Abstract
Yersiniosis is strongly associated with the consumption of pork contaminated with enteropathogenic Yersinia enterocolitica, which is harbored by domestic pigs without showing clinical signs of disease. In contrast to data on Y. enterocolitica isolated from conventionally reared swine, investigations into the occurrence of Y. enterocolitica in wild boars in Germany are rare. The objectives of the study were to get knowledge about these bacteria and their occurrence in wild boars hunted in northern Germany by isolation of the bacteria from the tonsils, identification of the bioserotypes, determination of selected virulence factors, macrorestriction analysis, multilocus sequence typing (MLST), and testing of antimicrobial susceptibility. Altogether, tonsils from 17.1% of 111 tested wild boars were positive for Y. enterocolitica by culture methods. All but two isolates belonged to biotype (BT) 1A, with the majority of isolates bearing a ystB nucleotide sequence which was revealed to have 85% identity to internal regions of Y. enterocolitica heat-stable enterotoxin type B genes. The remaining Y. enterocolitica isolates were identified to be BT 1B and did not carry the virulence plasmid. However, two BT 1A isolates carried the ail gene. Macrorestriction analysis and results from MLST showed a high degree of genetic diversity of the isolates, although the region where the samples were taken was restricted to Lower Saxony, Germany, and wild boars were shot during one hunting season. In conclusion, most Y. enterocolitica isolates from wild boars investigated in this study belonged to biotype 1A. Enteropathogenic Y. enterocolitica bioserotypes 4/O:3 and 2/O:9, usually harbored by commercially raised pigs in Europe, could not be identified.
INTRODUCTION
The genus Yersinia belongs to the bacterial family Enterobacteriaceae and is actually composed of 18 species (1), with 3 of these species being shown to be human pathogens (Yersinia pestis, Y. pseudotuberculosis, Y. enterocolitica). Y. enterocolitica has been found in a variety of food, animal, and environmental samples and comprises both pathogenic and nonpathogenic strains. It is traditionally subdivided into bioserotypes on the basis of the combination of biochemical and serological tests (2). Y. enterocolitica species have six biotypes (BTs), of which five (BTs 1B, 2, 3, 4, and 5) contain pathogenic strains, characterized by the presence of the 70-kb virulence plasmid, termed pYV (3, 4). Because strains of BT 1A lack pYV, they have been regarded as avirulent in the past, although patients with diarrhea and outbreaks of gastrointestinal infections due to BT 1A yersiniae have been reported (5–8). The virulence plasmid pYV carries genes encoding adhesin A (YadA), Yersinia outer membrane proteins (Yops) from the type III secretion system, and the transcriptional regulator gene (virF) (4). Apart from pYV itself, pYV-bearing strains of Y. enterocolitica require a number of chromosomally encoded factors to express full virulence, such as the inv gene, which encodes the primary invasion factor for Y. enterocolitica (9); the ail gene, which encodes an outer membrane protein that promotes attachment and invasion (10); the rfbC gene, which can be used to identify pathogenic Y. enterocolitica O:3 strains (11); and the ystA gene, which encodes a heat-stable enterotoxin (12).
The ystB gene, which codes for another heat-stable, mouse-reactive enterotoxin, is mainly present in BT 1A strains of Y. enterocolitica (13).
In Europe, Y. enterocolitica strains of BT 4 (serotype O:3) and BT 2 (serotype O:9) are often associated with clinical cases in humans (14). Swine are an important reservoir of these bioserotypes, and they usually carry the agent asymptomatically in the tonsils (15). Besides the consumption of raw minced pork, which was identified to be a risk factor for human yersiniosis in Germany, dogs and cats might also act as sources of Y. enterocolitica (3, 16). Reports on the occurrence of Y. enterocolitica in wild boars (Sus scrofa) are rare, and the epidemiological link between wild boars and domestic pigs is still unknown (17). Serological investigations in northeastern Germany and in Switzerland detected anti-Yersinia antibodies in more than 60% of wild boars (18, 19), which is comparable to the results obtained in finishing pigs (20).
In order to get insight into the occurrence of Y. enterocolitica in wild boars and to estimate the role of wild boars as a reservoir of pathogenic Yersinia species as well as the potential risk of wild boar meat for consumers, we investigated Yersinia isolates from the tonsils of 111 wild boars hunted in Lower Saxony, Germany, with respect to their virulence profiles and phylogenetic affiliation in relation to the sequence types (STs) from humans and pigs described so far.
