Abstract
Objective
To develop a natural language processing software inference algorithm to classify the content of primary care consultations using electronic health record Big Data and subsequently test the algorithm's ability to estimate the prevalence and burden of childhood respiratory illness in primary care.
Design
Algorithm development and validation study. To classify consultations, the algorithm is designed to interrogate clinical narrative entered as free text, diagnostic (Read) codes created and medications prescribed on the day of the consultation.
Setting
Thirty-six consenting primary care practices from a mixed urban and semirural region of New Zealand. Three independent sets of 1200 child consultation records were randomly extracted from a data set of all general practitioner consultations in participating practices between 1 January 2008–31 December 2013 for children under 18 years of age (n=754 242). Each consultation record within these sets was independently classified by two expert clinicians as respiratory or non-respiratory, and subclassified according to respiratory diagnostic categories to create three ‘gold standard’ sets of classified records. These three gold standard record sets were used to train, test and validate the algorithm.
Outcome measures
Sensitivity, specificity, positive predictive value and F-measure were calculated to illustrate the algorithm's ability to replicate judgements of expert clinicians within the 1200 record gold standard validation set.
Results
The algorithm was able to identify respiratory consultations in the 1200 record validation set with a sensitivity of 0.72 (95% CI 0.67 to 0.78) and a specificity of 0.95 (95% CI 0.93 to 0.98). The positive predictive value of algorithm respiratory classification was 0.93 (95% CI 0.89 to 0.97). The positive predictive value of the algorithm classifying consultations as being related to specific respiratory diagnostic categories ranged from 0.68 (95% CI 0.40 to 1.00; other respiratory conditions) to 0.91 (95% CI 0.79 to 1.00; throat infections).
Conclusions
A software inference algorithm that uses primary care Big Data can accurately classify the content of clinical consultations. This algorithm will enable accurate estimation of the prevalence of childhood respiratory illness in primary care and resultant service utilisation. The methodology can also be applied to other areas of clinical care.
Keywords: PRIMARY CARE
Strengths and limitations of this study.
This study developed and tested a natural language processing software inference algorithm to classify the content of clinical consultations using primary care Big Data.
Large, independent sets of 1200 gold standard consultation records in which respiratory conditions had been classified by two expert clinicians were used to train, test and validate the algorithm.
The algorithm demonstrated excellent specificity and positive predictive values for detecting respiratory conditions.
The algorithm is not reliant on predetermined clinical coding but is limited by the accuracy of information recorded by clinicians. It is designed to replicate clinical judgements rather than make independent classifications. It is also not able to classify conditions with very low prevalence.
The algorithm is able to analyse very large data sets including routinely recorded unstructured clinical narrative. These would be impractical to analyse manually. This will enable assessment of longitudinal trends and exploration of differences based on age, gender, geographical location, ethnicity and socioeconomic deprivation.
Background
Primary care influences child health outcomes by managing illness and by providing preventive and health promotion services. Primary care that is well organised and effectively delivered, can compensate for substantial social disadvantage and help to reduce inequalities in child health outcomes.1 2 Despite this evidence, population-wide approaches for children's health (with the exception of immunisation and the Well Child schedule3) have not been well developed or resourced in New Zealand primary care settings.4
Primary care is utilised by all New Zealand children,5 but there is currently little knowledge of morbidity and utilisation patterns. These patterns have been successfully mapped in adult populations to explore associations between general practice utilisation and ethnicity or socioeconomic deprivation.6 7 A better understanding of childhood illness presentation and service utilisation patterns in primary care will enable the development of more systematic approaches to care and improve resource allocation.
New Zealand is in a strong position to analyse patterns of childhood morbidity due to universal enrolment with a primary care provider at birth, every individual having a unique health number and a highly computerised primary care system containing detailed electronic consultation records. Few child health initiatives have made use of these existing enrolment bases and the rich data that exist on primary care Electronic Health Records (EHR). These data are considered to be Big Data given the volume of data available, the variety of structured and unstructured data including clinical consultation notes and the variable data veracity including multiple sources (clinicians, patients, caregivers) and ambiguities (spellings, abbreviations).8
The use of EHR Big Data presents several challenges. These include accessing data from individual primary care practices, the variety of software packages and systems used by practices, the mechanics of extracting and combining these data, and, most importantly, generating accurate and informative analyses from the plethora of data available.9 10 Novel software (Integrated Query Engine; IQE) that can extract EHR data directly from primary care practices has been developed by the local Primary Health Organisation (PHO).6 7 PHOs are responsible for coordinating the population health activities of general practice in New Zealand. The IQE is able to work with each of the three computerised medical records systems currently used by all practices within the PHO's practice network. It extracts line level data directly from each EHR database and, in real-time, transfers these data across a secure system to the PHO. The IQE is routinely used for extraction and transmission of EHR data sets from the general practices to the PHO for quality indicator and administrative purposes. Its ability to extract, transport and load data accurately and consistently has been established over a decade of use by the PHO.
