SUMMARY
Leukaemia inhibitory factor (LIF) has been shown to have an important role during muscle regeneration. The regenerative capacity of muscles after contusion injury in LIF-knockout mice is impaired compared with that of wild-type mice.
To clarify whether LIF modulates muscle regeneration by regulating myogenic precursor cell activity, we studied LIF expression and myogenic precursor cell activity in gastrocnemius muscles from Wistar rats at various times after contusion injury using immunohistochemistry and the direct effect of LIF on a rat myoblast cell line (L6).
After contusion injury, transient upregulation of the mRNA expression of LIF, LIF receptors and signal transducer and activator of transcription (STAT) 3, downstream of LIF and involved in enhanced cell proliferation, was observed. A marked increase in LIF protein in the cytosol of damaged myofibres was strongly correlated with a significant increase in the number of myogenic precursor cells (MyoD-positive cells) by 12 h after contusion. In addition, coexpression of LIF and MyoD protein in control and injured muscles after contusion injury from 3 h to 7 days was evident.
Treatment of L6 cells with LIF (1 ng/mL) in serum-free medium enhanced proliferation (bromodeoxyuridine incorporation) by 24 h. This was accompanied by increased expression of c-Myc protein within 12 h and was abolished by short interference RNA against c-Myc mRNA.
Together, the results of the present study suggest that LIF acts via paracrine and autocrine actions to regulate myogenic precursor cell activity during muscle regeneration after contusion injury and that the proliferative effect of LIF on L6 cells occurs via c-Myc signalling.
Keywords: c-Myc, L6 cells, leukaemia inhibitory factor, muscle contusion, MyoD, myogenic precursor cell, satellite cell
INTRODUCTION
Numerous growth factors have been shown to affect myogenic precursor cell activity during skeletal muscle regeneration. Many are positive modulators of myogenic precursor cell activity, such as hepatocyte growth factor, basic fibroblast growth factor, insulin-like growth factor-1, leukaemia inhibitory factor (LIF), platelet-derived growth factor and transforming growth factor-β.1 The response of myogenic precursor cells to LIF during the process of muscle regeneration is greater than the responses to the other growth factors.2
Leukaemia inhibitory factor is a poly functional cytokine of the interleukin (IL)-6 cytokine family. It shares the common gp130 receptor subunit together with IL-6, but has its own receptor, namely LIFR (a transmembrane signalling subunit). Leukaemia inhibitory factor mediates its signal via the heterodimerization of gp130 and LIFR.3 Leukaemia inhibitory factor mRNA expression is found at very low levels in intact muscles of mice4 and humans,5 but, in response to injury, expression is substantially upregulated at the site of muscle injury.5–7 Using in situ hybridization, Kami and Senba7 showed that expression of LIF mRNA in mononuclear cells within the damaged area of muscle was increased at 3 h and persisted for 7 days after contusion. Conversely, LIFR mRNA was upregulated in both mononuclear cells and the nuclei of muscle precursor cells in the injured area at 3 h to Day 2 after contusion.8 The relationship between LIF and myogenic precursor cell activity during skeletal muscle regeneration has been explored using LIF-knockout mice. Skeletal muscle regeneration in LIF-null mice was attenuated after crush injury, whereas exogenous administration of LIF increased the regenerative process and enlarged myofibres.9 The results suggest that LIF is important in modulating myogenic precursor cell activity after muscle injury. Studies in vivo in rats10 and in vitro in C2C12 cells (a mouse myoblast cell line)11 have shown that LIF induces the proliferation of myogenic precursor cells via the Janus tyrosine kinase (JAK) 2–signal transducer and activator of transcription (STAT) 3 pathway, whereas another study in C2C12 cells showed that early exposure to LIF inhibited differentiation of the proliferating myoblasts via an extracellular signal-regulated kinase (ERK) pathway.12 Very recently, Hunt et al.13 published results suggesting that LIF had an antiapoptotic effect, but not a proliferative effect, in C2C12 cells. Therefore, the in vitro effect of LIF on myoblast proliferation remains contentious.
Therefore, to better understand the action of LIF in the regulation of myogenic precursor cell activity during muscle repair after injury, we determined the cellular localization and expression pattern of LIF protein and myogenic precursor cell activity induced by contusion injury in the present study using double immunofluorescence staining specific to LIF and myogenic precursor cell markers, as well as examining the direct effects of LIF on a rat myoblast cell line (L6) in culture.
METHODS
Animals
Eight-week-old male Wistar rats, weighing approximately 270–290 g, were obtained from the National Laboratory Animal Centre of Thailand (Nakhon Pathom, Thailand). All animal experiments were performed in accordance with the animal use and care guidelines established by the Ethics Committee on the Use of Experimental Animals, Faculty of Science, Mahidol University. Rats were randomly allocated to either the control (n = 5) or contusion (n = 30) group. Rats in the contusion group were further subdivided into 3, 6 and 12 h and Day 1, 3 and 7 post-contusion groups (n = 5 in each).
Muscle contusion
An apparatus, based on the work of Kami et al.,10,14,15 was constructed to induce a reproducible physical trauma to the hind limb muscles of rats, as described previously.16 Briefly, rats were anaesthetized with thiopental (100 mg/kg, i.p.) and subjected to a single contusion of the hind limb muscle without skin incision. To achieve this, a 640 g cylinder was dropped by gravity from a stand at a height of 25 cm to the mid-belly of the gastrocnemius muscle, which was fixed in the posterolateral position.
Real-time polymerase chain reaction
Total RNA was isolated from 50 mg gastrocnemius muscle from control or injured rats using Trizol reagent. The first-strand cDNA was generated using 1 μg total RNA with the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA). Real-time polymerase chain reaction (PCR) was performed as described previously.16 Briefly, 1 μL cDNA template (50 ng), 6.25 μL PCR master mix, 1 μL of 5 μmol/L forward and reverse primers (Table 1) and 3.25 μL nuclease-free water were mixed, with the final volume being 12.5 μL. The reaction mixture was then subjected to 40 cycles of PCR using the following conditions: 95°C for 10 min, 95°C for 15 s and 60°C for 1 min. The relative quantification of LIFR, LIF and STAT3 mRNA expression was calculated using the 2−ΔΔCT method. In all these experiments, GAPDH was used as an endogenous control. Data were analysed using 7500 software version 2.03 (ABI, Foster City, CA, USA).
Table 1.
Primers used in the present study
| Gene | Accession no. | Primers (5′–3′) | Amplicon size (bp) |
|---|---|---|---|
| LIFR | NM_031048 | Forward: TCCATGCACCCCAAATCACGTCGAA Reverse: CGCCTGGTTCTCGGGGTCTGTCTCA |
133 |
| LIF | NM_022196 | Forward: TGCCCCTACTGCTCATTCTG Reverse: GGTTCCCCTTGAGCTGTGTA |
193 |
| STAT3 | NM_012747 | Forward: AGGTGTACCACCAAGGTCTCA Reverse: CCAATCGGAGGCTTAGTGAA |
166 |
| GAPDH | NM_017008 | Forward: ATCACTGCCACTCAGAAGACT Reverse: CATGCCAGTGAGCTTCCCGTT |
153 |
LIF, leukaemia inhibitory factor; LIFR, LIF receptor; STAT3, signal transducer and activator of transcription 3.
