Abstract
Human mitochondrial DNA (mtDNA) is replicated and repaired by the mtDNA polymerase gamma, polγ. Polγ is composed of three subunits encoded by two nuclear genes: (1) POLG codes for the 140-kilodalton (kDa) catalytic subunit, p140 and (2) POLG2 encodes the ∼110-kDa homodimeric accessory subunit, p55. Specific mutations are associated with POLG- or POLG2-related disorders. During DNA replication the p55 accessory subunit binds to p140 and increases processivity by preventing polγ's dissociation from the template. To date, studies have demonstrated that homodimeric p55 disease variants are deficient in the ability to stimulate p140; however, all patients currently identified with POLG2-related disorders are heterozygotes. In these patients, we expect p55 to occur as 25% wild-type (WT) homodimers, 25% variant homodimers and 50% heterodimers. We report the development of a tandem affinity strategy to isolate p55 heterodimers. The WT/G451E p55 heterodimer impairs polγ function in vitro, demonstrating that the POLG2 c.1352G>A/p.G451E mutation encodes a dominant negative protein. To analyze the subcellular consequence of disease mutations in HEK293 cells, we designed plasmids encoding p55 disease variants tagged with green fluorescent protein (GFP). P205R and L475DfsX2 p55 variants exhibit irregular diffuse mitochondrial fluorescence and unlike WT p55, they fail to form distinct puncta associated with mtDNA nucleoids. Furthermore, homogenous preparations of P205R and L475DfsX2 p55 form aberrant reducible multimers. We predict that abnormal protein folding or aggregation or both contribute to the pathophysiology of these disorders. Examination of mitochondrial bioenergetics in stable cell lines overexpressing GFP-tagged p55 variants revealed impaired mitochondrial reserve capacity.
Introduction
Multiple copies of the maternally inherited 16.569 kb mitochondrial DNA (mtDNA) reside within human mitochondria as circular, covalently closed molecules. Discrete regions of protein-mtDNA interactions known as nucleoids occur along the matrix-side of the mitochondrial inner membrane. Recent super-resolution microscopy techniques have revealed that each nucleoid harbors 1–3 mtDNA molecules (1,2) and each cell contains ∼300–800 nucleoids. The mtDNA genome is replicated and repaired by the mtDNA polymerase gamma, polγ. Polγ is composed of three subunits encoded by two nuclear genes: (1) POLG codes for the 140-kilodalton (KDa) catalytic subunit, p140 and (2) POLG2 encodes the ∼110-kDa homodimeric accessory subunit, p55. Underscoring the importance of mtDNA replication in human health, specific mutations are associated with POLG- or POLG2-related disorders (3). Mitochondrial diseases are devastating disorders that comprise a continuum of overlapping phenotypes and the age of onset of these diseases range from early childhood to late adulthood. The prevalence of autosomal recessive POLG-related disorders is estimated to be approximately 1 in 10 000 (4) and a recent study of newborn cord blood revealed that 1 in 200 infants harbored one of ten common pathogenic mtDNA mutations (5). POLG-related disorders are currently defined by six major phenotypes of neurodegenerative disease that include: Alpers–Huttenlocher syndrome (AHS), childhood myocerebrohepatopathy spectrum (MCHS), myoclonic epilepsy myopathy sensory ataxia (MEMSA), the ataxia neuropathy spectrum (ANS), autosomal recessive progressive external ophthalmoplegia (arPEO) and autosomal dominant progressive external ophthalmoplegia (adPEO) (4,6). In agreement with the requirement for mtDNA replication re-initiation between embryonic day (E)6 and 7.5 (7), the essential role of p140 in animal cells was demonstrated utilizing POLG knockout (KO) mice. The POLG KO resulted in embryonic lethality at E7.5–8.5 with subsequent depletion of mtDNA (8). Similarly, several studies have demonstrated the essential role of p55 in mtDNA replication: (i) two separate null mutations in the Drosophila melanogaster POLG2 gene caused lethality in the early pupal stage of fly development (9), (ii) homozygous POLG2 KO mice were embryonic lethal at E8–8.5 (10) and (iii) in a porcine oocyte knockdown model, oocyte maturation required POLG2 (11).
The p55 dimer is unique in comparison to other DNA polymerase processivity factors, as it shares primary amino acid sequence and tertiary structural conservation with prokaryotic aminoacyl-tRNA synthetases (12,13). The p55 accessory subunit has been well-characterized biochemically as a DNA-binding protein that regulates p140 activity (14–19). During DNA replication, p55 binds asymmetrically to the catalytic subunit and each monomer plays a distinct role in conferring processivity. By increasing processivity the p55 accessory subunit prevents polγ's dissociation from the template during DNA replication (20,21). The p140-proximal p55 subunit appears to strengthen the interaction with DNA while the distal p55 subunit is important for accelerating the nucleotide incorporation step (21). Evidence supporting tight association of p55 monomers within the homodimer comes from studies demonstrating that the affinity between monomers is extremely high with an estimated Kd of <0.1 nM (22) and the structure of the dimer interface is extensive at about 4000 Å2 (21). In the absence of genetic linkage and cosegregation analyses, four nuclear mutations encoding p55 disease variants that are biochemically defective at stimulating polγ activity have previously been identified, P205R, R369G, G451E and L475DfsX2 p55 (16,23–25) and Table 3. We predict that if homodimeric wild-type (WT) p55 molecules and defective variant p55 molecules could readily dissociate and re-associate, then polγ DNA synthesis reactions would fail or become defective. However, in support of the tight association between monomers, the presence of the G451E disease variant that by itself failed to stimulate polγ, had no effect on DNA synthesis when the ratio of homodimeric WT p55 to G451E was varied and then utilized in DNA synthesis reactions (16).
Table 3.
Summary of experimental data on human POLG2-related disease variantsa
| p55 Variant | Symptoms | Age at diagnosis (years) | Homodimer |
Heterodimer |
NGFP fusion | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| DNA synthesis | Affinity for p140 | Affinity for DNA | Protein stabilityb | DNA synthesis | Affinity for p140 | Affinity for DNA | Formation of puncta | |||
| WT | N/A | N/A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | + |
| P205R | Seizures, liver, hypotonia |
0.5 | ++ | +++ | ++ | ++ | +++ | N/A | N/A | – |
| R369G | PEO | 19, 55 | + | ++ | +++ | +++ | +++ | N/A | N/A | + |
| G451E | PEO | 60 | – | – | ++ | +++ | + | + | ++ | + |
| L475D fsX2 |
Seizures, blindness, hypotonia | 8 | – | – | – | – | +++ | N/A | N/A | – |
PEO, progressive external ophthalmoplegia.
aReferences for data contained in this table can be found in the text.
bAberrant protein stability was judged by analytical gel filtration chromatography and partially denaturing PAGE.
Because all currently identified patients with POLG2-disorders harbor heterozygous POLG2 mutations that alter evolutionary conserved residues (23,25), we hypothesize that affected individuals would harbor mixtures of variant and WT p55 molecules. As previous biochemical studies suggest that monomers within the homodimer do not readily dissociate and because we expect both POLG2 alleles to be expressed equally, p55 should occur as 50% heterodimers, 25% WT homodimers and 25% variant homodimers in vivo. In the literature to date, studies have only examined the biochemical consequences of p55 variant homodimers. To analyze the consequences of P205R, R369G, G451E and L475DfsX2 p55 heterodimeric disease variants, plasmids with one gene coding for WT p55 with an N-terminal Strep-tag (NStrep-p55) and a second gene encoding a WT or a disease variant with an N-terminal hexa histidine-affinity tag (NHis-p55) were constructed. Heterodimeric p55 disease variants harboring the two tags were purified utilizing a tandem affinity strategy and polγ biochemical analyses revealed a dominant negative p55 variant that impairs polγ activity. To complement the biochemical studies, WT and p55 disease variants were N-terminally tagged with green fluorescent protein (NGFP) and expressed in human embryonic kidney-293 (HEK293) cells to analyze the subcellular consequence of POLG2-disease mutations. In agreement with previous in vitro work suggesting protein misfolding or aggregation contributes to POLG2-related disorders, we have identified two variants that have altered intramitochondrial localization in vivo. Examination of mitochondrial bioenergetics revealed impaired mitochondrial reserve respiratory capacity in stable cell lines expressing NGFP-p55 disease variants.
Results
Purification of disease variant p55 heterodimers
In order to study heterodimeric p55 variants, a technique was developed to purify dual-tagged p55 dimers harboring one monomer with a His-tag and another monomer with a Strep-tag (Fig. 1A). Whole cell extracts (WCEs) were batch bound to Ni-NTA agarose followed by extensive washing and elution. Ni-NTA eluate was monitored for His-tagged protein by Bradford analysis and peak fractions were pooled. The eluted protein was then applied to a column containing Strep-Tactin (ST) resin to capture dual-tagged p55. The ST resin was extensively washed to remove homodimeric His-tagged protein then the dual-tagged p55 was eluted with desthiobiotin buffer. The ST eluate was subjected to an FPLC two-step approach to obtain highly purified p55 (Materials and Methods). Representative amount of each monomer in the purified dual-tagged WT p55 preparation were quantitated using a Western blot approach (Fig. 1B and C). To estimate the amount of each tagged monomer in the preparation the areas of the bands from 0.6 to 2.3 µg of homodimeric NHis-p55 and NStrep-p55 standards were plotted versus µg loaded to generate standard curves. The average amount of NHis-p55 monomer in the dimer was estimated to be 58 ± 11% (Fig. 1B) and that of the NStrep-p55 monomer to be 55 ± 20% (Fig. 1C), errors reported are standard deviation (SD). These results demonstrate that each monomeric species of the p55 dimeric preparation consist of a 1:1 ratio of NHis-p55 to NStrep-p55. The purification and Western blotting procedures were repeated for each of the p55 disease variant heterodimers and preparations were verified to contain 1:1 ratios of NHis-p55 variant:NStrep-p55.
Figure 1.
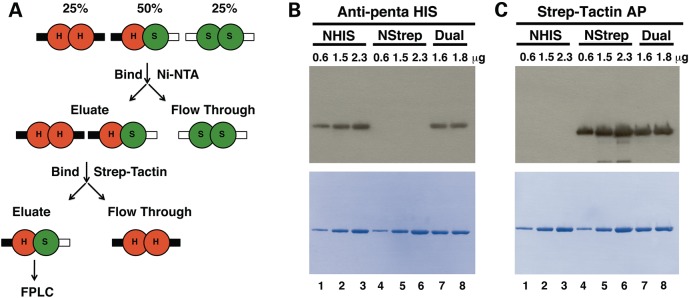
Purification of dual-tagged p55. (A) Purification scheme. See text for details. (B and C) Western blots of three forms of p55 probed with either (B) Anti-penta HIS antibody or (C) Strep-Tactin alkaline phosphatase (AP) conjugate. The amount of protein loaded per well in μg is indicated above each well of the blot. Lower panels of (B) and (C) are Coomassie stained immunoblots following autoradiography and quantitation, and emphasize proper sample loading, high specificity of each probe for its tag and for each tag for its resin. WT NHis-p55 dimers [H in (A) and NHIS in (B) and (C)]; WT NStrep-p55 dimers (S and NStrep); dual-tagged WT NHis-p55/NStrep-p55 dimers (H/S and dual).
