Abstract
Giardia is the most common parasitic cause of gastrointestinal infections worldwide, with transmission through surface water playing an important role in various parts of the world. Giardia duodenalis (synonyms: G. intestinalis and G. lamblia), a multispecies complex, has two zoonotic subtypes, assemblages A and B. When British Columbia (BC), a western Canadian province, experienced several waterborne giardiasis outbreaks due to unfiltered surface drinking water in the late 1980s, collection of isolates from surface water, as well as from humans and beavers (Castor canadensis), throughout the province was carried out. To better understand Giardia in surface water, 71 isolates, including 29 from raw surface water samples, 29 from human giardiasis cases, and 13 from beavers in watersheds from this historical library were characterized by PCR. Study isolates also included isolates from waterborne giardiasis outbreaks. Both assemblages A and B were identified in surface water, human, and beavers samples, including a mixture of both assemblages A and B in waterborne outbreaks. PCR results were confirmed by whole-genome sequencing (WGS) for one waterborne outbreak and supported the clustering of human, water, and beaver isolates within both assemblages. We concluded that contamination of surface water by Giardia is complex, that the majority of our surface water isolates were assemblage B, and that both assemblages A and B may cause waterborne outbreaks. The higher-resolution data provided by WGS warrants further study to better understand the spread of Giardia.
INTRODUCTION
Giardia causes large numbers of gastrointestinal infections worldwide; health impacts include diarrhea, as well as malabsorption and impaired growth in children (1). Despite its significance, transmission of Giardia is poorly understood. While other modes of transmission (person to person, foodborne) are possible, because of the persistence of the infectious cyst stage in surface water at 0 to 4°C for over 2 months and the resistance to chemical drinking water disinfection, the likelihood of transmission through unfiltered surface water is very high (1, 2).
Giardia was thought to consist of six species, including G. duodenalis (synonyms: G. intestinalis and G. lamblia; infecting humans and other mammals), G. agilis (infecting amphibians), G. ardeae, G. psittaci (infecting birds), G. muris, and G. microti (infecting rodents) (3). More recently, molecular tools have provided new information about taxonomy. Assemblages A and B are genetically distinct, and some have proposed naming assemblage A G. duodenalis and assemblage B G. enterica (4).
In British Columbia (BC), a western Canadian province (population, 4,113,487), drinking water sources are commonly surface supplies from unprotected watersheds (5). Many small drinking water systems are unfiltered, and some are treated by chlorination alone. Two decades ago, BC's high rate of giardiasis was highlighted by the occurrence of 13 confirmed or probable waterborne outbreaks (6). Investigations at that time included testing of raw drinking water samples collected from across BC and fecal samples from patients with no history of travel outside BC and from potential animal sources of Giardia drinking water contamination. Giardia isolates were retrieved by culture, and the trophozoites were characterized (7). Molecular characterization was limited by methods available at that time, but DNA-based characterization by PCR and Sanger sequencing has provided better tools to analyze and compare isolates. With the decreasing cost of whole-genome sequencing (WGS), more comprehensive comparisons between isolates can now be made.
Using a previously collected library of Giardia isolates, we genetically characterized 71 isolates (human, beaver, and water) collected from all parts of the province. These isolates were both outbreak associated and sporadic (not outbreak associated). Isolates were studied by PCR, with further information obtained by WGS of a subset of samples. We show that current and emerging molecular tools can be used to obtain new information to better understand the waterborne spread of giardiasis.
MATERIALS AND METHODS
Isolate information.
(i) Outbreak isolates (human, beaver, and water).
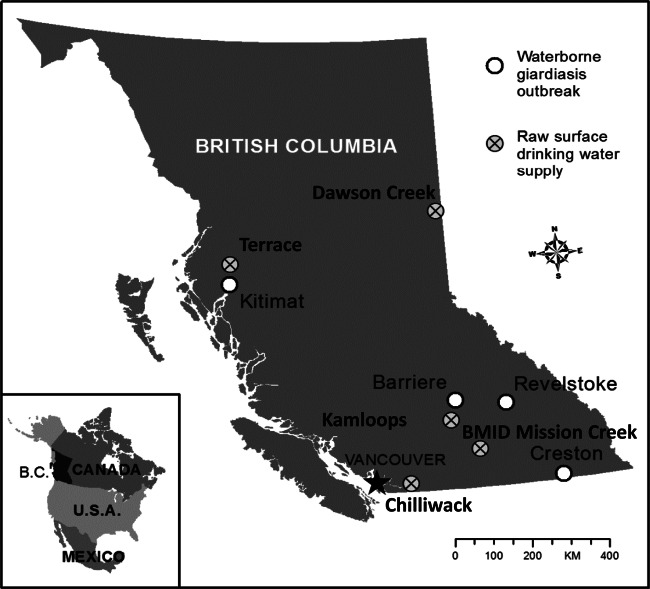
Between 1989 and 1995, four outbreaks of waterborne giardiasis occurred in small BC towns (Fig. 1). Public health investigations implicated untreated and contaminated drinking water as the source of the outbreaks; investigations were carried out by testing surface drinking water samples, human feces, and in two outbreaks, samples from beavers (Castor canadensis) epidemiologically implicated as the source of fecal contamination (Table 1). The first outbreak of giardiasis in Creston (southeastern BC) (Fig. 1) was investigated after the abrupt onset of an excess number of cases of gastrointestinal illness was reported to local public health officials. Over 70 town residents who drank the untreated (no chlorination or filtration) surface water were found to have Giardia cysts in their feces. When investigations implicated drinking water, a boil water notice was issued and two raw water samples were collected a week apart from a household tap in town. A third drinking water sample was collected at the drinking water intake of the town's surface supply. Giardia cysts were detected in all of the water samples, and three isolates were retrieved, one from each of the three separate water samples. The town's drinking water was from an undeveloped forested watershed (no human habitation, farming, or domestic animal use). At the time of this winter outbreak, the watershed was under deep snow and the creek (drinking water supply) was covered with ice. Investigations to locate the source of drinking water contamination found a beaver living in the water close to and upstream of the drinking water intake. No parasites were detected in water samples collected upstream of this beaver's location, and when the beaver was removed, Giardia cysts were no longer detected in drinking water samples and new human infections stopped. However, a Giardia isolate was retrieved from the beaver that was removed from the intake. Seventy-two laboratory-confirmed human infections were reported; eight different human source isolates were retrieved from human case fecal samples collected over a 4-day period.
