Abstract
The cellulosome is a supramolecular multienzyme complex formed by species-specific interactions between the cohesin modules of scaffoldin proteins and the dockerin modules of a wide variety of polysaccharide-degrading enzymes. Cellulosomal enzymes bound to the scaffoldin protein act synergistically to degrade crystalline cellulose. However, there have been few attempts to reconstitute intact cellulosomes due to the difficulty of heterologously expressing full-length scaffoldin proteins. We describe the synthesis of a full-length scaffoldin protein containing nine cohesin modules, CipA; its deletion derivative containing two cohesin modules, ΔCipA; and three major cellulosomal cellulases, Cel48S, Cel8A, and Cel9K, of the Clostridium thermocellum cellulosome. The proteins were synthesized using a wheat germ cell-free protein synthesis system, and the purified proteins were used to reconstitute cellulosomes. Analysis of the cellulosome assembly using size exclusion chromatography suggested that the dockerin module of the enzymes stoichiometrically bound to the cohesin modules of the scaffoldin protein. The activity profile of the reconstituted cellulosomes indicated that cellulosomes assembled at a CipA/enzyme molar ratio of 1/9 (cohesin/dockerin = 1/1) and showed maximum synergy (4-fold synergy) for the degradation of crystalline substrate and ∼2.4-fold-higher synergy for its degradation than minicellulosomes assembled at a ΔCipA/enzyme molar ratio of 1/2 (cohesin/dockerin = 1/1). These results suggest that the binding of more enzyme molecules on a single scaffoldin protein results in higher synergy for the degradation of crystalline cellulose and that the stoichiometric assembly of the cellulosome, without excess or insufficient enzyme, is crucial for generating maximum synergy for the degradation of crystalline cellulose.
INTRODUCTION
The cellulosome is a supramolecular multienzyme complex composed of a wide variety of polysaccharide-degrading enzymes and structural proteins and is displayed on the cell surface of anaerobic cellulolytic bacteria (1, 2) (Fig. 1). Clostridium thermocellum is one of the most investigated cellulosome-producing anaerobic bacteria. The formation of C. thermocellum cellulosomes is mediated by two specific interactions. One interaction is between the type I dockerin module at the C termini of polysaccharide-degrading enzymes and the nine internal cohesin modules of the primary scaffoldin protein, and the other interaction is mediated between the type II dockerin module at the C terminus of the primary scaffoldin protein and the internal cohesin modules of the cell surface-displayed secondary scaffoldin protein. The scaffold of the cellulosome complex assembles through the interaction of one primary scaffoldin protein containing nine type I cohesin modules with four different secondary scaffoldin proteins. The genome of C. thermocellum ATCC 27405 contains at least 79 cellulosomal genes, 8 of which encode the cohesin-containing scaffoldin protein while the remaining 71 partly encode the dockerin-containing cellulosomal components. Cellulosomes isolated from C. thermocellum cells grown on different carbon sources have different enzymatic compositions (3, 4).
FIG 1.
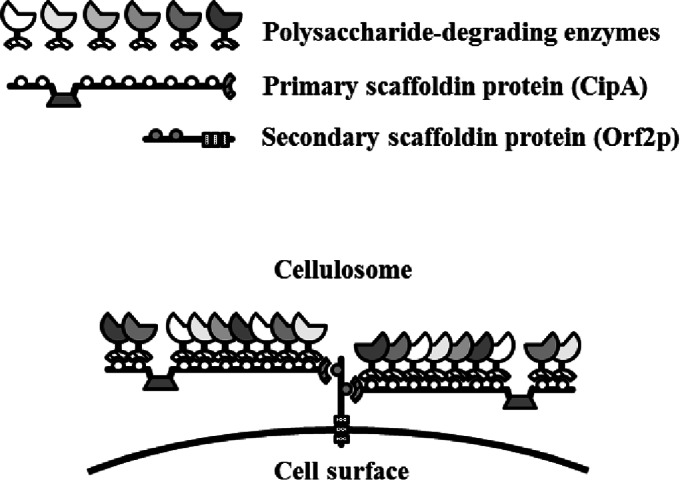
Schematic representation of the C. thermocellum cellulosome. The cellulosome is a cell surface-displayed supramolecular multienzyme complex containing a wide variety of polysaccharide-degrading enzymes. The formation of the C. thermocellum cellulosome is mediated by two specific interactions. One interaction is between the type I dockerin module at the C termini of polysaccharide-degrading enzymes and the internal nine cohesin modules of the primary scaffoldin protein, and the other interaction is mediated between the type II dockerin module at the C terminus of the primary scaffoldin protein and the internal cohesin modules of the cell surface-displayed secondary scaffoldin protein. The scaffold of the cellulosome complex assembles through the interaction of one primary scaffoldin protein (CipA, containing nine type I cohesin modules) and four secondary scaffoldin proteins (SdbA, which contains one type II cohesin module; Orf2p, which contains two type II cohesin modules; OlpB, which contains seven type II cohesin modules; and Cthe0736, which contains seven type II cohesin modules). In the diagram, Orf2p is used as a representative secondary scaffoldin protein.
The C. thermocellum cellulosomal enzymes Cel48S, Cel8A, and Cel9K were previously identified by proteomic analysis as major enzymatic components of the C. thermocellum cellulosome obtained from cells grown on crystalline cellulose and other cellulosic substrates (3, 4). The domain organization of these cellulosomal cellulases is shown in Fig. 2A. Cel48S contains a glycoside hydrolase (GH) family 48 (GH48) and a type I dockerin module and is one of the most abundant cellulosomal cellulases in the C. thermocellum cellulosome (3, 4). Cel48S is classified as a cellobiohydrolase and processively hydrolyzes the cellulose chain from the reducing end (5, 6). Cel9K, which contains a GH family 9 (GH9), a carbohydrate-binding module (CBM) family 4 (CBM4), and a type I dockerin module, is the major cellulosomal cellulase (3, 4) and is also classified as a cellobiohydrolase (Cel9K may be classified as a processive endoglucanase) but processively hydrolyzes the cellulose chain from the nonreducing end (2, 7). The third enzyme, Cel8A, contains a GH family 8 (GH8) and a type I dockerin module and is one of the most abundant cellulosomal endoglucanases in the cellulosome (3, 4). These three enzymes are predicted to degrade crystalline cellulose by synergistic actions between exo- and endomode glucanases: in this model, the Cel8A endoglucanase cleaves cellulose at the internal β-1,4-glucosidic bond, and the Cel48S and Cel9K exoglucanases processively hydrolyze the cellulose chain from the reducing and nonreducing ends, respectively.
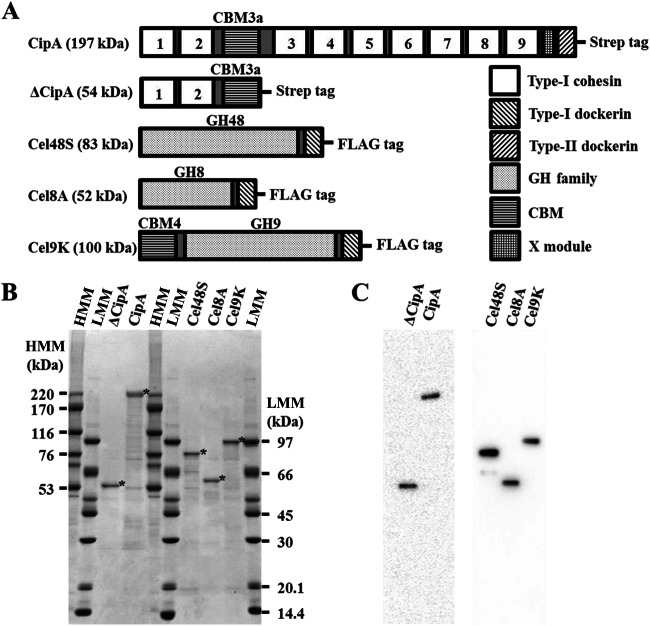
FIG 2.
Domain organization and SDS-PAGE analysis of the cellulosomal components. (A) The C. thermocellum cellulosomal scaffoldin protein, CipA, contains nine type I cohesin modules (cohesins 1 to 9), a CBM3a, an X module, and a type II dockerin module. The N-terminal segment of CipA, designated ΔCipA, contains two type I cohesin modules (cohesins 1 and 2) and a CBM3a. The model complex employed here contained three enzymatic components: Cel48S, a GH48 enzyme; Cel8A, a GH8 enzyme; and Cel9K, a GH9 enzyme that also contains a CBM4 module at its N terminus. All three enzymes contain a type I dockerin module at their C termini. The dockerin module specifically associates with the type I cohesin modules of the scaffoldin protein. Mature scaffoldin and enzymatic components were synthesized in a cell-free system as GST fusion proteins containing a Strep and a FLAG tag, respectively, at the C terminus and then purified by cleavage with PreScission protease in a column. (B) Purified samples were subjected to SDS-PAGE on a 4 to 20% gel and stained with Coomassie brilliant blue. The bands corresponding to the purified proteins are indicated by asterisks. HMM, high molecular mass; LMM, low molecular mass. (C) Purified proteins were detected by Western blotting with anti-Strep or anti-FLAG M2 monoclonal antibody that targeted the C-terminal Strep or FLAG tag of the scaffoldin or enzymatic component, respectively.
