Abstract
Although DNA methylation is now recognized as an important mediator of complex diseases, the extent to which the genetic basis of such diseases is accounted for by DNA methylation is unknown. In the setting of large, extended families representing a minority, high-risk population of the USA, we aimed to characterize the role of epigenome-wide DNA methylation in type 2 diabetes (T2D). Using Illumina HumanMethylation450 BeadChip arrays, we tested for association of DNA methylation at 446 356 sites with age, sex and phenotypic traits related to T2D in 850 pedigreed Mexican-American individuals. Robust statistical analyses showed that (i) 15% of the methylome is significantly heritable, with a median heritability of 0.14; (ii) DNA methylation at 14% of CpG sites is associated with nearby sequence variants; (iii) 22% and 3% of the autosomal CpG sites are associated with age and sex, respectively; (iv) 53 CpG sites were significantly associated with liability to T2D, fasting blood glucose and insulin resistance; (v) DNA methylation levels at five CpG sites, mapping to three well-characterized genes (TXNIP, ABCG1 and SAMD12) independently explained 7.8% of the heritability of T2D (vi) methylation at these five sites was unlikely to be influenced by neighboring DNA sequence variation. Our study has identified novel epigenetic indicators of T2D risk in Mexican Americans who have increased risk for this disease. These results provide new insights into potential treatment targets of T2D.
Introduction
Complex diseases are the result of intricate interactions between genetic and epigenetic mechanisms, involving important internal and external environmental triggers (1). Understanding these etiological processes requires systematic investigations into their various components, and epigenetic determinants are being increasingly recognized as central players (2–4). The last decade has seen pivotal technological advances aimed at characterizing DNA methylation on a genome-wide scale. Despite these developments, however, attempts to link alterations in the DNA ‘methylome’ to complex diseases are still in a nascent stage.
Type 2 diabetes (T2D) is a complex disease with global implications, necessitating the delineation of underlying pathogenic mechanisms. Although significantly heritable (5,6) with numerous genetic risk variants identified to date (7–9), the genetic basis of T2D remains largely unaccounted for. Studies in mice (10) and humans (11–13) have assessed the role of DNA methylation in T2D risk, providing preliminary insights into the methylation-T2D nexus. A recent study of twins in the MuTHER Consortium (14) has shown evidence that DNA methylation changes influenced by shared environmental effects may be important in biological functions relevant to T2D. However, large-scale, fine-resolution DNA methylation surveys have, to our knowledge, not been conducted in extended families, to examine the contribution of DNA methylation changes to the heritability of T2D.
We utilized phenotypic and genotypic data available from the San Antonio Family Heart Study (15,16), an ongoing project involving large and extended pedigrees of Mexican Americans, to assess the contribution of DNA methylation to the heritability of T2D. Here, we report the results of epigenome-wide associations between DNA methylation and T2D-related traits and document the genetic basis of this association by estimating the heritability of DNA methylation and investigating the role of local sequence variation.
Results
In this family-based study, we interrogated 485 577 genome-wide DNA methylation sites and analyzed a total of 446 356 autosomal CpG sites to investigate genetic and phenotypic associations. Selection criteria of these methylation probes are shown in Supplementary Material, Figure S1 and detailed information on the 446 356 CpG sites is shown in Supplementary Material, Table S1. The included sites contained 35 395 probes (7.9%) that had a known single nucleotide polymorphism (SNP) at the CpG site and 92 117 (20.64%) probes that contained at least one SNP that was not located at the site. We examined the association of these 446 356 sites with diabetes-related traits in 850 pedigreed Mexican Americans whose clinical characteristics are provided in Supplementary Material, Table S2. Participants were predominantly female (63%) and at the time of assessment ∼21% of individuals had T2D and another ∼17% had impaired fasting glucose. There was also a high prevalence of obesity (56%) and hypertension (33%). Familial relationships among study participants are shown in Supplementary Material, Table S3.
Heritability of DNA methylation levels
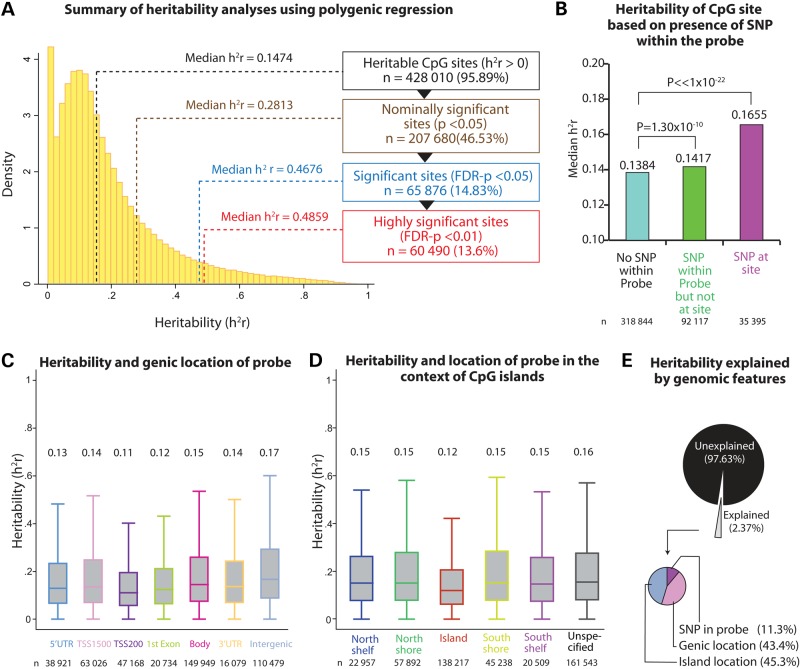
We first estimated the narrow-sense heritability of each normalized methylation score, identifying 65 876 (14.8%) significantly heritable CpG sites using a false discovery rate (FDR) of 0.05 (Fig. 1A, Supplementary Material, Table S1). The median heritability of all tested sites was 0.14, and for those that were significantly heritable (FDR < 0.05) it was 0.47.
Figure 1.
Heritability of DNA methylation levels in Mexican Americans. (A) Histogram of heritability (h2r) estimates for all CpG sites. Subgroups of CpG sites based on statistical significance and their corresponding median estimates of heritability are shown. (B) Median heritability based on the presence of DNA sequence variants within the probes targeting CpG sites. (C) Distribution of heritability of DNA methylation based on the gene context of the CpG site. Box plots show the distribution of heritability estimates based on gene region. Number above each box shows the median heritability value. UTR, untranslated region; TSS, transcription start site. (D) Distribution of DNA methylation heritability in the context of CpG islands. The box plots show distribution of heritability of CpG sites within each category of CpG island location. Number above each box shows the median heritability value. (E) Variability of heritability explained by genomic features. The results were obtained using multivariable analysis of variance.
We next examined if the estimated heritability was influenced by genomic features. We found (Fig. 1B) that probes containing a SNP and probes containing a SNP at the CpG site had significantly higher heritabilities (P = 1.30 × 10−10 and P < 1.0 × 10−22, respectively) compared with probes that did not contain a SNP. The genic location of the probes moderately influenced heritability (Fig. 1C), such that probes within 200 bp of the transcription start site and those within the first exon were least heritable while the median heritability was higher at locations more distal to the transcription start site. Also, probes within CpG islands were least heritable while those within shores and shelves were generally more heritable (Fig. 1D). Collectively, we observed (Fig. 1E) that these three features explained only 2.37% of the variability in heritability. Of this total explained variation, 45%, 43% and 11% was attributed to island location, genic location and the presence of probe-SNP, respectively.
Association of methylation levels with local sequence variants (meQTL mapping)
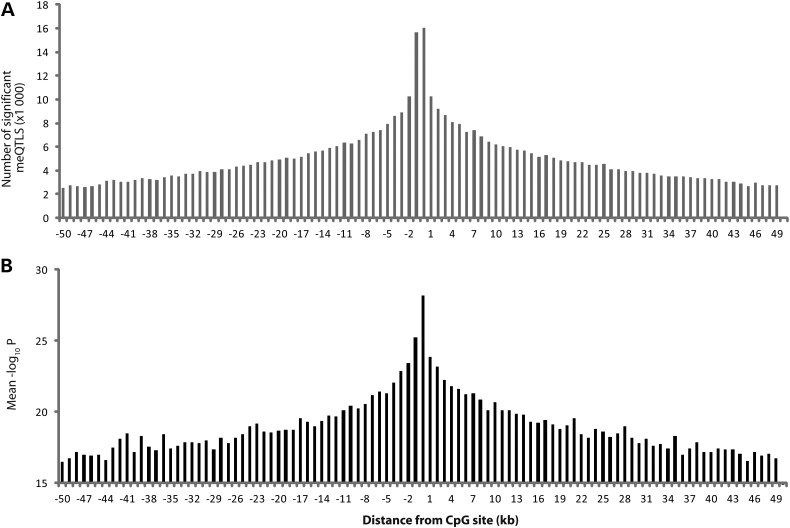
We examined if methylation levels were influenced by sequence variants in close proximity to the CpG site (CpG site ±50 kb), using genotype data from ∼1 million sequence variants. Of the 446 356 CpG sites analyzed, 444 535 (99.6%) had at least one local SNP for testing. We examined 18 518 148 associations between CpG sites and SNPs (average 41.67 SNPs per CpG site; detailed in Supplementary Material, Table S4). After correcting for local linkage disequilibrium as well as for the number of CpG sites tested, we found that DNA methylation levels at 61 799 (14.3%) CpG sites were significantly associated with at least one SNP. All statistically significant meQTL associations are provided in Supplementary Material, Table S5. The 61 799 CpG sites had DNA methylation levels that were significantly associated with a total of 633 918 SNPs (average 10.3 significant associations per CpG site). The number of significant associations as well as the strength of the meQTL associations was inversely related to the distance of the SNP from the CpG site (Fig. 2).