FIG 1.
Map of BC showing the locations of the four giardiasis outbreaks and the raw surface water supplies tested.
TABLE 1.
Giardia isolates from waterborne outbreaks in BC summarized by assemblage
| Town with outbreak and isolate source (no. of isolates) | Collection date(s) | Assemblage(s) (no. of isolates) |
|---|---|---|
| Creston | ||
| Drinking water (3) | February, March 1990 | B (3) |
| Beaver (1) | March 1990 | A (1) |
| Human (8) | February 1990 | B (4), A (4) |
| Kitimat | ||
| Drinking water (1) | March 1990 | B (1) |
| Human (3) | December 1989, February 1990 | A (1), B (2) |
| Barriere | ||
| Drinking water (3) | July, September 1990 | A (1), B (2) |
| Human (3) | December 1990, January 1991 | A (1), B (2) |
| Revelstoke | ||
| Human (3) | September 1995 | A (1), B (2) |
| Beaver (1) | August 1995 | B (1) |
The Kitimat waterborne outbreak (Fig. 1) also occurred in the winter of the same year but in northwestern BC. This town used unfiltered surface source drinking water; the water was chlorinated, but gravel infiltration gallery beds at the intake had been disrupted just prior to the outbreak. Although investigations were not as extensive as in the previously described outbreak, drinking water was implicated following a report to public health officials of an abrupt increase in excess numbers of laboratory-confirmed cases of giardiasis (n = 28). A boil water notice was issued, and drinking water samples tested were found to contain Giardia cysts. Isolates from the water source, as well as human isolates from this outbreak, were included for molecular characterization in this study. In the following year, physicians in a third BC town, Barriere, (Fig. 1), notified public health officials of an excess number of giardiasis cases (25 laboratory confirmed). Patients were interviewed by public health officials and drinking of untreated surface water was implicated as the probable source of infection. The drinking water surface supply was from a watershed known to be inhabited by wildlife. However, no watershed investigation was carried out and while isolates from cyst-positive surface water samples were retrieved, they were collected 3 months before the human isolates were retrieved. The fourth outbreak (Revelstoke, Fig. 1), 4 years after the Creston outbreak, occurred in yet another small BC town also using an untreated surface supply for drinking water. Its source was from an undeveloped (no human habitation, no farming or domestic animal activities) watershed. Epidemiological investigation implicated drinking water after the abrupt occurrence of an excess number of cases (62 laboratory confirmed) of gastrointestinal illness was reported, and a beaver was found living in a culvert in the drinking water intake from the creek. Giardia was found in its feces; when it was removed, the outbreak declined. Isolates were retrieved from this beaver and additional human patients.
(ii) Non-outbreak-associated water isolates.
Because of numerous waterborne outbreaks, water samples from various areas of the province were collected (during all seasons) over a 4-year period (1990 to 1994). In the present study, 29 surface water sample isolates were characterized, including 22 not associated with outbreaks and 7 associated with outbreaks. Of the 22 isolates from water samples not associated with outbreaks, 17 were from one mixed-use watershed (Table 2). Water samples from this watershed were collected over 3 years during the same 10-year period to assess the effects of settling ponds on Giardia cyst concentration (Table 2; Fig. 1). The remaining 12 isolates were retrieved from raw water samples collected at different times at the community drinking water intake; 5 additional isolates were collected from the reservoir/settling pond a short distance downstream of the intake. The town, which used drinking water settling ponds and a long contact time for chlorination of their surface drinking water supply, reported no excess cases of giardiasis during this time. Four other geographically separate surface water sites (Fig. 1) were also tested for cysts, and five water source isolates were retrieved (Table 2). No excess cases of giardiasis were reported in the four towns using these surface drinking water supplies.