The efficient degradation of recalcitrant crystalline cellulose by C. thermocellum is essentially dependent on the formation of a cellulosome complex mediated by the primary scaffoldin protein, CipA (8, 9). CipA is composed of four different types of modules: a CBM family 3a (CBM3a) that interacts with crystalline cellulose, nine type I cohesin modules that bind to the nine enzyme molecules containing a type I dockerin module, an X module that may participate in CipA dimer formation, and a type II dockerin module at the C terminus for binding the type II cohesin module of the secondary scaffoldin proteins (2) (Fig. 2A). Although the indispensability of CipA for the rapid degradation of crystalline cellulose by C. thermocellum has been demonstrated (6), the difficulty of isolating full-length CipA from recombinant Escherichia coli cells has hampered studies on cellulosomes reconstituted from full-length CipA. Genetic instability of the highly repeated DNA sequences encoding cohesins 3 to 8 also complicates the isolation of CipA (10). Furthermore, studies on cellulosomes face the common problem of the heterologous expression of a large scaffoldin protein. Therefore, most previous studies on cellulosomes focused on minicellulosomes assembled by deletion variants of scaffoldin proteins that contain only two or three cohesin modules. These deletion variants display two or three enzyme molecules (8, 11–13), whereas the number of cohesin modules in native cellulosomal scaffoldin proteins is typically between 6 and 11 (1). These minicellulosomes usually show significantly lower activity toward crystalline cellulose than do native cellulosomes (8, 12, 14). Thus, elucidating the mechanism responsible for the high activity for crystalline cellulose exhibited by native cellulosomes requires in vitro reconstitution of the cellulosome complexes from their large scaffoldin proteins.
The cellulolytic activity of the in vitro-reconstituted cellulosome, comprising full-length CipA produced by recombinant E. coli cells and the protein mixture secreted from cipA-deficient C. thermocellum cells, has recently been investigated (14). The reconstituted cellulosome was assembled by assuming that the secreted protein mixture has an average molecular mass of 80 kDa. The assembled cellulosome showed ∼80% of the activity of the native cellulosome from C. thermocellum toward crystalline cellulose and 12-fold-higher activity than the unassembled protein mixture. Furthermore, the heterologous expression of full-length CipA by the thermophilic anaerobic bacterium Thermoanaerobacterium saccharolyticum has recently been reported (10). That study demonstrated intercellular complementation of the functional cellulosome with full-length CipA secreted from recombinant T. saccharolyticum cells and the cellulosomal components secreted from cipA-deficient C. thermocellum cells. However, only these two attempts to reconstitute intact cellulosome complexes from recombinant full-length scaffoldin proteins have been reported to date, and there have been few attempts to reconstitute cellulosome complexes from purified components that include recombinant full-length scaffoldin proteins.
It has been reported that a wheat germ cell-free protein synthesis system using purified wheat embryos is suitable for synthesizing a large set of multidomain proteins simultaneously (15) and for producing large multidomain proteins that are difficult to produce using E. coli (15, 16). Recently, the high-throughput synthesis of the 42 components (excluding CipA) of the C. thermocellum cellulosome using the wheat germ cell-free system was reported (17). Therefore, while full-length CipA has been produced by recombinant E. coli cells (14), the wheat germ cell-free system appears to be a promising tool for reconstituting intact cellulosomes from full-length scaffoldin proteins and purified enzymatic components. Here, we report the synthesis of full-length CipA and the major cellulosomal enzymes of the C. thermocellum cellulosome using the wheat germ cell-free system and analysis of the stoichiometric assembly and cellulolytic activities of cellulosome complexes reconstituted from the purified full-length CipA protein and major cellulosomal cellulases.
MATERIALS AND METHODS
Materials.
The genomic DNA of C. thermocellum ATCC 27405 (NBRC 103400) was obtained from the National Institute of Technology and Evaluation, Japan; the nucleotide sequence is publicly available from the National Center for Biotechnology Information database under accession no. NC_009012.1. Plasmid pEUGST-GFP (16) was used as a cassette vector for the construction of pEU derivatives for the cell-free protein synthesis of a glutathione S-transferase (GST) fusion protein, and wheat germ extract was prepared as reported previously (18). E. coli DH5α (TaKaRa, Japan) was used as a cloning host. All PCR primers were synthesized by Greiner Japan and are listed in Table 1.
TABLE 1.
Primers used for DNA amplification
| Name | Nucleotide sequencea |
|---|---|
| pEUUSacI-5′ | 5′-GATCACGAGCTCATACATAACCTTATGTATCATACACATACG-3′ (SacI) |
| pEUDKpnI-3′ | 5′-CTACAGGGTACCGGAGAAAGGCGGACAGGTAT-3′ (KpnI) |
| pMWSacI-3′ | 5′-GATCACGAGCTCGTAATCATGGTCATAGCTG-3′ (SacI) |
| pMWKpnI-5′ | 5′-CTACAGGGTACCGGCACTGGCCGTCGTTTTAC-3′ (KpnI) |
| CipA-NPr5′ | 5′-TTCCAGGGGCCCCTGGGAAGATCTGCCACAATGACAGTCGAG-3′ (BglII) |
| CipA-CSt3′ | 5′-TGGGACGTCGACTTACTTTTCGAACTGCGGGTGGCTCCAGCTTGCCTGTGCGTCGTAATCACTTGATG-3′ (SalI) |
| ΔCipA-CSt3′ | 5′-TGGGACGTCGACGCGTTTACTTTTCGAACTGCGGGTGGCTCCAGCTTGCACTGCCACCGGGTTCTTTAC-3′ (SalI) |
| Cel48S-NPr5′ | 5′-TTCCAAGGTCCACTGGGATCCACTAGTGGTCCTACAAAGGCACCTAC-3′ |
| Cel48S-CF3′ | 5′-ACGTCGACGCGTTTACTTGTCATCGTCATCCTTGTAGTCGTCACCGTTCTTGTACGGCAATG-3′ |
| Cel8A-NPr5′ | 5′-TTCCAAGGTCCACTGGGATCCACTAGTGCAGGTGTGCCTTTTAACAC-3′ |
| Cel8A-CF3′ | 5′-ACGTCGACGCGTTTACTTGTCATCGTCATCCTTGTAGTCGTCACCATAAGGTAGGTGGGGTATGC-3′ |
| Cel9K-NPr5′ | 5′-TTCCAAGGTCCACTGGGATCCACTAGTTTGGAAGACAAGTCTCCAAAG-3′ |
| Cel9K-CF3′ | 5′-ACGTCGACGCGTTTACTTGTCATCGTCATCCTTGTAGTCGTCACCTTTATGTGGCAATACATCTATC-3′ |
| pEUGST-NPr3′ | 5′-CAGTGGACCTTGGAACAGAACCTCTAGATCCGATTTTGGAGG-3′ |
| pEU-C5′ | 5′-TAAACGCGTCGACGTCCCATGGTTTTG-3′ |
| Cohesin4-5′ | 5′-CACCGACAGATGATTCGA-3′ |
| Cohesin8-3′ | 5′-GTCAACTTGTTCGGAGTTAT-3′ |
| pEUUn | 5′-ACATACGATTTAGGTGACACTA-3′ |
| 2pEUDn | 5′-GAGAGCGCACGAGGGAGCTT-3′ |
Recognition sequences of restriction endonucleases used for the construction of plasmids in this study are shown in boldface, and the names of the restriction endonucleases are shown in parentheses following the nucleotide sequences of the primers.
DNA substrates for cell-free protein synthesis of GST fusion proteins.
Plasmid pMWGST-GFP, a low-copy-number vector for the construction of pMW derivatives for the cell-free protein synthesis of a GST fusion protein, was constructed as follows. A DNA fragment consisting of the 5′ untranslated region (UTR)-GST-green fluorescent protein (GFP)-3′UTR region in pEUGST-GFP, which contained the promoter sequence of SP6 RNA polymerase, the omega sequence, the coding region of GST-GFP fusion protein, and the 3′ UTR of tobacco mosaic virus, was amplified by PCR with primers pEUUSacI-5′ and pEUDKpnI-3′. A DNA fragment consisting of a low-copy-number plasmid was PCR amplified from plasmid pMW218 (Nippon Gene, Japan) using primers pMWSacI-3′ and pMWKpnI-5′. After digesting the two PCR products with SacI and KpnI, the two DNA fragments were ligated to yield pMWGST-GFP, which was then transformed into DH5α cells. Positive transformants were selected on Luria-Bertani (LB) agar plates containing 30 μg/ml kanamycin (Km) (LB-Km plates) and incubated at 30°C.
Plasmids pMWGST-CipA and pMWGST-ΔCipA for the cell-free protein syntheses of GST fusion scaffoldin proteins were constructed as follows. The CipA and ΔCipA genes were PCR amplified from C. thermocellum ATCC 27405 (NBRC 103400) genomic DNA using primers CipA-NPr5′ and CipA-CSt3′ and primers CipA-NPr5′ and ΔCipA-CSt3′, respectively. After digestion with BglII and SalI, the amplified DNA fragments were introduced into the BamHI-SalI sites of pMWGST-GFP to exchange the GFP and scaffoldin protein genes, yielding plasmid pMWGST-CipA or pMWGST-ΔCipA; DH5α cells were then transformed with either pMWGST-CipA or pMWGST-ΔCipA. Positive transformants were selected on LB-Km plates and incubated at 30°C.