Muscle histology
Muscle histology was processed as described previously.16 Briefly, gastrocnemius muscles were fixed in 10% neutral buffered formalin for 24 h at room temperature (25°C) and embedded in paraffin wax. Serial cross-sections were made at 4 μm for Mayer’s haematoxylin and eosin staining. Images were taken at a magnification of × 100 under a light microscope (CX21; Olympus, Tokyo, Japan).
Indirect immunofluorescence staining
Gastrocnemius muscles were fixed in 10% neutral buffered formalin overnight at 4°C and transferred to fix in 30% sucrose solution for cryoprotection for 24 h (Fluka Biochemika, Buchs, Switzerland). Muscle samples were cut into 10 μm sections using a cryostat (CM 1800; Leica, Wetzlar, Germany) and immersed in boiled (100°C) citric acid (Sigma, St Louis, MO, USA), pH 6.0, for 6 min to retrieve antigens. Sections were allowed to cool to room temperature (25°C) and were then immersed in cold methanol (Merck, Darmstadt, Germany) at −20°C for 10 min. The sections were then washed with phosphate-buffered saline (PBS) and incubated with 4% bovine serum albumin (BSA; Sigma) for 1 h at room temperature in a humidity chamber. Rabbit anti-rat polyclonal MyoD antibody (1 : 50; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was applied to each section and incubated in a humidity chamber at room temperature for 1 h and then at 4°C overnight. The sections were washed three times with PBS, after which fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit secondary antibody (1 : 300; Santa Cruz Biotechnology) was added and sections were incubated in the dark in a humidity chamber at room temperature for 2 h. Sections were then washed three times with PBS + 0.01% Tween 20 (Sigma). For the second staining, goat anti-rat polyclonal LIF antibody (1 : 50; Santa Cruz Biotechnology) was applied to each section and sections were incubated in a dark humidity chamber at RT for 2 h. The sections were washed three times with PBS, then Cy5-conjugated rabbit anti-goat secondary antibody (1 : 125; Zymax, Invitrogen, Carlsbad, CA, USA) was added, incubated in the dark in a humidity chamber at room temperature for 2 h before being washed three times with PBS + 0.01% Tween 20. Double immunostaining for LIF and MyoD was viewed under an Olympus confocal microscope (FV-1000) at × 400 magnification.
Cell proliferation assay
Rat myoblast cells (L6) from American Type Culture Collection (Manassas, VA, USA) were grown in growth medium consisting of Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS) and antibiotics (final concentration: 100 U/mL penicillin and 100 μg/mL streptomycin). When cells reached 60–70% confluence, they were trypsinized and seeded into a six-well plate at a density of 8 × 104 cells/well before being cultured in serum-free medium according to the methods of Florini and Roberts.17 Briefly, cells were cultured in DMEM supplemented with 5% FBS and antibiotics for 24 h, then washed twice with PBS. Serum-containing DMEM was then replaced with serum-free DMEM (without antibiotics) and the cells were cultured in serum-free medium for 24 h. Thereafter, the cells were cultured in serum-free medium (DMEM), serum-free medium (DMEM) + LIF at different concentrations (0.1, 1, 10 and 100 ng/mL) and growth medium (DMEM + 10% FBS) for 24 and 48 h. Changes in cell number were determined in triplicate experiments by counting in a haemocytometer. Serum-free medium (DMEM) and growth medium (DMEM + 10% FBS) were used as negative and positive controls, respectively. Data are presented as a percentage of the negative controls (DMEM).
To evaluate whether the effects of LIF on changes of L6 cell number are the result of cell proliferation, the bromodeoxyuridine (BrdU) incorporation assay was performed to determine the DNA replication state of the cells, which reflects cell division. Briefly, BrdU (Sigma), at a final concentration of 10 μmol/L, was added to the culture media and cells were incubated in the presence of BrdU for 1 h at 37°C and 5% CO2 (pulse labelling). Full details of the methodology used for BrdU labelling are given below.
Immunocytochemistry
The L6 cells were cultured in the presence of serum-free medium as described above with the exception that they were seeded in an eight-well chamber slide (Laboratory-Tek; Fisher Scientific, Loughborough, UK) at a density of 4 × 103 cells/well. For c-Myc and proliferating cell nuclear antigen (PCNA) immunostaining, cells were cultured in either DMEM, DMEM + LIF (1 ng/mL) or DMEM + 10% FBS for 12 h. Cells were then fixed in 4% para-formaldehyde at room temperature for 1 min, followed by −20°C methanol for 1 min and 2% BSA at room temperature for 30 min. For single c-Myc immunostaining, cells were incubated with rabbit anti-rat c-Myc antibody (1 : 100; Santa Cruz Biotechnology) at room temperature for 1 h. Cells were then washed twice with PBS. Thereafter, goat anti-rabbit Alexa Fluor 488 (1 : 500; Invitrogen, Molecular Probes, Carlsbad, CA, USA) was added to the cells and cells were incubated at room temperature for 1 h. For double immunostaining of c-Myc and PCNA, primary rabbit anti-rat c-Myc antibody (1 : 100; Santa Cruz Biotechnology) together with mouse anti-rat PCNA antibody (1 : 50; Santa Cruz Biotechnology) were used along with secondary antibodies (1 : 500), namely goat anti-rabbit Alexa Fluor 488 (Invitrogen, Molecular Probes) and goat anti-mouse Texas Red (Rockland Immunochemicals, Gilbertsville, PA, USA). To visualize nuclei, cells were counterstained with antifade solution containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA, USA). Images were captured using a Nikon fluorescence microscope (Eclipse E600, 11.2 Color Mosaic Model; Nikon, Tokyo, Japan) at × 200 magnification.
For BrdU labelling, cells were cultured either in DMEM, DMEM + LIF (1 ng/mL) or DMEM + 10% FBS for 24 and 48 h. After cells had been pulse labelled with BrdU for 1 h (incubation of BrdU was started at 23 and 47 h after treatment conditions were applied), cells were fixed with 4% paraformal-dehyde at room temperature for 10 min. Thereafter, DNA was denatured with 2 mol/L HCl (Fisher Scientific) in PBS containing 0.5% Triton X-100 at 37°C for 30 min and neutralized with 0.1 mol/L borate buffer (Sigma), pH 8.5, at room temperature for 10 min. Cells were washed with PBS and incubated with primary BrdU antibody, FITC conjugated (1 : 100; Abcam, Cambridge, MA, USA) at room temperature for 1 h. After three washes with PBS, cells were post-fixed with 4% paraformaldehyde at room temperature for 5 min and then washed twice with PBS. Cells were counterstained with antifade solution (Vector Laboratories). Images were captured using a Zeiss (Jena, Germany) laser scanning microscope (LSM 5 Live) at × 630 magnification.