TABLE 2.
Giardia isolates from nonoutbreak surface drinking water in BC summarized by assemblage
| Surface water site (no. of isolates) and collection date(s) | Assemblage (no. of isolates) |
|---|---|
| Mission Creek intake (10) | |
| September 1990 | B (1) |
| May, December 1991 | B (2) |
| April, July 1992 | B (2) |
| September 1992 | A (1) |
| October 1992 | B (1) |
| November 1992 | A (1) |
| September 1993 (2) | B (2) |
| Mission Creek settling pond (7) | |
| August, September 1992 | B (3) |
| August, September 1993 | B (3) |
| October 1993 | B (1) |
| Elk Creek (1), May 1994 | A (1) |
| Muskwa River (1), February 1991 | B (1) |
| South Thompson River (1), April 1991 | B (1) |
| Deep Creek (2), April 1991 | B (2) |
(iii) Non-outbreak-associated human isolates.
Non-outbreak-associated human isolates were also retrieved from cyst-positive feces during this study period. Of the human patients, none had traveled outside BC. Patients ranged in age from 3 to 72 years (median, 30 years); seven isolates were from patients less than 12 years of age. Isolates were from 13 males and 16 females. All but four human isolates were from symptomatic persons; the four asymptomatic persons were tested because other members of their family or work groups had been diagnosed with giardiasis. While there were 17 isolates from human infections associated with the outbreak, there were also 12 not associated with outbreaks.
(iv) Non-outbreak-associated beaver isolates.
Non-outbreak-associated beaver isolates were obtained from animals located at two geographically separate sites (Table 3). One group (four isolates from four beavers) was located 11 miles east of Creston (outbreak site), and the second group (three isolates from three beavers) was located in a farm pond hundreds of miles southwest of Creston.
TABLE 3.
Isolates from BC beavers not associated with outbreaks characterized by assemblage
| BC site location | Source (no. of isolates) | Collection date | Assemblage (no. of isolates) |
|---|---|---|---|
| Pond near Aldergrove | Beaver (3) | December 1987 | A (3) |
| Goat River | Beaver (4) | April 1990 | B (4) |
| Pantage Creek | Beaver (1) | November 1985 | A (1) |
| Slocan River | Beaver (1) | December 1987 | B (1) |
| Zimacord River | Beaver (1) | April 1985 | B (1) |
Isolate retrieval.
Large-volume raw surface water samples were collected, concentrated, and processed for the presence of Giardia cysts (8). Isolates were also purified from human and beaver fecal samples containing cysts (9). All study isolates were excysted by in an in vivo gerbil model (10). Giardia trophozoites were cultured and cryopreserved in 10% dimethyl sulfoxide (DMSO) at −80°C (7). For the present molecular characterization study, trophozoites were retrieved and DNA was obtained from the 71 Giardia study isolates described above.
DNA extraction.
Briefly, trophozoites were centrifuged at 10,000 rpm for 10 min at room temperature, washed twice with cold phosphate-buffered saline, centrifuged for 3 min, and then incubated at 58°C for 30 min with 20 μl of proteinase K and buffer ATL (Qiagen, Mississauga, ON, Canada). DNA was then extracted from trophozoites with a QIAamp DNA minikit (Qiagen, Mississauga, ON, Canada). Extracted DNA was quantified with a Qubit fluorometer (Invitrogen, Carlsbad, CA, USA), and quality was assessed with the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA).
PCR and Sanger sequencing.
A fragment of the 18S rRNA gene was amplified with primers RH11 (CATCCGGTCGATCCTGCC) and RH4 (AGTCGAACCCTGATTCTCCGCCAGG) (11). The PCR mixtures (50-μl volumes) consisted of 2 μl of sample, 1× GoTaq reaction buffer, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 5% DMSO, 0.2 μM each primer, and 1.25 U of Taq polymerase (Promega, Madison, WI, USA). The reactions were carried out in the GeneAmp PCR System 9700 (Life Technologies) as described by Appelbee et al. (11). PCR products were purified with the QIAquick PCR purification kit (Qiagen, Mississauga, ON, Canada). Purified amplicons were sequenced with the ABI PRISM BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) with the amplification primers and 2% DMSO on the ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, CA, USA). Sequences were aligned in Geneious v.6 (Biomatters, Inc., San Francisco, CA) (12), and their identities were verified by BLAST (http://www.ncbi.nlm.nih.gov) (13). The 18S rRNA gene alignment was analyzed, together with the 18S rRNA gene sequences of standard strains WB (assemblage A) and GS_B (assemblage B) (4, 14) retrieved from NCBI with Molecular Evolutionary Genetics Analysis version 6.0 (MEGA6) (15). Neighbor-joining (NJ) analyses were conducted with the Kimura two-parameter model. Bootstrap support was estimated by using 1,000 replicates. A maximum-likelihood (ML) analysis was also conducted by MEGA6 with the general time-reversible model under gamma distributed with invariant sites for rate among sites. The confidence of the node was estimated by using 500 bootstrap replicates.