Plasmids pEUGST-Cel48S, pEUGST-Cel8A, and pEUGST-Cel9K for the cell-free protein synthesis of GST fusion cellulosomal cellulases were constructed as follows. The Cel48S, Cel8A, and Cel9K genes were PCR amplified from C. thermocellum genomic DNA using primers Cel48S-NPr5′ and Cel48S-CF3′, primers Cel8A-NPr5′ and Cel8A-CF3′, and primers Cel9K-NPr5′ and Cel9K-CF3′, respectively. A plasmid DNA fragment corresponding to pEUGST was PCR amplified from pEUGST-GFP using primers pEUGST-NPr3′ and pEU-C5′. Each cellulase gene and the plasmid DNA fragment was ligated using an In-fusion HD cloning kit (Clontech, Japan) to yield plasmid pEUGST-Cel48S, pEUGST-Cel8A, or pEUGST-Cel9K, which was then transformed into DH5α cells. Positive transformants were selected on LB plates containing 50 μg/ml ampicillin (Amp) (LB-Amp plates) and incubated at 37°C.
The cellulosomal genes in the pMW and pEU derivatives were sequenced by the dideoxy chain termination method with fluorescent dye terminators (Eurofins Genomics, Japan). However, direct sequencing of the CipA gene in pMWGST-CipA was not possible because the CipA gene contains highly repeated nucleotide sequences encoding cohesins 3 to 8. Therefore, the CipA gene was sequenced as follows, using a modification of the method reported for sequencing the CipA gene in C. thermocellum genomic DNA (19). The region of the CipA gene in pMWGST-CipA encoding cohesins 3 to 9 was digested with PstI, which cleaves at the center of each cohesin sequence, to yield six ∼500-bp DNA fragments. Each ∼500-bp DNA fragment was subcloned into the PstI site of pUC19. The nucleotide sequence of each DNA fragment was confirmed by analysis of the subcloned sequences from 30 positive transformants. The nucleotide sequences of the PstI-digested DNA fragments encoding cohesins 4 and 5 and cohesins 5 and 6 were identical. Therefore, the length of the PCR product amplified from pMWGST-CipA with primers Cohesin4-5′ and Cohesin8-3′, which specifically amplify an ∼2,500-bp DNA fragment encoding cohesins 4 to 8, was confirmed to rule out the possibility that the CipA gene in pMWGST-CipA contains a deletion within the highly repeated DNA region.
The DNA substrates for cell-free protein synthesis of the GST fusion proteins were PCR amplified from pEUGST fusion derivatives using primers pEUUn and 2pEUDn. PCR was performed for 25 cycles under the following conditions: 96°C for 30 s, 50 or 55°C for 30 s, and 72°C for 60 s/1 kbp with PrimeStar HS or GXL DNA polymerase (TaKaRa), using a TaKaRa Dice TP-650 PCR Thermal Cycler.
Cell-free protein synthesis and purification.
Wheat germ cell-free protein synthesis and purification of the GST fusion proteins was performed as described previously (20). The synthesized fusion proteins were cleaved with PreScission protease in a glutathione-Sepharose 4B MicroSpin column (GE Healthcare, Japan). The flowthrough fraction contained proteins of the predicted sizes for FLAG tag- or Strep tag-fused mature proteins, as revealed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 4 to 20% gradient gels (ATTO, Japan) stained with Coomassie brilliant blue and Western blot analysis using anti-FLAG M2 (Sigma-Aldrich, Japan) or anti-Strep tag (the NWSHPQFEK tag antibody; Gen Script, USA) monoclonal antibody using an ECL Select Detection kit (GE Healthcare). The protein concentration was estimated by densitometric analysis with Image J software (National Institutes of Health, USA) with bovine serum albumin (BSA) as a standard.
Cellulosome assembly.
Cellulosomes were assembled using cell-free synthesized proteins as follows. The cellulosome complex was assembled by mixing the scaffoldin and enzymatic components at different molar ratios in buffer A (50 mM sodium acetate, pH 5.5, 2 mM CaCl2, 2 mM dithiothreitol, and 0.01% BSA) at 40°C for 30 min. The cohesin-dockerin interaction was analyzed by an electrophoretic mobility shift assay using native PAGE on 4 to 20% gradient gels with Western blot analysis using an anti-FLAG M2 or anti-Strep tag monoclonal antibody. To assay cellulase activity and analyze stoichiometric assembly, the cellulosome complex was assembled by mixing scaffoldin protein at different concentrations with the enzyme mixture at a fixed concentration at a Cel48S/Cel8A/Cel9K molar ratio of 4.06:1.82:0.72. Stoichiometric assembly of the cellulosome complex was analyzed by size exclusion chromatography. After incubating the cellulosome assembly in buffer A without 0.01% BSA at 40°C for 30 min, the assembled mixture was subsequently applied to a KW404-4F column (4.6-mm diameter by 300-mm length; Shodex, Japan) connected to a Prominence high-performance liquid chromatography (HPLC) system (Shimadzu, Japan) in buffer A without 0.01% BSA at 25°C. The assembled mixture was eluted at a flow rate of 0.35 ml/min and fractionated every minute. The fractions were concentrated using a Nanosep centrifugal device (10-kDa cutoff; Pall Corporation, USA) and assayed using phosphoric acid-swollen cellulose (PASC) as a substrate.
Cellulase assay.
The cellulolytic activities of the enzymes and the reconstituted cellulosomes were assayed at 55°C in buffer A, according to procedures described previously (16). Three assay substrates were used at a final concentration of 5 mg/ml (0.5%): Avicel (Avicel PH-101; Sigma-Aldrich), PASC prepared from Avicel as described previously (21), and carboxymethylcellulose (CMC) (Sigma-Aldrich). To assay cellulase activity toward recalcitrant substrates, the incubation time was extended from 30 min to 48 h. The amount of reducing sugars released from the substrate was quantified using 3′5′-dinitrosalicylic acid (DNS) reagent and measuring the absorbance at 535 nm with glucose as a standard (22). One unit of enzymatic activity was defined as the amount of enzyme producing 1 μmol of reducing sugar per minute, and the specific activity was defined as the enzymatic activity per milligram of enzyme. Assays were performed at different concentrations of enzyme to determine if the amount of product increased in proportion to the amount of enzyme.
RESULTS
Cell-free protein synthesis and purification of cellulosomal components.
To reconstitute the C. thermocellum cellulosome, we constructed plasmids encoding the full-length scaffoldin protein, CipA; its deletion derivative, ΔCipA; and the three major enzymatic components, Cel48S, Cel8A, and Cel9K, to synthesize these proteins as GST fusion proteins using a wheat germ cell-free protein synthesis system (Fig. 2). The CipA gene contains a highly repeated DNA sequence encoding cohesins 3 to 8. The CipA gene is often deleted spontaneously, even in recA-deficient E. coli cells when cloned into a high-copy number plasmid, such as pUC-derived pEU plasmid for wheat germ cell-free protein synthesis. Therefore, we constructed a low-copy-number plasmid containing the pSC101 ori for the wheat germ cell-free protein synthesis of GST fusion full-length CipA. The nucleotide sequence of the CipA gene in pMWGST-CipA was confirmed by subcloning the highly repeated DNA region encoding cohesins 3 to 9, as described for sequencing of the CipA gene in C. thermocellum genomic DNA (19). The cell-free synthesized GST fusion proteins were recovered in soluble form, purified by glutathione affinity chromatography, and cleaved to yield the cellulosomal proteins of the predicted sizes. The purified proteins were confirmed by SDS-PAGE (Fig. 2B) and Western blotting using an antibody against the C-terminal Strep or FLAG tag of the scaffoldin or enzymatic components, respectively (Fig. 2C). The amount of cellulosomal components purified from 1 ml of translation mixture was between 1 and 10 μg.
Substrate specificities of cellulosomal enzymes.
The substrate specificities of the three major cellulosomal enzymes, Cel48S, Cel8A, and Cel9K, of C. thermocellum were investigated. As shown in Table 2, Cel8A exhibited the highest activity for CMC, followed by PASC and then crystalline cellulose (Avicel). In contrast, Cel9K exhibited higher activity for PASC, followed by CMC and then Avicel. Cel48S also exhibited higher activity for PASC, followed by Avicel and then CMC. We next investigated the synergistic actions of these enzymes for degrading cellulosic substrates by mixing the enzymes at a Cel48S/Cel8A/Cel9K molar ratio of 4.06:1.82:0.72. This molar ratio was based on the reported ratio of normalized spectral abundance factors (NSAF) of cellulosomal components determined by mass spectrometry analysis of native cellulosome prepared from cells grown on crystalline cellulose (4); the NSAF values of proteins have been used for determining relative protein ratios in a multiprotein complex (23). The synergies for the degradation of cellulosic substrates generated by mixing these enzymes are shown in Table 2. The larger synergy observed for the degradation of the more recalcitrant substrates (recalcitrance was in the following order: Avicel > PASC > CMC) is explained by the synergy model between exo- and endoglucanases, as is the limited synergy for CMC degradation: endoglucanases efficiently cleave CMC, but cellobiohydrolases cannot processively hydrolyze the CMC chain from the cleavage site.