Screening for functional interaction between heterodimeric p55 disease variants and p140
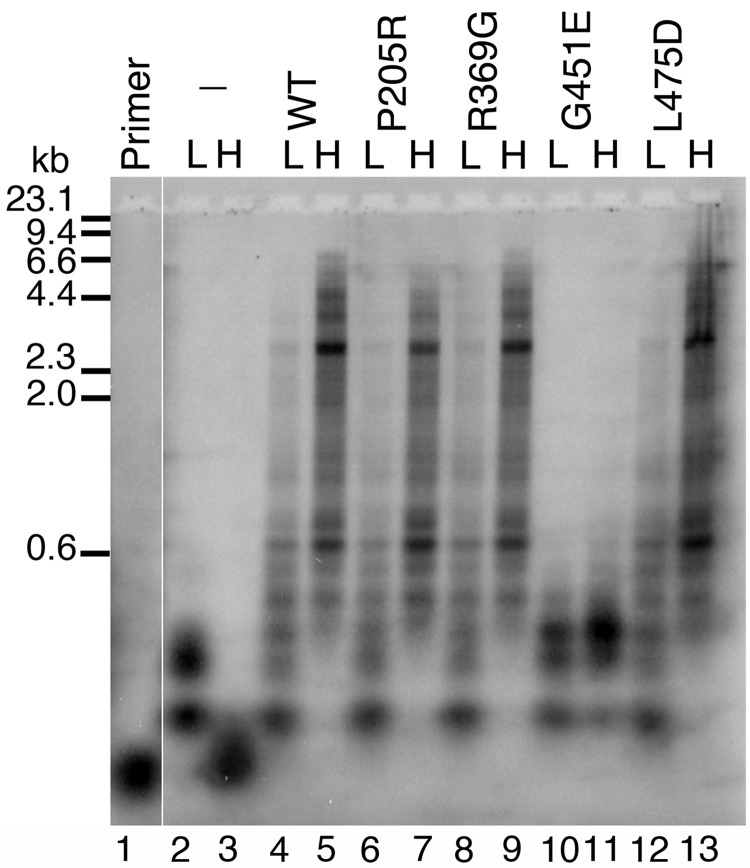
Previously we demonstrated that homogeneous preparations of P205R p55 and R369G p55 have a decreased ability to stimulate p140 while L475DfsX2 p55 and G451E p55 completely fail to stimulate the catalytic subunit (16,23). To screen heterodimeric variants for their ability to stimulate polγ in vitro we used an end-labelled primer extension functional assay (Fig. 2). As judged by alkaline gel electrophoresis and by denaturing polyacrylamide gel electrophoresis (PAGE) (Supplementary Material, Fig. S1), p140 alone can synthesize a maximum of ∼150 nucleotides past the primer in the absence of the accessory subunit and supplemental salt (Fig. 2, lane 2). The ability of the isolated catalytic subunit to extend primers becomes severely inhibited in reactions supplemented with 150 mm NaCl (Fig. 2, compare lane 3 and lane 2), and significant stalling and/or dissociation was also evident by denaturing PAGE (Supplementary Material, Fig. S1). Addition of the dual-tagged WT p55 accessory subunit in the reaction significantly stimulates primer extension at low salt (Fig. 2, compare lanes 4 and 2) and the higher physiological salt concentration further enhances both the quantity and the length of primer extension products to as much as ∼7 kb or the full length of the M13mp18 template (Fig. 2, compare lanes 5 and 4). P205R, R369G and L475DfsX2 p55 heterodimeric disease variants displayed the same degree of stimulation as WT p55 at both low and high salt (Fig. 2). These results suggest that when P205R, R369G and L475DfsX2 variants combine with a WT p55 subunit, they retain the ability to stimulate processive DNA synthesis.
Figure 2.
Functional interaction of p55 heterodimer variants with the catalytic subunit. Reactions were analyzed by alkaline gel electrophoresis and contained 2 nM singly primed M13mp18 DNA, 5 nM p140 and 5 nM dual-tagged WT p55 or heterodimeric variant. L, low salt (none added to the reaction); H, high salt (reactions supplemented with 150 mm NaCl); WT, dual-tagged WT p55 WT; P205R, WT/P205R; R369G, WT/R369G; G451E, WT/G451E; L475D, WT/L475DfsX2 p55 heterodimers. The hyphen (–) indicates the absence of p55 in these reactions (p140 only). Primer, no protein added to the reaction. Markers, λ-DNA/Hind III fragments labeled with [α-32P]dTMP. Log(λ-DNA/Hind III nucleotide fragments) was plotted against band migration distance (Millimeters), and linear regression generated a correlation coefficient of 0.98 over the range analyzed (23.1, 9.4, 6.6, 4.4, 2.3, 2.0 and 0.6 kilo-nucleotides). This standard curve was used to estimate the size of polγ products observed on the alkaline gel whereas shorter products were observed directly on denaturing PAGE (Supplementary Material, Fig. S1). Full-length primer extension products are 7249 nucleotides in length.
In contrast, at low and high salt concentrations polγ harboring the WT/G451E p55 heterodimer was severely inhibited in the ability to extend end-labelled primers (Fig. 2 lanes 10 and 11). Maximal synthesis of less than 600 nucleotide length products was observed at both salt concentrations. Interestingly, the inhibition by the heterodimer is distinct from the complete failure to interact with the mtDNA polymerase observed for the G451E homodimer, which synthesized DNA products less than 150 nucleotides in length at low salt and less than 50 nucleotides at high salt, identically to reactions containing the isolated catalytic subunit (16,23). Even though the lengths of the DNA products are slightly longer than those synthesized by the isolated catalytic subunit, the WT/G451E heterodimer fails to synthesize full-length WT holoenzyme extension products. This experiment demonstrates that one G451E monomer in the WT/G451E heterodimer is dominant negative and ‘poisons’ the functional interaction between the accessory subunit and the p140 catalytic subunit. Furthermore, the defective DNA synthesis associated with polγ harboring the WT/G451E p55 heterodimer supports previous observations that monomers within the p55 dimer are strongly associated and likely do not re-assort during the course of p55 purification or during biochemical reactions.
Polγ harboring the WT/G451E p55 heterodimer incorporates dTMP less efficiently than WT polγ
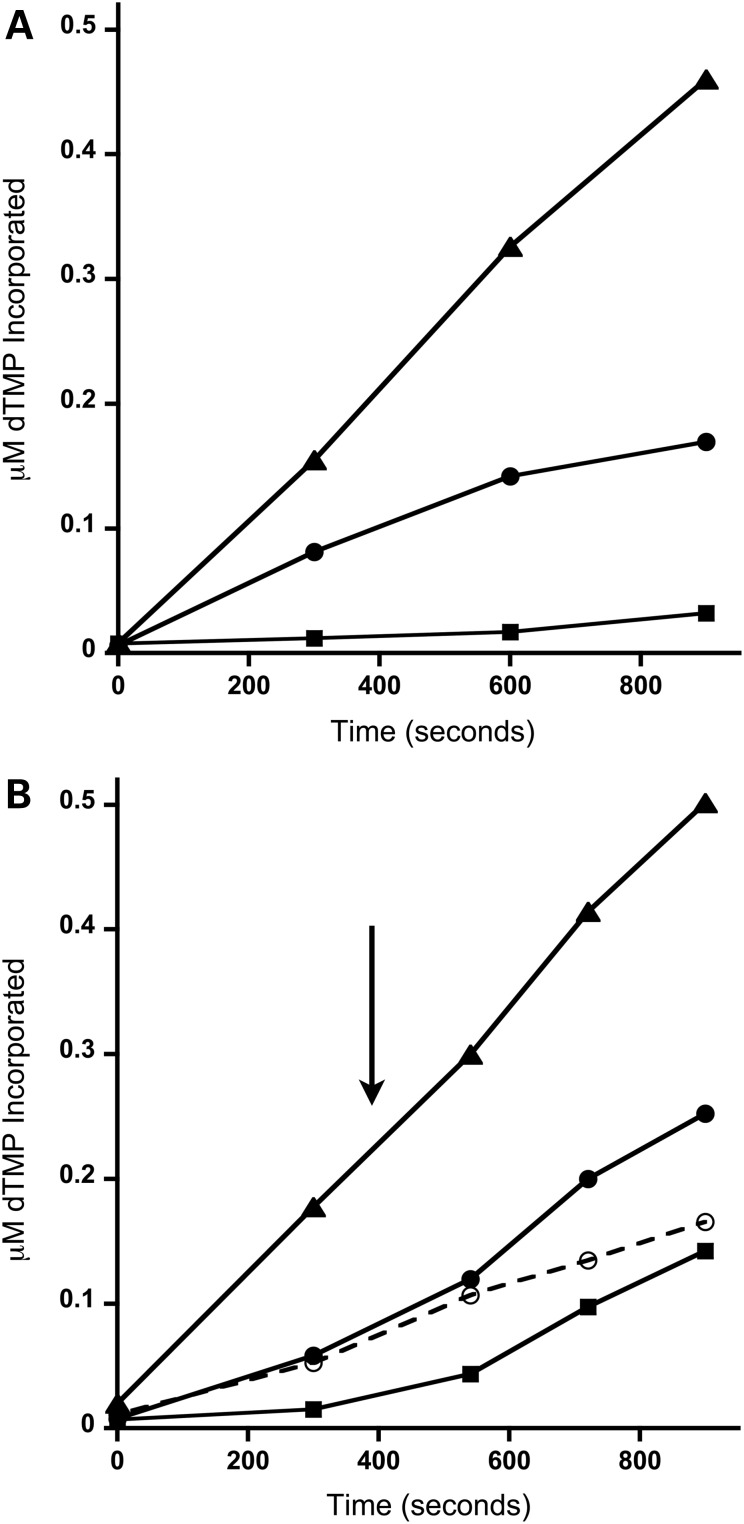
To further investigate the dominant negative effect observed for the PEO-associated WT/G451E p55 heterodimer we employed a steady-state DNA polymerase assay. In the absence of the accessory subunit the steady-state rate of dTMP incorporation into DNA by p140 was estimated to be ∼27-fold less than the holoenzyme (compare the kcat of 0.02 for p140 with the kcat of 0.53 for the holoenzyme, Table 1 and Fig. 3A). In agreement with the defective mtDNA polymerase activity observed in the functional assay, the observed kcat of the WT/G451E p55 heterodimer containing polγ was 2.3-fold less than that of WT p55 (Fig. 3A and Table 1). Additionally, supplementation of reactions containing the WT/G451E p55 heterodimer with a molar equivalent of WT/G451E p55 heterodimer had no effect on kcat (Fig. 3B, dashed line and Table 1). Interestingly, mtDNA polymerase activity could be restored to the WT holoenzyme level by supplementing the WT/G451E p55-containing polγ reaction with an equivalent of WT p55 (Fig. 3B) resulting in an increase of kcat by ∼2-fold (Table 1). The restoration of mtDNA polymerase activity by addition of WT p55 was also observed for reactions initially containing the catalytic subunit only.
Table 1.