Library preparation and WGS.
Paired-end (PE) DNA libraries were constructed with the Nextera XT DNA kit (Illumina, San Diego, CA) according to the manufacturer's protocol, with some modifications. To remove small inserts, DNA libraries were cleaned up twice with the Agencourt AMPure XP-PCR purification system (Beckman Coulter Inc., Brea, CA) at a 0.5× ratio. Samples were then normalized to equal molar concentrations, denatured with 0.1 N NaOH, and neutralized by dilution as recommended by Illumina. Library quality was assessed with the High Sensitivity DNA Chip on the Bioanalyzer 2011 (Agilent Technologies). DNA libraries were pooled in six to eight samples per run in accordance with the library pooling considerations described in the manufacturer's protocol. Samples were loaded onto a MiSeq flow cell with 10% PhiX spiked in and sequenced with MiSeq v2 reagent kits (Illumina, Inc., San Diego, CA) with a 250-bp PE output.
Whole-genome sequence assembly and mapping.
The quality of the reads was assessed by FastQC, and the adaptor sequences and sequences of poor quality were trimmed by Trim Galore v.0.3.1 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). All Giardia genomes were de novo assembled from the PE reads with the assembler SPAdes v3.1 (16). We assessed the assemblage identity of each isolate by estimating the percentage of PE reads from each isolate mapped to the reference genomes of Giardia (assemblages A, B, and E) available in GiardiaDB (http://giardiadb.org) with a cutoff of 85% (17). Before read mapping, low-quality raw reads were filtered and the remaining reads were mapped to the reference genomes with Bowtie 2 (18).
Multilocus phylogenetic analysis.
Makeblastdb (part of the BLAST+ package) was used to convert all of the assembled Giardia genomes into blast databases. The sequences of three loci, those for triose-phosphate isomerase (tpi), β-giardin (bg), and glutamate dehydrogenase (gdh), were retrieved from the assembled genomes with the command line fastacmd in the BLAST+ package. The alignments of these three genes were concatenated with the 18S rRNA gene alignment from the Sanger sequencing data for phylogenetic analysis. The phylogenetic relationships between the isolates of assemblages A and B were analyzed by MEGA6 (15). Parsimony analyses were done by using a heuristic search with random stepwise addition of data and tree bisection-reconnection in MEGA6 for 1,000 bootstrap replicates. An NJ (19) tree was also built with MEGA6 by using Jukes-Cantor distance. The confidence of the topology was assessed by using 1,000 bootstrap replicates.
Core genome phylogeny.
Over 4,000 protein orthologs (core gene set) common to Giardia assemblages A, B, and E (4) were downloaded from GiardiaDB (http://giardiadb.org). These orthologs were searched against and retrieved from the 12 draft assembled genomes generated in this paper by using tblastn under default conditions (13). Orthologs retrieved from the assembled genomes with <50% nucleotide sequence identity to and <70% coverage of the queries were filtered from the data matrix. All the orthologs were individually aligned by MUSCLE (20). Over 4,000 alignments were concatenated into a single alignment and analyzed by FastTree (21) and MEGA6. The tree was visualized with FigTree (available from http://tree.bio.ed.ac.uk/software/figtree/).
Nucleotide sequence accession numbers.
The new sequences determined in this study were deposited in GenBank under accession numbers KM190677 to KM190837, KM201106 to KM201175, and KP687755 to KP687790. The accession numbers of all of the sequences can be found via BioProject record no. PRJNA280606.
RESULTS
PCR and Sanger sequencing analysis.
Overall, the 18S rRNA gene PCR and sequence analysis of the 71 BC isolates showed that 26 (37%) were of assemblage A and 45 (63%) were of assemblage B. Of the 29 raw surface water isolates, 4 (14%) were of assemblage A while 25 (86%) were of assemblage B. This contrasted with both the 29 human source isolates, of which 16 (55%) were of assemblage A and 13 (45%) were of assemblage B, and the 13 beaver source isolates, of which 6 (50%) were of assemblage A and 7 (50%) were of assemblage B. Analysis by geographical sample sites was done to address possible overrepresentation by outbreak or ongoing water studies. Results of water isolates from eight different sites showed that almost all were of assemblage B. Three of the 4 of assemblage A water isolates of the 29 from raw surface water were from repeat sampling of the two water systems studied over time. Of the human isolates tested (from 16 different geographic sites), 16 (55%) were of assemblage A and 13 (45%) were of assemblage B. When all of the strains isolated from beavers were analyzed on the basis of their eight different geographical sites (duplicates from the same site or time were removed), four were of assemblage A and four were of assemblage B. Thus, the pattern of isolate assemblages in BC humans and beavers and in non-outbreak and outbreak settings contrasted with the predominantly assemblage B isolates from surface water.