Analysing morbidity patterns within these extracted data is problematic because primary care practices do not consistently or frequently use diagnostic labelling, and there is marked variability between clinicians and conditions.11–17 For example, in one New Zealand primary care sample, less than 10% of acute respiratory illness presentations were coded.18 This indicates that use of diagnostic coding is insufficient to provide an accurate estimation of illness prevalence. One way of addressing this challenge is to create a natural language processing algorithm that can automatically make diagnostic classifications based on the signs and symptoms recorded within the clinical narrative for each consultation, combined with medications prescribed and diagnostic labels. Such software (Pattern Recognition Over Standard Aesculapian Information Collections; PROSAIC) has previously been developed by the PHO to estimate the incidence of influenza-like illness.18
This study aimed to extend the use of PROSAIC to identify childhood respiratory conditions within primary care consultations by building an algorithm to classify the unstructured clinical narrative written by clinicians. This paper describes the development and validation of this inference algorithm.
Methods
Setting
All 60 primary care practices within the networks of two PHOs in the Greater Wellington region of New Zealand were invited to participate and 36 consented. The ‘Normal Hours’ cohort consisted of the 77 467 children (75% of the two PHO's child population; N=103 359) under 18 years of age enrolled in these 36 primary care practices between 1 January 2008 until 31 December 2013 (the study period). This Normal Hours cohort represented 270 576 person years; children both joined and left this cohort during the 6-year period of the study (eg, births, deaths, turning 18 years of age, or moving into or out of a consenting practice).
Two of the consenting primary care practices operated out-of-hours services. An additional two stand-alone out-of-hours clinics operated within the Greater Wellington region and one of these also consented to participate. The ‘Out-of-Hours’ cohort consisted of 28 776 children (16 098 from consenting primary care practices and 12 678 from other practices) who presented to these out-of-hours services during the study period.
The data set was extracted with IQE software and included all child-general practitioner (GP) consultations at consenting practices during the study period (n=754 242). Of these, 692 968 involved the Normal Hours cohort and 61 274 the Out-of-Hours cohort. Patient names were not included in the extracted data set, but each consultation record was associated with an individual's unique National Health Index (NHI) number. This enables records to be matched between data sets. All data sets were held and analysed within the PHO, which routinely handles identifiable health information and consequently has rigorous protocols in place to ensure patient confidentiality. No identifiable data was ever accessed by the research team external to the PHO.
Exploratory analysis and methodology development
A random sample of 1193 clinical records from seven general practitioners (GPs; 693 records), three practice nurses (300 records) and 200 out-of-hours consultations was extracted from the data set to enable exploratory analysis of the data and development of the methodology. Challenges identified within the exploratory analysis were discussed among the entire research team and consensus was achieved.
Creating the respiratory condition categories
A hierarchical classification system was developed (figure 1). The first level of the hierarchy divided all consultations into either ‘respiratory’ or ‘not respiratory’. The ‘not respiratory’ category included consultations for conditions such as injury or gastroenteritis, as well as consultations in which the respiratory system was examined as a screening test, but no signs, symptoms or diagnoses were recorded. These screening consultations were excluded so that the burden of respiratory illness estimate was not inflated by consultations that did not result from a respiratory illness.
Figure 1.
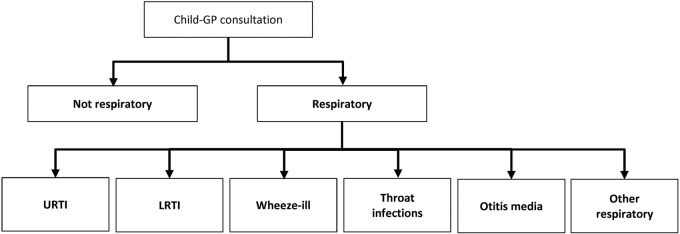
Hierarchy for classification of consultations, using free text notes, diagnostic Read codes and medication prescription. GP, general practitioner; URTI, upper respiratory tract infection; LRTI, lower respiratory tract infection; Wheeze-ill, wheeze-related illness.
The second level of the hierarchy subclassified consultations into one or more specific respiratory conditions. When selecting these conditions, consideration was given to the degree to which these could be mapped to conditions of high prevalence (those that are common) and/or conditions responsible for significant morbidity and hospitalisation (those that are important). Initially 14 categories were selected; however, exploratory analysis indicated that the prevalence of some of these conditions was very low (<4%), and insufficient to effectively train an algorithm. Consequently, conditions with low individual prevalence were combined within categories based on anatomical proximity (eg, pharyngitis and tonsillitis).
Ultimately, six condition categories were created: (1) upper respiratory tract infections; (2) lower respiratory tract infections; (3) wheeze-related illness; (4) throat infections; (5) otitis media; and (6) other respiratory conditions. The main conditions included within each category are presented in table 1.
Table 1.
