Abstract
Birds are the primary hosts of Chlamydia psittaci, a bacterium that can cause avian chlamydiosis in birds and psittacosis in humans. Wild seabirds are frequently admitted to wildlife rescue centers (WRC) at European Atlantic coasts, for example, in connection with oil spills. To investigate the extent of chlamydial shedding by these birds and the resulting risk for animals in care and the medical staff, seabirds from a French WRC were sampled from May 2011 to January 2014. By use of a quantitative PCR (qPCR), 195 seabirds belonging to 4 orders, 5 families and 13 species were examined, of which 18.5% proved to be Chlamydiaceae positive. The highest prevalence of shedders was found in northern gannets (Morus bassanus) (41%), followed by European herring gulls (Larus argentatus) (14%) and common murres (Uria aalge) (7%). Molecular characterization and phylogenetic analysis of qPCR-positive northern gannet samples revealed two variants of a strain closely related to C. psittaci. In European herring gulls and in one common murre, strains showing high sequence similarity to the atypical Chlamydiaceae-like C122 previously found in gulls were detected. Our study shows that seabirds from the northeastern Atlantic Ocean carry several chlamydial organisms, including C. psittaci-related strains. The staff in WRCs should take protective measures, particularly in the case of mass admissions of seabirds.
INTRODUCTION
The family Chlamydiaceae comprises a group of obligate intracellular Gram-negative bacteria that are widely distributed throughout the world, causing a wide range of diseases in humans and animals (1). It is composed of a single genus, Chlamydia, that thus far comprises 11 species: C. abortus, C. avium, C. caviae, C. felis, C. gallinacea, C. muridarum, C. pecorum, C. pneumoniae, C. psittaci, C. suis, and C. trachomatis (2, 3). C. psittaci is the primary avian pathogen that can infect a large number of bird species, as well as mammalian hosts (4, 5). Avian strains of C. psittaci are currently divided into at least 15 outer membrane protein A gene (ompA)-based genotypes (5, 6), each one tending to be associated with certain bird species.
Avian chlamydiosis has been known for centuries and is a major factor of economic loss to the poultry industry, as well as a permanent risk for zoonotic transmission to humans (7). Depending on the infecting chlamydial strain and the susceptibility of the avian hosts, avian chlamydiosis is mainly characterized by respiratory, ocular, enteric, or nervous disorders that may occasionally be fatal. In addition, latent infection has a potential of causing recurrent clinical disease resulting in chronicity (7, 8). Infected birds intermittently excrete chlamydial agents through feces and nasal secretions, thus representing an important reservoir of infection for birds and humans (9). Generally, transmission occurs through inhalation of aerosolized respiratory secretions or ingestion of contaminated dried dusts. Factors affecting transmission include the susceptibility of the avian host, dose of infection, virulence of the strain, stress, and environmental conditions (7, 8, 10).
The disease caused by C. psittaci in humans is called psittacosis and varies from inapparent or mild influenza-like symptoms to severe and even potentially fatal systemic disease with severe pneumonia (10, 11). Psittacosis is of concern for public health authorities and specific control measures have been recommended (12, 13).
Until recently, C. psittaci had been considered to be the sole causative agent of chlamydiosis in birds, but there is new evidence suggesting that avian chlamydiosis involves more chlamydial agents. In particular, two more avian chlamydial species, C. avium and C. gallinacea (14), and one Candidatus taxon, “Candidatus Chlamydia ibidis” (15), were recently described. The diversity of chlamydial agents in birds was further extended by studies reporting mammalian chlamydiae, such as C. abortus, C. pecorum, and C. trachomatis, also being capable of infecting avian hosts (16, 17).
