ABSTRACT
Describing the viral diversity of wildlife can provide interesting and useful insights into the natural history of established human pathogens. In this study, we describe a previously unknown picornavirus in harbor seals (tentatively named phopivirus) that is related to human hepatitis A virus (HAV). We show that phopivirus shares several genetic and phenotypic characteristics with HAV, including phylogenetic relatedness across the genome, a specific and seemingly quiescent tropism for hepatocytes, structural conservation in a key functional region of the type III internal ribosomal entry site (IRES), and a codon usage bias consistent with that of HAV.
IMPORTANCE
Hepatitis A virus (HAV) is an important viral hepatitis in humans because of the substantial number of cases each year in regions with low socioeconomic status. The origin of HAV is unknown, and no nonprimate HAV-like viruses have been described. Here, we describe the discovery of an HAV-like virus in seals. This finding suggests that the diversity and evolutionary history of these viruses might be far greater than previously thought and may provide insight into the origin and pathogenicity of HAV.
INTRODUCTION
The vast majority of emerging infectious diseases (EIDs) in humans have origins in wildlife (1). What is less well understood is how many well-established human viruses also have animal origins, either in the recent or distant past. Human immunodeficiency virus (HIV; family Retroviridae) is a good and relatively recent example, adapting from its precursor simian immunodeficiency virus (SIV) and eliminating any need for an animal host to sustain new human infections (2). Measles virus (family Paramyxoviridae) is another and much older example, which has been shown to share an ancestor with the now-eradicated rinderpest virus of cattle and is believed to have emerged in people following a long and close association between people and livestock (3). In both cases, a virus is believed to have emerged from an animal source, adapted, and ultimately become fully sustained by human-to-human transmission.
Here, we describe the discovery of a virus in seals that is related to human hepatitis A virus (HAV; genus hepatovirus, family Picornaviridae). We tentatively name the virus “phopivirus” reflecting a contraction of phocid (host taxa) and picornavirus, and present evidence that supports a common evolutionary ancestry with HAV.
RESULTS
Discovery and genetic characterization.
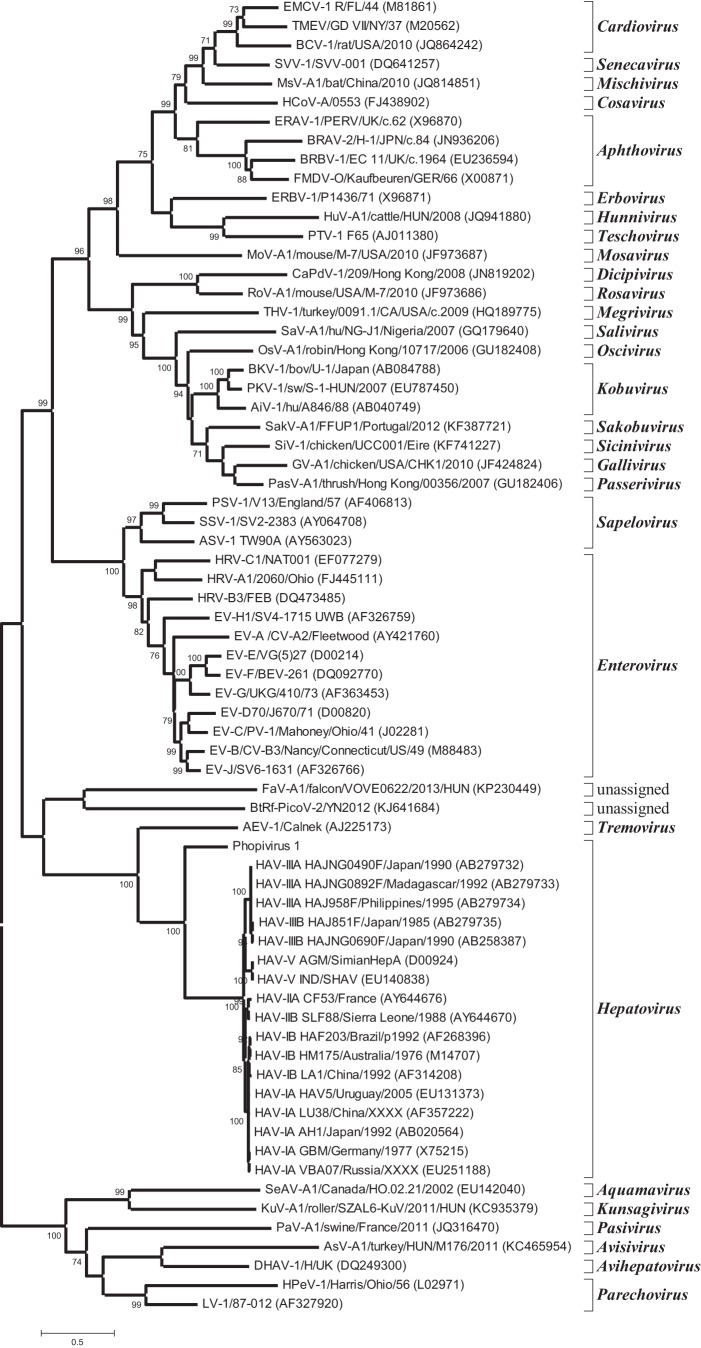
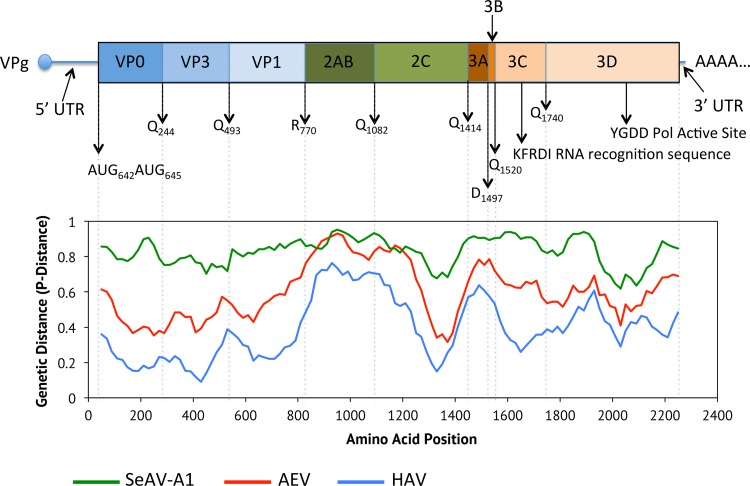
The lungs, livers, spleens, and oral mucosae from three harbor seals (Phoca vitulina vitulina) were assayed by high-throughput sequencing (HTS) for the presence of unknown viruses. These individuals all died in an unusual mortality event in 2011 caused by avian influenza virus subtype H3N8 (4) but were processed for additional viral discovery (described here) to look for viruses that might be cooccurring with the influenza. Sequences for a novel picornavirus with homology to HAV were identified in these samples, which were assembled into a putative genome in silico and then subsequently confirmed by PCR and dideoxy sequencing. The genome of phopivirus was shown to be 7,476 nucleotides (nt) long, including a single open reading frame (ORF) of 6,690 nt coding for the polyprotein, a long 5′ untranslated region (UTR) of 642 nt that includes the internal ribosome entry site (IRES), and a short 3′ UTR of 145 nt that terminates in a poly(A) tail. Maximum-likelihood phylogenetic reconstruction showed that phopivirus is most closely related to HAV (Fig. 1) and shares sufficient amino acid sequence identity in the P1 (74.8%), P2 (44.7%) and P3 (55.5%) regions to be considered a novel species in the genus hepatovirus (Fig. 1) (5, 6). A schematic of the genome with predicted polyprotein cleavage sites is presented in Fig. 2. No discernible cleavage site could be identified between the 2A and 2B peptides based on homology to known cleavage sites in other picornaviruses. Further experimental analyses will be required to investigate this feature and its potential cleavage products.
