ABSTRACT
Considerable evidence exists that bacteria detect eukaryotic communication molecules and modify their virulence accordingly. In previous studies, it has been demonstrated that the increasingly antibiotic-resistant pathogen Pseudomonas aeruginosa can detect the human hormones brain natriuretic peptide (BNP) and C-type natriuretic peptide (CNP) at micromolar concentrations. In response, the bacterium modifies its behavior to adapt to the host physiology, increasing its overall virulence. The possibility of identifying the bacterial sensor for these hormones and interfering with this sensing mechanism offers an exciting opportunity to directly affect the infection process. Here, we show that BNP and CNP strongly decrease P. aeruginosa biofilm formation. Isatin, an antagonist of human natriuretic peptide receptors (NPR), prevents this effect. Furthermore, the human NPR-C receptor agonist cANF4-23 mimics the effects of natriuretic peptides on P. aeruginosa, while sANP, the NPR-A receptor agonist, appears to be weakly active. We show in silico that NPR-C, a preferential CNP receptor, and the P. aeruginosa protein AmiC have similar three-dimensional (3D) structures and that both CNP and isatin bind to AmiC. We demonstrate that CNP acts as an AmiC agonist, enhancing the expression of the ami operon in P. aeruginosa. Binding of CNP and NPR-C agonists to AmiC was confirmed by microscale thermophoresis. Finally, using an amiC mutant strain, we demonstrated that AmiC is essential for CNP effects on biofilm formation. In conclusion, the AmiC bacterial sensor possesses structural and pharmacological profiles similar to those of the human NPR-C receptor and appears to be a bacterial receptor for human hormones that enables P. aeruginosa to modulate biofilm expression.
IMPORTANCE
The bacterium Pseudomonas aeruginosa is a highly dangerous opportunist pathogen for immunocompromised hosts, especially cystic fibrosis patients. The sites of P. aeruginosa infection are varied, with predominance in the human lung, in which bacteria are in contact with host molecular messengers such as hormones. The C-type natriuretic peptide (CNP), a hormone produced by lung cells, has been described as a bacterial virulence enhancer. In this study, we showed that the CNP hormone counteracts P. aeruginosa biofilm formation and we identified the bacterial protein AmiC as the sensor involved in the CNP effects. We showed that AmiC could bind specifically CNP. These results show for the first time that a human hormone could be sensed by bacteria through a specific protein, which is an ortholog of the human receptor NPR-C. The bacterium would be able to modify its lifestyle by favoring virulence factor production while reducing biofilm formation.
INTRODUCTION
Pseudomonas aeruginosa is a well-known opportunistic pathogen and a major cause of mortality among cystic fibrosis patients (1). The morbidity associated with this bacterium and its resistance to antibiotherapy are largely attributed to its transition in host tissues (particularly in lung, soft tissues, skin, and urinary bladder) from a planktonic to a biofilm lifestyle (2, 3). The present emergence of multiresistant strains is now critical and is prompting the need to find new therapeutic approaches (4).
The high adaptability of P. aeruginosa and its rapid response to the host environment imply that it can detect a large range of eukaryotic chemical signals (5). However, few bacterial sensors for eukaryotic molecules have been characterized to date (6). Studies on the Escherichia coli quorum-sensing regulator A (QseA) led to the identification of QseC/QseB, a two-component regulatory system (7, 8). This system is activated not only by the bacterial quorum-sensing signal autoinducer AI3 but also by eukaryotic neurohormones (epinephrine/norepinephrine) (9). It was suggested that QseC could be a bacterial ortholog of eukaryotic adrenergic receptors (10). Bioinformatic screening of E. coli QseC analogs has demonstrated the presence of proteins related to this sensor in a large number of bacterial pathogens, including P. aeruginosa (11), in which norepinephrine acts as a virulence inducer (12). It appears that P. aeruginosa is able to sense different eukaryotic communication molecules, including gamma interferon (13), dynorphin (14), gamma aminobutyric acid (GABA) (15), and natriuretic peptides (16, 17).
Natriuretic peptides are a family of eukaryotic hormones and neurohormones composed of three members: atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), and C-type natriuretic peptide (CNP). These peptides are mainly expressed in cardiomyocytes and endothelial cells (18) explaining their major role in cardiovascular homeostasis. However, ANP and CNP are also produced in significant amounts by the lung bronchial and alveolar epithelia and particularly by Clara (Club) cells (19). The circulating blood levels of natriuretic peptides are increased during community-acquired pneumonia (20), and the lipopolysaccharides (LPS) of Gram-negative bacteria induces the plasmatic release of both BNP (21) and CNP (22). In previous studies, we demonstrated that P. aeruginosa can detect BNP and CNP at micromolar concentrations and reacts with an overall increase of virulence (16, 17). In P. aeruginosa, the effect of CNP appears to be mediated by cyclic AMP (cAMP) synthesis, which subsequently activates the regulatory proteins Vfr (17) and PtxR (16), leading to a rise of HCN and exotoxin A production and a reorganization of LPS structure. For these reasons, it was suggested that CNP promotes the acute infection phenotype of P. aeruginosa (23), favoring the switch of P. aeruginosa to a planktonic lifestyle, and a decrease in biofilm formation. The effects of BNP and CNP on P. aeruginosa appear to be mediated by distinct mechanisms, since BNP, unlike CNP, did not modify intrabacterial cAMP levels (17) and was without effect on P. aeruginosa virulence for Caenorhabditis elegans (16). In humans, each type of natriuretic peptide acts through a particular receptor subtype, suggesting the expression of specific bacterial orthologs of the human natriuretic peptide receptors in P. aeruginosa.
In the present study, we extended our knowledge of the effect of natriuretic peptides on P. aeruginosa by investigating for the first time the effect of these peptides on the biofilm formation activity of the bacterium. Using in silico approaches, we identified the P. aeruginosa protein AmiC as a potential CNP binding sensor site and investigated in vitro the binding of natriuretic peptide receptor agonists to AmiC. The pharmacological profile of AmiC as a bacterial CNP receptor was completed by studying the effect of isatin, an antagonist of human CNP receptors that is also a known bacterial metabolite and signaling compound. We further determined the affinity of AmiC for several natriuretic peptide receptor agonists. Using an amiC-deficient mutant and its complemented derivative, we validated the crucial role of P. aeruginosa AmiC in the effect of CNP, but not BNP, on biofilm formation. This study provides an integrated explanation of the CNP sensing system in P. aeruginosa and opens up the way to develop a new strategy to control P. aeruginosa biofilm formation.
RESULTS
Effects of CNP and BNP on P. aeruginosa biofilm formation.
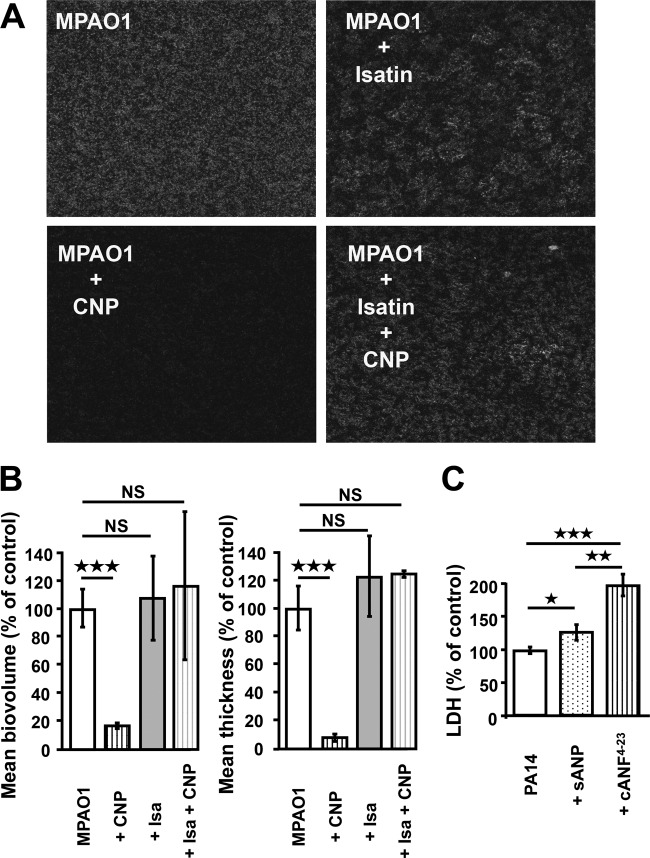
The influence of CNP and BNP on the biofilm formation activity of P. aeruginosa was investigated under dynamic and static conditions. Unless otherwise stated, peptides were used at a micromolar concentration, since (i) this corresponds to the concentration usually used for studying the effects of peptides on eukaryotic models (24), (ii) the peptides were previously shown to be active at this concentration on quorum-sensing molecules and toxin production in P. aeruginosa (16), and (iii) the duration of the biofilm experiment, which required a large amount of medium (continuous flow) containing the peptides, was not compatible with the realization of a whole dose-response study. When P. aeruginosa PA14 was exposed to CNP or BNP under dynamic conditions, we observed a highly significant decrease of the biofilm volume (−81.0% ± 6.6% and −80.3% ± 4.4%, respectively; P < 0.001) and of the biofilm average thickness (−72.3 ± 9.7% and −76.0 ± 4.8%, respectively; P < 0.001) (Fig. 1A). These effects were associated with a reduction of the number and mean size of mushroom-like structures in the biofilm (Fig. 1B). Exposure of P. aeruginosa MPAO1 to CNP for 24 h under static conditions led to similar results, with a marked decrease of biofilm formation (Fig. 2A) associated with a reduction of volume and thickness (−83.3 ± 1.9% and −91.7 ± 2.2%, respectively; P < 0.001) (Fig. 2B).