Estimated dissociation and rate constants
| Kd(DNA) (nM)a | kcat (s−1)b |
kcat (s−1)b Spiked with WT p55c |
|
|---|---|---|---|
| Isolated p140 | N/A | 0.02 ± 0.001 | 0.27 ± 0.01 |
| p140 + WT/WT p55 | 24 ± 9 | 0.53 ± 0.02 | 0.56 ± 0.04 |
| p140 + WT/G451E p55 | 63 ± 2 | 0.23 ± 0.01 | 0.37 ± 0.05d |
Kd(DNA) and kcat values were estimated as described under Materials and Methods.
aValues reported are averages with standard deviations.
bStandard error estimates were derived by linear regression analysis to calculate apparent kcat values.
cA 1 nMolar equivalent of WT p55 was added to the reaction at 6.5 min and kcat values were estimated using linear regression post WT p55 addition. See Figure 3 and Materials and Methods.
dWhen a 1 nMolar equivalent of G451E p55 heterodimer is added to the G451E p55 heterodimer reaction in place of WT p55 then the estimated kcat post G451E p55 heterodimer addition is 0.2 ± 0.01, standard error estimate derived by linear regression analysis.
Figure 3.
The WT/G451E p55 heterodimer is dominant negative in a polγ steady-state polymerase assay. (A) DNA polymerase time course analysis of WT polγ (triangles), polγ harboring WT/G451E heterodimers (circles) and p140 catalytic subunit only (squares). Samples were collected at 0, 5, 10 and 15 min. (B) Addition of WT p55 during the DNA polymerase time course stimulates polγ harboring WT/G451E p55 heterodimers in vitro. The arrow represents the addition of 1 nM WT p55 to the reactions at 6.5 min (solid lines) or the addition of 1 nM WT/G451E p55 (dashed line). Symbols are as in (A). Samples were collected at 0, 5, 9, 12 and 15 min. A representative experiment is shown in the figure and the analyses were performed twice.
The WT/G451E p55 heterodimer has weakened binding affinities for double-stranded DNA and for p140
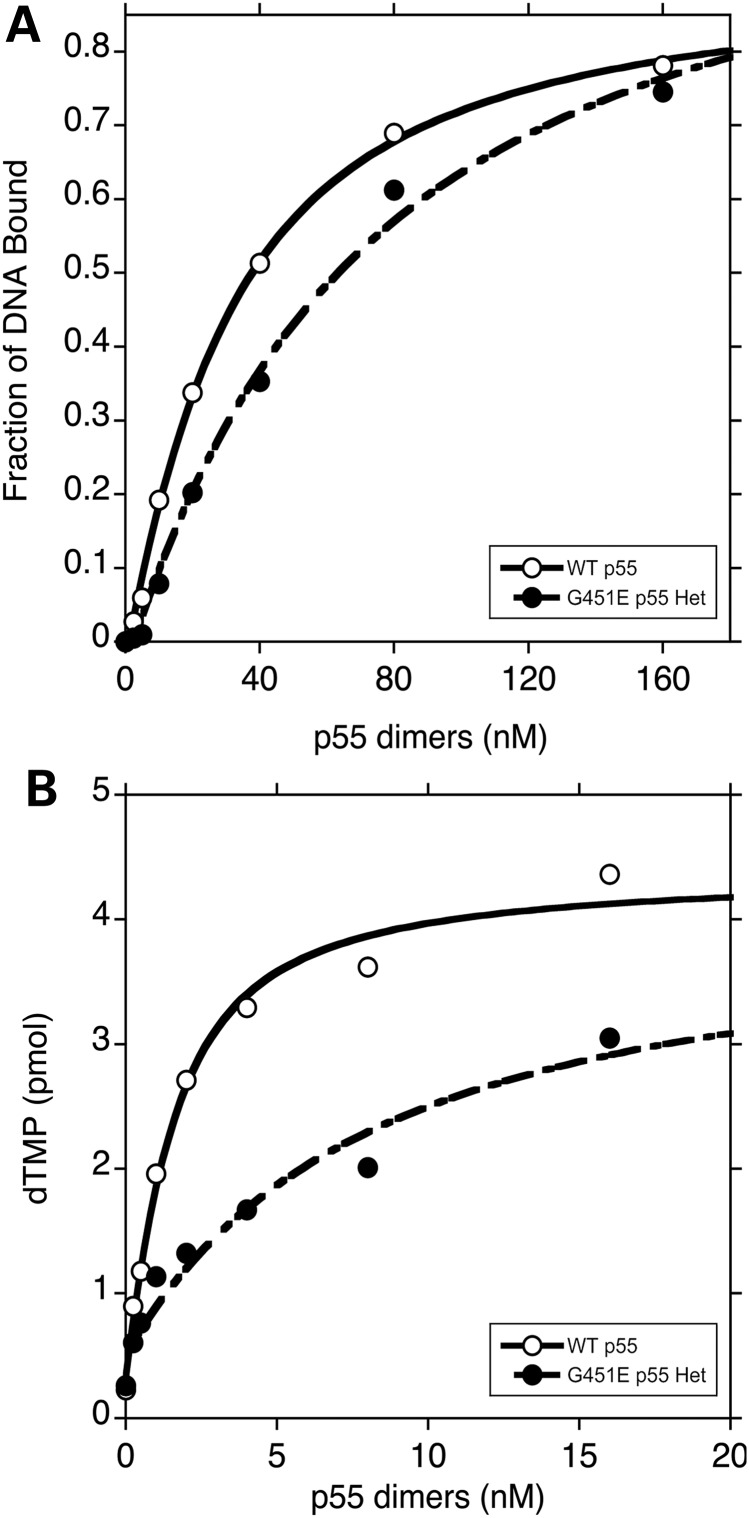
To determine whether or not the WT/G451E p55 heterodimer impairs polγ activity due to a weakened affinity for double-stranded DNA (dsDNA) or due to a weakened affinity for p140 we utilized two assays: (1) a synthetic dsDNA was employed in an electrophoretic mobility shift assay (EMSA) to estimate the dissociation constants for dsDNA binding, Kd(DNA), and (2) a mtDNA polymerase functional assay was exploited to estimate the dissociation constants for p140 binding, Kd(p140). We observed an apparent ∼3-fold decrease in DNA-binding affinity for the WT/G451E p55 heterodimer as compared with WT p55, 63 ± 2 and 24 ± 9 nM respectively (Fig. 4A and Table 1). However, the WT/G451E displayed a stronger affinity for DNA as compared with the G451E homodimer, which when compared with WT p55 was decreased by 6-fold (23).
Figure 4.
The WT/G451E p55 heterodimer has weakened binding affinities for dsDNA and for p140. (A) Determination of the dissociation constants for DNA binding, Kd(DNA). Representative binding isotherms for WT p55 (open circle) and WT/G451E (closed circle) p55 binding to end-labeled double-stranded oligonucleotides are presented. Reactions (20 µl) contained 15 nM dsDNA and varying amounts of accessory subunits. DNA-binding reactions were resolved by EMSAs as described in Materials and Methods. (B) Determination of the dissociation constants for p140 binding, Kd(p140). Representative binding isotherms for accessory subunits binding to p140 as measured by DNA polymerase activity on poly(dA)•p(dT)12–18 are presented. Reactions (50 µl) contained 1 nM p140 and varying amounts of WT (open circle) or WT/G451E (closed circle) p55 heterodimer. Experiments were repeated twice independently. The dissociation constants were calculated by quadratic curve fitting as described in Materials and Methods.
To determine the physical interaction between the heterodimeric WT/G451E accessory subunit and the catalytic subunit we performed a functional steady-state kinetic assay on poly(dA)•p(dT)12–18 at the physiological salt concentration of 150 mm NaCl. Under these conditions, no appreciable DNA synthesis is observed by the isolated catalytic subunit and DNA synthesis is only detected when p140 and p55 interact thereby increasing the affinity of polγ for DNA. This enhanced affinity was assessed over a range of protein ratios, permitting the construction of binding isotherms and the estimation of inter-subunit dissociation constants (Fig. 4B). In this assay we observed an ∼5-fold weaker Kd(p140) for the heterodimer in comparison to WT p55 dimers, 5.5 ± 2.2 and 1.2 ± 0.2 nM, respectively (Fig. 4B). However, the heterodimer displayed a much stronger affinity for p140 as compared with the G451E homodimer, which when compared with WT p55 was decreased by 316-fold (23). Therefore, in comparison to the WT p55, the WT/G451E p55 heterodimer has weaker binding affinities for both the catalytic subunit and for dsDNA.
WT NGFP p55 colocalizes with mtDNA-containing nucleoids within the mitochondrial reticulum of HEK293 cells
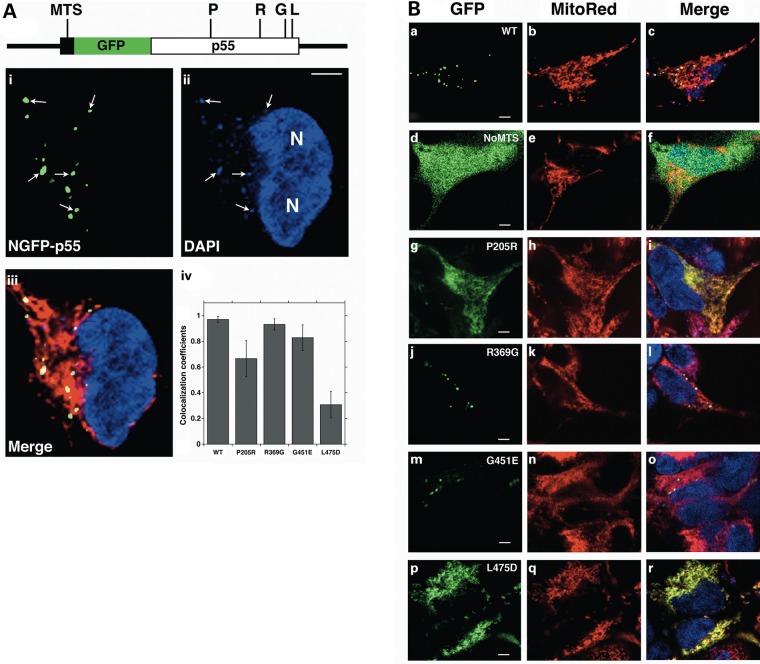
Immunocytochemistry analysis has demonstrated that in 143B osteosarcoma cells p55 colocalizes with the mitochondrial twinkle helicase to form puncta within the mitochondrial reticulum (26). To complement our biochemical studies and assess the in vivo effects of these disease variants, we transfected HEK293 cells with a WT p55 mammalian expression plasmid for live-cell imaging (Materials and Methods). The plasmid encodes p55 with an amino-terminal green fluorescent protein tag (WT NGFP p55). Cellular DNA was fluorescently labeled with 4′,6-diamidino-2-phenylindole (DAPI), and MitoTracker® Red CMXRos was employed to label mitochondria (Fig. 5). To quantify WT NGFP p55 localization to mitochondria and to mtDNA we calculated colocalization coefficients based on dual-color images for both green relative to red (p55 colocalized to mitochondria) and green relative to blue (p55 colocalized to mtDNA) fluorescence emissions according to Manders et al. (27) (Materials and Methods). The average weighted green relative to red colocalization coefficient was estimated to be 0.92 ± 0.04 (Table 2) providing strong evidence that NGFP p55 localizes to the mitochondrial reticulum. Furthermore, the average green relative to blue coefficient was estimated to be 0.98 ± 0.01 (error is SD) confirming localization to mtDNA. The green-to-blue coefficient was calculated from four WT NGFP p55 images from two experiments containing a total of 74 cells (cell number was determined by counting DAPI-stained nuclei). Interestingly, we observed that some DAPI-stained mtDNA do not localize with NGFP p55 and may represent mtDNA that are not bound by polγ or actively replicating. A representative image is shown in Figure 5A and arrows emphasize WT NGFP p55 molecules colocalized with DAPI-stained mtDNA. Taken together, our results support that following translation of the NGFP-POLG2 mRNA on cytoplasmic ribosomes, WT NGFP-p55 is correctly imported into mitochondria, folded correctly within the mitochondrial matrix and localized to DAPI-stained mtDNA-containing nucleoids.
