Abstract
Infection by human pathogens through the consumption of fresh, minimally processed produce and solid plant-derived foods is a major concern of the U.S. and global food industries and of public health services. Enterohemorrhagic Escherichia coli O157:H7 is a frequent and potent foodborne pathogen that causes severe disease in humans. Biofilms formed by E. coli O157:H7 facilitate cross-contamination by sheltering pathogens and protecting them from cleaning and sanitation operations. The objective of this research was to determine the role that several surface structures of E. coli O157:H7 play in adherence to biotic and abiotic surfaces. A set of isogenic deletion mutants lacking major surface structures was generated. The mutant strains were inoculated onto fresh spinach and glass surfaces, and their capability to adhere was assessed by adherence assays and fluorescence microscopy methods. Our results showed that filament-deficient mutants bound to the spinach leaves and glass surfaces less strongly than the wild-type strain did. We mimicked the switch to the external environment—during which bacteria leave the host organism and adapt to lower ambient temperatures of cultivation or food processing—by decreasing the temperature from 37°C to 25°C and 4°C. We concluded that flagella and some other cell surface proteins are important factors in the process of initial attachment and in the establishment of biofilms. A better understanding of the specific roles of these structures in early stages of biofilm formation can help to prevent cross-contaminations and foodborne disease outbreaks.
INTRODUCTION
Enterohemorrhagic Escherichia coli (EHEC) infection can cause a wide range of human diseases, including mild to severe diarrhea and hemorrhagic colitis, and life-threatening conditions, such as hemolytic-uremic syndrome (HUS) (1). Because of public health and food safety implications, E. coli O157:H7 is the most widely studied serotype (2, 3). Consumption of contaminated fresh produce is an important transmission pathway of this and other EHEC foodborne pathogens and significantly contributes to the rising numbers of foodborne outbreaks (4).
E. coli O157:H7 can adapt to stressful environmental conditions by forming biofilms (5). Biofilms can be defined as a sessile community of microorganisms that are attached to the surface and often embedded in an extracellular matrix. This matrix is formed by the cells themselves, and it consists of polysaccharides, proteins, and nucleic acids (6). A single microbial species can form biofilms on either biotic or abiotic surfaces, but in most cases, biofilms consist of multiple microbial species (7, 8).
The formation of biofilms starts with initial attachment events (6). Bacteria sense, recognize, and respond to various environmental conditions that trigger the transformation from planktonic to sessile growth. These conditions, such as a decrease in the levels of nutrients or temperature (9, 10), cause the bacteria to respond by altering their metabolism and gene expression profile. Some of the changes affect the proteins on the external surfaces of cells, and these proteins are primarily responsible for adhesion of bacteria to abiotic and biotic surfaces during the process of initial attachment (11). By screening a library of E. coli mutants for defects in biofilm formation, Genevaux and colleagues (12) revealed that half of the mutant strains were also defective in flagellum-mediated motility. The importance of pili, flagella, and other filamentous surface adhesins in the early stages of biofilm formation of E. coli was further demonstrated in studies by Pratt and Kolter (13) and Xicohtencatl-Cortes et al. (14).
The aim of the current study was to expand our knowledge of the roles of several major cell surface structures in the interactions of bacteria with food and environmental matrices during the process of initial attachment to biotic and abiotic surfaces. We investigated the roles of curli (csgA), the EspA filament (espA), a flagellar filament structural protein (fliC), the hemorrhagic coli fimbriae (hcpA), long polar filament A (lpfA), outer membrane protein A (ompA), a putative fimbria-like protein (yehD), and perosamine synthetase (per), an enzyme essential for the production of O antigen, in the adhesion of cells to spinach leaves and abiotic (glass) surfaces. The coding sequences of these genes were deleted by λ Red recombinase-mediated site-directed mutagenesis, and the effects of the mutations on E. coli O157:H7 cell morphology, growth kinetics, and ability to attach to abiotic and spinach leaf surfaces were examined. A better understanding of the process of initial attachment of enterohemorrhagic bacteria to produce leaves may contribute to the development of better and safer food handling practices and thus mitigate the risk of foodborne infection outbreaks.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1 and Table 2, respectively. Escherichia coli O157:H7 strain EDL933 was used as the parental strain for site-directed mutagenesis of targeted genes by using the λ Red recombinase system. Wild-type (WT) EDL933 and isogenic in-frame deletion mutants, along with selected complementation strains, with or without a plasmid encoding green (GFP) or red (mCherry) fluorescent protein, were used in different assays in this study. E. coli Top10 was used as the recipient strain for plasmid construction. The strains were routinely grown in tryptic soy broth (TSB), Luria-Bertani (LB) broth, or Hanahan's broth (SOB) or on TSB or LB agar (Becton Dickinson, Franklin Lakes, NJ, and Acumedia, Neogen, Lansing, MI) at 30°C or 37°C. When required, antibiotics were added to the media at the following concentrations: kanamycin (Kan), 50 μg/ml; ampicillin (Amp), 100 μg/ml; chloramphenicol (Cm), 5 and 20 μg/ml; and gentamicin (Gen), 7 μg/ml (Sigma, Saint Louis, MO).
TABLE 1.
Bacterial strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| EDL933 | Prototypical EHEC O157:H7 strain | 17 |
| Top10 | F− mcrA ΔlacX74 recA1 rpsL (Strr) endA1 λ− | Life Technologies |
| EDL933ΔcsgA | EDL933 with csgA (curli) gene deleted; Cmr | This study |
| EDL933ΔespA | EDL933 with espA (secreted protein EspA) gene deleted; Cmr | This study |
| EDL933ΔfliC | EDL933 with fliC (flagellin) gene deleted; Cmr | This study |
| EDL933ΔhcpA | EDL933 with hcpA (adhesive pilus) gene deleted; Cmr | This study |
| EDL933ΔlpfA | EDL933 with lpfA (long polar filament A) gene deleted; Cmr | This study |
| EDL933ΔompA | EDL933 with ompA (outer membrane protein) gene deleted; Cmr | This study |
| EDL933Δper | EDL933 with per (perosamine synthetase) gene deleted; Cmr | This study |
| EDL933ΔyehD | EDL933 with yehD (fimbria-like protein) gene deleted; Cmr | This study |
| EDL933ΔfliC/pETcoco-2 fliC+ | Complemented version of mutant strain EDL933ΔfliC | This study |
| EDL933ΔompA/pETcoco-2 ompA+ | Complemented version of mutant strain EDL933ΔompA | This study |
| EDL933Δper/pETcoco-2 per+ | Complemented version of mutant strain EDL933Δper | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pKD3 | Cm resistance gene cassette-containing plasmid; Ampr Cmr | 15 |
| pKD46 | λ Red recombinase expression plasmid; Ampr | 15 |
| pGFP | Cloning vector; Ampr | Clontech |
| pGFP-Gen | Gentamicin-resistant version of pGFP | This study |
| pmCherry-Gen | Gentamicin-resistant version of pmCherry | This study |
| pFastBac1 | Cloning vector, source of Genr cassette; Genr Ampr | Invitrogen |
| pmCherry-C1 | Cloning vector, source of mCherry cassette; Kanr Neor | Clontech |
| pETcoco-2 | Cloning vector, vector for mutation complementation assays; Ampr | Novagen |
| pETcoco-2 fliC+ | Complementation plasmid for ΔfliC mutant | This study |
| pETcoco-2 ompA+ | Complementation plasmid for ΔompA mutant | This study |
| pETcoco-2 per+ | Complementation plasmid for Δper mutant | This study |
Construction of isogenic mutants.