FIG 1 .
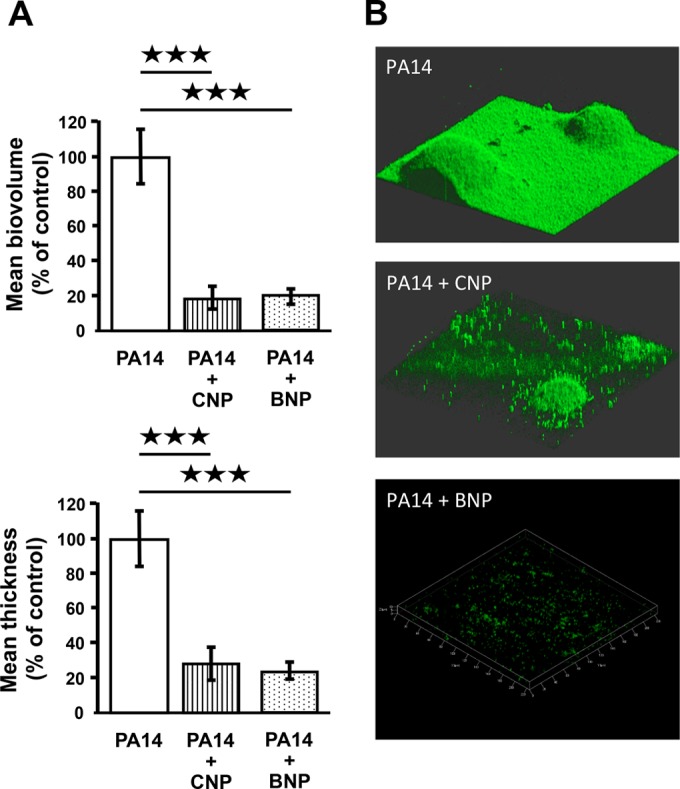
Biofilm formation by Pseudomonas aeruginosa PA14 exposed to C-type natriuretic peptide (CNP) or brain natriuretic peptide (BNP) in dynamic condition. (A) COMSTAT analyses of biofilms of P. aeruginosa PA14 control (PA14) or PA14 exposed to CNP (10−6 M) or BNP (10−6 M). Data are the means for twelve samples from six independent experiments for control bacteria, ten samples from six independent experiments for CNP-exposed bacteria, and six samples from two independent experiments for BNP-exposed bacteria. ***, P < 0.001. (B) 3D shadow representations of the biofilm structures developed under dynamic condition by P. aeruginosa PA14 control or PA14 exposed to CNP or exposed to BNP at 37°C for 24 h in LB broth. Biofilms were stained with Syto 61 red dye or SytoX dye and observed by confocal laser scanning microscopy.
FIG 2 .
Effect of antagonists and agonists of natriuretic peptide receptors. (A) 2D black-and-white images resulting from CLSM observations of MPAO1 24-h biofilms obtained under static conditions after growth under control conditions and exposed to CNP (10−6 M), isatin (10−5 M) or both isatin and CNP. (B) COMSTAT analyses of biofilms of P. aeruginosa MPAO1 exposed to CNP, to isatin, or both to isatin and CNP. Data are means from three independent experiments for CNP exposure and two independent experiments for isatin and isatin-plus-CNP exposure. ***, P < 0.001; NS, not significantly different. (C) Cytotoxic activity of PA14 alone (control), PA14 exposed to the NPR-C agonist cANF4-23 (10−6 M), or PA14 exposed to the NPR-A agonist sANP (10−6 M). The cytotoxic effect of the bacterium was determined by measuring lactate dehydrogenase (LDH) accumulation in the medium. Data are the means for nine samples from three independent experiments for cANF4-23 and means for six samples from two independent experiments for sANP. ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Effects of specific antagonist and agonists of natriuretic peptide receptors on P. aeruginosa biofilm formation and virulence activity.
The biofilm formation activity of P. aeruginosa MPAO1 was also studied in the presence of isatin, an antagonist of both A and C subtypes natriuretic peptide receptors (NPR-A and NPR-C). Administered alone for 24 h, isatin affected the organization of the biofilm, which appeared to be more irregular, with the presence of large water channels (Fig. 2A). We also noted a limited and nonsignificant increase of the biomass and thickness of the biofilm (Fig. 2B). However, when P. aeruginosa MPAO1 was exposed to isatin (10−5 M) for 20 min prior to the administration of CNP, the inhibitory effect of CNP on the biofilm formation activity of the bacterium was abolished (Fig. 2A and B).
As we had previously shown that natriuretic peptides enhance P. aeruginosa cytotoxicity activity and global virulence (16), we tested the effects of sANP, a specific NPR-A agonist, and the artificial peptide cANF4-23, a specific NPR-C agonist, on bacterial cytotoxicity for lung cells. The peptide cANF4-23 (an NPR-C agonist) caused a strong increase of P. aeruginosa cytotoxicity activity against A549 human lung epithelial cells (+98.3% ± 16.9%; P < 0.001) (Fig. 2C). In parallel, we observed that the sANP peptide (an NPR-A agonist) also enhanced the P. aeruginosa cytotoxicity in the same cell model, but to a lower extent (+25.9% ± 12.1%; P < 0.05) (Fig. 2C).
In silico study of a functional C-type natriuretic peptide receptor ortholog in P. aeruginosa.
In mammalian cells, CNP preferentially binds the NPR-C receptor and has a weak affinity for the NPR-A receptor (25), whereas BNP strongly binds NPR-A and has a weak affinity for NPR-C. The observations that the effects of CNP were blocked by the NPR-C antagonist isatin and mimicked by the NPR-C agonist cANF4-23 suggested the presence of a bacterial ortholog of NPR-C in P. aeruginosa. This hypothesis was investigated using in silico approaches. Searching bibliographic data, we noted a DALI structure similarity search (26) that identified a P. aeruginosa protein, AmiC, which shows structural homologies with the binding domain of natriuretic peptides receptors (27, 28). Using tools from the Protein Data Bank in Europe (http://www.ebi.ac.uk/pdbe/), we investigated in detail the structure of P. aeruginosa AmiC bound to its known ligand acetamide. We observed that it shares high structural similarity with the natriuretic peptide receptor subtype NPR-C bound to one of its agonists, the atrial natriuretic peptide (ANP) (data not shown). This supports the hypothesis that AmiC could act as a bacterial natriuretic peptide sensor.
The three-dimensional (3D) structure of P. aeruginosa AmiC (Protein Data Bank [PDB] ID: 1PEA) (29) was compared to that of the human NPR-C receptor (hNPR-C) (PDB ID: 1JDP) (30). Using the STAMP software (31), we superimposed an AmiC monomer onto each protomer of the structure of an hNPR-C dimer in complex with CNP. Although the sequences of these proteins share only 19% identity and 26% similarity, these two proteins present an overall similar 3D structure in the homodimeric form (Fig. 3A), particularly at the interface between monomers, which is considered the binding cleft of hNPR-C. The highest similarity was found in the helix of AmiC, corresponding to the region involved in the binding of CNP to hNPR-C. Next, we split the AmiC monomer into two parts, corresponding to the two lobes of the protein. The first lobe consists of amino acids 8 to 123 and 261 to 338, and the second lobe consists of amino acids 124 to 260 and 339 to 375 of AmiC. Then, we superimposed each lobe on the 3D structure of hNPR-C (Fig. 3B). Each lobe of hNPR-C and AmiC showed a high level of secondary structure conservation with similar 3D positions, reinforcing the concept of structural homology between these two proteins. Using GASH structural superposition software (32) on each lobe of hNPR-C and AmiC, we observed that each of these two proteins has 254 residues estimated to be involved in equivalent secondary structures, covering 61.7% of the AmiC sequence. The hinge between the two lobes was essentially formed with flexible loops. Once the structures were superimposed, we looked at the amino acids involved in the binding of CNP, supporting the hypothesis that AmiC and hNPR-C bind the peptide in a similar fashion. Table S1 in the supplemental material lists AmiC amino acids that are potentially involved in CNP binding and the corresponding amino acids involved in the binding of hNPR-C to CNP, as described by He et al. (33). Amino acids located within 4 Å of CNP in the binding sites of both hNPR-C and AmiC are shown schematically in Fig. S1 in the supplemental material. A total of 24 amino acids from AmiC and 21 residues from hNPR-C with equivalent physicochemical properties are located in the same regions and may stabilize the binding of CNP to AmiC and NPR-C equivalently.
FIG 3 .