FIG 1 .
Maximum-likelihood phylogenetic reconstruction of the picornaviruses, based on peptide 3D amino acid sequences. The model used was LG (34) complete deletion, gamma distributed with invariant sites (G+I). The tree shows that phopivirus is most closely related to HAV. Relationships are consistent when repeated for other peptides. Scale indicates number of amino acid substitutions per site per year. Bootstrap values are shown at nodes.
FIG 2 .
Schematic of the phopivirus genome, with predicted polyprotein cleavage sites, and sequence identities across the genome to HAV, avian encephalomyelitis virus (AEV), and seal aquamavirus A1 (SeAV-A1). The picornavirus genome consists of a single open reading frame (ORF) which produces a polyprotein that is cleaved into the structural (P1) and non-structural (P2 and P3) proteins.
Natural history of phopivirus.
To investigate whether seals are the natural host of phopivirus, we first looked for evidence of lineage-specific diversifying selection, as jumps between host species are frequently accompanied by bursts of adaptive evolution. No significant evidence of episodic selection was detected in either the VP1 or 3D gene (P < 0.05), suggesting that phopivirus was not recently introduced to seal hosts. We then looked for evidence of the virus in other individuals and host species, surveying a total of 113 samples collected opportunistically from 37 individuals by using PCR (targeting a 157-nt fragment of the VP1 gene). This cohort included 29 harbor seals, 6 harp seals (Phoca groenlandicus), and 2 gray seals (Halichoerus grypus). All were juveniles and all died between 2011 and 2013 along the New England coast (Table 1). Phopivirus was detected in 11/29 harbor seals and 1/6 harp seals but was not found in either of the two gray seals tested (likely a reflection of the limited sample number tested). Comparison of the partial VP1 sequence amplified by this PCR confirmed that all harbor seals were infected with the same virus, with 100% sequence identity between amplicons. This may reflect the more conserved nature of VP1 or may indicate constraints in viral diversity, as seen for HAV (7). The sequence identified in the harp seal was slightly different, however, with 97% identity to the harbor seal strain. These data may indicate the circulation of species-specific strains of phopivirus, and they further support the suggestion that phocid seals are the natural hosts of this virus.
TABLE 1 .
Results showing the presence/absence of phopivirus in different animals and tissues by both qPCR and conventional PCR
| Animala | Result of qPCR (conventional PCR) inb: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Adrenal gland | Dermal lesion | Liver | Lung | Oral mucosa | Spleen | Thoracic lymph node | Skin | |
| COA120801 Pv | Neg (neg) | |||||||
| IFAW11 279 Pv | Neg (neg) | |||||||
| IFAW11 280 Pv | Neg (neg) | |||||||
| IFAW11 283 Pv | Neg (neg) | |||||||
| IFAW12 258 Pv | Neg (neg) | |||||||
| MARC 12 046 Pv | Neg (neg) | |||||||
| MARC 12 052 Pv | Neg (neg) | |||||||
| MH 08 035 Pg | Neg (neg) | Neg (neg) | Neg (neg) | |||||
| MH 08 048 Pv | Neg (neg) | Neg (neg) | Neg (neg) | |||||
| NEAQ 11 006 Pgc | Pos (neg) | Pos (neg) | Pos (pos) | |||||
| NEAQ 11 175 Pv | Neg (neg) | Neg (neg) | Neg (neg) | |||||