Figure 5.
P205R and L475DfsX2 p55 disease variants fail to form discrete mitochondrial nucleoids. (A) Linear representation of the NGFP p55 primary amino acid sequence. The amino-terminal MTS is shown as a black rectangle while GFP and p55 are shown as green and white rectangles respectively. The location of the P205R, R369G, G451E and L475DfsX2 p55 disease variant substitutions are indicated by P, R, G and L on the linear map respectively. WT NGFP p55 forms i. green punctate structures that ii. colocalize with DAPI-stained mtDNA nucleoids within the iii. red mitochondrial reticulum of cultured HEK293 cells. Two DAPI-stained nuclei are labeled ‘N’ in ii. while in i. and ii. five mtDNA nucleoids are emphasized with white arrows. iv. Quantitation of p55 disease variant intramitochondrial mistargeting. Mito Red (MitoTracker® Red) to green GFP fluorescence colocalization coefficients quantifies the failure of P205R- and L475DfsX2-p55 to form puncta. Values reported are average weighted co-localization coefficients with standard deviations (error bars). WT, NGFP p55 (65 cells); P205R, NGFP P205R p55 (52 cells); R369G, NGFP R369G p55 (60 cells); G451E, NGFP G451E p55 (60 cells); L475D, NGFP L475DfsX2 p55 (80 cells). Three images of each variant were used with total number of cells indicated in parentheses. (B) Representative images of WT NGFP p55 (WT, a–c), NoMTS NGFP p55 (NoMTS, d–f), NGFP P205R p55 (P205R, g–i), NGFP R369G p55 (R369G, j–l), NGFP G451E p55 (G451E, m–o) and NGFP L475DfsX2 p55 (L475D, p–r). Merge, blue, green and red fluorescence channels are merged together in (A) iii. and (B) c, f, i, l, o and r. Scale bars in (A) and (B) are 4 µm.
Table 2.
Average weighted colocalization coefficients for WT NGFP p55 and disease variants
| NGFP constructa | Average weighted colocalization coefficients: Green col. redb | Average weighted colocalization coefficientb,c |
|---|---|---|
| WT | 0.92 ± 0.04 | 0.96 ± 0.03 |
| P205R | 0.89 ± 0.03 | 0.67 ± 0.14 |
| R369G | 0.76 ± 0.21 | 0.93 ± 0.04 |
| G451E | 0.76 ± 0.24 | 0.83 ± 0.10 |
| L475D | 0.78 ± 0.05 | 0.31 ± 0.10 |
| NoMTSd | 0.22 ± 0.05 | N/A |
aWT, NGFP p55 (55 cells); P205R, NGFP P205R p55 (52 cells); R369G, NGFP R369G p55 (60 cells); G451E, NGFP G451E p55 (60 cells); L475D, NGFP L475DfsX2 p55 (80 cells); NoMTS, NoMTS NGFP p55 (87 cells). Total number of cells analyzed from 18 separate pictures is indicated in parentheses.
bValues reported are averages with standard deviations.
cFor ease of visualization the red relative to green colocalization coefficient was set to 1 and represents the ability of NGFP p55 to form puncta. The coefficients for each image were calculated by subtracting the weighted red channel colocalized with green channel coefficient from 1 then calculating the average and standard deviation for each variant. Thus, the lower the average weighted red channel colocalized to green channel coefficient the weaker the ability to form puncta and vice versa. This datum is represented graphically in Figure 5 A panel iv.
dThe diffuse nature of NoMTS p55 construct within the cytoplasm precluded meaningful quantitation of the coefficient for the red channel colocalized with the green channel; however, we observed a 4.2-fold decrease relative to WT p55 in the green relative to red colocalization coefficient.
The native p55 mitochondrial targeting sequence is required for mitochondrial import
Computational methods predict a mitochondrial targeting sequence (MTS) of 25 amino acid residues for p55 (28). To determine the role of the presumed p55 MTS we utilized site-directed mutagenesis to construct a plasmid lacking the encoded MTS, which we named NoMTS NGFP p55 (Materials and Methods). When the NoMTS plasmid was transfected into HEK293 cells, the GFP-tagged accessory subunit lacking the MTS failed to localize within mitochondria as indicated by the diffuse green fluorescence observed within the cytoplasm (Fig. 5B panels d–f show a representative image). The failure of p55 lacking the MTS to localize within mitochondria was indicated by the calculated average green relative to red colocalization coefficient of 0.22 ± 0.05, which is 4.2-fold less than WT NGFP p55 (Table 2). This result confirms that in vivo the N-terminal 25 amino acid residues are required to direct p55 to the mitochondrion.
The NGFP-P205R and NGFP-L475DfsX2 p55 variants fail to form WT p55-like puncta
Site-directed mutagenesis was utilized to separately introduce POLG2-disease mutations into the WT NGFP p55 plasmid template (Materials and Methods). Similar to WT NGFP p55, all NGFP-tagged p55 disease variants expressed in the HEK293 cells localize to mitochondria as confirmed by the estimated green relative to red colocalization coefficients that ranged from 0.76 ± 0.24 to 0.89 ± 0.03 (Table 2). Both G451E and R369G variants form punctate nucleoid structures similar to WT p55 (Fig. 5A iv and B j–l and m–o). Strikingly, both the P205R and L475DfsX2 p55 variants did not form WT p55-like punctate structures, but instead had irregular diffuse green fluorescence throughout the mitochondrial reticulum (Fig. 5A iv. and B g–i and p–r and Table 2). Previously we observed an inability or weakened ability of these homodimeric variants to bind mtDNA and the L475DfsX2 p55 variant formed aberrant oligomeric complex when analyzed by analytical gel filtration chromatography (23).
Cell lines expressing NGFP p55 disease variants have impaired reserve respiratory capacity
Following plasmid transfection, geneticin (G418) was utilized to select stable cell lines expressing NGFP-tagged WT or disease variant p55s (Materials and Methods). Consistent with previous studies on other cell types (29–31), we utilized galactose-based medium to force ATP-linked oxidative phosphorylation (OXPHOS) over glycolysis in HEK293 cell lines. OXPHOS is the process of coupling substrate oxidation to ATP production. The Seahorse XF24 Extracellular Flux Analyzer tissue culture plate contains 24 wells to simultaneously monitor cellular oxygen consumption rates (OCR, a measure of OXPHOS) and extracellular acidification rates (ECAR, a measurement of intracellular lactate production and glycolysis) in 24 transient microchambers. In the standard XF24 bioenergetics assay, basal OCRs or basal respiration is initially measured followed by the addition of OXPHOS inhibitors.
The OCR profiles for HEK293 cell lines expressing NGFP WT p55, disease variants and age-matched HEK293 cells were determined (Fig. 6 and Supplementary Material, Fig. S2). First, oligomycin was delivered into the microchambers to inhibit ATP synthesis by blocking the proton channel of the mitochondrial ATP synthase. Oligomycin treatment resulted in a rapid decrease in OCR allowing measurement of ATP-linked respiration or ATP production. Next, the proton ionophore 2,4-dinitrophenol (DNP) was added to collapse the membrane potential (ΔΨ) by transporting hydrogen ions across the mitochondrial inner membrane. DNP treatment results in an accelerated consumption of oxygen without ATP synthesis and allows measurement of maximal OCR or maximal respiration. Last, rotenone and antimycin A were delivered, which inhibit complex I and III of the mitochondrial electron transport chain respectively. Following treatment with each inhibitor, OCRs are measured to determine bioenergetic parameters such as reserve respiratory capacity, proton leak and non-mitochondrial oxygen consumption (Materials and Methods). In comparison to HEK293 cells expressing NGFP WT p55, we detected no differences in basal OCR, maximal OCR, ATP-linked oxygen consumption or proton leak in cell lines expressing p55 disease variants (Fig. 6 and Supplementary Material, Fig. S3). However, 6- to 16-fold decreases in reserve respiratory capacity were detected in all NGFP-p55 disease variant cell lines, whereas reserve respiratory capacity remained virtually unchanged for NGFP WT p55 (Fig. 6D).
Figure 6.
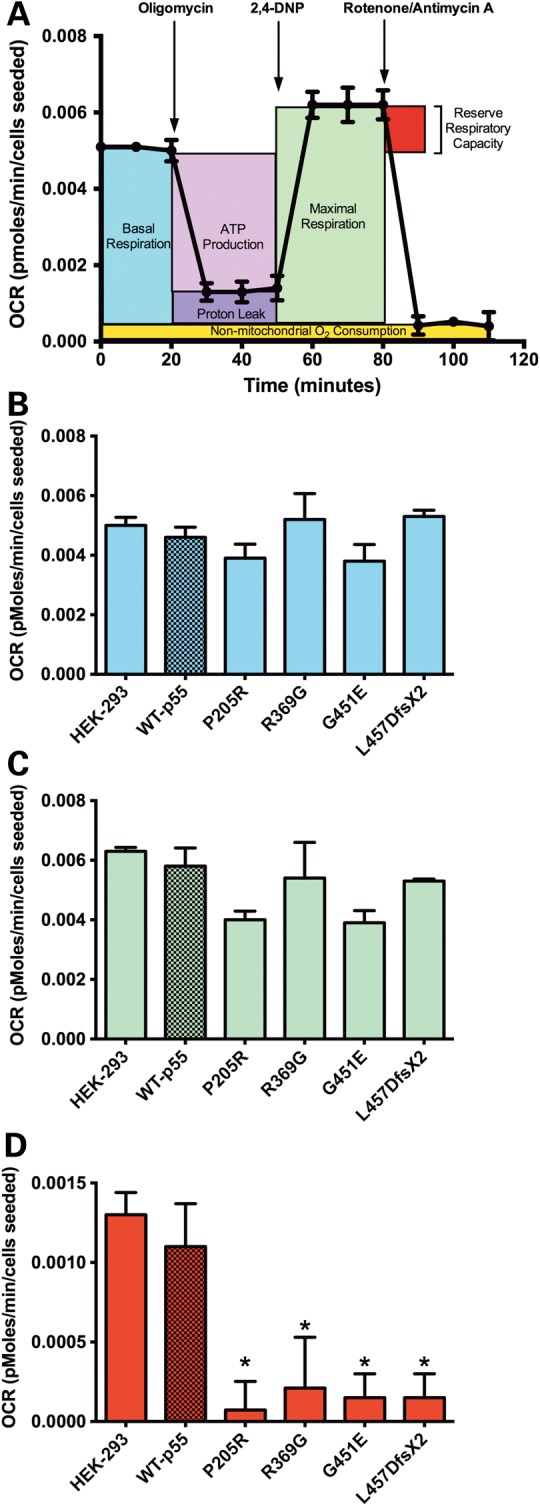
Cell lines harboring POLG2 variants have impaired reserve capacity. (A) Representative OCR profile of the mitochondrial bioenergetic parameters of WT NGFP p55 cells grown in galactose medium. By inhibiting ATP synthase with oligomycin, basal OCR can be differentiated into ATP-linked oxygen consumption and that which is required to overcome the natural proton leak. Maximal oxygen consumption is determined after the injection of the uncoupler, 2,4-DNP, while respiratory reserve capacity is the difference between maximal and basal OCR. Completely inhibiting respiration with the ETC inhibitors rotenone and antimycin A reveals non-mitochondrial oxygen consumption. The average (B) basal OCR, (C) maximal OCR and, (D) reserve respiratory capacity for untransfected HEK293 cells and cell lines expressing the NGFP p55 variants and WT NGFP p55 are shown. Data are the means ± SD, n = 7 per group, normalized to cells seeded, *P ≤ 0.05 versus WT NGFP p55 control as determined by Student's t-test.