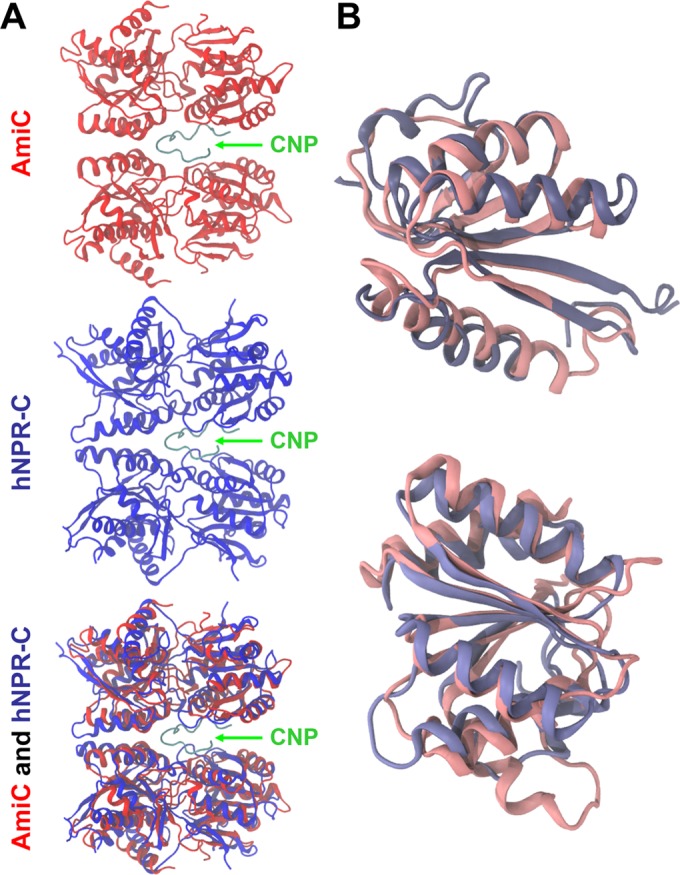
Cartoon representation of AmiC (PDB ID: 1PEA) (red) and hNPR-C (PDB ID: 1JDP) (blue) superimposed using the STAMP algorithm. (A) Overall superimposition of AmiC and NPR-C, shown in cartoon form. The CNP peptide is in green. (B) Superimposition by lobes of AmiC and hNPR-C. The lobe on the left corresponds to amino acids 8 to 123 and 261 to 338 of AmiC, and the lobe on the right corresponds to amino acids 124 to 260 and 339 to 375. These images were made using VMD (63).
We also determined the virtual capacity of P. aeruginosa AmiC to bind natriuretic peptides (BNP and CNP) by molecular docking using AutoDock (34). Docking revealed that CNP can bind to AmiC at the dimer interface of the bacterial protein (see Fig. S2A and B in the supplemental material). The most relevant calculated binding site of CNP on AmiC requires 13 amino acids: Arg64, Glu68, Ala90, Met92, Pro93, Glu96, Arg97, Asp99, Glu112, Tyr113, Pro115, Asn116, and Tyr366. Since these predictions were obtained by an in silico study, the involvement of each of these residues needs to be validated by constructing and studying the 13 correspondent mutant proteins, which will be addressed in a future work. In contrast, we observed in silico that BNP has no affinity for the AmiC protein. The NPR-C antagonist isatin also appeared capable of binding to the AmiC dimer interface (see Fig. S2C and D). In this case, the AmiC amino acids involved in the recognition of isatin are predicted to be Ile71, Arg72, Val94, and Arg97. These amino acids are located in the same region involved as the putative CNP binding site, and one residue (Arg97) could play a key role, since it potentially interacts with both CNP and isatin.
Effect of CNP on the expression of the amidase operon in P. aeruginosa.
In P. aeruginosa, AmiC is considered a sensor protein. In the absence of stimulation (basal conditions), AmiC binds to the AmiR protein, an antitermination factor, which down-regulates the transcription of the amiEBCRS operon (35). Upon binding of acetamide, AmiC releases AmiR, triggering the transcription of the whole ami operon (36). In order to verify that CNP could act in the same manner as acetamide on AmiC, we measured amiB, amiC, amiE, and amiR mRNA levels by quantitative reverse transcription-PCR (qRT-PCR) 1 h after exposure of the bacterium to CNP. The amiC, amiE, and amiR mRNA levels were significantly increased (2.97- ± 0.2-, 3.66- ± 0.39-, and 2.73- ± 0.67-fold, respectively; P < 0.001) after 1 h of exposure to CNP (10−6 M) (Fig. 4), whereas the amount of lecB mRNA, encoded by a gene adjacent to the ami operon, was not modified. In contrast, BNP (10−6 M) had no significant effect on the whole ami operon transcriptional activity (data not shown).
FIG 4 .
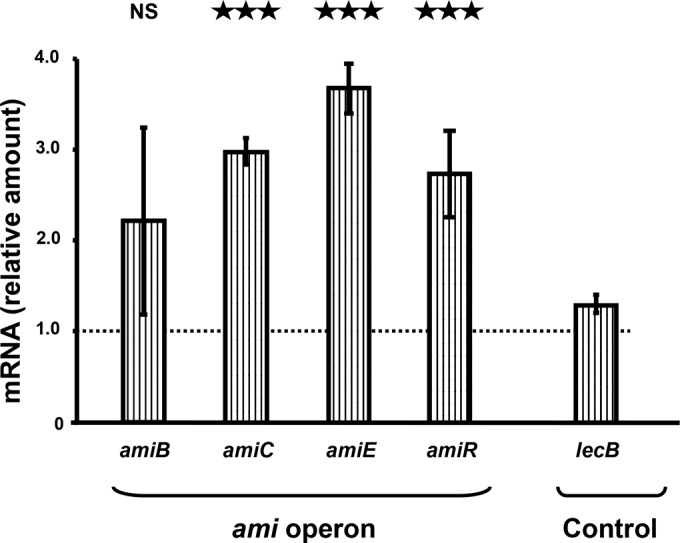
Effect of C-type natriuretic peptide (CNP) on expression level of genes from the ami operon. Expression levels of ami genes in PAO1-treated bacteria relative to those in non-PAO1-treated bacteria (control). RNAs were extracted 1 h after PAO1 exposure to physiologic solution (control) or CNP (10−6 M) and assayed by quantitative real-time reverse transcription-PCR (qRT-PCR). These data are the means from two independent experiments. ***, P < 0.001; NS, not significantly different.
AmiC binding with natriuretic peptide receptor agonists.
In order to confirm that the action of CNP and isatin on P. aeruginosa is mediated directly through AmiC and is not an indirect effect, we investigated their binding to AmiC in vitro. Recombinant AmiC was expressed in E. coli. We first confirmed the oligomeric state of AmiC using native gel electrophoresis and analytical gel filtration (see Fig. S3 in the supplemental material). These methods showed an apparent molecular mass of 120 ± 30 kDa and 136 ± 3 kDa, respectively. These results are most consistent with AmiC (protomer molecular mass, 45.5 kDa) forming a dimer with an irregular shape, as predicted by the model (Fig. S3). We then tested the interaction of AmiC with natriuretic peptide receptor agonists and isatin using microscale thermophoresis (Fig. 5A). AmiC bound to CNP tightly, showing a dissociation constant (KD) of 2.0 ± 0.3 µM (Fig. 5). In contrast, we observed that BNP possessed no affinity for AmiC (Fig. 5A). Isatin could also bind to AmiC, although in this case, the interaction was not so strong, showing a KD of 600 ± 200 µM (Fig. 5A and B). Additionally, we measured the interactions of other natriuretic peptide receptor agonists. sANP, a specific NPR-A agonist, showed a weak interaction with AmiC which had clearly not proceeded to completion at 900 µM (Fig. 5B), the highest concentration that we were able to assess. These data clearly predict that the KD is greater than 600 µM. Conversely, our initial experiments on osteocrin (a specific NPR-C agonist) showed a very strong interaction with AmiC, apparently tighter than we were able to determine using the available instrument. This indicates a KD value below 100 nM. In the same way, cANF4-23, another NPR-C specific agonist, presented a KD value of 40 ± 10 µM (Fig. 5).
FIG 5 .

CNP, BNP, and NPR agonist and isatin affinities for AmiC in vitro. (A) Recombinant AmiC was fluorescently labeled and incubated with various concentrations of natriuretic peptide receptor agonists and isatin. These mixtures were then analyzed using by microscale thermophoresis. The results were normalized to a fitted zero for unbound AmiC, and the thermophoresis signal is shown, in parts per thousand. Addition of cANF4-23 caused AmiC fluorescence to increase in a dose-dependent fashion; this increase was used as the signal rather than thermophoresis, as the effect confounds the thermophoresis signal. (B) The results were fitted to the dissociation constant formula from the law of mass action. The fitted values for KD are shown for isatin, BNP, sANP, cANF4-23, and osteocrin. We showed that these compounds either bound very weakly (sANP), bound more tightly (osteocrin), or exhibited no interaction (BNP). The data are the means from three independent experiments.
Involvement of AmiC in CNP-regulated P. aeruginosa biofilm formation.