In wild birds, C. psittaci is highly prevalent among Psittaciformes and Columbiformes (7), but representatives of other orders are also recognized as natural carriers (18), for instance, Anseriformes, Charadriiformes, Falconiformes, Passeriformes, Procellariformes, and Strigiformes (4, 18–25). Moreover, other Chlamydiaceae or nonclassified chlamydiae have been detected in Charadriiformes and Pelecaniformes (15, 22, 26). The reported prevalence of Chlamydiaceae in wild birds ranged from 1 to 74%, depending on avian hosts, the molecular tools used, and the sampling size (20, 27, 28). As in domestic birds, stress due to weather changes, nesting, migration, or food shortages may precipitate the disease in wild birds (10), even though the infection often remains inapparent. Outbreaks of disease with relatively high morbidity and mortality have been described (8, 18, 24, 29, 30). Although domestic birds are the most common source of infection in humans, wild birds have also been reported to be a source of C. psittaci infection in the wild (31–33) or in wildlife rescue centers (WRCs) (25).
Nowadays, injured or diseased wild birds can be submitted to WRCs by the public or by people from environmental protection associations for welfare reasons. Although raptors (i.e., Falconiformes and Strigiformes) and birds from the orders Columbiformes or Passeriformes are the main bird species admitted in WRCs in Europe (25, 34–37), seabirds (i.e., bird species with a life history linked to the marine environment) from the Anseriformes, Charadriiformes, Procellariformes, and Suliformes can also be taken into care, for instance, in connection with oil spills. In such disaster situations, seabirds can be admitted by hundreds or thousands (38–40), leading to overcrowding, close contact with the veterinary and/or nursing staff and an elevated risk of stress-induced chlamydial shedding. Until now, only a few studies about the shedding of Chlamydiaceae by seabirds admitted to WRCs have been published (25, 34, 35). Consequently, studying the epidemiology of chlamydial infections in seabirds represents a major challenge both in terms of biodiversity conservation and public health issues.
The purpose of the present study was to investigate the occurrence and the diversity of shedding of Chlamydiaceae by seabirds commonly admitted to WRCs at Europe's Atlantic coast, since this area is particularly at risk for oil spills (40, 41).
MATERIALS AND METHODS
Seabird population and sampling.
Birds included in the present study were individuals belonging to European seabird species, admitted to the Wildlife Health Centre (Centre Vétérinaire de la Faune Sauvage et des Ecosystèmes des Pays de la Loire [CVFSE]) of the Nantes Atlantic College of Veterinary Medicine, Food Science and Engineering (Oniris) from May 2011 to January 2014 inclusive. A total of 195 seabirds belonging to 4 orders, 5 families, and 13 species were sampled during this period (Table 1). Three species represented >80% of the birds: European herring gulls (Larus argentatus; 41.5%), northern gannets (Morus bassanus; 23.5%), and common murres (Uria aalge; 15.5%) in rank order. All seabirds sampled were found washed up on beaches or suspected sick or injured in harbors from the French North coast of the Bay of Biscay (Bretagne and Pays de Loire regions). They were collected as live casualties by the public or members of environmental protection associations and admitted to the CVFSE for appropriate medical care. Information regarding the location, date, species, and any relevant details about the incident history were collected at the time of admission by the CVFSE staff. The body weight and age of each seabird were also recorded at this stage. Each bird was classified either as a fledgling (bird with flight feathers not yet fully emerged), immature (bird with an adult body size but immature plumage), or adult (independent bird with mature plumage). After admission, a clinical examination was performed by a veterinarian, and any observed clinical signs were recorded. The causes for casualty admissions were determined on the basis of clinical signs and the incident history. Dry cloacal swabs were sampled from the birds during the initial clinical examination or in not more than the 48 h following admission. They were stored at −80°C during 1 week to up to 11 months before being transported under cooling conditions to the avian chlamydiosis National Reference Laboratory (NRL) for analysis.
TABLE 1.
