actionable, 1,156 patients (58%) had a mutation in a potentially actionable gene (Fig 2C). For the likely somatic mutations identified, only six genes were present at greater than 5% (with not all of these being druggable), six genes were present at a frequency of between 1% and 5%, and the remainder were present at less than 1% (Fig 1). On the basis of the mutation spectrum assessed, the percentage of patients with potentially actionable alterations varied from 5% to 79% by tumor type (Figs 2A to 2C). As we transitioned from the 11-gene Sequenom panel to the 46- and 50-gene Ampliseq panels, there were only modest increases in patients with potentially actionable alterations (Data Supplement).
Fig 2.
Frequency of actionable alterations. (A) Frequency of potentially actionable alterations among the 2,000 patients tested (left panel). For this analysis, patients were classified into mutually exclusive categories based on the highest category of alteration (actionable somatic mutation > nonactionable somatic mutation > germline variant; reporting each patient once). For the analysis, TP53, IDH1, and IDH2 were considered not actionable, and KRAS was considered actionable. Frequency of potentially actionable alterations by tumor type among tumor types with 10 or more patients tested (right panel). (B) Frequency of potentially actionable alterations with KRAS as well as TP53, IDH1, and IDH2 considered not actionable (left panel). Frequency of potentially actionable alterations by tumor type with this classification (right panel). (C) Frequency of potentially actionable alterations considering KRAS, TP53, IDH1, and IDH2 actionable (left panel). Frequency of potentially actionable alterations by tumor type with this classification (right panel).
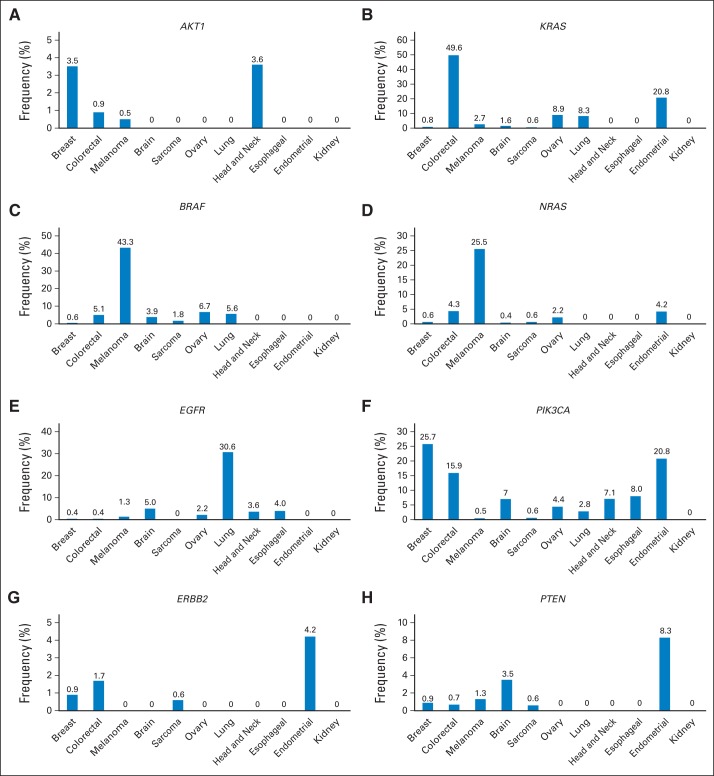
The frequency of mutations in specific genes is shown for different tumor types where 20 patients or more were tested (Table 1). In addition to the expected common disease-mutation associations (eg, KRAS–colon cancer, EGFR–lung cancer), we identified a set of potentially actionable genes frequently mutated in specific diseases but also uncommonly mutated in other cancer types (eg, BRAF, PIK3CA). Furthermore, some genes had a low incidence of mutations across several tumor types (eg, ERBB2; Fig 3).
Table 1.
Frequency of Alterations in All Tested Genes by Tumor Type
| Gene | Overall (N = 2,000) |
Breast (n = 655) |
CRC (n = 353) |
Melanoma (n = 224) |
Brain (n = 258) |
Sarcoma (n = 166) |
Ovary (n = 45) |
Lung (n = 36) |
Head and Neck (n = 28) |
Esophageal (n = 25) |
Endometrial (n = 24) |
Kidney (n = 20) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | No. of Patients | % | |
| Tested using Ion Torrent | 1,749 | 531 | 288 | 224 | 201 | 161 | 45 | 36 | 28 | 25 | 24 | 20 | ||||||||||||
| ABL1 | 5 | 0.3 | 2 | 0.4 | 1 | 0.4 | 1 | 0.5 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| AKT1* | 28 | 1.4 | 23 | 3.5 | 3 | 0.9 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 3.6 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| ALK | 2 | 0.1 | 0 | 0.0 | 1 | 0.4 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 4.2 | 0 | 0.0 |
| APC | 88 | 5.0 | 8 | 1.5 | 69 | 24.0 | 1 | 0.5 | 1 | 0.5 | 1 | 0.6 | 2 | 4.4 | 0 | 0.0 | 1 | 3.6 | 0 | 0.0 | 1 | 4.2 | 0 | 0.0 |
| ATM | 11 | 0.6 | 3 | 0.6 | 1 | 0.4 | 4 | 1.8 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 1 | 2.8 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| BRAF* | 140 | 7.0 | 4 | 0.6 | 18 | 5.1 | 97 | 43.3 | 10 | 3.9 | 3 | 1.8 | 3 | 6.7 | 2 | 5.6 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| CDH1 | 3 | 0.2 | 0 | 0.0 | 2 | 0.7 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| CDKN2A | 12 | 0.7 | 1 | 0.2 | 0 | 0.0 | 5 | 2.2 | 2 | 1.0 | 1 | 0.6 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 4.0 | 0 | 0.0 | 0 | 0.0 |
| CSF1R | 2 | 0.1 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 4.2 | 0 | 0.0 |
| CTNNB1 | 17 | 1.0 | 0 | 0.0 | 4 | 1.4 | 6 | 2.7 | 0 | 0.0 | 3 | 1.9 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 4.2 | 0 | 0.0 |
| EGFR | 33 | 1.9 | 2 | 0.4 | 1 | 0.4 | 3 | 1.3 | 10 | 5.0 | 0 | 0.0 | 1 | 2.2 | 11 | 30.6 | 1 | 3.6 | 1 | 4.0 | 0 | 0.0 | 0 | 0.0 |
| ERBB2 | 15 | 0.9 | 5 | 0.9 | 5 | 1.7 | 0 | 0.0 | 0 | 0.0 | 1 | 0.6 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 4.2 | 0 | 0.0 |
| ERBB4 | 10 | 0.6 | 1 | 0.2 | 5 | 1.7 | 3 | 1.3 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 4.0 | 0 | 0.0 | 0 | 0.0 |