Analysis of isolates from the raw surface drinking water sources not associated with outbreaks (Table 1) showed that of the samples (17 strains over 3 years) collected from a single small water system (Fig. 1) over time, 2 (12%) were of assemblage A and 15 (86%) were of assemblage B. When analyzed by the collection sites within this surface drinking water source system, most of the isolates from samples (10 of 12) collected at the drinking water intake were of assemblage B. We note that both assemblages A and B were detected in this system during the same month (September 1992).
Analysis of PCR and sequence data from isolates retrieved during waterborne outbreaks (Table 2) showed mixtures of both assemblages A and B. Further analysis of the Creston outbreak, which had the most isolates retrieved (n = 12), showed that during the outbreak period, all 3 water isolates retrieved included assemblage B isolates (2 from tap water in a Creston home, 1 from the drinking water intake at the creek). Four isolates, however, from human patients were of assemblage A and four were of assemblage B. As previously noted, when the watershed investigation showed evidence of beaver tracks in the snow (no other human or animal activity was observed) and after the animal was located living in the creek at the intake, the beaver was removed. The isolate retrieved from this beaver was of assemblage A.
Similarly, analysis of the four Kitimat outbreak isolates showed both assemblages; assemblage B was found in the implicated water supply, but one human patient was infected with an assemblage A isolate and two other human outbreak patients were infected with assemblage B isolates. Although the Barriere outbreak was less abrupt in presentation than the other three outbreaks, similar findings were observed, with both assemblages B and A in the implicated surface drinking water supply (samples collected within a 1-month period) and human isolates, including two of assemblage B and one of assemblage A. A sporadic human strain collected 3 years after the outbreak was of assemblage A and was identical to outbreak strains at the 18S rRNA gene locus. In the Revelstoke outbreak, a mixture of assemblages was detected, with the isolate from the beaver (residing in the drinking water intake and considered the source of the outbreak) being of assemblage B while isolates from three human patients were of assemblages A (one isolate) and B (two isolates). As shown in Table 3, there were two beaver groups; the isolates from all of the members of one group were assemblage A and those from all of the members of the other group were assemblage B isolates. Of the four isolates from other single beavers (not from groups but from widely separated locations in BC), two were assemblage A and two were assemblage B Giardia.
Multilocus phylogenetic analysis.
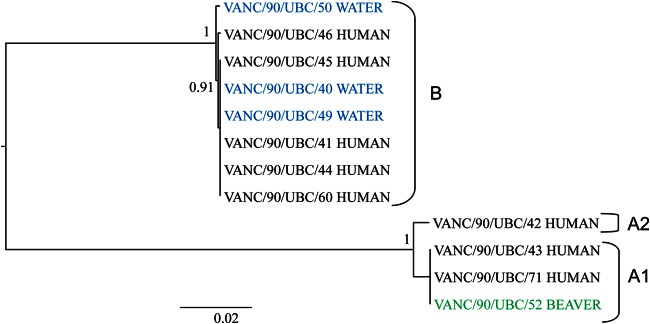
To confirm the PCR and sequencing assemblage data, multilocus phylogenetic analysis (gdh, bg, tpi, 18S rRNA gene) was carried out with sequences derived from WGS. Results corroborated the PCR and sequencing assemblage designations. A phylogenetic tree of the 12 Creston outbreak-associated isolates (water, human, and beaver) is shown in Fig. 2. Assemblage B isolates (three from water, five from humans) from the Creston outbreak clustered closely, and assemblage A isolates (one from a beaver, three from humans) nested in two groups. Isolates in Creston assemblage A were differentiated into assemblage subgroups A1 and A2, which had 99.1% nucleotide sequence identity (Fig. 2).
FIG 2.
Phylogeny of Giardia isolates from the Creston outbreak based on four genes (gdh, bg, tpi, and the 18S rRNA gene) and 500 bootstrap replicates. Isolates are colored according to their sources (blue, water; black, human; green, beaver).