MATERIALS AND METHODS
Sample collection and isolation of strains.
Tonsils were collected from the heads of 111 wild boars hunted in Lower Saxony, Germany, during one hunting season, between November 2012 and January 2013. The animals were shot at the weekends and immediately delivered to a butcher specialized for game. Heads were separated on the same day and stored in a cold storage room until sampling of tonsils on Monday mornings. The age of the animals was estimated according to their tooth development. Sampling of tonsils was performed according to the instructions for tonsil sampling given in the Technical Specifications for Harmonised National Surveys on Yersinia enterocolitica in Slaughter Pigs (21). After they were removed, the tonsils were stored in a sterile plastic bag at room temperature and sent to the laboratory within 1 h. For the isolation of presumptive Yersinia spp., the standardized ISO 10273:2003 method was used (22). In brief, approximately 10 g of lacerated tonsils was inoculated in nine times the mass of prewarmed peptone-sorbitol-bile-salt (PSB) broth (Sigma-Aldrich, Munich, Germany) to give a 1:10 dilution. After incubation for 2 to 3 days at 25°C with shaking, a volume of 500 μl of the enrichment broth was transferred into 4.5 ml of a 0.25% potassium hydroxide (KOH) solution and mixed for 20 s. Subsequently, 1 ml of the inoculated KOH solution and 1 ml of the PSB enrichment broth were spread over at least five cefsulodin-irgasan-novobiocin (CIN) agar plates (bioMérieux, Nürtingen, Germany). The agar plates were incubated at 30°C for 24 h. In parallel, 10 ml of the initial PSB enrichment broth was transferred into 90 ml of prewarmed irgasan-ticarcillin-potassium chlorate (ITC) broth (Sigma-Aldrich, Munich, Germany) to give a 1:100 dilution. After incubation at 25°C for 48 h, a total of 1 ml of the ITC enrichment broth was spread over five Salmonella-Shigella-deoxycholate-calcium chlorate (SSDC) agar plates (Merck, Darmstadt, Germany). The agar plates were incubated at 30°C for 24 to 48 h. Both the SSDC and the CIN agar plates were checked for characteristic colonies with a bull's-eye appearance using a stereomicroscope.
Species identification by MALDI-TOF MS and cultivation of isolates.
Five presumptive Yersinia colonies per sample were subcultured on Luria-Bertani (LB) agar plates. After incubation for 24 h at 30°C, one inoculation loop of bacteria was prepared as described earlier (23) and loaded into a matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) system (Microflex LT; Bruker Daltonics). The mass spectrometry fingerprint was generated with Bruker real-time classification software (version 3.0) and analyzed by using the MALDI biotyper database (last update, October 2012). The database contains the profiles from 11 Yersinia species; however, the profiles from Y. pestis strains are not included. For control purposes, the type and clinical strains Y. enterocolitica subsp. palearctica DSM 11502, Y. enterocolitica subsp. enterocolitica DSM 11503, Y. enterocolitica subsp. enterocolitica DSM 11504, and Y. pseudotuberculosis DSM 8992T were included in each MALDI-TOF MS experimental setup.
Biotyping and serotyping.
The isolates were serotyped by a slide agglutination test using Y. enterocolitica commercial antisera for the antigens O:3, O:5, O:8, O:9, and O:27 (Sifin, Berlin, Germany). To exclude autoagglutination, 1 drop of a 0.9% saline solution was mixed with the bacterial culture material prior to serotyping. Biotyping was performed at the National Reference Center for Salmonella and Other Bacterial Enteric Pathogens, Robert Koch Institute, Wernigerode, Germany. The isolates were biotyped on the basis of pyrazinamidase and Tween activity; esculin hydrolysis; indole production; and salicin, xylose, and trehalose fermentation tests according to the ISO 10273:2003 method (2, 22).
DNA isolation, 16S rRNA gene sequencing, and identification of virulence markers.
Genomic DNA was extracted by using a DNeasy tissue kit (Qiagen, Hilden, Germany). For species confirmation, a part of the 16S rRNA gene comprising 458 bp of the 533-bp PCR product was amplified by PCR and sequenced. For this, the FD1mod and r533 primer pair was used, and the cycling conditions were those used in a previously described protocol (24). For the detection of the chromosomal gene ail, a real-time PCR assay described by Maede et al. (25) was performed on a LightCycler 480 II instrument (Roche Diagnostics, Mannheim, Germany). In addition, the presence of further chromosomal virulence genes (ystA and ystB) and the plasmid-borne virulence genes yadA, virF, and yopT was investigated by PCR amplification as described earlier (26, 27).