| FBXW7 | 24 | 1.4 | 0 | 0.0 | 0 | 0.0 | 2 | 0.9 | 0 | 0.0 | 1 | 0.6 | 0 | 0.0 | 1 | 2.8 | 1 | 3.6 | 0 | 0.0 | 5 | 20.8 | 0 | 0.0 |
| FGFR1 | 2 | 0.1 | 1 | 0.2 | 0 | 0.0 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| FGFR2 | 9 | 0.5 | 2 | 0.4 | 1 | 0.4 | 2 | 0.9 | 2 | 1.0 | 0 | 0.0 | 1 | 2.2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| FGFR3 | 8 | 0.5 | 1 | 0.2 | 2 | 0.7 | 2 | 0.9 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 3.6 | 0 | 0.0 | 0 | 0.0 | 1 | 5.0 |
| FLT3 | 1 | 0.1 | 0 | 0.0 | 1 | 0.4 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| GNAS* | 13 | 0.7 | 0 | 0.0 | 5 | 1.4 | 4 | 1.8 | 0 | 0.0 | 1 | 0.7 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| HNF1A | 4 | 0.2 | 0 | 0.0 | 0 | 0.0 | 2 | 0.9 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 4.2 | 0 | 0.0 |
| HRAS | 12 | 0.7 | 5 | 0.9 | 1 | 0.4 | 3 | 1.3 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 2 | 7.1 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| IDH1* | 106 | 5.3 | 1 | 0.2 | 1 | 0.3 | 3 | 1.3 | 92 | 35.7 | 5 | 3.0 | 0 | 0.0 | 1 | 2.8 | 0 | 0.0 | 0 | 0.0 | 1 | 4.2 | 0 | 0.0 |
| JAK2 | 2 | 0.1 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| JAK3 | 3 | 0.2 | 2 | 0.4 | 0 | 0.0 | 0 | 0.0 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| KDR | 9 | 0.5 | 0 | 0.0 | 1 | 0.4 | 5 | 2.2 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 1 | 2.8 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| KIT | 15 | 0.9 | 1 | 0.2 | 0 | 0.0 | 10 | 4.5 | 1 | 0.5 | 2 | 1.2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| KRAS* | 226 | 11.3 | 5 | 0.8 | 175 | 49.6 | 6 | 2.7 | 4 | 1.6 | 1 | 0.6 | 4 | 8.9 | 3 | 8.3 | 0 | 0.0 | 0 | 0.0 | 5 | 20.8 | 0 | 0.0 |
| MET* | 7 | 0.4 | 2 | 0.4 | 2 | 0.6 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 2.8 | 1 | 3.6 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| MLH1 | 3 | 0.2 | 1 | 0.2 | 0 | 0.0 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| MPL | 3 | 0.2 | 2 | 0.4 | 0 | 0.0 | 0 | 0.0 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| NOTCH1 | 4 | 0.2 | 1 | 0.2 | 1 | 0.4 | 0 | 0.0 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 4.0 | 0 | 0.0 | 0 | 0.0 |
| NRAS* | 83 | 4.2 | 4 | 0.6 | 15 | 4.3 | 57 | 25.5 | 1 | 0.4 | 1 | 0.6 | 1 | 2.2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 4.2 | 0 | 0.0 |
| PDGFRA | 6 | 0.3 | 0 | 0.0 | 2 | 0.7 | 2 | 0.9 | 2 | 1.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| PIK3CA* | 259 | 12.9 | 168 | 25.7 | 56 | 15.9 | 1 | 0.5 | 18 | 7.0 | 1 | 0.6 | 2 | 4.4 | 1 | 2.8 | 2 | 7.1 | 2 | 8.0 | 5 | 20.8 | 0 | 0.0 |
| PTEN | 22 | 1.3 | 5 | 0.9 | 2 | 0.7 | 3 | 1.3 | 7 | 3.5 | 1 | 0.6 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 2 | 8.3 | 0 | 0.0 |
| PTPN11 | 7 | 0.4 | 1 | 0.2 | 0 | 0.0 | 2 | 0.9 | 4 | 2.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| RB1 | 11 | 0.6 | 0 | 0.0 | 3 | 1.0 | 1 | 0.5 | 2 | 1.0 | 1 | 0.6 | 0 | 0.0 | 1 | 2.8 | 0 | 0.0 | 2 | 8.0 | 0 | 0.0 | 0 | 0.0 |
| RET* | 7 | 0.4 | 0 | 0.0 | 2 | 0.6 | 1 | 0.5 | 1 | 0.4 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| SMAD4 | 51 | 2.9 | 6 | 1.1 | 30 | 10.4 | 1 | 0.5 | 3 | 1.5 | 0 | 0.0 | 1 | 2.2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 4.2 | 0 | 0.0 |
| SMARCB1 | 2 | 0.1 | 0 | 0.0 | 1 | 0.4 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| SMO | 4 | 0.2 | 0 | 0.0 | 1 | 0.4 | 1 | 0.5 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 1 | 2.8 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| SRC | 2 | 0.1 | 1 | 0.2 | 0 | 0.0 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| STK11 | 6 | 0.3 | 1 | 0.2 | 1 | 0.4 | 2 | 0.9 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 2 | 5.6 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| TP53 | 539 | 30.8 | 150 | 28.3 | 155 | 53.8 | 27 | 12.1 | 70 | 34.8 | 28 | 17.4 | 20 | 44.4 | 9 | 25.0 | 9 | 32.1 | 13 | 52.0 | 9 | 37.5 | 4 | 20.0 |
| VHL | 3 | 0.2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 1 | 0.5 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 2 | 10.0 |
NOTE. Two thousand patient underwent testing overall, 1,749 patients on Ion Torrent platform and 251 patients on Sequenom platform. Thirty-three patients underwent EZH2 and GNA11 mutation testing and 352 had GNAQ testing, and no mutations were found. In addition, 352 patients had IDH2 testing, and two mutations were found (in brain tumors).