Figure 2.
Association of meQTLs with CpG sites. The bar charts show the number of significantly associated meQTLs (A) and the average strength of evidence (−log10 P, B) based on distance between the meQTL and corresponding CpG site.
We also tested the hypothesis that heritability of CpG sites is determined by the number of significantly associated SNPs. For each additional significantly associated SNP, the heritability of a CpG site was improved by 0.64% (95% CI 0.63%–0.66%; P, inestimably low). The number of significant methylation-SNP associations explained 12.47% of the variability in heritability. Together, these results indicate that while the contribution of surrounding sequence variants to the heritability of CpG sites is highly statistically significant, most of the variability in heritability estimates cannot be explained solely by the sequence variation assessed in this study.
Association of methylation levels with age and sex
Age and sex are known confounders in many epidemiological studies of complex diseases and have been shown to influence DNA methylation levels. We therefore investigated whether there was any evidence of significant association of age and sex with methylation levels. At an FDR of 0.05, 99 487 (22.3%) and 12 432 (2.8%) CpG sites were significantly associated with age and sex, respectively (Table 1, Supplementary Material, Table S6). Using the BSMAP software (17), we found that none of the autosomal sites associated with sex could be mapped to the X chromosome.
Table 1.
Association of DNA methylation with age and sex based on genomic features
| Subgroups | Association with age |
Association with sex |
||||||
|---|---|---|---|---|---|---|---|---|
| Significant sites |
Hypermethylation with age | Significant sites |
Hypermethylation in females | |||||
| Significant/analyzed | %a | N | %b | Significant/analyzed | % | N | % | |
| All sites | 99 487/446 356 | 22.3 | 38 221 | 38.4 | 12 432/446 356 | 2.8 | 10 645 | 85.6 |
| By SNP position | ||||||||
| No SNP in probe | 72 448/318 884 | 22.7 | 29 412 | 40.6 | 9285/318 844 | 2.9 | 7971 | 85.9 |
| SNP in probe | 19 237/92 117 | 20.9 | 7574 | 39.4 | 2489/92 117 | 2.7 | 2132 | 85.7 |
| SNP at CpG site | 7802/35 395 | 22.0 | 1235 | 15.8 | 658/35 395 | 1.9 | 542 | 82.4 |
| By gene location | ||||||||
| 5′UTR | 8138/38 921 | 20.9 | 3844 | 47.2 | 899/38 921 | 2.3 | 764 | 85.0 |
| TSS1500 | 14 107/63 026 | 22.4 | 5938 | 42.1 | 2231/63 026 | 3.5 | 2074 | 93.0 |
| TSS200 | 7731/47 168 | 16.4 | 5232 | 67.7 | 1028/47 168 | 2.2 | 927 | 90.2 |
| First exon | 4625/20 734 | 22.3 | 3379 | 73.1 | 534/20 734 | 2.6 | 493 | 92.3 |
| Gene body | 32 170/149 949 | 21.5 | 9454 | 29.4 | 3419/149 949 | 2.3 | 2750 | 80.4 |
| 3′UTR | 3327/20 734 | 22.3 | 565 | 17.0 | 310/20 734 | 2.6 | 226 | 72.9 |
| Intergenic | 29 389/110 479 | 26.6 | 9809 | 33.4 | 4011/110 479 | 3.6 | 3411 | 85.0 |
| By CpG island | ||||||||
| North Shelf | 5184/22 957 | 22.6 | 446 | 8.6 | 369/22 957 | 1.6 | 302 | 81.8 |
| North Shore | 13 323/57 892 | 23.0 | 4960 | 37.2 | 2618/57 892 | 4.5 | 2301 | 87.9 |
| Island | 27 868/138 217 | 20.2 | 25 226 | 90.5 | 4055/138 217 | 2.9 | 3620 | 89.3 |
| South Shore | 9765/45 238 | 21.6 | 3429 | 35.1 | 2181/45 238 | 4.8 | 1944 | 89.1 |
| South Shelf | 4492/20 509 | 21.9 | 359 | 8.0 | 361/20 509 | 1.8 | 278 | 77.0 |
| Unspecified | 38 855/161 543 | 24.1 | 3801 | 9.8 | 2848/161 543 | 1.8 | 2 200 | 77.3 |
| By meQTL | ||||||||
| No significant meQTL | 83 746/384 557 | 21.8 | 33 503 | 40.0 | 8543/384 557 | 2.2 | 7244 | 84.8 |
| ≥1 Significant meQTLs | 15 741/61 799 | 25.5 | 4718 | 30.0 | 3889/61 799 | 6.3 | 3401 | 87.5 |
aOut of the total significant sites.
bOut of significant sites within the category.
We next investigated the potential contribution of genomic features to the strength of these associations. CpG sites associated with age and sex were highly likely to: demonstrate no SNP within the probe; lie within the gene body or within 1500 bp of the transcription start site (TSS); and be located within CpG Islands (Table 1). To quantify the contribution of genomic features to the association of DNA methylation with age and sex we conducted multivariable analysis of variance. The presence of a SNP within the probe, genic location, CpG island context, and association with local SNPs explained 0.1%, 0.3%, 12.7% and 0.1% of the variability in the association of CpG sites with age, respectively, and <0.01%, 0.1%, 0.9% and 0.1%, of the variability in the association of CpG sites with sex, respectively.
To identify common biological pathways that might be enriched for DNA methylation changes associated with age and sex, we performed pathway analyses on a reduced subset of genes whose DNA methylation levels were associated with each of these. Pathways associated with age (n = 68, Fisher's exact P < 0.05; Supplementary Material, Table S7) included several involved in age-related diseases or processes influenced by aging (neurological disorders/neuronal system/neuronal transmission/long-term potentiation, inhibition/regulation of insulin secretion, energy metabolism, cell communication and cell signaling pathways). Those related to sex (n = 17, Fisher's exact P < 0.05; Supplementary Material, Table S8) included the nuclear receptor transcription pathway, and pathways involving molecular hubs that show variation between the sexes (e.g. NOTCH and IGF1 pathways).
Association of DNA methylation with diabetes-related traits
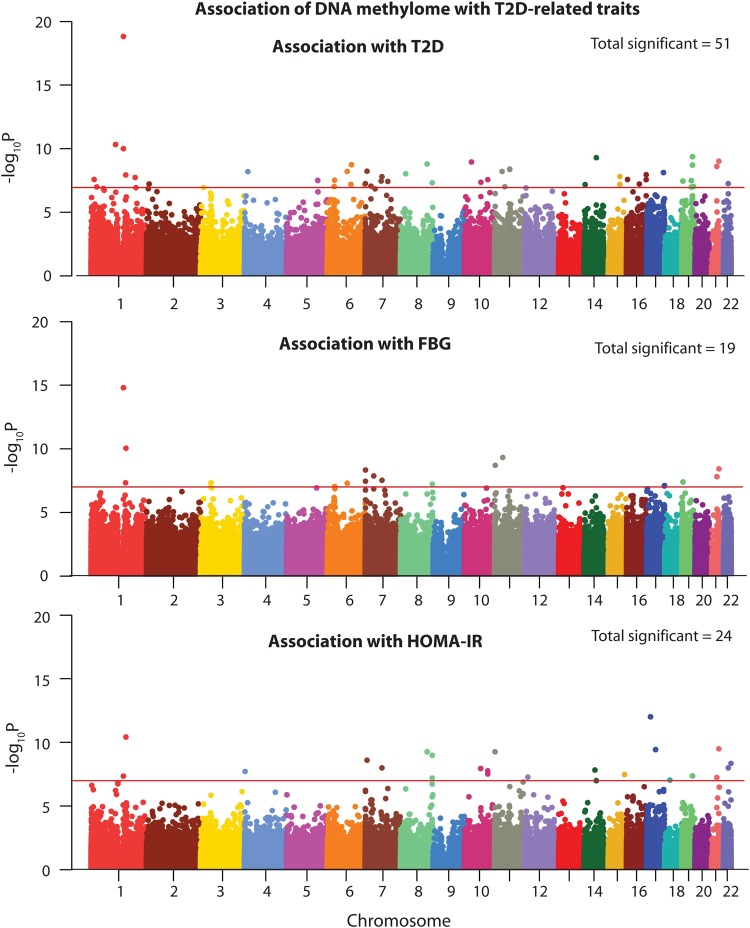
We studied the association of DNA methylation with three traits related to type 2 diabetes—liability to T2D (hereinafter referred to as T2D), fasting blood glucose (FBG) levels and homeostatic model of assessment-insulin resistance (HOMA-IR). After adjusting for potential confounders and correcting for multiple tests at a FDR of 0.05, we observed several significant associations (Fig. 3, Supplementary Material, Table S9). A total of 51 CpG sites were significantly associated with T2D, 19 with FBG and 24 with HOMA-IR. The association patterns for FBG and HOMA-IR were retained even after adjusting for the use of anti-diabetic medication (Supplementary Material, Fig. S2).
Figure 3.
Association of genome-wide methylation levels with diabetes-related traits. Manhattan plots depicting association of each CpG site studied with liability of T2D (top panel), FBG (middle panel) and HOMA-IR (bottom panel). Statistically significant associations are those that lie above the red colored line which indicates FDR-corrected epigenome-wide significance level.