Respiratory classification categories and the conditions included in each
| Classification category | Respiratory conditions included within category* |
|---|---|
| Upper respiratory tract infections |
|
| Lower respiratory tract infections |
|
| Wheeze-related illness |
|
| Throat infections |
|
| Otitis media |
|
| Other respiratory |
|
*These classifications are based purely on the information within the electronic health record including consultation notes, medications prescribed and diagnostic Read Codes created on the day of the consultation. It does not include subsequent laboratory tests.
GP, general practitioner.
Methodological decisions
Inclusion of both practice nurse and GP consultations was initially planned as these both contribute to the primary care burden, however, during exploratory analysis, it was found that many nursing clinical records were created as a result of telephone calls (including messages left), immunisation visits and general health and development checks. It was not possible to cross reference clinical records with appointment bookings to differentiate between clinical consultations and other non-consultation records because many nurses did not keep appointments in the same manner as did GPs. Consequently, nurse-only clinical records were excluded. If a nurse and GP both consulted the same child on the same day, PROSAIC merged these clinical records so that important information was not omitted (GPs often did not re-record information already captured within the nursing notes).
When interpreting the content of individual consultations, the following methodological decisions were made: (1) any directly declared and diagnosed condition by the GP was accepted at face value, even if the clinical experts within the research team disagreed with the GP's impression; (2) when the GP's clinical impression regarding symptoms differed from that of the child's (or their parent's) report, the GP's assessment was accepted at face value; (3) signs and symptoms reported within the consultation were deemed to be part of the current episode unless these were clearly delineated as being historical (ie, ‘pneumonia 3 years ago’) or absent (ie, ‘no wheeze’).
Algorithm development and training
The Child Respiratory Algorithm (‘the Algorithm’) was created to classify each consultation, based on the respiratory condition or conditions that were assessed or managed during the consultation, using PROSAIC software. The PROSAIC system was chosen: (1) as it had been successfully used previously to solve similar problems of classifying acute general practice presentations from clinical narrative; and (2) as the research team had expertise with this software; and also, (3) because it had been developed by the local PHO involved in the research, it was able to be further developed and refined for this study. PROSAIC used clinical narrative entered as free text, diagnostic (Read) codes created on the day of the consultation and medications prescribed on the day of the consultation, to process the algorithm and make this classification. PROSAIC is a collection of bespoke software subsystems providing tools to process natural language. These include software that: gathers clinical discourse into appropriate units (paragraphs, sentences, words); expands and disambiguates temporal expressions; and identifies pertinent expressions from within a specific lexicon and assesses the context of each expression's use (or negation). The tools include a variety of rule-processing classes that allow an algorithm to direct the way in which language is assessed, from basic pertinent expression existence tests, to the assignment and assessment of Bayesian probabilities. The algorithm defines lexicons specific to pertinent findings and directs PROSAIC on how to apply its tools and interpret results from these. Algorithms can be specifically developed for any given concept or content area using established natural language processing and linguistic concordance techniques.18–22 The way in which this algorithm processes data has been described with reference to identifying influenza-like illness (manuscript under review).
Gold standards
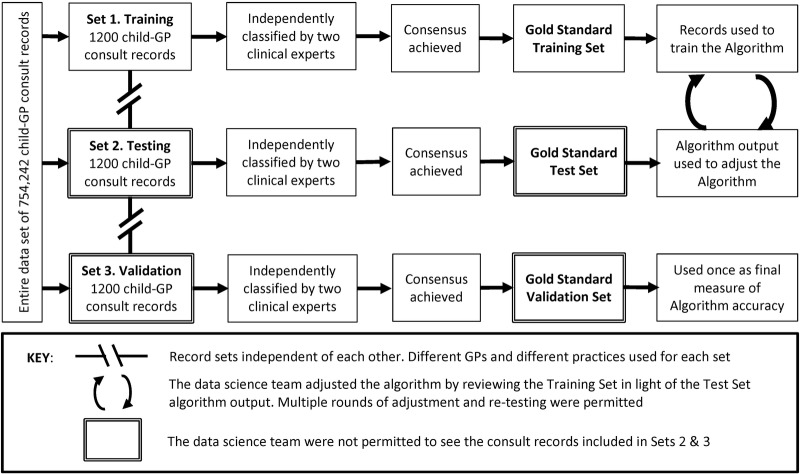
Three independent ‘gold standard’ sets of consultation records were used to train, test and validate the algorithm (figure 2). To create each gold standard set, 10 primary care practices were identified at random from the PHO network. One GP was selected at random from each of these 10 practices and 100 child clinical consultation records were selected at random from each of these GPs. The notes of GPs and practices used in any one particular set were explicitly excluded from selection in subsequent sets. Two hundred child clinical consultation records were also randomly selected from out-of-hours services. This process produced three independent subsets of 1200 consultation records from the entire data set of 754 242 records. Two clinical experts (TD and LM; each with over 20 years general practice experience) independently assessed each of the 3600 consultation records within these subsets. These experts used the same data fields as the algorithm to independently classify each consultation into its appropriate categories according to the hierarchical classification system (figure 1). These classification categories were not mutually exclusive, for example, a single consultation may have provided care for sinusitis as well as for a wheeze-related illness. Manual classification of clinical notes is very difficult and can be associated with substantial error.14 23 For this reason, the two expert clinicians independently classified each consultation using a coding interface that was custom-designed to make classification as easy as possible, reduce classification errors and ultimately improve concordance between clinicians. The blind agreement for each gold standard set is presented in table 2. When discordance occurred, consensus was reached by discussion. A third clinical expert (NT) was available to mediate if necessary, but was not required. Initial discordance primarily related to classification of secondary diagnoses.