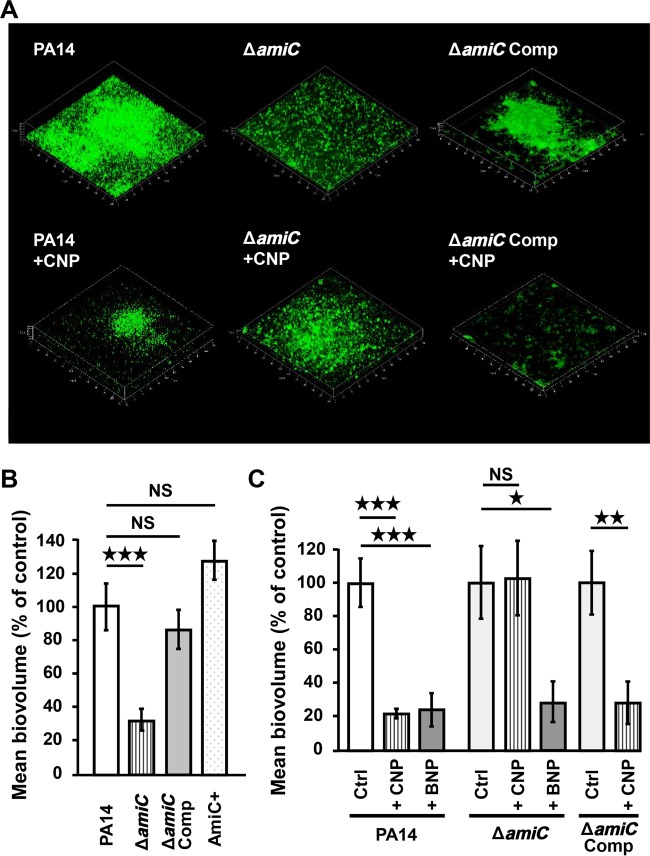
Under dynamic conditions, the biofilm formed by the PA14 ΔamiC strain appeared reduced in comparison to wild-type strain PA14 WT (Fig. 6A). Conversely, the morphology of the biofilm formed by the PA14 AmiC+ strain (PA14 containing the wild-type amiC gene plus extra amiC plasmid-borne copies) or by the PA14 EV strain carrying the empty vector was similar to that of the wild-type (data not shown). Complementation of the ΔamiC strain with the amiC gene carried by a vector restored entirely the ability of the bacteria to form a biofilm (Fig. 6A and B). The measure of the biofilm biovolumes revealed 64.5 ± 12.5% reduction for the PA14 ΔamiC strain in comparison to the PA14 wild type (P < 0.001), whereas the biovolume of the biofilm generated by the complemented strain is similar to that of wild-type PA14 (+13.7% ± 14.6%) (Fig. 6B). The biofilm formed by the strain PA14 AmiC+ was slightly, but nonsignificantly, enhanced (Fig. 6B).
FIG 6 .
Involvement of AmiC protein on P. aeruginosa biofilm formation. (A) 3D shadow representations of the biofilm structures developed by the P. aeruginosa PA14 control (PA14), the ΔamiC strain, and the amiC complemented strain (amiC Comp), not exposed to peptide or exposed to CNP (10−7 M) under dynamic conditions, at 37°C for 24 h in LB broth. Biofilms were stained with SytoX and observed by confocal laser scanning microscopy. (B) COMSTAT analyses of biofilms of P. aeruginosa PA14 control, the ΔamiC strain, the amiC complemented strain, and the AmiC+ strain. (C) COMSTAT analyses of biofilms of the P. aeruginosa PA14 control, the ΔamiC strain, and the amiC complemented strain exposed to CNP (10−7 M), to BNP (10−7 M), or to neither of them. Data are the means for 27 samples from 11 independent experiments for the PA14 control, 11 samples from four independent experiments for PA14 exposed to CNP, nine samples from three independent experiments for PA14 exposed to BNP, 19 samples from seven independent experiments for the ΔamiC control strain, 12 samples from four independent experiments for the ΔamiC strain exposed to CNP, eight samples from three independent experiments for the amiC strain exposed to BNP, 24 samples from six independent experiments for the amiC Comp control strain, and 12 samples from four independent experiments for the amiC Comp strain exposed to CNP. Note that the values for the wild-type strain (left), ΔamiC strain (middle), and amiC Comp strain (right) values are set to 100% independently. ***, P < 0.001; *, P < 0.05; NS, not significantly different.
We observed that the ability of CNP to reduce biofilm formation is abolished in the ΔamiC strain (Fig. 6A and C) and is restored in the complemented strain (−71.9% ± 12.7%) (P < 0.01) (Fig. 6C). In contrast, BNP, which strongly reduced biofilm formation as well (−75.1% ± 9.9%) (P < 0.001) (Fig. 6C), continued to have the same impact on biofilm formation in the ΔamiC mutant strain (−70.8% ± 12.1%) (P < 0.05) (Fig. 6C).
In parallel, since it had been shown previously that CNP also modifies P. aeruginosa virulence, we tested the effect of AmiC on cytotoxicity for lung cells. We observed that, whereas the PA14 wild-type strain showed an increase in cytotoxic activity after CNP exposure (+76.6% ± 17%; P < 0.001), the ΔamiC strain did not exhibit significantly modified cytotoxicity in the presence of CNP compared with controls (ΔamiC strain without CNP exposure) (+14.7% ± 6.0%) (see Fig. S4C in the supplemental material). In accordance with these data, we observed that the increased production of HCN triggered by CNP on wild-type PA14 disappeared in the ΔamiC mutant strain (Fig. S4D). It is interesting that the strain overexpressing AmiC (AmiC+) strongly lost its ability to kill lung cells (Fig. S4A), confirming a role for AmiC in the regulation of P. aeruginosa virulence.
DISCUSSION
We have shown that P. aeruginosa is sensitive to micromolar concentrations of natriuretic peptides (i.e., BNP and CNP) and responds to these eukaryotic factors by a major increase in virulence (16, 17). Previously, the intrabacterial cascade involving Vfr, PtxR, RhlR, and LasR proteins, leading to a remodeling of the lipopolysaccharide (LPS) structure and an increase of HCN and exotoxin A production, was identified (16), but the nature of the P. aeruginosa natriuretic peptide sensors remained a central question.
P. aeruginosa is an opportunistic pathogen, and in cystic fibrosis patients, it is known to form biofilms in lung alveoli (2). As an exchange surface between air and blood, the alveolar epithelium formed by pneumocytes is particularly thin (0.2 µm). If bacteria develop on this epithelium, they are certainly exposed to diffusible blood containing endocrine factors such as natriuretic peptides. These are also produced locally by endothelial cells (22), by lung airway epithelium cells, and by nonciliated Clara cells (19). The growth of bacteria in biofilms is generally associated with a decrease of diffusible virulence factor synthesis (37). This assists in establishing a chronic infectious status, which progressively weakens the host, leading to a potential lethal outcome (3). Conversely, an anticipated release of bacterial virulence factors, particularly when the bacterium is not protected by the biofilm, should trigger the host defense responses and the clearance of the pathogen and increase antibiotic sensitivity. Due to the effect on biofilm formation in P. aeruginosa, CNP and related molecules could potentially be interesting for therapy. CNP strongly decreased the biofilm formation activity of both P. aeruginosa PAO1 and PA14 strains, confirming the hypothesis that virulence factor production and biofilm formation are inversely related. This is consistent with our previous results showing that CNP enhances cAMP production in P. aeruginosa (17). Indeed, cAMP and c-di-GMP synthesis appear to be antagonistically regulated in bacteria. While c-di-GMP promotes the growth of biofilms (23), cAMP prevents P. aeruginosa biofilm formation (38). The present results are also in agreement with the observation that CNP induces a rearrangement of the Pseudomonas LPS structure (17) and enhances algC mRNA synthesis (16), since LPS contributes to bacterial surface properties and AlgC is involved in synthesis of the LPS core (39).
In order to identify the potential natriuretic peptide sensor in P. aeruginosa, we started from the hypothesis that this protein should show structural homology with the binding domain of eukaryotic natriuretic peptide receptors. van den Akker et al. (28) have shown that the binding domain of the eukaryotic natriuretic peptide receptor subtype A (NPR-A), a receptor which binds ANP and BNP, exhibits a type I periplasmic binding protein fold and that the closest structural neighbor of this ANP binding domain is the AmiC protein from P. aeruginosa. Analysis of the crystal structure of the dimerized hormone binding domain of NPR-A and of the structure of the AmiC-AmiR complex showed that in NPR-A and AmiC, the same residues in helices α3 and α7 interact, respectively, with ANP and AmiR. Furthermore, the pocket forming the ANP binding site on NPR-A overlaps with the recognition site of AmiR on AmiC (27, 28). In eukaryotes, NPR-A is known as a receptor enzyme coupled to cGMP synthesis. However, CNP does not lead to an increase of cGMP in P. aeruginosa, whereas it stimulates cAMP synthesis (17). Then, we looked for possible similarities between AmiC and human NPR-C (hNPR-C), the sole ANP and CNP eukaryotic receptor coupled to an adenylate cyclase activity (40). We observed in silico that hNPR-C and P. aeruginosa AmiC proteins present 19% identity and 26% similarity, confirming the structural homology of eukaryotic natriuretic peptide receptors and AmiC (27). The AmiC binding domain helix α3 is delineated by amino acids 85 to 99 (41). In the present study, we noted that the binding of CNP to AmiC occurred in the same region; 6 of the 13 CNP binding amino acids (Ala90, Met92, Pro93, Glu96, Arg97, and Asp99) are in helix α3, suggesting that CNP could compete with AmiR for binding on AmiC. We compared all the amino acids putatively involved in the AmiC-CNP interaction to those observed in the interaction of CNP with NPR-C in eukaryotes (33). The physicochemical properties of amino acids involved in these two interactions (i.e., CNP/AmiC in bacteria and CNP/NPR-C in eukaryotes) showed a remarkably high level of conservation (this study). We determined that AmiC (like NPR-C) (40) is functionally expressed as a dimer, reinforcing the hypothesis that AmiC could have a CNP binding activity.