Western blot analysis
Protein from L6 cells was extracted with lysis buffer containing 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS), 25 mmol/L β-glycerophosphate, 50 mmol/L NaCl, 50 mmol/L NaF, 5 mmol/L benzamidine, 20 mmol/L Tris–HCl (pH 7.6), 1 mmol/L EDTA, 1 mmol/L sodium orthovanadate, 5 mmol/L N-ethylmaleimide, 1 mmol/L phenylmethylsulphonyl fluoride and 1 × protease inhibitor. Cells were washed twice with PBS, placed on ice, 200 μL lysis buffer was added to the cells and they were incubated on ice with gentle shaking for 15 min. The lysate was then centrifuged at 10 621 g for 10 min at 4°C. The supernatant was transferred to a precooled Eppendorf tube and kept at −20°C for protein assay. Protein concentrations in the extract were measured using the Lowry protein assay at a wavelength of 750 nm. This assay has been shown to yield more valid and reliable results for protein concentrations than the Bradford assay.18
For immunoblotting, 25 μg protein was loaded in a 4% stacking and 10% separating gel. A protein ladder (Magicalmark; Invitrogen) was used to determine molecular weight. Protein was transferred from the gel to polyvinylidene difluoride membranes in transfer buffer at 100 V for 1 h, then blocked with 5% fat-free milk in Tris-buffered saline with 0.1% Tween 20 (TBS-T) for 1 h on a shaker with gentle shaking at room temperature. Primary rabbit anti-rat c-Myc antibody (1 : 500; Santa Cruz Biotechnology) was applied in TBS-T + 5% milk overnight at 4°C. After washing with TBS-T, goat anti-rabbit peroxidase labelled secondary antibody (1 : 10 000; Vector Laboratories) was applied in 10 mL of 5% milk in TBS-T and membranes were incubated at room temperature for 1 h. After washing in TBS-T, membranes were incubated with ECL Plus Western Blotting Detection Reagents (GE Health-care Biosciences, Pittsburgh, PA, USA) for 5 min at room temperature. Protein bands were detected with a Kodak Film Processor X-OMAT 2000A (Eastman Kodak, Rochester, NY, USA). Membranes were then stripped using stripping buffer consisting of 62.5 mmol/L Tris, pH 6.8, 2% SDS, 100 mmol/L β-mercaptoethanol at 50°C for 30 min and then washed with TBS-T for detecting α-tubulin using mouse anti-rat α-tubulin antibody (1 : 2000; Sigma).
Short interference RNA transfection
In these experiments, L6 cells were seeded in DMEM + 5% FBS (no antibiotics) at density of 8 × 104 cells/well on a six-well plate. Lipofectamine 2000 (Invitrogen) was diluted with OPTI-MEM I (Invitrogen) and the mixture incubated at room temperature for 5 min. Thereafter, either scrambled short interference (si) RNA (medium GC content; Invitrogen) or c-Myc siRNA Pre-designed (Invitrogen, Ambion, Austin, TX, USA) was added to the diluted Lipofectamine, mixed gently and incubated at room temperature for 20 min. Then, the Lipofectamine–siRNA (scrambled/c-Myc) complexes were added to each well immediately after L6 cells had been plated to enhance the transfection efficiency of this cell line. The final concentration of scrambled and c-Myc siRNA was 20 nmol/L. The L6 cells were incubated with Lipofec-tamine–siRNA complexes for 5 h, followed by incubation with DMEM + 5% FBS (no antibiotics) for 19 h. Thereafter, DMEM + 5% FBS was replaced with serum-free medium (DMEM) for 24 h. Finally, the serum-free medium was replaced with DMEM + LIF 1 ng/mL (the effective dose for L6 cell proliferation) to determine the direct effect of LIF on the regulation of c-Myc protein expression. Changes in c-Myc protein expression 12 h after LIF treatment were investigated using western blot analysis, as described above. Band densities of western blot results were measured using IMAGEJ software version 1.42q (National Institutes of Health, Bethesda, MD, USA; available from http://rsb.info.nih.gov/ij/, accessed 11 June 2009).
Statistical analysis
All data are presented as the mean ± SEM. Differences between groups were analysed using Kruskal–Wallis and Dunn’s tests, one-way ANOVA and Tukey’s post hoc test or independent Student’s t-test as appropriate. P < 0.05 was considered significant.
RESULTS
Rapid upregulation of LIFR, LIF and STAT3 mRNA expression in response to contusion
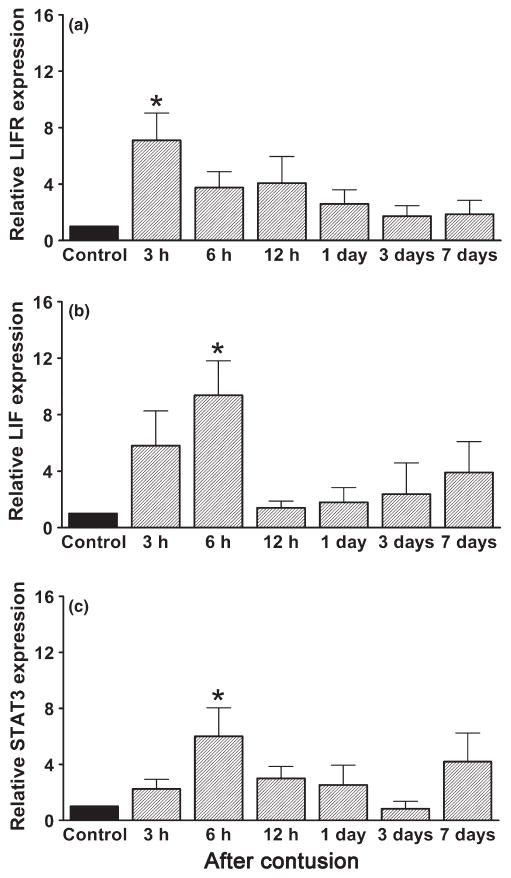
Investigation of the expression pattern of LIFR, LIF and STAT3 mRNA over the time-course of muscle regeneration after contusion revealed that within 3 h of injury, LIFR mRNA levels were significantly upregulated by approximately sevenfold compared with control (P < 0.05; Fig. 1a), followed by upregulation of LIF mRNA by approximately ninefold (P < 0.05; Fig. 1b) 6 h after injury. In addition, STAT3 mRNA (a downstream signalling molecule in the LIF pathway) was increased significantly by approximately sixfold at 6 h (P < 0.05; Fig. 1c).
Fig. 1.