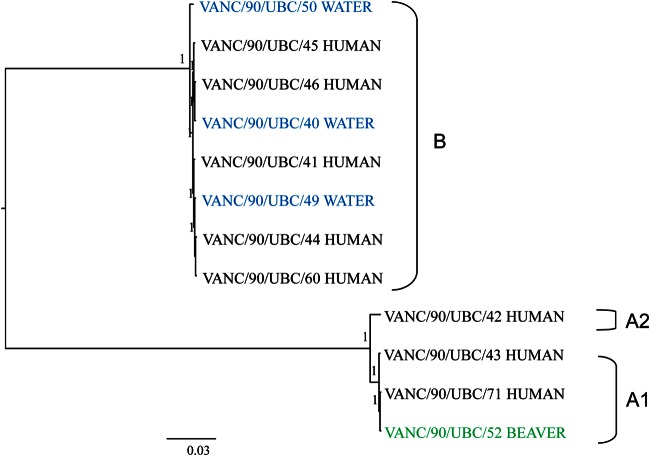
The average insert size of samples was ca. 750 bp. The average sequencing depth obtained for the outbreak isolates was 85× (range, 33× to 138×). The genome size ranged from 10 to 12 Mb with a GC content of ca. 48% (see Table S1 in the supplemental material). No mixed genomes were observed among the 12 isolates sequenced. The phylogenetic tree based on 4,086 core genes with over 2.5 million amino acids (Fig. 3) for strains from the Creston outbreak was congruent with the four-locus tree, but it revealed greater resolution among assemblage B isolates. For instance, there were at least three well-supported clades found within the assemblage B isolates. Isolates of assemblage A are divergent from those of assemblage B (ca. 77% amino acid identity). Within each assemblage, most of the outbreak isolates had very high percentages of identity in their genomes. There was less than 1% amino acid sequence variation among the isolates of assemblages A1 (99.2% identity) and B (99.1% identity), respectively, except an isolate from a water source (VANC/90/UBC/50) that was 98.5% identical to the other seven assemblage B isolates. Isolates of assemblage A2 were differentiated from those of A1 by a ca. 3% sequence dissimilarity (Fig. 3). The core gene tree also indicated close connections among human and water source isolates of assemblage B, for instance, the cluster of VANC/90/UBC/46 and VANC/90/UBC/40, as well as the cluster of VANC/90/UBC/44, VANC/90/UBC/60, and VANC/90/UBC/49. Similarly, one human isolate, VANC/90/UBC/71, clustered with one beaver isolate, VANC/90/UBC/52, with strong support in assemblage A1 (Fig. 3).
FIG 3.
ML tree of the Giardia isolates from the Creston outbreak constructed with 4,086 core genes and 100 bootstrap replicates. Isolates are colored according to their sources (blue, water; black, human; green, beaver).
DISCUSSION
Many small towns in the western Canadian province of BC use surface sources for drinking water. Most of the water was treated only by chlorination, and almost none was filtered at the time of the waterborne giardiasis outbreaks. Although no further waterborne outbreaks (suspected or confirmed) have been identified since the original work (resulting in the isolate library), Giardia remains one of the province's most common enteric pathogens. This study used the unique library of Giardia isolates collected during the period of public health focus to obtain new information by using recently developed molecular tools to characterize isolates.
Although the library of isolates may have selection bias from in vivo retrieval and culture of trophozoites, we also note some advantages of the characterization of isolates by culture-based methods rather than cyst-based PCR (22, 23). First, axenic culture is more likely to support the growth of a pure culture, which aids in the interpretation of WGS data. It should be noted that while it is possible to interpret WGS data from mixed populations, the limited availability of G. duodenalis genomes (both complete and draft) impacts the interpretation of mixed data sets at this time. Furthermore, PCR used to characterize cultured trophozoites is not subject to issues of inhibitory compounds such as heme and humic acids concentrated during the DNA extraction process. Lastly, the large amount of DNA available from culture needed to prepare libraries for WGS enables a more comprehensive and higher-resolution characterization than cyst-based PCR.
While new molecular tools are increasing the understanding of the biology of Giardia, it is not yet fully understood. WGS will contribute to more extensive knowledge of G. duodenalis. For example, the genomes of the five Giardia isolates sequenced (4, 14, 24, 25) to date support the proposal that G. duodenalis (assemblage A) and G. enterica (assemblage B) are, in fact, two distinct species.
This work characterized a large number of surface water isolates retrieved from samples from all parts of BC by sequencing commonly used genetic markers (the 18S rRNA gene, the β-giardin gene, gdh, and tpi). Overall, most of the water isolates contained assemblage B, despite geographic and temporal differences in isolate collection. It is worth noting that despite the consumption of unfiltered drinking water in the five non-outbreak-associated communities studied, no excess numbers of human cases of giardiasis were reported.
It was an unexpected finding that both assemblages A and B were present in all four waterborne outbreaks. The best-characterized and -understood outbreak (in Creston) was previously thought to be a point source outbreak attributed to a single beaver residing in the drinking water intake (at a time of year when the watershed was under deep snow, the water supply was under ice, there was no human activity and no evidence of other wildlife, and no overland water transport or animal grazing was possible). Notably, the outbreak (no new cases) and detection of cysts in the drinking water stopped after the beaver was removed, clearly implicating the beaver. While the isolates from drinking water were assemblage B organisms, both assemblage A and B cases were found in the human patients and the imputed source for drinking water (fecal contamination by a beaver) was assemblage A.
To further study this finding, WGS was performed to better understand the relatedness among Creston isolates. This could not be accomplished with traditional genotyping approaches (sequencing of key genetic markers), as they do not afford sufficient resolution, nor do PCR-based methods allow this level of characterization. Figures 2 and 3 show multilocus and core genome phylogenetic analyses by WGS of the Creston outbreak isolates, with assemblage A clustering into A1 and A2; assemblage B was even more tightly clustered by WGS of the core genome.