Abbreviation: CRC, colorectal cancer.
Genes that were on both Sequenom and Ion Torrent platforms. Information on these genes was available in all 2,000 patients. Frequency of mutations in all other genes was calculated based on the 1,749 patients tested.
Fig 3.
Frequency of selected alterations in different tumor types. Frequency of (A) AKT1, (B) KRAS, (C) BRAF, (D) NRAS, (E) EGFR, (F) PIK3CA, (G) ERBB2, and (H) PTEN mutations in breast cancer, colorectal cancer, melanoma, brain tumors, sarcoma, ovarian cancer, lung cancer, head and neck cancer, esophageal cancer, endometrial cancer, and kidney tumors.
Enrollment Onto Clinical Trials
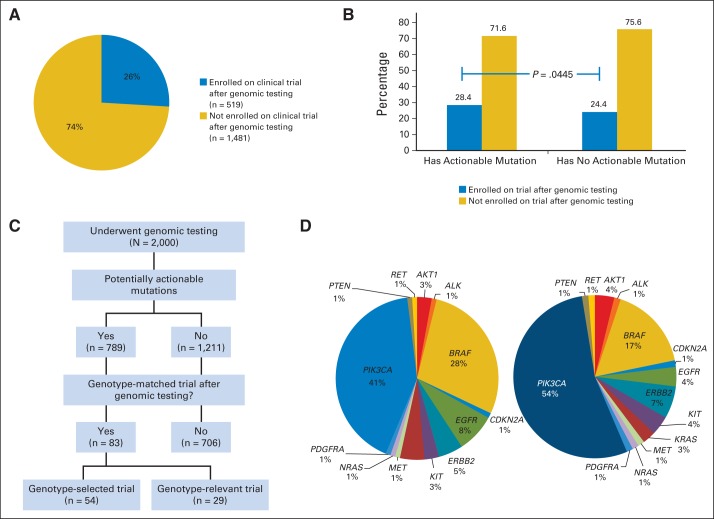
Five hundred nineteen patients (26%) enrolled onto therapeutic clinical trials at MD Anderson Cancer Center after genomic testing results were available (Fig 4A). Patients with mutations in potentially actionable genes were marginally more likely to be treated on therapeutic clinical trials after test results were available than those without actionable mutations (28.4% v 24.4%, respectively; P = .0445; Fig 4B).
Fig 4.
Clinical trial enrollment. (A) Proportion of patients enrolled onto a therapeutic clinical trial after results of genomic testing were available. (B) Proportion of patients enrolled onto a therapeutic clinical trial among patients with actionable mutations and those without actionable alterations (28.4% v 24.4%, respectively; P = .0445). (C) CONSORT diagram of patients who went on genotype-matched trials. (D) Key genomic alterations of patients who were enrolled onto genotype-matched trials (left panel). The right panel depicts the key genomic alterations of patients who were enrolled onto genotype-matched trials excluding alterations detectable by standard-of-care assays (EGFR, n = 5; BRAF, n = 16; and KRAS mutant, n = 3).
Among the 789 patients with potentially actionable alterations (defined as shown in Fig 2A), 83 (11%) went on genotype-matched trials after genomic testing; 54 (7%) were treated on a genotype-selected trial (requiring a mutation for eligibility), and 29 (4%) were treated on a genotype-relevant trial (ie, a trial without biomarker selection but with an agent that targeted the gene product or downstream signaling). In addition, 121 patients (15%) were treated on other clinical trials (Fig 3C). Figure 4D demonstrates the key genomic alterations of patients who were enrolled onto genotype-matched trials. The right panel in Figure 4D depicts the key genomic alterations of patients who were enrolled onto genotype-matched trials excluding 24 patients who had alterations detectable by standard-of-care assays (EGFR in lung cancer, BRAF in melanoma, and KRAS in colon cancer).