MLST and macrorestriction analysis.
The sequence types of all Y. enterocolitica isolates were determined by multilocus sequence typing (MLST) and by use of the Yersinia MLST website (http://pubmlst.org/yersinia/) (28). New MLST types were assigned by the MLST database curators. The affiliation to groups of related genotypes was determined by using the software eBURST (29). Macrorestriction analysis was performed to investigate the clonality of the isolates. NotI (Roche Diagnostics, Mannheim, Germany)-digested fragments of genomic DNA were separated in a CHEF DR II system (Bio-Rad, Munich, Germany) as described by Liang and coworkers (30). The total run time was 19 h, and 0.5× Tris-borate-EDTA buffer was used as the running buffer. The pulse time was increased from 2 to 20 s over the entire run time. The gels were stained with ethidium bromide and analyzed using BioNumerics software (version 7.0; Applied Maths, Sint-Martens-Latem, Belgium). The band patterns were analyzed using the Dice coefficient with 0.5% optimization and 1% position tolerance.
Antimicrobial susceptibility testing.
Testing of susceptibility to a panel of 14 antibiotics/antibiotic combinations was performed by broth microdilution following the recommendations given in Clinical and Laboratory Standards Institute (CLSI) documents M07-A9 (31), M100-S22 (32), and VET01-S2 (33). For quality control purposes, Escherichia coli ATCC 25922 was used as a reference strain.
RESULTS AND DISCUSSION
In recent years, a couple of studies on the presence of bacterial zoonotic pathogens, including Yersinia spp., in wild boars aimed at estimating the role of wild boars as a reservoir for pathogens have become available (18, 19, 34–36). It could be shown that wild boars might act as reservoirs for various pathogens, e.g., Brucella suis, nontoxigenic tox-bearing Corynebacterium ulcerans, and hepatitis E virus (37–39). The purposes of those investigations were to evaluate the risk of transmission of pathogens to domestic pigs or to estimate the risk that consumers would become infected, considering the increased popularity of wild boar meat. However, a relationship between enteropathogenic Yersinia isolates from wild boars and isolates from domestic pigs or humans was not identified.
In order to gain knowledge on the occurrence of Y. enterocolitica in wild boars, the tonsils of 111 wild boars hunted in Lower Saxony, Germany, were investigated. (In total, the number of wild boars shot during the 2012-2013 hunting season in Lower Saxony was 49,881 [40], far greater than our sample size.). Altogether 17.1% of the wild boars' tonsils were positive for Y. enterocolitica, while two boars (1.8%) carried isolates identified to be Y. frederiksenii. All isolates were identified by MALDI-TOF MS, and results were confirmed by 16S rRNA gene sequencing (Table 1). The sequenced 458-bp regions corresponded exactly (or showed a single-base-pair exchange) to Y. enterocolitica 16S rRNA gene sequences deposited in the GenBank database (accession no. CP009838.1 and FN812722.1). For 18 out of 19 positive tonsils, Y. enterocolitica colonies were identified using CIN agar plates, while SSDC agar plates contained a large amount of contaminating flora. It has recently been reported that members of the genera Morganella, Proteus, Serratia, and Aeromonas in particular might interfere with the differentiation of Yersinia on SSDC agar plates (41). Y. pseudotuberculosis was not detected in any of the samples; however, CIN and SSDC agar plates were not designed for isolation of this species (19). Antimicrobial susceptibility testing revealed that all Yersinia isolates showed ampicillin MIC values of ≥32 mg/liter, and therefore, these isolates were classified as resistant, while the isolates were susceptible to or exhibited low MIC values for all other antimicrobial agents tested. Hence, the resistance profiles of the Yersinia isolates tested differed from those of the livestock isolates, which, in some cases, exhibit resistance to sulfonamides, tetracycline, or streptomycin, in addition to ampicillin (42, 43).
TABLE 1.