To identify the most relevant CpG sites associated with all three T2D-related traits, we generated a composite significance (CS) score, which combined the log-transformed significance values for the association of a CpG site with each of the three T2D-related traits into a single metric. We observed that 277 sites had a CS score exceeding zero and 78 and 53 CpG sites had a CS score corresponding to a combined P < 0.05 and <0.017 (additional correction for three traits), respectively (Supplementary Material, Table S10). Of the 277 observed CpG sites with CS score greater than zero, 46 had probes containing at least one SNP (49 SNPs total). In a subset of 197 individuals for whom whole-genome sequence data was available, we found: for 26 SNPs (across 25 probes) there was no evidence of genetic variation in our cohort subset at the SNP; for one SNP there was an insufficient call rate for analysis; for 17 SNPs (across 16 probes) there was no significant association between the genetic variation and DNA methylation levels; and for five probes genetic variation influenced DNA methylation levels (Bonferroni-corrected P < 0.05; Supplementary Material, Table S11). Of the five probes for which there was evidence of genetic variation influencing DNA methylation levels, all contained a SNP at the CpG site. Overall, this suggests that sequence variation at the CpG site may be measured as a change in DNA methylation levels, but not all genetic variation will influence detection of the probe, particularly if the variation is more distal to the CpG site.
Details of the five CpG sites that were most significantly associated with T2D-related traits are given in Table 2. Three of these CpG sites mapped to well-characterized genes: thioredoxin interacting protein (TXNIP), ATP-binding cassette sub-family G member 1 (ABCG1) and Sterile Alpha Motif Domain Containing 12 (SAMD12). Of note, seemingly small differences in the median methylation levels (2–5%) between participants with and without T2D were still associated with statistical significance indicating that minor variations in the extent of methylation for these five sites could yield critical information regarding T2D status. To ensure that our findings were not unduly influenced by low variance of methylation, we ran simulations on 1000 randomly generated phenotypes for the two most significant CpG sites and found that our associations are unlikely to be artifactual (Supplementary Material, Fig. S3). Lastly, we observed that with the exception of the intergenic site cg08309687, the remaining CpG sites were not associated with nearby SNPs (CpG ± 50 kb), suggesting that most of the observed associations are unlikely to be due to known localized sequence variation.
Table 2.
Top five CpG sites most significantly associated with T2D, FBG or insulin resistance
| Characteristic | CpG site |
||||
|---|---|---|---|---|---|
| cg19693031 | cg06500161 | cg25217710 | cg07960624 | cg08309687 | |
| Chromosome | 1 | 21 | 1 | 8 | 21 |
| Coordinate | 145 441 552 | 43 656 587 | 156 609 523 | 119 208 486 | 35 320 596 |
| Heritability | 0.42 | 0.47 | 0.45 | 0.72 | 0.46 |
| q_heritability | 8.16 × 10−10 | 9.28 × 10−10 | 3.83 × 10−6 | 3.06 × 10−31 | 3.22 × 10−8 |
| b_age | 0.0028 | 0.0052 | −0.0046 | −0.0196 | 0.0028 |
| q_age | 1.0000 | 1.0000 | 0.5067 | 1.82 × 10−25 | 1.0000 |
| b_sex | 0.5852 | −0.1536 | 0.0123 | −0.2058 | 0.2688 |
| q_sex | 5.06 × 10−15 | 1.0000 | 1.0000 | 1.0000 | 5.02 × 10−3 |
| b_T2Da | −0.5788 | 0.3875 | 0.3164 | −0.4508 | −0.4705 |
| q_T2D | 6.83 × 10−14 | 4.21 × 10−4 | 1.0000 | 6.92 × 10−4 | 1.09 × 10−3 |
| b_FBG | −0.2699 | 0.1969 | 0.3099 | −0.2080 | −0.2419 |
| q_FBG | 6.89 × 10−10 | 1.65 × 10−3 | 3.95 × 10−5 | 0.1538 | 6.67 × 10−3 |
| b_HOMA-IR | −0.1963 | 0.2197 | 0.3310 | −0.2684 | −0.2285 |
| q_HOMA-IR | 0.0194 | 0.0001 | 1.69 × 10−5 | 2.42 × 10−4 | 2.56 × 10−2 |
| CS score | 24.04 | 10.00 | 9.17 | 7.59 | 6.73 |
| Combined P-value | 9.15 × 10−25 | 9.91 × 10−11 | 6.66 × 10−10 | 2.58 × 10−8 | 1.87 × 10−7 |
| Median methylation score | |||||
| Participants with T2D | 0.67 | 0.59 | 0.60 | 0.44 | 0.17 |
| Participants without T2D | 0.72 | 0.57 | 0.60 | 0.42 | 0.19 |
| Gene symbol | TXNIP | ABCG1 | – | SAMD12 | – |
| Gene context | 3′UTR | Body | Intergenic | 3′UTR | Intergenic |
| Significant meQTLs | 0 | 0 | 0 | 0 | 2 |
b, regression coefficient; q, significance after controlling for FDR of 0.05; CS score, composite significance score; T2D, type 2 diabetes; FBG, fasting blood glucose; HOMA-IR, homeostatic model of assessment-insulin resistance.
aFor discrete traits, SOLAR returns a negative regression coefficient if a variable is associated with an increased risk. For consistency of presentation and interpretation, the coefficients have been multiplied by −1 here.
To directly estimate the contribution of the top five CpG sites to the heritability of T2D, we estimated the shrinkage in the estimated heritability due to inclusion of the five methylation levels as covariates (in addition to age, age2, sex, age × sex interaction, age2 × sex interaction and the use of antihypertensive or lipid lowering drugs) in a polygenic regression model. We found that the estimated heritability of T2D in the base model was 0.6086 (SE 0.1412), P = 2.0 × 10−7 but this estimate shrunk to 0.5610 (SE 0.1705), P = 1.2 × 10−4 in the alternative model. Thus, the five top significant CpG sites accounted for 7.8% of the heritability of T2D.
We also tested the null hypothesis that the most significant CpG site associations with T2D-related traits are confounded by body mass index (BMI). For this we further adjusted the polygenic regressions shown in Table 2 by including BMI as a covariate. The results of these analyses are shown in Table 3. These results show that even after accounting for BMI, all significantly associated CpG sites retained the strength and significance of the associations. We therefore rejected the null hypothesis that our results are confounded by BMI.
Table 3.
Association of the top five CpG sites before and after adjusting for BMI
| CpG site | Trait |
|||||
|---|---|---|---|---|---|---|
| T2Da | FBG |
HOMA-IR | ||||
| Beforeb b(p) | Afterc b(p) | Beforeb b(p) | Afterc b(p) | Beforeb b(p) | Afterc b(p) | |
| cg19693031 | −0.58 (1.53 × 10−19) | −0.91 (1.9 × 10−14) | −0.27 (1.55 × 10−15) | −0.26 (1.52 × 10−15) | −0.20 (4.39 × 10−8) | −0.18 (1.09 × 10−8) |
| cg06500161 | 0.39 (9.43 × 10−10) | 0.32 (0.0006) | 0.20 (3.70 × 10−9) | 0.16 (2.86 × 10−6) | 0.22 (3.23 × 10−10) | 0.13 (4.25 × 10−5) |
| cg25217710 | 0.32 (0.0018) | 0.42 (0.0006) | 0.31 (8.88 × 10−11) | 0.28 (2.72 × 10−9) | 0.33 (3.81 × 10−11) | 0.24 (4.46 × 10−8) |
| cg07960624 | −0.45 (1.55 × 10−9) | −0.33 (0.0068) | −0.21 (3.46 × 10−7) | −0.16 (6.88 × 10−5) | −0.27 (5.47 × 10−10) | −0.16 (5.82 × 10−5) |
| cg08309687 | −0.47 (2.45 × 10−9) | −037 (0.0007) | −0.24 (1.50 × 10−8) | −0.21 (2.31 × 10−7) | −0.23 (5.78 × 10−8) | −0.18 (7.13 × 10−6) |
b, regression coefficient; p, significance after adjusting for BMI; T2D, type 2 diabetes; FBG, fasting blood glucose; HOMA-IR, homeostatic model of assessment- insulin resistance.
aFor discrete traits, SOLAR returns a negative regression coefficient if a variable is associated with an increased risk. For consistency of presentation and interpretation, the coefficients have been multiplied by −1 here.
bBased on polygenic regression models adjusting for age, age2, sex, age × sex interaction, age2 × sex interaction, Sentrix id, Sentrix position, the use of antihypertensive and lipid lowering drugs and cellular heterogeneity.
cBased on polygenic regression models adjusting for age, age2, sex, age × sex interaction, age2 × sex interaction, Sentrix id, Sentrix position, the use of antihypertensive and lipid lowering drugs, cellular heterogeneity and BMI.
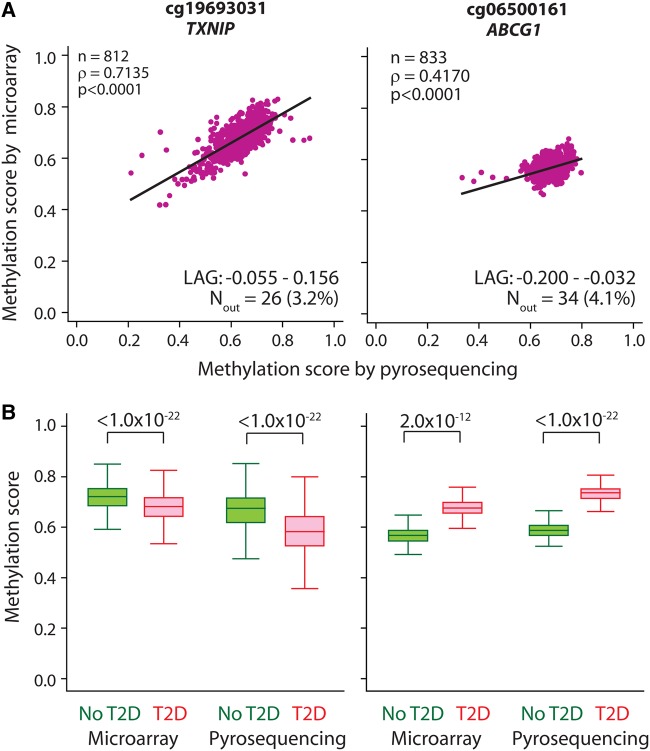
We used pyrosequencing to validate our results for the top two CpG sites (cg19693031 in TXNIP and cg06500161 in ABCG1). We found (Fig. 4) that the pyrosequencing data were highly correlated with the microarray data (Fig. 4A) and that even when the estimates from pyrosequencing were used these sites were significantly associated with T2D (P < 1.0 × 10−22 for both sites, Fig. 4B). When the methylation levels measured by pyrosequencing were used as covariates in the polygenic regression models, we found that the TXNIP and ABCG1 sites were significantly associated with T2D (P = 1.2 × 10−7 and 1.1 × 10−5, respectively) even after adjusting for age, age2, sex, BMI, batch effects, plate effects and well effects. The TXNIP site was also strongly associated with FBG and HOMA-IR (P = 1.5 × 10−15 and 1.2 × 10−7, respectively), while the ABCG1 site was associated with HOMA-IR (P = 0.0107). All the models used the same covariates mentioned above.
