Abstract
Enterococcus faecalis F4-9 isolated from Egyptian salted-fermented fish produces a novel bacteriocin, termed enterocin F4-9. Enterocin F4-9 was purified from the culture supernatant by three steps, and its molecular mass was determined to be 5,516.6 Da by mass spectrometry. Amino acid and DNA sequencing showed that the propeptide consists of 67 amino acid residues, with a leader peptide containing a double glycine cleavage site to produce a 47-amino-acid mature peptide. Enterocin F4-9 is modified by two molecules of N-acetylglucosamine β-O-linked to Ser37 and Thr46. The O-linked N-acetylglucosamine moieties are essential for the antimicrobial activity of enterocin F4-9. Further analysis of the enterocin F4-9 gene cluster identified enfC, which has high sequence similarity to a glycosyltransferase. The antimicrobial activity of enterocin F4-9 covered a limited range of bacteria, including, interestingly, a Gram-negative strain, Escherichia coli JM109. Enterocin F4-9 is sensitive to protease, active at a wide pH range, and moderately resistant to heat.
INTRODUCTION
Food spoilage bacteria and food-borne pathogens are public health concerns worldwide; therefore, food preservation techniques have received considerable attention (1). However, consumers have become aware of the hazardous effects of using chemical preservatives, increasing the demand for natural foods with fewer preservatives and unimpaired sensory quality (2). To achieve these aims, food science researchers have developed a novel approach to food preservation, biopreservation using bacteriocins from lactic acid bacteria (LAB).
Bacteriocins are antimicrobial peptides ribosomally synthesized by prokaryotes, which exhibit bactericidal activity against species closely related to the producer strain (3, 4, 5). In general, the effect of bacteriocins on bacterial cells is exerted by increasing the permeability of the cytoplasmic membrane, ultimately destabilizing membrane function (6). LAB bacteriocins have numerous desirable features, including safety, easy digestion by human proteases, stability at wide pH and temperature ranges, antimicrobial activity against a wide spectrum of food-borne pathogens and food spoilage bacteria, and no cross-resistance with antibiotics. These features have prompted food scientists to assess the potential for bacteriocins as biopreservatives.
Bacteriocins produced by Gram-positive bacteria are classified into three classes as proposed by Cotter et al. (7): class I (lantibiotics), comprising lanthionine-containing peptides; class II, including non-lanthionine-containing bacteriocins; and bacteriolysins (previously class III bacteriocins). Bacteriolysins are large, heat-labile antimicrobial peptides that possess a domain-type structure that functions in translocation, receptor binding, and lethal activity. Moreover, Klaenhammer (8) has suggested that bacteriocins that are modified with carbohydrate or lipid should be classified as a separate class IV, as in the cases of glycocin F (formerly plantaricin KW30) (9) and sublancin 168 (10).
Bacteriocins produced by Enterococcus spp. are termed enterocins. Most enterocins belong to class II bacteriocins, which is subdivided into four subcategories. Each class is represented by the following enterocins: class IIa, enterocin A (11); class IIb, enterocin 1071 (12); class IIc, enterocin AS-48 (13); and class IId, enterocin B (14).
Some bacteriocins undergo unusual posttranslational modifications (PTMs) such as glycosylation (9). In glycosylation, a carbohydrate group is covalently linked to a peptide or protein. The most common patterns of glycosylation are linkage through the hydroxyl group of serine/threonine residues (O-linked) or the side chain nitrogen of asparagine (N-linked); a less common linkage forms via the sulfur atom of a cysteine residue (S-linked) (15). Glycosylation confers more diverse biological functions and greater proteomic dissimilarities than other PTMs (16). To date, only two bacteriocins were reported as glycosylated bacteriocins: glycocin F produced by Lactobacillus plantarum KW30 (9) and sublancin 168 produced by Bacillus subtilis 168 (10).
Here we present the purification and characterization of a novel class IV bacteriocin, enterocin F4-9, which is posttranslationally modified by glycosylation. To our knowledge, this is the first report of a glycosylated bacteriocin produced by Enterococcus spp.
MATERIALS AND METHODS
Bacterial strains and media.
Strain F4-9 was isolated from Egyptian salted-fermented fish and identified as Enterococcus faecalis by the 16S rRNA gene sequence as described previously (17). E. faecalis F4-9 was stored at −80°C in MRS medium (Oxoid, Basingstoke, United Kingdom) with 15% glycerol; it was propagated in MRS medium at 30°C for 16 h before use. For determination of antimicrobial spectra, indicator strains were propagated at the appropriate temperature (30°C or 37°C), as recommended by culture collections. Indicator strains of LAB were cultured in MRS medium, and non-LAB indicator strains, except those mentioned below, were cultured in tryptic soy broth (BD, Sparks, MD) supplemented with 0.6% yeast extract (Nacalai Tesque, Kyoto, Japan) (TSBYE). Cupriavidus necator and Pseudomonas putida were cultured in nutrient broth at 25°C for 48 h and Luria-Bertani (LB) medium (BD) at 37°C for 24 h, respectively. Escherichia coli JM109 was grown in LB medium at 37°C. E. coli JM109 transformants were cultivated in LB agar medium containing 1.5% agar (Nacalai Tesque), 20 μg ml−1 ampicillin, and 50 μg ml−1 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (Nacalai Tesque).
Bacteriocin activity assay.
The spot-on-lawn method was used to assess bacteriocin activity (17). Briefly, 10-μl portions of twofold dilutions of a bacteriocin preparation were spotted onto a double layer comprised of 5 ml of Lactobacilli Agar AOAC (BD) or TSBYE with 1% agar and inoculated with an overnight culture of an indicator strain as the upper layer and 10 ml of MRS medium with 1.2% agar as the bottom layer. After incubating overnight at the appropriate temperature for the indicator strains, the bacterial lawns were checked for inhibition zones. In the purification steps, the activity titer was defined as the reciprocal of the highest dilution that yields a clear zone of inhibition in the indicator lawn; this value was expressed in arbitrary activity units per milliliter of bacteriocin preparation. The MICs of the bacteriocins against various indicator strains were determined using the above-mentioned spot-on-lawn method with the purified bacteriocin solution. The MIC was defined as the minimum bacteriocin concentration to yield a clear zone of growth inhibition in the indicator lawn.
Purification of enterocin F4-9.