Figure 2.
Use of independent consultation record sets to train, test and validate the Algorithm. GP, general practitioner.
Table 2.
Gold standard consultation record sets
| Blind agreement by GP clinical experts* |
||||
|---|---|---|---|---|
| Gold standard | Records included | Respiratory consultations identified | Agreement if consultation is respiratory or not respiratory | Complete agreement for all respiratory classifications included in consultations |
| Training set (set 1) | 1200 | 529 (0.44; 0.40 to 0.48) | 1139 (0.95; 0.93 to 0.97) | 1037 (0.86; 0.84 to 0.89) |
| Test set (set 2) | 1200 | 556 (0.46; 0.42 to 0.51) | 1146 (0.96; 0.94 to 0.97) | 1060 (0.88; 0.85 to 0.91) |
| Validation set (set 3) | 1200 | 553 (0.46; 0.42 to 0.51) | 1151 (0.96; 0.94to 0.98) | 1046 (0.87; 0.84 to 0.90) |
Data are n (proportion; 95% CI).
*Consensus was reached for all records with discordant classifications following initial independent coding.
GP, general practitioner.
This process resulted in three independent ‘gold standard’ sets of 1200 child consultation records in which all respiratory conditions present had been accurately classified. The aim of algorithm development was to enable the algorithm to make the same judgements as expert clinicians (the gold standard) when assessing clinical notes. The first of these sets was used to train the algorithm, the second set to test the algorithm and the third set was used to validate the algorithm.
Training and testing the algorithm
The training set gold standard (set 1) was used to train the algorithm to replicate the judgements made by the clinical experts. For each classification category, the sensitivity, specificity, positive predictive value and F-measure of individual symptoms and combinations of symptoms were calculated within the training set. These data were assessed by three GP clinical experts and compared with existing evidence (identified by way of a systematic search of the literature) to ensure no anomalies were present. These data were then used to inform the weight given to each symptom or group of symptoms by the algorithm. In order to provide a conservative estimate of the burden of respiratory illness, training aimed to keep the total number of false positives to a minimum (ie, maximise specificity). Algorithm performance was analysed to identify portions of the algorithm that were performing poorly. These were then modified to improve algorithm specificity, while maintaining or improving sensitivity. Beyond an optimal point, specificity is gained at the expense of sensitivity as the algorithm struggles to disambiguate ever increasing subtleties in language. Consultation records within the training set were able to be viewed by the data science team (JM and OJ) who created and adjusted the algorithm.
When the algorithm was considered to be performing well against the training set (set 1), its performance was tested on a new independent data set (the test set gold standard; set 2). The use of an independent test set ensured that the results of this test were not biased by features or anomalies associated with the notes of specific GPs or practices included in the training set used to inform the algorithm, and to increase confidence that the algorithm would perform well across the validation set and ultimately the entire data set that was comprised of consultation records from a wide range of GPs and practice contexts. Following each round of testing, the algorithm was further refined to improve its performance. The data science team personnel were able to see the algorithm output (results) from the test set (set 2) but were not permitted to see any of the clinical records, to avoid the introduction of training bias.
The clinical records within the validation set (set 3) were not viewed by the data science team. The training and test sets were able to be used repetitively as the algorithm was adjusted, whereas the validation set was used only once to provide final measures of accuracy.
Analysis
Data extracted from primary care practices were stored in a Microsoft SQL Server Database. All analyses were conducted using R statistical programming language.
