Abstract
Mutations in alpha- and beta-tubulins are increasingly recognized as a major cause of malformations of cortical development (MCD), typically lissencephaly, pachygyria and polymicrogyria; however, sequencing tubulin genes in large cohorts of MCD patients has detected tubulin mutations in only 1–13%. We identified patients with a highly characteristic cerebellar dysplasia but without lissencephaly, pachygyria and polymicrogyria typically associated with tubulin mutations. Remarkably, in seven of nine patients (78%), targeted sequencing revealed mutations in three different tubulin genes (TUBA1A, TUBB2B and TUBB3), occurring de novo or inherited from a mosaic parent. Careful re-review of the cortical phenotype on brain imaging revealed only an irregular pattern of gyri and sulci, for which we propose the term tubulinopathy-related dysgyria. Basal ganglia (100%) and brainstem dysplasia (80%) were common features. On the basis of in silico structural predictions, the mutations affect amino acids in diverse regions of the alpha-/beta-tubulin heterodimer, including the nucleotide binding pocket. Cell-based assays of tubulin dynamics reveal various effects of the mutations on incorporation into microtubules: TUBB3 p.Glu288Lys and p.Pro357Leu do not incorporate into microtubules at all, whereas TUBB2B p.Gly13Ala shows reduced incorporation and TUBA1A p.Arg214His incorporates fully, but at a slower rate than wild-type. The broad range of effects on microtubule incorporation is at odds with the highly stereotypical clinical phenotype, supporting differential roles for the three tubulin genes involved. Identifying this highly characteristic phenotype is important due to the low recurrence risk compared with the other (recessive) cerebellar dysplasias and the apparent lack of non-neurological medical issues.
Introduction
Since the first discovery of TUBA1A in 2007 (OMIM *602529) as a novel gene for lissencephaly, genes encoding alpha- and beta-tubulins, the main components of microtubule polymers, are now being recognized for their major involvement in malformations of cortical development (MCD) (1,2), and these disorders are now collectively termed tubulinopathies. Mutations in TUBA1A are predominantly associated with a variable spectrum of lissencephaly, pachygyria and polymicrogyria-like cortical malformations, with or without microcephaly and abnormalities of other brain structures (particularly the basal ganglia and corpus callosum) (1,3–11). TUBB2B (OMIM *612850) mutations frequently cause polymicrogyria-like abnormalities, as well as lissencephaly, cortical dysplasia, schizencephaly and microcephaly to a lesser extent (12–17). Two major subgroups of TUBB3 (OMIM *602661) mutations have been described previously: those associated with mildly abnormal gyral pattern (microgyria and gyral disorganization) and those with congenital fibrosis of the extraocular muscles type 3 (CFEOM3) with neurological symptoms (2,18,19). Mutations in TUBB (OMIM *191130, previously referred to as TUBB5) have been associated with microcephaly with structural brain abnormalities, TUBB4A (OMIM *602662) variants with hypomyelination (20,21) and, most recently, mutations in TUBB2A (OMIM *615101) have been identified in individuals with simplified gyral patterning or unaffected cerebral cortex but with infantile seizures (22). In addition to alpha- and beta-tubulins, mutations in a gamma-tubulin gene, TUBG1 (OMIM *191135), have also been associated with pachygyria (10), further highlighting the importance of tubulin and microtubule function during brain development.
Clinically, individuals with tubulin mutations display a range of outcomes, from severe intellectual disability and intractable seizures to learning difficulties and absence of epilepsy (12,16,18). Screening tubulin genes in large cohorts of MCD patients has yielded a mutation detection rate of only 1–13.3% (6,8,9,12,13). Selection of MCD patients for sequencing of these genes is difficult because of the variable clinical and imaging features.
It is clear, however, that the overall brain development is affected in these patients, and besides predominant cortical malformations, the basal ganglia, corpus callosum, brainstem and cerebellum are frequently affected. The cerebellum and vermis are often described as hypoplastic or dysplastic, without further specification of the abnormality (6,19). The range of overlapping phenotypes stemming from tubulin gene variation has been examined extensively; however, all studies published to date have ascertained patients based exclusively on the presence of MCD.
In this study, we present 10 patients ascertained due to their characteristic cerebellar dysplasia combined with basal ganglia dysplasia and frequent brainstem asymmetry. Although lissencephaly, pachygyria and polymicrogyria-like malformations were absent, this combination of imaging features prompted us to sequence tubulin genes. Strikingly, this yielded a high mutation detection rate, as seven out of nine patients carry tubulin mutations (78%, six of eight families).
Results
From the large cohort of patients with hindbrain imaging abnormalities referred to the University of Washington Hindbrain Research Program, we identified 10 patients, including two siblings, with a distinct combination of brainstem asymmetry, superior cerebellar dysplasia and basal ganglia dysplasia and without a diagnosis of MCD (e.g. pachygyria and polymicrogyria).
Clinical features
All individuals displayed delayed psychomotor development, with a broad range of severities (Table 1). Motor development was usually more affected than speech. Four patients had documented seizures. Four were reported to have significant behavioural problems: attention deficit hyperactivity disorder and aggression being most common. Occipital frontal circumference was available for nine individuals, five of whom had microcephaly (>2 standard deviations below the mean) and two of whom had macrocephaly (>2 standard deviations above the mean). Of note, abnormal eye movements were recorded in seven individuals, including oculomotor apraxia (OMA) in four. Strabismus was present in five. Polyneuropathy and/or CFEOM were not reported in any of the patients. No patients had dysmorphic features, non-central nervous system malformations or hearing loss.
Table 1.
Summary of clinical data
| Patient gender | Mutation | Motor development | Cognition and speech | Seizures | OFC | Behaviour | Eye movements |
|---|---|---|---|---|---|---|---|
| UW168-3 F | TUBA1A c.645G>A (p.R214H) | 4yr 3m sitting, not crawling | Few single words at 4yr 3m | ‘Startle sz’ and ‘absence sz’ | <−2.5 SD | NA | Roving eye movements |
| UW167-3 F | TUBA1A c.655A>G (p.I219V) | 10yr walks with assistance, ataxia | No speech | IS, recurrence of sz 4yr | 0 SD | NA | Wandering gaze, nystagmoid movements; greatly improved (11yr) |
| UW169-3 F | TUBB2B c.38G>C (p.G13A) | NA | NA | No (11yr) | −4 SD | Asperger's, ADHD, temper tantrums | Strabismus |
| UW169-4 F | TUBB2B c.38G>C (p.G13A) | NA | NA | No (5yr) | −4 SD | ADHD symptoms, temper tantrums | Strabismus |
| UW164-3 M | TUBB3 c.1070C>T (p.P357L) | Sitting 14m, walking 28m, no pincer grasp 5yr | First word 14m IQ ‘low end of normal’, dysarthric 5yr | Partial complex | +3 SD | ADHD-like | OMA, strabismus |
| UW165-3 F | TUBB3 c.1070C>T (p.P357L) | Hypotonia, sitting 10m, walking 26m | First words 18m, normal speech 2yr 6m | No | 0 SD | Normal | OMA, abnormal saccades |
| UW170-3 M | TUBB3 c.862G>A (p.E288K) | Hypotonia, sitting if placed in position at 13m | No words 14m | No | −2 SD | NA | Mild OMA, strabismus |
| UW161-3 F | Not tested | Sitting 12m, walking 34m | NA | No (9yr) | +2 SD | Temper tantrums | OMA |
| UW166-3 F | No | 28m: drools, rolls front to back | No words 28m | sz partially responsive to AED (28m) | −3.2 SD | Normal | Abnormal saccades disconjugate gaze |
| UW146-3 M | No | Sitting 10m, walking 26m | 10 words and 10 signs 2yr 2m | No | +0.5 SD | Temper tantrums (2yr) | Nystagmus, bilateral ptosis, strabismus |
ADHD, attention deficit hyperactivity disorder; OMA, oculomotor apraxia; IS, infantile spasms; yr, year(s); m, month(s); NA, not assessed; SD, standard deviation; sz, seizures.