Standard DNA procedures followed well-established protocols and specific recommendations from relevant manufacturers. The λ Red recombination technology (15) was used for in-frame deletion of the individual coding sequences of 8 target genes (csgA, espA, fliC, hcpA, lpfA, ompA, per, and yehD). Plasmid pKD46 was transformed into strain EDL933 by electroporation using a Gene Pulser transfection apparatus with a pulse controller (Bio-Rad, Hercules, CA). Linear mutagenesis DNA products with target sequences flanking the Cm resistance gene (cat) were generated by PCR, using pKD3 as the template and oligonucleotides (Integrated DNA Technologies, Coralville, IA, and Eurofins, Huntsville, AL) designed as described by Datsenko and Wanner (15). Mutagenesis was carried out as described by Datsenko and Wanner (15) and Serra-Moreno et al. (16). The deletion of the gene of interest was verified by PCR, using the downstream oligonucleotide CTR-Cm-R, which anneals to a sequence internal to the cat gene, and oligonucleotides upstream of the deleted target sequences to allow the amplification of the sequences encompassing the deletion junctions 5′ to the deleted genes. These oligonucleotides were designed in such a way that the PCR products for individual target genes differed by 100 bp (ranging from 0.4 to 1.1 kb), allowing easy differentiation of mutants with gene deletions (see Fig. S1 in the supplemental material). All oligonucleotides used in this study are listed in Table S1.
Construction of plasmids.
Gen-resistant GFP- and mCherry-expressing plasmids were constructed for labeling and simultaneous observation of WT and mutant EDL933 strains by fluorescence microscopy techniques. The Gen-resistant version of the plasmid pGFP (Clontech, Mountain View, CA) was previously constructed in our laboratory to accommodate experimental needs involving E. coli strains with Amp resistance (our unpublished data). The plasmid pGFP-Gen was constructed by replacing the Amp resistance gene (bla) in pGFP with the Gen resistance gene (accC1) from pFastBac1 (Life Technologies, Carlsbad, CA). The pmCherry-Gen plasmid was constructed from pGFP-Gen by replacing the GFP coding sequence with the mCherry coding sequence from pmCherry-C1 (Clontech). The single-copy complementation plasmid pETcoco-2 (Novagen, Billerica, MA), used for fliC, ompA, and per deletions, was constructed by in-frame subcloning of the respective deleted coding sequences in pETcoco-2 under the transcriptional control of the T7 promoter. Additional information on plasmid construction is provided in Fig. S2 and S3 in the supplemental material.
Cell growth kinetics.
Individual bacterial strains were grown in 200 μl TSB or 10% TSB in 96-well microplates at 37°C with shaking. A Synergy 4 hybrid microplate reader controlled by Gen5 software (BioTek Instruments, Winooski, VT) was used for microplate incubation and to obtain information on bacterial growth kinetics by measuring the optical density at 600 nm (OD600) at 10-min intervals during the incubation. The experiment was repeated three times, and data were analyzed using OriginPro 7.5 (OriginLab Corporations, Northampton, MA).
Adherence and detachment assays.
Packaged baby spinach leaves were purchased from a local store and rinsed with sterile phosphate-buffered saline (PBS; pH 7.4). Spinach leaves were cut into 1-cm-diameter circular sections and placed in 24-well tissue culture microplates (Becton Dickinson, Franklin Lakes, NJ) containing 1 ml of sterile PBS. WT and mutant EDL933 strains carrying pmCherry-Gen were used as inoculums to examine attachment to spinach by spiral plating following deattachment from the leaves. The WT EDL933 strain carrying pGFP-Gen was also included in each inoculum as an internal reference for binding assays intended for analyses using confocal laser scanning microscopy (CLSM). Approximately 107 bacterial cells of WT or mutant EDL933 carrying pmCherry-Gen and equal amounts of the WT EDL933 strain carrying pGFP-Gen were added individually or in combination to each well to allow adherence of bacterial cells to the leaves for 24 h at 4°C and 25°C. After incubation, the leaves were washed three times with PBS. Three discs were homogenized together to recover bacteria for enumeration. E. coli O157:H7 cells in the suspension, rinse, and homogenate fractions were spiral plated onto LB agar plates containing 7 μg/ml Gen by using an Eddy Jet 2 spiral plater (Thermo Fisher Scientific, Pittsburgh, PA). Data were analyzed using OriginPro 7.5. A separate set of samples was observed by CLSM after washing.
In another experiment, 1-cm-diameter circular sections of fresh spinach leaves were incubated with GFP-labeled WT and mCherry-labeled mutant bacterial cells (in sterile PBS) as described above for 10 min, followed by air drying for 60 min in a biosafety hood and storage at 4°C overnight. The leaf samples were then washed and attached cells enumerated as described above.
Microscopic observations.
The attachment of GFP- and mCherry-labeled E. coli O157:H7 cells to spinach leaves was examined using a Zeiss LSM710 CLSM system (Thornwood, NY). Samples were prepared and washed as described above. The images were observed using a Zeiss Axio Observer inverted microscope with a 63× 1.4-numerical-aperture (NA) Plan-Apochromat oil-immersion objective, with a 488-nm argon laser and a 561-nm diode laser with a pin hole of 45 μm passing through an MBS 488/561 beam splitter filter with limits set at 490 to 535 nm for GFP and 570 to 640 nm for mCherry. The Zeiss Zen 2012 (Thornwood, NY) 64-bit software was used to obtain single images, or if the tissue was not flat, 10 to 20 z-stack images with a 1-μm thickness were obtained to produce three-dimensional renderings, which were used to develop the two-dimensional maximum-intensity projections for publication. Data were analyzed using OriginPro 7.5.