Discussion
Characterization of the role of POLG2 mutations in mitochondrial disease has been complicated by a lack of family genetics; however, an understanding of mutation pathogenicity has come from several reports analyzing biochemical defects in p55 disease variants (16,23–25). Previously we identified novel POLG2 mutations by examining a cohort of 112 patients that exhibited heterogeneous symptoms resembling POLG-related disease but lacking POLG gene mutations and demonstrated that p55 homodimer disease variants are defective in their ability to stimulate the polγ catalytic subunit, p140 (23). To address the consequence of heterodimer disease variants, we developed a tandem affinity strategy to isolate p55 heterodimeric disease variants for biochemical characterization. To complement the biochemical studies we also designed mammalian expression plasmids encoding WT NGFP p55 and NGFP disease variants to analyze the subcellular consequence of POLG2-disease mutations. Furthermore, we examined the bioenergetics of cell lines expressing NGFP p55 disease variants.
The POLG2 exon 8 c.1352G>A mutation encoding G451E p55 is dominant negative
Previously a 60-year-old female with adPEO caused by a heterozygous POLG2 mutation was identified (16). At the time of the study, her two siblings were unaffected and her mother was reported affected but deceased. Biochemical analysis revealed that the G451E p55 homodimer completely failed to stimulate p140 (16,23). The loss of WT p55 function, i.e. failure to stimulate p140, does not easily predict a dominant mutation in vivo unless a single copy of POLG2 is haploinsufficient. Recently we reported the contrary. In our vertebrate model POLG2 is haplosufficient, as mice heterozygous for a POLG2 KO allele lacked any detectable mitochondrial dysfunction or phenotype over a two-year time period (10). However, the critical importance of p55 in vertebrate embryogenesis was evident by the embryonic lethality of homozygous POLG2 KO mice.
In this report, we describe a method to study the consequences of p55 heterodimeric disease variants (Fig. 1). Polγ DNA synthesis was inhibited in reactions containing the WT/G451E p55 heterodimer (Fig. 2). The inhibition of DNA synthesis by the WT/G451E p55 heterodimer was distinct from the G451E p55 homodimer. In this study, the defective activity associated with polγ containing the WT/G451E p55 heterodimer, was likely not due to thermal instability, as steady-state reactions supplemented with a molar equivalent of heterodimer had no effect on kcat when comparing values pre- and post-addition (Table 1 and Fig. 3). The observed polγ defect is largely explained by the ∼5-fold weaker ability of WT/G451E p55 to bind to p140 (Fig. 4). Previously, the homodimeric R369G p55 variant was characterized as having an identical ∼5-fold decrease in affinity for p140 and an identical pattern of DNA synthesis inhibition when reactions at physiological salt concentration (150 mm NaCl) were visualized via alkaline gel electrophoresis (Table 3) (23). However, at lower salt concentrations, R369G p55 homodimers were able to synthesis longer stretches of DNA up to ∼5 kb, although the activity was still defective compared with WT polγ, and DNA binding remained unaltered. Thus, the defect associated with the WT/G451E p55 heterodimer variant is unique in that DNA synthesis reactions remain ‘poisoned’ at both low and high salt concentrations (Fig. 2).
The ∼3-fold weaker affinity for DNA (Fig. 4), may help to explain an additive or synergistic inability to synthesize products greater than ∼600 nts at both salt concentrations. Relative to WT p55, the DNA-binding affinity of the heterodimer was 2-fold stronger than homodimeric G451E p55, which was previously shown to be ∼6-fold weaker than WT p55 (23). This suggests that the single WT p55 monomer in the heterodimer was capable of increasing the affinity of p55 dimers to DNA. Interestingly, in the steady-state assay the inhibition of polγ DNA synthesis by WT/G451E p55 is lifted by adding a molar equivalent of WT p55 to the reaction (Fig. 3B). This rescuing effect is likely explained by the higher affinities of the WT p55 for p140 and for DNA, which would allow the WT accessory subunit to ‘bump off’ and replace the heterodimer. The displacement of the heterodimer by WT p55 would allow continuation of polγ DNA synthesis. Based on these biochemical analyses we predict that in vivo the G451E monomer impairs polγ activity when associated with the WT monomer. In the absence of genetic linkage and co-segregation analyses our biochemical findings suggest that c.1352G>A/p.G451E allele is dominant negative in vivo leading to both G451E homodimeric loss (25%) and WT/G451E heterodimeric impairment (50%) of polγ function. Therefore, in a heterozygous patient only 25% of the POLG2 gene products (WT homodimers) would remain competent for polγ activity. The rescuing effect noted above is intriguing and suggests that overexpression of WT p55 in a dominant negative c.1352G>A mutant cell could potentially rescue the disease phenotype in vivo.
Structural implications of p55 heterodimeric disease variants
Within the polγ holoenzyme it was demonstrated that the p55 dimer binds asymmetrically to the p140 catalytic subunit with the majority of the intersubunit interactions occurring between the proximal p55 monomer and p140 (20). Our experiments demonstrated that P205R, R369G and L475DfsX2 p55 heterodimers do not have noticeable biochemical defects (Fig. 2). Previously we demonstrated that homogeneous preparations of these three variants are deficient or defective in the ability to stimulate p140 (23). Based on the crystal structure of the holoenzyme, the location of the P205R p55 substitution would not appear to disrupt the interface between the catalytic subunit and the proximal p55 monomer; however, both the R369G and L475DfsX2 substitutions would. The lack of biochemical defects associated with these heterodimeric variants suggests that areas of the p140-proximal p55 subunit interface are maintained by simple re-alignment of the polγ subunits. In these cases the p55 dimer may serve an evolutionarily redundant purpose. We predict that a WT/R369G or WT/L475DfsX2 p55 with a variant monomer positioned proximally to p140 cannot bind and therefore, must ‘roll over’ to adjust the variant into the distal position. As a result the WT protomer aligns in the proximal position and the critically important subunit interactions are maintained allowing WT-like polγ activity. One caveat to this functional redundancy is that p55 disease variants may undergo aberrant protein folding (see below).
Within the holoenzyme the G451E p55 substitution is expected to disrupt the interaction made between the proximal G451 p55 residue and the p140 L559 residue (23), while the distal G451 p55 residue is not predicted to contact the catalytic subunit (20). One might expect that simple re-alignment of the WT/G451E p55 heterodimer within the polγ holoenzyme would allow for WT-like DNA polymerase activity as was observed for the other variants mentioned above; however, our in vitro biochemistry revealed that polγ harboring WT/G451E p55 is indeed defective. Our results suggest that interactions made between both the proximal and distal G451 p55 residues are critically important for maintenance of polγ activity. Based on these findings we predict that polγ binding to DNA induces conformational changes that result in increased interactions between the distal p55 monomer and the catalytic subunit.
P205R and L475DfsX2 p55 variants have altered intramitochondrial localization
To analyze the subcellular localization of p55 we separately transfected HEK293 cells with a mammalian expression plasmids encoding WT or NGFP p55 variants. Precursor proteins synthesized on cytoplasmic ribosomes and destined for the mitochondrial matrix must translocate both the mitochondrial outer and inner membranes via the translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) complexes, respectively (32). Here we demonstrated that the p55 MTS is required for proper recognition by the TOM and TIM machinery for mitochondrial import, correct p55 folding within mitochondria and localization with mtDNA-containing nucleoids (Fig. 5A). Previously we determined that recombinant homogeneous preparations of L475DfsX2 p55 formed aberrant reducible multimers (23). Consistent with the notion of protein misfolding or aggregation in vivo, live-cell confocal fluorescence microscopy revealed that amino-terminally tagged GFP P205R and L475DfsX2 p55 variants are indeed targeted to mitochondria but relative to WT p55, have irregular diffuse green fluorescence throughout the reticulum (Fig. 5). Thus, the defects associated with P205R and L475DfsX2 variants are not likely the result of severely reduced protein levels. Aberrant protein folding was re-examined utilizing homogeneous preparations of P205R p55 via partially denaturing PAGE. Similar to L475DfsX2 we discovered that P205R p55 also contains laddered bands and smears not present in the WT p55 sample indicative of aberrant reducible multimers (Supplementary Material, Fig. S4). Protein misfolding is implicated in many p53-mediated cancers and has been reported to possibly be involved in up to half of all human diseases (33). Following mitochondrial import the failure of P205R and L475DfsX2 p55 to form puncta likely results from abnormal submitochondrial protein folding; therefore, we predict that P205R and L475DfsX2 p55 disease variant folding or aggregation or both contributes to the pathophysiology of these POLG2-related disorders. For POLG2 c.1352G>A/p.G451E we hypothesize that late-onset adPEO results from dominant negative loss of function in vivo. In general, the failure of G451E homo- and heterodimers to enhance polγ processivity would cause the replication complexes to stall during mtDNA replication and is consistent with the accumulation of mtDNA deletions detected in the patients with PEO (34). Although P205R, R369G and L475DfsX2 heterodimers displayed the same degree of stimulation as WT p55 we previously observed biochemical defects associated with the homodimer. Within these particular heterodimers, the WT p55 monomer may serve as a type of intermolecular chaperone to stabilize the variant monomer. In vivo the 25% misfolded p55 may negatively affect a range of mitochondrial-associated metabolic functions by aberrantly associating with mitochondrial proteins or enzymes. For example, mtDNA-containing nucleoid proteins or metabolic enzymes found in submitochondrial compartments, such as enzymes required for the tricarboxylic acid cycle or beta-oxidation of fatty acids, could be disrupted by misfolded p55 disease variants.
Mitochondrial reserve respiratory capacity is impaired in NGFP p55 disease variant cell lines
Cell lines expressing NGFP p55 disease variants had 6- to 16-fold less mitochondrial reserve capacity in comparison to the WT NGFP p55 cell line (Fig. 6). Mitochondrial reserve capacity is the difference between maximal OCR and basal OCR. It is widely hypothesized that when a cell is under stress, mitochondria draw upon this reserve capacity to meet cellular energy demands required for organ functions, cellular repair processes and detoxification of reactive oxygen species (29). In other words, reserve capacity refers to the amount of ATP that can be produced by OXPHOS in response to a sudden increase in energy demand, such as stress or increased workload (35). These results demonstrate that, although each p55 variant may display different biochemical defects (e.g. binding and stimulating the p140 catalytic subunit, protein folding and co-localization) the overall effect on the mitochondria is to reduce the reserve capacity. In the body, a reduction of this capacity would compromise the ability of muscle tissue to deal with stress (exercise) and cause general weakness of skeletal muscles and exercise intolerance, which is a hallmark symptom of PEO (4).