Enterocin F4-9 was purified from 1 liter of the culture supernatant of E. faecalis F4-9 in MRS medium at 30°C for 16 h based on the method of Sawa et al. (18, 19) with minor modifications. The cells were removed by centrifugation at 6,000 × g for 15 min; 25 g of activated Amberlite XAD-16 resin (Sigma-Aldrich, St. Louis, MO) was then mixed with the culture supernatant and slowly shaken overnight at 4°C. The resin matrix was washed with 50 ml of Milli-Q water and 500 ml of 20% (vol/vol) ethanol. Enterocin F4-9 was eluted with 150 ml of 70% (vol/vol) 2-propanol (Nacalai Tesque) containing 0.1% trifluoroacetic acid (TFA) (Nacalai Tesque). To remove the 2-propanol, the eluted active fraction was evaporated. Then, enterocin F4-9 was diluted with 55 ml of 50 mM sodium phosphate buffer (pH 5.6) (PB). Enterocin F4-9 was loaded on an SP Sepharose Fast Flow cation exchange chromatography column (length, 70 mm; internal diameter, 10 mm) (GE Healthcare, Uppsala, Sweden) equilibrated with 50 ml of PB; the column was then washed with PB. Enterocin F4-9 was eluted with 50 ml of 0.1 M NaCl in PB. This active fraction was applied to a 3-ml Resource RPC column (GE Healthcare) in an LC-2000 Plus high-performance liquid chromatography (HPLC) system (JASCO, Tokyo, Japan). The active fraction was eluted using the following program with a gradient of Milli-Q–acetonitrile containing 0.1% TFA at a flow rate of 1.0 ml min−1: 0 to 45 min, 0 to 60% (vol/vol) acetonitrile. Purified enterocin F4-9 was stored at −30°C. The antibacterial activity of the fractions obtained in each purification step was determined as described above using E. faecalis JCM 5803T as the indicator strain. The peptide concentration of each fraction was measured by the Pierce bicinchoninic acid (BCA) protein assay kit (TaKaRa Bio, Shiga, Japan). Purified bacteriocin was concentrated using a Speed Vac concentrator (Savant, Farmingdale, NY) and dissolved at suitable concentrations with Milli-Q water.
Mass spectrometry and amino acid sequencing.
The molecular masses of bacteriocin peptides were determined by electrospray ionization time of flight mass spectrometry (ESI-TOF MS) with the JMS-T100LC mass spectrometer (JEOL, Tokyo, Japan). The amino acid sequence analysis of enterocin F4-9 was conducted by Edman degradation with the PPSQ-31 protein sequencer (Shimadzu, Kyoto, Japan).
Identification of enterocin F4-9 structural gene and the biosynthetic gene cluster.
Genomic DNA of E. faecalis F4-9 was extracted with MagExtractor Genome (Toyobo, Osaka, Japan). To identify the structural gene (enfA49) encoding enterocin F4-9, we used the primers, In-enfA49-F (F stands for forward), In-enfA49-R (R stands for reverse), Out-enfA49-F, and Out-enfA49-R (Table 1) based on a nucleotide sequence coding a hypothetical protein from Enterococcus faecalis TX1346 (GenBank accession no. WP_002396198). These primers and genomic DNA were employed to amplify enfA49 with Ex Taq DNA polymerase (Promega, Madison, WI) according to standard methodology (20). The amplified fragments were cloned into the pGEM-T vector (Promega) and sequenced by FASMAC (Kanagawa, Japan). The fragment containing enfA49 was amplified by PCR using newly designed specific primers (F49-F and F49-R) (Table 1) based on the nucleotide sequence obtained above; this fragment was sequenced directly for confirmation.
TABLE 1.
Oligonucleotide primers used to amplify and analyze the structural gene and the biosynthetic genes of enterocin F4-9
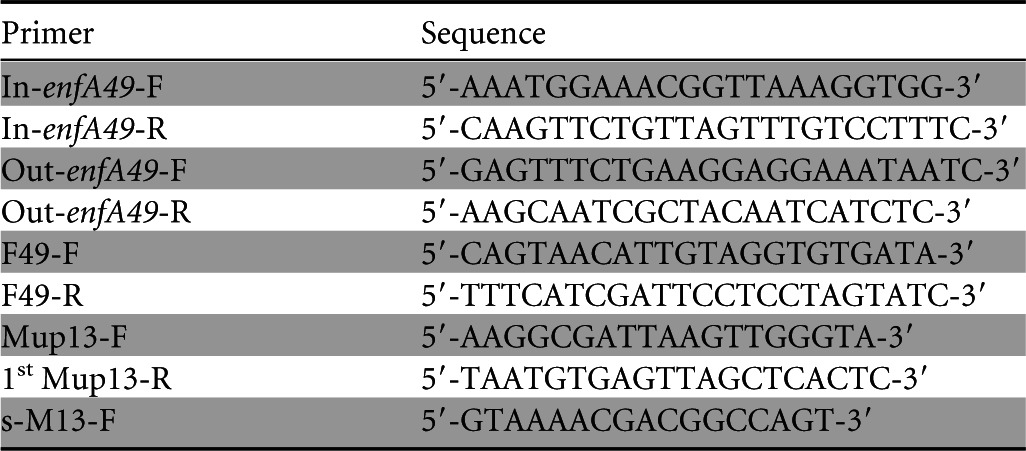
To obtain the DNA sequences flanking enfA49, various primers were designed based on the available genome sequence of E. faecalis TX1346 (GenBank accession no. AEBI01000099). The sequence of the further upstream region was also identified by anchored PCR by the method of Ishibashi et al. (21) with minor modifications. Briefly, total DNA from E. faecalis F4-9 was digested with KpnI, EcoRI, and BamHI (Nippon Gene, Tokyo, Japan); the digested DNA was ligated into a pUC18 cloning vector (Toyobo) that had been treated with the corresponding restriction enzymes and then dephosphorylated. Each of the various ligation products was used as the template for ligation-anchored and nested PCR using enfA49-specific primers and vector-specific primers (Mup13-F, s-M13-F, and 1st Mup13-R). The resulting products were purified and cloned into the pGEM-T vector, and these plasmids were sequenced.
Deglycosylation reaction.
The deglycosylation reaction was performed with β-N-acetylglucosaminidase (4,000 U ml−1; Biolabs, Ipswich, MA) according to the manufacturer's instructions. Briefly, 12 μg of enterocin F4-9 was dissolved in 12 μl of 10× G1 reaction buffer (5 mM CaCl2 and 50 mM sodium acetate), 12 μl of 10× bovine serum albumin (BSA), and 95 μl of Milli-Q water. Then, it was O-deglycosylated with 2 μl of β-N-acetylglucosaminidase (extracted from Xanthomonas manihotis). The reactant was incubated at 37°C for 40 h. The O-deglycosylated peptide was purified by reverse-phase HPLC (RP-HPLC) and analyzed using ESI-TOF MS.
Determination of the role of disulfide bonds on the antimicrobial activity of enterocin F4-9.
Enterocin F4-9 was reduced and alkylated by the method of Bhugaloo-Vial et al. (22) with slight modifications. Briefly, 12 μg of peptide was dissolved in 10 μl of 100 mM Tris-HCl (pH 8.5), and 0.2 μl of 1 M dithiothreitol (DTT) (Sigma-Aldrich) was added. The reaction was allowed to proceed overnight at room temperature in a nitrogen atmosphere. Then, 0.3 μl of 1 M iodoacetic acid (Nacalai Tesque) was added, and the mixture was placed in the dark for 1 h. The reaction was quenched by 0.2 μl of 1 M DTT. The reactant was desalted and fractionated by RP-HPLC and analyzed using ESI-TOF MS. The antimicrobial activity for the reduced and alkylated peptide was determined by the spot-on-lawn method using E. faecalis JCM 5803T as an indicator strain.