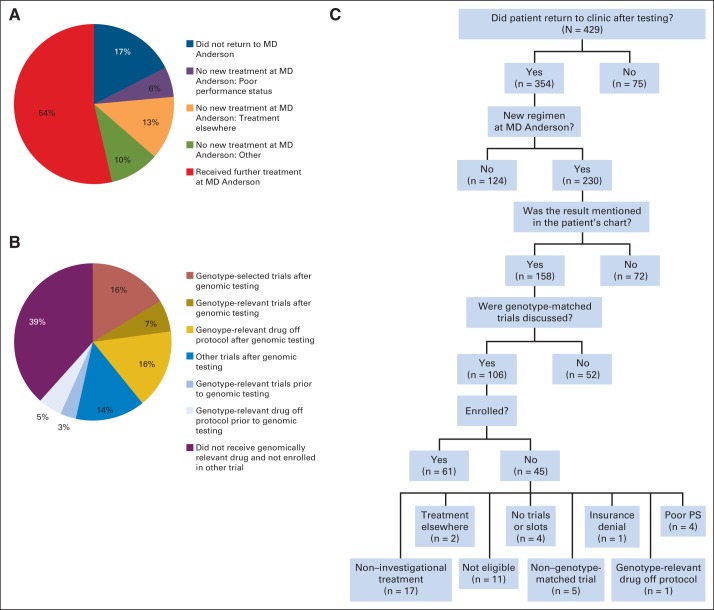
To gain insights into obstacles for trial enrollment, we reviewed the records of the 429 patients with PIK3CA/AKT1/PTEN/BRAF mutations. The median time from CLIA test results to last follow-up was 257 days (range, 4 to 749 days for patients who returned to clinic). Surprisingly, 199 (46%) of 429 patients with PIK3CA/AKT1/PTEN/BRAF mutations did not undergo a new regimen of therapy at our institution after testing (Fig 5A); 75 (17%) of 429 patients did not return to the institution after testing, 55 (13%) returned but elected to be treated elsewhere (usually locally; ie, closer to home), 26 (6%) did not initiate new treatment as a result of declining performance status, and 43 (10%) did not start new treatment for other reasons (eg, stable disease). Of the 230 patients who received a new treatment at MD Anderson Cancer Center, 40 (17%) were treated on genotype-selected trials after testing, 16 (7%) were treated on genotype-relevant trials, 35 (15%) were treated on other trials, and 40 (17%) received a genotype-relevant drug (as standard of practice or off protocol) after testing (Fig 5B). In addition, eight patients (2%) had been treated on a genotype-relevant trial before testing, and 12 patients (5%) received a genotype-relevant drug off protocol before testing. Thus, of the 230 patients with PIK3CA/AKT1/PTEN/RAF mutations who received subsequent therapy at MD Anderson Cancer Center, 116 (50%) received a genotype-matched drug (96 patients after testing, 42%).
Fig 5.
Genotype-matched trial enrollment in patients who had tumors with PIK3CA/AKT1/PTEN/BRAF mutations. (A) Subsequent treatment of 429 patients who had tumors with PIK3CA/AKT1/PTEN/BRAF mutations. (B) Treatments given to the 230 patients with PIK3CA/AKT1/PTEN/BRAF–mutant tumors who received a subsequent new treatment regimen at The University of Texas MD Anderson Cancer Center. (C) Reasons patients who had tumors with PIK3CA/AKT1/PTEN/BRAF mutations were not enrolled onto trials. PS, performance status.
The clinical course of the 429 patients with PIK3CA/AKT/PTEN/BRAF mutations is shown in Figure 5C. Of the 354 patients who returned to the institution after testing, 230 went on to subsequent new regimens. One hundred fifty-eight (68%) of 230 patients had the genomic testing result noted in the transcribed clinical notes, 106 (46%) had documentation of discussion of genotype-matched trials, and 61 (27%) were enrolled onto a genotype-matched trial (Fig 5C). Trials were discussed with 45 patients who subsequently were not treated on a genotype-matched trial but rather elected noninvestigational therapy, a non–genotype-matched trial, or treatment elsewhere or received genotype-relevant drug off protocol. In addition, slots could not be identified for four patients, four patients had too poor performance status, 11 patients were ineligible for trials for other reasons, and one patient had insurance denial for trial participation.
Also of note, 13 patients ultimately went on more than one genotype-matched trial after genomic testing. Fourteen patients had more than one actionable genomic alteration, and two patients went on trials simultaneously targeting two alterations (one patient with KRAS and PIK3CA and the other with KIT and PIK3CA).
We also evaluated the time required for genomic testing. Of the 429 patients with PIK3CA/AKT/PTEN or BRAF mutations enrolled onto the study, 103 patients already had CLIA genomic testing (usually single-gene testing) done before multiplex testing. For the other 326 patients, time from consent to test results was an average of 31 days and median of 26 days. Notably, 102 patients (23.8%) with PIK3CA/AKT1/PTEN/BRAF mutations had another treatment started before test results were received; 13 of these were genotype-relevant choices.
DISCUSSION
Broad implementation of multiplex hotspot testing across an institution is feasible. However, overall, only a small portion of patients with actionable alterations were enrolled onto genotype-matched trials. Notably, 46% of patients with PIK3CA, AKT, PTEN, or BRAF alterations did not receive subsequent treatment at the institution; however, of patients who received subsequent treatment, 23% were treated on a genotype-matched trial after genomic testing. One hundred fifteen (48%) of 242 patients who received additional treatment after multiplex testing received a genotype-relevant drug. Thus, for patients who receive additional treatment, multiplex testing was used to drive genomically informed clinical decision making.
Seven hundred eighty-nine patients (39%) had a mutation in a potentially actionable gene. However, the frequency of actionable alterations differs widely (from 5% to 79%) between different tumor lineages; thus, the utility of this platform may differ based on tumor type. Furthermore, likely actionable mutations at a frequency of greater than 5% were present in only four genes, and likely actionable mutations at a frequency between 1% and 5% were also present in only four genes. The low frequency of mutations in most actionable genes could present a challenge for validating the actionability of the targets. In addition, to identify sufficient patients for genotype-selected trials, it will be necessary to test large numbers of patients with multiplexed testing.