This suggests that the dynamics of Giardia transmission, particularly of assemblage A, through contaminated untreated drinking water are complex. Since point source outbreaks are often considered to be clonal, caused by a single strain of a contaminating microbe, the detection in this study of different assemblages (A and B) was unexpected. However, it is probable that surface water is dynamically contaminated by different host sources, given the range of animal hosts known to carry zoonotic assemblages (1). Furthermore, it is well documented that mixed Giardia infections in both humans and animals (dogs, cattle, wildlife) can occur (26–29). Given the mixture of assemblages detected within a short time frame (days to weeks) in all four outbreaks, it is possible that the water source was dynamically contaminated by multiple animals or that a single host was infected with a mixture of G. duodenalis assemblages. The detection of only one assemblage by genotyping and PCR may be attributed to the retrieval method; it is known that axenic culture of strains from a mixed infection may preferentially support one strain over the other (29).
Outbreaks of waterborne disease may result from coinciding circumstances, including inadequate drinking water treatment, failure of treatment following severe weather events (30), increased virulence of contaminating agents, and the level of population (herd) immunity to the contaminating agent(s) (31). Waterborne outbreaks caused by polymicrobial contamination have also been reported; this is not surprising, given that water sources may be impacted by multiple pollution sources or that one host could be infected with multiple pathogens (32). Given the multitude of potential fecal contamination sources and the use of unfiltered drinking water in many small communities in BC during the 1980s and early 1990s, more waterborne outbreaks may have been expected. However, excess giardiasis cases in a human population are mostly likely to occur when large numbers of cysts from a contaminated surface water source overcome the herd immunity of a population drinking inadequately treated surface water (31).
While it was observed that both assemblages A and B (water as well as human and beaver isolates) were present in all regions of BC and while both assemblages were detected within single water systems and within outbreaks, this study also shows a clear predominance of assemblage B among water isolates (compared to both human and beaver isolates collected during the same period in BC). This may mean that the overall ecology and transmission of Giardia is dominated by assemblage B, and this may not be ascertained given the isolates available for this study. It may also be that, phenotypically, this assemblage is better able to persist in the environment, but that has not been supported by the scientific literature to date. Further study of this assemblage (designated by some recently as a separate species) is warranted.
Given the findings of this study, further studies of Giardia in the environment should consider an extensive sampling strategy with testing done in replicates to capture the diversity of assemblages and to avoid testing and sampling biases. Furthermore, the degree of variation among isolates seemingly identical by single- or multilocus genotyping (and specifically for assemblage A, which appears to be more clonal) may obscure the detection of mixed infections or mixed contamination of water. Newer tools such as WGS have already been used to study outbreaks due to other microbes. WGS will probably be a useful tool to better understand source attribution and to better understanding pathogen transmission dynamics. WGS, when used to examine the core genome phylogeny of an outbreak, confirmed findings generated by multilocus genotyping. Higher-resolution WGS analysis methods such as single nucleotide polymorphism analyses may be used to develop a more comprehensive understanding of the phylogenetic relationships, genomic strategies, and adaptation strategies of this poorly understood but ubiquitous waterborne pathogen.
Supplementary Material
ACKNOWLEDGMENTS
The funding support of the Canadian Institutes for Health Research (CIHR) in retrieving the isolate library is acknowledged.
We are grateful for the support of the province's public health and local officials and members of the Environmental Microbiology Program at the British Columbia Public Health Microbiology Reference Laboratory (BCPHMRL). In particular, we thank Loan Hoang, Anna Li, Renata Zanchettin, Brian Auk, C. P. Fung, Justin Dirk, and Sara Tan for their ongoing dedication and assistance. We also thank Sunny Mak at the British Columbia Centre for Disease Control for his assistance with generating maps.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00524-15.
REFERENCES
- 1.Feng YY, Xiao LH. 2011. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev 24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson RCA. 2000. Giardiasis as a re-emerging infectious disease and its zoonotic potential. Int J Parasitol 30:1259–1267. doi: 10.1016/S0020-7519(00)00127-2. [DOI] [PubMed] [Google Scholar]
- 3.van Keulen H, Feely DE, Macechko PT, Jarroll EL, Erlandsen SL. 1998. The sequence of Giardia small subunit rRNA shows that voles and muskrats are parasitized by a unique species Giardia microti. J Parasitol 84:294–300. doi: 10.2307/3284485. [DOI] [PubMed] [Google Scholar]