Effects of pH and heat treatment on enterocin F4-9 activity.
The purified enterocin F4-9 solution, at a concentration of 6 μM, was adjusted to pH values between 2.0 and 10.0 using the following buffers (50 mM): glycine-HCl (pH 2.0), sodium acetate (pH 4.0), sodium phosphate (pH 6.0), Tris-HCl (pH 8.0), and glycine NaOH (pH 10.0). The bacteriocin preparations at different pH values were heated at 80°C, 100°C, and 121°C for 15 min. The bacteriocin preparations were also left at room temperature (25°C) overnight. After that, they were readjusted to pH 6.0 by mixing with 0.5 M sodium phosphate buffer (pH 6.0) before the activity test. The residual activities were determined by the spot-on-lawn method using E. faecalis JCM 5803T as an indicator strain. The untreated bacteriocin preparation adjusted to pH 6.0 by the 0.5 M sodium phosphate buffer was used as a control with 100% activity.
Effects of proteolytic enzymes on enterocin F4-9 activity.
The purified enterocin F4-9 at a concentration of 10 μM was treated with the following enzymes: trypsin (Sigma-Aldrich), α-chymotrypsin (Sigma-Aldrich), and proteinase K (TaKaRa). All enzymes were dissolved in suitable buffers and filter sterilized. Each enzyme solution was mixed with purified enterocin F4-9 solution at an enzyme-to-peptide molar ratio of 1:100, 1:1, and 5:1. After incubation at 37°C for 6 h, the enzymes were inactivated by heating at 80°C for 10 min. The residual activities were determined as described above. Enterocin F4-9 without enzyme was incubated at 37°C for 6 h and then heated at 80°C for 10 min and considered a control with 100% activity.
Mode of action of enterocin F4-9.
The mode of action of enterocin F4-9 was analyzed by the method described previously according to the killing manner toward the indicator strain (14, 23). Briefly, a culture of E. faecalis JCM 5803T was incubated at 37°C until it reached an optical density at 620 nm (OD620) of 0.14, and enterocin F4-9 at final concentrations of 3 μM (1.5× MIC) and 40 μM (22× MIC) was added to the culture. The culture without enterocin F4-9 served as a control. Then, to monitor the effect of enterocin F4-9 against the indicator strain, bacterial growth was measured at OD620 for 8 h at 37°C by the Infinite F200 Pro spectrophotometer (Tecan, Männedorf, Switzerland).
Computer analysis of DNA and amino acid sequences.
The obtained DNA and amino acid sequences were analyzed using the GENETYX-WIN software version 8.0.1 (Genetyx, Tokyo, Japan). Database searches were carried out using the BLAST program of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/).
Nucleotide sequence accession number.
The DNA sequence obtained has been deposited into the DNA Data Bank of Japan (DDBJ) with accession number LC029806.
RESULTS
Purification, mass spectrometry, and amino acid sequence of enterocin F4-9.
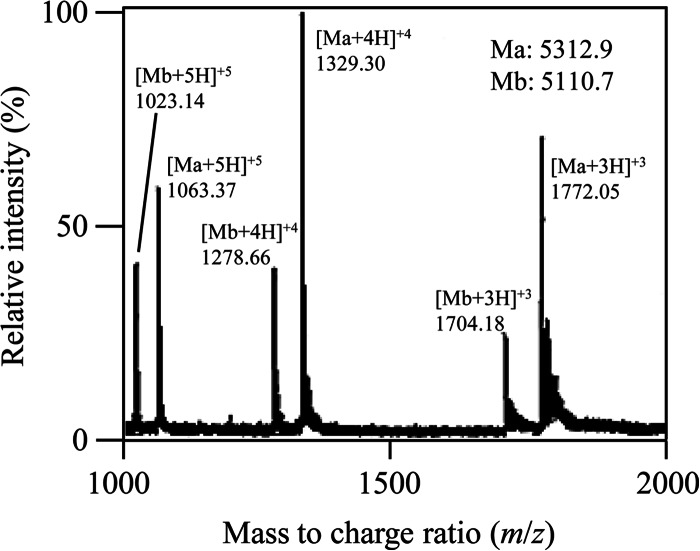
The bacteriocin produced by E. faecalis F4-9 was purified from the culture supernatant in 3 steps as shown in Table 2. The antimicrobial peptide was obtained as a single peak by reverse-phase HPLC. The molecular mass of the purified bacteriocin was determined to be 5,516.6 Da by ESI-TOF MS (Fig. 1). Because this molecular mass has not been reported for bacteriocins, this result demonstrates that enterocin F4-9 is a novel bacteriocin.
TABLE 2.
Purification of enterocin F4-9 produced by E. faecalis F4-9
| Step | Vol (ml) | Total activity (AU)a | Total amt of protein (mg)b | Sp act (AU mg−1) | Yield (%) |
|---|---|---|---|---|---|
| Supernatant | 1,000 | 2.00 × 105 | 8.34 × 103 | 23.9 | 100 |
| Amberlite XAD-16 | 100 | 1.60 × 105 | 3.20 × 102 | 5.00 × 102 | 80 |
| SP Sepharose | 50 | 8.00 × 104 | 1.67 × 10 | 4.80 × 103 | 40 |
| RP-HPLC | 15 | 2.40 × 104 | 0.779 | 3.08 × 104 | 12 |
Antimicrobial activity (in arbitrary units [AU]) was assayed by the spot-on-lawn method using E. faecalis JCM 5803T as the indicator strain.
The protein concentration (in milligrams per milliliter) was measured with the Pierce BCA protein assay kit.
FIG 1.
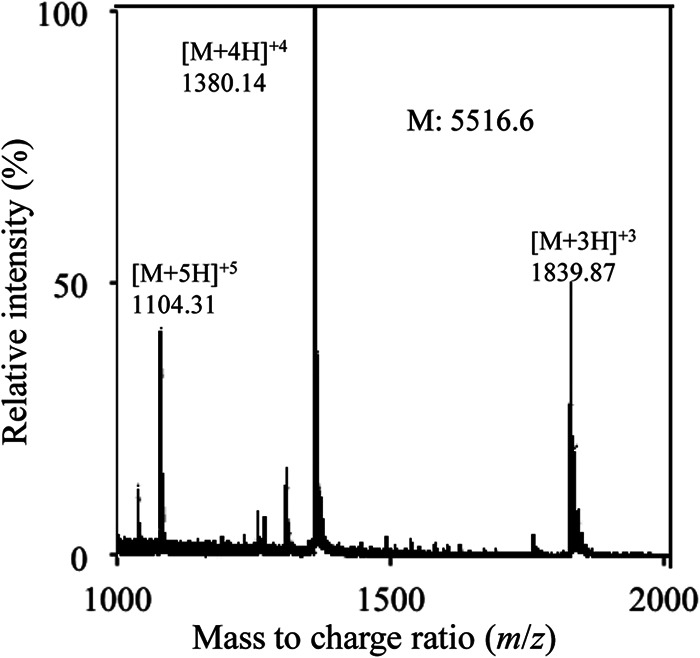
ESI-TOF mass spectrometry of purified enterocin F4-9. Multiple charged molecular ions of enterocin F4-9 were detected and are indicated. The molecular mass (M) of enterocin F4-9 was calculated based on the most abundant peak (quadrivalent charged peaks).