In P. aeruginosa, the amiC gene appears highly conserved (99.57% sequence homology and total preservation of the amino acid sequence between PAO1 and PA14 strains), suggesting that a crucial role is played by this protein in this species. The amiC gene is encoded in an operon including the amiL, amiE, amiB, amiC, amiR, and amiS genes (36) (http://www.pseudomonas.com). AmiC activation by acetamide, its classical ligand in bacteria, is known to provoke the release of AmiR, leading to the transcriptional activation of the whole ami operon (35, 41). We therefore hypothesized that if CNP acts on AmiC in a fashion similar to that of acetamide, it should also induce the ami operon expression. This was confirmed by our observations. CNP actually induced amiERC transcription, and the specificity of this action was reinforced by the lack of effect of BNP on the ami operon transcription (data not shown). As mentioned elsewhere, P. aeruginosa AmiC appears to be functionally related to hNPR-C, and it is interesting that, in eukaryotic cells, NPR-C preferentially binds to CNP with a higher affinity than to BNP (33). Such highly conserved specific recognition modes between eukaryotic and bacterial sensors were observed previously in Helicobacter pylori for the human hormone somatostatin, whose bacterial sensor can recognize only one agonist subtype (42).
The issue of how CNP contacts AmiC in P. aeruginosa needs to be addressed, as AmiC is predicted to be located in the bacterial cytoplasm (http://www.pseudomonas.com). This means that CNP must enter the bacterium before binding to AmiC. Such an assumption is plausible, since another eukaryotic peptide, dynorphin, was shown able to freely cross the membrane of P. aeruginosa, inducing increased virulence (14). This behavior was explained by the high percentage of basic and hydrophobic amino acid residues present in dynorphin (76%) (43). Similarly, CNP contains 73% basic or hydrophobic amino acids. Moreover, natriuretic peptides have been shown to form pores in phospholipid bilayers (44) and could therefore manage their own entrance into the bacterium. However, we cannot exclude the possibility that AmiC protein could, under some conditions, reach the P. aeruginosa inner membrane. Indeed, AmiC belongs to the type I periplasmic binding fold protein family, and the Uniprot database claims that AmiC is located in the periplasmic space (http://www.uniprot.org; reference P27017). One protein being located in multiple bacterial compartments to undertake unrelated functions is not unprecedented. Indeed, the EF-Tu protein, which acts in the bacterial cytoplasm as a ribosomal elongation factor, was shown to migrate upon stress to the plasma membrane and acquire new functions as a sensor molecule (45). In P. aeruginosa, EF-Tu, after migration into bacteria membrane, also behaves as a plasminogen-binding protein (46). To support this hypothesis, we observed, using AmiC antibodies, that while AmiC protein is mainly located in the bacterial cytosol, a proportion of AmiC is associated with the inner membrane compartment (see Fig. S6 in the supplemental material). In this case, we assumed that CNP is delivered to AmiC proteins that have reached the inner membrane through protein transporters, for example. To reinforce this hypothesis, we observed that a protein encoded by the gene PA14_64270 (ortholog of PA4858 in PAO1) possesses a high structural similarity with AmiC, could virtually bind CNP, and is necessary for the CNP effect on bacterial biofilm (see Fig. S5 in the supplemental material).
Although six bacterial sensors for eukaryotic messengers have been identified (6), to date, none of the bacterial sensors structurally identified possesses similarity to the eukaryotic counterpart receptor. For example, E. coli QseC, the sensor protein for epinephrine, is a histidine sensor kinase which crosses the bacterial membrane twice and which appears to be structurally very different from the G-coupled protein receptor (9, 47). In the same vein, gamma interferon, which binds the eight-transmembrane-region P. aeruginosa OprF protein (48), in mammals activates a single transmembrane receptor. Since our data suggest that AmiC and hNPR-C possess a homologous 3D structure and that CNP acts as an agonist on P. aeruginosa, we undertook to study the pharmacological profile of AmiC, as happens in eukaryotic receptor studies.
Results from our pharmacological studies carried out using cANF4-23, an NPR-C agonist, and isatin, an NPR-A/NPR-C antagonist (49) also support the hypothesis that AmiC acts as an ortholog of eukaryotic NPR-C. In silico molecular modeling revealed that isatin binding to AmiC would require only 4 amino acids, but, as expected for an antagonist, these amino acids are located in the potential CNP binding pocket of AmiC. The antagonist effect of isatin was confirmed experimentally. Isatin had no intrinsic activity on the biofilm formation activity of P. aeruginosa but inhibited the effect of CNP. Conversely, the artificial peptide cANF4-23, a specific NPR-C agonist in eukaryotes, was able to reproduce the effects of CNP on P. aeruginosa global virulence on lung cells, as observed by Blier et al. (16). These data reinforced the hypothesis that AmiC acts as a sensor for natriuretic peptides but do not exclude the possibility that these peptides bind to another protein that in turn affects AmiC. In order to validate our hypothesis, we purified P. aeruginosa AmiC recombinantly produced in E. coli and checked the interactions of CNP, BNP, isatin, osteocrin (an NPR-C agonist) (50), cANF4-23 (an NPR-C agonist), and sANP (an NPR-A agonist) (51) with AmiC. As expected, AmiC bound strongly to CNP, showing a KD of 2.0 ± 0.3 µM, consistent with the effective concentrations of CNP on P. aeruginosa. In contrast, BNP does not interact with AmiC, suggesting that BNP acts on P. aeruginosa through another bacterial target, a hypothesis strongly reinforced by the fact that BNP retains an effect on biofilm formation in the amiC mutant strain (this study). Isatin also showed a specific interaction, although at a significantly higher concentration (600 ± 200 µM). As normal blood isatin concentrations are usually on the order of 0.3 to 1.3 µM (52), this implies that in vivo, isatin is unlikely to be at a sufficiently high concentration to antagonize the effects of CNP on AmiC. Finally, we attempted to test specific agonists of NPR-A and NPR-C, to confirm that AmiC reflects NPR-C specificity. sANP showed some interaction with AmiC, but this interaction was weak and indeed was not complete at the highest ligand concentration available, indicating a KD (likely considerably) in excess of 600 µM, probably explaining the weak effect of this peptide on bacterial cytotoxicity (this study). In contrast, initial experiments on osteocrin showed that it bound sufficiently strongly that it was beyond the limit of detection of our experiment (100 nM), and cANF4-23 possesses a KD in the same range as that of CNP. These observations suggest that AmiC does indeed show the same specificity as NPR-C and is selective against NPR-A agonists. Finally, using an amiC-deficient mutant strain and its complemented derivative, we physiologically validated the crucial role of AmiC sensor in CNP effects on P. aeruginosa virulence expression and biofilm formation.
The last puzzling question is the reason why a bacterium such as P. aeruginosa should express a sensor system for peptides which are naturally labile molecules present only in vertebrates. The fact that the CNP sensor should also be a binding site for isatin provides some clues on a possible explanation. Isatin, or indole-2,3-dione, is an interspecies signal molecule produced in large amounts by bacteria such as Escherichia coli (53). Until now, pseudomonads were not thought to synthesize isatin or isatin-related compounds, but as shown by Lee et al. (54) and in the present study, P. aeruginosa can detect this molecule. Isatin is also produced in large amounts by plant tissues, where it is a precursor in auxin biosynthesis (55), and by mammalian tissues, including lungs (56, 57). Therefore, in the host, P. aeruginosa is regularly exposed to significant amounts of isatin. As is the case for GABA (15), it will gain an advantage by adapting its behavior in response to the host physiology. Nevertheless, even if this explanation is correct, the fact that AmiC showed common properties with the eukaryotic NPR-C, including CNP recognition, remains to be explained.
In conclusion, the present work provides the first demonstration that bacteria not only are sensitive to eukaryotic messengers but also could express eukaryotic receptor-like structures to detect human hormones. The AmiC sensor protein acts as an ortholog of the eukaryotic receptor NPR-C, acting as a CNP/isatin sensor in P. aeruginosa that modulates bacterial biofilm formation. Since the CNP hormone is produced in the lung, the effects of CNP on the biofilm formation activity of P. aeruginosa could represent a natural defense mechanism against lung colonization. Furthermore, a natriuretic peptide receptor agonist has been used as a bronchodilator drug (58). On one hand, this could pose a risk of enhancing P. aeruginosa virulence, and on the other, it may prevent biofilm formation. This opens up the tantalizing prospect that compounds in development for other indications could be retargeted for use against biofilms.
MATERIALS AND METHODS
Reagents and test substances.
The CNP peptide was obtained from Polypeptide (Strasbourg, France). Isatin and hBNP were purchased from Sigma-Aldrich (St Quentin Fallavier, France). Osteocrin, sANP, and cANF4-23 were synthesized by J. Leprince (PRIMACEN platform, University of Rouen, France). Dimethyl sulfoxide (DMSO) was purchased from Acros Organics (Geel, Belgium).
Bacterial strains and bacterial cultures.
Pseudomonas aeruginosa PAO1 was obtained from an international collection (17). P. aeruginosa MPAO1 was originally from the Iglewski lab. A transposon library was constructed from the MPAO1 strain at the University of Washington (59). The P. aeruginosa PA14 was from Harvard Medical School (Boston, MA) (60) and kindly provided by the Biomerit Research Center (University Cork, Ireland). All strains used in this work are listed in Table S2 in the supplemental material. All strains were grown at 37°C in Luria Bertani medium (LB). For treatment with CNP, an overnight culture was diluted to an optical density at 600 nm (OD600) of 0.08 using a ThermoSpectronics (Cambridge, United Kingdom) spectrophotometer. CNP was added 2 h after the onset of the culture, a time corresponding to the middle of the exponential growth phase. For all studies performed using isatin, because of its hydrophobicity, this molecule was initially dissolved in a solution of DMSO in water (5/95). The final concentration of DMSO in bacterial culture medium was kept under 0.2%. In this case, control experiments were conducted in the presence of the same final concentration of DMSO. The final bacterial density and the absence of contamination were controlled by plating.