Occurrence of Y. enterocolitica and Y. frederiksenii in 111 wild boars shot in Lower Saxony, Germany
| Strain | No. (%) of positive wild boars by age |
|||
|---|---|---|---|---|
| <12 mo (n = 48) | 12 to 24 mo (n = 47) | >24 mo (n = 16) | Total (n = 111) | |
| Y. frederiksenii | 2 (4.1) | 0 | 0 | 2 (1.8) |
| Y. enterocolitica BT 1A | 5 (10.4) | 11 (23.4) | 1 (6.3) | 17 (15.3) |
| Y. enterocolitica BT 1B | 0 | 1 (2.1) | 1 (6.3) | 2 (1.8) |
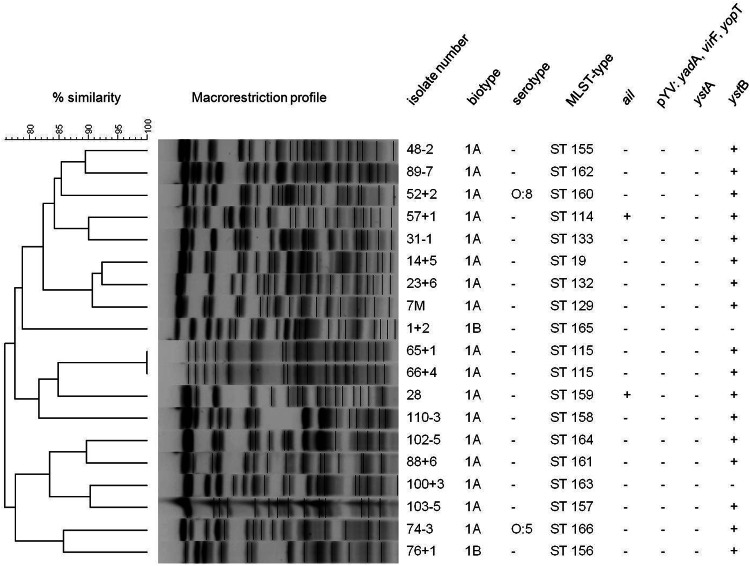
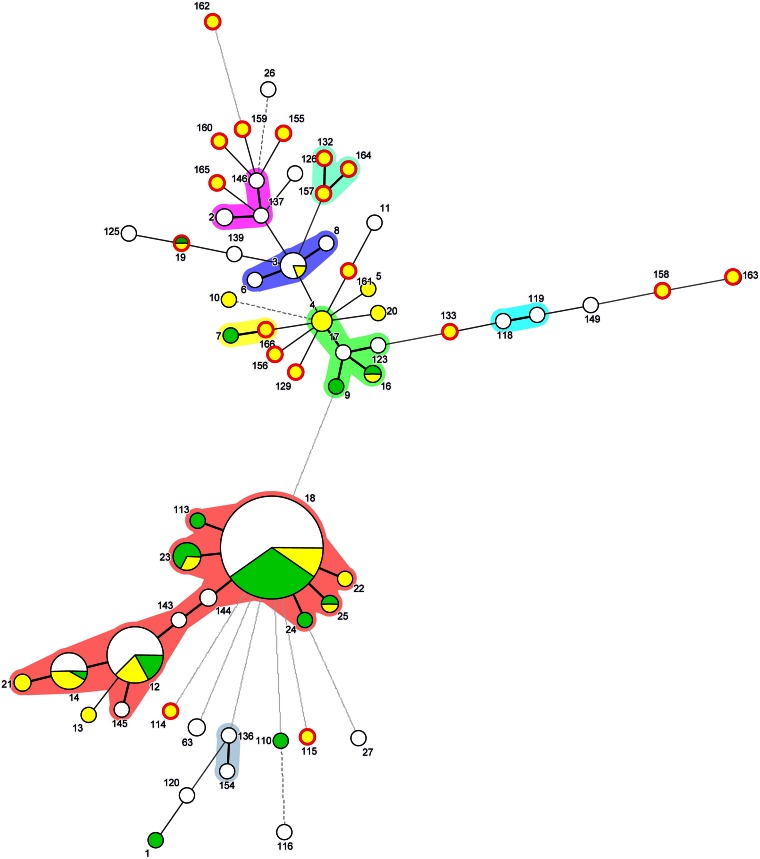
The majority of the isolates belonged to BT 1A (89.5%); the exceptions were two isolates identified to be BT 1B (Table 1). The absence of the plasmid-borne genes (yadA, yopT, and virF) confirmed the lack of plasmid pYV and the results of biotyping (Fig. 1). Among all Y. enterocolitica biotypes, BT 1A is regarded as the most heterogeneous biotype and includes a wide range of serotypes. Of these, strains of serotypes O:5, O:6,30, O:6,31, O:7,8, and O:10 and O-nontypeable strains are isolated the most often (5). In the present study, the isolates were tested only for the most relevant serotypes causing gastroenteritis in humans in Europe, and serotyping succeeded for only two isolates, belonging to serotypes O:5 and O:8 (Fig. 1). Because of the restricted availability of commercially available sera, most isolates had to be referred to as nontypeable by serotyping. Molecular analysis for virulence markers detected the ail gene in two Y. enterocolitica isolates of BT 1A, and these isolates lacked other classical virulence genes besides ystB (Fig. 1). Regarding the minimum spanning tree, both STs of the ail-positive isolates (ST114 and ST159) were genetically distant from each other (Fig. 2). The ail gene is often used to differentiate pathogenic from nonpathogenic Yersinia strains, even though ail-positive BT 1A isolates were recently reported, leading to the conclusion that methods which are exclusively based on the detection of ail genes are insufficient to distinguish between pathogenic and nonpathogenic strains (17, 44, 45). In contrast to our results, 14 out of 17 enteropathogenic Yersinia isolates recovered from wild boars shot between 2007 and 2008 in Switzerland were identified to be Y. enterocolitica and carriers of the chromosomal virulence gene marker ail (19). Of these, 5, 4, and 3 isolates were assigned to bioserotypes 4/O:3, 2/O:9, and 2/O:5,27, respectively (19). Whether the deviating findings of the Swiss study (19) and the present study are based on differences in the methodologies used for the isolation of Y. enterocolitica or represent an uneven geographical distribution of isolates remains to be clarified.
FIG 1.
NotI macrorestriction patterns and characteristics of Y. enterocolitica isolates from wild boars.
FIG 2.