Demographic characteristics
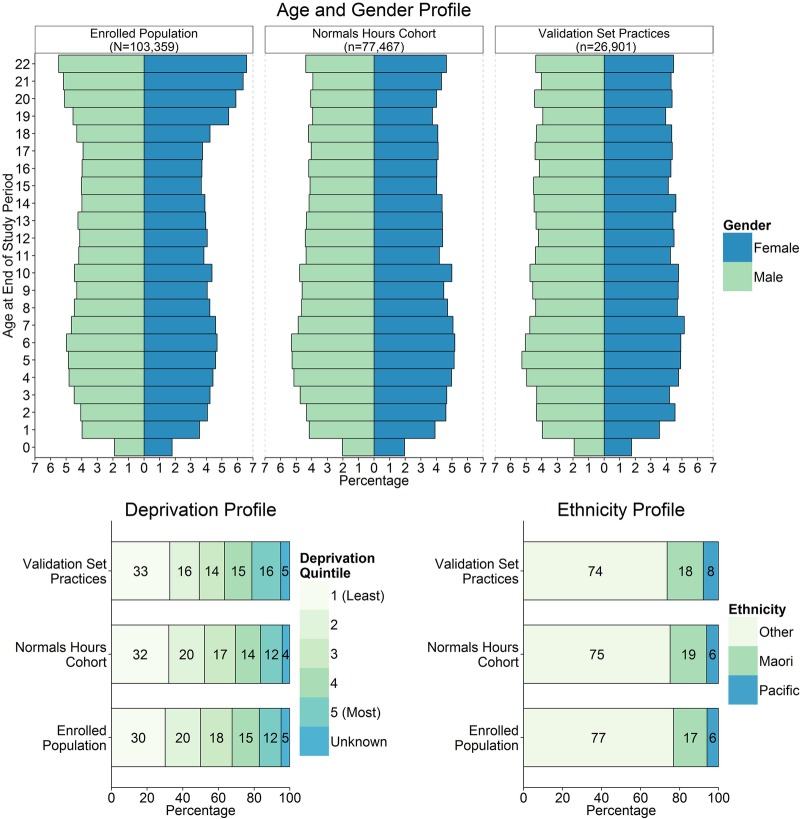
The demographic characteristics of age, gender, ethnicity (other, Māori, Pacific) and New Zealand Deprivation Index (a measure of socioeconomic deprivation24 25) of children enrolled in practices included in the gold standard validation set (n=26 901) were compared with those of the Normal Hours cohort (n=77 467) and all children enrolled within the two PHOs (N=103 359).
Algorithm validation
The performance of the algorithm in classifying clinical notes was assessed against the validation set gold standard. Bootstrapping was used to create measures of accuracy with CIs. This involved simple random sampling with replacement to create 10 000 samples of n=500 from the validation set (set 3; n=1200). Incidence, sensitivity, specificity, positive predictive value and F-measure were calculated from each sample providing 10 000 measures from which to calculate the 95% CIs. Positive predictive values >0.7 were defined a priori as being acceptable.26
Results
The demographic characteristics of the children enrolled in practices included in the validation set (set 3) were broadly comparable with children in all practices included in the Normal Hours cohort (figure 3).
Figure 3.
Demographic characteristics of the funded population, the Normal Hours cohort and children enrolled in the practices that were included in the validation set (set 3).
Within the 1200 record validation set, 355 records (30%) were Read coded by the treating GP at the time of the consultation. There were 133 records (11%) with a respiratory Read code, representing 24% of the 553 respiratory consultations identified by expert clinicians.
The algorithm identified a total of 555 consultations as containing one or more respiratory conditions. Of these, 408 consultations (74%) were identified as containing only one respiratory condition, 131 (24%) were identified as containing two respiratory conditions, 12 (2%) were identified as containing three respiratory conditions and two consultations (0.3%) were identified as containing four separate respiratory conditions.
The performance of the Algorithm in classifying consultations within the validation gold standard set (set 3) is presented in table 3. The algorithm classified 46% (95% CI 42% to 50%) of consultations within the validation set as being related to the assessment or management of respiratory illness. The algorithm was able to identify respiratory consultations with a sensitivity of 0.72 (95% CI 0.67 to 0.78) and a specificity of 0.95 (95% CI 0.93 to 0.98). The positive predictive value of algorithm respiratory classification was 0.93 (95% CI 0.89 to 0.97).
Table 3.
Automated software inference algorithm measures of performance in the validation set (set 3)
| Diagnostic category | Incidence (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | Positive predictive value (95% CI) | Negative predictive value (95% CI) | F-measure (95% CI) |
|---|---|---|---|---|---|---|
| Respiratory | 0.46 (0.42 to 0.50) | 0.72 (0.67 to 0.78) | 0.95 (0.93 to 0.98) | 0.93 (0.89 to 0.97) | 0.80 (0.76 to 0.84) | 0.81 (0.77 to 0.85) |
| LRTI | 0.04 (0.02 to 0.06) | 0.61 (0.39 to 0.83) | 0.99 (0.98 to 1.00) | 0.76 (0.55 to 0.95) | 0.98 (0.97 to 0.99) | 0.67 (0.47 to 0.85) |
| URTI | 0.21 (0.18 to 0.25) | 0.54 (0.45 to 0.64) | 0.98 (0.96 to 0.99) | 0.86 (0.78 to 0.94) | 0.89 (0.86 to 0.92) | 0.66 (0.57 to 0.74) |
| Wheeze ill | 0.09 (0.06 to 0.12) | 0.96 (0.90 to 1.00) | 0.96 (0.94 to 0.98) | 0.70 (0.59 to 0.82) | 1.00 (0.99 to 1.00) | 0.81 (0.73 to 0.89) |
| Throat infections | 0.10 (0.08 to 0.13) | 0.50 (0.37 to 0.64) | 0.99 (0.99 to 1.00) | 0.91 (0.79 to 1.00) | 0.95 (0.92 to 0.96) | 0.64 (0.51 to 0.76) |
| Otitis media | 0.12 (0.10 to 0.15) | 0.58 (0.45 to 0.71) | 0.99 (0.98 to 1.00) | 0.90 (0.81 to 1.00) | 0.94 (0.92 to 0.96) | 0.71 (0.59 to 0.81) |
| Other | 0.02 (0.01 to 0.04) | 0.66 (0.38 to 0.92) | 0.99 (0.98 to 1.00) | 0.68 (0.40 to 1.00) | 0.99 (0.98 to 1.00) | 0.66 (0.42 to 0.87) |
LRTI, lower respiratory tract infections; Other, other respiratory condition; URTI, upper respiratory tract infections; Wheeze ill, wheeze-related illness.