Brain imaging
Brain magnetic resonance imaging (MRI) studies were performed between 8 months and 8 years of age for all 10 individuals (Table 2).
Table 2.
Summary of MRI
| Patient | Age MRI | Vermis | Pons | Medulla | Cortex | Corpus callosum | Enlarged LV | Basal ganglia DYS | Thalami | Hippocampus | CN V HYPO | Others |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UW168-3 | 4yr 3m | HYPO | ASYM | N | Diffuse irregular gyration and sulcation | Partial ACC | + | Y | Globular | N | ++ | Deficient falx, bilat coloboma |
| UW167-3 | 2yr 1m | HYPO | ASYM | DYSP | Diffuse (L > R) irregular gyration and sulcation | Partial ACC | ++ | Y | Globular | N | +++ | Decreased WM |
| UW169-3 | 8yr 7m | N | N | N | Irregular gyration and sulcation, multiple shallow sulci | N | + | Y | Globular | Bilaterally malrotated, left » right | N | Cavum septum pellucidum |
| UW169-4 | 2yr 2m | N | N | N | Diffuse irregular gyration and sulcation | N | + | Y | Globular | Left malrotated, right normal | + | Cavum septum pellucidum |
| UW164-3 | 6yr 5m | N | ASYM | DYSP | Diffuse irregular gyration and sulcation, multiple shallow sulci | Mildly shortened | N | Y | Globular | Incompletely folded | N | |
| UW165-3 | 2yr 6m | HYPO | ASYM | DYSP | Diffuse irregular gyration and sulcation | Mildly shortened | + | Y | Globular | N | + | Deficient falx, prominent perivascular spaces, slightly decreased WM |
| UW170-3 | 9m | HYPO | ASYM | N | Irregular gyral pattern | Thin, hypopl splenium | ++ | Y | Globular | N | ++ | Low-resolution MRI |
| UW161-3 | 8m | HYPO | ASYM | DYSP | Asymmetric gyral pattern, multiple shallow sulci | Thin | + | Y | Globular | NA (lack of coronal images) | + | |
| UW166-3 | 1yr 3m | HYPO | ASYM | DYSP | Irregular gyral pattern | ACC | +++ | Y | Globular | Incompletely folded (HYD) | ++ | Progressive WM loss, communicating HYD |
| UW146-3 | 2yr 2m | HYPO | ASYM | N | Diffuse irregular gyration and sulcation | Thin, hypoplastic rostrum | ++ | Y | Globular | N | ++ | Decreased WM |
+, mild; ++, moderate; +++, severe; ACC, agenesis of the corpus callosum; ASYM, asymmetric; bilat, bilateral; CN, cranial nerve; DYS, dysplasia; HYD, hydrocephalus; HYPO, hypoplasia; LV, lateral ventricles; m, months; N, normal; WM, white matter; yr, years.
Cerebellum and cerebellar vermis
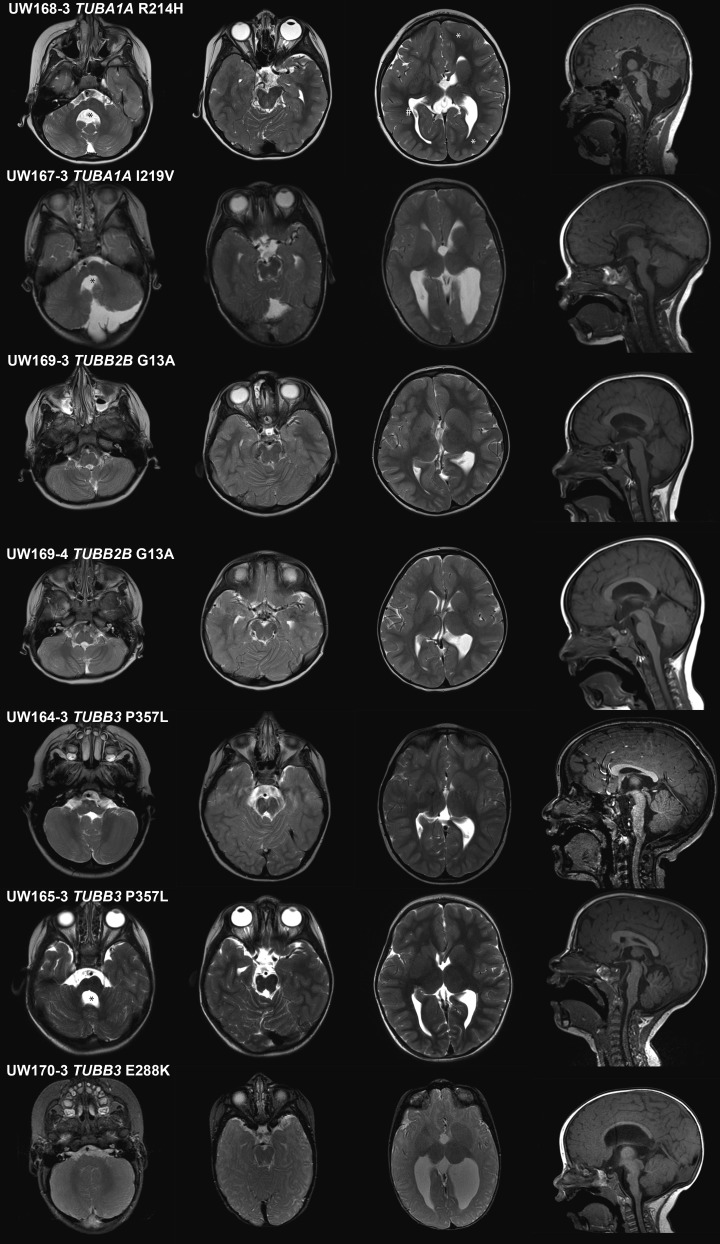
All patients have a distinct dysplasia of the superior cerebellum, especially the vermis (with ‘diagonal’ folia, i.e. folia crossing the midline at an oblique angle), best visible at the midline on axial views (Supplementary Material, Fig. S1, see also Fig. 1 and Supplementary Material, Fig. S2, second column). The vermis is hypoplastic in all but three individuals (7/10), with the anterior vermis more severely affected. The cerebellar hemispheres are either normal size or mildly hypoplastic with mild asymmetry.
Figure 1.