Minimum spanning tree based on the MLST profiles obtained from the MLST database (last accessed 18 December 2014; http://pubmlst.org/yersinia/) and generated using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). The Arabic numbers indicate the ST assignment. The colored areas around the circles represent single-locus variants. The length of the connecting lines is proportional to the number of different MLST alleles. Green, isolates of human origin; yellow, isolates of animal origin; white, the origins of the isolates were not defined in the MLST database. The sizes of the circles are proportional to the number of isolates. The position of the Y. enterocolitica isolates within the population from wild boars collected during this study (including all MLSTs) is indicated by a red edge.
The ystB gene was found in all Y. enterocolitica isolates except two, one of which belonged to BT 1B and one of which belonged to BT 1A (Fig. 1). To exclude the possibility of mispriming, PCR experiments and subsequent sequencing of the ystB PCR amplicons were conducted. Sequence analysis was performed with the program nucleotide BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The ystB nucleotide sequence revealed 86% and 85% identities to internal regions of ystB genes from beavers (GenBank accession no. KJ592627 and KJ592626.1) and from a human clinical isolate (GenBank accession no. D88145.1), respectively. However, the presence of the complete gene and its functionality still need to be examined.
Two Y. enterocolitica isolates were identified to be BT 1B (Fig. 1), which is considered highly pathogenic for humans (46). However, neither isolate carried the virulence plasmid, calling their pathogenic potential into question.
Among the 19 Y. enterocolitica isolates, 18 different macrorestriction patterns were detected (Fig. 1). Only two isolates obtained from the same quarry showed indistinguishable NotI restriction endonuclease digestion profiles. As described previously, macrorestriction analysis and results from MLST showed a high degree of genetic diversity of the Y. enterocolitica BT 1A isolates from wild boars (47), even though all samples were taken in a circumscribed geographical region during the same hunting season. Only a single isolate (ST19) shared an MLST sequence pattern previously detected in a single Y. enterocolitica BT 1A strain of human origin, which was isolated in the year 2003 in the United Kingdom (http://pubmlst.org/perl/bigsdb/bigsdb.pl?page=info&db=pubmlst_yersinia_isolates&id=126). For the remaining Y. enterocolitica isolates, 17 novel MLST types were assigned.
In conclusion, most Y. enterocolitica isolates from wild boars belonged to biotype 1A, while enteropathogenic Y. enterocolitica bioserotypes 4/O:3 and 2/O:9, usually harbored by commercially raised pigs in Europe, could not be identified.
ACKNOWLEDGMENTS
We thank Elke Siever, Inna Pahl, and Vera Nöding for excellent technical assistance.