TABLE 2.
Enzymatic activities of cellulosomal enzymes for cellulosic substrates
| Enzyme | Sp act (U/mg) (±SE)a |
||
|---|---|---|---|
| CMC | PASC | Avicel | |
| Cel48S | 0.0031 ± 0.000016 | 0.087 ± 0.0088 | 0.0038 ± 0.000092 |
| Cel8A | 14 ± 1.7 | 3.7 ± 0.68 | 0.0013 ± 0.00029 |
| Cel9K | 0.030 ± 0.0027 | 0.37 ± 0.052 | 0.0029 ± 0.000098 |
| Cel48S + Cel8A + Cel9Kb | 3.1 ± 0.22 (1.2) | 1.3 ± 0.076 (1.6) | 0.0066 ± 0.00030 (2.0) |
The enzymatic activities were determined by measuring the amount of reducing sugars released from the substrate (0.5%), as described in Materials and Methods. Assays were performed at different concentrations of enzyme to determine if the amount of product increased in proportion to the amount of enzyme. The data are presented as the means from the results of two to eight independent experiments.
Cel48S, Cel8A, and Cel9K were mixed at a Cel48S/Cel8A/Cel9K molar ratio of 4.06:1.82:0.72, which corresponds to a weight ratio of 0.67:0.18:0.14. Synergies generated by mixing Cel48S, Cel8A, and Cel9K for the degradation of each substrate are indicated in parentheses.
Stoichiometric assembly of the cellulosome.
Before reconstituting the cellulosome complex, we examined whether the cellulosome complex stoichiometrically assembled by interactions between the cohesin and dockerin modules at equimolar concentration. Assembly of the minicellulosome was analyzed by electrophoretic mobility shift assays (Fig. 3A). Specifically, a fixed concentration of ΔCipA was mixed with different amounts of each of the three enzymes and then analyzed by native PAGE and Western blotting using an anti-Strep tag antibody that targeted the C-terminal Strep tag of the scaffoldin protein. Here, “ΔCipA/enzyme” denotes the molar ratio of ΔCipA containing two cohesin modules to the enzyme containing one dockerin module, “CipA/enzyme” denotes the molar ratio of full-length CipA containing nine cohesin modules to the enzyme containing one dockerin module, and “cohesin/dockerin” denotes the molar ratio of the cohesin module to the dockerin module. The results showed that the bands corresponding to the minicellulosome complex converged to a single band when the complexes assembled at a ΔCipA/enzyme molar ratio of 1/2 (cohesin/dockerin = 1/1), suggesting that in cellulosomes formed at this molar ratio, the minicellulosome complex comprises two cohesin modules of ΔCipA saturated with enzyme molecules. These results indicated that minicellulosomes comprise two cohesin modules of ΔCipA displaying two enzyme molecules when the cohesin and dockerin modules are available in stoichiometric (equimolar) amounts during minicellulosome formation. Next, we analyzed the assembly of the cellulosome complex reconstituted from full-length CipA (Fig. 3B). Bands corresponding to the cellulosome complexes were not observed in the gels. This is likely due to smearing of the bands or the inability of the cellulosome complexes with a high molecular weight to enter the gel, which made it difficult to analyze their stoichiometric assembly. Thus, we next analyzed cellulosome assembly with an anti-FLAG tag antibody that targeted the C-terminal FLAG tag of the enzymes (Fig. 3C). The bands corresponding to the cellulosome complexes assembled at a CipA/enzyme ratio of 1/2.25 (cohesin/dockerin = 1/0.25) shifted to a low molecular weight compared with the bands corresponding to the complexes assembled at CipA/enzyme ratios of 1/36 and 1/9 (cohesin/dockerin = 1/4 and 1/1), suggesting that the complexes assembled at a CipA/enzyme ratio of 1/2.25 (cohesin/dockerin = 1/0.25), which are predicted to display fewer than nine enzyme molecules, exhibited lower molecular weights than the enzyme-saturated complexes assembled at CipA/enzyme ratios of 1/36 and 1/9 (cohesin/dockerin = 1/4 and 1/1). On the other hand, if the cellulosome complex assembled stoichiometrically, then at a CipA/enzyme ratio of 1/9 (cohesin/dockerin = 1/1), the bands corresponding to the unassembled enzymes should disappear and shift to a high-molecular-weight band corresponding to the complex. Surprisingly, the bands corresponding to the unassembled enzymes completely shifted to a higher molecular weight even when the complexes were assembled at a CipA/enzyme ratio of 1/36 (cohesin/dockerin = 1/4); at this ratio, three-quarters of the enzyme was predicted to be in an unassembled form. This shift to apparently higher molecular weight is probably not due to the formation of a nonspecific aggregate with CipA, since CipA alone did not form an aggregate during electrophoresis (i.e., the band corresponding to CipA remained a single band [Fig. 3B]). Therefore, these results suggested that the electrophoretic mobility shift assay was not suitable for analyzing the stoichiometric assembly of the cellulosome complex reconstituted from full-length CipA.
FIG 3.
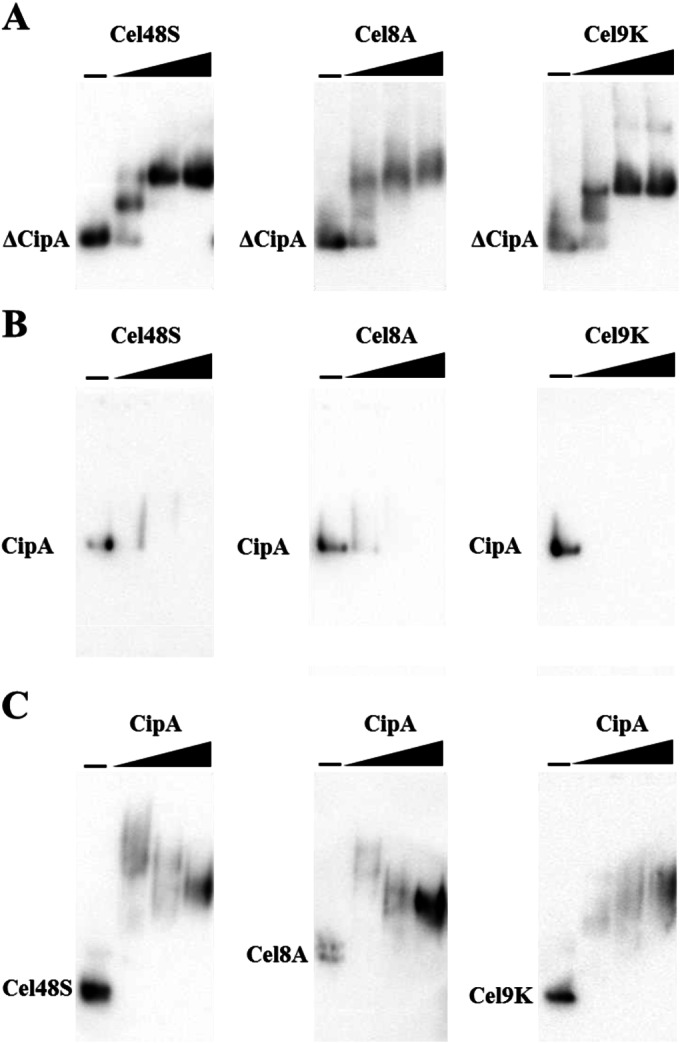
Electrophoretic mobility shift assay of the minicellulosome and cellulosome assembly. (A) Minicellulosome complexes assembled by mixing ΔCipA at a fixed concentration (0.050 μM) and each enzyme at various concentrations. The molar ratio of ΔCipA containing two cohesin modules to the enzyme containing one dockerin module is designated “ΔCipA/enzyme,” and the molar ratio of the cohesin module to the dockerin module is designated “cohesin/dockerin.” The complexes were assembled at ΔCipA/enzyme molar ratios of 1/0, 1/1, 1/2, and 1/4 (cohesin/dockerin = 1/0, 1/0.5, 1/1, and 1/2) and analyzed by native PAGE on 4 to 20% gradient gels with Western blot analysis using an anti-Strep tag monoclonal antibody. The bands corresponding to ΔCipA are indicated. (B) Cellulosome complexes assembled by mixing CipA at a fixed concentration (0.025 μM) and each enzyme at various concentrations. The molar ratio of CipA containing nine cohesin modules to the enzyme containing one dockerin module is designated “CipA/enzyme.” The complexes were assembled at CipA/enzyme molar ratios of 1/0, 1/4.5, 1/9, and 1/18 (cohesin/dockerin = 1/0, 1/0.5, 1/1, and 1/2) and detected by Western blotting with an anti-Strep tag monoclonal antibody. The bands corresponding to CipA are indicated. (C) Cellulosome complexes assembled by mixing Cel48S, Cel8A, or Cel9K at a fixed concentration (0.050 μM) and CipA at various concentrations. The complexes were assembled at CipA/enzyme molar ratios of 0, 1/36, 1/9, and 1/2.25 (cohesin/dockerin = 0, 1/4, 1/1, and 1/0.25) and detected by Western blotting with anti-FLAG M2 tag monoclonal antibody. The bands corresponding to Cel48S, Cel8A, and Cel9K are indicated.