| NEAQ 11 239 Pv | Neg (neg) | Neg (neg) | Neg (neg) | |||||
| NEAQ 11 250 Pv | Neg (neg) | |||||||
| NEAQ 11 254 Pv | Neg (neg) | |||||||
| NEAQ 11 263 Pv | Neg (neg) | Neg (neg) | Neg (neg) | |||||
| NEAQ 11 278 Pv | Pos (neg) | Neg (neg) | Pos (neg) | |||||
| NEAQ 11 280 Pv | Neg (neg) | Pos (neg) | Pos (neg) | |||||
| NEAQ 11 286 Pvd | Pos (pos) | Pos (pos) | Pos (pos) | Pos (pos) | Pos (pos) | Pos (pos) | ||
| NEAQ 11 294 Pvd | Pos (pos) | Pos (pos) | Pos (pos) | Pos (pos) | Pos (pos) | Pos (pos) | ||
| NEAQ 11 295 Pvd | Pos (pos) | Pos (pos) | Pos (pos) | Pos (pos) | Pos (pos) | Pos (pos) | ||
| NEAQ 11 300 Pv | Neg (neg) | Neg (neg) | Neg | |||||
| NEAQ 11 303 Pv | Pos (pos) | Pos (pos) | Pos (pos) | |||||
| NEAQ 11 306 Pv | Pos (pos) | Pos (pos) | ||||||
| NEAQ 11 318 Pv | Pos (pos) | Pos (pos) | Pos (neg) | Pos (pos) | ||||
| NEAQ 11 352 Pv | Neg (neg) | Neg (neg) | Neg (neg) | Neg (neg) | ||||
| NEAQ 11 354 Pv | Pos (pos) | Pos (neg) | Pos (neg) | Pos (neg) | ||||
| NEAQ 11 355 Pg | Neg (neg) | Neg (neg) | Neg (neg) | |||||
| NEAQ 12 015 Pg | Neg (neg) | Neg (neg) | Neg (neg) | |||||
| NEAQ 12 045 Hg | Neg (neg) | Neg (neg) | Neg (neg) | Neg (neg) | ||||
| NEAQ 12 068 Hg | Neg (neg) | Neg (neg) | Neg (neg) | |||||
| NEAQ 12 156 Pv | Neg (neg) | Neg (neg) | Neg (neg) | Neg (neg) | ||||
| NEAQ 12 159 Pv | Neg (neg) | Neg (neg) | Neg (neg) | Neg (neg) | ||||
| NEAQ 12 180 Pv | Pos (pos) | Pos (pos) | Pos (pos) | |||||
| NEAQ 12 199 Pv | Pos (pos) | Pos (pos) | Pos (pos) | Pos (pos) | ||||
| NEAQ 13 022 Pv | Neg (neg) | Neg (neg) | Neg (neg) | Neg (neg) | Neg (neg) | |||
| NEAQ 13 039 Pg | Neg (neg) | Neg (neg) | Neg (neg) | Neg (neg) | ||||
| NEAQ 13 070 Pg | Neg (neg) | Neg (neg) | Neg (neg) | Neg (neg) | ||||
Pg, Phoca groenlandicus (harp seal); Pv, Phoca vitulina (harbor seal); Hg, Halichoerus grypus (gray seal).
Results for samples with a positive qPCR are shown in bold font. Pos, positive; Neg, negative.
The sequence found in this animal is slightly different (97% identical) from the one found in harbor seals.
Samples from the indicated animals were used for fluorescent in situ hybridization (FISH).
Pathogenesis.
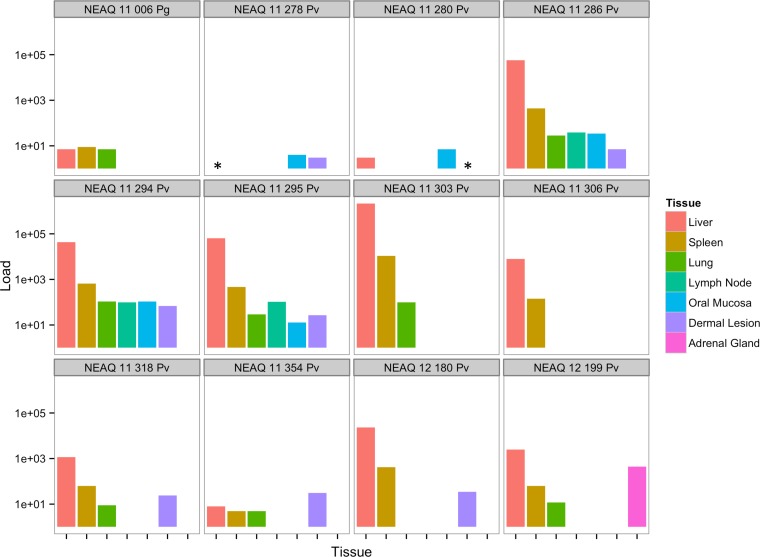
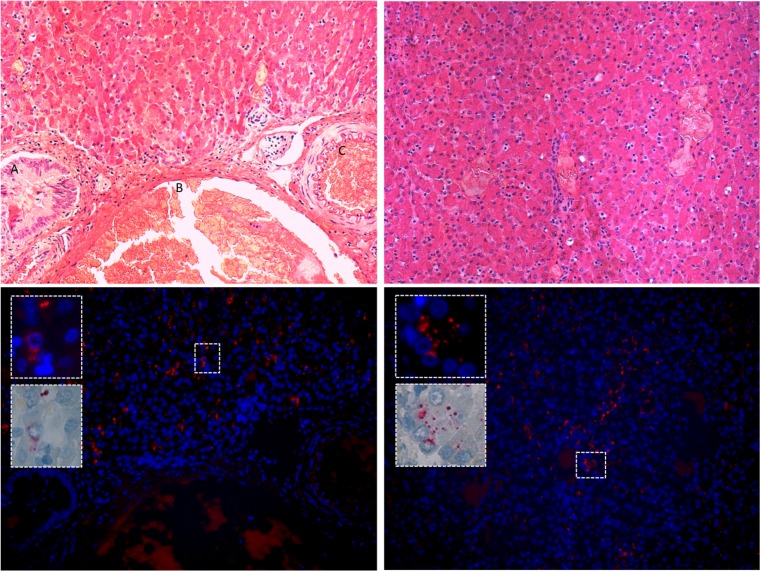
The distribution of phopivirus in different tissues was assessed using quantitative PCR and in situ hybridization (ISH). Viral loads, expressed as genome copy numbers per 250 ng of nucleic acid, were consistently highest in the liver, followed by the spleen (Fig. 3). ISH was then used to assay liver, spleen, lung, lymph node, and oral mucosa samples from three positive animals and one negative animal to confirm infection. Only the livers (hepatocytes) of the three PCR-positive animals were truly infected (Fig. 4), despite the detection of viral RNA in other tissues by qPCR. No observable pathological changes were associated with phopivirus infection (slides reviewed by veterinary pathologist J. A. St. Leger, as well as by medical hepatopathologist J. H. Lefkowitch for comparison with HAV infection).
FIG 3 .
Phopivirus viral loads for all 12 positive animals identified in this study. Viral loads were determined by qPCR and are expressed as genome copy numbers per 250 ng. Pg, Phoca groenlandicus (harp seal); Pv, Phoca vitulina (harbor seal); *, tissue not available for testing.