Relative mRNA expression of the (a) leukaemia inhibitory factor receptor (LIFR), (b) leukaemia inhibitory factor (LIF) and (c) signal transducer and activator of transcription (STAT) 3 in control and injured rat gastrocnemius muscles at different times after contusion. Expression was normalized against that of GAPDH and is expressed in the fold change relative to the mean control value (no SEM). Data are presented as the mean ± SEM (n = 5). * P < 0.05 compared with control (Kruskal–Wallis and Dunn’s (non-parametric) tests).
Correlation between upregulation of LIF protein in damaged myofibres and increased myogenic precursor cell number
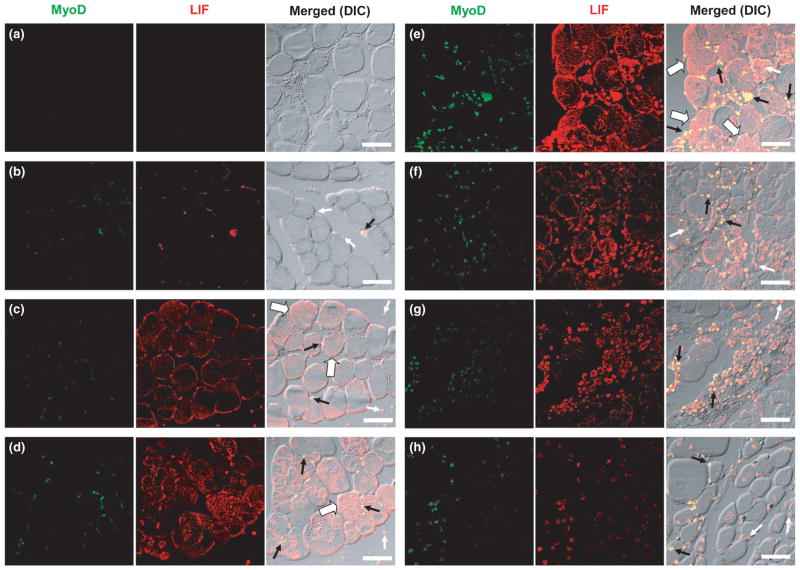
To demonstrate that LIF could act as a mitogenic factor on myogenic precursor cell activity after contusion injury, we used double immunostaining of MyoD (a marker of myogenic precursor cells, including myoblasts derived from satellite cells and committed myogenic cells) and LIF to determine the localization and expression pattern of myogenic precursor cell activity and LIF protein expression. Based on the results of the mRNA expression study, we expected to observe concurrent increases in LIF protein expression and myogenic precursor cell number after contusion injury. When primary antibodies were omitted during the staining process (negative control), no signals for either MyoD or LIF were observed (Fig. 2a). However, in the control gastrocnemius muscle, signals for MyoD and LIF were seen in the perimeter of myofibres, but not in the cytoplasm (Fig. 2b). At 3 h after contusion injury, immunostaining for LIF was clearly seen in the mononuclear cells and cytoplasm of myofibres (Fig. 2c). However, a marked increase in LIF staining on damaged myofibres and MyoD-positive cells was observed 6 h after contusion (Fig. 2d). At 12 h after contusion, pronounced LIF protein expression in damaged myofibres and the number of MyoD-positive cells in the damaged area were apparently maximal (Fig. 2e). From Day 1 to Day 7, LIF staining in intact myofibres decreased towards control levels (Fig. 2f–h). Interestingly, colocalization of MyoD and LIF signals was also noted in control and injured muscles from 3 h to Day 7 after contusion injury, suggesting the expression of LIF protein in the myogenic precursor cells prior to and after injury (Fig. 2b–h). Corresponding images of muscle histology from haematoxylin and eosin staining compared with the double-immuno-staining images are shown in Fig. S1, available online as supplemenatry material for this paper.
Fig. 2.
Localization and expression pattern of MyoD (green) and leukaemia inhibitory factor (LIF; red) proteins in control and injured rat gastrocnemius muscles at different times after contusion. Colocalized MyoD and LIF are stained yellow in the merged images. (a) No primary MyoD and LIF antibodies; (b) control muscle; (c) 3 h, (d) 6 h, (e) 12 h, (f) 1 day, (g) 3 days and (h) 7 days after contusion. Small white arrows indicate LIF-positive cells. Small black arrows indicate cells coexpressing MyoD and LIF. Large white arrows (c–e) indicate myofibres that express LIF in their cytoplasm. Immunostaining of MyoD and LIF proteins was performed on four different animals. Bars, 50 μm. DIC, differential interference contrast.
Effects of LIF on L6 cell proliferation in serum-free medium
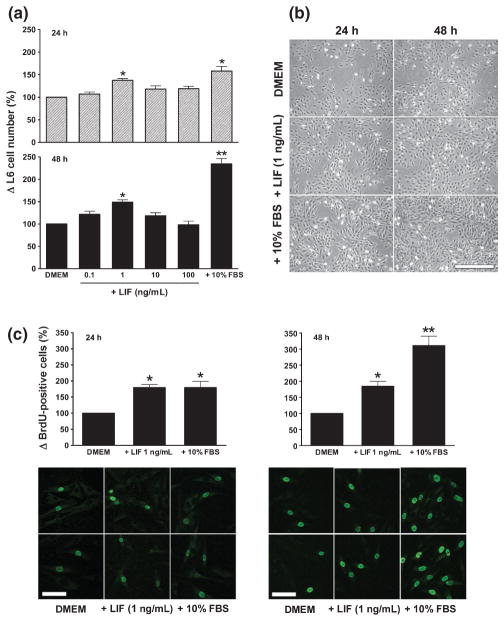
Although the in vivo study clearly showed that LIF was strongly expressed in myofibres and MyoD-positive cells soon after contusion injury, the causal relationship between LIF and the proliferation of myogenic precursor cells cannot be made. We therefore chose to study the direct effect of LIF in vitro using L6 cells, a rat myoblast cell line. Preliminary experiments showed that, in conventional growth medium (i.e. DMEM + 10% FBS), L6 cells proliferated rapidly such that any effect of LIF was masked. Therefore, subsequent experiments were performed under serum-free conditions (DMEM), using DMEM + 10% FBS and DMEM media as positive and negative controls, respectively. As shown in Fig. 3a, by 24 h after culture in serum-free medium, the addition of 1 ng/mL LIF significantly increased cell number (P < 0.05; Fig. 3a) and enhanced the proliferation of L6 cells, as determined by increased BrdU incorporation (P < 0.05; Fig. 3c), compared with serum-free medium alone. At this time, the effect of LIF (1 ng/mL) was comparable to that of 10% FBS. Of note, at higher concentrations of LIF (from 10 to 100 ng/mL), L6 cell numbers were not increased significantly (Fig. 3a). Prolonged culture in the medium containing LIF (1 ng/mL) for 48 h did not further increase L6 cell number (Fig. 3a) or enhance cell proliferation (Fig. 3c) compared with its effect at 24 h. In contrast, the number and proliferation of L6 cells in medium containing 10% FBS were almost double at 48 h (Fig. 3a,c). Representative images of changes in L6 cell number and BrdU incorporation after treatment with DMEM, DMEM + LIF 1 ng/mL and DMEM + 10% FBS are shown in Fig. 3b,c.