Biofilm formation under dynamic conditions.
After 3 h of preculture in LB at 37°C, P. aeruginosa PA14 was inoculated at an OD600 of 0.08 in LB medium and subcultured for 2 h. CNP (1 µM final concentration) was added, and bacteria were grown for additional 3 h. Bacteria were then washed and adjusted to an OD600 of 0.1 in 0.9% NaCl supplemented with CNP (1 µM). The bacterial suspensions were then used to study biofilm formation under dynamic conditions at 37°C in a three-channel flow cell as described by Bazire et al. (61). Briefly, each bacterial suspension was injected into a flow cell channel, and bacteria were allowed to adhere to the glass coverslip for 2 h. A flow (2.5 ml/h) of LB medium containing CNP (1 µM) was then applied for 24 h. At the end of the experiments, the biofilms were stained with 5 µM Syto 61 red dye or 5 µM SytoX (Molecular Probes). Observations were made using a confocal laser scanning microscope (LSM 710 confocal laser-scanning microscope; Zeiss). The biofilm thicknesses and corresponding biovolumes were estimated by measuring field samples from at least 3 independent experiments using COMSTAT software (62).
Biofilm formation under static conditions.
The static bacterial biofilm formation activity was determined after 24 h of development. Precultures of P. aeruginosa, containing the pSMC2.1 plasmid harboring the green fluorescent protein (GFP) gene, were grown overnight at 37°C. An aliquot of preculture (100 µl) was then added to 5 ml of LB medium supplemented with 400 µg/ml kanamycin. Tested molecules or the corresponding excipient were added after 2 h of subculture. Three hours later, bacteria were collected by centrifugation (7,000 × g, 10 min). The supernatant was then removed, and the pellets were resuspended in 25 ml of physiological solution (0.9% NaCl) and adjusted to an OD600 of 0.1. Sterile glass slides, previously cleaned with 4% TFD4 in water (Dutscher, Brumath, France), were layered in petri dishes and covered by the bacterial suspension. The slides were then incubated for 2 h at 37°C under static conditions. At the end of the attachment period, the physiological solution was replaced by LB medium and incubated for 24 h at 37°C in static condition. The slides were then rinsed twice in 25 ml sterile phosphate-buffered saline (PBS; 0.1 M; pH 7.4), and bacteria were heat fixed. Biofilms were observed using a confocal laser scanning microscope (LSM 710; Zeiss). Images were treated using a 3-median filter and segmented using Zen 2009 software (Zeiss). The volumes and thicknesses of the biofilms were estimated as described elsewhere (62). All 2D images are representative of 15 observations.
In silico studies.
STAMP software (31) was used within VMD (63) on a single computer. GASH structural superposition software (32) was used on the web server (http://sysimm.ifrec.osaka-u.ac.jp/gash/). Potential ligand/protein interactions were investigated in silico by the molecular docking technique using the amidase sensor protein of P. aeruginosa AmiC (PDB ID: 1QO0). Essential hydrogen atoms, Kollman united atom type charges, and solvation parameters were added with the aid of AutoDock tools (34). Affinity (grid) maps of 20- by 20- by 20-Å grid points and 0.375-Å spacing were generated using the Autogrid program (34). AutoDock parameter set- and distance-dependent dielectric functions were used in the calculation of the van der Waals and electrostatic terms, respectively. Docking simulations were made using the Lamarckian genetic algorithm (LGA). Initial position, orientation, and torsions of the ligand molecules were set randomly. Each docking experiment was derived from three different runs that were set to terminate after a maximum of 1,000,000 energy evaluations. The population size was set to 100.
qRT-PCR mRNA assays.
RNA extraction and quantification were realized as previously described (61) using bacteria grown for 3 h in LB. The primers employed are presented in Table S3 in the supplemental material. The transcription level of 16S rRNA was used as the endogenous control. PCRs were performed in triplicate, and the standard deviations were lower than a CT (cycle threshold) of 0.15. Relative quantification of mRNA was obtained according to Bazire et al. (64) using the comparative CT (2−ΔΔCT) method (65).
Lung epithelial cell cytotoxicity tests.
The human A549 lung epithelial cell type II line (ATCC-CCL185TM) (American Type Culture Collection, Manassas, VA) was grown at 37°C in 5% CO2 atmosphere in Dulbecco’s modified Eagle’s medium (DMEM; Lonza) supplemented with 10% fetal calf serum (Lonza) and 1% of penicillin and streptomycin (Penistrep; Lonza). For cytotoxicity assays, cells were seeded in 24-well plates at a final density of 3 × 105 cells per well and grown for 48 h before use. A minimum of 24 h before infection assays, cells were deprived of antibiotics and fetal calf serum by the addition of a fresh serum- and antibiotic-free medium. For the assays, A549 cells were incubated for 2 h with control or pretreated P. aeruginosa PA14 at a multiplicity of infection of 10. Bacterial cytotoxicity was determined by measurement of lactate dehydrogenase (LDH) release. LDH is a stable cytosolic enzyme that diffuses into the culture medium upon cell lysis. This marker of cytotoxicity was assayed using the Cytotox 96 enzymatic assay (Promega, Charbonnières, France) as previously described (17).
P. aeruginosa AmiC purification.
To express P. aeruginosa AmiC in E. coli, a synthetic codon-optimized gene was prepared (MWG Operon) with a leader sequence of MHHHHHHSSGVDLGTENLYFQS to provide a cleavable 6×His tag. This was cloned into the NcoI and HindIII restriction sites of the pET-Duet1 vector (Novagen). This construct was transformed into Rosetta2 (DE3) cells (Novagen) and grown in 500 ml of ZYM-5052 medium (66) at 37°C until mid-log phase and then at 20°C overnight. Cells were harvested by centrifugation at 4,750 × g for 30 min, and resuspended in 20 mM Tris-HCl, 0.5 M NaCl, 20 mM imidazole (pH 8.0) (buffer A). Cells were lysed using a sonicator and clarified by centrifugation at 20,000 × g. The protein was purified using a 1-ml HisTrap crude FF column and a Superdex 200 16/600 high-resolution column (GE Healthcare), using an ÄKTAxpress system (GE Healthcare) at 4°C. Lysate was loaded over the IMAC column at 0.25 ml min−1 and washed with buffer A. AmiC was eluted in buffer A supplemented with 250 mM imidazole (pH 8.0) and loaded onto the gel filtration column. The protein was then eluted isocratically in 10 mM HEPES-NaOH, 0.5 M NaCl (pH 7.0). DMSO was added to the purified protein to 1% (vol/vol), and the protein was concentrated using a Vivaspin 20 centrifugal concentrator (Sartorius; 10-kDa molecular mass cutoff) at 4°C.
Microscale thermophoresis.
AmiC was labeled using the RED-NHS labeling kit (NanoTemper Technologies). The labeling reaction was performed according to the manufacturer’s instructions in the supplied labeling buffer applying a concentration of 20 µM protein (molar dye-to-protein ratio ~ 2:1) at room temperature for 30 min. Unreacted dye was removed with the supplied dye removal columns equilibrated with MicroScale thermophoresis (MST) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10 mM MgCl2). The label/protein ratio was determined using photometry at 650 and 280 nm. Typically, a ratio of 0.8 was achieved.
AmiC, either labeled (CNP, isatin, BNP, and cANF4-23) or unlabeled (sANP and osteocrin), was adjusted to 100 nM with MST buffer supplemented with 0.05% Tween 20. CNP, isatin, sANP, and osteocrin were dissolved in MST buffer supplemented with 0.05% Tween 20. A series of 1:1 dilutions were prepared in the identical buffer, producing ligand concentrations ranging from 13 nM to 434 µM (CNP), 3.12 µM to 3 mM (isatin), 892 nM to 1.8 mM (sANP), 28 nM to 450 µM (BNP), 68 nM to 1.1 mM (cANF4-23), and 120 pM to 4 µM (osteocrin). For thermophoresis, each ligand dilution was mixed with 1 volume of AmiC, which leads to a final concentration of AmiC of 50 nM and final ligand concentrations at half of the ranges above. After a 5-min incubation, followed by centrifugation at 10,000 × g for 10 min, approximately 4 µl of each solution was filled into Monolith NT standard treated capillaries (NanoTemper Technologies GmbH). Thermophoresis was measured using a Monolith NT.115 or Monolith NT.LabelFree instrument (NanoTemper Technologies GmbH) at an ambient temperature of 25°C with 5-s/30-s/5-s laser off/on/off times, respectively. Instrument parameters were adjusted with 50% LED power and 20 to 40% MST power. Data from three independently pipetted measurements were analyzed (NT.Analysis software version 1.5.41; NanoTemper Technologies) using the signal from thermophoresis and temperature jump. Following data analysis, the thermophoresis signals were fitted to the formula for KD from the law of mass action:
where f(c) is the observed thermophoresis signal, “unbound” is the signal in the absence of ligand (normalized to 0), “bound” is the signal at infinite ligand concentration, c is the concentration of ligand in the same units as AmiC concentration, and KD is the dissociation constant. The data were fitted using GraphPad Prism version 5, and figures for thermophoresis were generated using this software. For cANF4-23, incubation with AmiC resulted in a dose-dependent increase in AmiC fluorescence, which confounds the MST signal. This increase was therefore measured using the MST device, and the data were fitted to the above equation. To confirm that the fluorescence increase was related to binding, samples were heated to 95°C for 10 min with 2% (wt/vol) SDS and 20 mM dithiothreitol (DTT) and retested: addition of cANF4-23 no longer increased fluorescence.