FIG 4 .
Harbor seal infected liver tissue. (Top) Two liver sections from animal NEAQ-11-295-Pv, stained with hematoxylin; left-hand image includes bile duct (A), hepatic vein (B), and hepatic artery (C). No inflammation is observed in the infected tissue. (Bottom) Distribution of phopivirus in liver using fluorescent in situ hybridization (FISH), with higher-magnification insets demonstrating clear cytoplasmic infection.
A common ancestry.
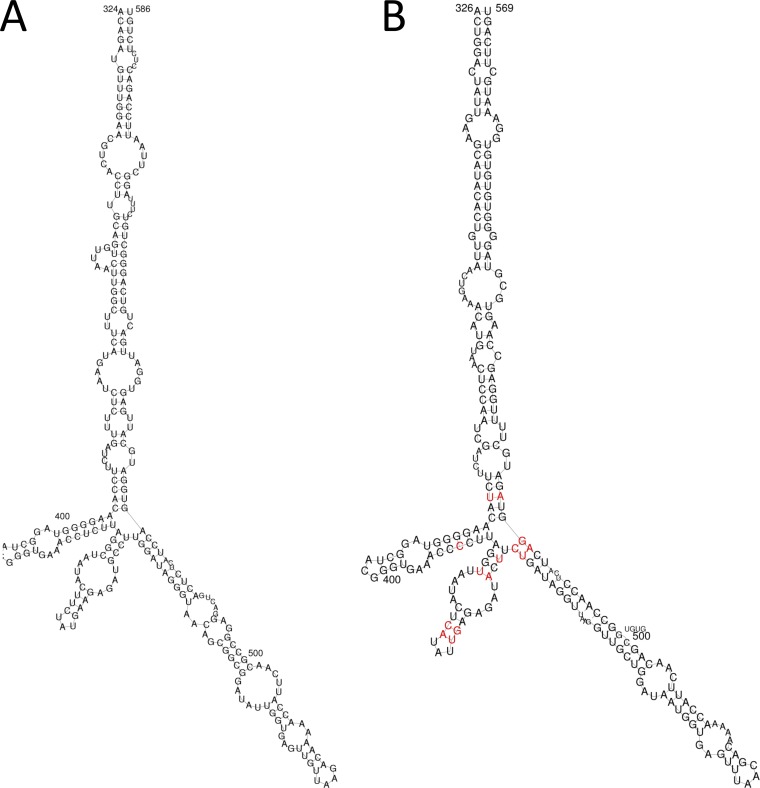
The genetic and phenotypic data presented above support a common ancestry between phopivirus and HAV. Our data show that these viruses also share several additional characteristics that are generally considered to be unique to HAV. For example, a comparison of the phopivirus and HAV 5′ UTR sequences (containing the unique type III IRES) showed substantial conservation in the secondary structure of domain IV. This domain is critical for the initiation of translation in HAV (8, 9), and the selection of complementary mutations that preserve base pairing and secondary structure in the phopivirus sequence suggests a shared critical role in translation (Fig. 5). Both viruses also share important unique/uncommon similarities in codon use. Relative occurrence measures the frequency (%) with which synonymous codons occur relative to the occurrence of the codon that is used most often for each amino acid (7). When assessed for HAV, it was shown that the codons most often preferred by the host were rarely used by the virus—a trait that has to date not been observed in any other known picornavirus (7). Here, we show that phopivirus uses this same strategy and that both HAV and phopivirus display a strong codon bias that is complementary to that of the host cell for many different amino acids (Table 2 and Table 3). We confirmed that this feature does not extend to related viruses, such as avian encephalomyelitis virus (AEV; genus Tremovirus), thus highlighting the evolutionary distinction of phopivirus and HAV.
FIG 5 .
Structural comparison of the type III IRES, domain IV (which corresponds to nucleotides 320 to 570 of the phopivirus 5′ UTR). Highlighted nucleotides represent pairings where complementary mutations have been selected in order to preserve structural integrity. (A) HAV. (B) Phopivirus.
TABLE 2 .
Relative occurrence of synonymous codons for each amino acid
| Amino acid (symbols) | Codon(s) | Occurrence (%) of codon ina: |
||||||
|---|---|---|---|---|---|---|---|---|
| Humans | HAV | Poliovirus | Seals | Phopivirus | Chickens | AEV | ||
| Phenylalanine (Phe, F) | UUU | 86.7 | 100.00 | 100.00 | 87.8 | 100.00 | 83.17 | 100.00 |
| UUC | 100.0 | 28.20 | 95.43 | 100.0 | 22.09 | 100.00 | 65.34 | |
| Leucine (Leu, L) | UUA | 19.4 | 49.32 | 63.98 | 19.8 | 98.30 | 18.18 | 31.89 |
| UUG | 32.6 | 100.00 | 100.00 | 20.6 | 100.00 | 32.73 | 100.00 | |
| CUU | 33.3 | 44.66 | 52.15 | 30.3 | 57.63 | 32.21 | 41.18 | |
| CUC | 49.5 | 8.22 | 68.28 | 46.8 | 16.95 | 43.64 | 24.15 | |
| CUA | 18.2 | 9.32 | 71.51 | 18.5 | 35.59 | 15.58 | 16.10 | |
| CUG | 100.0 | 26.03 | 77.42 | 100.0 | 22.03 | 100.00 | 43.03 | |
| Isoleucine (Ile, I) | AUU | 76.9 | 100.00 | 100.00 | 42.0 | 100.00 | 76.36 | 100.00 |
| AUC | 100.0 | 16.63 | 80.48 | 100.0 | 23.08 | 100.00 | 49.06 | |
| AUA | 36.1 | 31.62 | 60.56 | 26.0 | 39.56 | 40.00 | 80.52 | |
| Valine (Val, V) | GUU | 39.1 | 100.00 | 46.01 | 23.5 | 100.00 | 46.45 | 91.10 |
| GUC | 51.6 | 14.81 | 47.15 | 81.3 | 37.88 | 48.23 | 46.32 | |
| GUA | 25.3 | 16.99 | 46.39 | 17.4 | 33.33 | 27.66 | 34.97 | |