Brain imaging of mutation-positive patients: representative images from brain MRI, each row represents one patient. First to third column, axial T2 images and fourth column, sagittal midline T1 images. First column, level of the cerebellum and brainstem; brainstem asymmetry (except UW169-3 and UW169-4) and enlarged fourth ventricle (black asterisks), normal-sized cerebellum (except UW167-3). Second column, level of superior cerebellum showing distinct ‘diagonal’ dysplasia of the foliar pattern. Third column, level of basal ganglia; dysplastic and amorphous basal ganglia in all, also note the irregularities in sulcal depth and organization (dysgyria); representative examples in UW168-3 of shallow sulci (marked by white asterisks) and deep sulci (#). Fourth column, thinning of brainstem in all, hypoplastic vermis (except UW169-3, UW169-4 and UW164-3) and abnormal corpus callosum (CC) in three (partial agenesis in UW168-3 and UW167-3 and thin CC in UW170-3). See Table 2 for details of imaging by individual.
Brainstem
In eight of the 10 individuals, the pons is asymmetrically hypoplastic with a midline ventral indentation and asymmetrical inferior and middle cerebellar peduncles. Additional diffusion tension imaging studies of individual UW165-3 showed asymmetry of the corticospinal tracts at the level of the pons (Supplementary Material, Fig. S3). In five of 10, the medulla has a globular contour, with indistinct demarcation between the pyramids and the olivary nuclei.
Basal ganglia, thalami and corpus callosum
The basal ganglia are asymmetrically dysplastic with bulbous appearance in all individuals, with diffuse, branched or absent anterior limb of the internal capsule (Fig. 1 and Supplementary Material, Fig. S2, third column). The lateral ventricles have an irregular contour and abnormal rounding of the frontal horns (10/10), likely related to the basal ganglia dysplasia. The thalami are globular-shaped in all. The corpus callosum is variably affected, ranging from almost complete agenesis to normal (Fig. 1 and Supplementary Material, Fig. S2, fourth column).
Cortex
As cortical malformations are considered a key feature in the tubulinopathies, but not reported in our cohort, we carefully re-reviewed the cortex in all individuals. Lissencephaly, pachygyria and polymicrogyria were not observed; however, patchy and asymmetric abnormalities in gyral size and orientation and varying sulcal depths are present in all individuals (Supplementary Material, Fig. S1). A pattern involving a cluster of multiple shallow sulci was frequently observed, especially in the frontal lobes. This pattern differs from polymicrogyria (normal cortical thickness, no microgyri, no blurring of the grey–white matter boundaries) and from simplified gyral pattern, in which the gyri are too few in number (23).
Variants in TUBA1A, TUBB2B and TUBB3
DNA was available for nine of 10 patients, and we identified mutations in seven (78%). In six individuals, these were novel, unreported variants: TUBB2B c.38G>C (p.Gly13Ala) in two siblings; TUBB3 c.1070C>T (p.Pro357Leu) in two unrelated patients and TUBB3 c.862G>A (p.Glu288Lys) and TUBA1A c.655A>G (p.Ile219Val) in sporadic patients. In addition, we identified the previously published missense change TUBA1A c.641G>A (p.Arg214His) in one patient (12). All variants are de novo, except TUBB2B p.Gly13Ala, which was inherited by two affected sisters from their mosaic, apparently asymptomatic father (Supplementary Material, Fig. S4). Variants were determined to be absent from 1000 genomes, the Exome Variant Server, dbSNP142 and ExAC databases. We did not identify mutations in TUBA8, TUBB2A or TUBB4A.
CADD scores were generated for each mutation and for previously published disease-associated tubulin gene variants (Supplementary Material, Table S1) (12). The TUBA1A mutations associated with the mild phenotype (p.Arg214His, p.Ile219Val and p.Val353Ile) have a lower average CADD score than those associated with microlissencephaly, consistent with the difference in severity of the phenotypes (Student's t-test, P = 0.04). In contrast, CADD scores for TUBB2B and TUBB3 variants did not differ significantly across phenotypic groups, although the sample sizes are small.
Homology modelling
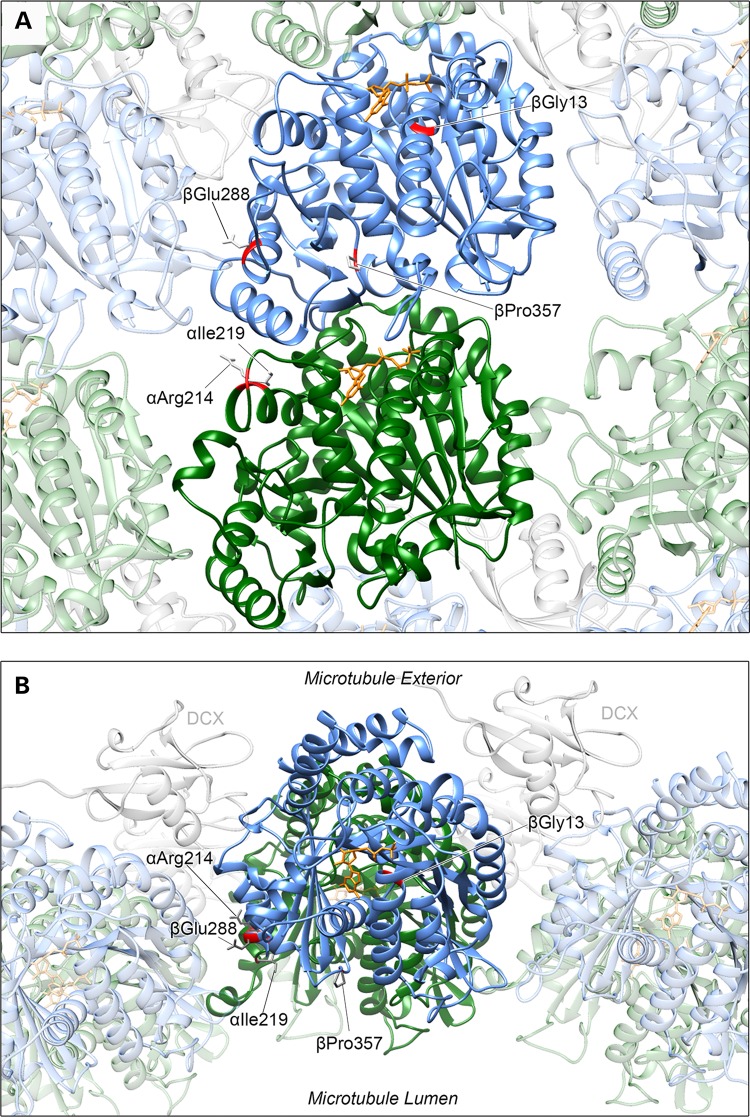
Altered amino acids are located in a variety of positions throughout the alpha-/beta-tubulin heterodimer (Fig. 2A and B and Table 3), with each substitution affecting highly conserved residues (Supplementary Material, Fig. S5). None of the substitutions are predicted to affect interactions with microtubule-associated proteins (MAPs) that predominantly associate with the external face of microtubule polymers.
Figure 2.