For this analysis, we categorized TP53, IDH1, and IDH2 as not actionable, although IDH-targeted therapies are now in trials, and recently trials have started using TP53 as a selection marker. Also notable is that we considered KRAS as an actionable gene, because there are KRAS genotype-selected trials. However, these trials may not be applicable for several tumor types, and available slots are limited; thus, this is an overestimation of true actionability. Although we categorized mutations as being in a potentially actionable gene, it should be noted that during clinical trial consideration, the level of evidence for the functional impact of each mutation also needs to be considered. Therefore, the frequency of actionable mutations may be less than the frequency of mutations in potentially actionable genes. Also of note, there is growing interest in using larger panels, such as whole-exome sequencing or targeted full-length sequencing of actionable genes. These approaches not only cover nonhotspot alterations, but they may also yield copy number information and, depending on design, gene rearrangements. Such larger panels may demonstrate potentially actionable alterations in many more patients.3–6
We noted several challenges in linking genomic testing to genomically matched trials. First, 17% of patients did not return to MD Anderson Cancer Center after testing, and 13% elected to be treated closer to home. Therefore, in nearly a third of the patients, the genomic information from testing was likely not used for therapy planning. This is at least in part attributable to MD Anderson Cancer Center being a referral center. Although one option would be to limit testing to patients who are local, genomic testing remains an important tool to deliver the most informative consultations. However, before testing, it is important to determine whether the patient is interested in clinical trials. Second, 6% of patients did not initiate new treatment as a result of declining performance status, and several were ineligible for trials because of their performance status. It remains difficult to predict health deterioration in patients with advanced cancer, and unfortunately, 105 patients (5%) died within 3 months of testing. Thus, careful assessment of performance status and comorbidities is needed. Testing for non–standard-of-care markers is most likely to benefit patients with adequate performance status to participate in clinical trials in the next two lines of treatment.
Many patients underwent genomic characterization to guide future treatments rather than point-of-care testing to decide on next-line therapy. This approach allows for the most expedited delivery of the next line of therapy but represents an opportunity lost for enrollment onto genotype-matched trials. In our study, time from consent to genomic report was a median of 26 days; this delay, at least in part, is attributable to the time to locate archival tissue blocks. Point-of-care tumor biopsies for molecular profiling or liquid biopsy approaches may not only immediately obtain samples for testing, but may also overcome potential challenges as a result of genomic evolution.
Even studies that have performed point-of-care testing have demonstrated the challenges to offering a genotype-matched treatment option to all patients. In the Lung Cancer Mutation Consortium, an oncogenic driver was found in 64% of patients who underwent genomic testing; however, only 28% of patients were enrolled onto a marker-selected trial.7 In the SAFIR01 breast cancer trial, a targetable alteration was identified in 46% of patients and therapy was personalized in 13% of patients, with 28% of patients with a targetable alteration receiving a matched therapy.8 In the institutional series from Vanderbilt, Lovly et al9 reported that among 150 patients with melanoma who underwent SNaPSHOT testing, 60% had at least one mutation and 43% of patients harboring metastatic disease with actionable mutations were treated in genomically matched trials. In our study, genomic analysis was not point-of-care testing and was not linked to a prespecified treatment algorithm. Keeping that in mind, with widespread implementation of genomic testing, enrollment of 11% of patients who underwent genomic testing onto genomically matched trials overall and nearly 40% of patients with PIK3CA/AKT/PTEN/BRAF mutations receiving genomically relevant drugs after testing establish the potential value of genomic testing for therapy selection. However, a major obstacle was the paucity of genomically matched trials, especially for less common tumor types and for the less commonly mutated genes. Tracking frequency of alterations (Table 1) can help determine feasibility of genotype-selected trials in each disease type.
Over the study period, there was a steady increase in number of genes for which we have genotype-selected trials. We have leveraged our genomic testing to design genotype-selected investigator-initiated trials. We have also recruited several industry-sponsored trials and activated genomically selected histology-independent basket trials. Novel trial approaches to explore antitumor efficacy in rare molecular subtypes and novel trial access mechanisms such as just-in-time trial access activating genomically matched trials appropriate for individual patient genotype are needed.
Our study was observational in design; we relied on retrospective assessment of impact of testing on treatment choices. Notably, only 69% patients had the genomic testing results acknowledged in the transcribed clinic notes, and 44% had documentation of discussion of genotype-matched trials. It is unclear whether test results were indeed not appreciated and trial options not discussed or whether these were simply not documented. However, it is possible that alerting treating physicians that genomic testing results are available may improve trial accrual. To facilitate this, we have activated a clinical trial alert system. Physicians now receive an e-mail alert when actionable genomic test results are received, with a list of genotype-matched trials. It has been recently reported that even oncologists at leading cancer centers have variable comfort levels with their knowledge of genomics.10 To assist in decision making, we have put together a Precision Oncology Decision Support Team. For common actionable genes, variant-level information on mutations that have been experimentally characterized based on published literature is included on our Web site PersonalizedCancerTherapy.org. We expect that by increasing education and streamlining decision making, we can improve the implementation of genomically informed cancer therapy.
Acknowledgment
We thank Elena Vess for secretarial assistance. We thank Wanlin Wang, Chetna Wathoo, Rabiul Islam, Lilibeth Castillo, Blessy Sajan, Constance Dennis, Faith Minglana, and Kristin Hargraves for assistance in data acquisition.
Appendix
Table A1.