The positive predictive value of the algorithm classifying consultations as being related to specific respiratory diagnostic categories ranged from 0.68 (95% CI 0.40 to 1.00; other respiratory conditions) to 0.91 (95% CI 0.79 to 1.00; throat infections; table 3).
Discussion
Principal findings
The algorithm demonstrated excellent performance for classifying consultations as being respiratory-related. The high specificity for all classifications indicated that the goal of generating a conservative estimate of the burden of respiratory illness by keeping false positives to a minimum had been achieved. Nearly all respiratory consultation records were able to be subclassified according to diagnostic categories with acceptable positive predictive values (with the exception of ‘other respiratory conditions’). Only 2% of consultations were classified as ‘other respiratory conditions’, indicating that the classification categories included all of the prevalent respiratory conditions and that the algorithm was able to make a classification decision for nearly all respiratory consultations.
The application of the algorithm using PROSAIC will enable considerably more accurate estimation of respiratory condition prevalence and patterns of service utilisation in primary care than relying on diagnostic codes. In the validation set, less than a quarter of the respiratory consultations identified by expert clinicians had respiratory Read codes. Diagnostic coding is often absent or inaccurate,11–18 27 particularly for secondary diagnoses.13 Two or more conditions were identified in over a quarter of the validation set respiratory consultations, reinforcing the need for these to be classified when estimating prevalence and burden of illness.
Strengths
The specificity of the algorithm in classifying consultations as being respiratory-related exceeds that previously reported.28 The algorithm developed in this study had higher sensitivity and specificity than that developed by Wu et al17 to diagnose asthma. It is likely that the algorithm had both high specificity and sensitivity for classifying Wheeze illness because the medications prescribed and symptoms described were highly predictive for Wheeze illness and for few other conditions. The algorithm had lower sensitivity (0.50 compared with 0.96) but much higher specificity (0.99 compared with 0.34) than a data abstraction approach using search term methods for pharyngitis as reported by Benin et al.29 The current study's results are consistent with the aim of minimising the occurrence of false positives. The algorithm is intended to be used to estimate the prevalence of these conditions in primary care; it is not designed to make or inform clinical diagnoses. An additional strength is that PROSAIC can interpret multiple data types (consultation records, diagnostic codes, medications) and can be used on large sets of unstructured notes gathered from multiple practitioners in multiple primary care practices with multiple EHR systems rather than being confined to a single data type, a single institution or a single EHR system.17 29
Two clinical experts were used to create the three gold standard sets of 1200 clinical records with which the algorithm was trained and tested, and against which the algorithm was validated. This minimised the chance of error found previously when research assistants or individual coders have performed this task,14 and increased confidence in the performance data reported. The use of these three distinct sets of gold standard consultation records, which included records of 30 different clinicians at 30 different practices, minimised the introduction of training bias. Care was taken to ensure that the sample of clinical narrative used for algorithm training, testing and validation taken from the study population was random and avoided contributing to overtraining or bias. This sample included practitioners from urban and rural settings, as well as normal hours and out-of-hours practices, to account for differences in clinical notation. The high number of records within these sets matches or exceeds those reported previously.17 28 29 The demographic characteristics of children from practices included in the gold standard validation set were very similar to those of all children in the Normal Hours cohort, indicating that this randomly selected data set was broadly representative of the entire data set.
Natural language processing programmes have previously been found to function more effectively for variables that are narrowly and consistently defined.30 The 93% accuracy found in the current study is notable given the enormous variability in style, structure and content of consultation record keeping by individual practitioners, and the presence of notations and spelling errors.30–32 The algorithm's concordance with the consensus of two expert clinicians was only slightly less than the clinicians’ concordance with each other (93% vs 96%). This is consistent with previous research, which has found that automated coding is at least, if not more, accurate than expert coding,19 23 and it can be applied to very large data sets, whereas expert coding can only be applied to relatively small samples.
Limitations
This study used the treating GP's stated diagnosis, or experts’ assessment of the presumed diagnosis, based on clinical information and prescriptions recorded, as the gold standard. Consequently, this gold standard includes potentially erroneous diagnoses made by the treating GPs,13 16 and is limited by the information that the GPs determined was pertinent to record. However, the goal of this study was to estimate the burden of illness within primary care as defined by the care received. The GP perception of the conditions being managed is of prime importance in assessing health service utilisation, and hence this limitation does not affect the algorithm's ability to provide important and useful data.