Tubulin variants affect amino acids located in varying regions of the heterodimer. Affected TUBA1A residues (Arg214 and Ile219) are highlighted on an alpha-tubulin protein subunit (green) and TUBB2B Gly13 and TUBB3 Glu288 and Pro357 on a beta-tubulin subunit (blue) as part of a heterodimer incorporated into a microtubule polymer lattice depicted from (A) within the microtubule polymer and (B) from above. TUBA1A Arg214 and Ile219 and TUBB3 Glu288 and Pro357 residues are located on the surface of the heterodimer, whereas TUBB2B Gly13 is buried within the beta-tubulin subunit. αArg214 is positioned towards, but not predicted to directly interact with, both the longitudinal intradimer (between residues of the same heterodimer) and lateral interprotofilament (adjacent subunits) interfaces. βGlu288 is also positioned at an interface between an adjacent protofilament subunit but with no direct interaction predicted. When incorporated into a polymer, the βPro357 residue faces into the microtubule lumen. It is not predicted to have lateral or longitudinal contact with neighbouring subunits. Due to their location, none of the affected residues is predicted to affect interaction with MAPs that predominantly associate with the exterior surface of the polymer lattice, as demonstrated by DCX in the model (grey).
Table 3.
Summary of homology modelling and functional analysis
| Gene | Variant | In silico | Conservation | In vitro consequences |
|---|---|---|---|---|
| TUBA1A | p.Arg214His (c.641G>A) | Alpha-helix 6. Within intradimer and interprotofilament interfaces | αArg214 conserved in most alpha-tubulin isoforms | Incorporates into microtubules but at a reduced rate following cold-induced depolymerization |
| p.Ile219Val (c.655A>G) | Loop between alpha-helices 6 and 7 | αIle219 conserved throughout alpha-tubulin isoforms | Not determined | |
| TUBB2B | p.Gly13Ala (c.38G>C) | Alpha-helix 1. Within beta-tubulin core. Three predicted H-bonds between β-subunit and GDP disrupted | βGly13 hyperconserved in both beta- and alpha-tubulin isoforms | Reduced incorporation into microtubules |
| TUBB3 | p.Glu288Lys (c.862G>A) | Alpha-helix 9. Within interprotofilament interface. Loss of two H-bonds with Thr285 (intrasubunit) predicted | βGlu288 conserved in beta- and most alpha-tubulin isoforms | Does not incorporate into microtubules |
| p.Pro357Leu (c.1070C>T) | Loop between beta-strands 9 and 10. Facing microtubule lumen | βPro357 hyperconserved in both beta- and alpha-tubulin isoforms | Does not incorporate into microtubules |
TUBA1A
p.Arg214His affects an arginine that is conserved in most human alpha-tubulin isoforms. Positioned on alpha-helix 6 of the alpha-tubulin subunit, Arg214 is on the surface of the protein, within the interprotofilament interface between an adjacent alpha-tubulin subunit when incorporated within a microtubule polymer. It is also in relative proximity to (but not predicted to associate with) helix 10 of the beta-tubulin within the same heterodimer. p.Ile219Val affects an isoleucine conserved throughout alpha-tubulins. Five amino acids from Arg214, this residue is located within a short loop between two alpha-helices at the interface with an adjacent alpha-subunit of a neighbouring microtubule protofilament. The branched isoleucine side chain does not extend into this interface, but is instead orientated towards the centre of the protein. Ile219 is predicted to form a hydrogen bond with Cys213; however, this is preserved when substituted with a valine.
TUBB2B
p.Gly13Ala affects a glycine located on the first alpha-helix, within the core of the beta-tubulin subunit. Hyper conserved through alpha- and beta-tubulins, this residue is positioned within a tight groove between the alpha-helix and the preceding beta-strand, facilitated by glycine's lack of side chain (Supplementary Material, Fig. S6). As a result of the substitution, the side chain of the resulting alanine is predicted to protrude into the groove. The surrounding residues at the end of the first beta-strand and beginning of the helix are involved in direct interaction with a guanosine nucleotide (GTP/GDP) bound at the exchangeable (E)-site of the beta-tubulin (24). Consequently, further subtle changes are predicted in the orientation of additional side chains involved with nucleotide binding, especially Gln11 and Cys12.
TUBB3
p.Glu288Lys is located within the ninth alpha-helix of the beta-tubulin subunit. When incorporated within a microtubule polymer, the acidic glutamate side chain is orientated laterally towards a beta-tubulin subunit of an adjacent protofilament. This residue is conserved through both alpha- and beta-tubulins, and substitution with a basic lysine residue is predicted to disrupt two putative hydrogen bonds between Glu288 and Thr285 within the same subunit (Supplementary Material, Fig. S7). TUBB3 p.Pro357Leu affects one of the two neighbouring prolines in a loop between beta-strands 9 and 10 of the beta-tubulin subunit. When incorporated within the polymer lattice, Pro357 faces into the microtubule lumen (Fig. 2B). This residue is not predicted to interact with longitudinal or laterally adjacent tubulin subunits.
Effect of tubulin gene variants on microtubule incorporation and polymerization in vitro
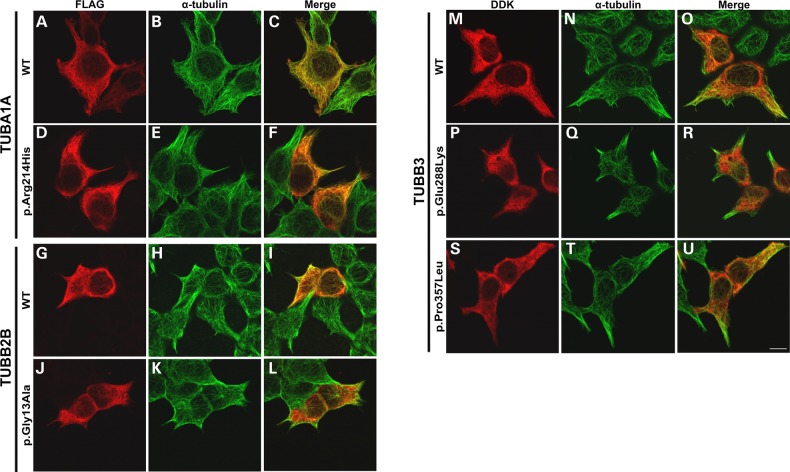
To investigate the in vitro functional consequences of tubulin gene variants identified, wild-type and variant TUBA1A, TUBB2B and TUBB3 constructs were transiently expressed in HEK293 cells and immunocytochemically stained with antibodies specific to endogenous and transgenic tubulin. Mutations had variable effects on co-assembly with endogenous alpha- or beta-tubulin subunits and incorporation into microtubule polymers (Fig. 3 and Table 3).
Figure 3.