Codons Tested on Sequenom 11-Gene Assay and the Ampliseq 46-Gene (CMS46) and 50-Gene (CMS50) Assays
| Gene | Codons Tested in Sequenom Panel | Codons Tested in CMS46 | Codons Tested in CMS50 |
|---|---|---|---|
| ABL1* | 237-260, 275-283, 303-319, 350-362, 387-412 | 232-260, 275-279, 314-360, 380-412 | |
| AKT1* | 17,173, 179 | 16-59 | 16-52, 154-183 |
| ALK* | 1172-1177, 1259-1277 | 1172-1204, 1270-1279 | |
| APC | 865-886, 1105-1122, 1289-1322, 1349-1382, 1430-1467, 1487-1509 | 860-891, 1089-1125, 1284-1326, 1342-1384, 1426-1471, 1483-1524, 1543-1582 | |
| ATM* | 343-355, 395-412, 601-614, 837-862, 1307-1324, 1674-1693, 1733-1758, 1785-1802, 1935-1957, 2436-2445, 2650-2667, 2693-2715, 2721-2739, 2888-2891, 2937-2950, 2996-3016, 3037-3052 | 326-355, 407-412, 601-626, 834-865, 1292-1325, 1674-1707, 1726-1757, 1790-1815, 1926-1946, 2436-2454, 2650-2667, 2682-2711, 2718-2736, 2865-2891, 2933-2950, 2996-3026, 3041-3057 | |
| BRAF* | 464, 466, 469, 594, 597, 600, 601 | 439-471, 581-605 | 439-473, 581-611 |
| CDH1 | 69-92, 351-373, 395-415 | 65-96, 337-374, 380-408 | |
| CDKN2A* | 51-76 | 51-90, 98-140 | |
| CSF1R* | 299-318, 952-973 | 297-319, 953-973 | |
| CTNNB1 | 12-45 | 9-48 | |
| EGFR* | 89-125, 280-297, 575-601, 698-722, 729-761, 766-790, 803-823, 830-866 | 96-123, 279-297, 575-601, 695-726, 729-796, 807-823, 855-875 | |
| ERBB2* | 753-769, 772-797, 832-852, 875-883 | 752-797, 839-882 | |
| ERBB4* | 136-141, 177-186, 234-247, 272-289, 303-322, 343-363, 588-619, 923-943 | 136-141, 167-186, 225-247, 254-290, 295-323, 333-367, 580-623, 919-948 | |
| EZH2 | 625-649 | ||
| FBXW7 | 264-279, 381-400, 450-472, 478-506, 566-583 | 264-287, 378-403, 434-473, 478-509, 567-594 | |
| FGFR1* | 121-139, 247-268 | 120-148, 247-273 | |
| FGFR2* | 250-268, 297-313, 367-395, 546-558 | 250-275, 296-313, 362-399, 546-558 | |
| FGFR3* | 247-268, 377-409, 634-653, 681-712, 790-807 | 247-277, 367-402, 631-653, 690-719, 771-807 | |
| FLT3* | 441-458, 569-575, 589-613, 662-682, 828-846 | 437-466, 570-610, 663-685, 828-847 | |
| GNA11* | 202-219 | ||
| GNAQ* | 209 | 206-245 | |
| GNAS | 201 | 196-218 | 196-240 |
| HNF1A | 198-217, 253-282 | 192-221, 253-282 | |
| HRAS* | 5-23, 48-79 | 5-35, 42-82 | |
| IDH1 | 132 | 118-134 | 101-135 |
| IDH2 | 172 | 133-177 | |
| JAK2* | 604-622 | 603-622 | |
| JAK3* | 568-578, 709-729 | 128-140, 568-580, 709-733 | |
| KDR* | 240-258, 267-280, 472-490, 872-892.959-985, 1138-1161, 1192-1216, 1301-1321, 1336-1356 | 244-291, 471-480, 872-894, 961-988, 1135-1156, 1192-1221, 1283-1310, 1324-1357 | |
| KIT* | 47-69, 501-514, 536-549, 550-585, 641-684, 714-728, 807-828, 836-854 | 23-58, 494-514, 525-587, 627-661, 664-684, 714-724, 802-828, 832-858 | |
| KRAS* | 12,13,61,146 | 5-28, 40-67, 136-150 | 5-66, 114-150 |
| MET* | 375,848,988,1010, 1112,1124,1248,1253, 1268 | 160-187, 362-379, 992-1017, 1105-1126, 1247-1268 | 159-188, 339-378, 816-856, 981-1012, 1105-1132, 1246-1274 |
| MLH1 | 373-393 | 373-415 | |
| MPL* | 499-522 | 501-522 | |
| NOTCH1* | 1566-1605, 1673-1697 | 1566-1602, 1673-1680, 2536-2476 | |
| NPM1* | 283-295 | 283-295 | |
| NRAS* | 12,13,61,146 | 6-22, 53-69 | 3-31, 43-69, 124-150 |
| PDGFRA* | 552-570, 647-688, 819-847 | 552-583, 644-668, 671-709, 819-854 | |
| PIK3CA* | 60,88,110,111,345,405, 418,420,453,539,542, 545,546,909,1021,1025, 1043,1046,1047,1049 | 77-98, 328-351, 418-422, 533-551, 688-716, 1019-1049, 1065-1069 | 54-90, 106-118, 316-351, 390-422, 449-468, 522-549, 677-720, 898-924, 1017-1051, 1065-1069 |
| PTEN* | 5-24, 55-70, 167-184, 212-222, 240-266, 282-300, 316-342 | 1-25, 55-70, 99-135, 165-184, 212-215, 231-267, 282-300, 312-342 | |
| PTPN11* | 53-82, 486-506 | 46-82, 485-527 | |
| RB1 | 132-154, 195-203, 350-371, 549-565, 566-585, 655-680, 703-724, 743-765 | 130-159, 196-203, 314-345, 350-366, 452-463, 547-582, 655-691, 703-724, 743-770 | |
| RET* | 918 | 609-627, 630-654, 762-774, 880-901, 914-931 | 608-654, 762-786, 875-924 |
| SMAD4 | 109-128, 167-184, 228-247, 304-319, 330-363, 385-404, 444-472, 497-526 | 98-136, 142-146, 165-202, 242-263, 307-319, 326-365, 384-424, 443-474, 494-532 | |
| SMARCB1 | 39-55, 154-167, 182-203, 376-386 | 35-72, 144-206, 373-386 | |
| SMO* | 186-218, 310-340, 399-418, 516-542, 626-646 | 186-228, 307-331, 391-419, 511-542, 608-646 | |
| SRC* | 514-534 | 499-533 | |
| STK11* | 30-62, 174-199, 253-281, 325-360 | 22-64, 155-181, 191-207, 253-285, 317-361 | |
| TP53 | 1-18, 81-114, 126-135, 149-181, 187-223, 230-253, 269-306, 332-344 | 1-20, 68-113, 126-138, 149-223, 225-258, 263-307, 332-367 | |
| VHL | 88-110, 120-149, 147-175 | 78-108, 114-150, 155-174 |
Genes considered actionable in the clinical trial enrollment analysis.