Prevalence of Chlamydiaceae shedders among seabird species admitted to the CVFSE from May 2011 to January 2014
| Order | Family | Common species name (scientific name) | 23S-qPCR Chlamydiaceae |
||
|---|---|---|---|---|---|
| No. positivea | Mean Cq | Cq range | |||
| Anseriformes | Anatidae | Common scoter (Melanitta nigra) | 2/5 | 31.1 | 30.4–31.8 |
| Charadriiformes | Alcidae | Common murre (Uria aalge) | 2/30 | 35.1 | 32.1–38.1 |
| Alcidae | Razorbill (Alca torda) | 0/3 | |||
| Laridae | Black-headed gull (Chroicocephalus ridibundus) | 0/10 | |||
| Laridae | Common gull (Larus canus) | 0/1 | |||
| Laridae | European herring gull (Larus argentatus) | 11/81 | 35.6 | 28.3–39.0 | |
| Laridae | Great black-backed gull (Larus marinus) | 0/5 | |||
| Laridae | Lesser black-backed gull (Larus fuscus) | 1/7 | 34.7 | ||
| Laridae | Little gull (Hydrocoloeus minutus) | 0/2 | |||
| Laridae | Mediterranean gull (Ichthyaetus melanocephalus) | 0/1 | |||
| Laridae | Yellow-legged gull (Larus michahellis) | 1/2 | 37.7 | ||
| Procellariformes | Procellaridae | Northern fulmar (Fulmarus glacialis) | 0/2 | ||
| Suliformes | Sulidae | Northern gannet (Morus bassanus) | 19/46 | 27.5 | 19.6–37.3 |
| Total | 36/195 | 31.0 | |||
For the number of positive results, the values are expressed as follows: the number of positive birds/the total number of birds examined.
Molecular analysis of samples: detection of Chlamydiaceae-specific DNA in cloacal swab samples.
A first screening was performed with whole genomic DNAs extracted (QIAamp DNA minikit; Qiagen, France) from the cloacal swab samples and by using a 23S rRNA-based Chlamydiaceae-specific quantitative PCR targeting a 23S rRNA gene fragment conserved in all Chlamydiaceae (23S-qPCR Chlamydiaceae), according to the method of Ehricht et al. (42). An internal control for potential PCR inhibition (TaqMan exogenous internal positive control; Applied Biosystems) and a positive control (C. psittaci strain Loth) were systematically included. All samples with a quantitative cycle (Cq) above 39 were considered negative.
Chlamydial species molecular characterization.
(i) Species-specific qPCR assays.
DNA samples positive in the 23S-qPCR Chlamydiaceae were further analyzed to determine the chlamydial species by using two C. psittaci-specific qPCR systems (referred to here as incA-qPCR C. psittaci and ompA-qPCR C. psittaci, respectively) (16, 43), as well as using specific qPCRs for the detection of C. abortus and C. pecorum (16), C. avium (44), C. gallinacea (K. Laroucau et al., unpublished data), and the taxon “Candidatus Chlamydia ibidis” (NRL, unpublished data). C. psittaci strain Loth, C. abortus strain AB7, C. pecorum strain iB1, C. gallinacea strain 08-1274/3, C. avium strain 10-743 SC13, and “Candidatus Chlamydia ibidis” strain 10-1398/6 were used as controls.
(ii) MLST analysis on qPCR C. psittaci-positive samples.
Multilocus sequence typing analysis (MLST) was carried out on incA-qPCR C. psittaci-positive samples according to the scheme developed by Pannekoek et al. (45, 46), targeting seven housekeeping genes, namely, gatA, oppA, hflX, gidA, enoA, hemN, and fumC. Target genes were amplified and sequenced using primers described on the Chlamydiales MLST website (http://pubmlst.org/chlamydiales/), except for the fumC locus, for which new forward fumC-CpsiF1 (5′-TTCCTGGGCTCCTGAGGTTA-3′) and reverse fumC-CpsiR1 (5′-CTCTCCGGTTTCTTGACGCA-3′) primers had to be designed. PCR-amplified segments were sequenced on both DNA strands by Eurofins Genomics (Ebersberg, Germany). Sequences for each locus were queried against the online Chlamydiales MLST databases to determine allelic designations and a subsequent allelic profile was used to determine the sequence type (ST). The new allele sequences are accessible via the Chlamydiales MLST website. Multiple alignments of the seven concatenated housekeeping gene fragments were conducted using the Bionumerics software package (version 4.6; Applied-Maths, Belgium). A dendrogram was constructed using the UPGMA (unweighted pair-group method with arithmetic averages) method.
(iii) Analysis of chlamydial 16S rRNA, 23S rRNA, and ompA genes.