To avoid unexpected behavior of the cellulosome complex in the electrophoretic mobility shift assay, we next tried to analyze the stoichiometric assembly of the cellulosome complex by size exclusion chromatography. The cellulosome complexes, assembled by mixing full-length CipA at various concentrations with the mixture of Cel48S, Cel8A, and Cel9K at a fixed concentration, were fractionated by size exclusion chromatography. Because the cellulosome complexes and the unassembled enzymes could not be detected by UV due to their small amounts, we detected them by the activity for PASC substrate; the activity for this substrate was predicted to reflect the amount of the enzyme mixture, because the enzyme mixture synergistically and efficiently hydrolyzed the substrate (Table 2) and the cellulosome formation did not substantially affect the activity for the substrate (Table 3). The unassembled enzymes were eluted after 11 min (Fig. 4A), which was consistent with the elution time corresponding to the molecular masses of the enzymes, ranging from 52 to 100 kDa. In contrast, the mixture assembled at a CipA/enzyme ratio of 1/9 (cohesin/dockerin = 1/1), which was predicted to comprise an enzyme-saturated complex displaying nine enzyme molecules, was almost eluted after 6 min (Fig. 4D), which was consistent with the elution time of Blue Dextran (2,000 kDa), suggesting that the enzymes were sufficiently integrated into the cellulosome complex with a high molecular weight during the incubation at 40°C for 30 min before being subjected to size exclusion chromatography. Furthermore, because the mixtures assembled at a CipA/enzyme ratio of <1/9 (cohesin/dockerin < 1/1) were predicted to comprise a small amount of the enzyme-saturated complex and an excess amount of the free enzymes, successive changes of the elution profiles between the complexes assembled at CipA/enzyme ratios of 0, 1/36, 1/18, and 1/9 (cohesin/dockerin = 0, 1/4, 1/2, and 1/1) (Fig. 4A, B, C, and D) supported the stoichiometric assembly of the cellulosome complex. Therefore, the size exclusion chromatographic analysis demonstrated the stoichiometric assembly of the cellulosome complex reconstituted from full-length CipA, and the results suggested that the dockerin module of the enzymes stoichiometrically bound to the cohesin modules of full-length CipA.
TABLE 3.
Enzymatic activities of reconstituted minicellulosome and cellulosome complexes for cellulosic substrates
| Scaffold/enzymea | Sp act (U/mg) (± SE)b |
||
|---|---|---|---|
| CMC | PASC | Avicel | |
| 0 | 3.1 ± 0.22 (1.0) | 1.3 ± 0.076 (1.0) | 0.0066 ± 0.00030 (1.0) |
| ΔCipA/enzyme | |||
| 1/2 | ND | ND | 0.011 ± 0.00037 (1.7) |
| 1/1 | ND | ND | 0.014 ± 0.0012 (2.2) |
| 1/0.5 | ND | ND | 0.012 ± 0.0010 (1.9) |
| CipA/enzyme | |||
| 1/9 | 2.4 ± 0.26 (0.8) | 1.0 ± 0.016 (0.8) | 0.026 ± 0.0018 (4.0) |
| 1/2.25 | 3.2 ± 0.50 (1.0) | 1.0 ± 0.080 (0.8) | 0.016 ± 0.0016 (2.5) |
The scaffold/enzyme values indicate the molar ratio of the scaffoldin protein to the enzyme mixture; for example, the cellulosome complex assembled with 0.030 μM CipA and 0.27 μM enzyme mixture is indicated as 1/9.
Minicellulosome or cellulosome complexes were assembled by mixing ΔCipA or CipA at various concentrations and the enzyme mixture at a fixed concentration, as described in Materials and Methods. The enzyme mixture comprised the Cel48S/Cel8A/Cel9K molar ratio of 4.06:1.82:0.72. Assays were performed at different concentrations of enzyme to determine if the amount of product increased in proportion to the amount of enzyme. The data are presented as the means from two to eight independent experiments. The activities of the minicellulosome and cellulosome complexes relative to the unassembled enzyme mixture are indicated in parentheses. ND, activity was not determined.
FIG 4.
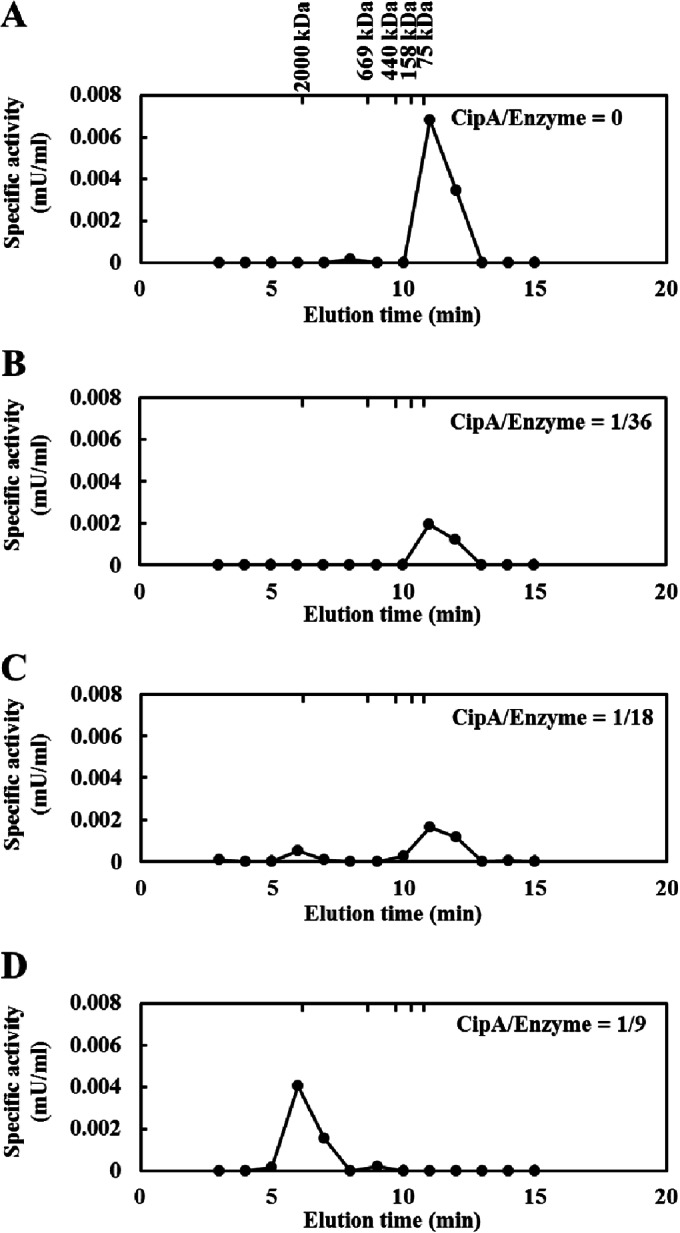
Size exclusion chromatography of reconstituted cellulosome complexes. Stoichiometric assembly of the reconstituted cellulosome complex was analyzed by size exclusion chromatography as described in Materials and Methods. The cellulosome complexes were assembled by mixing CipA at various concentrations with the enzyme mixture at a fixed concentration (0.41 μM). The enzyme mixture comprised a Cel48S/Cel8A/Cel9K molar ratio of 4.06:1.82:0.72. The CipA/enzyme values indicate the molar ratio of CipA to the enzyme mixture; for example, a cellulosome complex assembled with 0.045 μM CipA and 0.41 μM enzyme mixture is shown as “CipA/enzyme = 1/9.” The complexes were assembled at CipA/enzyme molar ratios of 0 (cohesin/dockerin = 0) (A), 1/36 (cohesin/dockerin = 1/4) (B), 1/18 (cohesion/dockerin = 1/2) (C), and 1/9 (cohesin/dockerin = 1/1) (D). The enzymatic activity was determined by measuring the amount of reducing sugars released from 0.5% PASC. The molecular mass markers (2,000-kDa Blue Dextran, 669-kDa thyroglobulin, 440-kDa ferritin, 158-kDa aldolase, and 75-kDa conalbumin) are indicated.
Cellulolytic activity of the reconstituted cellulosome.