Fig. 3.
Changes in (a, b) the number of L6 cells and (c) bromodeoxyuridine (BrdU) incorporation in response to leukaemia inhibitory factor (LIF). (a) The number of L6 cells 24 and 48 h after LIF treatment with different concentrations, as indicated, was compared with that in serum-free medium (Dulbecco’s modified Eagle’s medium; DMEM) and growth medium (DMEM + 10% fetal bovine serum; FBS). Graphs were plotted from data obtained from three separate experiments and show the mean ± SEM. (b) Representative images of cells cultured in DMEM, DMEM + LIF 1 ng/mL and DMEM + 10% FBS at 24 and 48 h. Bars, 200 μm. (c) The number of BrdU-positive cells at 24 and 48 h after treatment with DMEM + LIF 1 ng/mL was compared with that in cells cultured in serum-free medium (DMEM) and growth medium (DMEM + 10% FBS). The number of BrdU-positive cells was determined in 10 random images for each treatment group in each experiment. Graphs were plotted from data obtained from three separate experiments and show the mean ± SEM of values expressed as a percentage of values in the control (DMEM) group. *P < 0.05, **P < 0.01 compared with DMEM (one-way ANOVA and Tukey’s post hoc test). Two representative images of BrdU-positive cells from each treatment group at 24 and 48 h are shown beneath the graphs. Bars, 50 μm.
Effects of LIF on nuclear c-Myc expression
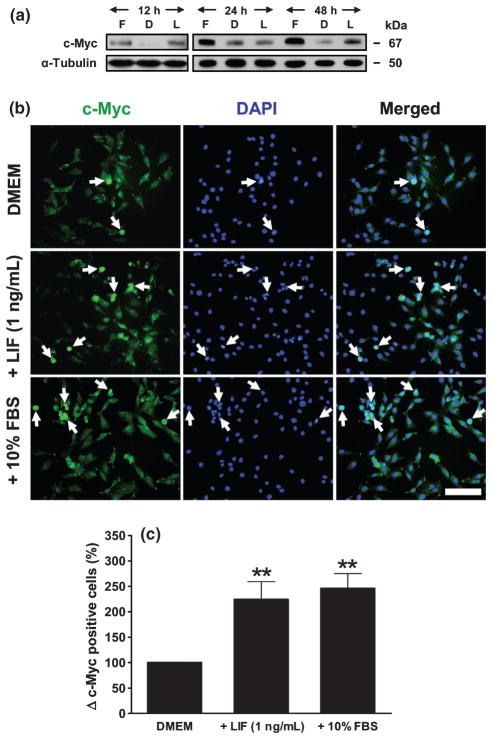
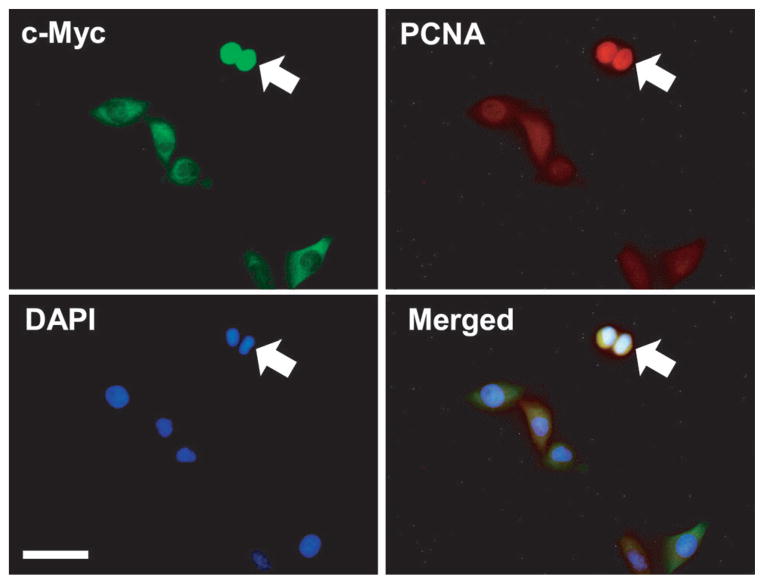
Because our in vivo results (Fig. 1) as well as previous studies10,11 have implicated STAT3 signalling in LIF-enhanced muscle regeneration and myoblast proliferation, we further explored whether LIF treatment upregulated expression of c-Myc protein, a downstream mediator of STAT3 signalling in the regulation of cell cycle progression (G1 to S phase). Indeed, both LIF and growth medium upregulated c-Myc expression, as demonstrated by western blotting (Fig. 4a). Immunofluorescence staining (Fig. 4b) extended the data obtained using western blot analysis that nuclear, but not cytoplasmic, c-Myc protein was upregulated by LIF. Quantitative analysis of nuclear c-Myc expression revealed that the number of nuclear c-Myc-positive cells (cells showing costaining with c-Myc and DAPI) after LIF treatment was increased significantly (P < 0.01) and comparable to that in growth medium condition at 12 h (Fig. 4c). To confirm that LIF had a mitotic activity via upregulation of nuclear c-Myc protein, triple immunostaining with c-Myc, PCNA and DAPI was performed. Figure 5 shows that LIF treatment is associated with increased nuclear expression of c-Myc in the dividing cells (PCNA-positive cells).
Fig. 4.
Effects of leukaemia inhibitory factor (LIF) on the proliferation of L6 cells in serum-free medium are mediated by increased c-Myc protein expression. (a) Representative western blot from three separate experiments showing c-Myc protein expression in L6 cells after different times of culture (12, 24 and 48 h) Dulbecco’s modified Eagle’s medium (DMEM; D), DMEM + LIF 1 ng/mL (L) and DMEM + 10% fetal bovine seum (FBS; F). (b) Immunofluorescence staining showing c-Myc protein expression in serum-starved L6 cells after 12 h culture in the presence of DMEM, DMEM + LIF 1 ng/mL and DMEM + 10% FBS. Immunostaining of c-Myc protein was performed in three separate experiments. Images were merged with 4′,6′-diamidino-2-phenylindole (DAPI) to show nuclear localization. Arrows indicate the nuclear localization of c-Myc protein. Bar, 100 μm. (c) Quantitative analysis of nuclear c-Myc-positive cells (cells costained with c-Myc and DAPI) after 12 h LIF treatment, determined in 10 random images for each treatment for each experiment. Graphs were plotted from data obtained from three separate experiments and show the mean ± SEM of values expressed as a percentage of values in the control (DMEM) group. **P < 0.01 compared with DMEM (one-way ANOVA and Tukey’s post hoc test).
Fig. 5.