Tubulin gene variants demonstrate varying levels of microtubule incorporation in vitro. Wild-type and variant FLAG-tagged TUBA1A (A–F), TUBB2B (G–L) and DDK-tagged TUBB3 (M–U) expression constructs were transfected into HEK293 cells to observe incorporation into microtubule polymers during interphase. Cells were fixed for immunocytochemical examination 24 h post-transfection and stained using anti-FLAG (A, D, G and J) or anti-DDK (M, D and G) antibodies to detect the transgenic tubulin and anti-α-tubulin (B, E, H, K, N, Q and T) to detect the endogenous microtubule network. TUBA1A p.Arg214His successfully incorporates into microtubule polymers, as evident by the presence of fibre-like arrangement in the anti-FLAG channel (D). In comparison to wild-type TUBB2B, p.Gly13Ala also demonstrates integration into microtubules but at a reduced level (J–L); polymer fibres are notably less well defined in the anti-FLAG channel in comparison to endogenous alpha-tubulin and an increased proportion of the visible p.Gly13Ala beta-tubulin remains unpolymerized in the cell cytoplasm. This is supported by predominantly green polymers observed in the merged cell image (L). Neither TUBB3 p.Glu288Lys nor p.Pro357Leu demonstrates any evidence of microtubule incorporation in vitro as indicated by diffuse cytoplasmic staining instead of defined microtubules (M–U). Although wild-type TUBB3 can be seen to co-assemble with endogenous alpha-tubulin into microtubule polymers, both variants remain unpolymerized throughout the cell cytoplasm. Images were acquired blind to construct type and are representative of >95% of the cells observed. Scale bar = 10 μm.
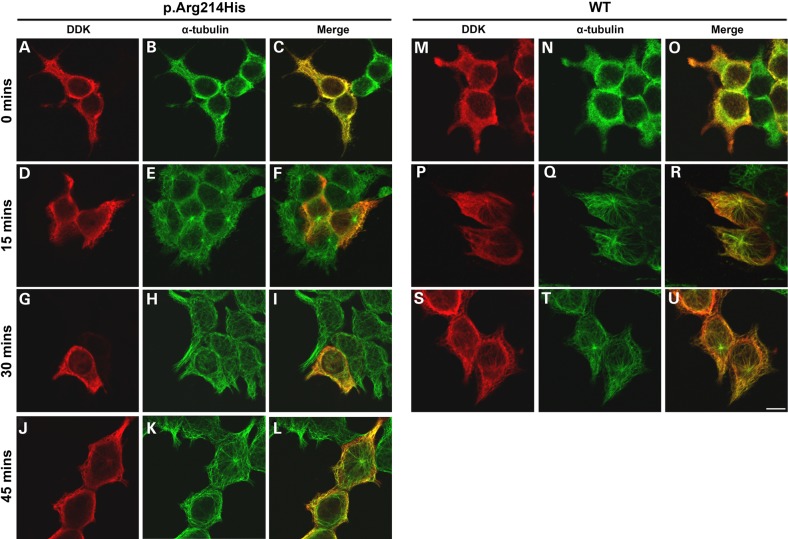
TUBA1A p.Arg214His demonstrates a diminished rate of microtubule reintegration, following cold-induced depolymerization in comparison to wild-type protein (Fig. 4). Despite its ability to polymerize, this suggests that the arginine-to-histidine substitution subtly perturbs the rate that affected heterodimers integrate into growing microtubules. TUBB2B p.Gly13Ala demonstrates a notable reduction in microtubule incorporation in vitro, with very faint polymers visible and the majority of variant beta-tubulin remaining unpolymerized throughout the cell cytoplasm. Neither of the TUBB3 variants demonstrates any signs of microtubule incorporation, as transgenic protein remains diffuse throughout the cell with no polymer arrangement observed.
Figure 4.
TUBA1A p.Arg214His demonstrates slowed microtubule reincorporation following cold-induced depolymerization. To examine the effect of the p.Arg214His amino acid substitution on microtubule dynamics in vitro, HEK293 cells expressing wild-type and variant TUBA1A were incubated at 4°C to induce microtubule polymer depolymerization. After 30 min, cover slips were returned to 37°C to promote microtubule re-polymerization. To observe the rate of microtubule growth in the presence of each construct, cells were removed after 0 (A–C and M–O), 15 (D–F and P–R), 30 (G–I and S–U) and 45 (J–L) min of polymer rescue and methanol-fixed prior to immunocytochemical staining. Wild-type TUBA1A (M–U) demonstrates reincorporation at a rate similar to endogenous tubulin. After 15 and 30 min at 37°C (D–I), microtubule repolymerization in TUBA1A p.Arg214His-expressing cells can be observed in the endogenous alpha-tubulin channel (E and H), whereas transgenic protein remains unpolymerized with no visible microtubules (D and G). TUBA1A p.Arg214His incorporation is evident after 45 min at 37°C (J); however, the proportion of variant tubulin appears to be at a markedly lower level than that of endogenous alpha-tubulin (L). Images were acquired blind to construct type and are representative of >95% of the cells observed. Scale bar = 10 μm.
Discussion
This study describes a highly recognizable pattern of cerebellar dysplasia, brainstem asymmetry and basal ganglia dysplasia without major cortical malformation in 10 patients with developmental delays and eye movement abnormalities. In the majority of the tested patients (78%), we identified mutations in TUBA1A, TUBB2B or TUBB3 that were either de novo or inherited from a mosaic parent. Four out of the five mutations identified are novel. Our data add a distinct and recognizable phenotype to the tubulinopathy spectrum, with relatively mild cortical involvement (12). Although the cohort presented is small, the mutation detection rate suggests that the described pattern of brain abnormalities is highly predictive for tubulin mutations. This detection rate is much higher than any previously published in patients with cortical malformations, ranging from 1 to 13.3% (6,8,9,12,13). We expect this number to be higher in selected cohorts of patients with cortical malformations combined with other abnormalities, (e.g. basal ganglia and corpus callosum), and indeed a detection rate of 30% has been found for the combination of lissencephaly with cerebellar hypoplasia (6).
Clinical implications
The Baraitser–Winter Neurogenetics Database (www.lmdatabases.com, v 1.0.32) includes 336 conditions encompassing cerebellar vermis hypoplasia or aplasia, making it challenging to identify specific genetic causes in patients with cerebellar malformations. A more detailed understanding of the imaging abnormalities associated with each genetic condition is essential for accurately diagnosing these patients. For example, identifying the characteristic cerebellar dysplasia of tubulinopathies distinguishes them from other disorders with cerebellar dysplasia that are often autosomal recessive and associated with other medical issues such as hearing loss (Chudley–McCullough syndrome), retinal dystrophy (Poretti–Boltshauser syndrome), muscular dystrophy (dystroglycanopathies), as well as progressive retinal, kidney and liver disease (Joubert syndrome).
Although previous studies have not focused on cerebellar abnormalities in tubulinopathies, cerebellar dysplasia has been mentioned in multiple patients (all with cortical malformations), confirming its association with tubulin gene mutations (11,12,15,16,19). In another study, up to 30% of the patients with lissencephaly with cerebellar hypoplasia had mutations in TUBA1A, as opposed to only 1% in the classic lissencephaly cohort (6). It would be interesting to investigate whether cerebellar dysplasia (without hypoplasia) is also a useful diagnostic marker for tubulin mutations in patients with lissencephaly.
Although OMA is commonly present in patients with cerebellar abnormalities, OMA has rarely been associated with tubulinopathies, so it is surprising that the majority of our patients presented with gross motor delays and OMA or other impairments of eye movement. In contrast, OMA has only been described in two patients with the distinct tubulinopathy H-ABC (hypomyelination with atrophy of the basal ganglia and cerebellum) due to TUBB4A mutations and never in patients with TUBA1A, TUBB2B or TUBB3 mutations (21). This may be due to under ascertainment of eye movement abnormalities in patients with severe disability.