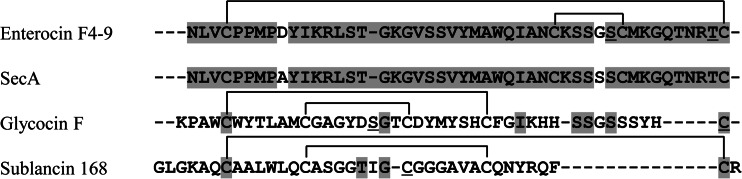
The amino acid sequence of enterocin F4-9 was identified by Edman degradation, and the sequence was NLVXPPMPDYIKRLSTGKGVSSVYMAWQIANXKSSGXXMMGQTNRXX, where X represents an unidentified residue and the underlining indicates that the residue was identified with low reliability. The sequence obtained shows high homology with the predicted product of secA, a hypothetical protein of unknown function encoded in the E. faecalis TX1346 genome (GenBank accession no. WP_002396198), as shown in Fig. 2.
FIG 2.
Amino acid sequence of enterocin F4-9 and comparison with the sequences of the hypothetical protein (SecA) of E. faecalis TX1346, glycocin F, and sublancin 168. Identical residues (gray background), glycosylation sites (underlined), and hypothetical disulfide bonds (indicated by bars above the sequence) are indicated. Gaps introduced to maximize alignment of the sequences are indicated by dashes.
Antimicrobial spectrum of enterocin F4-9.
The antimicrobial spectrum of enterocin F4-9 is shown in Table 3. Enterocin F4-9 exhibited a narrow antimicrobial spectrum against some Gram-positive food spoilage bacteria, including Enterococcus strains and Bacillus coagulans. Interestingly, enterocin F4-9 exhibited a strong inhibitory activity against E. coli JM109. In keeping with other bacteriocin-producing bacteria, E. faecalis F4-9 exhibits self-immunity against enterocin F4-9.
TABLE 3.
Antimicrobial spectrum of the purified enterocin F4-9
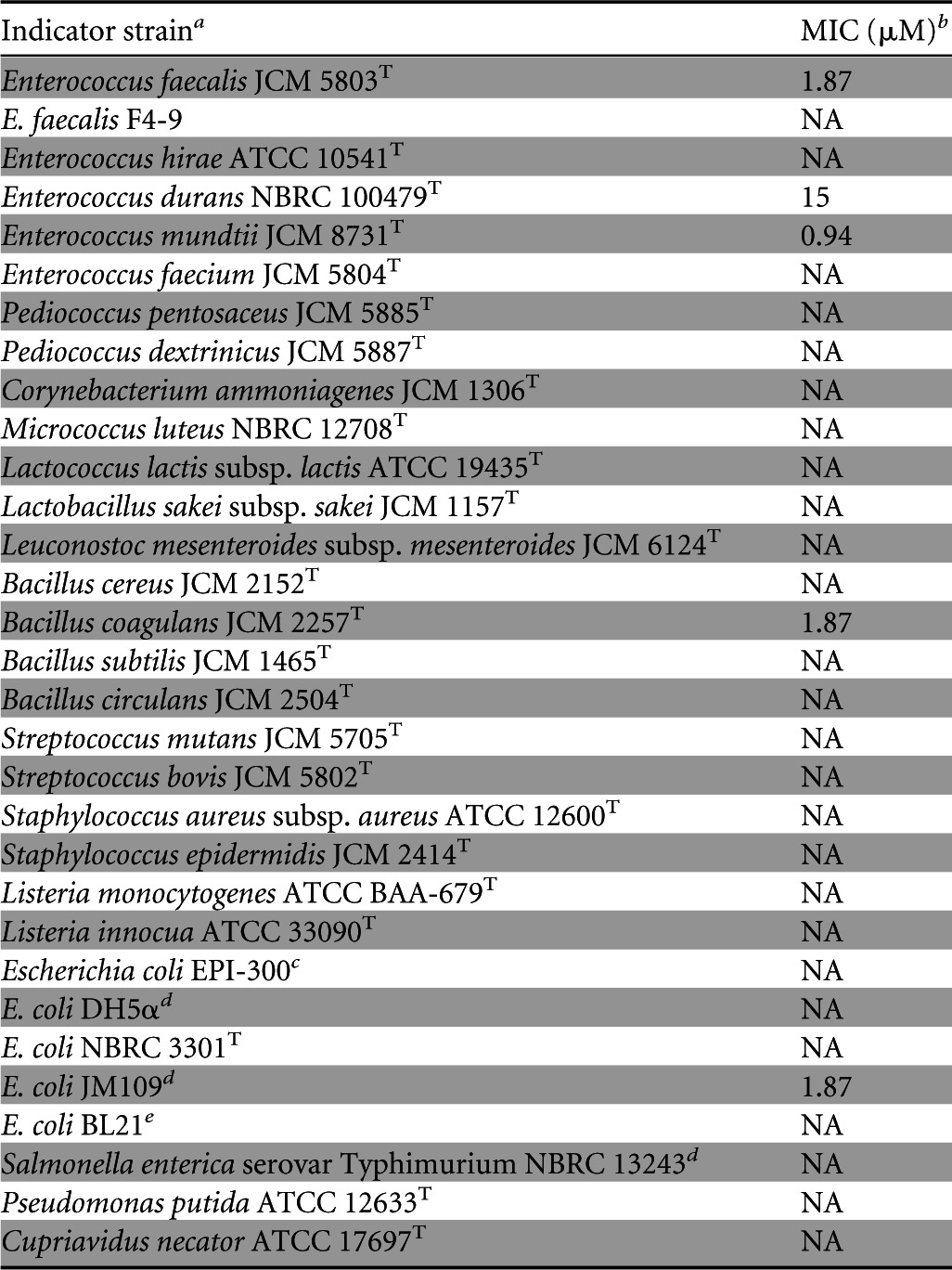
JCM, Japan Collection of Microorganisms (Wako, Japan); ATCC, American Type Culture Collection (Manassas, VA); NBRC, National Institute of Technology and Evaluation Biological Resource Center (Chiba, Japan).
NA, no activity (>30 μM).
Epicentre (Madison, WI).
Promega (Madison, WI).
GE Healthcare (Uppsala, Sweden).
Effects of pH, heat treatment, and enzymes on enterocin F4-9 activity.