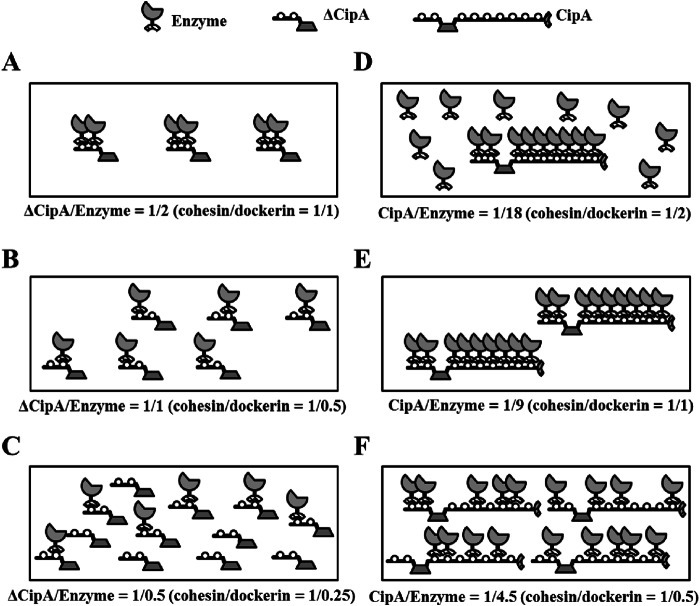
To investigate the effect of the stoichiometry of cellulosome assembly on cellulolytic activity, we assembled cellulosome complexes by mixing different concentrations of the scaffoldin protein with fixed concentrations of the three enzymes and then measured the activities toward cellulosic substrates (Fig. 5 and Table 3). The activity profile of the minicellulosome complexes reconstituted from ΔCipA indicated that the complex assembled at a ΔCipA/enzyme molar ratio of 1/1 (cohesin/dockerin = 1/0.5) exhibited the highest activity toward Avicel and showed 2.2-fold-higher activity toward Avicel than the unassembled enzyme mixture (Fig. 5A and Table 3). This was despite the fact that the enzyme-saturated minicellulosome complex was assembled at a ΔCipA/enzyme ratio of 1/2 (cohesin/dockerin = 1/1), as confirmed by the electrophoretic mobility shift assay (Fig. 3A). Moreover, the complex assembled at a ΔCipA/enzyme ratio of >1/2 (cohesin/dockerin > 1/1) was slightly more efficient at Avicel degradation than the complex assembled at a ΔCipA/enzyme ratio of 1/2 (cohesin/dockerin = 1/1) (Table 3). This result suggested that the minicellulosome complex displaying only one enzyme molecule, such as the complexes shown in Fig. 6B and C, was slightly more efficient at Avicel degradation than the enzyme-saturated complex shown in Fig. 6A. Since ΔCipA contains CBM3a, which targets the cellulosome complex to the crystalline substrate (24), these results suggested that the CBM3a-mediated targeting of the minicellulosome complex to the crystalline substrate contributed more to the degradation of the substrate than the synergistic action of the two enzyme molecules on ΔCipA.
FIG 5.
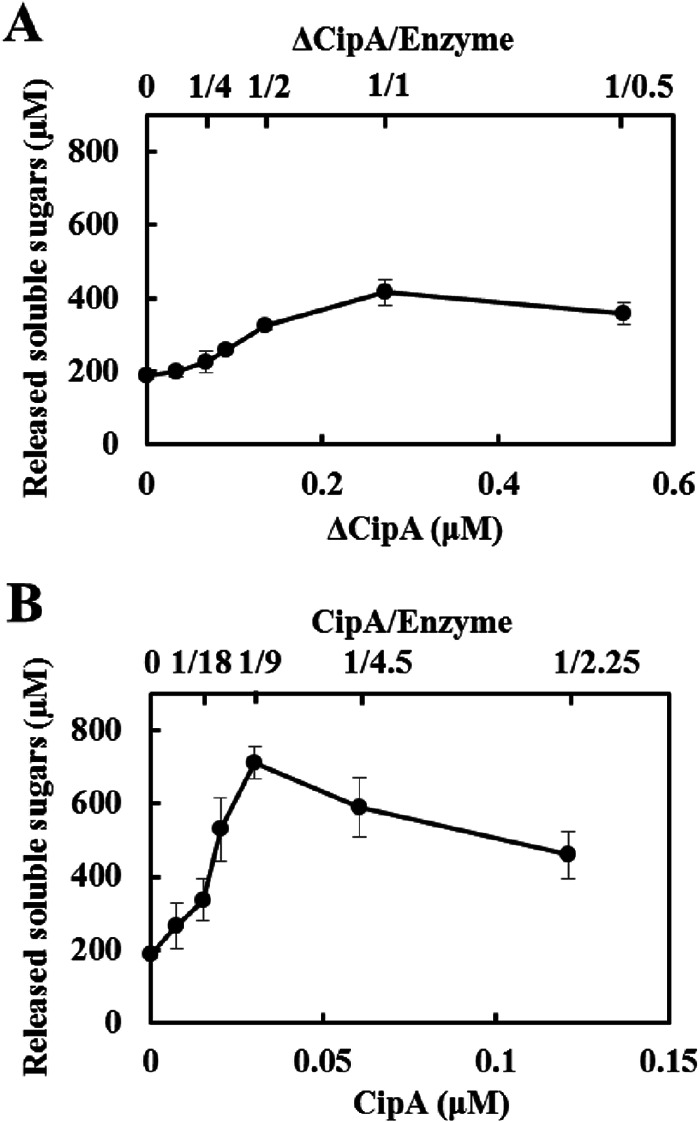
Enzymatic activities of reconstituted minicellulosome and cellulosome complexes for crystalline cellulose. The enzymatic activities of minicellulosome (A) and cellulosome (B) complexes were determined by measuring the amount of reducing sugars released from 0.5% Avicel at 55°C in 24 h. Minicellulosome or cellulosome complexes were assembled by mixing ΔCipA or CipA at various concentrations with the enzyme mixture at a fixed concentration (0.27 μM). The enzyme mixture comprised a Cel48S/Cel8A/Cel9K molar ratio of 4.06:1.82:0.72. The scaffold/enzyme values indicate the molar ratio of scaffoldin protein to the enzyme mixture; for example, a cellulosome complex assembled with 0.030 μM CipA and 0.27 μM enzyme mixture is indicated as 1/9. The data are presented as the means from three independent experiments ± standard errors (SE).
FIG 6.
Schematic models of minicellulosome and cellulosome complexes. The minicellulosome or cellulosome complexes were assembled by mixing ΔCipA or CipA at various concentrations with an enzyme mixture at a fixed concentration. (A) The minicellulosome complex assembled at a ΔCipA/enzyme ratio of 1/2 (cohesin/dockerin = 1/1) is predicted to comprise an enzyme-saturated complex displaying two enzyme molecules. (B and C) The complexes assembled at ΔCipA/enzyme ratios of 1/1 (cohesin/dockerin = 1/0.5) (B) and 1/0.5 (cohesin/dockerin = 1/0.25) (C) are predicted to display only one enzyme molecule. (D) In contrast, the cellulosome complex assembled at a CipA/enzyme ratio of 1/18 (cohesin/dockerin = 1/2) is predicted to comprise an enzyme-saturated complex displaying nine enzyme molecules and the free enzymes. (E) The complex assembled at a CipA/enzyme ratio of 1/9 (cohesin/dockerin = 1/1) is predicted to comprise the enzyme-saturated complex. (F) The complex assembled at a CipA/enzyme ratio of 1/4.5 (cohesin/dockerin = 1/0.5) is predicted to display fewer than nine enzyme molecules.
On the other hand, the activity profile of the cellulosome complexes reconstituted from CipA indicated that the complex assembled at a CipA/enzyme ratio of 1/9 (cohesin/dockerin = 1/1) exhibited the highest activity toward Avicel of the assembled cellulosomes and 4-fold-higher activity toward Avicel than the unassembled enzyme mixture (Fig. 5B and Table 3), whereas its activities for CMC and PASC were similar to those of the unassembled mixture (Table 3). Analysis of the cellulosome assembly using size exclusion chromatography suggested that the dockerin module of the enzymes stoichiometrically bound to the cohesin modules of CipA (Fig. 4); therefore, the complex assembled at a CipA/enzyme ratio of 1/9 (cohesin/dockerin = 1/1) was predicted to comprise cellulosome complexes displaying nine enzyme molecules, as shown in Fig. 6E. In contrast, the complexes assembled at CipA/enzyme ratios of <1/9 (cohesin/dockerin < 1/1) were predicted to comprise a small amount of enzyme-saturated complex and an excess amount of the free enzyme (Fig. 6D), and the complexes assembled at CipA/enzyme ratios of >1/9 (cohesin/dockerin > 1/1) would display fewer than nine enzyme molecules (Fig. 6F). Therefore, these results suggested that stoichiometric cellulosome assembly, without excess or insufficient enzyme, exhibited maximum synergy for the degradation of crystalline cellulose, whereas cellulosome formation was not essential for the degradation of noncrystalline cellulose. Moreover, because the activities of the cellulosome complexes assembled at a CipA/enzyme ratio of >1/9 (cohesin/dockerin > 1/1) toward Avicel gradually decreased as the molar ratio of CipA increased (Fig. 5B), the synergy of the nine enzyme molecules on CipA contributed more to the degradation of crystalline cellulose than the CBM3a-mediated targeting effect. Furthermore, since the cellulosome complex assembled at a CipA/enzyme ratio of 1/9 (cohesin/dockerin = 1/1) showed ∼2.4-fold-greater synergy for the degradation of Avicel than the minicellulosome complex assembled at a ΔCipA/enzyme ratio of 1/2 (cohesin/dockerin = 1/1) (Table 3), it appears that displaying more enzyme molecules on a single scaffoldin protein generates greater synergy for the degradation of crystalline cellulose.