Low-temperature scanning electron microscopy (LT-SEM) was used to compare cell morphologies of the WT and mutant EDL933 strains. Cells were placed as spots on carbon adhesive tabs (Electron Microscopy Sciences, Incorporated, Hatfield, PA) secured to 15-mm by 30-mm copper plates and air dried, followed by rapid freezing in liquid nitrogen and transfer to a Quorum PP2000 cryo-prep chamber (Quorum Technologies, East Sussex, United Kingdom). The specimens were etched inside the cryotransfer system and coated with a 10-nm layer of platinum before being observed using an S-4700 field emission scanning electron microscope (Hitachi High Technologies America, Inc., Dallas, TX). An accelerating voltage of 5 kV was used to view the specimens. Images were captured using a 4 pi analysis system (Durham, NC). Images were analyzed with ImageJ (NIH, Bethesda, MD). Data were analyzed using OriginPro 7.5.
Adherence of cells to glass slides was observed with a Nikon N-Storm microscope (Melville, NY). One-hundred-microliter aliquots of 107 CFU/ml EDL933 WT and mutant cells harboring the pGFP-Gen plasmid were suspended in sterile PBS and layered on top of a microscope coverslip. After 24 h of incubation at 25°C, the coverslips were rinsed with sterile PBS and attached to a microscope slide. The GFP was excited with a 488-nm AOTF modulated laser line and imaged using an Apo total internal-reflection fluorescence (TIRF) 100× 1.49-NA oil-immersion objective. Images were recorded with an iXon DU897 electron microscopy charge-coupled device (EM-CCD) camera (Andor Technologies, South Windsor, CT). We captured at least 10 field-of-view areas of 80 by 80 μm for each of the wild-type and mutant cells, with larger numbers of views for mutants with diminished bindings. The experiment was repeated three times, and data were analyzed using OriginPro 7.5 and ImageJ.
Statistical analysis.
Statistical analysis of the data for the bacterial enumeration was performed using GraphPad software (GraphPad Software Inc., La Jolla, CA). The statistical significance of the differences in sample means of the mutant and wild-type strains was calculated using unpaired Student's t test. Results were considered significant if the P value was <0.05.
RESULTS
Generation of isogenic deletion mutants.
Bacterial surface structures play a vital role in attachment of bacterial cells to biotic and abiotic surfaces. To gain better insight into the role of major cell surface components in cellular attachment, we generated knockout null mutants in enterohemorrhagic E. coli O157:H7 EDL933 (17) by using λ Red recombinase-mediated mutagenesis protocols (15, 16, 18). The targeted genes were those encoding the curli filament (csgA), the EspA filament (espA), a flagellar structural protein that carries the antigenic determinant for the H antigen (fliC), the hemorrhagic coli fimbriae (hcpA), the long polar filament A (lpfA), the abundant outer membrane protein A (ompA), perosamine synthetase (per), and a predicted fimbria-like adhesion protein (yehD).
A functional chloramphenicol resistance gene (cat) was expressed in all of the intended isogenic mutants, indicating successful replacement of the target genes with cat. The deletion of these genes was confirmed by PCRs using forward primers specific to sequences upstream of the deleted genes and a common reverse primer specific to a sequence in the knocked-in cat gene. The sizes of the PCR fragments encompassing the deletion junctions were consistent with those expected for each of the resultant mutants (400 to 1,100 bp long, with 100-bp increments, for EDL933 ΔcsgA, ΔespA, ΔfliC, ΔhcpA, ΔlpfA, ΔompA, Δper, and ΔyehD mutants) (see Fig. S1 in the supplemental material), indicating the successful deletion of the targeted genes and insertion of the cat gene in place of the deleted target genes.
Phenotypic characterization of isogenic mutants.
The colony morphologies of the mutants grown on LB agar plates were visually inspected, and no significant alteration compared to the WT was observed, except for EDL933Δper, which gave rise to slightly smaller colonies after overnight incubation at 37°C. EDL933Δper did not exhibit a “rough” phenotype under those growth conditions. The cellular morphologies of EDL933 and the isogenic mutants were examined using LT-SEM. LT-SEM was applied because this technique does not require any chemical fixation, and utilization of liquid nitrogen to freeze the bacteria, with observation at low temperatures, results in images of bacteria without distortion by chemical treatment. Our results showed that the deletion mutations affected the cell length and width to various degrees (Fig. 1A). The mutants that showed the most remarkable changes in cell shape were EDL933ΔompA and EDL933Δper. It has been shown that the OmpA protein contributes to the structural integrity of the outer membrane and that mutation of the ompA gene affects the shape and size of various bacteria (19–21). The EDL933ΔompA strain, most likely due to compromised outer membrane integrity, exhibited a spherical morphology (Fig. 1B, top right panel). The EDL933Δper mutant, like EDL933ΔompA, had a reduced size (Fig. 1A and B, bottom left panel). Probably due to deficiency in the lipopolysaccharides, the Δper cells seemed to aggregate in a linear fashion, forming bacterial filaments that were tens of micrometers long (Fig. 1B, bottom right panel). The perosamine synthetase is required for synthesis of the O157 antigen, and deletion of the per gene likely renders the EDL933 strain deficient in the O antigen (22–25). This was confirmed by a negative reaction in an agglutination assay (see Fig. S4 in the supplemental material) using multiclonal antibodies targeting the E. coli O157 O antigen (Oxoid DrySpot E. coli O157 latex agglutination assay kit; Thermo Fisher Scientific). The complemented mutant strain EDL933Δper/pETcoco-2per+ exhibited a positive reaction indistinguishable from that of the wild-type strain in the agglutination assay. Negative-staining transmission electron microscopy (NS-TEM) confirmed that the deletion of the fliC gene resulted in a complete loss of flagella in the EDL933ΔfliC mutant. The lack of flagella did not seem to result in any other changes in cellular morphology. NS-TEM images showed that the complemented EDL933ΔfliC/pETcoco-2fliC+ cells had fully restored flagella (see Fig. S4).
FIG 1.