The mutations encoding G451E and R369G p55 are associated with late-onset adPEO. We have demonstrated that a G451E monomer is dominant negative in vitro and when reconstituted with the remaining two WT subunits of the trimeric polγ holoenzyme, poisons DNA polymerase activity. Consistent with the notion of blocking or competing with WT gene products in vivo both R369G and G451E p55 retain the ability to associate with mtDNA and form punctate mitochondrial nucleoids. R369G and G451E homodimers were previously demonstrated to display defective DNA synthesis and weakened binding affinity to p140 (23) and Table 3. The biochemical results from both of these mutant proteins are consistent with the late-onset autosomal dominant PEO seen in the patients (16,23,24). In contrast to G451E and R369G p55 proteins, the P205R and L475DfsX2 p55 variants were targeted to mitochondria but misfold and fail to form discrete puncta. The mild biochemical defects of the P205R homodimer and the lack of biochemical defects associated with the heterodimer are somewhat inconsistent with the observed early onset of disease (Table 3). Likewise, the early presentation of symptoms associated with the heterozygous c.1423_1424delTT/p.L475DfsX2 mutation is inconsistent with an autosomal dominant disorder and more closely resembles phenotypes associated with autosomal recessive mitochondrial disease. One explanation is that the age of onset in individuals harboring these mutations is influenced by other genetic or environmental factors. Despite the experimental differences we have observed among the p55 disease variants, all POLG2 mutations cause defective mitochondrial reserve capacity when overexpressed in HEK293 cell lines. Future studies in our cell-based system will investigate if defective bioenergetics associated with P205R and L475DfsX2 p55 result from an increased activation of the mitochondrial unfolded protein response (UPRmt). Interestingly, studies in worms and rat hepatoma cells have demonstrated that perturbation of mtDNA maintenance activate the UPRmt (36). While other unidentified factors may influence mitochondrial disease phenotypes, it is clear that the c.1352G>A/p.G451E mutation is dominant negative and causes PEO. Based on our findings we propose that a knock-in G451E p55 mouse model would best replicate human POLG2-related disease. Transgenic mouse models of Twinkle-related disorders have faithfully replicated late-onset disease phenotypes such as cytochrome c oxidase deficiency and mtDNA deletions in postmitotic tissues (37). Additionally, mouse models of orthologous nuclear mutations exploiting gene knock-in or a tissue-specific promoter have contributed to our understanding of the role of mtDNA maintenance in human disease (38). The design of an animal model of POLG2-related disease would be significant and aid in the determination of the physiological consequences of disease mutations including tissue-specific defects. Furthermore, a POLG2-related disease animal model could be exploited to examine potential therapeutic-interventions.
In summary, we developed two robust systems to analyze dominant negative effects and altered intramitochondrial localization of p55 disease variants. Our in vitro biochemical analysis of the WT/G451E p55 heterodimer demonstrates a greatly reduced processivity due to decreased affinity for p140 and for DNA (Table 3). In the absence of family genetics our discovery that G451E p55 forms a dominant negative heterodimer clarifies how the POLG2 exon 8 c.1352G>A mutation disturbs the mtDNA replication machinery thereby causing adPEO. Furthermore our mitochondrial localization studies indicate that P205R and L475DfsX2 are not folded correctly upon mitochondrial import and thus, do not likely interact with the p140 (Table 3). Finally, our bioenergetics analysis revealed that mitochondrial reserve capacity is impaired by these disease variants. Taken together, this study provides further understanding of the mechanisms contributing to these complex and devastating disorders.
Materials and Methods
Construction of plasmids expressing heterodimeric p55 disease variants
Two amino-terminal (N-terminal) tags were separately engineered into the human POLG2 gene. The pET-p55CHIS homodimer parent plasmid and plasmids derived from the parent plasmid harboring homodimeric disease variants (23) were used as DNA templates in separate PCR reactions to amplify the POLG2 and variant open reading frames (ORFs). The N-terminal hexa histidine tag (NHis-p55) was fused in-frame to WT, P205R, R369G and G451E p55 ORFs via PCR amplification with the p55NHIS_F_NdeI (5′-AAA AAA AAC ATA TGG CTA GCA GAG GAT CGC ATC ACC ATC ACC ATC ACG GCG CCG ATG CGG GGC AGC CG) and p55NHIS_R_KpnI (5′-AAA GGT ACC TTA AAC ATT CTT AGC TGA TGA TA) primer set. To generate the NHis-L475DfsX2 p55 truncation variant, the p55NHIS_F_NdeI and p55L475DfsX2NHIS_R_KpnI (5′-AAA GGT ACC TTA ATC AAA GTC TTT TAA TTT GGA T) primer set was used. PCR products were separately cloned into the NdeI and KpnI sites (MCS-2) of the pETDuet-1 dual expression vector (EMD Millipore) to generate plasmids encoding N-terminal His tagged WT, P205R, R369G, G451E and L475DfsX2 p55. Next, the POLG2 gene coding for WT p55 with an N-terminal Strep tag (NStrep-p55) was PCR amplified from pET-p55CHIS using the p55NStrepNcoI_F (5′-GAG ATA TAC CAT GGC TAG CTG GAG CCA CCC GCA GTT CGA AAA AGG CGC CGA TGC GGG GCA GCC G) and p55NStrepEcoRI_R (5′-AAA GAA TTC TTA AAC ATT CTT AGC TGA TGA TA) primer set. The NStrep-p55 PCR product was cloned into the pETDuet-1 MCS-1 using NcoI and EcoRI restriction sites. All of the plasmids containing p55-tagged ORFs were confirmed by DNA sequencing and the two WT plasmids were separately transformed into BL21(DE3) RIPL to express homodimeric NStrep-p55 and NHis-p55. The WT NStrep-p55 ORF was subcloned into the NcoI and EcoRI sites of all NHIS containing pETDuet-1 plasmids generating the dual-tagged WT p55 expression plasmid and the p55 heterodimer disease variant expression plasmids.
Expression and purification of disease variant heterodimeric p55 and WT p140
WT and heterodimeric human p55 disease variants were expressed in BL21-CodonPlus® (DE3)-RIPL E. coli. One-liter cultures were grown to an OD600 of 1 and then chilled to 20°C followed by induction with 1 mm IPTG for 17 h. Cells were pelleted at 250g, washed with 20 ml 10 mm Tris, pH7.5, 1 mm EDTA, 50 mm NaCl then pellets were snap frozen in liquid nitrogen and stored at −80°C. Cell pellets from 1 l of culture were thawed on wet ice, then suspended in 60 ml lysis buffer containing 50 mm Tris–HCl pH 8.0, 500 mm NaCl, 25 mm imidazole, 1% Triton X-100, 3 mg/ml lysozyme and 1:1000 protease inhibitor cocktail (Sigma P8849). The cell suspension was passed through a 40 ml French Pressure Cell (Amicon) at 18 000 psi and 200 µl of 0.5 mg/ml avidin was added to the resulting WCE and mixed. The WCE was centrifuged at 30 000g for 15 min at 4°C to obtain a soluble lysate. The lysate was mixed end-over-end at 4°C for 30 min with 2 ml of Ni-NTA agarose slurry (Qiagen) pre-equilibrated in lysis buffer. The Ni-NTA resin was batch washed two times with 40 ml lysis buffer, followed by pouring the resin into a disposable column and further washed in the column with 13 column volumes of the same buffer except the Triton X-100 was replaced with 0.01% NP-40. Then HIS-tagged proteins were eluted from the Ni-NTA resin by 50 mm Tris–HCl, pH 8.0, 0.25 M Imidazole. The Ni-NTA eluate was pooled and applied to a disposable column containing 2 ml of ST slurry (IBA GmbH) pre-equilibrated in ST-wash buffer containing 100 mm Tris–HCl, pH 8.0, 500 mm NaCl, 1 mm EDTA, 0.01% NP-40 and further washed in the column with 10 column volumes of ST-wash buffer. Heterodimeric p55 was eluted from the ST resin by 100 mm Tris–HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA and 2.5 mm desthiobiotin. The ST eluate was processed further by fast protein liquid chromatography as previously described (39). To purify homodimeric NStrep-p55 the Ni-NTA resin step was excluded and to purify NHis-p55 the ST resin step was excluded. Specificity of both the ST and the Ni-NTA resins were tested by separately applying 250 ng of purified homodimeric NHis-p55 or NStrep-p55 to 0.4 ml of resin, respectively. Columns were extensively washed followed by the release of protein with elution buffer. Washed and eluted fractions were visualized by PAGE followed by Coomassie staining. The high specificity of the resins was confirmed by the presence of NHis-p55 only in the unbound fractions of the ST column and by the presence of NStrep-p55 only in the unbound fractions of the Ni-NTA column.
For this study, the exonuclease-deficient (Exo–) pol γ lacking the N-terminal 25 amino acid residue MTS was denoted as WT p140. Overexpression of the N-terminal hexa-histidinyl form of this protein, in Spodoptera frugiperda (Sf9) insect cells, and purification was carried out as described (39).
Determining protein concentration
The concentration of purified protein was determined in relation to a range of BSA standards (0.75–3 µg) via Coomassie staining. Images of the Coomassie stained gels were collected with the SYNGENE G:BOX gel doc system and band areas were quantitated using ImageJ64. The areas of the BSA standards were plotted against µg loaded and linear regression generated correlation coefficients of 0.98–0.99 over the range analyzed. The BSA standard curves were run on each gel and the concentration of purified protein was then estimated.
HIS-tag detection
After Western transfer to PVDF membranes, the membranes were incubated with 0.2 µg/ml mouse anti-penta-His monoclonal antibody (Qiagen) in TN buffer (50 mm Tris–HCl, pH 7.5, 500 mm NaCl) plus 1 mg/ml BSA for 1.5 h. Next, the blot was washed three times with TN buffer for 8 min followed by incubation in a 1/3000 dilution of goat anti-mouse alkaline phosphatase-conjugated antibody (Bio-Rad) in TN buffer with 1 mg/ml BSA for 1 h 25 min. After three 8-min washes in TNT (50 mm Tris–HCl, pH 7.5, 500 mm NaCl and 0.1% Triton X-100) and three 8-min washes in TN, bands were visualized by chemiluminescence with CDP-Star (Roche) on Kodak Scientific Imaging Film.
Strep-tag detection
After Western transfer, the membrane was first washed in phosphate buffered saline (PBS) buffer (4 mm KH2PO4, 16 mm Na2HPO4, 115 mm NaCl, pH 7.4) for 15 min and then blocked with PBS blocking buffer (PBS buffer with 3% BSA and 0.5% v/v Tween 20), at room temperature for 1 h. The blot was washed three times with PBS-Tween buffer (PBS buffer with 0.1% v/v Tween 20) for 5 min followed by incubation in a 1/4000 dilution of StrepTactin-alkaline phosphatase conjugate (Bio-Rad) in PBS-Tween for 1 h. After two 1-min washes in PBS-Tween and two 1-min washes in PBS buffer, bands were visualized with CDP-Star (Roche) on Kodak Scientific Imaging Film. Autoradiograms of three to five second exposures were scanned and bands were quantitated in ImageJ64. The areas of the NHis-p55 and NStrep-p55 homodimer standards were plotted against µg loaded and linear regression generated correlation coefficients of 0.94–0.96 over the range analyzed. Standard curves were run on each blot and the concentration of heterodimer was then estimated. Following HIS- and Strep-tag visualization with CDP-Star, the PVDF membranes were washed in ddH2O for 5 min and then stained with 0.1% Coomassie Blue for 5 min. Next, the membranes were destained in 50% methanol, 10% acetic acid for 5 min and then rinsed twice with ddH2O followed by air-drying to visualize the efficiency of sample loading and electrotransfer.