DISCUSSION
Here, we synthesized the full-length scaffoldin protein and the three major enzymatic components of the C. thermocellum cellulosome as GST fusion proteins using a wheat germ cell-free protein synthesis system and purified the proteins by glutathione affinity chromatography (Fig. 2). The enzymatic properties of the three enzymes, such as optimum pH and temperature and substrate specificity, have been reported (5, 7, 25). Specifically, Cel8A exhibited the highest activity for CMC, followed by acid-swollen cellulose (ASC) and then crystalline cellulose (Avicel) (25). In contrast, Cel48S exhibited higher activity for ASC, followed by Avicel and then CMC (5). The specific activities of Cel9K for these substrates have not been reported. As shown in Table 2, the substrate specificities of the enzymes synthesized by in vitro translation were comparable to the previous findings described above, suggesting that the cell-free synthesized enzymes exhibited enzymatic properties similar to those produced in recombinant E. coli cells, as reported previously (16). Next, we investigated the stoichiometric assembly of cellulosome complexes reconstituted from the purified full-length CipA and major cellulosomal cellulases (Fig. 3 and 4). The stoichiometric assembly of cellulosomes has been suggested by many biochemical analyses of minicellulosome assemblies (11–13, 26) and structural analyses of cohesin-dockerin complexes (27–29), supporting the view that all native cellulosomes stoichiometrically assemble at a cohesin/dockerin molar ratio of 1/1. However, direct evidence using native cellulosomes or full-length scaffoldin proteins has never been reported. In this study, analysis of the cellulosome assembly using size exclusion chromatography demonstrated the stoichiometric assembly of the cellulosome complex reconstituted from full-length CipA (Fig. 4), but this could not be confirmed by the electrophoretic mobility shift assay due to unexpected migration of the bands corresponding to the unassembled enzymes on a native PAGE gel (Fig. 3C). Crystallographic study of the C. thermocellum cohesin-dockerin complex showed a dual binding mode of the dockerin module to the cohesin module, but this crystallographic study also revealed that two dockerin modules cannot simultaneously bind to one cohesin module (27), suggesting a strict stoichiometric assembly. Thus, the unexpected migration of the bands may be due to the higher concentration of protein in the native PAGE gel during electrophoresis compared to the concentration of the assembled complex in solution. Size exclusion chromatographic analysis of the reconstituted cellulosomes supported this notion, because the elution profiles of the complexes assembled at a CipA/enzyme ratio of <1/9 (cohesin/dockerin < 1/1) indicated that the unassembled enzymes did not associate with the high-molecular-mass complex in the elution buffer (Fig. 4B and C). One possible explanation for the unexpected behavior of the unassembled enzymes on native PAGE gels is as follows. Both type I and type II cohesin-dockerin interactions exhibited high affinity, with a dissociation constant of <10−9 M (30, 31). However, it has been reported that CipA on the cell surface localized by the type II cohesin-dockerin interaction can be replaced by free type II dockerin-containing proteins, suggesting that dockerin exchange occurs (32). Therefore, it is predicted that assembled enzymes can be replaced with a high concentration of free enzymes. If dockerin exchange occurs in the gel at high frequency during electrophoresis, it may retard the migration of bands corresponding to the unassembled enzymes. Dockerin exchange is also predicted to occur in the minicellulosome complex. However, the reduced number of cohesin modules in the minicellulosome complex may prevent decreased band mobility caused by dockerin exchange in the gel.
The activity profiles of the minicellulosome and cellulosome complexes suggested that displaying more enzyme molecules on a single scaffoldin protein generates more synergy for the degradation of crystalline cellulose and that the generated synergy contributes more to the degradation of crystalline cellulose than the CBM3a-mediated targeting effect. This conclusion is consistent with that of a previous study on the reconstituted cellulosome (14). In the paper by Krauss et al., the cellulosome reconstituted from full-length CipA was stoichiometrically assembled by assuming that the unassembled enzyme mixture had an average molecular mass of 80 kDa. This reconstituted cellulosome exhibited 12-fold-higher activity for Avicel than the unassembled enzyme mixture secreted from cipA-deficient C. thermocellum cells. In contrast, in the current study, the cellulosome complex stoichiometrically assembled at a CipA/enzyme molar ratio of 1/9 (cohesin/dockerin = 1/1) exhibited 4-fold-higher activity for Avicel than the unassembled enzyme mixture (Table 3). Thus, these two different in vitro-reconstituted cellulosomes exhibited different synergies for Avicel degradation (12- versus 4-fold synergy). Because the enzyme mixture prepared from cipA-deficient C. thermocellum cells contains a larger variety of cellulosomal cellulases, in addition to noncellulosomal cellulases, such as Cel9I and Cel48Y (33), the 3-fold difference in the observed synergy may be attributable to the difference in the variety of enzymatic components in the cellulosome complex (<70 versus 3 components). These findings are also supported by previous results showing that a mixture of C. cellulolyticum cellulosome complexes with different enzymatic compositions, separated by anion-exchange chromatography, synergistically acted to degrade Avicel (34). Furthermore, it is estimated that the activity for Avicel of the purified C. thermocellum cellulosome comprising a large variety of cellulosomal cellulases, reported by Krauss et al. (14), is ∼7-fold higher than that of the cellulosome complex comprising three enzymes reconstituted in this study (0.19 versus 0.026 U/mg). Taken together, these results suggest that the inclusion of a larger variety of enzymatic components in a cellulosome may generate more synergy and exhibit greater activity for the degradation of crystalline cellulose.
The mechanisms by which cellulosomes are assembled by various anaerobic bacteria have been investigated. The cellulosomal components are generally thought to bind randomly to each cohesin module in the scaffoldin protein. For example, C. thermocellum CipA contains nine cohesin modules with roughly four sequence diversities: the amino acid sequences of cohesins 3 to 8 are almost identical, but those of cohesins 1, 2, 3 to 8, and 9 are dissimilar. However, the cellulosomal components of C. thermocellum indiscriminately bound to recombinant cohesins 1 and 6 (14) and to cohesins 2 and 3 (35), even though the sequences of each cohesin pair are dissimilar. This indiscriminate binding was demonstrated by SDS-PAGE analysis of the cohesin-binding fractions of the whole cellulosomal components. However, nonrandomized integration of cellulosomal enzymes into a scaffoldin protein was recently proposed based on the results of electrophoretic mobility shift assays of the assembly of an artificial minicellulosome (36). These data suggested that preferential binding of cellulosomal enzymes to the cohesin modules did not result from slight differences in the binding affinity between the cohesin and dockerin modules, but rather, resulted from differences in the length of the intercohesin linker: a shorter intercohesin linker promoted preferential binding. Furthermore, computational simulations of the C. thermocellum cellulosome assembled with three cellulosomal enzymes (Cbh9A, Cel5B, and Cel48S) suggested that large multimodular enzymes (Cbh9A) may bind more frequently to CipA than smaller, single-modular enzymes (Cel5B or Cel48S) (37). Because CipA contains intercohesin linkers of various lengths and the three enzymes used in this study have different modular structures (Fig. 2A), these enzymes may also bind preferentially to certain cohesin modules in CipA. If so, this preferential binding may affect the ratio of these enzymes displayed on one CipA molecule (Cel48S/Cel8A/Cel9K = 5.5:2.5:1.0), estimated by assuming that these enzymes indiscriminately bind to the nine cohesin modules in CipA with similar binding affinities. However, it seems that this preferential binding does not affect the conclusion of this study, i.e., that the stoichiometric assembly of the cellulosome is crucial for generating maximum synergy for the degradation of crystalline cellulose. Further studies on cellulosome assembly using full-length CipA may provide a clue to solving the puzzle of cellulosome formation.
ACKNOWLEDGMENTS
We thank S. Tomitsuka for technical support and S. Shinoda for helpful discussions.
This work was supported by PRESTO from the Japan Science and Technology Agency; the Japan Association for Chemical Innovation; Challenging Exploratory Research grant 26550103 from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan; Grant-in-Aid for Young Scientists (B) 22760612 from the MEXT of Japan; and the Strategic Research Foundations at Private Universities 2013–2017 and 2014–2018 from the MEXT of Japan.