Enterocin F4-9 stability was assayed under a wide range of pH and temperature conditions. The activity of enterocin F4-9 decreased with exposure to high temperature and pH (Table 4). After autoclaving at 121°C for 15 min, activity was completely lost, whereas enterocin F4-9 retained full activity at 100°C in the middle acidic condition only. Enterocin F4-9 also retained 100% activity at 80°C in a wide pH range. However, it retained full activity at room temperature overnight only in the pH range from 2 to 6 and completely lost activity at pH 8 at room temperature both overnight and even after just 15 min (data not shown). Enterocin F4-9 was completely inactivated by all proteolytic enzymes employed at different molar ratios.
TABLE 4.
Effects of pH and temperature on the activity of enterocin F4-9
| Temperature treatment | Residual activity (%) after treatmenta |
||||
|---|---|---|---|---|---|
| pH 2 | pH 4 | pH 6 | pH 8 | pH 10 | |
| 25°C, overnight | 100b | 100 | 100 | 0 | 0 |
| 80°C, 15 min | 100 | 100 | 100 | 100 | 0 |
| 100°C, 15 min | 0 | 100 | 0 | 0 | 0 |
| 121°C, 15 min | 0 | 0 | 0 | 0 | 0 |
Data represent activity after the indicated treatments (temperature and pH). Data represent the observations for three independent experiments.
The untreated bacteriocin preparation adjusted to pH 6.0 by the 0.5 M sodium phosphate buffer was used as a control with 100% activity.
Mode of action of enterocin F4-9.
The optical density of E. faecalis JCM 5803T without enterocin F4-9 rose from 0.14 to 0.23. Addition of enterocin F4-9 at a high concentration (40 μM) maintained cell density at the same level. Further, cells treated with a lower concentration (3 μM) of enterocin F4-9 also exhibited a similar inhibition pattern (Fig. 3). These results demonstrate that enterocin F4-9 inhibits multiplication of E. faecalis JCM 5803T, which is proposed to be the bacteriostatic effect.
FIG 3.
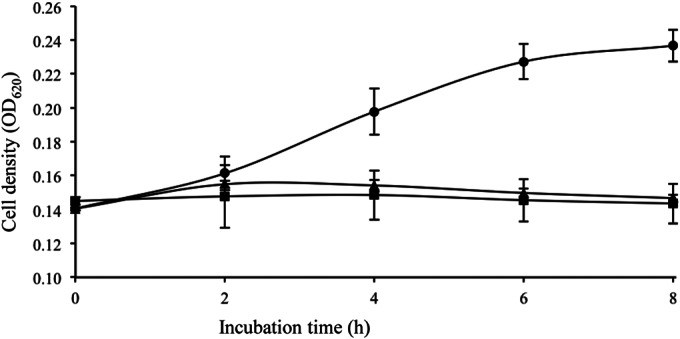
Effect of enterocin F4-9 on E. faecalis JCM 5803T growth. Enterocin F4-9 at final concentrations of 0 μM (circles), 3 μM (triangles), and 40 μM (squares) were added to the cultures of the indicator strain, E. faecalis JCM 5803T. Values are means ± standard deviations (error bars) derived from triplicate experiments.
Determination of the structural gene for elucidation of the structure of enterocin F4-9.
Subsequently, PCR was conducted using primer sets designed based on the DNA sequence of E. faecalis TX1346. The amplified fragment containing the structural gene of enterocin F4-9 was obtained and named enfA49. enfA49, which encodes the propeptide of enterocin F4-9, corresponds to 67 amino acid residues (Fig. 4). This propeptide possesses a double glycine site for cleavage of the leader sequence; the mature peptide is composed of 47 amino acid residues. The residues unidentified by Edman degradation were identified by DNA sequencing and were Cys4, Cys32, Ser37, Cys38, Thr46, and Cys47; the residue identified with low reliability was confirmed to be Lys40. Based on this sequence, the calculated molecular mass of enterocin F4-9 (5,113.0 Da) does not agree with the observed molecular mass (5,516.6 Da). This discrepancy indicates that this bacteriocin likely undergoes some modifications.
FIG 4.
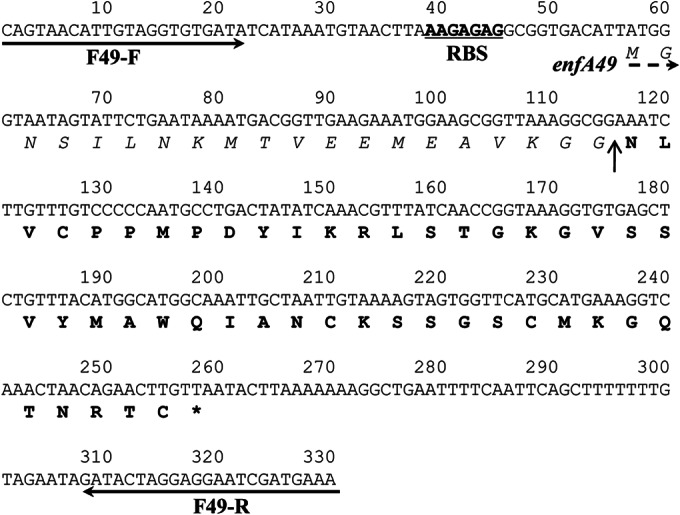
Nucleotide sequence of the region containing the enterocin F4-9 structural gene (enfA49). The starting point of the propeptide is indicated by the dashed arrow. The position of the enterocin-specific primer is indicated with a solid black arrow. The single peptide cleavage site is indicated by a vertical arrow. The putative ribosome binding site (RBS) and stop codon are underlined and indicated by an asterisk, respectively. The amino acids corresponding to the leader and mature peptides are shown in italic type and boldface type, respectively.
Determination of enterocin F4-9 biosynthetic gene cluster.
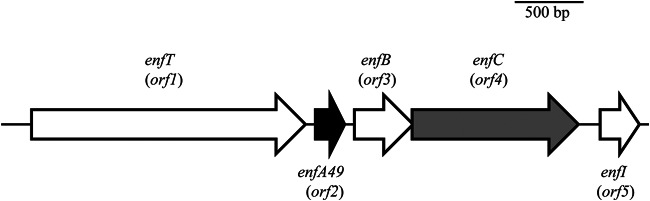
The enfA49 gene cluster (approximately 4.7 kb) was predicted to contain five protein-coding genes (Fig. 5). Database analysis of the identified open reading frames (ORFs) was used to propose their functions. The structural gene (enfA49) is located in the same locus with the other putative genes. enfT (ORF1) exhibited 82% identity to an ABC transporter and ATP-binding protein of E. faecalis (GenBank accession no. WP_002396197). enfB (ORF3) exhibited 38% identity to two different putative proteins, a thiol-disulfide isomerase of Paenibacillus peoriae (GenBank accession no. WP_010348043), which facilitates disulfide bond formation, and a putative bacteriocin transport accessory protein of E. faecalis 13-SD-W-01 (GenBank accession no. EPI01858). enfC (ORF4) exhibited 80% and 33% identities to glycosyltransferase, the group 2 family protein of E. faecalis TX1346 (GenBank accession no. EFU16565), and to SunS, the S-glycosyltransferase that transfers a sugar onto the precursor of the glycopeptide sublancin (GenBank accession no. WP_004399050), respectively. enfI (ORF5) exhibited 66% identity to the bacteriocin immunity protein for mundticin KS of Enterococcus mundtii (GenBank accession no. BAB88213).