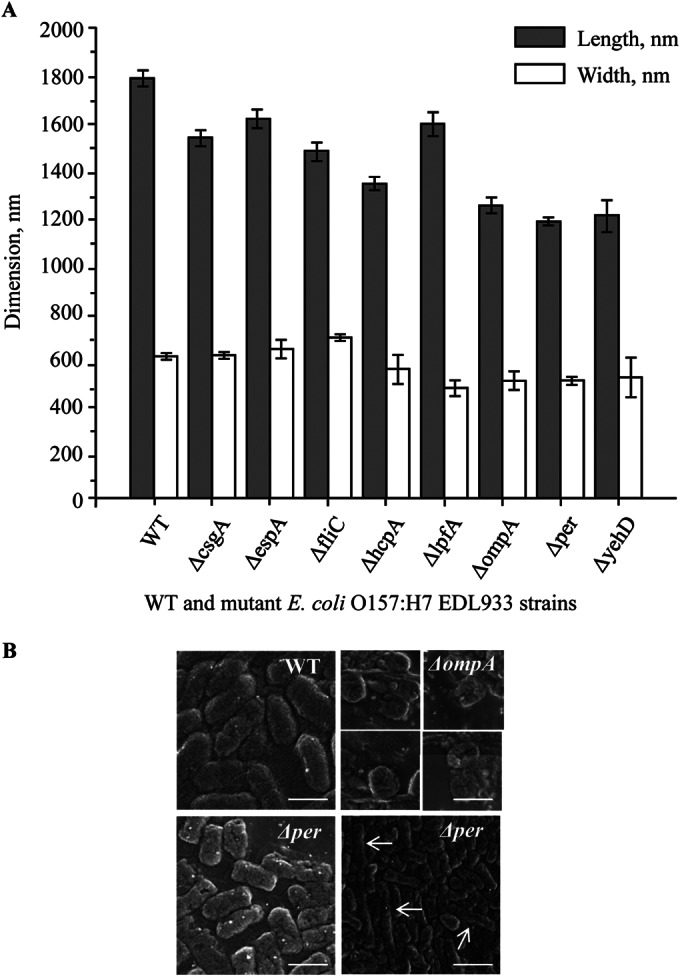
LT-SEM characterization of E. coli O157:H7 EDL933 wild-type and mutant strains. (A) Average two-dimensional measurements of E. coli O157:H7 EDL933 and the isogenic deletion mutant strains. Gray bars represent cell lengths (longer dimension), and white bars represent cell widths (shorter dimension). Data are means ± standard errors (SE) of results from three independent experiments. (B) Typical cell morphologies of EDL933 and selected isogenic mutants. Note the cell rounding in the ΔompA strain (top right) and the filamentation in the Δper strain (bottom right; examples of filamentous cells are marked with white arrowheads). Bars, 1 μm and 3 μm (bottom right).
Deleting major surface components of bacterial cells might influence the viability of the strain. To characterize the viability of cells, we compared the growth kinetics of wild-type EDL933 and the individual isogenic mutant strains. As shown in Fig. 2, the growth kinetics of the deletion mutants were highly comparable to that of the WT strain, with identical optical doubling times (∼38 min), except for the EDL933Δper mutant, which exhibited a slightly extended optical doubling time (∼42 min) and reduced total growth (Fig. 2A). The complemented mutant strain EDL933Δper/pETcoco-2per+ exhibited an optical doubling time and total growth comparable to those of the wild-type strain (see Fig. S5 in the supplemental material). In 10% TSB, a medium with reduced nutrient value and ionic strength, the WT and mutant strains grew at the same rate, with an optical doubling time of approximately 125 min, with the exception of the EDL933ΔompA mutant, which exhibited irregular and slow growth (Fig. 2B). We speculate that this strain, lacking the major outer membrane protein A, which is responsible for outer membrane stability, was more susceptible to the stress of the lower ionic strength of the growth medium. The growth rate of the complementary mutant strain EDL933ΔompA/pETcoco-2ompA+ was restored to a level comparable to that of the wild-type strain (see Fig. S5).
FIG 2.
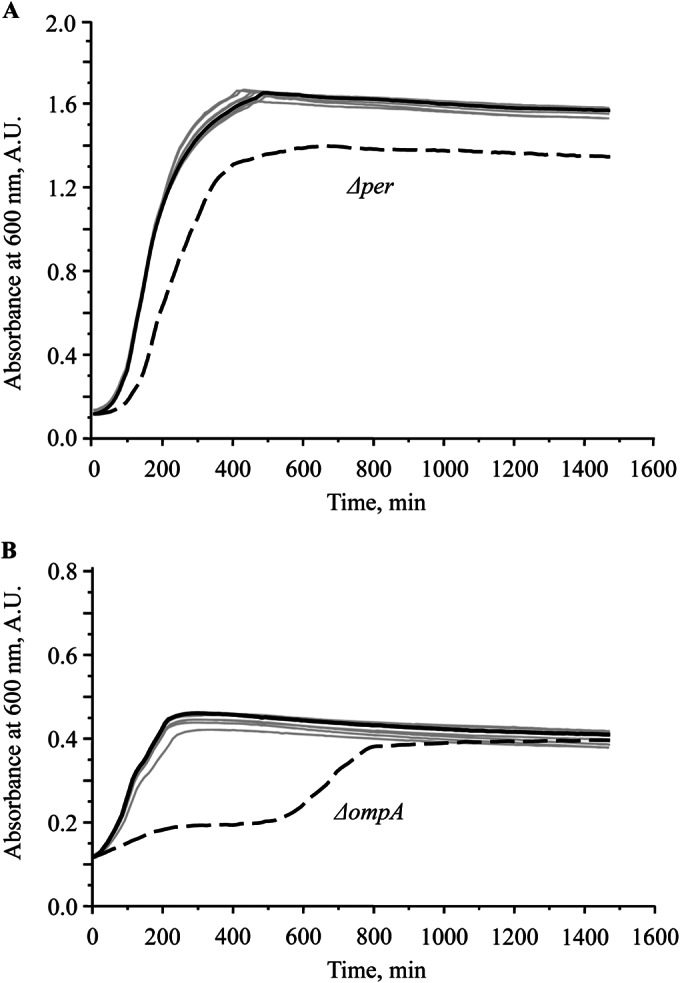
Growth kinetics of wild-type and mutant E. coli O157:H7 EDL933 strains. (A) In TSB, the growth curves of the wild type (dark solid line) and the mutant strains (gray solid lines) were nearly identical, with the exception of mutant strain EDL933Δper (dashed line), which exhibited a slightly shorter optical doubling time (42 min) than that of the wild type and the other deletion mutants (38 min). (B) In 10% TSB, the EDL933ΔompA mutant strain (dashed line) exhibited biphasic and slower growth than the wild-type strain (dark solid line) and the other mutant strains (gray solid lines). The experiment was repeated three times. A.U., absorbance units.