REFERENCES
- 1.Savin C, Martin L, Bouchier C, Filali S, Chenau J, Zhou Z, Becher F, Fukushima H, Thomson NR, Scholz HC, Carniel E. 2014. The Yersinia pseudotuberculosis complex: characterization and delineation of a new species, Yersinia wautersii. Int J Med Microbiol 304:452–463. doi: 10.1016/j.ijmm.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Wauters G, Kandolo K, Janssens M. 1987. Revised biogrouping scheme of Yersinia enterocolitica. Contrib Microbiol Immunol 9:14–21. [PubMed] [Google Scholar]
- 3.Stamm I, Hailer M, Depner B, Kopp PA, Rau J. 2013. Yersinia enterocolitica in diagnostic fecal samples from European dogs and cats: identification by Fourier transform infrared spectroscopy and matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 51:887–893. doi: 10.1128/JCM.02506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelis GR, Boland A, Boyd AP, Geuijen C, Iriarte M, Neyt C, Sory MP, Stainier I. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev 62:1315–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tennant SM, Grant TH, Robins-Browne RM. 2003. Pathogenicity of Yersinia enterocolitica biotype 1A. FEMS Immunol Med Microbiol 38:127–137. doi: 10.1016/S0928-8244(03)00180-9. [DOI] [PubMed] [Google Scholar]
- 6.Stephan R, Joutsen S, Hofer E, Säde E, Björkroth J, Ziegler D, Fredriksson-Ahomaa M. 2013. Characteristics of Yersinia enterocolitica biotype 1A strains isolated from patients and asymptomatic carriers. Eur J Clin Microbiol Infect Dis 32:869–875. doi: 10.1007/s10096-013-1820-1. [DOI] [PubMed] [Google Scholar]
- 7.Mallik S, Virdi JS. 2010. Genetic relationships between clinical and non-clinical strains of Yersinia enterocolitica biovar 1A as revealed by multilocus enzyme electrophoresis and multilocus restriction typing. BMC Microbiol 10:158. doi: 10.1186/1471-2180-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenwood MH, Hooper WL. 1990. Excretion of Yersinia spp. associated with consumption of pasteurized milk. Epidemiol Infect 104:345–350. doi: 10.1017/S0950268800047361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pepe JC, Miller VL. 1993. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc Natl Acad Sci U S A 90:6473–6477. doi: 10.1073/pnas.90.14.6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller VL, Farmer JJ III, Hill WE, Falkow S. 1989. The ail locus is found uniquely in Yersinia enterocolitica serotypes commonly associated with disease. Infect Immun 57:121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weynants V, Jadot V, Denoel PA, Tibor A, Letesson JJ. 1996. Detection of Yersinia enterocolitica serogroup O:3 by a PCR method. J Clin Microbiol 34:1224–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delor I, Cornelis GR. 1992. Role of Yersinia enterocolitica Yst toxin in experimental infection of young rabbits. Infect Immun 60:4269–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant T, Bennett-Wood V, Robins-Browne RM. 1998. Identification of virulence-associated characteristics in clinical isolates of Yersinia enterocolitica lacking classical virulence markers. Infect Immun 66:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.European Food Safety Authority. 2007. Monitoring and identification of human enteropathogenic Yersinia spp. EFSA J 595:1–30. doi: 10.2903/j.efsa.2007.595. [DOI] [Google Scholar]
- 15.Fredriksson-Ahomaa M, Bucher M, Hank C, Stolle A, Korkeala H. 2001. High prevalence of Yersinia enterocolitica 4:O3 on pig offal in southern Germany: a slaughtering technique problem. Syst Appl Microbiol 24:457–463. doi: 10.1078/0723-2020-00055. [DOI] [PubMed] [Google Scholar]
- 16.Rosner BM, Stark K, Höhle M, Werber D. 2012. Risk factors for sporadic Yersinia enterocolitica infections, Germany 2009-2010. Epidemiol Infect 140:1738–1747. doi: 10.1017/S0950268811002664. [DOI] [PubMed] [Google Scholar]