For further characterization chlamydial 16S rRNA, 23S rRNA, and/or ompA gene fragments were amplified by using previously described representative primer pairs (47–50) (Table 2). Both forward and reverse strands of each PCR amplicon were sequenced with the respective PCR primers. Nucleotide sequences were subjected to BLAST analysis against the NCBI database to identify related sequences and aligned. Phylogenetic reconstruction was conducted on Mega6 by using the maximum-likelihood method based on the Jukes-Cantor model and applying neighbor-joining and BioNJ algorithms. Reliability of the generated phylogenetic tree was evaluated by 500 replications of bootstrap resampling (51).
TABLE 2.
Primers used for sequencing
| Targeted sequence | Primer | Sequence (5′-3′) | PCR product size (bp) | Reference |
|---|---|---|---|---|
| 16S rRNA | 16S1 | CGGATCCTGAGAATTTGATC | 1,400 | 47 |
| rp2 | CTACCTTGTTACGACTTCAT | 1,400 | 48 | |
| 23S rRNA | 16SF2 | CCGCCCGTCACATCATGG | 1,000 | 49 |
| 23SIGR | TGGCTCATCATGCAAAAGGCA | 1,000 | 49 | |
| ompA | CTU | ATGAAAAAACTCTTGAAATCGG | 1,000 | 50 |
| CTL | CAAGATTTTCTAGA(T/C)TTCAT(C/T)TTGTT | 1,000 | 50 |
Data analysis.
The cloacal shedding prevalence of Chlamydiaceae was first determined in each of the seabird species admitted to the CVFSE during the study period. Moreover, all data from the chlamydial detection and genotyping and questionnaire were analyzed using statistical methods. The correlation and differences among variables of interest (the prevalence of cloacal shedding of Chlamydiaceae, the species and genotype of the Chlamydiaceae, the shedding level [all dependent variables]) and independent variables expressing the potential risk factors (seabird species, season, age, and admission causes) were tested using a chi-square test or a Student t test. Specifically, for each Chlamydiaceae-positive seabird species, the proportion of positive birds and the related Chlamydiaceae detected were determined for four seasons. Seasons were defined according to the biology features of the species studied: spring and fall in February/March and in September/October, respectively, corresponding to the migratory movements; April to August for the summer breeding season; and November to January for the wintering period (52).
Ethical standards.
The live animals in this study were admitted as sick or injured wild bird casualties to the Pays de la Loire Regional Wildlife and Ecosystem Veterinary Centre for appropriate clinical care. Samples were collected according to French legislation.
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited at the European Nucleotide Archive (ENA) under accession numbers LN810440 to LN810449, LN810454 to LN810464, and LN810468 to LN810483 for 16S rRNA, 23S rRNA, and ompA gene sequences, respectively.
RESULTS
Prevalence of Chlamydiaceae shedders among different seabird species.
Using the 23S-qPCR, cloacal shedding of Chlamydiaceae was detected in 18.5% of the birds, belonging to 6 species and 4 families (Table 1). In species covered with sufficient sample numbers (i.e., n ≥ 30 birds), the prevalence of Chlamydiaceae shedders was significantly higher among northern gannets (41%; chi-square test, P < 0.01) than among European herring gulls (14%) and common murres (7%). Although the number of samples from common scoters was low (n = 5), the proportion of shedders was high (40%). Furthermore, the mean Cq values observed in northern gannets and common scoters were 27.5 and 31.1, respectively, thus indicating a sizeable level of chlamydial excretion. This contrasted with lower mean values from European herring gulls and common murres (with a mean Cq of around 35), but variations among individual birds were considerable (Table 1).
While the prevalence was not significantly different from one season to another in European herring gulls, the prevalence in northern gannets was significantly higher in summer (62%) than in autumn and winter taken together (25%; chi-square test, P < 0.02) (Table 3). Interestingly, in northern gannets, the summer period with a high Chlamydiaceae excretion level (mean Cq of 26.1) contrasted with the moderate (mean Cq of 29.6) and low (mean Cq of 36.4) levels during autumn and winter, respectively (Table 3). No age-related significant difference of proportion (chi-square test) or of excretion level (mean Cq; Student t test) of positive birds were noticed in any of the sampled seabird species (Table 3). No specific cause of admission was linked to a Chlamydiaceae-positive seabird species neither to the higher seasonal Chlamydiaceae shedding observed in northern gannets in the summer period (Table 3). None of the seabirds sampled during this survey showed signs of clinical avian chlamydiosis.