Coexpression of nuclear c-Myc and proliferating cell nuclear antigen (PCNA) in proliferating L6 cells. Images show triple immunostaining for c-Myc, PCNA and 4′,6′-diamidino-2-phenylindole (DAPI) in serum-starved L6 cells 12 h after leukaemia inhibitory factor (LIF) treatment. Immunostaining of c-Myc and PCNA proteins was performed in three separate experiments. Representative images were merged with DAPI to show the nuclear localization of the c-Myc and PCNA proteins. Arrows indicate dividing cells. Bar, 50 μm.
c-Myc acts downstream of LIF to enhance L6 cell proliferation
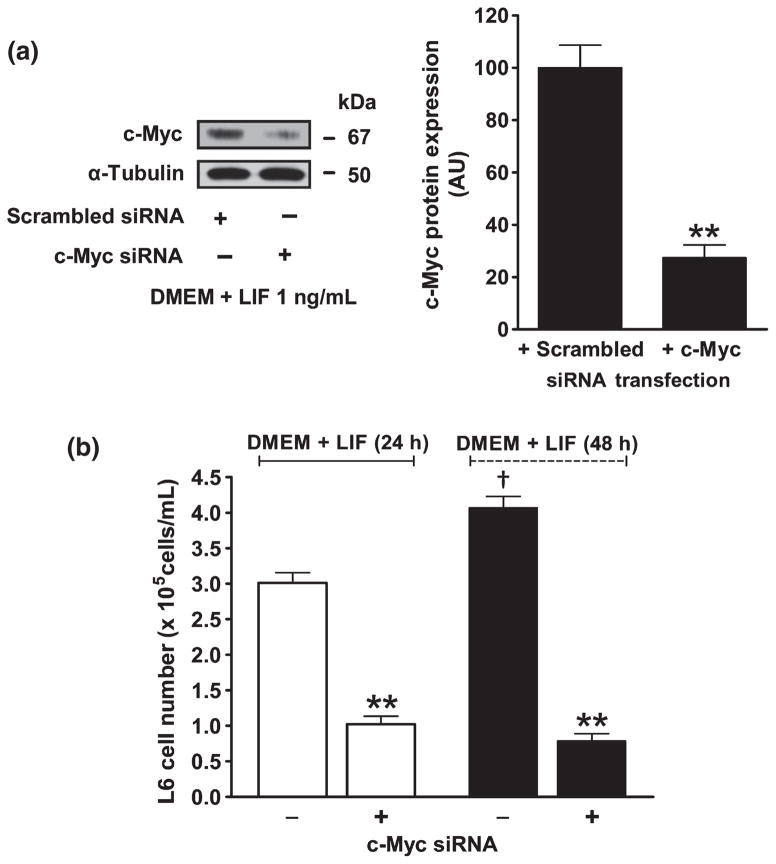
To confirm whether c-Myc was a mediator of the effect of LIF on myoblast proliferation, c-Myc siRNA was added to the culture medium before LIF treatment to block c-Myc mRNA transcription. After c-Myc siRNA was pretransfected, western blot analysis revealed that c-Myc protein expression after LIF treatment was suppressed by approximately 75% (27.4 ± 4.8 AU) compared with scrambled siRNA transfection (100.0 ± 8.5 AU; Fig. 6a). In addition, when the expression of c-Myc protein was blocked by siRNA, LIF failed to induce cell proliferation at 24 and 48 h after treatment (Fig. 6b).
Fig. 6.
Effects of leukaemia inhibitory factor (LIF) on the proliferation of L6 cells are abolished by c-Myc knockdown. (a) Western blot of c-Myc protein expression 12 h after LIF treatment after transfection with either scrambled or c-Myc short interference (si) RNA and graphs showing band density normalized against α-tubulin. Quantitative analysis (arbitrary units) was determined using ImageJ software version 1.42q (National Institutes of Health, Bethesda, MD, USA; available from http://rsb.info.nih.gov/ij/, accessed 11 June 2009). Data are the mean ± SEM of three separate experiments. **P < 0.01 compared with scrambled siRNA. (b) Changes in L6 cell numbers after c-Myc siRNA transfection. Data were collected at 24 and 48 h after serum-free medium (DMEM) was replaced with DMEM + LIF 1 ng/mL. Data are the mean ± SEM of three separate experiments. †P < 0.01 compared with DMEM + LIF (24 h); **P < 0.01 compared with no c-Myc siRNA transfection (independent Student’s t-test).
DISCUSSION
The key findings of the present study are as follows: (i) expression of LIFR, LIF and STAT3 mRNA is rapidly and significantly upregulated during the early phase of muscle regeneration, as demonstrated using real-time PCR; (ii) LIF protein is expressed in damaged myofibres and the pattern of expression after contusion injury is strongly correlated with increases in myogenic precursor cell number, as evidenced by double immunostaining; (iii) in addition to its expression in damaged myofibres, LIF protein was expressed in myogenic precursor cells and mononuclear cells after contusion injury; and (iv) the direct effect of LIF on the proliferation of the rat myoblast cell line (L6) in vitro was mediated via c-Myc signalling.
Several lines of evidence suggest a role for LIF in muscle regeneration after injury. Thus, LIF mRNA is expressed at low levels in adult muscle, but is upregulated by inflammation,4 crush injury7,19 or mechanical overloading.20,21 Impaired muscle regeneration after crush injury in LIF-null mice is prevented by administration of LIF.2 In addition, LIF has a direct proliferative effect on isolated satellite cells from rats11 and C2C12 myoblasts.11 Because LIF has a short half-life in the circulation22 and high renal clearance,23 it is believed to function as a paracrine factor.4
The most likely source of LIF is the injured/inflamed muscle. Indeed, results from the present and other studies support this notion. Data from real-time PCR in the present study showed that after contusion injury, the expression of LIF and its receptors was upregulated, which corroborates well with results from in situ hybridization studies.7,8,19 In addition, there was a strong positive correlation between LIF protein expression in damaged myofibres and myogenic precursor cell activity in the present study, as assessed by the concurrent increase in the intensity of LIF staining in the cytoplasm of damaged myofibres and the increase in the number of MyoD-positive nuclei. On the basis of these findings, myogenic precursor cell proliferation is possibly regulated via the paracrine action of LIF by its interaction with its receptor through a heterodimer receptor (gp130 and LIFR-β) on the myogenic precursor cell membrane, thereby activating the translocation of STAT3 from the cytoplasm to the nucleus to initiate myogenic precursor cell proliferation.10
A previous study showed that LIF was produced by cultured primary myoblasts from wild-type and mdx mice.6 However, there is no information available regarding the expression of LIF mRNA and protein in myogenic precursor cells. Although the work of Kami et al.7,8 revealed expression of LIF and LIFR mRNA in mononucleated cells after contusion, these did not identify the nature of the cells. In the present study, colocalization of LIF and MyoD protein expression was observed in both control and injured muscles at 3 h to Day 7 after contusion, suggesting the production of LIF in the myogenic precursor cells prior to and after injury. This finding leads us to propose that LIF may also act as an autocrine factor in regulating myogenic precursor cell activity after contusion injury.