| GUG | 100.0 | 45.15 | 100.00 | 100.0 | 69.70 | 100.00 | 100.00 | |
| Serine (Ser, S) | UCU | 77.9 | 100.00 | 46.38 | 57.2 | 80.00 | 69.80 | 100.00 |
| UCC | 90.8 | 29.87 | 71.01 | 93.5 | 26.67 | 77.72 | 67.53 | |
| UCA | 62.6 | 88.93 | 100.00 | 42.8 | 100.00 | 57.43 | 74.23 | |
| UCG | 22.6 | 4.70 | 25.12 | 30.3 | 0.00 | 25.74 | 32.99 | |
| AGU | 62.1 | 33.56 | 49.28 | 58.7 | 51.67 | 55.45 | 82.99 | |
| AGC | 100.0 | 6.71 | 39.61 | 100.0 | 16.67 | 100.00 | 44.33 | |
| Proline (Pro, P) | CCU | 88.4 | 100.00 | 49.39 | 68.2 | 100.00 | 90.00 | 100.00 |
| CCC | 100.0 | 21.74 | 40.08 | 100.0 | 55.26 | 100.00 | 46.95 | |
| CCA | 85.4 | 81.64 | 100.00 | 68.2 | 100.00 | 92.35 | 70.73 | |
| CCG | 34.8 | 2.42 | 27.53 | 27.7 | 0.00 | 45.88 | 20.73 | |
| Threonine (Thr, T) | ACU | 69.3 | 100.00 | 82.31 | 37.1 | 87.30 | 80.61 | 91.34 |
| ACC | 100.0 | 20.74 | 100.00 | 100.0 | 15.87 | 100.00 | 66.23 | |
| ACA | 79.9 | 92.59 | 80.00 | 47.9 | 100.00 | 97.58 | 100.00 | |
| ACG | 32.3 | 4.44 | 29.62 | 24.7 | 3.17 | 46.67 | 40.69 | |
| Alanine (Ala, A) | GCU | 66.4 | 100.00 | 80.92 | 55.6 | 100.00 | 90.83 | 100.00 |
| GCC | 100.0 | 28.37 | 66.41 | 100.0 | 23.08 | 100.00 | 56.87 | |
| GCA | 57.0 | 58.51 | 100.00 | 49.7 | 98.08 | 82.97 | 81.52 | |
| GCG | 26.7 | 1.42 | 32.06 | 20.8 | 5.77 | 39.74 | 35.55 | |
| Tyrosine (Tyr, Y) | UAU | 79.7 | 100.00 | 70.00 | 48.1 | 100.00 | 66.29 | 100.00 |
| UAC | 100.0 | 28.41 | 100.00 | 100.0 | 58.82 | 100.00 | 64.47 | |
| Histidine (His, H) | CAU | 72.2 | 100.00 | 64.03 | 59.7 | 100.00 | 65.97 | 100.00 |
| CAC | 100.0 | 22.11 | 100.00 | 100.0 | 24.32 | 100.00 | 39.43 | |
| Glutamine (Gln, Q) | CAA | 36.0 | 92.86 | 97.87 | 31.8 | 100.00 | 37.12 | 67.49 |
| CAG | 100.0 | 100.00 | 100.00 | 100.0 | 35.00 | 100.00 | 100.00 | |
| Asparagine (Asn, N) | AAU | 89.0 | 100.00 | 79.58 | 85.0 | 100.00 | 75.11 | 100.00 |
| AAC | 100.0 | 21.75 | 100.00 | 100.0 | 33.33 | 100.00 | 69.38 | |
| Lysine (Lys, K) | AAA | 76.5 | 100.00 | 100.00 | 71.5 | 100.00 | 79.59 | 72.29 |
| AAG | 100.0 | 58.58 | 91.84 | 100.0 | 45.87 | 100.00 | 100.00 | |
| Aspartic acid (Asp, D) | GAU | 86.9 | 100.00 | 92.73 | 66.7 | 100.00 | 100.00 | 100.00 |
| GAC | 100.0 | 21.04 | 100.00 | 100.0 | 25.23 | 98.42 | 73.82 | |
| Glutamic acid (Glu, E) | GAA | 73.2 | 100.00 | 100.00 | 73.0 | 100.00 | 75.79 | 59.27 |
| GAG | 100.0 | 76.51 | 80.00 | 100.0 | 39.56 | 100.00 | 100.00 | |
| Cysteine (Cys, C) | UGU | 84.1 | 100.00 | 100.00 | 100.0 | 100.00 | 66.17 | 100.00 |
| UGC | 100.0 | 26.58 | 81.90 | 97.6 | 19.45 | 100.00 | 76.42 | |
| Tryptophan (Trp, W) | UGG | 100.0 | 100.00 | 100.00 | 100.0 | 100.00 | 100.00 | 100.00 |
| Arginine (Arg, R) | CGU | 36.9 | 4.21 | 17.39 | 38.1 | 8.20 | 44.26 | 38.51 |
| CGC | 85.2 | 2.11 | 15.46 | 61.9 | 3.28 | 85.25 | 34.78 | |
| CGA | 50.8 | 3.51 | 8.70 | 54.2 | 3.28 | 43.44 | 20.50 | |
| CGG | 93.4 | 2.11 | 13.53 | 72.9 | 3.28 | 79.51 | 11.80 | |
| AGA | 100.0 | 100.00 | 100.00 | 100.0 | 100.00 | 100.00 | 67.70 | |
| AGG | 98.4 | 31.58 | 53.14 | 92.4 | 26.23 | 95.90 | 100.00 | |
| Glycine (Gly, G) | GGU | 48.6 | 60.87 | 88.24 | 36.0 | 100.00 | 57.87 | 100.00 |
| GGC | 100.0 | 27.90 | 58.82 | 100.0 | 25.93 | 100.00 | 63.56 | |
| GGA | 74.3 | 100.00 | 100.00 | 56.5 | 100.00 | 89.34 | 60.17 | |
| GGG | 69.8 | 34.78 | 72.06 | 61.2 | 24.07 | 81.22 | 58.05 | |
The most abundant codon in each amino acid family is assigned as 100%; the frequencies of other codons are then expressed relative to this most abundant synonym (for example, human UUU is used 86.7% as often as UUC).
TABLE 3.