Subtle cortical dysgenesis: dysgyria
No findings of lissencephaly, pachygyria, cobblestone cortex or polymicrogyria (abnormal cortical thickness, irregular cortical surface and grey–white boundary and/or microgyri) were present in the individuals recruited for this study (23). However, we observed abnormal sulci, the majority being shallow, with less frequent areas of sulci extending too deeply into the white matter, the latter predominantly in the perisylvian areas. We therefore consider the terms simplified gyral pattern, polymicrogyria and polymicrogyria-like cortical dysplasia to be potentially confusing. This is also supported by fetal pathology studies showing atypical features in polymicrogyria-like cortical dysplasia, suggesting a different pathophysiological mechanism from classic polymicrogyria (11,13). We propose using the term tubulinopathy-associated dysgyria for the subtle abnormalities of gyral shape without imaging evidence of lissencephaly, pachygyria, cobblestone cortex, polymicrogyria or other cortical abnormalities.
Recurrence risk
We identified two affected siblings with a TUBB2B mutation, inherited from an asymptomatic father who was found to be mosaic for this mutation. Familial recurrence has previously been described in two sisters with polymicrogyria and a TUBA1A mutation with mosaicism in the maternal line (5). These findings emphasize the importance of careful analysis of the parental sequencing results for evidence of mosaicism and adequate counselling about possible recurrence, as germline mosaicism in a parent can never be fully excluded.
Genotype–phenotype correlations
The same tubulin mutations are frequently observed in unrelated individuals sharing similar brain malformations, e.g. TUBA1A p.Arg264Cys in individuals with characteristic central pachygyria (3,9,12). Substitutions affecting identical residues but resulting in different amino acids have also produced comparable phenotypes (e.g. TUBA1A p.Arg402Cys and p.Arg402His) (6,12). Consistent with this, in our cohort of patients with strikingly similar MRI findings, we identified one (TUBB3 p.Pro357Leu) mutation in two unrelated patients and a second mutation (TUBB2B p.Gly13Ala) in two siblings. Contrary to this trend, the TUBA1A p.Arg214His variant found in UW168-3 was previously identified in a fetus with a more severe phenotype (11,12). Other phenotypic dissimilarities have also been described, e.g. TUBA1A p.Arg390Cys associated with both tubulinopathy-associated dysgyria (6) and asymmetrical perisylvian polymicrogyria (10). Although these instances are relatively uncommon, they limit the utility of genotype–phenotype correlations in clinical practice.
The large number of TUBA1A mutations that have been identified to date (49 at present), the majority of which are associated with severe brain malformations, makes it possible to evaluate whether in silico predictions such as CADD scores correlate with severity of the brain malformation. In fact, the TUBA1A variants associated with subtle cortical abnormalities have lower CADD scores than those associated with microlissencephaly, so it may someday be possible to use in silico predictions to improve prognostic information for patients and families.
Functional effects of mutations
Our data indicate that mutations in three different tubulin genes lead to a very consistent clinical phenotype, implicating a shared biological mechanism. Surprisingly, the mutations also affect diverse regions of the alpha-/beta-tubulin heterodimer, resulting in potential effects on microtubule dynamics, polymer incorporation or guanine nucleotide binding. TUBA1A p.Arg214His was associated with the mildest functional deficit in our in vitro pipeline, incorporating into microtubule polymers at comparable levels to wild-type but at a reduced rate. This mild deficit is consistent with the predicted subtle structural effects of TUBA1A p.Arg214His on the alpha-tubulin subunit. Despite residing at the interprotofilament interface, closer in silico inspection suggests that neither TUBA1A p.Arg214His nor p.Ile219Val directly affects protein–protein interactions. Interestingly, both variants are in close proximity to a p.Asp218Tyr substitution, previously identified in an individual with lissencephaly (6). This demonstrates how little one can predict of the resulting phenotype based purely on the affected region of tubulin proteins, as substitutions affecting adjacent residues can result in notably different brain abnormalities.
Structural predictions indicate that TUBB2B p.Gly13Ala affects conserved residues that are part of the nucleotide binding site. GTP hydrolysis at the beta-tubulin subunit is responsible for both polymer incorporation and dynamics (25). This is consistent with the reduced microtubule incorporation observed in vitro and relatively high CADD score, but would be the first association of a mutation potentially affecting guanine nucleotide binding with a mild cerebrocortical phenotype (12).
As with the TUBA1A variants, TUBB3 p.Glu288Lys and p.Pro357Leu substitutions are not predicted to affect specific intra- or intersubunit interactions; however, this observation may be irrelevant, as neither mutant protein incorporates into microtubules in vitro. We therefore hypothesize that the p.Glu288Lys and p.Pro357Leu substitutions prevent correct protein processing through one of several possible mechanisms including protein folding, alpha-/beta-tubulin heterodimerization or subsequent integration into growing microtubule polymers. This may additionally support the observation by Bahi-Buisson et al. (12) that impaired tubulin folding is generally associated with milder cortical phenotypes, while also emphasizing the necessity to support in silico predictive modelling with in vitro cellular analysis.
Based on our in vitro assays, TUBA1A p.Arg214His has the least effect on tubulin polymerization, whereas TUBB2B p.Gly13Ala has an intermediate effect, and the TUBB3 variants abolish polymerization entirely. The fact that all these mutations lead to the same brain phenotype could be due to a number of different factors, including: (i) a differential requirement for the tubulin proteins during brain development, with TUBA1A being more critical than the beta-tubulins. This may be due to functional redundancy between the different beta-tubulins which are expressed in overlapping patterns during brain development (26); (ii) similar functional effects for all of the mutations. Complete lack of mutant TUBB3 polymerization might result in a functional consequence similar to reduced polymerization of mutant TUBA1A, possibly due to negative effects of mutant TUBA1A incorporated into microtubules and (iii) genetic or environmental modifiers contributing to tubulin function and the resulting brain phenotype.
Conclusion
In summary, we have identified a highly characteristic combination of cerebellar, brainstem and basal ganglia abnormalities with subtle cortical involvement associated with mutations in three different tubulin genes TUBA1A, TUBB2B or TUBB3. Although tubulin mutations have been commonly associated with severe cortical malformations, this work emphasizes that the cortical involvement can be subtle in patients with more obvious brainstem and basal ganglia involvement. We confirm that a pipeline of in silico and in vitro work is necessary to add contextual evidence in tubulinopathies, and future work will focus on biochemical and cellular mechanisms by which tubulin mutations disrupt the development of the cortex, basal ganglia, cerebellum and brainstem.
Materials and Methods
Patient selection
From the large cohort of patients with hindbrain imaging abnormalities referred to the University of Washington Hindbrain Research Program, we identified patients with a distinct combination of brainstem asymmetry, superior cerebellar dysplasia and basal ganglia dysplasia and without a diagnosis of MCD (e.g. pachygyria and polymicrogyria). Clinical information was collected through chart review and structured interviews with the parents. All MRIs were systematically reviewed for abnormalities of the cortex, basal ganglia, white matter, ventricles, corpus callosum, hippocampus, brainstem, cerebellum, cerebellar vermis and cranial nerves (by R.O., G.E.I. and D.D.). When possible, DNA was collected from blood and/or saliva. Patients were enrolled through a human subjects research protocol approved at the University of Washington. Informed consent was provided by the patients, their parents or their legal guardians.