TABLE 3.
Epidemiological characteristics and results for chlamydia detection by using qPCR in seabird species shedders admitted to the CVFSE from May 2011 to January 2014
| Common species name (n)a | Epidemiological datab |
23S-qPCR resultc | |||
|---|---|---|---|---|---|
| Season of admission | No. positive | Age | Admission cause(s) | ||
| Common scoter (2) | Summer | 1/1 | 1 ad | Moult default | 1 pos/neg (30.4) |
| Fall | 1/1 | 1 ad | Unknown cause | 1 pos/neg (31.8) | |
| Common murre (2) | Summer | 2/9 | 1 ad | Moult default | 2 pos/neg (35.1) |
| European herring gull (11) | Spring | 1/4 | 1 ad | Trauma | 1 pos/neg (33.7) |
| Summer | 7/59 | 6 ad, 1 im | Trauma, unknown cause | 6 pos/neg (34.0) + 1 pos/pos (38.1) | |
| Winter | 3/10 | 3 im | Trauma, unknown cause | 3 pos/pos (38.4) | |
| Lesser black-backed gull (1) | Fall | 1/1 | 1 ad | Botulism suspected | 1 pos/neg (34.7) |
| Yellow-legged gull (1) | Fall | 1/2 | 1 ad | Trauma | 1 pos/neg (37.7) |
| Northern gannet (19) | Summer | 13/21 | 8 ad, 5 im | Oiled, trauma, unknown cause | 13 pos/pos (26.1) |
| Fall | 5/14 | 1 ad, 4 im | Trauma, unknown cause | 5 pos/pos (29.6) | |
| Winter | 1/10 | 1 ad | Unknown cause | 1 pos/pos (36.4) | |
n, number of seabird shedders.
For the number of positive results, the values are expressed as follows: the number positive/the total number of seasonal admissions. Age is indicated as either “ad” for adult or “im” for immature.
The 23S-qPCR results are expressed as follows: 23S-qPCR Chlamydiaceae/incA-qPCR C. psittaci, with the qPCR Chlamydiaceae mean Cq value indicated in parentheses. pos, positive; neg, negative.
Molecular characterization of Chlamydiaceae-positive samples.
Using the incA-based C. psittaci-specific qPCR, all of the Chlamydiaceae-positive northern gannet samples were positive, as well as 4 of 11 European herring gull samples (Table 3). In contrast, Chlamydiaceae-positive DNA samples from common scoters, common murres, and the two other gull species were all negative. All of these samples were also tested negative for C. abortus, C. pecorum, C. avium, C. gallinacea, and the taxon “Candidatus Chlamydia ibidis.”
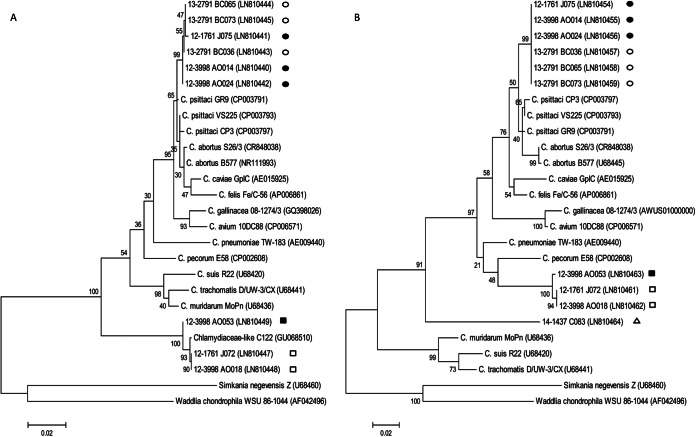
Due to low DNA content, no genotype could be assigned to the four incA-qPCR C. psittaci-positive samples detected in European herring gulls. The seven Chlamydiaceae-positive but incA-qPCR C. psittaci-negative European herring gull samples were also negative using the C. psittaci ompA-qPCR. In the case of two samples with high DNA content, i.e., 12-1761_J072 and 12-3998_AO018, partial sequencing of 16S and 23S rRNA genes was achieved. The resulting phylogenetic trees, which included sequences from representative species of Chlamydiales, showed those samples to form a distinct line of descent separated from those of C. psittaci and all other hitherto-established Chlamydiaceae species, but clearly grouped together in a cluster within Chlamydiaceae and included the “Chlamydiaceae-like” C122 from the seabird species Larus glaucescens (26), as well as sample 12-3998_AO053 recovered from a common murre in the present study (Fig. 1).