Attachment of E. coli O157:H7 cells to an abiotic surface.
Previous work from our and other laboratories demonstrated that E. coli O157:H7 is capable of attaching to glass to form biofilms by itself or in dual-species cocultures with other bacteria (5, 7, 26, 27). To assess the effects of deletion and, through it, the possible roles of the products of the csgA, espA, fliC, hcpA, lpfA, ompA, per, and yehD genes, we used TIRF microscopy to observe bacteria that were tightly bound to the surface. Our results showed that the mutant strains' ability to adhere to an abiotic surface was moderately to severely compromised (Fig. 3). The average cell counts for the WT and the isogenic mutants in an 80-μm by 80-μm square view ranged from ∼1.5 (ΔompA mutant) to ∼13 (WT). The surface adherence abilities of the wild type and the ΔfliC or ΔompA mutant strain were significantly different (P < 0.05). However, the microscopic enumeration might have overestimated the ΔfliC and ΔompA mutant counts, as views without attached cells were intentionally omitted. The EDL933ΔfliC and EDL933ΔompA mutant strains were the least able to bind to glass (Fig. 3); thus, our results suggest that the products of the ompA and fliC genes play an important role in attachment to abiotic surfaces.
FIG 3.
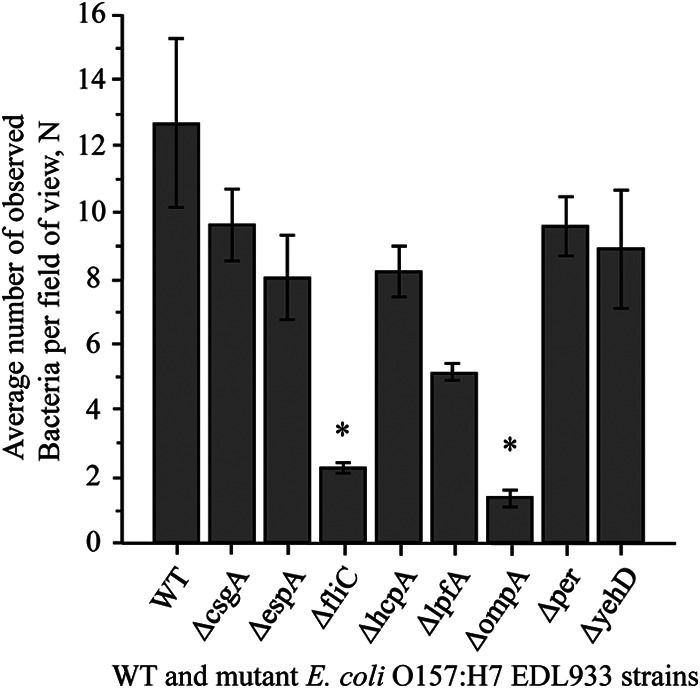
Attachment of GFP-labeled E. coli O157:H7 EDL933 wild-type and mutant strains to glass. The graphs shows the average E. coli O157:H7 EDL933 cell counts per field of view. Data are means ± SE of results from three independent experiments. Unpaired Student's t test was used to determine whether differences were statistically significant (WT versus mutant). Asterisks indicate significant differences from the binding ability of the wild type.
Attachment of E. coli O157:H7 cells to spinach leaves.
To assess the effects of the mutations on the colonization ability of cells on fresh produce, surface attachment assays were conducted using WT and isogenic mutant EDL933 strains. GFP-labeled WT EDL933 and mCherry-labeled WT EDL933 (as a control) or the individual isogenic mutants were mixed at a 1:1 ratio as inoculums. Figure 4 shows that certain mutant EDL933 cells attached to spinach leaves at a significantly lesser degree than the WT EDL933 strain during the 24-h incubation at either 25°C or 4°C (Fig. 4). The reduction in attachment of mutant strains was especially prominent in the cases of the EDL933ΔfliC and EDL933ΔompA mutant strains. The numbers of attached EDL933ΔfliC mutant bacteria were decreased to 20.4% (at 4°C) and 16.7% (at 25°C) of the WT levels (Fig. 4). The deletion of the gene encoding the OmpA protein also severely hindered the ability of EDL933 cells to attach to spinach leaves. The numbers of EDL933ΔompA mutant cells were 15.1% and 11.4% of the WT levels at 4°C and 25°C, respectively (Fig. 4). The deficiencies of the mutant strains in binding to spinach leaves were fully complemented in the strains carrying the complementary plasmids, achieving levels of 90.7% (EDL933Δfli/pETcoco-2fliC+) and 95.3% (EDL933ΔompA/pETcoco-2ompA+) of the WT level at 25°C (see Fig. S6 in the supplemental material).
FIG 4.
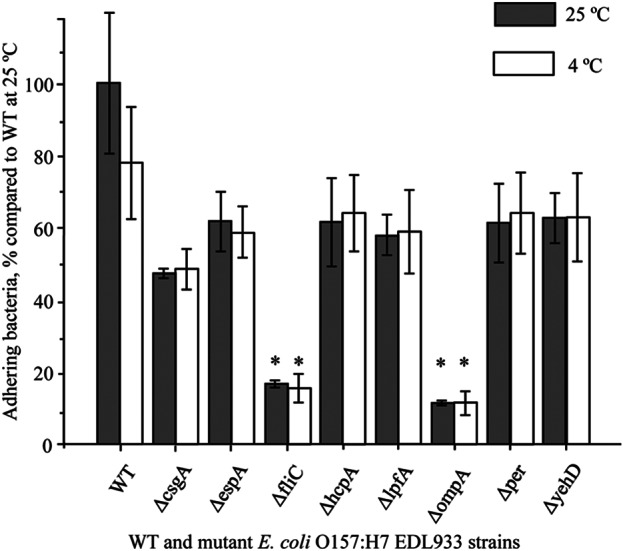
Adhesion of WT and mutant E. coli O157:H7 EDL933 strains on spinach. After incubation with spinach leaves for 24 h at 25°C (gray bars) and 4°C (white bars) and rinsing off weakly bound cells, strongly bound E. coli cells were enumerated by spiral plating. Data are means ± SE of results from three independent experiments. Unpaired Student's t test was used to determine whether differences were statistically significant (WT versus mutant). Asterisks indicate significant differences from the binding ability of the wild type.