Genetic studies
TUBA1A (RefSeq accession: NC_000012.12), TUBB3 (NC_000016.10), TUBB2B (NC_000006.12) and TUBA8 (NC_000022.11) were sequenced using molecular inversion probe (MIP) capture and sequencing on an MiSeq, as described previously (27). The MIPs targeted all coding exons and 10 bp of exon-flanking intronic sequence. In patients without mutations, we performed Sanger sequencing of TUBB2A (NC_000006.12) and TUBB4A (NC_000019.10). All variants were confirmed by Sanger sequencing (primer sequences available on request) and checked against four online polymorphism databases: 1000 genomes (www.1000genomes.org), the Exome Variant Server (http://evs.gs.washington.edu/EVS/), dbSNP (http://www.ncbi.nlm.nih.gov/snp) and ExAC (http://exac.broadinstitute.org).
Bioinformatics analysis of tubulin gene variants
Combined Annotation-Dependent Depletion 1.0 (CADD; http://cadd.gs.washington.edu) bioinformatics software was used to analyse predicted deleterious effects of identified mutations on protein function (28). In addition, well-phenotyped TUBA1A, TUBB2B and TUBB3 variants included in a study by Bahi-Buisson et al. (12) were also analysed to enable comparison between mutations associated with unaffected/simplified gyration and with other, more severe, cortical abnormalities: microlissencephaly, lissencephaly, central pachygyria and central and general polymicrogyria-like cortical malformations.
Homology modelling
In silico structural predictions of wild-type and variant TUBA1A, TUBB2B and TUBB3 proteins were generated via a previously described homology modelling pipeline (29). The best homologies for these models were based on 99, 100 and 94% identities of alpha- and beta-tubulin templates (PDB: 4I4T) (30). Microtubule lattice architecture was based on a previously published template (PDB: 2XRP) (31).
Cell culture
Human embryonic kidney (HEK293) cell culturing, seeding and transfection were performed as described previously (22).
Expression construct generation
In vitro functional analysis of TUBA1A and TUBB2B variants was performed using expression constructs derived from a C-terminally FLAG-tagged TUBA1A (pRK5TUBA1A-CFLAG) clone, as published previously (6). pRK5TUBA1A-FLAG was subsequently modified to reflect wild-type TUBB2B (pRK5TUBB2B-CFLAG) by directional cloning. Tubulin gene variants were introduced into wild-type TUBA1A and TUBB2B, as well as a C-terminally DDK-tagged human TUBB3 expression construct (OriGene Technologies, USA; pCMV6 entry), using QuickChange site-directed mutagenesis (Stratagene, UK). Entire wild-type and variant transgene coding regions were sequence-validated following maxiprep yields (QIAgen, UK). The TUBA1A p.Ile219Val variant was only identified in the final stages of this study, and in vitro analysis was therefore not available. We included this patient due to the position of the affected amino acid, its de novo inheritance and the similarity of the resulting phenotype with that of others in the cohort.
Immunocytochemistry
HEK293 cells were fixed in −20°C methanol for 3 min and cell membranes subsequently permeabilized with phosphate-buffered saline (Sigma, UK) plus 0.5% Triton X-100 (Sigma) to facilitate immunocytochemical staining of intracellular epitopes. Cells expressing TUBA1A and TUBB2B constructs were stained using primary antibodies specific to FLAG (Sigma, Cat. F1804; 1:500 dilution), alpha-tubulin (Sigma, Cat. T6199; 1:500 dilution) and fluorescent-conjugated secondary antibodies raised against rabbit and mouse (Life Technologies, UK, Cat. A-11011 and A-11001). TUBB3-expressing cells were sequentially stained for transgenic and endogenous tubulin protein, as outlined previously (22). Immunostained cells were mounted onto glass slides using ProLong Gold Antifade Reagent (Life Technologies) and images acquired using confocal microscopy (Zeiss LSM 710 & Zen Software, Germany).
Microtubule polymerization assay
HEK293 cells expressing wild-type and variant tubulin gene constructs were incubated at 4°C to induce depolymerization of the microtubule polymer network. After 30 min, HEK293 cells were returned to 37°C. At specified times following their return to optimal temperature, cells were removed, methanol-fixed and immunostained to observe the rate of microtubule re-polymerization in the presence of each tubulin construct.
Supplementary Material
Funding
To M.I.R. lab: National Institute of Social Care and Health Research (NISCHR to M.I.R.), Epilepsy Research UK (ERUK to S.-K.C. and M.I.R.) and the Waterloo Foundation (to M.I.R.). To R.O.: EMBO short-term fellowship and the Simonsfonds from the Dutch Human Genetics Society (NVHG). To D.D.: University of Washington Pediatrics Department funds and private donations from families of children with brain malformations. The work was also supported by the University of Washington Intellectual and Developmental Disabilities Research Center Genetics Core (National Institutes of Health U54HD083091).
Supplementary Material
Acknowledgements
We express our deep appreciation to the patients and families who participated in this work. We also thank Jim Barkovich (University of California, San Francisco, CA, USA) for helpful discussions of MRI findings, the cortical abnormalities in particular.
Conflict of Interest statement. None declared.