Figure 4.
Validation of Illumina Human Methylation 450k array results with pyrosequencing for top two sites significantly associated with T2D-related traits. (A) Correlation plots with regression line fits for the methylation levels of top two CpG sites (with the mapping genes). n, number of samples that passed the quality check in pyrosequencing; ρ, Spearman's correlation coefficient; P, statistical significance of ρ; LAG, limits of agreement determined using Bland–Altman's method; Nout, number of samples outside LAG. (B) Box plots showing distribution of methylation levels obtained by microarray and pyrosequencing for each site in subjects with (red) and without (green) T2D. Numbers at the top show significance values obtained by Mann–Whitney U test for the indicated comparisons.
Finally, we examined whether genes identified using our association studies were enriched within diabetes-related pathways. We collated a list of 42 genes to which 53 CpG sites associated with T2D traits (CS score ≥1.77, combined P < 0.017) mapped. Even in this small dataset, pathway analysis (Supplementary Material, Table S12) indicated significant enrichment in 31 pathways (Fisher's exact P < 0.05), including those related to circadian clock (P = 0.005), adipocytokine signaling (P = 0.009), leptin pathway (P = 0.023), HDL-mediated lipid transport (P = 0.031) and insulin signaling (P = 0.033).
Epigenome-wide inflation of significance values
We observed that there is substantial epigenome-wide inflation in various analyses presented here. Table 4 shows that the epigenome-wide inflation factor (λmedian) was very high for all analyses with the exception of association with T2D and HOMA-IR. We investigated the inflation using two strategies. First, we examined if accounting for population admixture reduces the inflation factors. For this, we used top four principal components (PCs) derived from SNP-based ancestry markers [procedure detailed in (18)] as covariates in the polygenic regression models. Table 4 further shows that accounting for population admixture somewhat reduced the inflation factors but with the exception of association with T2D and HOMA-IR the inflation factors continued to be high.
Table 4.
Epigenomic inflation factor for significance values
| Analysis |
λmedian |
|
|---|---|---|
| Before correction With PCs | After correction With PCs | |
| Heritability | 6.74 | 4.14 |
| Association with | ||
| Age | 10.24 | 1.95 |
| Sex | 2.03 | 1.84 |
| T2D | 1.01 | 0.32 |
| FBG | 1.95 | 1.65 |
| HOMA-IR | 1.13 | 0.98 |
PC, top four principal components reflecting population admixture; T2D, type 2 diabetes; FBG, fasting blood glucose; HOMA-IR, homeostatic model of assessment- insulin resistance.
Second, we conducted rigorous simulation studies. We simulated 10 000 quantitative trait loci using the observed distribution of T2D-related traits and generated an empirical distribution of likelihood ratio χ2. These analyses were conducted for the 53 significantly associated CpG sites. The results of the simulation studies are shown in Supplementary Material, Table S13. These results clearly demonstrated that the inflation observed in our analyses was not a statistical artefact. For each site, there was no evidence for a systematic departure from the expected likelihood ratio χ2 values.
Discussion
There are some striking observations from this study. We found that ∼15% of CpG sites assayed were significantly heritable, which is less than that recently observed by McRae et al. (19) (48.5%) in their family-based study. Using peripheral blood cells, our average heritability estimate of 0.14 was similar to that observed previously using adipose tissue (h2 = 0.19) (14), to that using peripheral blood corrected for cellular heterogeneity (h2 = 0.18) (19) and also to the estimates observed in peripheral blood by our group and others, using earlier Illumina platforms (20,21). Typically, mean heritability estimates of DNA methylation levels in other tissues are lower, and have been estimated at 0.12, 0.07, 0.05 and 0.03 in human umbilical vein epithelial cells, cord blood mononuclear cells, placenta and brain, respectively (22–24). In comparing our data to that previously published, we found that the Pearson's correlation coefficient between estimates of heritability reported by us and McRae et al. (19) was 0.79, suggesting a reasonably good agreement.
Only 14.3% of CpG sites assessed were associated with common genetic variants within 50 kb of the site, which was similar to but slightly higher than previous reports using different Illumina platforms (4%–9%) (20,21,24–26). This variation only accounted for ∼12.5% of the heritability, which is somewhat lower than the estimate of 19% given by Grundberg et al. (14). Despite a potential role for the contribution of genetic variation on DNA methylation levels, our results suggest that the heritability of DNA methylation is likely influenced by, as yet, unknown genetic mechanisms, a phenomenon also supported by various other human and animal studies (27–29). Indeed, there is now scientific evidence to believe that Darwinian evolution may have taken place due to epigenetic inheritance independently of sequence alterations and that this may drive variation within complex traits (30). Although our data lend further support to the role of epigenetic inheritance, additional studies are needed to better understand this.
We observed that 22.4% of the studied CpG sites are significantly associated with age, which is higher than that reported by Hannum et al. (31) who found 15% of sites on the 450k array were associated with age. Several studies show that DNA methylation levels are mostly positively correlated with age, which is particularly evident in CpG islands (32–34), a finding which we also replicate (Table 1). We found that 38.4% of CpG sites showed increased methylation with age but this was even more noticeable (90.5%) in CpG islands. In our study, the relationship between hypermethylation and age is not as strong as what has been seen in other studies (32,33), which may be due to the detection of a large number of associations with CpG sites outside of CpG islands, whose methylation is generally decreased with age (32,33). We found that CpG sites outside of CpG islands were more likely to be heritable and it might be that the powerful extended pedigree-based design of our study has identified additional heritable age-related CpG sites, which may not have been identified by other studies. Several studies (31,32,35) have identified key genes whose level of methylation strongly associates with age, and some studies have included development of prediction models, which show correlations between age and predicted age of up to 96% (based on 71 CpG sites). Our most significant associations with age included several CpG sites within the genes FHL2, ELOVL2 and KLF14, which have previously been implicated in several other studies (31–33,35), and in general our study confirmed associations of age with DNA methylation in previously reported genes. Also, even using a reduced subset approach, our pathway analysis identified enrichment of genes involved in cellular communication, neuronal systems, insulin secretion and metabolism, disorders and processes that are very relevant to aging.
We found ∼3% of autosomal CpG sites to be significantly associated with sex and of these ∼86% were hypermethylated in females. Few studies have globally examined the association of DNA methylation with sex, particularly with respect to autosomal methylation levels. Two recent studies utilizing the Illumina 450k DNA methylation platform identified sex biases in 0.15–0.18% of autosomal CpG sites with hypermethylation in females seen for 55–77% of probes, which is less than what we see, likely due to the use of small cohorts, pooling methods and differential tissue used in these studies (36,37). We found that many of our most significantly associated CpG sites (e.g. those in KRT77, RNASEH2C, RFTN1 and ARID1B) had also been identified in the study by Xu et al. (37) (analyzing human prefrontal cortex of Caucasians), which suggests that CpG sites associated with sex are not necessarily tissue- or cohort-specific. However, the role of these CpG sites, or the genes in which they reside, in sex biases is not an obvious one. Our pathway analysis found enrichment of genes associated with the nuclear receptor transcription pathway, involving several nuclear receptors associated with sex development and sex-biased disorders. We also identified pathways in which the central molecule may be involved in sex-specific crosstalk [e.g. NOTCH (38)] or may be influenced by sex steroids (e.g. IGF1 39) and might contribute to sex-biases in disease (40–42). These observations are important because in studies attempting to unravel the association between differential methylation and complex disease, a correction for sex will be necessary even if only autosomal CpG sites are selected for analyses.
We observed biologically relevant associations between DNA methylation levels and diabetes-related traits independently of BMI. Most genome-wide association studies performed to date investigating the role of DNA methylation in T2D have focused on pooled DNA strategies or analyzed a small number of samples, both of which reduce the power of the study and make it difficult to identify statistically significant associations. Using a pooled DNA approach, Toperoff et al. (12) identified significant differences in DNA methylation levels at CpG sites within SLC30A8, TCF7L2 and FTO, although DNA methylation at the FTO locus appears to be driven by sequence variation (43). Hidalgo et al. (11), identified a genome-wide significant association of a CpG site within ABCG1 with insulin and HOMA-IR, using the same microarray platform utilized in this study, a finding which we also replicate.
In our dataset, we found many statistically significant associations between DNA methylation levels and T2D, FBG and HOMA-IR (51, 19 and 24, respectively). Using a statistical approach to combine association with three related phenotypes (generation of a CS score and combined P-value), we identified five CpG sites that accounted for 7.8% of the heritability of T2D, three of which localized to candidate genes whose DNA methylation levels may be strongly indicative of T2D pathophysiology: TXNIP, ABCG1 and SAMD12. Two of these (TXNIP and ABCG1) are well characterized and stand out as prime candidates for T2D pathophysiology due to their known biological functions. Although the ABCG1 probe each contained a known SNP, there was no evidence to suggest that it significantly influenced DNA methylation levels in our cohort. Furthermore, pyrosequencing analysis validated our findings for TXNIP and ABCG1.