- 17.Fredriksson-Ahomaa M, Wacheck S, Bonke R, Stephan R. 2011. Different enteropathogenic Yersinia strains found in wild boars and domestic pigs. Foodborne Pathog Dis 8:733–737. doi: 10.1089/fpd.2010.0711. [DOI] [PubMed] [Google Scholar]
- 18.Al Dahouk S, Nockler K, Tomaso H, Splettstoesser WD, Jungersen G, Riber U, Petry T, Hoffmann D, Scholz HC, Hensel A, Neubauer H. 2005. Seroprevalence of brucellosis, tularemia, and yersiniosis in wild boars (Sus scrofa) from north-eastern Germany. J Vet Med 52:444–455. doi: 10.1111/j.1439-0450.2005.00898.x. [DOI] [PubMed] [Google Scholar]
- 19.Fredriksson-Ahomaa M, Wacheck S, Koenig M, Stolle A, Stephan R. 2009. Prevalence of pathogenic Yersinia enterocolitica and Yersinia pseudotuberculosis in wild boars in Switzerland. Int J Food Microbiol 135:199–202. doi: 10.1016/j.ijfoodmicro.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 20.von Altrock A, Roesler U, Waldmann KH. 2011. Herd factors associated with the serological Yersinia prevalence in fattening pig herds. Foodborne Pathog Dis 8:1249–1255. doi: 10.1089/fpd.2011.0883. [DOI] [PubMed] [Google Scholar]
- 21.European Food Safety Authority. 2009. Technical specifications for harmonised national surveys on Yersinia enterocolitica in slaughter pigs. EFSA J 7:1374. doi: 10.2903/j.efsa.2009.1374. [DOI] [Google Scholar]
- 22.Anonymous. 2003. Horizontal method for the detection of presumptive pathogenic Yersinia enterocolitica. ISO 10273:2003 International Organization for Standardization, Geneva, Switzerland. [Google Scholar]
- 23.Ayyadurai S, Flaudrops C, Raoult D, Drancourt M. 2010. Rapid identification and typing of Yersinia pestis and other Yersinia species by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. BMC Microbiol 10:285. doi: 10.1186/1471-2180-10-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sihvonen LM, Haukka K, Kuusi M, Virtanen MJ, Siitonen A, YE Study Group . 2009. Yersinia enterocolitica and Y. enterocolitica-like species in clinical stool specimens of humans: identification and prevalence of bio/serotypes in Finland. Eur J Clin Microbiol Infect Dis 28:757–765. doi: 10.1007/s10096-008-0696-y. [DOI] [PubMed] [Google Scholar]
- 25.Maede D, Reiting R, Strauch E, Ketteritzsch K, Wicke A. 2008. A real-time PCR for detection of pathogenic Yersinia enterocolitica in food combined with an universal internal amplification control system. J Verbr Lebensm 3:141–151. doi: 10.1007/s00003-008-0341-9. [DOI] [Google Scholar]
- 26.Arnold T, Hensel A, Hagen R, Aleksic S, Neubauer H, Scholz HC. 2001. A highly specific one-step PCR—assay for the rapid discrimination of enteropathogenic Yersinia enterocolitica from pathogenic Yersinia pseudotuberculosis and Yersinia pestis. Syst Appl Microbiol 24:285–289. doi: 10.1078/0723-2020-00040. [DOI] [PubMed] [Google Scholar]
- 27.Thoerner P, Bin Kingombe CL, Bogli-Stuber K, Bissig-Choisat B, Wassenaar TM, Frey J, Jemmi T. 2003. PCR detection of virulence genes in Yersinia enterocolitica and Yersinia pseudotuberculosis and investigation of virulence gene distribution. Appl Environ Microbiol 69:1810–1816. doi: 10.1128/AEM.69.3.1810-1816.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang J, Wang X, Xiao Y, Cui Z, Xia S, Hao Q, Yang J, Luo L, Wang S, Li K, Yang H, Gu W, Xu J, Kan B, Jing H. 2012. Prevalence of Yersinia enterocolitica in pigs slaughtered in Chinese abattoirs. Appl Environ Microbiol 78:2949–2956. doi: 10.1128/AEM.07893-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 32.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing, 22nd informational supplement. M100-S22 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. 2013. performance standards of antimicrobial disk and dilution susceptibility tests of bacteria isolated from animals; second informational supplement. VET01-S2 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 34.Wacheck S, Fredriksson-Ahomaa M, Konig M, Stolle A, Stephan R. 2010. Wild boars as an important reservoir for foodborne pathogens. Foodborne Pathog Dis 7:307–312. doi: 10.1089/fpd.2009.0367. [DOI] [PubMed] [Google Scholar]