Numbers of rare codons in hosts and viruses
| Source | No. ofa: |
||
|---|---|---|---|
| RCs | RCs abundant (100%) in host | Amino acids with RCs | |
| Humans | 5 | NA | 4 |
| HAV | 25 | 12 | 15 |
| Poliovirus | 7 | 0 | 4 |
| Seals | 9 | NA | 6 |
| Phopivirus | 20 | 9 | 12 |
| Chickens | 4 | NA | 3 |
| AEV | 5 | 0 | 3 |
RCs, rare codons; NA, not applicable.
DISCUSSION
Most zoonotic viral diseases occur only periodically, when conditions permit or promote the spillover of viruses from wildlife into humans (10). The impact they have on public health and economic stability can be substantial (11–13); however, they seldom persist for very long and rarely (it would seem) adapt sufficiently to become human viruses, sustained entirely by human-to-human transmission. Identifying those viruses that do appear to have adapted to humans or, indeed, to other animals, may provide insights into the frequency with which spillover leads to complete host adaptation, the extent to which contact with animals has shaped human virodiversity, and perhaps even an understanding of the pathways or viral families that are most likely to contribute new human pathogens in the future.
Here, we describe a novel picornavirus within the genus hepatovirus that is the closest known relative of HAV yet discovered. We propose the virus be named phopivirus and present evidence that supports a common evolutionary ancestry with HAV. First, phylogenetic analysis of the P1, P2, and P3 regions all confirm relatedness of the two viruses. Second, both phopivirus and HAV have a principal tropism for hepatocytes, with seemingly quiescent infection in juveniles (we note that no pathological changes were observed in the juvenile seals examined here and suggest this is perhaps analogous to HAV infection of children, where disease rarely manifests [14, 15]). Third, we demonstrate conservation in a key region of the type III IRES (8, 15), which has been shown to preserve function in HAV replication (8, 9) and which is different from the hepatitis C virus-like IRES used by the phylogenetically related AEV (16). Finally, we show that both phopivirus and HAV utilize a strategy of highly biased codon usage that is complementary to that of the host and unique among the picornaviruses (again, not used by AEV).
We suggest that these data support the hypothesis that HAV may have zoonotic origins but qualify that we do not assume phopivirus to be the direct ancestor of HAV. Humans have certainly been hunting seals for thousands of years (17, 18), providing a classical pathway of emergence; however, we cannot discern from current data whether HAV evolved following a spillover of phopivirus from seals into humans or vice versa or whether it evolved from a quite unrelated host species.
It has previously been suggested that Old World nonhuman primates (NHPs) might be the source of HAV (19). Indeed, both human and NHP strains of HAV are known to exist; however, the data have never been entirely clear, and it is not really known whether HAV emerged into primates from an unrelated species (for example, seals) and then dispersed throughout the primate family or whether HAV strains have coevolved with primates. The limited HAV diversity in NHPs might argue against coevolution (20), as would the detection of identical HAV sequences in different NHP species from entirely different continents (HAV strain AGM-27 from an African green monkey in Kenya and Indian simian HAV strain IND-SHAV from a rhesus macaque in India [21, 22]) or the sharing of HAV strains (genotype III) between NHPs and several human populations around the world (19, 23–27). We also note that the known diversity of HAV in NHPs is almost exclusively limited to species or individuals that have substantial contact with humans in natural settings (e.g., macaques) or captive colonies (21–24, 27–30). Therefore, while we do not reject the possibility that HAV originated in NHPs, we suggest that additional information on the global diversity and distribution of HAVs in NHPs is needed to better clarify their role in its evolutionary history and current epidemiology—particularly those in species with little or no contact with humans.
Our finding of phopivirus in seals is significant because it is the first nonprimate hepatovirus to be discovered. It suggests that several such viruses could exist in wildlife (many of which may be closer to HAV than phopivirus) and that the evolutionary history of HAV may be far more complex than previously considered. Increased surveillance and discovery for related viruses in wildlife will now be required to better understand the course of events leading to the diversification of phopivirus and HAV.
MATERIALS AND METHODS
Viral discovery and genome expansion.
The lungs, livers, spleens, and oral mucosae from three harbor seals (Phoca vitulina) were processed for viral discovery. Tissue samples were placed in NucliSens lysis buffer (bioMérieux) with 2- by 3-mm tungsten carbide beads (Qiagen), disrupted using the TissueLyser (2 times for 60 s at 25 Hz; Qiagen), incubated for 2 h at 55°C with 4 µl 600 mAU/ml proteinase K (Qiagen), and centrifuged for 3 min at 4°C and 12,000 rpm to collect the supernatant. Total nucleic acids (TNAs) were then extracted on the NucliSens easyMAG (bioMérieux) platform according to the manufacturer’s instructions and DNase treated using 2 U/ml Turbo DNase (catalog number AM2238; Ambion). The remaining nucleic acids, now enriched for RNA templates, were assayed by high-throughput sequencing using Ion Torrent technology (PGM platform; 1 million reads per sample) according to the manufacturer’s instructions. Sequence reads were aligned against host reference databases to remove host background using the Bowtie2 mapper and then primer trimmed and filtered based on quality, GC content, and sequence complexity. The remaining reads were de novo assembled using Newbler (version 2.6), and contigs and unique singletons subjected to homology search using MegaBlast against the GenBank nucleotide database. Sequences that showed poor or no homology at the nucleotide level were searched against the GenBank viral protein database using BLASTx. Viral sequences from the BLASTx analysis were subjected to a homology search against the GenBank protein database to correct for biased E values. The putative phopivirus genome, assembled as described above, was subsequently confirmed by PCR and traditional dideoxy sequencing of 500-nt overlapping fragments. Rapid amplification of cDNA ends (RACE) was used to confirm the sequences of the 5′ and 3′ termini, using the SMARTer PCR cDNA synthesis kit (Clontech) and LA Taq DNA polymerase (TaKaRa) according to the manufacturer’s instructions.
Phylogenetic and genetic analyses.