For each CpG site nominally associated with T2D liability (CSS > 0), the affiliated loci did not overlap with any of the 63 autosomal loci that have been detected by previous GWAS, as described in the most recent meta-analysis (9). This is in contrast to a small MeDIP-Seq study (44), which found ∼20% of differentially methylated regions overlapped with loci previously identified by GWAS. A similar finding was observed for obesity in a small 450k array dataset examining lean versus obese African Americans (45). The discrepancy in overlap with GWAS loci between our study and others is likely due to different detection methods (for example, Yuan et al. 44 found that only ∼60% of 450k probes were nominally significant with the same direction of effect as their corresponding loci from MeDIP-Seq experiments), significantly smaller sample sizes (each of the other two studies examined ≤46 cases) or inadequate correction for experimental biases (Xu et al. 45 did not correct for differences in probe types, all batch effects or cellular heterogeneity). More recently, the insulin resistance study by Hidalgo et al. (11), which used a much larger sample size did not identify any CpG loci that overlapped with loci from GWAS studies of insulin resistance. Overall, our results suggest that independent genetic and epigenetic loci may exert influence of T2D liability, although it is possible that the genes involved in genetic and epigenetic mechanisms are part of the same or interacting biological pathways.
The TXNIP protein regulates intra- and extra-cellular reduction–oxidation cycles (46,47) and is an important determinant of glucose metabolism, glucose tolerance and T2D (48–50). Although a modest association has previously been shown between TXNIP variants and diabetes (48), to our knowledge, altered DNA methylation within this gene has not been shown to contribute to T2D disease risk. ABCG1 plays a role in lipid and glucose homeostasis (reviewed in 51), and gene and protein expression is reduced in macrophages of diabetic subjects (52). A recent study of 40 600 subjects on 14 SNPs within or surrounding ABCG1 failed to reveal any significant associations with T2D (53). However, Hidalgo et al. (11) identified associations with methylation levels in a CpG site within the ABCG1 gene (the same one identified in this study) and insulin resistance in non-diabetic patients; our results are concordant with this but also add a new dimension, extending the association to FBG and T2D. It is important to note that for each of these genes, our DNA methylation results were consistent with the direction of effect expected, based on previously published studies, suggesting that DNA methylation is a novel mechanism by which these genes may be regulated to influence T2D pathophysiology.
In addition to these highly promising candidate genes, several other genes with potential roles related to T2D (including those with cholesterol, lipid, obesity or adipogenesis-related functions) also contained CpG sites whose DNA methylation levels had combined P < 0.05. Of note, SREBF1 controls cholesterol homeostasis (reviewed by Sato 54) and is required for regulation of carbohydrate metabolism (55), decreased expression is also seen in adipose and skeletal muscle tissue of diabetic subjects (56). Several studies have reported modest associations of genetic variants in and around the SREBF1 gene with T2D and hyperglycemia (57–61), however, the role of DNA methylation in this gene as a determinant of T2D is also novel. Other interesting candidate genes included LOXL2 (62), CPT1A (63), SOCS3 (64), CALHM1 (65,66), ICA1 (67), ZBTB7A (68), CUX1 (69), NFE2L3 (70) and LDLRAP1 (71,72). Not surprisingly, using pathways analysis, we detected gene enrichment in insulin signaling, as well as in pathways related to lipid transport, which is often dysregulated in individuals with T2D.
Of the 49 SNP-containing probes that had a CS score > 0, the SNPs located directly at the CpG site were more likely to significantly influence DNA methylation levels and explained a higher proportion of the variance in methylation levels detected than SNPs elsewhere within the probe. Importantly, the CpG site within ABCG1 that was strongly associated with T2D liability was within a SNP-containing probe (rs9982016) but our analysis indicated a lack of significant association with methylation levels (uncorrected P = 0.1895). Also, pyrosequencing validation suggested that this SNP is unlikely to strongly affect DNA methylation levels. Our findings warrant careful consideration in studies where probes are excluded on the basis of SNP presence alone, as important candidate genes could be excluded from analyses.
A key issue raised by our study is the extent of observed epigenome-wide inflation of the significance values in association studies. Three findings need to be considered. First, our analyses showed that correction for population admixture using SNP-based PCs (as suggested by 73) only partly and inadequately accounts for this inflation. Second, inflation has been previously observed in polygenic regression analyses suited for family data essentially due to dense relatedness among individuals (74,75). However, our analytical procedures explicitly accounted for relatedness among participants. Third, our robust simulation studies failed to demonstrate the presence of a systematic shift in significance values for the most significant CpG sites. Together, these findings strongly support the possibility of likely real associations. It is conceivable that given the wave-like changes that characterize methylation signals (76), many CpG sites, unlike SNPs, may show a simultaneous and real association with disease. It is therefore likely that the inflation seen, reflects true variation of methylation levels and that such variation may be under strong genomic control.
A limitation of this study is that we did not have data on glycated hemoglobin and as such, some undiagnosed cases of diabetes may be missed. In the case of such misclassification, it can be expected that the results would be biased towards the null. The fact that we were still able to extract biologically coherent associations indicates that our results would have been only minimally influenced by a possible misclassification bias.
In summary, our study used a high-resolution genome-wide methylation survey to identify genes associated with age, sex and T2D. The methylation profiles of only the top five significant CpG sites accounted for 7.8% of the heritability of T2D in our cohort, and the full contribution of the DNA methylome to the genetic basis of T2D is likely to be much larger. This appears to be largely independent of the contribution of sequence variants [which has previously been estimated to be ∼10% (7)]. Thus, our results provide an additive and independent explanation to the genetic basis of T2D. These results are also notable because the study participants represent a minority population in the USA that has a high prevalence of T2D. Epigenetic, personalized treatment regimens are already on the clinical horizon for cancers (77). Our results, combined with those of others, point to similar treatment options for T2D in the near future.
Materials and Methods
Study participants
Participants in this study were from the San Antonio Family Heart Study (SAFHS) (15,16), an ongoing, prospective evaluation of Mexican-American families living in San Antonio. We included a total of 859 participants from 39 families, who were assessed between 2002 and 2006. The methods for recruitment of participants and their phenotyping and genotyping have been extensively described elsewhere (16,78). Written consent was obtained for all individuals in this study. Approval for this study was received from the Institutional Review Board at The University of Texas Health Science Center at San Antonio.
Diabetes-related phenotypic traits
We used three phenotypic traits related to T2D: presence of T2D (defined using the ADA criteria of fasting glucose ≥ 126 mg/dL (79) or those participants already receiving anti-diabetic medication), FBG (mg/dl) and HOMA-IR (defined as FBG in mg/dl × fasting plasma insulin in μU/ml/405) (80).
Profiling of genome-wide DNA methylation
DNA samples (500 ng) underwent bisulfite conversion using the EZ-96 DNA Methylation™ Kit (Zymo Research, Irvine, CA) and were subjected to genome-wide DNA methylation profiling using the Illumina Infinium HumanMethylation450 BeadChip assay (Illumina, San Diego, CA), as per the manufacturers' protocol. The methylation array targeted 485 577 methylation sites across the genome and incorporated both Infinium I and Infinium II bead types. BeadChips were scanned with the Illumina iScan and raw data were imported into GenomeStudio (V2011.1) to extract image intensities. We rejected nine samples for which the inbuilt SNP data did not match genome-wide SNP data that were available for these individuals (described later) leaving a total of 850 samples for analyses.
Methylation at each CpG site was represented as a methylation score (β) that ranged from 0 (unmethylated) to 1 (methylated), which represents a ratio of the quantile-normalized intensity of methylated to combined (methylated + unmethylated) locus intensity. Probes that were located on the sex chromosomes (n = 11 648), that were non-CpG loci (n = 2994) or that analyzed SNPs (n = 65) were excluded (Supplementary Material, Fig. S1). Since the distribution of β values is different based on Infinium I and Infinium II probe designs, we used the BMIQ method implemented in the R-based software, BMIQ (81) to correct for design-based differences. Finally, to ensure that the β values are amenable to variance component framework, we inverse normalized them as described below. We thus included a total of 470 870 probes for analyses, of which heritability analyses could be successfully completed for 446 356 sites. This paper is based on these 446 356 sites (detailed in Supplementary Material, Table S1). To determine if any probes contained a known SNP, we used R packages IlluminaHumanMethylation450kprobe (82) (for probe listing) and SNPlocs.Hsapiens.dbSNP.20120608 (83) (for SNP mapping based on dbSNP build 137) under the Bioconductor platform.
The exclusion of SNP-containing probes is a common quality control measure in many studies using DNA methylation arrays, however this global exclusion does not take into account the likelihood of a SNP being present (based on minor allele frequencies, which may be cohort specific), the effect that currently unknown SNPs may have on DNA methylation levels, or whether the genetic variant elicits an effect on DNA methylation levels. We therefore elected to include all probes, regardless of the presence of a SNP to ensure the inclusion of all true positive results. However, to ensure that genetic variation within the probe sequence did not influence the methylation levels of CpG sites that were associated with T2D, we then utilized whole-genome sequence data that were available for a subset (n = 197) of the population and tested for association between the known probe SNPs and DNA methylation levels.
Pyrosequencing
For validation of microarray data, 500 ng of genomic DNA was bisulfite converted as described earlier. Each bisulfite-treated sample was PCR-amplified for 40 cycles with primers designed using Pyromark Assay Design 2.0 software and subjected to pyrosequencing with the PyroMark96 MD (Qiagen, Valencia, CA); primer and probe sequences, as well as annealing temperatures are provided in Supplementary Material, Table S14. Percentage DNA methylation was determined at each CpG site using PyroMark CpG software 1.0.11.14 and group differences analyzed using the Mann–Whitney U test.