FIG 1.
Phylogenetic reconstruction based on 16S rRNA (A) and 23S rRNA (B) genes. Sequences obtained from northern gannet, European herring gull, common murre, and/or common scoter samples and representative Chlamydiaceae species, as well as two outgroup species comprising W. chondrophila and S. genevensis were used. Symbols: ●, northern gannet (group 1); ○, northern gannet (group 2); ■, common murre; □, European herring gull; △, common scoter.
Low DNA content was also the reason for our failure to sequence Chlamydiaceae-positive but incA-qPCR C. psittaci-negative samples obtained from the two other gull species (lesser black-backed gull and yellow-legged gull). Only from one common scoter sample, namely, 14-1437_C083, was the 23S rRNA sequence successfully obtained. Phylogenetic analysis revealed sample 14-1437_C083 to form another distinct line of descent separated from all other hitherto-established Chlamydia species within Chlamydiaceae (Fig. 1B).
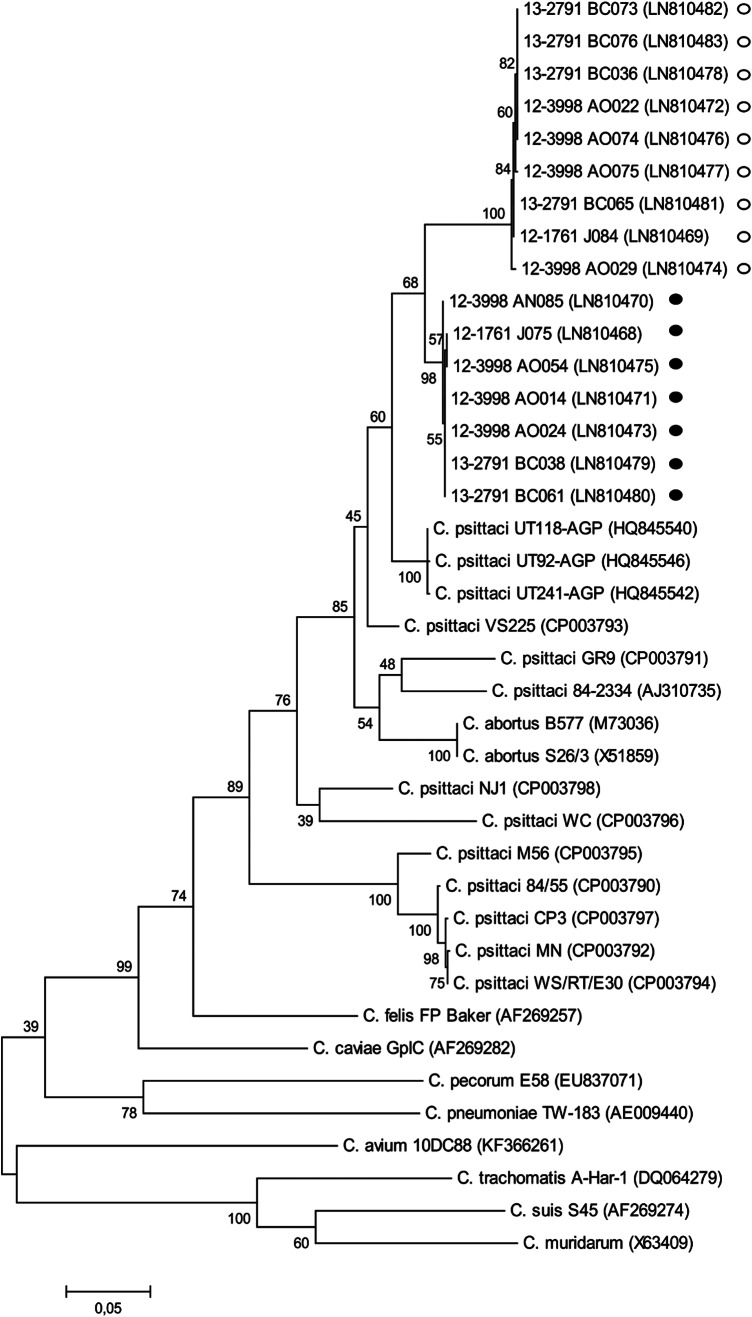
All northern gannet samples positive in C. psittaci-specific incA-qPCR showed Cq values similar to those in the Chlamydiaceae-specific 23S-qPCR, but a panel of samples (identified as group 1, Table 4) had higher Cq values when examined with ompA-based C. psittaci-specific qPCR. In contrast, similar Cq values were obtained from the two qPCR analyses for a second panel of samples (identified as group 2). Analysis of the ompA sequences of 16 of 19 northern gannet samples confirmed the existence of two distinct sequence groups and identified mutations at primer binding sites of the ompA-qPCR (see Fig. S1 in the supplemental material). Multiple alignments revealed that the ompA sequence of 12-3998_AO024 sample, arbitrarily defined as reference