Phylogenetic analyses of sequence data were performed using MUSCLE (31). Maximum-likelihood trees were reconstructed using MEGA version 6.0.6 (32), and the best-fitting models selected using the same program. To look for evidence of adaptive selection in phopivirus, lineage-specific diversifying selection was assessed in VP1 and 3D alignments using the codon-based branch site random-likelihood-effects algorithm, a model specifically developed to evaluate evidence of episodic diversifying selection within individual branches of a phylogeny (33), available in the HYPHY package on Datamonkey (http://www.datamonkey.org). Codon usage was assessed both for viruses and their hosts. For HAV, poliovirus, humans, seals, chickens, and avian encephalomyelitis virus (AEV), codon usage tables were obtained from a database based on genomes in NCBI GenBank (http://www.kazusa.or.jp), accessed on 1 March 2015. For phopivirus, codon usage frequencies were determined using the Cusp program (European molecular biology open software suite, http://emboss.sourceforge.net/apps/cvs/emboss/apps/cusp.html). From these tables, relative occurrence was determined by calculating the frequency of each codon, assigning 100% to the codon with the highest frequency for each amino acid, and calculating the occurrence (%) of all other codons for each amino acid relative to the occurrence of this most abundant synonym (7). An IRES structural alignment between the HAV 5′ UTR (X75216) and the phopivirus 5′ UTR was performed using the RNApaln program from ViennaRNA suite version 2.1.2. The secondary structure for HAV was obtained from Rfam (http://rfam.sanger.ac.uk/family/RF00228#tabview=tab3).
Screening PCR.
Quantitative real-time PCR (qPCR) using SYBR green was performed to assay samples for the presence of phopivirus. All samples were extracted as described above and reverse transcribed into cDNA by using SuperScript III (random hexamer protocol; Invitrogen). Primers were designed to target a 153-nt fragment of the confirmed VP1 sequence (FWD, 5′ AGTTGGGTCGAAGGAGCGTCA 3′, and RVS, 5′ TCCTTGTCCTGGTAAGGATGCTGC 3′). PCR was performed using 12.5 µl SYBR green PCR master mix (Applied Biosystems), 300 nM each primer, FWD and RVS, and 2 µl cDNA template (standardized to 250 ng/µl) added to a final volume of 25 µl. Standard controls (plasmid containing the target polyprotein sequence) were prepared at 1.5 × 105 to 1.5 × 100 per µl in a background of 2.5 ng/µl human placental DNA, to allow quantification of viral copy numbers. All samples, standards, and nontemplate controls were run in triplicate over 45 cycles. Conventional PCR was also performed on all samples in order to generate templates for sequencing to confirm positive results. PCR was performed with a fast-cycling PCR kit (Qiagen) according to the manufacturer’s instructions. The primers were the same as described above for qPCR.
Nucleotide sequence accession number.
The sequence for phopivirus has been assigned GenBank accession number KR703607.
ACKNOWLEDGMENTS
This study was funded by NIH/NIAID Centers for Research in Diagnostics and Discovery (grant U19AI109761 to W.I.L.). Sample collection was partially funded by NOAA’s John H. Prescott Marine Mammal Rescue Assistance Grant Program award no. NA07NMF4390220 and NOAA’s Marine Mammal Health and Stranding Response Program Unusual Mortality Event Contingency Fund. N.K. was supported by core funding provided by the Biotechnology and Biological Sciences Research Council under The Pirbright Institute Livestock Viral Diseases Programme.
The study benefitted from intellectual developments (or contributions) from the PREDICT project of the United States Agency for International Development (USAID) Emerging Pandemic Threats Program. Response to stranded marine mammals was authorized through Stranding Agreements between National Oceanic and Atmospheric Administration (NOAA), National Marine Fisheries Service Northeast Region, and Stranding Network participants New England Aquarium, International Fund for Animal Welfare, Marine Animal Rehabilitation Center, and College of the Atlantic.
We thank Salvatore Frasca, Jr., and Kelly Marcheterre of the Connecticut Veterinary Medical Diagnostic Laboratory for sharing samples that contributed to the investigation and Alexander Solovyov for help with structural analysis of the 5′ RNA (IRES).
Samples were collected by K.P. and T.R. Laboratory experiments were performed by S.J.A., E.L., M.D.S.-L., I.N.-M., H.S.I. and T.G. Pathology was performed by J.A.S.L. and J.H.L. Data analysis and interpretation were performed by S.J.A., J.A.S.L, J.H.L., E.L., M.D.S.-L., K.J., N.K., and W.I.L. The paper was written by S.J.A., J.A.S.L., T.R., T.G., K.P., K.N., and W.I.L. (with additional comments from all other coauthors).
The contents are the responsibility of the authors and do not necessarily reflect the views of USAID or the United States Government.
Footnotes
Citation Anthony SJ, St. Leger JA, Liang E, Hicks AL, Sanchez-Leon MD, Jain K, Lefkowitch JH, Navarrete-Macias I, Knowles N, Goldstein T, Pugliares K, Ip HS, Rowles T, Lipkin WI. 2015. Discovery of a novel hepatovirus (Phopivirus of seals) related to human hepatitis A virus. mBio 6(4):e01180-15. doi:10.1128/mBio.01180-15.