Confocal laser scanning microscopy was also used to assess the ability of WT and isogenic mutant strains to attach to spinach leaves (Fig. 5A and B). We observed that the WT EDL933 and mutant bacteria attached to spinach leaves with different efficacies. The mutant bacteria were less successful at establishing strong physical bonds with the epidermis of the spinach leaves and were more readily detached during the washing process. The EDL933ΔfliC and EDL933ΔompA strains showed an especially weak propensity to bind to spinach leaves (Fig. 5B). In each of these cases, the mCherry-labeled mutants counted for less than 13% of the cells in the mixed population of attached WT and mutant cells.
FIG 5.
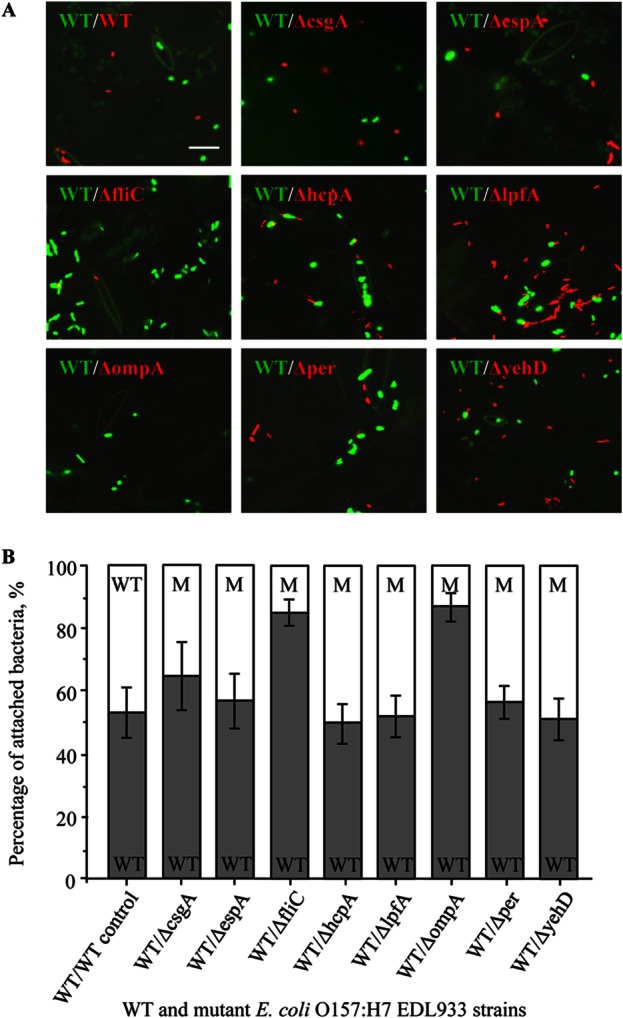
Attachment of WT and mutant E. coli O157:H7 EDL933 strains to spinach. GFP-labeled WT E. coli O157:H7 EDL933 and mCherry-labeled WT or mutant strains were coincubated with spinach leaves for 24 h at 25°C. (A) Strongly bound E. coli cells were imaged using confocal microscopy. Bar, 5 μm. (B) Average percentages of coinoculated WT and mutant attached cells. The gray bars represent the percentages of GFP-labeled WT bacteria (WT), and the corresponding white bars represent the percentages of mCherry-labeled WT (as a control) or mutant (M) bacteria. The percentage values were calculated from the cumulative results of three independent experiments, and data are means ± SE.
We were interested to see whether the mutant strains, especially the EDL933ΔfliC and EDL933ΔompA strains, would show a decreased ability to adhere to spinach leaves in a scenario where the bacterial cells were forced to physically interact with the surfaces of the leaves. The previous experiments showed that during the 24-h period of incubation in an aqueous environment, the bacteria had plenty of time to explore the surfaces of the leaves and establish stable contact with the epidermis. In this experiment, the spinach leaf specimens were incubated with WT EDL933 and individual isogenic deletion mutants for a short time (10 min) and then air dried and stored at 4°C overnight before being washed.
CLSM was employed to assess the detachment of WT and mutant E. coli O157:H7 cells after the inoculum was allowed to dry on spinach leaves (Fig. 6A). In contrast to the observations for attachment in an aqueous environment, E. coli O157:H7 cells were rarely seen attached to the examined underside of the leaves; instead, they were present at a high density in the stomata. Figure 6A shows GFP-labeled WT EDL933 bacteria mixed with mCherry-labeled EDL933ΔfliC, EDL933ΔhcpA, and EDL933ΔompA bacteria filling in the stomata of the spinach leaves.
FIG 6.
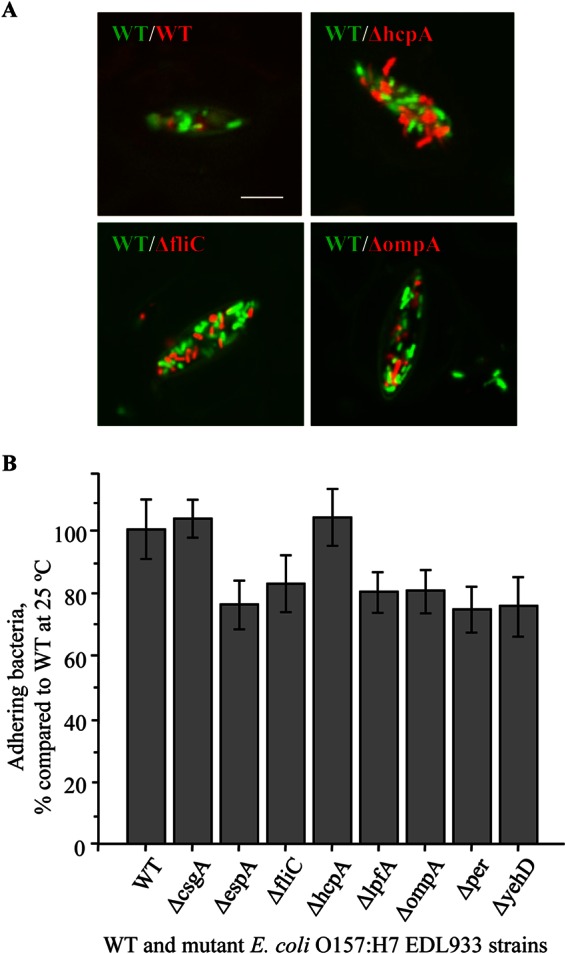
Binding of WT and mutant E. coli O157:H7 EDL933 strains to spinach leaves after air drying. (A) GFP-labeled WT and mCherry-labeled WT or mutant strains were incubated with spinach leaves for 10 min at 25°C and then air dried for 1 h before washing and retention analysis using confocal microscopy. Images of WT and selected mutants are shown. (B) Bacteria trapped in stomata were released in a stomacher and enumerated by spiral plating. Data are means ± SE of results from three independent experiments.