Determination of p55 affinity to DNA
The affinities of WT p55 and variants for DNA were determined by EMSA, and equilibrium binding constants (Kd(DNA)) were calculated as previously described (23).
Stimulation of activity of the catalytic subunit by the p55 variants
In vitro 5′-32P-end-labeled 35-mer primer extension assays were carried out as described previously (23) with the exception that 5 nM p140 and 5 nM heterodimeric p55 or variant was utilized and reactions were adjusted to 150 mm NaCl unless otherwise indicated. Samples were resolved by 1% alkaline agarose gel electrophoresis overnight in 30 mm NaOH/1 mm EDTA at 24 V. Gels were neutralized with one molar equivalent of HCl, dried at 60°C for 3 h, and imaged with a phosphor screen on a Typhoon 9400 phosphorimager (GE Healthcare).
Affinity of p55 to p140
Functional assay enzyme dilution buffer (FEDB) consisted of 25 mm Tris–HCl, pH 7.5, 10% glycerol, 1 mm EDTA, 0.5 mm β-mercaptoethanol, 0.005% NP-40, 160 mm NaCl, 25 µg/ml BSA. Samples (24 µl) of purified p140 (10 nM) and p55 variant (varying concentrations) were premixed in FEDB, and 10 µl was added to a cocktail to make reactions (50 µl) containing 25 mm HEPES-KOH, pH 7.5, 2 mm β-mercaptoethanol, 5 mm MgCl2, 0.05 µg/ml BSA, 40 µM dTTP, 13.32 nM [α-32P] dTTP (3000 Ci/mmol, PerkinElmer Life Sciences), 150 mm NaCl, 50 µg/ml poly(dA)•p(dT)12–18 (The Midland Certified Reagent Company, Incorporated), 1 nM p140 and 0–16 nM p55 or variant heterodimer protein. Reactions were assembled on ice, then incubated at 37°C for 4 min. Reactions were stopped by placing in an ice bath followed by the immediate addition of 0.2 ml stop solution (1 mg/ml BSA, 0.1 M sodium pyrophosphate tetrabasic decahydrate) and 1 ml of 10% trichloroacetic acid (TCA). TCA-insoluble radioactivity was determined by liquid scintillation counting. DNA polymerase activity at 150 mm NaCl indicated functional association of p140 with the p55 accessory subunit in the presence of nucleic acid. Heterodimeric G451E p55 and dual-tagged WT p55 containing polγ were each measured twice independently and the average incorporation of dTMP was plotted against the variable concentration of p55 dimers. Using a quadratic equation (23) standard error estimates were derived by nonlinear regression analysis to calculate apparent Kd(p140) values.
DNA polγ steady-state assay
Reactions were set up as in the affinity of p55 to p140 section with the exception that the final concentration of WT p55 or G451E p55 heterodimer was 1 nM. For the basic time course 220 µl reactions were prepared and incubated at 37°C. Next, 50 µl samples were collected at 0, 5, 10 and 15 min and added to test tubes on ice with 0.2 ml of stop solution then 1 ml of 10% TCA was added to each. TCA-insoluble radioactivity was determined by liquid scintillation counting. Linear regression was performed on data points obtained from 0 to 10 min time points to estimate kcat values. In separate experiments, the kcat was also determined over a timeframe of 0–6 min with samples taken every 30 s. The values obtained by linear regression analysis from these experiments (0.55 ± 0.02 for polγ harboring WT p55 and 0.2 ± 0.01 for polγ harboring heterodimeric G451E p55) were nearly identical to values obtained over the 0- to 10-min timeframe (Table 1). For time course analysis with p55 additions, 275 µl reactions were prepared and incubated at 37°C. At 6.5 min 8.75 µl of 21 nM WT p55 or G451E p55 heterodimer was added to the remaining 175 µl and mixed. Samples of 50 µl were collected at 0, 5, 9, 12 and 15 min and added to test tubes containing stop solution and TCA as described above. Linear regression was performed on data points obtained from 9 to 15 min to estimate Kcat values following addition of WT p55.
NGFP p55 fusion protein
The mammalian expression plasmid pJ603 expressing the human p55 fusion protein with an amino-terminal Aequorea coerulescens green fluorescent protein (NGFP) tag (NGFP p55) was synthesized by DNA 2.0. The GFP ORF (AB255038.1) was fused in-frame between the N-terminal 25 codons of POLG2, encoding the predicted p55 MTS, and the C-terminal remaining POLG2 cDNA (NM_007215.3). The pJ603-NGFP-POLG2 plasmid harbors the cytomegalovirus promoter that controls the expression of the NGFP-POLG2 gene and carries the neomycin-resistance cassette for selection of stable cell lines using geneticin (G418) after transfection. POLG2-disease mutations were introduced separately into pJ603-NGFP-POLG2 utilizing the QuikChange™ Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions. Primers set for each QuikChange™ reaction consisted of a reverse primer with the following complementary forward primers: (1) P205R, 5′-GGA TCT GGT AAA CAA GAG GCT ACG TTA TGG CCT TGC TCA GAT TGG AG; (2) R369G, 5′-TAC AAG AAA GAA AAA TCT TCA TGG AAA GGT ACT TAA ACT TCA CCC; (3) G451E, 5′-CTG AAA CTA CTT TGG AGA ATG AAT TAA TAC ATC TGA GAA GCA G; (4) L475DfsX2, 5′-TAT ATC CAA ATT AAA AGA CTT TGA TTA AGT ATA TAT CAT CAG CT. (5) The NoMTS construct lacking the p55 MTS was introduced into pJ603-NGFP-POLG2 utilizing the QuikChange™ Kit with the reverse primer and the complementary forward primer: 5′-ACC CAA GCT GGC TAG CGA CAC CAT GGT GAG CAA GGG CGC CGA GC primer.
Cell culture and transfection
HEK293 cells were obtained from ATCC and initially cultured in EMEM, Eagle's Minimum Essential Medium (ATCC) containing 5.6 mm glucose, 4 mm L-glutamine, 2 mm sodium pyruvate, 10% fetal bovine serum (FBS), 100 U/ml Penicillin and 100 µg/ml Streptomycin (Sigma-Aldrich). Cells were grown at 37°C in a humidified atmosphere with 5% CO2 in air. Cells were routinely detached by washing with PBS, pH 7.4, then incubated with 0.25% trypsin, 1 mm EDTA (Life Technologies, Inc.) for 2 min, followed by neutralization with media. The Lonza Nucleofector™ 2b device along with the corresponding kit (Amaxa™ cell line Kit V) was utilized to transfect HEK293 cells according to the manufacturer's protocol. For transient transfections HEK293 cells were transfected with the Lonza Nucleofector™ 2b device then grown overnight in Minimum Essential Medium Eagle (MEM) prior to analysis by confocal fluorescence microscopy as described below. Growing transfected cells in the presence of 800 µg/ml G418 selected for stable integration of plasmids. After 4–5 days of selection to eliminate untransfected cells, expression of WT and variant NGFP p55 transgenes were confirmed by fluorescence microscopy. G418-resistant clones were picked and regrown in multiwell plates, then expanded in tissue culture flasks. At this stage stable cell lines were maintained in EMEM containing 50 µg/ml G418. Vials of cells were prepared in freezing medium consisting of 40% EMEM, 50% FBS, 10% DMSO and stored in the gas-phase of a Nitrogen freezer for long-term storage. Stable cell lines were also grown in 10 mm Galactose, 6 mm L-glutamine, 10 mm HEPES, 1 mm sodium pyruvate, 100 U/ml Penicillin, 100 µg/ml Streptomycin, 50 µg/ml G418, 10% FBS and Dulbecco's Modified Eagle Medium (DMEM) deprived of Glucose (Life Technologies, Inc.) to force cells to rely on mitochondrial OXPHOS rather than glycolysis as previously described (40). Cells were grown at 37°C in a humidified atmosphere with 10% CO2 in air to compensate for the increased concentration of sodium bicarbonate in the galactose medium. Cell lines were grown for 7 months in galactose medium. When cultures became ∼70% confluent, cells were washed with PBS and trypsinized as described above. Each cell line was routinely re-seeded at 1:10, 1:20 and 1:40 dilutions to maintain cells in the replicative growth phase.
Live-cell confocal fluorescence microscopy
The day before microscopy cells suspended in MEM, Minimum Essential Medium Eagle (SIGMA-ALDRICH), 5.6 mm glucose, 10% FBS, 100 U/ml Penicillin, 100 µg/ml Streptomycin, 4 mm L-Glutamine, 1 mm sodium pyruvate without phenol red, were plated onto 35 mm glass bottom dishes No. 1.5 (MatTek Corporation) according to the manufacturer's instructions. Dishes were incubated at 37°C, 5% CO2 overnight. The following day cells were stained in fresh MEM containing 0.05 µg/ml DAPI for 90 min at 37°C, 5% CO2. MitoTracker® Red CMXRos (Life Technologies, Inc.) was added at a final concentration of 25 nM to visualize mitochondria and plates were incubated at 37°C, 5% CO2 for 4 min. Plates were washed and overlaid with Hank's balanced salt solution (Life technologies, Inc.) buffered with 25 mm HEPES, pH 7.3 prior to live-cell imaging. Confocal images were taken on a Zeiss LSM710 (Carl Zeiss Inc, Oberkochen, Germany) using a C-Apochromat 40X/1.2 Water DIC objective. For the red channel, a 561 nm diode laser at 9% power was used for excitation of the MitoTracker Red while a 566–691 filter collected the emission using a pinhole setting of 1 airy unit. For the green channel, a 488 nm ArKr laser line at 7% power was used for excitation of GFP while a bandpass 503–571 filter was used for collection of the emission signal. For the Blue channel, a 405 nm diode laser line at 20% power was used for excitation of DAPI while a bandpass 410–463 filter was used for the emission. The PMT voltage gain was held constant for all images with a setting of 620 V for the DAPI channel, 675 V for GFP and 530 V for the MitoTracker Red. Furthermore, all images were taken with a zoom of 1.4, a 3.2 µs pixel dwell time, a 0.29 µm pixel size and with line averaging set to 4.
Determining colocalization coefficients
The MitoTracker, GFP and DAPI fluorophores were chosen due to reliable separation of their emission spectra in the three detection channels of interest. Using Zeiss ZEN image-processing software, we estimated colocalization by matching the gray-values of individual pixels in tandem images generated from two fluorescent molecules and calculated the average weighted colocalization coefficient according to Manders et al. (27). We utilized a stringent threshold of 3-fold above background autofluorescence for all measurements. For example, the average sum of intensities of the WT NGFP p55 green and MitoTracker red colocalizing pixels as compared with the overall sum of the pixel intensities in the green channel was 92% colocalized (0.92 ± 0.04). Therefore, colocalization strongly supports that NGFP p55 is targeted to the mitochondrial reticulum in vivo.