- 35.Nikolova S, Tzvetkov Y, Najdenski H, Vesselinova A. 2001. Isolation of pathogenic yersiniae from wild animals in Bulgaria. J Vet Med 48:203–209. doi: 10.1046/j.1439-0450.2001.00448.x. [DOI] [PubMed] [Google Scholar]
- 36.Hayashidani H, Kanzaki N, Kaneko Y, Okatani AT, Taniguchi T, Kaneko K, Ogawa M. 2002. Occurrence of yersiniosis and listeriosis in wild boars in Japan. J Wildl Dis 38:202–205. doi: 10.7589/0090-3558-38.1.202. [DOI] [PubMed] [Google Scholar]
- 37.European Food Safety Authority. 2009. Porcine brucellosis (Brucella suis)—scientific opinion of the Panel on Animal Health and Welfare. EFSA J 1144:1–112. doi: 10.2903/j.efsa.2009.1144. [DOI] [Google Scholar]
- 38.Eisenberg T, Kutzer P, Peters M, Sing A, Contzen M, Rau J. 2014. Nontoxigenic tox-bearing Corynebacterium ulcerans infection among game animals, Germany. Emerg Infect Dis 20:448–452. doi: 10.3201/eid2003.130423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira-Filho EF, Bank-Wolf BR, Thiel HJ, König M. 2014. Phylogenetic analysis of hepatitis E virus in domestic swine and wild boar in Germany. Vet Microbiol 174:233–238. doi: 10.1016/j.vetmic.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 40.Gräber R, Keuling O. 2013. Schalenwild, p 25–41. In Gräber R, Strauß E, Johanshon S, Niedersächsisches Ministerium f̈ur Ernährung, Landwirtschaft und Verbraucherschutz (ed), Wild und Jagd—Landesjagdbericht 2012/13 Niedersächsisches Ministerium für Ernährung, Landwirtschaft und Verbraucherschutz, Hannover, Germany. [Google Scholar]
- 41.Denis M, Houard E, Labbé A, Fondrevez M, Salvat G. 2011. A selective chromogenic plate, YECA, for the detection of pathogenic Yersinia enterocolitica: specificity, sensitivity, and capacity to detect pathogenic Y. enterocolitica from pig tonsils. J Pathog 2011:296275. doi: 10.4061/2011/296275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baumgartner A, Kuffer M, Suter D, Jemmi T, Rohner P. 2007. Antimicrobial resistance of Yersinia enterocolitica strains from human patients, pigs and retail pork in Switzerland. Int J Food Microbiol 115:110–114. doi: 10.1016/j.ijfoodmicro.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 43.von Altrock A, Roesler U, Merle R, Waldmann KH. 2010. Prevalence of pathogenic Yersinia enterocolitica strains on liver surfaces of pigs and their antimicrobial susceptibility. J Food Prot 73:1680–1683. doi: 10.1016/j.jprot.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 44.Sihvonen LM, Hallanvuo S, Haukka K, Skurnik M, Siitonen A. 2011. The ail gene is present in some Yersinia enterocolitica biotype 1A strains. Foodborne Pathog Dis 8:455–457. doi: 10.1089/fpd.2010.0747. [DOI] [PubMed] [Google Scholar]
- 45.Kraushaar B, Dieckmann R, Wittwer M, Knabner D, Konietzny A, Made D, Strauch E. 2011. Characterization of a Yersinia enterocolitica biotype 1A strain harbouring an ail gene. J Appl Microbiol 111:997–1005. doi: 10.1111/j.1365-2672.2011.05112.x. [DOI] [PubMed] [Google Scholar]
- 46.Schubert S, Bockemuhl J, Brendler U, Heesemann J. 2003. First isolation of virulent Yersinia enterocolitica O8, biotype 1B in Germany. Eur J Clin Microbiol Infect Dis 22:66–68. doi: 10.1007/s10096-002-0859-1. [DOI] [PubMed] [Google Scholar]
- 47.Sihvonen LM, Jalkanen K, Huovinen E, Toivonen S, Corander J, Kuusi M, Skurnik M, Siitonen A, Haukka K. 2012. Clinical isolates of Yersinia enterocolitica biotype 1A represent two phylogenetic lineages with differing pathogenicity-related properties. BMC Microbiol 12:208. doi: 10.1186/1471-2180-12-208. [DOI] [PMC free article] [PubMed] [Google Scholar]