FIG 5.
Putative biosynthetic gene cluster of enterocin F4-9. A total of 5 ORFs were identified and illustrated. The structural gene and the putative glycosyltransferase gene of enterocin F4-9 are indicated by black and gray arrows, respectively.
Structural characterization of enterocin F4-9.
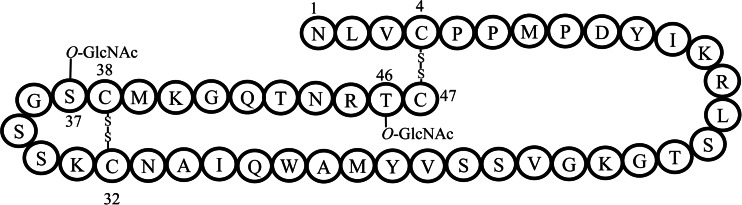
The presence of enfC suggests that enterocin F4-9 is subject to glycosylation, as in cases of sublancin 168 and glycocin F. Because of the difference between the observed and calculated molecular masses, we hypothesized that two molecules of N-acetylglucosamine (GlcNAc) were linked to enterocin F4-9. Many reports have shown that β-N-acetylglucosaminidase has strong selectivity for the cleavage of O-GlcNAc (GlcNAc attached to serine or threonine) (9, 24). Treatment of enterocin F4-9 with β-N-acetylglucosaminidase decreased the molecular mass to 5,312.9 Da, corresponding to partially deglycosylated peptides, and 5,110.7 Da, corresponding to completely deglycosylated peptides, and almost identical to the calculated mass with two disulfide bonds (Fig. 6) as illustrated in Table 5. The deOGlcNAcenterocin F4-9 (O-deglycosylated enterocin F4-9) was inactive, pinpointing that O-linked GlcNAc is essential for antimicrobial activity, as shown in Fig. 7. Stepper et al. (9) reported selectivity of β-N-acetylglucosaminidase for the O-GlcNAc substrate. On the basis of the enzyme-substrate specificity and Edman sequencing results, we confirmed that two molecules of GlcNAc are linked to Ser37 and Thr46 of enterocin F4-9, and it is therefore classified as an O-linked glycopeptide (Fig. 8).
FIG 6.
ESI-TOF mass spectrometry of the deglycosylated peptide. The masses of 1deOGlcNAcenterocin F4-9 and 2deOGlcNAcenterocin F4-9 were 5,312.9 and 5,110.7 Da, respectively.
TABLE 5.
Experimental and theoretical masses of the native and chemically modified enterocin F4-9
| Peptide | Molecular mass (Da) from ESI-TOF MS | Theoretical mass (Da) | Modification | Antimicrobial activity against E. faecalis JCM 5803Ta |
|---|---|---|---|---|
| Native | 5,516.6 | 5,113.0b | + | |
| Reduced and alkylated | 5,752.9 | 5,752.6 | Reduction of SS bonds and formation of S-CMCc | − |
| 1deOGlcNAc | 5,312.9 | 5,313.6 | One molecule of GlcNAc was released | − |
| 2deOGlcNAc | 5,110.7 | 5,110.6 | Two molecules of GlcNAc were released | − |
Symbols: +, ability to inhibit the growth of E. faecalis JCM 5803T; −, loss of the ability to inhibit the growth of E. faecalis JCM 5803T.
This shows the mass of the amino acid sequence deduced from the DNA sequence.
S-Carboxymethylated cysteine group.
FIG 7.
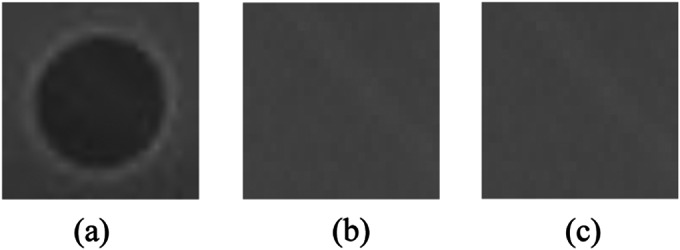
Antimicrobial activity assay of the native (a), reduced and alkylated (b), and deglycosylated (c) enterocin F4-9. The antimicrobial properties of each sample were assessed by its ability to inhibit the growth of E. faecalis JCM 5803T. As indicated, the reduced and alkylated peptide and the deglycosylated peptide did not exhibit antimicrobial activities.
FIG 8.
Proposed structure of enterocin F4-9. The residues modified with GlcNAc and the two putative disulfide bonds (Cys4-Cys47 and Cys32-Cys38) are indicated.
Further, treatment of enterocin F4-9 with DTT generated four free thiols that reacted with iodoacetic acid (Fig. 9 and Table 5). Edman degradation and chemical modification results all supported the conclusion that enterocin F4-9 contains no free thiols and has two disulfide bonds that are important for bioactivity (Fig. 7). The same role was reported for sublancin 168 and glycocin F (9). We propose that enterocin F4-9 has two nested disulfide bonds (Cys4-Cys47 and Cys32-Cys38); this was deduced from sublancin 168 and glycocin F structures, which contain inner and outer disulfide bonds.
FIG 9.
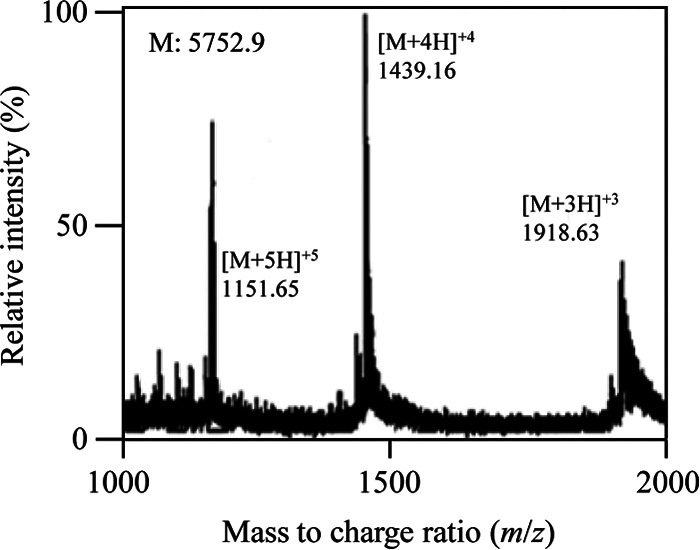
Analysis of the reduced and alkylated enterocin F4-9 by ESI-TOF mass spectrometry. Pure enterocin F4-9 was reduced and alkylated using DTT and iodoacetic acid. The four Cys residues were modified with an S-carboxymethylated cysteine (S-CMC) group. The expected molecular mass was observed.