REFERENCES
- 1.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451:990–993. doi: 10.1038/nature06536(2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharp PM, Hahn BH. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med 1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furuse Y, Suzuki A, Oshitani H. 2010. Origin of measles virus: divergence from rinderpest virus between the 11th and 12th centuries. Virol J 7:52. doi: 10.1186/1743-422x-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony SJ, St Leger JA, Pugliares K, Ip HS, Chan JM, Carpenter ZW, Navarrete-Macias I, Sanchez-Leon M, Saliki JT, Pedersen J, Karesh W, Daszak P, Rabadan R, Rowles T, Lipkin WI. 2012. Emergence of fatal avian influenza in New England harbor seals. mBio 3(4):e00166-12. doi: 10.1128/mBio.00166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams MJ, Lefkowitz EJ, King AMQ, Carstens EB. 2013. Recently agreed changes to the International Code of Virus Classification and Nomenclature. Arch Virol 158:2633–2639. doi: 10.1007/s00705-013-1749-9. [DOI] [PubMed] [Google Scholar]
- 6.Adams MJ, Lefkowitz EJ, King AM, Carstens EB. 2013. Recently agreed changes to the International Code of Virus Classification and Nomenclature. Arch Virol 158:2633–2639. doi: 10.1007/s00705-013-1749-9. [DOI] [PubMed] [Google Scholar]
- 7.Sánchez G, Bosch A, Pintó RM. 2003. Genome variability and capsid structural constraints of hepatitis A virus. J Virol 77:452–459. doi: 10.1128/JVI.77.1.452-459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown EA, Zajac AJ, Lemon SM. 1994. In-vitro characterization of an internal ribosomal entry site (Ires) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J Virol 68:1066–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown EA, Day SP, Jansen RW, Lemon SM. 1991. The 5′ nontranslated region of hepatitis-A virus RNA: secondary structure and elements required for translation in vitro. J Virol 65:5828–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morse SS, Mazet JA, Woolhouse M, Parrish CR, Carroll D, Karesh WB, Zambrana-Torrelio C, Lipkin WI, Daszak P. 2012. Prediction and prevention of the next pandemic zoonosis. Lancet 380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, Soropogui B, Sow MS, Keïta S, De Clerck H, Tiffany A, Dominguez G, Loua M, Traoré A, Kolié M, Malano ER, Heleze E, Bocquin A, Mély S, Raoul H. 2014. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med 371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 12.The World Bank 2014. The economic impact of the 2014 Ebola epidemic: short and medium term estimates for West Africa. The World Bank, Washington, DC. [Google Scholar]
- 13.WHO Ebola Response Team 2014. Ebola virus disease in West Africa—the first 9 months of the epidemic and forward projections. N Engl J Med 371:1481–1495. doi: 10.1056/NEJMoa1411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matheny SC, Kingery JE. 2012. Hepatitis A. Am Fam Physician 86:1027–1034. [PubMed] [Google Scholar]
- 15.Pintó RM, D’Andrea L, Pérez-Rodriguez FJ, Costafreda MI, Ribes E, Guix S, Bosch A. 2012. Hepatitis A virus evolution and the potential emergence of new variants escaping the presently available vaccines. Future Microbiol 7:331–346. doi: 10.2217/fmb.12.5. [DOI] [PubMed] [Google Scholar]
- 16.Bakhshesh M, Groppelli E, Willcocks MM, Royall E, Belsham GJ, Roberts LO. 2008. The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J Virol 82:1993–2003. doi: 10.1128/JVI.01957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffecker JF. 2005. A prehistory of the north: human settlement of the higher latitudes. Rutgers University Press, Piscataway, NJ. [Google Scholar]
- 18.Ray GC, McCormick-Ray J. 2014. Marine conservation—science, policy, and management. Wiley Blackwell, Chichester, United Kingdom. [Google Scholar]
- 19.Robertson BH. 2001. Viral hepatitis and primates: historical and molecular analysis of human and nonhuman primate hepatitis A, B and the GB related viruses. J Viral Hepat 8:233–242. doi: 10.1046/j.1365-2893.2001.00295.x. [DOI] [PubMed] [Google Scholar]
- 20.Cristina J, Costa-Mattioli M. 2007. Genetic variability and molecular evolution of hepatitis A virus. Virus Res 127:151–157. doi: 10.1016/j.virusres.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Arankalle VA, Ramakrishnan J. 2009. Simian hepatitis A virus derived from a captive rhesus monkey in India is similar to the strain isolated from wild African green monkeys in Kenya. J Viral Hepat 16:214–218. doi: 10.1111/j.1365-2893.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 22.Tsarev SA, Emerson SU, Balayan MS, Ticehurst J, Purcell RH. 1991. Simian hepatitis A virus (HAV) strain Agm-27: comparison of genome structure and growth in cell culture with other HAV Strains. J Gen Virol 72:1677–1683. doi: 10.1099/0022-1317-72-7-1677. [DOI] [PubMed] [Google Scholar]
- 23.Brown EA, Jansen RW, Lemon SM. 1989. Characterization of a simian hepatitis A virus (HAV): antigenic and genetic comparison with human HAV. J Virol 63:4932–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemon SM, Leduc JW, Binn LN, Escajadillo A, Ishak KG. 1982. Transmission of hepatitis A virus among recently captured Panamanian owl monkeys. J Med Virol 10:25–36. doi: 10.1002/jmv.1890100105. [DOI] [PubMed] [Google Scholar]
- 25.Jansen RW, Siegl G, Lemon SM. 1990. Molecular epidemiology of human hepatitis A virus defined by an antigen capture polymerase chain reaction method. Proc Natl Acad Sci U S A 87:2867–2871. doi: 10.1073/pnas.87.8.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson BH, Khanna B, Nainan OV, Margolis HS. 1991. Epidemiologic patterns of wild-type hepatitis A virus determined by genetic variation. J Infect Dis 163:286–292. doi: 10.1093/infdis/163.2.286. [DOI] [PubMed] [Google Scholar]
- 27.Nainan OV, Margolis HS, Robertson BH, Balayan M, Brinton MA. 1991. Sequence analysis of a new hepatitis A virus naturally infecting cynomolgus macaques (Macaca fascicularis). J Gen Virol 72:1685–1689. doi: 10.1099/0022-1317-72-7-1685. [DOI] [PubMed] [Google Scholar]
- 28.Andzhaparidze AG, Balaian MS, Savinov AP, Kazachkov YA, Titova IP. 1987. Spontaneous hepatitis similar to hepatitis A in African green monkeys. Vopr Virusol 32:681–686. (In Russian.) [PubMed] [Google Scholar]
- 29.Leduc JW, Lemon SM, Keenan CM, Graham RR, Marchwicki RH, Binn LN. 1983. Experimental infection of the New World owl monkey (Aotus trivirgatus) with hepatitis A virus. Infect Immun 40:766–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sa-nguanmoo P, Thawornsuk N, Rianthavorn P, Sommanustweechai A, Ratanakorn P, Poovorawan Y. 2010. High prevalence of antibodies against hepatitis A virus among captive nonhuman primates. Primates 51:167–170. doi: 10.1007/s10329-009-0172-z. [DOI] [PubMed] [Google Scholar]
- 31.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340(2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosakovsky Pond SL, Murrell B, Fourment M, Frost SD, Delport W, Scheffler K. 2011. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol 28:3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]