The numbers of bacteria after washing were determined by spiral plating (Fig. 6B) using mCherry-labeled WT and mutant cells. Our results showed that the difference in binding ability of WT and mutant bacterial cells was less pronounced in this scenario (Fig. 6B). The forced interaction (air drying) resulted in smaller numbers of attached bacteria recovered from the leaves. When the leaves were incubated overnight in a similarly concentrated bacterial suspension, 1 to 2% of cells, on average, remained bound to leaves after washing, while in this experiment, only 0.2 to 0.3% of cells were recovered after homogenization. There were no significant differences between the cell counts on the leaves for the wild type and the mutants, including the fliC and ompA mutants.
DISCUSSION
Upon exiting the gastrointestinal tract, bacteria face new challenges in the natural environment. The temperature is lower than 37°C, nutrients are less readily available, and in some cases the bacteria have to adapt to antibiotics or disinfectants. One of the survival strategies for E. coli is the formation of biofilms. Bacteria embedded in the matrices of biofilms exhibit increased tolerance to antibiotics, stress, and environmental factors (28–31). In order to form a biofilm, the bacteria first have to reach the surface in a liquid environment. Brownian motion, fluid dynamics, and gravity obviously play important roles in this process, but active motility increases the volume that the bacteria can explore and helps them to overcome obstacles, increasing the probability of successful surface adherence (32). The E. coli cell is propelled through water by its flagella, so it is not a surprise that E. coli strains with mutant flagellar apparatuses also exhibit deficiencies in biofilm formation (13). The earliest step of biofilm formation, which is the first contact of bacteria with a surface, is a reversible process. The bacteria probe the available surface, and if the physical (e.g., topographic) and chemical (electrostatic) interactions between the envelope of the bacterial cell and the abiotic or biotic surface are favorable and potent enough to overcome the repulsive forces, the bacteria will adhere permanently (32).
Our goal was to shed light on the process underlying the first event of biofilm formation and the role of selected surface structures in this process. By decreasing the temperature to 25°C and 4°C, we partially simulated the environmental conditions that bacteria face during cultivation, harvest, transportation, processing, and storage while they interact with fresh produce. We showed that deletion of the flagellin gene fliC (Fig. 3 to 5) severely impairs the ability of E. coli O157:H7 to attach to spinach leaves or to glass. The deletion of the outer membrane protein A gene (ompA) had a similarly strong effect (Fig. 3 to 5), and based on LT-SEM imaging (Fig. 1) and growth kinetics (Fig. 2), we speculate that the severe disruption of the membrane structure of the E. coli cell causes problems not only with adhesion but also with the physiology of the cell at a more fundamental level.
It has been shown that the product of the curli gene (csgA) is a key player in adherence of E. coli cells to spinach leaves (33–35). We showed here that deletion of the csgA gene had a moderate effect on adherence to spinach leaves (Fig. 4 and 5). However, it is possible that curli play a more significant role in the process of forming irreversible bonds with biotic surfaces at later stages of biofilm formation. The deletion of the long polar filament also had a moderate effect on adherence of E. coli O157:H7 EDL933 (Fig. 3 to 5). The ΔlpfA ΔlpfB double mutant likely will shed more light on the role of long polar filaments in adhesion, and we are planning to conduct experiments with this strain in the future.
An unexpected observation was the relatively small number of E. coli O157:H7 cells tightly bound to spinach leaf surfaces after allowing the inoculums to be in contact with leaves for a short time and to dry on the leaf surfaces (Fig. 6). In an aqueous environment, the wild-type E. coli O157:H7 cells seemed to need ample time to dock to the leaf surface and to establish strong or irreversible attachment to the leaves, and hence were retained on the epidermis in larger numbers. This process seemed to be severely hindered in the mutants with impaired motility (ΔfliC) or a defective outer membrane (ΔompA). In the case of inoculums drying on the leaf surface, the bacterial cells might not have had enough time to interact with the leaf surface and establish an irreversible attachment before the liquid evaporated, resulting in a loose attachment that was easily removed by rinsing. However, in the experiments where the bacterial suspension was allowed to evaporate, most of the cells remaining on leaves were found in the stomata of the leaves, where they were shielded from the rinsing liquid (Fig. 6). Although the mechanism for the bacterial cells accumulating in the stomata was not clear, this phenomenon may explain how bacteria survive decontamination treatments. In such a scenario, the effects of deletion of the fliC and ompA genes on the binding ability of O157:H7 EDL933 cells were less pronounced (Fig. 6B).
In conclusion, our research shows that E. coli EDL933 cells form specific interactions with the epidermis of the leaves of fresh produce and that the products of the fliC and ompA genes play crucial roles in the establishment of these specific interactions. We also showed that the stomata of cells can act as a trap under certain circumstances, allowing the accumulation of bacterial cells. By this mechanism, the bacteria can reach the internal tissues and intercellular spaces of the fresh produce, where they can survive in a sheltered microenvironment during produce processing and can present an increased risk for foodborne disease outbreaks.
Supplementary Material
ACKNOWLEDGMENTS
We thank Neil Billington, NIH, NHLBI, Laboratory of Molecular Physiology, for providing help with electron and TIRF microscopy and Marie-Paule Strub, NIH, NHLBI, Laboratory of Structural Biophysics, for providing the plasmids used in this study.
Lydia Nichols-Russell is a student intern from the University of Maryland, College Park, MD.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00215-15.