REFERENCES
- 1.Bayer EA, Belaich JP, Shoham Y, Lamed R. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol 58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 2.Demain AL, Newcomb M, Wu JH. 2005. Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev 69:124–154. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gold ND, Martin VJ. 2007. Global view of the Clostridium thermocellum cellulosome revealed by quantitative proteomic analysis. J Bacteriol 189:6787–6795. doi: 10.1128/JB.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raman B, Chongle P, Hurst GB, Rodriguez M, McKeown CM, Lankford PK, Samatova NF, Mielenz JR. 2009. Impact of pretreated switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis. PLoS One 4:e5271. doi: 10.1371/journal.pone.0005271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruus K, Wang WK, Ching J, Wu JH. 1995. Exoglucanase activities of the recombinant Clostridium thermocellum CelS, a major cellulosome component. J Bacteriol 177:1641–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guimaraes BG, Souchon H, Lytle BL, Wu JH, Alzari PM. 2002. The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum cellulosome. J Mol Biol 320:587–596. doi: 10.1016/S0022-2836(02)00497-7. [DOI] [PubMed] [Google Scholar]
- 7.Kataeva I, Li XL, Chen H, Choi SK, Ljungdahl LG. 1999. Cloning and sequence analysis of a new cellulose gene encoding CelK, a major cellulosome component of Clostridium thermocellum: evidence for gene duplication and recombination. J Bacteriol 181:5288–5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zverlov VV, Klupp M, Krauss J, Schwarz WH. 2008. Mutations in the scaffoldin gene, cipA, of Clostridium thermocellum with impaired cellulosome formation and cellulosome hydrolysis: insertions of a new transposable element, IS1447, and implications for cellulase synergism on crystalline cellulose. J Bacteriol 190:4321–4327. doi: 10.1128/JB.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson DG, Giannone RJ, Hettich RL, Lynd LR. 2013. Role of the CipA scaffoldin protein in cellulose solubilization, as determined by targeted gene deletion and complementation in Clostridium thermocellum. J Bacteriol 195:733–739. doi: 10.1128/JB.02014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Currie DH, Herring CD, Guss AM, Olson DG, Hogsett DA, Lynd LR. 2013. Functional heterologous expression of an engineered full length CipA from Clostridium thermocellum in Thermoanaerobacterium saccharolyticum. Biotechnol Biofuels 6:32. doi: 10.1186/1754-6834-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fierobe HP, Mechaly A, Tardif C, Belaich A, Lamed R, Shoham Y, Belaich JP, Bayer EA. 2001. Design and production of active cellulosome chimeras. Selective incorporation of dockerin-containing enzymes into defined functional complexes. J Biol Chem 276:21257–21261. doi: 10.1074/jbc.M102082200. [DOI] [PubMed] [Google Scholar]
- 12.Fierobe HP, Bayer EA, Tardif C, Czjzek M, Mechaly A, Belaich A, Lamed R, Shoham Y, Belaich JP. 2002. Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J Biol Chem 277:49621–49630. doi: 10.1074/jbc.M207672200. [DOI] [PubMed] [Google Scholar]
- 13.Fierobe HP, Mingardon F, Mechaly A, Belaich A, Rincon MT, Pages S, Lamed R, Tardif C, Belaich JP, Bayer EA. 2005. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J Biol Chem 280:16325–16334. doi: 10.1074/jbc.M414449200. [DOI] [PubMed] [Google Scholar]
- 14.Krauss J, Zverlov VV, Schwarz WH. 2012. In vitro reconstitution of the complete Clostridium thermocellum cellulosome and synergistic activity on crystalline cellulose. Appl Environ Microbiol 78:4301–4307. doi: 10.1128/AEM.07959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirano N, Sawasaki T, Tozawa Y, Endo Y, Takai K. 2006. Tolerance for random recombination of domains in prokaryotic and eukaryotic translation systems: limited interdomain misfolding in a eukaryotic translation system. Proteins 64:343–354. doi: 10.1002/prot.21008. [DOI] [PubMed] [Google Scholar]
- 16.Hirano N, Hasegawa H, Nihei S, Haruki M. 2013. Cell-free protein synthesis and substrate specificity of full-length endoglucanase CelJ (Cel9D-Cel44A), the largest multi-enzyme subunit of the Clostridium thermocellum cellulosome. FEMS Microbiol Lett 344:25–30. doi: 10.1111/1574-6968.12149. [DOI] [PubMed] [Google Scholar]
- 17.Deng K, Takasuka TE, Heins R, Cheng X, Bergeman LF, Shi J, Aschenbrener R, Deutsch S, Singh S, Sale KL, Simmons BA, Adams PD, Singh AK, Fox BG, Northen TR. 2014. Rapid kinetic characterization of glycosyl hydrolases based on oxime derivatization and nanostructure-initiator mass spectrometry (NIMS). ACS Chem Biol 9:1470–1479. doi: 10.1021/cb5000289. [DOI] [PubMed] [Google Scholar]
- 18.Takai K, Sawasaki T, Endo Y. 2010. Practical cell-free protein synthesis system using purified wheat embryos. Nat Protoc 5:227–238. doi: 10.1038/nprot.2009.207. [DOI] [PubMed] [Google Scholar]
- 19.Gerngross UT, Romaniec MP, Kobayashi T, Huskisson NS, Demain AL. 1993. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol Microbiol 8:325–334. doi: 10.1111/j.1365-2958.1993.tb01576.x. [DOI] [PubMed] [Google Scholar]
- 20.Hirano N, Ohshima H, Takahashi H. 2006. Biochemical analysis of the substrate specificity and sequence preference of endonuclease IV from bacteriophage T4, a dC-specific endonuclease implicated in restriction of dC-substituted T4 DNA synthesis. Nucleic Acids Res 34:4743–4751. doi: 10.1093/nar/gkl553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood TM. 1988. Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol 160:19–25. doi: 10.1016/0076-6879(88)60103-0. [DOI] [Google Scholar]
- 22.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 23.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. 2006. Quantitative proteomic analysis of distinct mammalian mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A 103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morag E, Lapidot A, Govorko D, Lamed R, Wilchek M, Bayer EA, Shoham Y. 1995. Expression, purification, and characterization of the cellulose-binding domain of the scaffoldin subunit from the cellulosome of Clostrdium thermocellum. Appl Environ Microbiol 61:1980–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz WH, Grabnitz F, Staudenbauer WL. 1986. Properties of a Clostridium thermocellum endoglucanase produced in Escherichia coli. Appl Environ Microbiol 51:1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lytle B, Myers C, Kruus K, Wu JH. 1996. Interaction of the CelS binding ligand with various receptor domains of the Clostridium thermocellum cellulosomal scaffoldin protein, CipA. J Bacteriol 178:1200–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho AL, Dias FM, Nagy T, Prates JA, Proctor MR, Smith N, Bayer EA, Davies GJ, Ferreira LM, Romao MJ, Fontes CM, Gilbert HJ. 2007. Evidence for a dual binding mode of dockerin modules to cohesins. Proc Natl Acad Sci U S A 104:3089–3094. doi: 10.1073/pnas.0611173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Currie MA, Adams JJ, Faucher F, Bayer EA, Jia Z, Smith SP. 2012. Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome. J Biol Chem 287:26953–26961. doi: 10.1074/jbc.M112.343897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Currie MA, Cameron K, Dias FM, Spencer HL, Bayer EA, Fontes CM, Smith SP, Jia Z. 2013. Small angle X-ray scattering analysis of Clostridium thermocellum cellulosome N-terminal complexes reveals a highly dynamic structure. J Biol Chem 288:7978–7985. doi: 10.1074/jbc.M112.408757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mechaly A, Fierobe HP, Belaich A, Belaich JP, Lamed R, Shoham Y, Bayer EA. 2001. Cohesin-dockerin interaction in cellulosome assembly: a single hydroxyl group of a dockerin domain distinguishes between nonrecognition and high affinity recognition. J Biol Chem 276:9883–9888. doi: 10.1074/jbc.M009237200. [DOI] [PubMed] [Google Scholar]
- 31.Jindou S, Kajino T, Inagaki M, Karita S, Beguin P, Kimura T, Sakka K, Ohmiya K. 2004. Interaction between a type-II dockerin domain and a type-II cohesin domain from Clostridium thermocellum cellulosome. Biosci Biotechnol Biochem 68:924–926. doi: 10.1271/bbb.68.924. [DOI] [PubMed] [Google Scholar]
- 32.Waller BH, Olson DG, Currie DH, Guss AM, Lynd LR. 2013. Exchange of type II dockerin-containing subunits of the Clostridium thermocellum cellulosome as revealed by SNAP-tags. FEMS Microbiol Lett 338:46–53. doi: 10.1111/1574-6968.12029. [DOI] [PubMed] [Google Scholar]
- 33.Berger E, Zhang D, Zverlov VV, Schwarz WH. 2007. Two noncellulosomal cellulases of Clostridium thermocellum, Cel9I and Cel48Y, hydrolyze crystalline cellulose synergistically. FEMS Microbiol Lett 268:194–201. doi: 10.1111/j.1574-6968.2006.00583.x. [DOI] [PubMed] [Google Scholar]
- 34.Fendri I, Tardif C, Fierobe HP, Lignon S, Valette O, Pages S, Perret S. 2009. The cellulosomes from Clostridium cellulolyticum: identification of new components and synergies between complexes. FEBS J 276:3076–3086. doi: 10.1111/j.1742-4658.2009.07025.x. [DOI] [PubMed] [Google Scholar]
- 35.Yaron S, Morag E, Bayer EA, Lamed R, Shoham Y. 1995. Expression, purification and subunit properties of cohesins 2 and 3 of the Clostridium thermocellum cellulosome. FEBS Lett 360:121–124. doi: 10.1016/0014-5793(95)00074-J. [DOI] [PubMed] [Google Scholar]
- 36.Borne R, Bayer EA, Pages S, Perret S, Fierobe HP. 2013. Unraveling enzyme discrimination during cellulosome assembly independent of cohesin-dockerin affinity. FEBS J 280:5764–5779. doi: 10.1111/febs.12497. [DOI] [PubMed] [Google Scholar]
- 37.Bomble YJ, Beckham GT, Matthews JF, Nimlos MR, Himmel ME, Crowley MF. 2011. Modeling the self-assembly of the cellulosome enzyme complex. J Biol Chem 286:5614–5623. doi: 10.1074/jbc.M110.186031. [DOI] [PMC free article] [PubMed] [Google Scholar]