The need to have conditions with sufficient prevalence to train the algorithm meant that a number of less prevalent conditions needed to be combined within single categories. As a result, this study will not be able to give estimations of the burden of some illnesses that, although rare, have considerable morbidity. These include croup, pertussis and pneumonia.
Nurse-only consultations were unable to be included due to the very large number of nursing records made, only a small proportion of which related to clinical consultations for an acute illness presentation. Including these records may have provided a more accurate estimate of the absolute number of respiratory consultations, but also an artificially low estimate of the proportion of primary care consultations that are respiratory-related because of the inclusion of non-consultation records.
Implications for clinical practice and future research
The algorithm will be used to provide an accurate estimate of the prevalence of respiratory condition consultations in primary care for children under 18 years of age. The utilisation of services for these six conditions will be analysed by age, gender, ethnicity, geographical location and New Zealand Deprivation Index. Trends in these data over time will also be examined. These analyses will provide valuable information that may be used to develop more systematic approaches to care.
This study has shown that using a natural language processing algorithm can allow very large numbers of consultation records to be analysed and categorised with greater accuracy than relying on diagnostic coding alone.18 The same approach will be applied to classifying other condition groups for skin infection and injury. Although this will require new classification hierarchies to be developed and new algorithm programming, the methodology described in this paper will be reused. It can also be used to gather similar data from different patient groups and populations. This approach has further and wider implications. It is already being used to monitor presentations of influenza-like illness on a daily basis from general practice, and could be extended for screening of other illnesses. The ability to analyse retrospective data as well as cross-sectional data allows comparison between specific time periods. Finally, this algorithm may be integrated into future versions of EHR software so that appropriate classification codes are suggested to clinicians in real time, thereby improving the quality and completeness of diagnostic coding.
Conclusions
A natural language processing software inference algorithm that analyses the content of clinical consultation records, diagnostic classifications and prescription information, is able to classify child-GP consultations related to respiratory conditions with similar accuracy to clinical experts. This algorithm will enable accurate estimation of the prevalence of childhood respiratory illnesses in primary care and the resultant service utilisation.
Acknowledgments
The authors gratefully acknowledge the primary care practices that consented to their consultation records being included in the study data set, and the primary health organisations that permitted use of their proprietary software and resources.
Footnotes
Contributors: AD, JMR, MS, LMB and NT conceived the study. All the authors contributed to the development of the overall study methodology. AD, LMB and NT provided clinical input into the algorithm design. JMR designed and built the natural language processing tools. OJ and JM programmed and trained the algorithm. AD and LMB classified the consultation records in the gold standard sets. BD was the principal writer of the manuscript. All the authors reviewed and revised the manuscript, and approved its final version. All the authors had full access to all of the data (including statistical reports and tables) in the study and can take responsibility for the integrity of the data, and the accuracy of the data analysis.
Funding: This work was supported by a New Zealand Lotteries Health Research Grant. The funding body had no role in the collection or analysis of data or the preparation of this manuscript.
Competing interests: OJ is an employee and LMB is a director of Compass Health Wellington Trust, which might have an interest in the submitted work.
Ethics approval: This study was approved by the University of Otago Ethics Committee (H13/044).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- 1.National Health Committee. Improving health for New Zealanders by investing in primary health care. Wellington, NZ: National Health Committee, 2000. [Google Scholar]
- 2.Ashworth M, Seed P, Armstrong D et al. . The relationship between social deprivation and the quality of primary care: a national survey using indicators from the UK Quality and Outcomes Framework. Br J Gen Pract 2007;57:441–8. [PMC free article] [PubMed] [Google Scholar]
- 3.Ministry of Health. Well child/tamariki ora national schedule. Wellington: Ministry of Health, 2013. [Google Scholar]
- 4.Dowell A, Turner N. New Zealand: the urgent need to focus on child health. NZ Fam Physician 2007;35:152–7. [Google Scholar]
- 5.Ministry of Health. New Zealand Health Survey: annual update of key findings 2012/13. Wellington, NZ: Ministry of Health, 2013. [Google Scholar]
- 6.Health Utilisation Research Alliance (HURA). Ethnicity data and primary care in New Zealand: lessons from the Health Utilisation Research Alliance (HURA) study. NZ Med J 2006;119:U1917. [PubMed] [Google Scholar]