References
- 1.Keays D.A., Tian G., Poirier K., Huang G.J., Siebold C., Cleak J., Oliver P.L., Fray M., Harvey R.J., Molnar Z. et al. (2007) Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell, 128, 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tischfield M.A., Cederquist G.Y., Gupta M.L., Jr., Engle E.C. (2011) Phenotypic spectrum of the tubulin-related disorders and functional implications of disease-causing mutations. Curr. Opin. Genet. Dev., 21, 286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahi-Buisson N., Poirier K., Boddaert N., Saillour Y., Castelnau L., Philip N., Buyse G., Villard L., Joriot S., Marret S. et al. (2008) Refinement of cortical dysgenesis spectrum associated with TUBA1A mutations. J. Med. Genet., 45, 647–653. [DOI] [PubMed] [Google Scholar]
- 4.Fallet-Bianco C., Loeuillet L., Poirier K., Loget P., Chapon F., Pasquier L., Saillour Y., Beldjord C., Chelly J., Francis F. (2008) Neuropathological phenotype of a distinct form of lissencephaly associated with mutations in TUBA1A. Brain, 131, 2304–2320. [DOI] [PubMed] [Google Scholar]
- 5.Jansen A.C., Oostra A., Desprechins B., De Vlaeminck Y., Verhelst H., Regal L., Verloo P., Bockaert N., Keymolen K., Seneca S. et al. (2011) TUBA1A mutations: from isolated lissencephaly to familial polymicrogyria. Neurology, 76, 988–992. [DOI] [PubMed] [Google Scholar]
- 6.Kumar R.A., Pilz D.T., Babatz T.D., Cushion T.D., Harvey K., Topf M., Yates L., Robb S., Uyanik G., Mancini G.M. et al. (2010) TUBA1A mutations cause wide spectrum lissencephaly (smooth brain) and suggest that multiple neuronal migration pathways converge on alpha tubulins. Hum. Mol. Genet., 19, 2817–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lecourtois M., Poirier K., Friocourt G., Jaglin X., Goldenberg A., Saugier-Veber P., Chelly J., Laquerriere A. (2010) Human lissencephaly with cerebellar hypoplasia due to mutations in TUBA1A: expansion of the foetal neuropathological phenotype. Acta Neuropathol., 119, 779–789. [DOI] [PubMed] [Google Scholar]
- 8.Morris-Rosendahl D.J., Najm J., Lachmeijer A.M., Sztriha L., Martins M., Kuechler A., Haug V., Zeschnigk C., Martin P., Santos M. et al. (2008) Refining the phenotype of alpha-1a tubulin (TUBA1A) mutation in patients with classical lissencephaly. Clin. Genet., 74, 425–433. [DOI] [PubMed] [Google Scholar]
- 9.Poirier K., Keays D.A., Francis F., Saillour Y., Bahi N., Manouvrier S., Fallet-Bianco C., Pasquier L., Toutain A., Tuy F.P. et al. (2007) Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A). Hum. Mutat., 28, 1055–1064. [DOI] [PubMed] [Google Scholar]
- 10.Poirier K., Saillour Y., Fourniol F., Francis F., Souville I., Valence S., Desguerre I., Marie Lepage J., Boddaert N., Line Jacquemont M. et al. (2013) Expanding the spectrum of TUBA1A-related cortical dysgenesis to polymicrogyria. Eur. J. Hum. Genet., 21, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fallet-Bianco C., Laquerriere A., Poirier K., Razavi F., Guimiot F., Dias P., Loeuillet L., Lascelles K., Beldjord C., Carion N. et al. (2014) Mutations in tubulin genes are frequent causes of various foetal malformations of cortical development including microlissencephaly. Acta Neuropathol. Commun., 2, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bahi-Buisson N., Poirier K., Fourniol F., Saillour Y., Valence S., Lebrun N., Hully M., Bianco C.F., Boddaert N., Elie C. et al. (2014) The wide spectrum of tubulinopathies: what are the key features for the diagnosis? Brain, 137, 1676–1700. [DOI] [PubMed] [Google Scholar]
- 13.Cushion T.D., Dobyns W.B., Mullins J.G., Stoodley N., Chung S.K., Fry A.E., Hehr U., Gunny R., Aylsworth A.S., Prabhakar P. et al. (2013) Overlapping cortical malformations and mutations in TUBB2B and TUBA1A. Brain, 136, 536–548. [DOI] [PubMed] [Google Scholar]
- 14.Guerrini R., Mei D., Cordelli D.M., Pucatti D., Franzoni E., Parrini E. (2012) Symmetric polymicrogyria and pachygyria associated with TUBB2B gene mutations. Eur. J. Hum. Genet., 20, 995–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaglin X.H., Poirier K., Saillour Y., Buhler E., Tian G., Bahi-Buisson N., Fallet-Bianco C., Phan-Dinh-Tuy F., Kong X.P., Bomont P. et al. (2009) Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat. Genet., 41, 746–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romaniello R., Arrigoni F., Bassi M.T., Borgatti R. (2015) Mutations in alpha- and beta-tubulin encoding genes: implications in brain malformations. Brain Dev., 37, 273–280. [DOI] [PubMed] [Google Scholar]
- 17.Romaniello R., Tonelli A., Arrigoni F., Baschirotto C., Triulzi F., Bresolin N., Bassi M.T., Borgatti R. (2012) A novel mutation in the beta-tubulin gene TUBB2B associated with complex malformation of cortical development and deficits in axonal guidance. Dev. Med. Child Neurol., 54, 765–769. [DOI] [PubMed] [Google Scholar]
- 18.Chew S., Balasubramanian R., Chan W.M., Kang P.B., Andrews C., Webb B.D., MacKinnon S.E., Oystreck D.T., Rankin J., Crawford T.O. et al. (2013) A novel syndrome caused by the E410K amino acid substitution in the neuronal beta-tubulin isotype 3. Brain, 136, 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirier K., Saillour Y., Bahi-Buisson N., Jaglin X.H., Fallet-Bianco C., Nabbout R., Castelnau-Ptakhine L., Roubertie A., Attie-Bitach T., Desguerre I. et al. (2010) Mutations in the neuronal ss-tubulin subunit TUBB3 result in malformation of cortical development and neuronal migration defects. Hum. Mol. Genet., 19, 4462–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breuss M., Heng J.I., Poirier K., Tian G., Jaglin X.H., Qu Z., Braun A., Gstrein T., Ngo L., Haas M. et al. (2012) Mutations in the beta-tubulin gene TUBB5 cause microcephaly with structural brain abnormalities. Cell Rep., 2, 1554–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simons C., Wolf N.I., McNeil N., Caldovic L., Devaney J.M., Takanohashi A., Crawford J., Ru K., Grimmond S.M., Miller D. et al. (2013) A de novo mutation in the beta-tubulin gene TUBB4A results in the leukoencephalopathy hypomyelination with atrophy of the basal ganglia and cerebellum. Am. J. Hum. Genet., 92, 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cushion T.D., Paciorkowski A.R., Pilz D.T., Mullins J.G., Seltzer L.E., Marion R.W., Tuttle E., Ghoneim D., Christian S.L., Chung S.K. et al. (2014) De novo mutations in the beta-tubulin gene TUBB2A cause simplified gyral patterning and infantile-onset epilepsy. Am. J. Hum. Genet., 94, 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barkovich A.J., Raybaud C. (2011) Pediatric Neuroimaging. Lipponcott Williams & Wilkins, Philadelphia. [Google Scholar]
- 24.Lowe J., Li H., Downing K.H., Nogales E. (2001) Refined structure of alpha beta-tubulin at 3.5 A resolution. J. Mol. Biol., 313, 1045–1057. [DOI] [PubMed] [Google Scholar]
- 25.Desai A., Mitchison T.J. (1997) Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol., 13, 83–117. [DOI] [PubMed] [Google Scholar]
- 26.Tischfield M.A., Baris H.N., Wu C., Rudolph G., Van Maldergem L., He W., Chan W.M., Andrews C., Demer J.L., Robertson R.L. et al. (2010) Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell, 140, 74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K. et al. (2012) Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science, 338, 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet., 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullins J.G. (2012) Structural modelling pipelines in next generation sequencing projects. Adv. Prot. Chem. Struct. Biol., 89, 117–167. [DOI] [PubMed] [Google Scholar]
- 30.Prota A.E., Bargsten K., Zurwerra D., Field J.J., Diaz J.F., Altmann K.H., Steinmetz M.O. (2013) Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science, 339, 587–590. [DOI] [PubMed] [Google Scholar]
- 31.Fourniol F.J., Sindelar C.V., Amigues B., Clare D.K., Thomas G., Perderiset M., Francis F., Houdusse A., Moores C.A. (2010) Template-free 13-protofilament microtubule-MAP assembly visualized at 8 A resolution. J. Cell Biol., 191, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.