High-density SNP genotyping
Study participants were previously genotyped for ∼1 million SNP markers using several Illumina genotyping arrays, including the HumanHap550v3, HumanExon510Sv1, Human1Mv1 and Human1M-Duov3. The Infinium Whole-Genome Genotyping Assay was employed according to manufacturers' instructions. Details of the data cleaning and imputing steps for this genotypic data have been described previously (20). A total of 995 320 SNPs were available for meQTL analysis.
Statistical analysis
Normalization of β-values and phenotypic traits
Since the heritability and association analyses (described later) were conducted under a variance components framework, it required that the methylation levels and the phenotypic traits exhibit a normal distribution. For this, we used an inverse-normalization process that involved ranking the values, generating cumulative density functions and determining z-values based on the cumulative densities.
Heritability analyses
Under the variance components framework, we used the pair-wise kinship matrix that quantified the extent of genetic similarity between individuals. To estimate the narrow-sense heritability (h2r) of a methylation site, we ran a polygenic model as follows
where, i(β) is the inverse-normalized methylation score, µ is the overall mean methylation score, b is the regression coefficient vector corresponding to the covariate matrix a, gi is the polygenic effect (used to estimate heritabilities) and ei is the measurement error. In each model, we used age, age2, sex, age × sex interaction, age2 × sex interaction, Sentrix id, Sentrix position, the use of antihypertensive and lipid lowering drugs and cellular heterogeneity as covariates. These models also directly permitted estimation of the association between inverse-normalized methylation levels with these covariates. We tested for potential overlap of sex-associated CpG sites with the sequence of the X chromosome (genome build Hg19) using in silico bisulfite conversion and mapping software BSMAP (17). We aligned the probe sequence for CpG sites significantly associated with sex against four reference genomes: reference, reverse complement, fully methylated and fully unmethylated.
Association analyses
We conducted three types of association analyses. First, we used a liability threshold modeling approach under a polygenic regression setting to test the association between discrete traits (T2D status) and methylation levels. Second, to test the association between quantitative traits (FBG, and HOMA-IR) and methylation levels, we used inverse-normalization followed by polygenic regression as described earlier. Third, we examined the association between methylation levels and local sequence variation (meQTL mapping defined as CpG ± 50 kb, based on an optimum window size identified by Quon et al. 23) using a measured genotype approach. In all association analyses, the statistical significance was tested by constraining the relevant regression coefficients to zero and comparing the log-likelihoods of the constrained and unconstrained models. For heritability analyses and association analyses, we used the SOLAR software package (84).
Correction for cellular heterogeneity
Our methylation studies were conducted on blood which contains a mixture of cell types but is more accessible than other tissues like adipocytes or islets. It is however, possible that differential methylation patterns may reflect underlying differences in the distribution of various cell types. Reinius et al. (85) and Houseman et al. (86) demonstrated the influence of differential cell proportions on DNA methylation signatures and Jaffe et al. (87) extended this procedure to the Illumina Infinium HumanMethylation450 array. We estimated the proportion of CD4+ T cells, CD8+ T cells, B cells, natural killer cells and granulocytes in each sample using this procedure (87) and adjusted all the polygenic regression models for these estimated cell counts as covariates.
Correction for multiple testing
For heritability analyses as well as association analyses, we corrected P-values using Benjamini and Hochberg's (88) method of controlling the FDR. For the meQTL association analyses, we used a two-step procedure to correct for multiple testing. First, to account for the potential linkage disequilibrium among closely spaced SNPs we used the method of Li and Ji (89) to estimate the effective number of SNPs. This method generates the eigenvalues of the genotype correlation matrix and then uses a Sidak type correction based on the sum of informative eigenvalues. Second, we used a Bonferroni correction for the number of CpG sites included in the analyses and multiplied the effective local α by the Bonferroni correction. To quantify the epigenome-wide inflation, we used the genomic inflation factor (75).
CS score
To quantify the association of each methylation site with all the T2D-related traits in a single metric, we added the log-transformed, inflation corrected and FDR-corrected significance values. This method is principally similar to Fisher's method (90) of combining P values with the difference that it uses logarithm to the base 10 (consistent with Manhattan plot representation of significance values).
Other statistical methods
We used Mann–Whitney tests to estimate the significance of non-normally distributed continuous variables across two groups and analysis of variance to estimate the significance of normally distributed variables across three or more groups. To ensure that low variation in methylation is not fallaciously responsible for observed associations, we ran the top two highly significant T2D CpG sites against 1000 simulated phenotypes, incorporating the same transformations and using the same covariates as for other polygenic analyses. To quantify the agreement between microarray and pyrosequencing techniques, we used the Bland–Altman procedure and estimated the regression-based limits of agreement. These analyses were done using the Stata 12.0 software package.
Pathway analyses
To test for pathway enrichment among our top methylation sites associated with age (reduced set of n = 2767 mapping to 2038 unique genes), sex (reduced set of n = 960 mapping to 772 unique genes) and diabetes (n = 53 mapping to 42 unique genes), Fisher's exact tests were performed in the R software package (91) on observed and permuted (1000X) data, with randomized gene lists drawn from the 20 261 genes covered by our genome-wide methylation data (20 260 genes were used for T2D analysis). The pathways tested in these enrichment analyses (n = 1273) are ones curated by KEGG, Biocarta, Pathway Interaction Database (PID) and Reactome, with the information downloaded from the Gene Set Enrichment Analysis (GSEA) website (92).
For pathway analyses of the methylation sites associated with age and sex, we first reduced the number of informative CpG sites since the number of significantly associated sites (99 487 for age and 12 432 for sex) was very high. For this, we used the procedure of non-negative matrix factorization (NMF) implemented in the R package NMF (93). The parameters used for NMF were in line with those used by Gaujoux and Seoighe (93) and are described in Supplementary Material, Table S15. After running the NMF, we used Kim and Park's algorithm (94) to extract the most informative subset of CpG sites.
Simulation studies for epigenome-wide inflation
Computer simulation was used to examine the distribution of the likelihood ratio test statistic. For each focal phenotype for which we obtained putative epigenome-wide significance, we performed 10 000 simulations under the null hypothesis to see whether the test distribution for the significant observed CpG sites (n = 53) conformed to that expected asymptotically. Simulations were performed in SOLAR (84) using the fast polygenic simulation method of Konigsberg and Blangero. (95) Each simulated phenotype was normally distributed (since the original phenotypes were inverse Gaussian transformed) and was simulated using the observed estimated heritability for the relevant observed phenotype (liability of T2D, FBG and HOMA-IR). For each covariate test, the null test distribution was accumulated and checked against its asymptotic expectations. We tested the difference in the distribution of the observed likelihood ratio statistics from their expected counterparts using three strategies: (i) inflation factor 1—this was defined as χ2 emp/3.84 where χ2 emp is the empirically derived χ2 at a P-value of 0.05; (ii) Inflation factor 2—this was defined as median (χ2 emp)/inverse χ (0.5, 1) where the denominator returns the expected median χ2 value; (iii) Proportion of the χ2 values exceeding 3.84. The results of simulation studies for the top 53 significant studies are shown in Supplementary Material, Table S13.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
Financial support for this study was provided by the NIDDK (DK087749). SNP genotyping and SOLAR is supported by NIMH (MH078143, MH078111 MH083824, MH059490, EB015611). Collection of family data and blood samples in the San Antonio Family Heart Study was supported by a NHLBI program project grant (HL045522). This investigation was conducted in facilities constructed with support from Research Facilities Improvement Program Grant Number C06 RR017515 from the National Center for Research Resources, National Institutes of Health.