In addition to damaged myofibres and myogenic precursor cells, mononucleated cells in the extracellular matrix (ECM) also expressed LIF protein. The cell type that expressed LIF in the ECM is possibly resident and/or phagocytic macrophages. Macrophages may serve as a source of LIF in the activation of myogenic precursor cell proliferation after they infiltrate the damaged myofibres.24 Evidence supporting this proposal came from an in vitro study that revealed that the number of myoblasts was increased when macrophages were included in the culture medium.25
To provide more mechanistic support for our in vivo results, we studied the effect of LIF on the proliferation of rat myoblast L6 cells in vitro. This cell line, established from the newborn rat thigh muscle, has the characteristics of myogenic precursor cells26 and has been used as a cell model for proliferation and differentiation by several investigators.27–29 We found that LIF increased the proliferation of L6 cells and the effects of LIF appeared to be maximal at a concentration of 1 ng/mL. These results are consistent with those of a previous study in C2C12 mouse myoblasts, in which LIF failed to further increase cell number at concentrations higher than 1 or 10 ng/mL.11,13 This phenomenon could be explained in terms of the binding kinetics between LIF and its membrane receptor (LIFR), which is saturated at a ligand concentration of 1 ng/mL. At higher concentrations, downregulation of LIFR probably occurs.30 The increase in L6 cell numbers is most likely due to enhanced cell proliferation rather than an increase in cell survival because the changes in cell number at 24 and 48 h were paralleled by increases in BrdU incorporation. This effect is concordant with previous work on LIF in cultured mouse and human myoblasts,31,32 as well as in primary rat satellite cells.11 In their study, Spangenberg and Booth11 delineated the signalling mechanism for LIF on cell proliferation through the JAK2–STAT3 pathway. Because the downstream signalling of STAT3 has not yet been elaborated, we pursued the potential of LIF activation of L6 cell proliferation occurring via the upregulating of nuclear c-Myc protein. We found that nuclear c-Myc was upregulated by LIF and that it was coexpressed with PCNA (a specific marker for dividing cells) and that the expression of c-Myc and cell proliferation were virtually abolished after c-Myc was knocked down by siRNA, supporting a critical link between LIF treatment, c-Myc and cell proliferation. Conversely, more recent studies have reported that LIF produces a dose-dependent inhibition of C2C12 myoblast cell differentiation. This effect of LIF occurred as a result of ERK activation.12 Furthermore, Hunt et al.13 have shown that LIF has an anti-apoptotic effect and that it increases C2C12 cell numbers by reducing staurosporine-induced apoptotic DNA fragmentation and decreasing the proteolytic activation of caspase 3. Thus, in addition to it effects enhancing myoblast proliferation via upregulation of c-Myc, LIF may have different modes of action on myoblasts because both inhibition of myoblast differentiation and prevention of apoptosis would also result in increased cell numbers.
In conclusion, we have shown in an in vivo study that, in addition to damaged myofibres, LIF is expressed in myogenic precursor cells after contusion injury. This finding suggests that LIF possibly acts in a paracrine and autocrine manner to regulate myogenic precursor cell activity. In addition, an in vitro study using L6 cells, we demonstrated a direct effect of LIF on cell proliferation by activation of nuclear c-Myc expression. These results may lead to alternative therapeutic strategies using LIF to enhance the process of skeletal muscle regeneration after trauma from sports-related injuries or accidents.
Supplementary Material
Acknowledgments
This work was supported, in part, by the Office of the Higher Education Commission, Thailand, under the program Strategic Scholarships for Frontier Research Network for the Joint PhD Program Thai Doctoral degree (to RS). Support was also provided by National Institutes of Health grant AR43349 (to KAE). The authors thank Professor Duncan Richard Smith (Institute of Molecular Biosciences, Mahidol University) for kindly providing secondary antibodies for LIF and MyoD double immunostaining, Miss Yuvadee Siriyasub (Center of Nanoimaging, Faculty of Science, Mahidol University) for her helpful assistance with the confocal microscopy and to Associate Professor Ok-Kyong Park Sarge (Department of Physiology, College of Medicine, University of Kentucky) and Professor Kim E Barrett (Department of Medicine, School of Medicine, University of California, San Diego, CA, USA) for critical comments on the manuscript.
Footnotes
Additional Supporting Information may be found in the online version of this article:
Figure S1. Effects of contusion injury on muscle degeneration and regeneration. (a–g) Haematoxylin and eosin staining of 4 μm sections of control (a) and injured gastrocnemius muscle at different times after contusion: 3 h (b), 6 h (c), 12 h (d), 1 day (e), 3 days (f) and 7 days (g).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Hawke TJ, Garry DJ. Myogenic satellite cells: Physiology to molecular biology. J Appl Physiol. 2001;91:534–51. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- 2.Kurek JB, Bower JJ, White JD, Muldoon CM, Austin L. Leukemia inhibitory factor and other cytokines as factors influencing regeneration of skeletal muscle. Basic Appl Myol. 1998;8:347–60. [Google Scholar]
- 3.Gearing DP, Thut CJ, VandeBos T, et al. Leukemia inhibitory factor receptor is structurally related to the IL-6 signal transducer, gp130. EMBO J. 1991;10:2839–48. doi: 10.1002/j.1460-2075.1991.tb07833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown MA, Metcalf D, Gough NM. Leukemia inhibitory factor and interleukin 6 are expressed at very low levels in the normal adult mouse and are induced by inflammation. Cytokine. 1994;6:300–9. doi: 10.1016/1043-4666(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 5.Reardon KA, Kapsa RM, Davis J, et al. Increased levels of leukemia inhibitory factor mRNA in muscular dystrophy and human muscle trauma. Muscle Nerve. 2000;23:962–6. doi: 10.1002/(sici)1097-4598(200006)23:6<962::aid-mus18>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 6.Kurek JB, Nouri S, Kannourakis G, Murphy M, Austin L. Leukemia inhibitory factor and interleukin-6 are produced by diseased and regenerating skeletal muscle. Muscle Nerve. 1996;19:1291–301. doi: 10.1002/(SICI)1097-4598(199610)19:10<1291::AID-MUS6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 7.Kami K, Senba E. Localization of leukemia inhibitory factor and inter-leukin-6 messenger ribonucleic acids in regenerating rat skeletal muscle. Muscle Nerve. 1998;21:819–22. doi: 10.1002/(sici)1097-4598(199806)21:6<819::aid-mus20>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 8.Kami K, Morikawa Y, Sekimoto M, Senba E. Gene expression of receptors for IL-6, LIF, and CNTF in regenerating skeletal muscles. J Histochem Cytochem. 2000;48:1203–13. doi: 10.1177/002215540004800904. [DOI] [PubMed] [Google Scholar]