Table A2.
List of Likely Germline Variants
| Gene | Codon | Wild Type | Variant Type |
|---|---|---|---|
| ABL1 | 247 | K | R |
| APC | 870 | P | S |
| APC | 1317 | E | Q |
| ATM | 410 | V | A |
| ATM | 604 | P | S |
| ATM | 858 | F | L |
| ATM | 1309 | A | T |
| ATM | 1691 | S | R |
| FGFR3 | 384 | F | L |
| JAK3 | 132 | P | T |
| JAK3 | 722 | V | I |
| KDR | 482 | C | R |
| KDR | 1356 | V | A |
| KIT | 541 | M | L |
| MET | 168 | E | D |
| MET | 362 | M | T |
| MET | 375 | N | S |
| MET | 1010 | T | I |
| MLH1 | 384 | V | D |
| PIK3CA | 391 | I | M |
| STK11 | 354 | F | L |
| TP53 | 273 | R | H |
Table A3.
Selected Therapeutic Implications of Potentially Actionable Genes on 50-Gene Panel
| Gene | Potential Therapeutic Implications |
|---|---|
| ABL1 | Treatment with ABL or BCR-ABL inhibitors |
| AKT1 | Treatment with AKT or mTOR inhibitors |
| ALK | Treatment with ALK inhibitors |
| ATM | Treatment with PARP inhibitors |
| BRAF | Treatment with BRAF inhibitors |
| CSF1R | Treatment with CSF1R monoclonal antibody and inhibitors |
| EGFR | Treatment with EGFR inhibitors |
| ERBB4 (HER4) | Treatment with HER4 inhibitors |
| FGFR1 | Treatment with FGFR1 inhibitors |
| FGFR2 | Treatment with FGFR2 inhibitors |
| FGFR3 | Treatment with FGFR3 inhibitors |
| FLT3 | Treatment with FLT3 inhibitors |
| GNA11 | Treatment with PKC and MEK inhibitors |
| GNAQ | Treatment with PKC and MEK inhibitors |
| HRAS | Treatment with MEK inhibitors |
| JAK2 | Treatment with JAK inhibitors |
| JAK3 | Treatment with JAK inhibitors |
| KDR | Treatment with KDR inhibitors |
| KIT | Treatment with KIT inhibitors |
| KRAS | Treatment with MEK inhibitors |
| MET | Treatment with MET inhibitors |
| MPL* | Treatment with JAK2 inhibitors |
| NOTCH1 | Treatment with γ-secretase inhibitors |
| NPM1 | Correlate with positive response to all-trans-retinoic acid therapy and chemotherapy in AML |
| NRAS | Treatment with MEK inhibitors |
| PDGFRA | Treatment with PDGFRA inhibitors |
| PIK3CA | Treatment with PI3K, AKT, or mTOR inhibitors |
| PTEN | Treatment with p110β, AKT, or mTOR inhibitors |
| PTPN11 | Treatment with MEK inhibitors |
| RET | Treatment with Ret inhibitors |
| SMO | Treatment with SMO inhibitors |
| SRC | Treatment with SRC inhibitors |
| STK11 | Treatment with mTOR or AMPK inhibitors |
NOTE. Genes were classified as potentially actionable if there is at least preclinical evidence suggesting genomic alteration may affect function and the gene can be targeted with an approved or investigational agent or if the gene is being used as an enrollment criterion for ongoing genotype-selected trials.
Abbreviations: AML, acute myeloid leukemia; EGFR, epidermal growth factor receptor; HER4, human epidermal growth factor receptor 4; mTOR, mammalian target of rapamycin; PARP, poly (ADP-ribose) polymerase; PDGFRA, platelet-derived growth factor receptor alpha; PI3K, phosphatidylinositol 3-kinase.
Borderline classification as actionable.
Footnotes
See accompanying editorial on page 2725
Supported in part by the Sheikh Khalifa Al Nahyan Ben Zayed Institute for Personalized Cancer Therapy, National Cancer Institute Grant No. U01 CA180964, National Center for Advancing Translational Sciences Grant No. UL1 TR000371, the Nellie B. Connally Breast Cancer Research Endowment, Cancer Prevention Research Institute of Texas Grant No. RP110584, National Institutes of Health Grant No. R21 CA159270, and The University of Texas MD Anderson Cancer Center Support Grant No. P30 CA016672.
Authors' disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Disclosures provided by the authors are available with this article at www.jco.org.