References
- 1.Youngson N.A., Morris M.J. (2013) What obesity research tells us about epigenetic mechanisms. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 368, 20110337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papait R., Greco C., Kunderfranco P., Latronico M.V., Condorelli G. (2013) Epigenetics: a new mechanism of regulation of heart failure? Basic Res. Cardiol., 108, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo Y., Wang Y., Wang Q., Xiao R., Lu Q. (2013) Systemic sclerosis: genetics and epigenetics. J. Autoimmun., 41, 161–167. [DOI] [PubMed] [Google Scholar]
- 4.Udali S., Guarini P., Moruzzi S., Choi S.W., Friso S. (2013) Cardiovascular epigenetics: from DNA methylation to microRNAs. Mol. Aspects Med., 34, 883–901. [DOI] [PubMed] [Google Scholar]
- 5.Mamtani M., Kulkarni H., Dyer T.D., Almasy L., Mahaney M.C., Duggirala R., Comuzzie A.G., Blangero J., Curran J.E. (2014) Waist circumference is genetically correlated with incident Type 2 diabetes in Mexican-American families. Diabet. Med., 31, 31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mamtani M., Kulkarni H., Dyer T.D., Almasy L., Mahaney M.C., Duggirala R., Comuzzie A.G., Blangero J., Curran J.E. (2013) Waist circumference independently associates with the risk of insulin resistance and type 2 diabetes in Mexican American families. PLoS One, 8, e59153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billings L.K., Florez J.C. (2010) The genetics of type 2 diabetes: what have we learned from GWAS? Ann . N. Y. Acad. Sci., 1212, 59–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwak S.H., Park K.S. (2013) Genetics of type 2 diabetes and potential clinical implications. Arch. Pharm. Res., 36, 167–177. [DOI] [PubMed] [Google Scholar]
- 9.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segre A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A. et al. (2012) Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet., 44, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akirav E.M., Lebastchi J., Galvan E.M., Henegariu O., Akirav M., Ablamunits V., Lizardi P.M., Herold K.C. (2011) Detection of beta cell death in diabetes using differentially methylated circulating DNA. Proc. Natl. Acad. Sci. U. S. A., 108, 19018–19023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hidalgo B., Irvin M.R., Sha J., Zhi D., Aslibekyan S., Absher D., Tiwari H.K., Kabagambe E.K., Ordovas J.M., Arnett D.K. (2014) Epigenome-wide association study of fasting measures of glucose, insulin, and HOMA-IR in the genetics of lipid lowering drugs and diet network study. Diabetes, 63, 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toperoff G., Aran D., Kark J.D., Rosenberg M., Dubnikov T., Nissan B., Wainstein J., Friedlander Y., Levy-Lahad E., Glaser B. et al. (2012) Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum. Mol. Genet., 21, 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volkmar M., Dedeurwaerder S., Cunha D.A., Ndlovu M.N., Defrance M., Deplus R., Calonne E., Volkmar U., Igoillo-Esteve M., Naamane N. et al. (2012) DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J., 31, 1405–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundberg E., Meduri E., Sandling J.K., Hedman A.K., Keildson S., Buil A., Busche S., Yuan W., Nisbet J., Sekowska M. et al. (2013) Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am. J. Hum. Genet., 93, 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacCluer J.W., Stern M.P., Almasy L., Atwood L.A., Blangero J., Comuzzie A.G., Dyke B., Haffner S.M., Henkel R.D., Hixson J.E. et al. (1999) Genetics of atherosclerosis risk factors in Mexican Americans. Nutr. Rev., 57, S59–S65. [DOI] [PubMed] [Google Scholar]
- 16.Voruganti V.S., Lopez-Alvarenga J.C., Nath S.D., Rainwater D.L., Bauer R., Cole S.A., Maccluer J.W., Blangero J., Comuzzie A.G. (2008) Genetics of variation in HOMA-IR and cardiovascular risk factors in Mexican-Americans. J. Mol. Med. (Berl.), 86, 303–311. [DOI] [PubMed] [Google Scholar]
- 17.Xi Y., Li W. (2009) BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics, 10, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melton P.E., Kent J.W. Jr., Dyer T.D., Almasy L., Blangero J. (2011) Genetic signal maximization using environmental regression. BMC Proc., 5(Suppl 9), S72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McRae A.F., Powell J.E., Henders A.K., Bowdler L., Hemani G., Shah S., Painter J.N., Martin N.G., Visscher P.M., Montgomery G.W. (2014) Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol., 15, R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carless M.A., Kulkarni H., Kos M.Z., Charlesworth J., Peralta J.M., Goring H.H., Curran J.E., Almasy L., Dyer T.D., Comuzzie A.G. et al. (2013) Genetic effects on DNA methylation and its potential relevance for obesity in Mexican Americans. PLoS One, 8, e73950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boks M.P., Derks E.M., Weisenberger D.J., Strengman E., Janson E., Sommer I.E., Kahn R.S., Ophoff R.A. (2009) The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One, 4, e6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon L., Joo J.E., Powell J.E., Ollikainen M., Novakovic B., Li X., Andronikos R., Cruickshank M.N., Conneely K.N., Smith A.K. et al. (2012) Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res., 22, 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quon G., Lippert C., Heckerman D., Listgarten J. (2013) Patterns of methylation heritability in a genome-wide analysis of four brain regions. Nucleic Acids Res., 41, 2095–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D., Cheng L., Badner J.A., Chen C., Chen Q., Luo W., Craig D.W., Redman M., Gershon E.S., Liu C. (2010) Genetic control of individual differences in gene-specific methylation in human brain. Am. J. Hum. Genet., 86, 411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell J.T., Tsai P.C., Yang T.P., Pidsley R., Nisbet J., Glass D., Mangino M., Zhai G., Zhang F., Valdes A. et al. (2012) Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet., 8, e1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbs J.R., van der Brug M.P., Hernandez D.G., Traynor B.J., Nalls M.A., Lai S.L., Arepalli S., Dillman A., Rafferty I.P., Troncoso J. et al. (2010) Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet., 6, e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaminsky Z.A., Tang T., Wang S.C., Ptak C., Oh G.H., Wong A.H., Feldcamp L.A., Virtanen C., Halfvarson J., Tysk C. et al. (2009) DNA methylation profiles in monozygotic and dizygotic twins. Nat. Genet., 41, 240–245. [DOI] [PubMed] [Google Scholar]
- 28.Roemer I., Reik W., Dean W., Klose J. (1997) Epigenetic inheritance in the mouse. Curr. Biol., 7, 277–280. [DOI] [PubMed] [Google Scholar]
- 29.Morgan H.D., Sutherland H.G., Martin D.I., Whitelaw E. (1999) Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet., 23, 314–318. [DOI] [PubMed] [Google Scholar]
- 30.Cortijo S., Wardenaar R., Colome-Tatche M., Gilly A., Etcheverry M., Labadie K., Caillieux E., Hospital F., Aury J.M., Wincker P. et al. (2014) Mapping the epigenetic basis of complex traits. Science, 343, 1145–1148. [DOI] [PubMed] [Google Scholar]
- 31.Hannum G., Guinney J., Zhao L., Zhang L., Hughes G., Sadda S., Klotzle B., Bibikova M., Fan J.B., Gao Y. et al. (2013) Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell, 49, 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Florath I., Butterbach K., Muller H., Bewerunge-Hudler M., Brenner H. (2014) Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum. Mol. Genet., 23, 1186–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steegenga W.T., Boekschoten M.V., Lute C., Hooiveld G.J., de Groot P.J., Morris T.J., Teschendorff A.E., Butcher L.M., Beck S., Muller M. (2014) Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age (Dordr.), 36, 9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christensen B.C., Houseman E.A., Marsit C.J., Zheng S., Wrensch M.R., Wiemels J.L., Nelson H.H., Karagas M.R., Padbury J.F., Bueno R. et al. (2009) Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet., 5, e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garagnani P., Bacalini M.G., Pirazzini C., Gori D., Giuliani C., Mari D., Di Blasio A.M., Gentilini D., Vitale G., Collino S. et al. (2012) Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell, 11, 1132–1134. [DOI] [PubMed] [Google Scholar]
- 36.Sun L., Lin J., Du H., Hu C., Huang Z., Lv Z., Zheng C., Shi X., Zhang Y., Yang Z. (2014) Gender-specific DNA methylome analysis of a Han Chinese longevity population. BioMed Res. Int., 2014, 396727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu H., Wang F., Liu Y., Yu Y., Gelernter J., Zhang H. (2014) Sex-biased methylome and transcriptome in human prefrontal cortex. Hum. Mol. Genet., 23, 1260–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreno C.S. (2010) The Sex-determining region Y-box 4 and homeobox C6 transcriptional networks in prostate cancer progression: crosstalk with the Wnt, Notch, and PI3K pathways. Am. J. Pathol., 176, 518–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanbur-Oksuz N., Derman O., Kinik E. (2004) Correlation of sex steroids with IGF-1 and IGFBP-3 during different pubertal stages. Turk. J. Pediatr., 46, 315–321. [PubMed] [Google Scholar]
- 40.Martocchia A., Sigala S., Proietti A., D'Urso R., Spano P.F., Missale C., Falaschi P. (2002) Sex-related variations in serum nerve growth factor concentration in humans. Neuropeptides, 36, 391–395. [DOI] [PubMed] [Google Scholar]
- 41.Paccou J., Dewailly J., Cortet B. (2012) Reduced levels of serum IGF-1 is related to the presence of osteoporotic fractures in male idiopathic osteoporosis. Joint Bone Spine, 79, 78–82. [DOI] [PubMed] [Google Scholar]
- 42.Key T.J., Appleby P.N., Reeves G.K., Roddam A.W. (2010) Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol, 11, 530–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bell C.G., Finer S., Lindgren C.M., Wilson G.A., Rakyan V.K., Teschendorff A.E., Akan P., Stupka E., Down T.A., Prokopenko I. et al. (2010) Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS One, 5, e14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan W., Xia Y., Bell C.G., Yet I., Ferreira T., Ward K.J., Gao F., Loomis A.K., Hyde C.L., Wu H. et al. (2014) An integrated epigenomic analysis for type 2 diabetes susceptibility loci in monozygotic twins. Nat. Commun., 5, 5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X., Su S., Barnes V.A., De Miguel C., Pollock J., Ownby D., Shi H., Zhu H., Snieder H., Wang X. (2013) A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics, 8, 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshihara E., Masaki S., Matsuo Y., Chen Z., Tian H., Yodoi J. (2014) Thioredoxin/Txnip: redoxisome, as a redox switch for the pathogenesis of diseases. Front. Immunol., 4, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sano R., Reed J.C. (2013) ER stress-induced cell death mechanisms. Biochim. Biophys. Acta, 1833, 3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreira N.E., Omae S., Pereira A., Rodrigues M.V., Miyakawa A.A., Campos L.C., Santos P.C., Dallan L.A., Martinez T.L., Santos R.D. et al. (2012) Thioredoxin interacting protein genetic variation is associated with diabetes and hypertension in the Brazilian general population. Atherosclerosis, 221, 131–136. [DOI] [PubMed] [Google Scholar]