Measurement of cellular bioenergetics
All materials and reagents for the Extracellular Flux assays except 2,4-dinitrophenol (Sigma-Aldrich) were from Seahorse Biosciences (Billerica, MA, USA). The Seahorse Biosciences XF24 extracellular flux analyzer (Billerica, MA, USA) was used to measure mitochondrial function in stable cell lines expressing NGFP p55 disease variants, WT NGFP p55 and age-matched HEK-293 controls. The XF24 allows for the determination of OCR and ECAR in real time (29,41). To maintain cell adherence, the assay plates were pre-coated with poly-D-lysine (0.01% solution), rinsed 2× with PBS and stored for up to 2 weeks at 4°C. For all bioenergetic measurements, cells grown in tissue culture for 7 months in galactose medium, as described above, were seeded to a density of 40 000 cells/well, which was pre-determined to be in the linear OCR range, and grown for 24 h in a 37°C humidified incubator with 10% CO2. For all experiments, the cells were checked for even seeding and 90–95% confluency. One hour prior to the start of the assay, the plate was rinsed 2× with unbuffered modified DMEM XF Assay Medium (Seahorse Biosciences, cat # 102365-106) containing 2 mm GlutaMax, 1 mm sodium pyruvate and 10 mm galactose and adjusted to a pH of 7.4. To each of the wells was added a final volume of XF assay medium (675 µl) then the plate was incubated at 37°C without CO2. Galactose was chosen as the sole sugar source to force the cells to rely on glutamine-driven mitochondrial OXPHOS over aerobic glycolysis as previously demonstrated (29–31,42). Mitochondrial function was assessed via the bioenergetic OCR profile for each transformant or control. Following equilibration, OCR was measured 3 times for 3 min following a 3 min mixing and 2 min waiting period. This cycle was repeated following the sequential injection of each inhibitor to a final concentration of 1.0 µM oligomycin, 150 µM 2,4-DNP and a mixture of 1.0 µM roteneone and 1.0 µM antimycin A. Parameters evaluated included the basal level of oxygen consumption, the oxygen consumption linked to ATP production and non-ATP production (proton leak), the maximal level of oxygen consumption, non-mitochondrial oxygen consumption and the reserve respiratory capacity.
Supplementary Material
Funding
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ES 065078 to WCC) and by a NIH Pathway to Independence Award to M.J.Y. (1K99ES022638-01).
Supplementary Material
Acknowledgements
We would like to thank Mr Jeff Tucker and Dr Agnes Janoshazi of the NIEHS Fluorescence Microscopy and Imaging Center for their expertise in confocal imaging. We would also like to thank Drs Scott Lujan, Kin Chan and Matthew Longley for critically reading this manuscript and Dr Robert Petrovich for insightful discussions regarding protein biochemistry.
Conflict of Interest statement. None.
References
- 1.Kukat C., Wurm C.A., Spahr H., Falkenberg M., Larsson N.G., Jakobs S. (2011) Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U S A, 108, 13534–13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown T.A., Tkachuk A.N., Shtengel G., Kopek B.G., Bogenhagen D.F., Hess H.F., Clayton D.A. (2011) Super-resolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol. Cell. Biol., 24, 4994–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Copeland W.C. (2014) Defects of mitochondrial DNA replication. J. Child Neurol. 29, 1216–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen B.H., Chinnery P.F., Copeland W.C. (2010) POLG-Related Disorders, GeneReviews at GeneTests: Medical Genetics Information Resource [database online] . Copyright, University of Washington, Seattle, 1997–2010. Available at http://www.genetests.org. [Google Scholar]
- 5.Wallace D.C., Chalkia D. (2013) Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol., 5, a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong L.J., Naviaux R.K., Brunetti-Pierri N., Zhang Q., Schmitt E.S., Truong C., Milone M., Cohen B.H., Wical B., Ganesh J. et al. (2008) Molecular and clinical genetics of mitochondrial diseases due to POLG mutations. Hum. Mutat., 29, E150–E172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart J.B., Larsson N.G. (2014) Keeping mtDNA in shape between generations. PLOS Genet., 10, e1004670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hance N., Ekstrand M.I., Trifunovic A. (2005) Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum. Mol. Genet., 14, 1775–1783. [DOI] [PubMed] [Google Scholar]
- 9.Iyengar B., Luo N., Farr C.L., Kaguni L.S., Campos A.R. (2002) The accessory subunit of DNA polymerase gamma is essential for mitochondrial DNA maintenance and development in Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A, 99, 4483–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humble M.M., Young M.J., Foley J.F., Pandiri A.R., Travlos G.S., Copeland W.C. (2013) Polg2 is essential for mammalian embryogenesis and is required for mtDNA maintenance. Hum. Mol. Genet., 22, 1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S.K., Zhao M.H., Zheng Z., Kwon J.W., Liang S., Kim S.H., Kim N.H., Cui X.S. (2015) Polymerase subunit gamma 2 affects porcine oocyte maturation and subsequent embryonic development. Theriogen., 83, 121–130. [DOI] [PubMed] [Google Scholar]
- 12.Carrodeguas J.A., Theis K., Bogenhagen D.F., Kisker C. (2001) Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer. Mol. Cell, 7, 43–54. [DOI] [PubMed] [Google Scholar]
- 13.Fan L., Kim S., Farr C.L., Schaefer K.T., Randolph K.M., Tainer J.A., Kaguni L.S. (2006) A novel processive mechanism for DNA synthesis revealed by structure, modeling and mutagenesis of the accessory subunit of human mitochondrial DNA polymerase. J. Mol. Biol., 358, 1229–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim S.E., Longley M.J., Copeland W.C. (1999) The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J. Biol. Chem., 274, 38197–38203. [DOI] [PubMed] [Google Scholar]
- 15.Carrodeguas J.A., Pinz K.G., Bogenhagen D.F. (2002) DNA binding properties of human pol gamma B. J. Biol. Chem., 277, 50008–50014. [DOI] [PubMed] [Google Scholar]
- 16.Longley M.J., Clark S., Yu Wai Man C., Hudson G., Durham S.E., Taylor R.W., Nightingale S., Turnbull D.M., Copeland W.C., Chinnery P.F. (2006) Mutant POLG2 disrupts DNA polymerase gamma subunits and causes progressive external ophthalmoplegia. Am. J. Hum. Genet., 78, 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson A.A., Tsai Y., Graves S.W., Johnson K.A. (2000) Human mitochondrial DNA polymerase holoenzyme: reconstitution and characterization. Biochem., 39, 1702–1708. [DOI] [PubMed] [Google Scholar]
- 18.Pinz K., Bogenhagen D. (2006) The influence of the DNA polymerase γ accessory subunit on base excision repair by the catalytic subunit. DNA Repair (Amst.), 5, 121–128. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y.S., Molineux I.J., Yin Y.W. (2010) A single mutation in human mitochondrial DNA polymerase pol gammaA affects both polymerization and proofreading activities, but only as a holoenzyme. J. Biol. Chem., 285, 28105–28116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y.S., Kennedy W.D., Yin Y.W. (2009) Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell, 139, 312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y.S., Lee S., Demeler B., Molineux I.J., Johnson K.A., Yin Y.W. (2010) Each monomer of the dimeric accessory protein for human mitochondrial DNA polymerase has a distinct role in conferring processivity. J. Biol. Chem., 285, 1490–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yakubovshaya E., Chen Z., Carrodeguas J.A., Kisker C., Bogenhagen D.F. (2006) Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J. Biol. Chem., 281, 374–382. [DOI] [PubMed] [Google Scholar]
- 23.Young M.J., Longley M.J., Li F.Y., Kasiviswanathan R., Wong L.J., Copeland W.C. (2011) Biochemical analysis of human POLG2 variants associated with mitochondrial disease. Hum. Mol. Genet., 20, 3052–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craig K., Young M.J., Blakely E.L., Longley M.J., Turnbull D.M., Copeland W.C., Taylor R.W. (2012) A p.R369G POLG2 mutation associated with adPEO and multiple mtDNA deletions causes decreased affinity between polymerase gamma subunits. Mitochondrion, 12, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young M.J., Copeland W.C. (2013) Mitochondrial disorders associated with the mitochondrial DNA polymerase γ: a focus on intersubunit interactions. In Wong L.J.C. (ed), Mitochondrial Disorders Caused by Nuclear Genes. Springer Science+Business Media, New York, pp. 49–72. [Google Scholar]
- 26.Garrido N., Griparic L., Jokitalo E., Wartiovaara J., Van Der Bliek A.M., Spelbrink J.N. (2003) Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell, 14, 1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manders E.M.M., Verbeek F.J., Aten J.A. (1993) Measurement of co-localization of objects in dual-colour confocal images. J. Microsc., 169, 375–382. [DOI] [PubMed] [Google Scholar]
- 28.Claros M.G., Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem., 241, 779–786. [DOI] [PubMed] [Google Scholar]
- 29.Hill B.G., Dranka B.P., Zou L., Chatham J.C., Darley-Usmar V.M. (2009) Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem. J., 424, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gohil V.M., Sheth S.A., Nilsson R., Wojtovich A.P., Lee J.H., Perocchi F., Chen W., Clish C.B., Ayata C., Brookes P.S., Mootha V.K. (2010) Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nat. Biotechnol., 28, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dott W., Mistry P., Wright J., Cain K., Herbert K.E. (2014) Modulation of mitochondrial bioenergetics in a skeletal muscle cell line model of mitochondrial toxicity. Redox Biol., 2, 224–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stojanovski D., Bohnert M., Pfanner N., van der Laan M. (2012) Mechanisms of protein sorting in mitochondria. Cold Spring Harb. Perspect. Biol., 4, a011320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaudhuri T.K., Paul S. (2006) Protein-misfolding diseases and chaperone-based therapeutic approaches. FEBS J., 273, 1331–1349. [DOI] [PubMed] [Google Scholar]
- 34.Krishnan K.J., Reeve A.K., Samuels D.C., Chinnery P.F., Blackwood J.K., Taylor R.W., Wanrooij S., Spelbrink J.N., Lightowlers R.N., Turnbull D.M. (2008) What causes mitochondrial DNA deletions in human cells? Nat. Genet., 40, 275–279. [DOI] [PubMed] [Google Scholar]
- 35.Desler C., Hansen T.L., Frederiksen J.B., Marcker M.L., Singh K.K., Juel Rasmussen L. (2012) Is there a link between mitochondrial reserve respiratory capacity and aging? J. Aging Res., 2012, 192503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haynes C.M., Ron D. (2010) The mitochondrial UPR—protecting organelle protein homeostasis. J. Cell Sci., 123, 3849–3855. [DOI] [PubMed] [Google Scholar]
- 37.Tyynismaa H., Mjosund K.P., Wanrooij S., Lappalainen I., Ylikallio E., Jalanko A., Spelbrink J.N., Paetau A., Suomalainen A. (2005) Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc. Natl. Acad. Sci. U S A, 102, 17687–17692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyynismaa H., Suomalainen A. (2009) Mouse models of mitochondrial DNA defects and their relevance for human disease. EMBO Rep., 10, 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasiviswanathan R., Longley M.J., Young M.J., Copeland W.C. (2010) Purification and functional characterization of human mitochondrial DNA polymerase gamma harboring disease mutations. Methods, 51, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marroquin L.D., Hynes J., Dykens J.A., Jamieson J.D., Will Y. (2007) Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol. Sci., 97, 539–547. [DOI] [PubMed] [Google Scholar]
- 41.Ferrick D.A., Neilson A., Beeson C. (2008) Advances in measuring cellular bioenergetics using extracellular flux. Drug Discov. Today, 13, 268–274. [DOI] [PubMed] [Google Scholar]
- 42.Reitzer L.J., Wice B.M., Kennell D. (1979) Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem., 254, 2669–2676. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.