DISCUSSION
In this paper, we have described the identification and characterization of a bacteriocin produced by E. faecalis F4-9. This novel bacteriocin-producing strain was isolated from Egyptian salted-fermented fish, a popular fish product in Egypt containing high-quality protein, vitamins, and minerals. Klein (25) previously reported that E. faecalis is frequently isolated from fish, crustaceans, and meat. Although various bacteriocin-producing LAB strains, including enterococci, have been isolated from fish, this is the first reported case, according to our knowledge, of a bacteriocin-producing LAB strain isolated from Egyptian salted-fermented fish.
Amino acid and DNA sequencing analyses revealed that enterocin F4-9 is comprised of 47 amino acid residues, with a precursor bacteriocin containing a double-glycine-type leader peptide (Fig. 4). The N-terminal leader sequence of class II bacteriocins has a role in the recognition of the precursor bacteriocin by transport and maturation components of bacteriocin-dependent secretion systems (26). The calculated mass of enterocin F4-9 was smaller than the observed mass by 403.5 Da. There have been numerous reports in which the calculated and observed masses of bacteriocins are different. In many cases, the bacteriocins were identified as lantibiotics (class I), where the observed mass was smaller than the calculated mass. Conversely, the observed mass of glycosylated bacteriocins is higher than the calculated mass, as in the case of enterocin F4-9. Wong-Madden and Landry (27) isolated and characterized β-N-acetylglucosaminidase from Xanthomonas manihotis and further reported its specificity for GlcNAc. Moreover, treatment of glycocin F by β-N-acetylglucosaminidase released O-GlcNAc from Ser18 (9). Ser37 and Thr46 residues of enterocin F4-9 could not be detected by Edman degradation, but the other candidate residues for glycosylation were detected, indicating that these residues are glycosylated. On the basis of our experimental findings and previous reports, we confirmed that enterocin F4-9 is modified by 2 molecules of GlcNAc, linked to Ser37 and Thr46 (Table 5 and Fig. 6). Enterocin F4-9 gene cluster analysis (Fig. 5), Edman degradation, and the β-N-acetylglucosaminidase assay all support the conclusion that enterocin F4-9 is a glycopeptide. This interpretation sufficiently accounts for the difference between the observed and calculated molecular masses.
Enterocin F4-9 remained active within a wide pH range, but it showed moderate resistance to heat treatment (Table 4), indicating that its stability is comparable to those of other reported glycopeptides. Glycocin F is sensitive to alkali treatment and is resistant to heat at 100°C, but its activity was destroyed completely by autoclaving (28). Sublancin 168 is stable at a broad pH range of 1.5 to 9.5 and can withstand autoclaving for 3 min at 121°C (29). Additionally, the antimicrobial activity of enterocin F4-9 was completely abolished by proteolytic enzymes. Similarly, glycocin F activity is destroyed by protease treatment (28), and sublancin 168 is sensitive to chymotrypsin (29).
Interestingly, enterocin F4-9 exhibits antimicrobial activity against E. faecalis and E. coli JM109, but not Enterococcus faecium and other E. coli (Table 3). This suggests that enterocin F4-9 utilizes a unique target molecule(s) to initiate its antimicrobial activity. The antimicrobial activity of glycocin F is also narrow and restricted to the Lactobacillus genus (28). Sublancin 168 exhibits antimicrobial activity against some Gram-positive bacteria, specifically Bacillus species, but it does not inhibit E. coli JM101 (29). It is clear that the antimicrobial spectrum of enterocin F4-9 differs from those of glycocin F and sublancin 168. This is not surprising, based on low sequence similarity between the three peptides, as shown in Fig. 2. Enterocin F4-9 inhibited proliferation of E. faecalis JCM 5803T without inducing cell lysis, implying a bacteriostatic effect (Fig. 3). A similar mode of action was also reported for glycocin F (28). Enterocin F4-9 is cationic (net charge, +4.63), and this is vital for interaction between the bacteriocin and negatively charged phospholipids in target bacterial cells (30).
O-Glycosylation is the most prevalent and diverse PTM in eukaryotes, but it is less widespread in bacteria. Serine and threonine are the preferred residues for O-glycosylation. Voisin et al. reported threonine-linked α-arabinofuranoside oligomers in type IV pilins of Pseudomonas aeruginosa (31). Glycosylated threonine, however, appears to be extremely rare in bacterial glycopeptides. To the best of our knowledge, enterocin F4-9 is the first reported bacteriocin with a glycosylated threonine residue. The loss of bioactivity of deOGlcNAcenterocin F4-9 and deOGlcNAcglycocin F strongly supports the hypothesis proposed by Stepper et al. (9) that O-GlcNAc reacts with a target molecule(s) in susceptible bacteria.
A lack of experimental evidence for class IV bacteriocins has led to withdrawal of this class from some classification schemes, such that proposed by Cotter et al. (7). However, the discovery of glycocin F, sublancin 168, and our novel glycopeptide enterocin F-49 demonstrate the existence of this class of bacteriocins.
In summary, enterocin F4-9 is the first glycosylated enterocin and the third glycosylated bacteriocin after glycocin F and sublancin 168. Because enterocin F4-9 has some unique properties, further studies on its mode of action could prove valuable for the application of enterocin F4-9 in the control of undesirable bacteria.
ACKNOWLEDGMENTS
This work was supported in part by the Egyptian Channel System Program (CSP) to Mohamed Abdelfattah Maky and JSPS KAKENHI grant 24380051. We are grateful to T. Nakashima of Kyushu University for kindly giving us access to the protein sequencer.