Statistical analysis.
The nonparametric Mann-Whitney test was used to compare the means within the same set of experiments.
SUPPLEMENTAL MATERIAL
2D ligand interaction diagrams. 2D representation of the CNP surrounded by amino acids of hNPR-C (a) and AmiC (b) located within 4 Å of the peptide. Download
Modelization of AmiC sensor and CNP or isatin interactions by the molecular docking approach. Download
Analytical native gel electrophoresis and size exclusion chromatography. Download
Involvement of AmiC protein in PA14 cytotoxicity. Download
Involvement of the transporter PA14_64270 (gene PA14_64270) orthologs of the protein PA4858 in PAO1 in the effect of CNP on both biofilm and virulence activity. Download
AmiC location using dot blot analysis. Download
Amino acids potentially involved in the binding of CNP to NPR-C and AmiC.
Strains and plasmids used in this study.
Primers used in this study.
ACKNOWLEDGMENTS
We thank Magalie Barreau and Olivier Maillot for technical assistance. We thank Christine Farmer for linguistic insight for the manuscript.
T. Rosay is a recipient of a doctoral fellowship from the French Ministry of Research (MRE). This work was supported by grants from the Communauté d’Agglomération d’Evreux, the Conseil Général de l’Eure, European Union (FEDER no. 31970), the French Association “Vaincre la Mucoviscidose” and the InterReg IVA PeReNE project.
Footnotes
Citation Rosay T, Bazire A, Diaz S, Clamens T, Blier A-S, Mijouin L, Hoffmann B, Sergent J-A, Bouffartigues E, Boireau W, Vieillard J, Hulen C, Dufour A, Harmer NJ, Feuilloley MGJ, Lesouhaitier O. 2015. Pseudomonas aeruginosa expresses a functional human natriuretic peptide receptor ortholog: involvement in biofilm formation. mBio 6(4):e01033-15. doi:10.1128/mBio.01033-15.
REFERENCES
- 1.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Bjarnsholt T, Jensen PØ, Fiandaca MJ, Pedersen J, Hansen CR, Andersen CB, Pressler T, Givskov M, Høiby N. 2009. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol 44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- 3.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Ashish A, Shaw M, Winstanley C, Ledson MJ, Walshaw MJ. 2012. Increasing resistance of the Liverpool epidemic strain (LES) of Pseudomonas aeruginosa (Psa) to antibiotics in cystic fibrosis (CF)—a cause for concern? J Cyst Fibros 11:173–179. doi: 10.1016/j.jcf.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Camilli A, Bassler BL. 2006. Bacterial small-molecule signaling pathways. Science 311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lesouhaitier O, Veron W, Chapalain A, Madi A, Blier AS, Dagorn A, Connil N, Chevalier S, Orange N, Feuilloley M. 2009. Gram-negative bacterial sensors for eukaryotic signal molecules. Sensors (Basel) 9:6967–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sperandio V, Li CC, Kaper JB. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect Immun 70:3085–3093. doi: 10.1128/IAI.70.6.3085-3093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sperandio V, Torres AG, Kaper JB. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol 43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 9.Hughes DT, Sperandio V. 2008. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol 6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. 2003. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci U S A 100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasko DA, Moreira CG, Li DR, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, Hughes DT, Huntley JF, Fina MW, Falck JR, Sperandio V. 2008. Targeting QseC signaling and virulence for antibiotic development. Science 321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hegde M, Wood TK, Jayaraman A. 2009. The neuroendocrine hormone norepinephrine increases Pseudomonas aeruginosa PA14 virulence through the las quorum-sensing pathway. Appl Microbiol Biotechnol 84:763–776. doi: 10.1007/s00253-009-2045-1. [DOI] [PubMed] [Google Scholar]
- 13.Wu L, Estrada O, Zaborina O, Bains M, Shen L, Kohler JE, Patel N, Musch MW, Chang EB, Fu YX, Jacobs MA, Nishimura MI, Hancock RE, Turner JR, Alverdy JC. 2005. Recognition of host immune activation by Pseudomonas aeruginosa. Science 309:774–777. doi: 10.1126/science.1112422. [DOI] [PubMed] [Google Scholar]
- 14.Zaborina O, Lepine F, Xiao G, Valuckaite V, Chen Y, Li T, Ciancio M, Zaborin A, Petrof EO, Turner JR, Rahme LG, Chang E, Alverdy JC. 2007. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog 3:e35. doi: 10.1371/journal.ppat.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dagorn A, Chapalain A, Mijouin L, Hillion M, Duclairoir-Poc C, Chevalier S, Taupin L, Orange N, Feuilloley MG. 2013. Effect of GABA, a bacterial metabolite, on Pseudomonas fluorescens surface properties and cytotoxicity. Int J Mol Sci 14:12186–12204. doi: 10.3390/ijms140612186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blier AS, Veron W, Bazire A, Gerault E, Taupin L, Vieillard J, Rehel K, Dufour A, Le Derf F, Orange N, Hulen C, Feuilloley MG, Lesouhaitier O. 2011. C-type natriuretic peptide modulates quorum sensing molecule and toxin production in Pseudomonas aeruginosa. Microbiology 157:1929–1944. doi: 10.1099/mic.0.046755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veron W, Lesouhaitier O, Pennanec X, Rehel K, Leroux P, Orange N, Feuilloley MG. 2007. Natriuretic peptides affect Pseudomonas aeruginosa and specifically modify lipopolysaccharide biosynthesis. FEBS J 274:5852–5864. doi: 10.1111/j.1742-4658.2007.06109.x. [DOI] [PubMed] [Google Scholar]
- 18.Drewett JG, Garbers DL. 1994. The family of guanylyl cyclase receptors and their ligands. Endocr Rev 15:135–162. doi: 10.1210/edrv-15-2-135. [DOI] [PubMed] [Google Scholar]
- 19.Nakanishi K, Tajima F, Itoh H, Nakata Y, Hama N, Nakagawa O, Nakao K, Kawai T, Torikata C, Suga T, Takishima K, Aurues T, Ikeda T. 1999. Expression of C-type natriuretic peptide during development of rat lung. Am J Physiol 277:L996–L1002. [DOI] [PubMed] [Google Scholar]
- 20.Nowak A, Breidthardt T, Christ-Crain M, Bingisser R, Meune C, Tanglay Y, Heinisch C, Reiter M, Drexler B, Arenja N, Twerenbold R, Stolz D, Tamm M, Müller B, Müller C. 2012. Direct comparison of three natriuretic peptides for prediction of short- and long-term mortality in patients with community-acquired pneumonia. Chest 141:974–982. doi: 10.1378/chest.11-0824. [DOI] [PubMed] [Google Scholar]
- 21.Vila G, Resl M, Stelzeneder D, Struck J, Maier C, Riedl M, Hülsmann M, Pacher R, Luger A, Clodi M. 2008. Plasma NT-proBNP increases in response to LPS administration in healthy men. J Appl Physiol 105:1741–1745. doi: 10.1152/japplphysiol.90442.2008. [DOI] [PubMed] [Google Scholar]
- 22.Suga S, Itoh H, Komatsu Y, Ogawa Y, Hama N, Yoshimasa T, Nakao K. 1993. Cytokine-induced C-type natriuretic peptide (CNP) secretion from vascular endothelial cells—evidence for CNP as a novel autocrine/paracrine regulator from endothelial cells. Endocrinology 133:3038–3041. doi: 10.1210/endo.133.6.8243333. [DOI] [PubMed] [Google Scholar]
- 23.Coggan KA, Wolfgang MC. 2012. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr Issues Mol Biol 14:47–70. [PubMed] [Google Scholar]
- 24.Klinger JR, Tsai SW, Green S, Grinnell KL, Machan JT, Harrington EO. 2013. Atrial natriuretic peptide attenuates agonist-induced pulmonary edema in mice with targeted disruption of the gene for natriuretic peptide receptor-A. J Appl Physiol 114:307–315. doi: 10.1152/japplphysiol.01249.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potter LR, Abbey-Hosch S, Dickey DM. 2006. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr Rev 27:47–72. [DOI] [PubMed] [Google Scholar]
- 26.Holm L, Sander C. 1995. Dali: a network tool for protein structure comparison. Trends Biochem Sci 20:478–480. doi: 10.1016/S0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 27.van den Akker F. 2001. Structural insights into the ligand binding domains of membrane bound guanylyl cyclases and natriuretic peptide receptors. J Mol Biol 311:923–937. doi: 10.1006/jmbi.2001.4922. [DOI] [PubMed] [Google Scholar]
- 28.van den Akker F, Zhang X, Miyagi M, Huo X, Misono KS, Yee VC. 2000. Structure of the dimerized hormone-binding domain of a guanylyl-cyclase-coupled receptor. Nature 406:101–104. doi: 10.1038/35017602. [DOI] [PubMed] [Google Scholar]
- 29.Pearl L, O’Hara B, Drew R, Wilson S. 1994. Crystal structure of AmiC: the controller of transcription antitermination in the amidase operon of Pseudomonas aeruginosa. EMBO J 13:5810–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He X, Chow D, Martick MM, Garcia KC. 2001. Allosteric activation of a spring-loaded natriuretic peptide receptor dimer by hormone. Science 293:1657–1662. doi: 10.1126/science.1062246. [DOI] [PubMed] [Google Scholar]