REFERENCES
- 1.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin Microbiol Rev 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karch H, Tarr PI, Bielaszewska M. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int J Med Microbiol 295:405–418. doi: 10.1016/j.ijmm.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Tarr PI, Gordon CA, Chandler WL. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2013. Surveillance for foodborne disease outbreaks—United States, 2009–2010. MMWR Morb Mortal Wkly Rep 62:41–47. [PMC free article] [PubMed] [Google Scholar]
- 5.Wang R, Bono JL, Kalchayanand N, Shackelford S, Harhay DM. 2012. Biofilm formation by Shiga toxin-producing Escherichia coli O157:H7 and non-O157 strains and their tolerance to sanitizers commonly used in the food processing environment. J Food Prot 75:1418–1428. doi: 10.4315/0362-028X.JFP-11-427. [DOI] [PubMed] [Google Scholar]
- 6.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 7.Liu NT, Nou X, Lefcourt AM, Shelton DR, Lo YM. 2014. Dual-species biofilm formation by Escherichia coli O157:H7 and environmental bacteria isolated from fresh-cut processing facilities. Int J Food Microbiol 171:15–20. doi: 10.1016/j.ijfoodmicro.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Archibald LK, Gaynes RP. 1997. Hospital-acquired infections in the United States. The importance of interhospital comparisons. Infect Dis Clin North Am 11:245–255. doi: 10.1016/S0891-5520(05)70354-8. [DOI] [PubMed] [Google Scholar]
- 9.Dewanti R, Wong AC. 1995. Influence of culture conditions on biofilm formation by Escherichia coli O157:H7. Int J Food Microbiol 26:147–164. doi: 10.1016/0168-1605(94)00103-D. [DOI] [PubMed] [Google Scholar]
- 10.Palmer RJ Jr, White DC. 1997. Developmental biology of biofilms: implications for treatment and control. Trends Microbiol 5:435–440. doi: 10.1016/S0966-842X(97)01142-6. [DOI] [PubMed] [Google Scholar]
- 11.Genevaux P, Bauda P, DuBow MS, Oudega B. 1999. Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch Microbiol 172:1–8. doi: 10.1007/s002030050732. [DOI] [PubMed] [Google Scholar]
- 12.Genevaux P, Bauda P, DuBow MS, Oudega B. 1999. Identification of Tn10 insertions in the dsbA gene affecting Escherichia coli biofilm formation. FEMS Microbiol Lett 173:403–409. doi: 10.1111/j.1574-6968.1999.tb13532.x. [DOI] [PubMed] [Google Scholar]
- 13.Pratt LA, Kolter R. 1999. Genetic analyses of bacterial biofilm formation. Curr Opin Microbiol 2:598–603. doi: 10.1016/S1369-5274(99)00028-4. [DOI] [PubMed] [Google Scholar]
- 14.Xicohtencatl-Cortes J, Sanchez Chacon E, Saldana Z, Freer E, Giron JA. 2009. Interaction of Escherichia coli O157:H7 with leafy green produce. J Food Prot 72:1531–1537. [DOI] [PubMed] [Google Scholar]
- 15.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serra-Moreno R, Acosta S, Hernalsteens JP, Jofre J, Muniesa M. 2006. Use of the lambda Red recombinase system to produce recombinant prophages carrying antibiotic resistance genes. BMC Mol Biol 7:31. doi: 10.1186/1471-2199-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen ML. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 18.Kannan P, Dharne M, Smith A, Karns J, Bhagwat AA. 2009. Motility revertants of opgGH mutants of Salmonella enterica serovar Typhimurium remain defective in mice virulence. Curr Microbiol 59:641–645. doi: 10.1007/s00284-009-9486-8. [DOI] [PubMed] [Google Scholar]
- 19.Sonntag I, Schwarz H, Hirota Y, Henning U. 1978. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol 136:280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y. 2002. The function of OmpA in Escherichia coli. Biochem Biophys Res Commun 292:396–401. doi: 10.1006/bbrc.2002.6657. [DOI] [PubMed] [Google Scholar]
- 21.Abe T, Murakami Y, Nagano K, Hasegawa Y, Moriguchi K, Ohno N, Shimozato K, Yoshimura F. 2011. OmpA-like protein influences cell shape and adhesive activity of Tannerella forsythia. Mol Oral Microbiol 26:374–387. doi: 10.1111/j.2041-1014.2011.00625.x. [DOI] [PubMed] [Google Scholar]
- 22.Shimizu T, Yamasaki S, Tsukamoto T, Takeda Y. 1999. Analysis of the genes responsible for the O-antigen synthesis in enterohaemorrhagic Escherichia coli O157. Microb Pathog 26:235–247. doi: 10.1006/mpat.1998.0253. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Reeves PR. 1998. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect Immun 66:3545–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilge SS, Vary JC Jr, Dowell SF, Tarr PI. 1996. Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect Immun 64:4795–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Awram P, Smit J. 2001. Identification of lipopolysaccharide O antigen synthesis genes required for attachment of the S-layer of Caulobacter crescentus. Microbiology 147:1451–1460. [DOI] [PubMed] [Google Scholar]
- 26.Uhlich GA, Chen CY, Cottrell BJ, Nguyen LH. 2014. Growth media and temperature effects on biofilm formation by serotype O157:H7 and non-O157 Shiga toxin-producing Escherichia coli. FEMS Microbiol Lett 354:133–141. doi: 10.1111/1574-6968.12439. [DOI] [PubMed] [Google Scholar]
- 27.Wang R, Kalchayanand N, Bono JL, Schmidt JW, Bosilevac JM. 2012. Dual-serotype biofilm formation by Shiga toxin-producing Escherichia coli O157:H7 and O26:H11 strains. Appl Environ Microbiol 78:6341–6344. doi: 10.1128/AEM.01137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson GG, O'Toole GA. 2008. Innate and induced resistance mechanisms of bacterial biofilms. Curr Top Microbiol Immunol 322:85–105. doi: 10.1007/978-3-540-75418-3_5. [DOI] [PubMed] [Google Scholar]
- 29.Murga R, Forster TS, Brown E, Pruckler JM, Fields BS, Donlan RM. 2001. Role of biofilms in the survival of Legionella pneumophila in a model potable-water system. Microbiology 147:3121–3126. [DOI] [PubMed] [Google Scholar]
- 30.Donlan RM. 2000. Role of biofilms in antimicrobial resistance. ASAIO J 46:S47–S52. doi: 10.1097/00002480-200011000-00037. [DOI] [PubMed] [Google Scholar]
- 31.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donlan RM. 2002. Biofilms: microbial life on surfaces. Emerg Infect Dis 8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macarisin D, Patel J, Bauchan G, Giron JA, Ravishankar S. 2013. Effect of spinach cultivar and bacterial adherence factors on survival of Escherichia coli O157:H7 on spinach leaves. J Food Prot 76:1829–1837. doi: 10.4315/0362-028X.JFP-12-556. [DOI] [PubMed] [Google Scholar]
- 34.Macarisin D, Patel J, Bauchan G, Giron JA, Sharma VK. 2012. Role of curli and cellulose expression in adherence of Escherichia coli O157:H7 to spinach leaves. Foodborne Pathog Dis 9:160–167. doi: 10.1089/fpd.2011.1020. [DOI] [PubMed] [Google Scholar]
- 35.Macarisin D, Patel J, Sharma VK. 2014. Role of curli and plant cultivation conditions on Escherichia coli O157:H7 internalization into spinach grown on hydroponics and in soil. Int J Food Microbiol 173:48–53. doi: 10.1016/j.ijfoodmicro.2013.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