- 9.Kurek JB, Bower JJ, Ramanella M, Koentgen F, Murphy M, Austin L. The role of leukemia inhibitory factor in skeletal muscle regeneration. Muscle Nerve. 1997;20:815–22. doi: 10.1002/(sici)1097-4598(199707)20:7<815::aid-mus5>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 10.Kami K, Senba E. In vivo activation of STAT3 signaling in satellite cells and myofibers in regenerating rat skeletal muscles. J Histochem Cytochem. 2002;50:1579–89. doi: 10.1177/002215540205001202. [DOI] [PubMed] [Google Scholar]
- 11.Spangenberg EE, Booth FW. Multiple signaling pathways mediate LIF-induced skeletal muscle satellite cell proliferation. Am J Physiol Cell Physiol. 2002;283:C204–11. doi: 10.1152/ajpcell.00574.2001. [DOI] [PubMed] [Google Scholar]
- 12.Jo C, Kim H, Jo I, et al. Leukemia inhibitory factor blocks early differentiation of skeletal muscle cells by activating ERK. Biochim Biophys Acta. 2005;1743:187–97. doi: 10.1016/j.bbamcr.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Hunt LC, Tudor EM, White JD. Leukemia inhibitory factor-dependent increase in myoblast cell number is associated with phosphotidylinositol 3-kinase-mediated inhibition of apoptosis and not mitosis. Exp Cell Res. 2010;316:1002–9. doi: 10.1016/j.yexcr.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 14.Kami K, Masuhara M, Kashiba H, Kawai Y, Nokuchi K, Senba E. Changes of vinculin and extracellular matrix components following blunt trauma to rat skeletal muscle. Med Sci Sports Exerc. 1993;25:832–40. doi: 10.1249/00005768-199307000-00012. [DOI] [PubMed] [Google Scholar]
- 15.Kami K, Nokuchi K, Senba E. Localization of myogenin, c-fos, c-jun, and muscle specific gene mRNAs in regenerating rat skeletal muscle. Cell Tissue Res. 1995;280:11–19. doi: 10.1007/BF00304506. [DOI] [PubMed] [Google Scholar]
- 16.Srikuea R, Phopramool C, Kitiyanant Y, Yimlamai T. Satellite cell activity in muscle regeneration after contusion in rats. Clin Exp Pharmacol Physiol. 2010;37:1078–86. doi: 10.1111/j.1440-1681.2010.05439.x. [DOI] [PubMed] [Google Scholar]
- 17.Florini JR, Roberts SB. A serum-free medium for the growth of muscle cells in culture. In Vitro Cell Dev Biol Plant. 1979;15:983–92. doi: 10.1007/BF02619157. [DOI] [PubMed] [Google Scholar]
- 18.Lu TS, Yiao SY, Lim K, Jensen RV, Hsiao LL. Interpretation of biological and mechanical variations between the Lowry versus Bradford method for protein quantification. North Am J Med Sci. 2010;2:325–8. doi: 10.4297/najms.2010.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kami K, Morikawa Y, Kawai Y, Senba E. Leukemia inhibitory factor, glial cell line-derived neurotrophic factor, and their receptor expressions following muscle crush injury. Muscle Nerve. 1999;22:1576–86. doi: 10.1002/(sici)1097-4598(199911)22:11<1576::aid-mus14>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 20.Sakuma K, Watanabe K, Totsuka T, Uramato I, Sano M, Sakamoto K. Differential adaptations of insulin-like growth factor-I, basic fibroblast growth factor, and leukemia inhibitory factor in the plantaris muscle of rats by mechanical overloading: An immunohistochemical study. Acta Neuropathol. 1998;95:123–30. doi: 10.1007/s004010050775. [DOI] [PubMed] [Google Scholar]
- 21.Sakuma K, Watanabe K, Sano M, Uramato I, Totsuka T. Differential adaptation of growth and differentiation factor 8/myostatin, fibroblast growth factor 6 and leukemia inhibitory factor in overloaded, regenerating and denervated rat muscle. Biochim Biophys Acta. 2000;1497:77–88. doi: 10.1016/s0167-4889(00)00044-6. [DOI] [PubMed] [Google Scholar]
- 22.Metcalf D, Nicola NA, Gearing DP. Effects of injected leukemia inhibitory factor on hematopoietic and other tissues in mice. Blood. 1990;76:50–6. [PubMed] [Google Scholar]
- 23.Hilton DJ, Nicola NA, Waring PM, Metcalf D. Clearance and fate of leukemia inhibitory factor (LIF) after injection into mice. J Cell Physiol. 1991;148:430–9. doi: 10.1002/jcp.1041480315. [DOI] [PubMed] [Google Scholar]
- 24.Tidball JG. Inflammatory processes in muscle injury and repair. Am J Physiol Regul Integr Comp Physiol. 2005;288:R345–53. doi: 10.1152/ajpregu.00454.2004. [DOI] [PubMed] [Google Scholar]
- 25.Merley F, Lescaudron L, Rouaud T, Crossin F, Gardahuat MF. Macrophages enhance muscle satellite cell proliferation and delay differentiation. Muscle Nerve. 1999;22:724–32. doi: 10.1002/(sici)1097-4598(199906)22:6<724::aid-mus9>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 26.Yaffe D. Retention of differentiation potentialities during prolonged cultivation of myogenic cells. Proc Natl Acad Sci USA. 1968;61:477–83. doi: 10.1073/pnas.61.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riuzzi F, Sorci G, Donato R. S100B stimulates myoblast proliferation and inhibits myoblast differentiation by independently stimulating ERK1/2 and inhibiting p38 MAPK. J Cell Physiol. 2006;207:461–70. doi: 10.1002/jcp.20580. [DOI] [PubMed] [Google Scholar]
- 28.Launay T, Hagstrom L, Lottin-Divoux S, et al. Blunting effect of hypoxia on the proliferation and differentiation of human primary and rat L6 myoblasts is not counteracted by Epo. Cell Prolif. 2010;43:1–8. doi: 10.1111/j.1365-2184.2009.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu R, Liu J, Fan J, et al. Novel evidence that testosterone promotes cell proliferation and differentiation via G protein-coupled receptors in the rat L6 skeletal muscle myoblast cell line. J Cell Physiol. 2011;226 doi: 10.1002/jcp.22710. Epub 3 March 2011. [DOI] [PubMed] [Google Scholar]
- 30.Bower J, Vakakis N, Nicola NA, Austin L. Specific binding of leukemia inhibitory factor to murine myoblasts in culture. J Cell Physiol. 1995;164:93–8. doi: 10.1002/jcp.1041640112. [DOI] [PubMed] [Google Scholar]
- 31.Austin L, Burgess AW. Stimulation of myoblast proliferation in culture by leukemia inhibitory factor and other cytokines. J Neurol Sci. 1991;101:193–7. doi: 10.1016/0022-510x(91)90045-9. [DOI] [PubMed] [Google Scholar]
- 32.Austin L, Bower J, Kurek J, Vakakis N. Effects of leukemia inhibitory factor and other cytokines on murine and human myoblast proliferation. J Neurol Sci. 1992;112:185–91. doi: 10.1016/0022-510x(92)90149-f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.