- 4.Adam RD, Dahlstrom EW, Martens CA, Bruno DP, Barbian KD, Ricklefs SM, Hernandez MM, Narla NP, Patel RB, Porcella SF, Nash TE. 2013. Genome sequencing of Giardia lamblia genotypes [sic] A2 and B isolates (DH and GS) and comparative analysis with the genomes of genotypes A1 and E (WB and pig). Genome Biol Evol 5:2498–2511. doi: 10.1093/gbe/evt197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Statistics Canada. 2012. The Canadian population in 2011: population counts and growth. Statistics Canada, Ottawa, Ontario, Canada. [Google Scholar]
- 6.British Columbia Provincial Health Officer. 2001. A report on the health of British Columbians. Provincial Health Officer's annual report 2000. Drinking water quality in British Columbia: The public health perspective. Ministry of Health Planning, Victoria, British Columbia, Canada. [Google Scholar]
- 7.Sarafis K, Isaac-Renton J. 1993. Pulsed-field gel-electrophoresis as a method of biotyping of Giardia duodenalisis. Am J Trop Med Hyg 48:134–144. [DOI] [PubMed] [Google Scholar]
- 8.Environmental Protection Agency. 1999. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. EPA 821-R-99-006. Office of Water, Environmental Protection Agency, Washington, DC. [Google Scholar]
- 9.Garcia LS. 2007. Diagnostic medical parasitology, 5th ed ASM Press, Washington, DC. [Google Scholar]
- 10.Isaac-Renton JL, Shahriari H, Bowie WR. 1992. Comparison of an in vitro method and an in vivo method of Giardia excystation. Appl Environ Microbiol 58:1530–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Appelbee AJ, Frederick LM, Heitman TL, Olson ME. 2003. Prevalence and genotyping of Giardia duodenalis from beef calves in Alberta, Canada. Vet Parasitol 112:289–294. doi: 10.1016/S0304-4017(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 12.Kearse M, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, TLM . 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, Davids BJ, Dawson SC, Elmendorf HG, Hehl AB, Holder ME, Huse SM, Kim UU, Lasek-Nesselquist E, Manning G, Nigam A, Nixon JEJ, Palm D, Passamaneck NE, Prabhu A, Reich CI, Reiner DS, Samuelson J, Svard SG, Sogin ML. 2007. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 15.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aurrecoechea C, Brestelli J, Brunk BP, Carlton JM, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Morrison HG, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ, Sullivan S, Treatman C, Wang HM. 2009. GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res 37:D526–D530. doi: 10.1093/nar/gkn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357-U354. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 20.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews RH, Chilton NB, Mayrhofer G. 1992. Selection of specific genotypes of Giardia intestinalis by growth in vitro and in vivo. Parasitology 105:375–386. doi: 10.1017/S0031182000074540. [DOI] [PubMed] [Google Scholar]
- 23.Bénéré E, Van Assche T, Cos P, Maes L. 2011. Variation in growth and drug susceptibility among Giardia duodenalis assemblages A, B, and E in axenic in vitro culture and in the gerbil model. Parasitology 138:1354–1361. doi: 10.1017/S0031182011001223. [DOI] [PubMed] [Google Scholar]
- 24.Franzén O, Jerlstrom-Hultqvist J, Castro E, Sherwood E, Ankarklev J, Reiner DS, Palm D, Andersson JO, Andersson B, Svard SG. 2009. Draft genome sequencing of Giardia intestinalis assemblage B isolate GS: is human giardiasis caused by two different species? PLoS Pathog 5:e1000560. doi: 10.1371/journal.ppat.1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jerlström-Hultqvist J, Franzén O, Ankarklev J, Xu FF, Nohýnková E, Andersson JO, Svärd SG, Andersson B. 2010. Genome analysis and comparative genomics of a Giardia intestinalis assemblage E isolate. BMC Genomics 11:543. doi: 10.1186/1471-2164-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geurden T, Geldhof P, Levecke B, Martens C, Berkvens D, Casaert S, Vercruysse J, Claerebout E. 2008. Mixed Giardia duodenalis assemblage A and E infections in calves. Int J Parasitol 38:259–264. doi: 10.1016/j.ijpara.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 27.Helmy YA, Klotz C, Wilking H, Krücken J, Nöckler K, Von Samson-Himmelstjerna G, Zessin KH, Aebischer T. 2014. Epidemiology of Giardia duodenalis infection in ruminant livestock and children in the Ismailia province of Egypt: insights by genetic characterization. Parasites Vectors 7:321. doi: 10.1186/1756-3305-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopkins RM, Meloni BP, Groth DM, Wetherall JD, Reynoldson JA, Thompson RCA. 1997. Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J Parasitol 83:44–51. doi: 10.2307/3284315. [DOI] [PubMed] [Google Scholar]
- 29.Upcroft JA, Upcroft P. 1994. Two distinct varieties of Giardia in a mixed infection from a single human patient. J Eukaryot Microbiol 41:189–194. doi: 10.1111/j.1550-7408.1994.tb01495.x. [DOI] [PubMed] [Google Scholar]
- 30.Hrudey SE, Hrudey EJ. 2006. Safe drinking water: lessons from recent outbreaks, p 333–336. In Thompson KC, Gray J (ed), Water contamination emergencies: enhancing our response. RSC Publishing, Cambridge, United Kingdom. doi: 10.1039/9781847552426. [DOI] [Google Scholar]
- 31.Isaac-Renton JL, Lewis LF, Ong CSL, Nulsen MF. 1994. A second community outbreak of waterborne giardiasis in Canada and serological investigation of patients. Trans R Soc Trop Med Hyg 88:395–399. doi: 10.1016/0035-9203(94)90397-2. [DOI] [PubMed] [Google Scholar]
- 32.Anonymous. 2000. Waterborne outbreak of gastroenteritis associated with a contaminated municipal water supply, Walkerton, Ontario, May-June 2000. Can Commun Dis Rep 26:170–173. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.