AUTHOR CONTRIBUTIONS
Conception and design: Funda Meric-Bernstam, Kenna Shaw, Yisheng Li, Keyur Patel, John Mendelsohn, Gordon B. Mills
Financial support: Funda Meric-Bernstam, John Mendelsohn, Gordon B. Mills
Administrative support: Funda Meric-Bernstam, Kenna Shaw, John Mendelsohn, Gordon B. Mills
Provision of study materials or patients: Funda Meric-Bernstam, Scott Kopetz, Michael A. Davies, Sarina A. Piha-Paul, Filip Janku, Naoto Ueno, David Hong, John De Groot, Vinod Ravi, Raja Luthra, Keyur Patel, Russell Broaddus
Collection and assembly of data: Funda Meric-Bernstam, Lauren Brusco, Chacha Horombe, Scott Kopetz, David Hong, Raja Luthra, Keyur Patel, Russell Broaddus
Data analysis and interpretation: Funda Meric-Bernstam, Lauren Brusco, Kenna Shaw, Scott Kopetz, Michael A. Davies, Mark Routbort, Sarina A. Piha-Paul, Filip Janku, Naoto Ueno, David Hong, John De Groot, Vinod Ravi, Yisheng Li, John Mendelsohn, Gordon B. Mills
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Funda Meric-Bernstam
Honoraria: Genentech, Roche Diagnostics, Sysmex
Consulting or Advisory Role: Novartis, Roche, Genentech
Research Funding: Novartis (Inst), AstraZeneca (Inst), Taiho (Inst), Genentech (Inst), Calithera (Inst), Debiopharma (Inst), Bayer (Inst)
Lauren Brusco
No relationship to disclose
Kenna Shaw
No relationship to disclose
Chacha Horombe
No relationship to disclose
Scott Kopetz
Consulting or Advisory Role: Amgen, Roche, GlaxoSmithKline, Jannsen, Bristol-Myers Squibb, Agendia, Merrimack, Sysmex, Bayer, Taiho, Sanofi, Array BioPharma
Research Funding: Roche, Amgen, GlaxoSmithKline, Sanofi, Sysmex, Biocartis, Guardant, Agendia
Michael A. Davies
Consulting or Advisory Role: GlaxoSmithKline, Genentech/Roche, Novartis, Sanofi
Research Funding: GlaxoSmithKline (Inst), Genentech/Roche (Inst), AstraZeneca (Inst), Merck (Inst), Oncothyreon (Inst), Myriad Genetics (Inst), Sanofi (Inst)
Mark Routbort
No relationship to disclose
Sarina A. Piha-Paul
No relationship to disclose
Filip Janku
No relationship to disclose
Naoto Ueno
No relationship to disclose
David Hong
No relationship to disclose
John De Groot
Consulting or Advisory Role: Deciphera Pharmaceuticals, Novartis, Roche/Genentech, Celldex, VBL Therapeutics, Foundation Medicine
Research Funding: Sanofi-Aventis, AstraZeneca, EMD-Serono, Eli Lilly, Novartis, Deciphera Pharmaceuticals
Vinod Ravi
No relationship to disclose
Yisheng Li
No relationship to disclose
Raja Luthra
No relationship to disclose
Keyur Patel
No relationship to disclose
Russell Broaddus
No relationship to disclose
John Mendelsohn
Leadership: Merrimack Pharmaceuticals-Director
Stock or Other Ownership: Merrimack Pharmaceuticals
Patents, Royalties, Other Intellectual Property: Royalties from University of California, San Diego
Gordon B. Mills
Honoraria: AstraZeneca/MedImmune, Critical Oncology Technologies, GlaxoSmithKline, Blend, Nuevolutino
Consulting or Advisory Role: AstraZeneca, Nuevolution, Critical Oncology Technologies
Research Funding: Critical Oncology technologies, GlaxoSmithKline, AstraZeneca/MedImmune
Patents, Royalties, Other Intellectual Property: Patent on HRD, licensed to Myriad Genetics
REFERENCES
- 1.Routbort MJ, Handal BA, Patel KP, et al. OncoSeek: A versatile annotation and reporting system for next generation sequencing-based clinical mutation analysis of cancer specimens. J Mol Diagn. 2012;14:747. (abstr) [Google Scholar]
- 2.Singh RR, Patel KP, Routbort MJ, et al. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15:607–622. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DB, Dahlman KH, Knol J, et al. Enabling a genetically informed approach to cancer medicine: A retrospective evaluation of the impact of comprehensive tumor profiling using a targeted next-generation sequencing panel. Oncologist. 2014;19:616–622. doi: 10.1634/theoncologist.2014-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meric-Bernstam F, Frampton GM, Ferrer-Lozano J, et al. Concordance of genomic alterations between primary and recurrent breast cancer. Mol Cancer Ther. 2014;13:1382–1389. doi: 10.1158/1535-7163.MCT-13-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagemann IS, Devarakonda S, Lockwood CM, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer. 2015;121:631–639. doi: 10.1002/cncr.29089. [DOI] [PubMed] [Google Scholar]
- 7.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.André F, Bachelot T, Commo F, et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: A multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15:267–274. doi: 10.1016/S1470-2045(13)70611-9. [DOI] [PubMed] [Google Scholar]
- 9.Lovly CM, Dahlman KB, Fohn LE, et al. Routine multiplex mutational profiling of melanomas enables enrollment in genotype-driven therapeutic trials. PLoS One. 2012;7:e35309. doi: 10.1371/journal.pone.0035309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray SW, Hicks-Courant K, Cronin A, et al. Physicians' attitudes about multiplex tumor genomic testing. J Clin Oncol. 2014;32:1317–1323. doi: 10.1200/JCO.2013.52.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]