- 49.Parikh H., Carlsson E., Chutkow W.A., Johansson L.E., Storgaard H., Poulsen P., Saxena R., Ladd C., Schulze P.C., Mazzini M.J. et al. (2007) TXNIP regulates peripheral glucose metabolism in humans. PLoS Med., 4, e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Greevenbroek M.M., Vermeulen V.M., Feskens E.J., Evelo C.T., Kruijshoop M., Hoebee B., van der Kallen C.J., de Bruin T.W. (2007) Genetic variation in thioredoxin interacting protein (TXNIP) is associated with hypertriglyceridaemia and blood pressure in diabetes mellitus. Diabet. Med., 24, 498–504. [DOI] [PubMed] [Google Scholar]
- 51.Tarling E.J. (2013) Expanding roles of ABCG1 and sterol transport. Curr. Opin. Lipidol., 24, 138–146. [DOI] [PubMed] [Google Scholar]
- 52.Mauldin J.P., Nagelin M.H., Wojcik A.J., Srinivasan S., Skaflen M.D., Ayers C.R., McNamara C.A., Hedrick C.C. (2008) Reduced expression of ATP-binding cassette transporter G1 increases cholesterol accumulation in macrophages of patients with type 2 diabetes mellitus. Circulation, 117, 2785–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schou J., Tybjaerg-Hansen A., Moller H.J., Nordestgaard B.G., Frikke-Schmidt R. (2012) ABC transporter genes and risk of type 2 diabetes: a study of 40,000 individuals from the general population. Diabetes Care, 35, 2600–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sato R. (2010) Sterol metabolism and SREBP activation. Arch. Biochem. Biophys., 501, 177–181. [DOI] [PubMed] [Google Scholar]
- 55.Ruiz R., Jideonwo V., Ahn M., Surendran S., Tagliabracci V.S., Hou Y., Gamble A., Kerner J., Irimia-Dominguez J.M., Puchowicz M.A. et al. (2014) Sterol regulatory element-binding protein-1 (SREBP-1) is required to regulate glycogen synthesis and gluconeogenic gene expression in mouse liver. J. Biol. Chem., 289, 5510–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sewter C., Berger D., Considine R.V., Medina G., Rochford J., Ciaraldi T., Henry R., Dohm L., Flier J.S., O'Rahilly S. et al. (2002) Human obesity and type 2 diabetes are associated with alterations in SREBP1 isoform expression that are reproduced ex vivo by tumor necrosis factor-alpha. Diabetes, 51, 1035–1041. [DOI] [PubMed] [Google Scholar]
- 57.Felder T.K., Oberkofler H., Weitgasser R., Mackevics V., Krempler F., Paulweber B., Patsch W. (2007) The SREBF-1 locus is associated with type 2 diabetes and plasma adiponectin levels in a middle-aged Austrian population. Int. J. Obes. (Lond.), 31, 1099–1103. [DOI] [PubMed] [Google Scholar]
- 58.Harding A.H., Loos R.J., Luan J., O'Rahilly S., Wareham N.J., Barroso I. (2006) Polymorphisms in the gene encoding sterol regulatory element-binding factor-1c are associated with type 2 diabetes. Diabetologia, 49, 2642–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eberle D., Clement K., Meyre D., Sahbatou M., Vaxillaire M., Le Gall A., Ferre P., Basdevant A., Froguel P., Foufelle F. (2004) SREBF-1 gene polymorphisms are associated with obesity and type 2 diabetes in French obese and diabetic cohorts. Diabetes, 53, 2153–2157. [DOI] [PubMed] [Google Scholar]
- 60.Laudes M., Barroso I., Luan J., Soos M.A., Yeo G., Meirhaeghe A., Logie L., Vidal-Puig A., Schafer A.J., Wareham N.J. et al. (2004) Genetic variants in human sterol regulatory element binding protein-1c in syndromes of severe insulin resistance and type 2 diabetes. Diabetes, 53, 842–846. [DOI] [PubMed] [Google Scholar]
- 61.Grarup N., Stender-Petersen K.L., Andersson E.A., Jorgensen T., Borch-Johnsen K., Sandbaek A., Lauritzen T., Schmitz O., Hansen T., Pedersen O. (2008) Association of variants in the sterol regulatory element-binding factor 1 (SREBF1) gene with type 2 diabetes, glycemia, and insulin resistance: a study of 15,734 Danish subjects. Diabetes, 57, 1136–1142. [DOI] [PubMed] [Google Scholar]
- 62.Coral K., Madhavan J., Pukhraj R., Angayarkanni N. (2013) High glucose induced differential expression of lysyl oxidase and its isoform in ARPE-19 cells. Curr. Eye Res., 38, 194–203. [DOI] [PubMed] [Google Scholar]
- 63.Nyman L.R., Tian L., Hamm D.A., Schoeb T.R., Gower B.A., Nagy T.R., Wood P.A. (2011) Long term effects of high fat or high carbohydrate diets on glucose tolerance in mice with heterozygous carnitine palmitoyltransferase-1a (CPT-1a) deficiency: Diet influences on CPT1a deficient mice. Nutr. Diab., 1, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ueki K., Kondo T., Kahn C.R. (2004) Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol. Cell. Biol., 24, 5434–5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sclafani A., Marambaud P., Ackroff K. (2014) Sucrose-conditioned flavor preferences in sweet ageusic T1r3 and Calhm1 knockout mice. Physiol. Behav., 126, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taruno A., Vingtdeux V., Ohmoto M., Ma Z., Dvoryanchikov G., Li A., Adrien L., Zhao H., Leung S., Abernethy M. et al. (2013) CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature, 495, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barkalifa R., Yagil Y., Yagil C. (2010) Sex-specific genetic dissection of diabetes in a rodent model identifies Ica1 and Ndufa4 as major candidate genes. Physiol. Genomics, 42, 445–455. [DOI] [PubMed] [Google Scholar]
- 68.Laudes M., Christodoulides C., Sewter C., Rochford J.J., Considine R.V., Sethi J.K., Vidal-Puig A., O'Rahilly S. (2004) Role of the POZ zinc finger transcription factor FBI-1 in human and murine adipogenesis. J. Biol. Chem., 279, 11711–11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stratigopoulos G., LeDuc C.A., Cremona M.L., Chung W.K., Leibel R.L. (2011) Cut-like homeobox 1 (CUX1) regulates expression of the fat mass and obesity-associated and retinitis pigmentosa GTPase regulator-interacting protein-1-like (RPGRIP1L) genes and coordinates leptin receptor signaling. J. Biol. Chem., 286, 2155–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwenk R.W., Vogel H., Schurmann A. (2013) Genetic and epigenetic control of metabolic health. Mol. Metab., 2, 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harada-Shiba M., Takagi A., Marutsuka K., Moriguchi S., Yagyu H., Ishibashi S., Asada Y., Yokoyama S. (2004) Disruption of autosomal recessive hypercholesterolemia gene shows different phenotype in vitro and in vivo. Circ. Res., 95, 945–952. [DOI] [PubMed] [Google Scholar]
- 72.Soufi M., Rust S., Walter M., Schaefer J.R. (2013) A combined LDL receptor/LDL receptor adaptor protein 1 mutation as the cause for severe familial hypercholesterolemia. Gene, 521, 200–203. [DOI] [PubMed] [Google Scholar]
- 73.Barfield R.T., Almli L.M., Kilaru V., Smith A.K., Mercer K.B., Duncan R., Klengel T., Mehta D., Binder E.B., Epstein M.P. et al. (2014) Accounting for population stratification in DNA methylation studies. Genet. Epidemiol., 38, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hemani G., Yang J., Vinkhuyzen A., Powell J.E., Willemsen G., Hottenga J.J., Abdellaoui A., Mangino M., Valdes A.M., Medland S.E. et al. (2013) Inference of the genetic architecture underlying BMI and height with the use of 20,240 sibling pairs. Am. J. Hum. Genet., 93, 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang J., Weedon M.N., Purcell S., Lettre G., Estrada K., Willer C.J., Smith A.V., Ingelsson E., O'Connell J.R., Mangino M. et al. (2011) Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet., 19, 807–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Faulk C., Dolinoy D.C. (2011) Timing is everything: the when and how of environmentally induced changes in the epigenome of animals. Epigenetics, 6, 791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ho A.S., Turcan S., Chan T.A. (2013) Epigenetic therapy: use of agents targeting deacetylation and methylation in cancer management. Onco Targets Ther., 6, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitchell B.D., Kammerer C.M., Blangero J., Mahaney M.C., Rainwater D.L., Dyke B., Hixson J.E., Henkel R.D., Sharp R.M., Comuzzie A.G. et al. (1996) Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation, 94, 2159–2170. [DOI] [PubMed] [Google Scholar]
- 79.(2003) Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care, 26(Suppl 1), S5–S20. [DOI] [PubMed] [Google Scholar]
- 80.Hanley A.J., Williams K., Stern M.P., Haffner S.M. (2002) Homeostasis model assessment of insulin resistance in relation to the incidence of cardiovascular disease: the San Antonio Heart Study. Diabetes Care, 25, 1177–1184. [DOI] [PubMed] [Google Scholar]
- 81.Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S. (2013) A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics, 29, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Triche T., Jr. IlluminaHumanMethylation450kprobe: Probe sequence data for microarrays of type IlluminaHumanMethylation450k. R package version 2.0.6. [Google Scholar]
- 83.Pages H. SNPlocs.Hsapiens.dbSNP.20120608: SNP locations for Homo sapiens (dbSNP Build 137). R package version 0.99.9.
- 84.Almasy L., Blangero J. (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet., 62, 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reinius L.E., Acevedo N., Joerink M., Pershagen G., Dahlen S.E., Greco D., Soderhall C., Scheynius A., Kere J. (2012) Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One, 7, e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Houseman E.A., Accomando W.P., Koestler D.C., Christensen B.C., Marsit C.J., Nelson H.H., Wiencke J.K., Kelsey K.T. (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics, 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jaffe A.E., Irizarry R.A. (2014) Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol., 15, R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B, 57, 289–300. [Google Scholar]
- 89.Li J., Ji L. (2005) Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb.), 95, 221–227. [DOI] [PubMed] [Google Scholar]
- 90.Dai H., Leeder J.S., Cui Y. (2014) A modified generalized Fisher method for combining probabilities from dependent tests. Front. Genet., 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luo W., Brouwer C. (2013) Pathview: an R/bioconductor package for pathway-based data integration and visualization. Bioinformatics, 29, 1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A., 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gaujoux R., Seoighe C. (2010) A flexible R package for nonnegative matrix factorization. BMC Bioinformatics, 11, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim H., Park H. (2007) Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis. Bioinformatics, 23, 1492–1502. [DOI] [PubMed] [Google Scholar]
- 95.Konigsberg L.W., Blangero J. (1993) Multivariate quantitative genetic simulations in anthropology with an example from the South Pacific. Hum. Biol., 65, 897–915. [PubMed] [Google Scholar]