- 31.Russell RB, Barton GJ. 1992. Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins 14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 32.Standley DM, Toh H, Nakamura H. 2005. GASH: an improved algorithm for maximizing the number of equivalent residues between two protein structures. BMC Bioinformatics 6:221. doi: 10.1186/1471-2105-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He XL, Dukkipati A, Garcia KC. 2006. Structural determinants of natriuretic peptide receptor specificity and degeneracy. J Mol Biol 361:698–714. doi: 10.1016/j.jmb.2006.06.060. [DOI] [PubMed] [Google Scholar]
- 34.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19:1639–1662. doi:. [DOI] [Google Scholar]
- 35.Wilson S, Drew R. 1991. Cloning and DNA sequence of amiC, a new gene regulating expression of the Pseudomonas aeruginosa aliphatic amidase, and purification of the amiC product. J Bacteriol 173:4914–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson SA, Wachira SJ, Drew RE, Jones D, Pearl LH. 1993. Antitermination of amidase expression in Pseudomonas aeruginosa is controlled by a novel cytoplasmic amide-binding protein. EMBO J 12:3637–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balasubramanian D, Schneper L, Kumari H, Mathee K. 2013. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res 41:1–20. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono K, Oka R, Toyofuku M, Sakaguchi A, Hamada M, Yoshida S, Nomura N. 2014. cAMP signaling affects irreversible attachment during biofilm formation by Pseudomonas aeruginosa PAO1. Microbes Environ 29:104–106. doi: 10.1264/jsme2.ME13151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldberg JB, Coyne MJ Jr., Neely AN, Holder IA. 1995. Avirulence of a Pseudomonas aeruginosa algC mutant in a burned-mouse model of infection. Infect Immun 63:4166–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anand-Srivastava MB, Sehl PD, Lowe DG. 1996. Cytoplasmic domain of natriuretic peptide receptor-C inhibits adenylyl cyclase. Involvement of a pertussis toxin-sensitive G protein. J Biol Chem 271:19324–19329. doi: 10.1074/jbc.271.32.19324. [DOI] [PubMed] [Google Scholar]
- 41.O’Hara BP, Norman RA, Wan PT, Roe SM, Barrett TE, Drew RE, Pearl LH. 1999. Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J 18:5175–5186. doi: 10.1093/emboj/18.19.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita K, Kaneko H, Yamamoto S, Konagaya T, Kusugami K, Mitsuma T. 1998. Inhibitory effect of somatostatin on Helicobacter pylori proliferation in vitro. Gastroenterology 115:1123–1130. doi: 10.1016/S0016-5085(98)70083-6. [DOI] [PubMed] [Google Scholar]
- 43.Marinova Z, Vukojevic V, Surcheva S, Yakovleva T, Cebers G, Pasikova N, Usynin I, Hugonin L, Fang W, Hallberg M, Hirschberg D, Bergman T, Langel U, Hauser KF, Pramanik A, Aldrich JV, Gräslund A, Terenius L, Bakalkin G. 2005. Translocation of dynorphin neuropeptides across the plasma membrane. A putative mechanism of signal transmission. J Biol Chem 280:26360–26370. doi: 10.1074/jbc.M412494200. [DOI] [PubMed] [Google Scholar]
- 44.Kourie JI. 1999. Synthetic mammalian C-type natriuretic peptide forms large cation channels. FEBS Lett 445:57–62. doi: 10.1016/S0014-5793(99)00081-2. [DOI] [PubMed] [Google Scholar]
- 45.Dallo SF, Kannan TR, Blaylock MW, Baseman JB. 2002. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol Microbiol 46:1041–1051. doi: 10.1046/j.1365-2958.2002.03207.x. [DOI] [PubMed] [Google Scholar]
- 46.Kunert A, Losse J, Gruszin C, Hühn M, Kaendler K, Mikkat S, Volke D, Hoffmann R, Jokiranta TS, Seeberger H, Moellmann U, Hellwage J, Zipfel PF. 2007. Immune evasion of the human pathogen Pseudomonas aeruginosa: elongation factor Tuf is a factor H and plasminogen binding protein. J Immunol 179:2979–2988. doi: 10.4049/jimmunol.179.5.2979. [DOI] [PubMed] [Google Scholar]
- 47.Xie W, Dickson C, Kwiatkowski W, Choe S. 2010. Structure of the cytoplasmic segment of histidine kinase receptor QseC, a key player in bacterial virulence. Protein Pept Lett 17:1383–1391. doi: 10.2174/0929866511009011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugawara E, Nestorovich EM, Bezrukov SM, Nikaido H. 2006. Pseudomonas aeruginosa porin OprF exists in two different conformations. J Biol Chem 281:16220–16229. doi: 10.1074/jbc.M600680200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Medvedev A, Crumeyrolle-Arias M, Cardona A, Sandler M, Glover V. 2005. Natriuretic peptide interaction with [3H]isatin binding sites in rat brain. Brain Res 1042:119–124. doi: 10.1016/j.brainres.2005.02.051. [DOI] [PubMed] [Google Scholar]
- 50.Moffatt P, Thomas G, Sellin K, Bessette MC, Lafrenière F, Akhouayri O, St-Arnaud R, Lanctôt C. 2007. Osteocrin is a specific ligand of the natriuretic peptide clearance receptor that modulates bone growth. J Biol Chem 282:36454–36462. doi: 10.1074/jbc.M708596200. [DOI] [PubMed] [Google Scholar]
- 51.Olson LJ, Lowe DG, Drewett JG. 1996. Novel natriuretic peptide receptor/guanylyl cyclase A-selective agonist inhibits angiotensin II- and forskolin-evoked aldosterone synthesis in a human zona glomerulosa cell line. Mol Pharmacol 50:430–435. [PubMed] [Google Scholar]
- 52.Mawatari K, Segawa M, Masatsuka R, Hanawa Y, Iinuma F, Watanabe M. 2001. Fluorimetric determination of isatin in human urine and serum by liquid chromatography postcolumn photoirradiation. Analyst 126:33–36. doi: 10.1039/b006484j. [DOI] [PubMed] [Google Scholar]
- 53.Lee J, Jayaraman A, Wood TK. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol 7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J, Bansal T, Jayaraman A, Bentley WE, Wood TK. 2007. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl Environ Microbiol 73:4100–4109. doi: 10.1128/AEM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou P, Koh HL. 2007. Determination of indican, isatin, indirubin and indigotin in Isatis indigotica by liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom 21:1239–1246. doi: 10.1002/rcm.2954. [DOI] [PubMed] [Google Scholar]
- 56.Watkins P, Clow A, Glover V, Halket J, Przyborowska A, Sandler M. 1990. Isatin, regional distribution in rat brain and tissues. Neurochem Int 17:321–323. doi: 10.1016/0197-0186(90)90154-L. [DOI] [PubMed] [Google Scholar]
- 57.Medvedev A, Buneeva O, Glover V. 2007. Biological targets for isatin and its analogues: implications for therapy. Biologics Targets Ther 1:151–162. [PMC free article] [PubMed] [Google Scholar]
- 58.Edelson JD, Makhlina M, Silvester KR, Vengurlekar SS, Chen X, Zhang J, Koziol-White CJ, Cooper PR, Hallam TJ, Hay DW, Panettieri RA Jr. 2013. In vitro and in vivo pharmacological profile of PL-3994, a novel cyclic peptide (hept-cyclo(Cys–His–Phe-d-Ala-Gly-Arg-d-Nle-Asp-Arg-Ile-Ser-Cys)-Tyr-[Arg mimetic]-NH(2)) natriuretic peptide receptor-A agonist that is resistant to neutral endopeptidase and acts as a bronchodilator. Pulm Pharmacol Ther 26:229–238. doi: 10.1016/j.pupt.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain pa14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bazire A, Shioya K, Soum-Soutéra E, Bouffartigues E, Ryder C, Guentas-Dombrowsky L, Hémery G, Linossier I, Chevalier S, Wozniak DJ, Lesouhaitier O, Dufour A. 2010. The sigma factor AlgU plays a key role in formation of robust biofilms by nonmucoid Pseudomonas aeruginosa. J Bacteriol 192:3001–3010. doi: 10.1128/JB.01633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. [DOI] [PubMed] [Google Scholar]
- 63.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 64.Bazire A, Diab F, Taupin L, Rodrigues S, Jebbar M, Dufour A. 2009. Effects of osmotic stress on rhamnolipid synthesis and time-course production of cell-to-cell signal molecules by Pseudomonas aeruginosa. Open Microbiol J 3:128–135. doi: 10.2174/1874285800903010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔCT) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 66.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2D ligand interaction diagrams. 2D representation of the CNP surrounded by amino acids of hNPR-C (a) and AmiC (b) located within 4 Å of the peptide. Download
Modelization of AmiC sensor and CNP or isatin interactions by the molecular docking approach. Download
Analytical native gel electrophoresis and size exclusion chromatography. Download
Involvement of AmiC protein in PA14 cytotoxicity. Download
Involvement of the transporter PA14_64270 (gene PA14_64270) orthologs of the protein PA4858 in PAO1 in the effect of CNP on both biofilm and virulence activity. Download
AmiC location using dot blot analysis. Download
Amino acids potentially involved in the binding of CNP to NPR-C and AmiC.
Strains and plasmids used in this study.
Primers used in this study.