- 7.Health Utilisation Research Alliance (HURA). Ethnicity, socioeconomic deprivation and consultation rates in New Zealand general practice. J Health Serv Res Policy 2006;11:141 10.1258/135581906777641721 [DOI] [PubMed] [Google Scholar]
- 8.Wang W, Krishnan E. Big data and clinicians: a review on the state of the science. JMIR Med Inform 2014;2:e1 10.2196/medinform.2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benin AL, Fenick A, Herrin J et al. . How good are the data? Feasible approach to validation of metrics of quality derived from an outpatient electronic health record. Am J Med Qual 2011;26:441–51. 10.1177/1062860611403136 [DOI] [PubMed] [Google Scholar]
- 10.Strauss JA, Chao CR, Kwan ML et al. . Identifying primary and recurrent cancers using a SAS-based natural language processing algorithm. J Am Med Inform Assoc 2013;20:349–55. 10.1136/amiajnl-2012-000928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordan K, Porcheret M, Croft P. Quality of morbidity coding in general practice computerized medical records: a systematic review. Fam Pract 2004;21:396–412. 10.1093/fampra/cmh409 [DOI] [PubMed] [Google Scholar]
- 12.Gray J, Orr D, Majeed A. Use of Read codes in diabetes management in a south London primary care group: implications for establishing disease registers. BMJ 2003;326:1130 10.1136/bmj.326.7399.1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peabody JW, Luck J, Jain S et al. . Assessing the accuracy of administrative data in health information systems. Med Care 2004;42:1066–72. 10.1097/00005650-200411000-00005 [DOI] [PubMed] [Google Scholar]
- 14.Gorelick MH, Knight S, Alessandrini EA et al. . Lack of agreement in pediatric emergency department discharge diagnoses from clinical and administrative data sources. Acad Emerg Med 2007;14:646–52. 10.1111/j.1553-2712.2007.tb01852.x [DOI] [PubMed] [Google Scholar]
- 15.Studney DR, Hakstian AR. A comparison of medical record with billing diagnostic information associated with ambulatory medical care. Am J Public Health 1981;71:145–9. 10.2105/AJPH.71.2.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Malley KJ, Cook KF, Price MD et al. . Measuring diagnoses: ICD code accuracy. Health Serv Res 2005;40(5pt 2):1620–39. 10.1111/j.1475-6773.2005.00444.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu ST, Sohn S, Ravikumar KE et al. . Automated chart review for asthma cohort identification using natural language processing: an exploratory study. Ann Allergy Asthma Immunol 2013;111:364–9. 10.1016/j.anai.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacRae J, Love T, McBain L et al. . Categorising onset of influenza-like illness demand in general practice using automated classification of free text daily consultation records. Health Informatics New Zealand 11th Annual Health Informatics Conference and Exhibition; Rotorua: HINZ, 2012. http://www.hinz.org.nz/uploads/file/2012conference/Papers/P18_MacRae.pdf. [Google Scholar]
- 19.Friedman C, Shagina L, Lussier Y et al. . Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004;11:392–402. 10.1197/jamia.M1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meystre S, Haug PJ. Automation of a problem list using natural language processing. BMC Med Inf Decis Mak 2005;5:30 10.1186/1472-6947-5-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popping R. Computer-assisted text analysis. Sage, 2000. [Google Scholar]
- 22.Stubbs M. Words and phrases: corpus studies of lexical semantics. Blackwell Publishers, 2001. [Google Scholar]
- 23.McColm D, Karcz A. Comparing manual and automated coding of physicians quality reporting initiative measures in an ambulatory EHR. J Med Pract Manag 2010;26:6–12. [PubMed] [Google Scholar]
- 24.Salmond C, Crampton P. NZDep2001 index of deprivation. Department of Public Health, Wellington School of Medicine and Health Science, 2002. [Google Scholar]
- 25.Salmond C, Crampton P, King P et al. . NZiDep: a New Zealand index of socioeconomic deprivation for individuals. Soc Sci Med 2006;62:1474–85. 10.1016/j.socscimed.2005.08.008 [DOI] [PubMed] [Google Scholar]
- 26.de Vet HCW, Knol DL, Mokkink LB et al. . Measurement in medicine a practical guide. Cambridge: Cambridge University Press, 2011. [Google Scholar]
- 27.Arous EJ, McDade TP, Smith JK et al. . Electronic medical record: research tool for pancreatic cancer? J Surg Res 2014;187:466–70. 10.1016/j.jss.2013.10.036 [DOI] [PubMed] [Google Scholar]
- 28.Chapman WW, Christensen LM, Wagner MM et al. . Classifying free-text triage chief complaints into syndromic categories with natural language processing. Artif Intell Med 2005;33:31–40. 10.1016/j.artmed.2004.04.001 [DOI] [PubMed] [Google Scholar]
- 29.Benin AL, Vitkauskas G, Thornquist E et al. . Validity of using an electronic medical record for assessing quality of care in an outpatient setting. Med Care 2005;43:691–8. 10.1097/01.mlr.0000167185.26058.8e [DOI] [PubMed] [Google Scholar]
- 30.Chan KS, Fowles JB, Weiner JP. Review: electronic health records and the reliability and validity of quality measures: a review of the literature. Med Care Res Rev 2010;67:503–27. 10.1177/1077558709359007 [DOI] [PubMed] [Google Scholar]
- 31.Pakhomov SV, Jacobsen SJ, Chute CG et al. . Agreement between patient-reported symptoms and their documentation in the medical record. Am J Manag Care 2008;14:530–9. [PMC free article] [PubMed] [Google Scholar]
- 32.Meystre SM, Savova GK, Kipper-Schuler KC et al. . Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008;3:128–44. [PubMed] [Google Scholar]