REFERENCES
- 1.Gálvez A, Abriouel H, López RL, Ben Omar N. 2007. Bacteriocin-based strategies for food biopreservation. Int J Food Microbiol 120:51–70. doi: 10.1016/j.ijfoodmicro.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Castro M, Palavecino N, Herman C, Garro O, Campos C. 2011. Lactic acid bacteria isolated from artisanal dry sausages: characterization of antibacterial compounds and study of the factors affecting bacteriocin production. Meat Sci 87:321–329. doi: 10.1016/j.meatsci.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Ray B. 2004. Fundamental food microbiology, 3rd ed CRC Press, Washington, DC. [Google Scholar]
- 4.Zendo T. 2013. Screening and characterization of novel bacteriocins from lactic acid bacteria. Biosci Biotechnol Biochem 77:893–899. doi: 10.1271/bbb.130014. [DOI] [PubMed] [Google Scholar]
- 5.Perez R, Zendo T, Sonomoto K. 2014. Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb Cell Fact 13:S3. doi: 10.1186/1475-2859-13-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jack R, Wan J, Gordon J, Harmark K, Davidson B, Hillier A, Wettenhall R, Hickey M, Conventry M. 1996. Characterization of the chemical and antimicrobial properties of pisccolin 126, a bacteriocin produced by Carnobacterium piciola JG 126. Appl Environ Microbiol 62:2897–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter P, Hill C, Ross R. 2005. Bacteriocins: developing innate immunity for foods. Nat Rev Microbiol 3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 8.Klaenhammer TR. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev 12:39–85. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 9.Stepper J, Shastri S, Loo T, Preston J, Novark P, Man P, Moore C, Havlicek V, Patchett M, Norris G. 2011. Cysteine S-glycosylation, a new post-translational modification found in glycopeptide bacteriocins. FEBS Lett 584:645–650. doi: 10.1016/j.febslet.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Oman T, Boettcher J, Wang H, Okalibe X, Van der Donk W. 2011. Sublancin is not a lantibiotic but an S-linked glycopeptide. Nat Chem Biol 7:78–80. doi: 10.1038/nchembio.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aymerich T, Holo H, Havarstein L, Hugas M, Garriga M, Nes I. 1996. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl Environ Microbiol 62:1676–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franz C, Grube A, Herrmann A, Abriouel H, Stärke J, Lombardi A, Tauscher B, Holzapfel W. 2002. Biochemical and genetic characterization of the two-peptide bacteriocin enterocin 1071 produced by Enterococcus faecalis FAIR-E 309. Appl Environ Microbiol 68:2550–2554. doi: 10.1128/AEM.68.5.2550-2554.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gálvez A, Giménez-Gallego G, Maqueda M, Valdivia E. 1989. Purification and amino acid composition of peptide antibiotic AS-48 produced by Streptococcus (Enterococcus) faecalis subsp. liquefaciens S-48. Antimicrob Agents Chemother 33:437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casaus P, Nilsen T, Cintas L, Nes I, Hernandez P, Holo H. 1997. Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology 143:2287–2294. doi: 10.1099/00221287-143-7-2287. [DOI] [PubMed] [Google Scholar]
- 15.Venugopal H, Edwards P, Schwalbe M, Claridge J, Libich D, Stepper J, Loo T, Patchett M, Norris G, Pascal S. 2011. Structural, dynamic, and chemical characterization of a novel S-glycosylated bacteriocin. Biochemistry 50:2748–2755. doi: 10.1021/bi200217u. [DOI] [PubMed] [Google Scholar]
- 16.Spiro R. 2002. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12:43R–56R. doi: 10.1093/glycob/12.4.43R. [DOI] [PubMed] [Google Scholar]
- 17.Zendo T, Eungruttanagorn N, Fujioka S, Tashiro Y, Nomura K, Sera Y, Kobayashi G, Nakayama J, Ishizaki A, Sonomoto K. 2005. Identification and production of a bacteriocin from Enterococcus mundtii QU 2 isolated from soybean. J Appl Microbiol 99:1181–1190. doi: 10.1111/j.1365-2672.2005.02704.x. [DOI] [PubMed] [Google Scholar]
- 18.Sawa N, Wilaipun P, Kinoshita S, Zendo T, Leelawatcharamas V, Nakayama J, Sonomoto K. 2012. Isolation and characterization of enterocin W, a novel two-peptide lantibiotic produced by Enterococcus faecalis NKR-4-1. Appl Environ Microbiol 78:900–903. doi: 10.1128/AEM.06497-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawa N, Koga S, Okamura K, Ishibashi N, Sonomoto K. 2013. Identification and characterization of novel multiple bacteriocins produced by Lactobacillus sakei D98. J Appl Microbiol 115:61–69. doi: 10.1111/jam.12226. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 21.Ishibashi N, Himeno K, Fujita K, Masuda Y, Perez R, Zendo T, Wilaipun P, Leeawatcharamas V, Nakayama J, Sonomoto K. 2012. Purification and characterization of multiple bacteriocins and an inducing peptide produced by Enterococcus faecium NKR-5-3 from Thai fermented fish. Biosci Biotechnol Biochem 76:947–953. doi: 10.1271/bbb.110972. [DOI] [PubMed] [Google Scholar]
- 22.Bhugaloo-Vial P, Douliez J-P, Mollé D, Dousset X, Boyaval P, Marion D. 1999. Delineation of key amino acid side chains and peptide domains for antimicrobial properties of divercin V41, a pediocin-like bacteriocin secreted by Carnobacterium divergens V41. Appl Environ Microbiol 65:2895–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu C-B, Zendo T, Nakayama J, Sonomoto K. 2008. Description of enterocin TW-49M, a novel enterocin B-homologous bacteriocin in carrot isolated Enterococcus durans QU 49. J Appl Microbiol 105:681–690. doi: 10.1111/j.1365-2672.2008.03798.x. [DOI] [PubMed] [Google Scholar]
- 24.Dong D, Hart G. 1994. Purification and characterization of an O-GlcNAc selective N-acetyl-β-d-glucosaminidase from rat spleen cytosol. J Biol Chem 269:19321–19330. [PubMed] [Google Scholar]
- 25.Klein G. 2003. Taxonomy, ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. Int J Food Microbiol 88:123–131. doi: 10.1016/S0168-1605(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 26.Izquierdo E, Wagner C, Marchioni E, Werner D, Ennahar S. 2009. Enterocin 96, a novel class II bacteriocin produced by Enterococcus faecalis WHE96, isolated from Munster cheese. Appl Environ Microbiol 75:4273–4276. doi: 10.1128/AEM.02772-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong-Madden S, Landry L. 1995. Purification and characterization of novel glycosidases from the bacterial genus Xanthomonas. Glycobiology 5:19–28. doi: 10.1093/glycob/5.1.19. [DOI] [PubMed] [Google Scholar]
- 28.Kelly W, Asmundson R, Huang C. 1996. Characterization of plantaricin KW30, a bacteriocin produced by Lactobacillus plantarum. J Appl Bacteriol 81:657–662. doi: 10.1111/j.1365-2672.1996.tb03561.x. [DOI] [Google Scholar]
- 29.Paik S, Chakicherla A, Hansen J. 1998. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J Biol Chem 273:23134–23142. doi: 10.1074/jbc.273.36.23134. [DOI] [PubMed] [Google Scholar]
- 30.Fujita K, Ichimasa S, Zendo T, Koga S, Yoneyama F, Nakayama J, Sonomoto K. 2007. Structural analysis and characterization of lacticin Q, a novel bacteriocin belonging to a new family of unmodified bacteriocins of Gram-positive bacteria. Appl Environ Microbiol 73:2871–2877. doi: 10.1128/AEM.02286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voisin S, Kus J, Houliston S, St-Michael F, Watson D, Cvitkovitch D, Kelly J, Brisson J, Burrows L. 2007. Glycosylation of Pseudomonas aeruginosa strain Pa5196 type IV pilins with mycobacterium-like α-1,5-linked d-Araf oligosaccharides. J Bacteriol 189:151–159. doi: 10.1128/JB.01224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]