type for group 1, exhibited a highest degree of similarity to three sequences from parrot isolates, namely, UT118-AGP, UT92-AGP, and UT241-AGP (GenBank accession numbers HQ845540, HQ845546, and HQ845542, respectively) (Fig. 2). For sample 12-3998_AO075, arbitrarily defined as reference type for group 2, BLAST analysis revealed that its ompA sequence exhibited a highest degree of similarity with two C. psittaci strains of genotype F (VS225 and 7778B15). In the ompA-based dendrogram shown in Fig. 2, northern gannet samples were distributed in two groups, in correlation with qPCR differences and ompA sequence analysis, forming, nevertheless, a separate clade in comparison with the established C. psittaci genotypes.
TABLE 4.
Detection of Chlamydiaceae in northern gannet samples using qPCR
| ompA sequence group and DNA sample identificationa | Cq |
||
|---|---|---|---|
| 23S-qPCR Chlamydiaceae | incA-qPCR C. psittaci | ompA-qPCR C. psittaci | |
| Group 1 | |||
| 12-1761_J075 | 23.6 | 25.3 | 32.1 |
| 12-3998_AN085 | 26.6 | 28.4 | 36.5 |
| 12-3998_AO014 | 23.5 | 25.3 | 34.4 |
| 12-3998_AO024* | 19.6 | 21.2 | 29.9 |
| 12-3998_AO054 | 23.2 | 24.9 | 33.4 |
| 13-2791_BC038 | 20.3 | 21.8 | 30.3 |
| 13-2791_BC061 | 28.5 | 29.3 | 39.4 |
| 13-2791_BC064† | 34.0 | 35.1 | Negb |
| 14-1437_C075† | 37.3 | 39.0 | Neg |
| 14-1437_C098† | 36.4 | 38.0 | Neg |
| Group 2 | |||
| 12-1761_J084 | 31.8 | 32.7 | 29.5 |
| 12-3998_AO022 | 24.6 | 26.4 | 25.2 |
| 12-3998_AO029 | 25.4 | 26.9 | 25.7 |
| 12-3998_AO074 | 30.0 | 31.9 | 30.3 |
| 12-3998_AO075* | 23.7 | 25.7 | 24.4 |
| 13-2791_BC036 | 27.9 | 28.8 | 29.5 |
| 13-2791_BC063† | 35.1 | 35.2 | 36.3 |
| 13-2791_BC065 | 26.2 | 26.9 | 27.4 |
| 13-2791_BC073‡ | 25.0 | 26.2 | 26.5 |
*, arbitrarily designed type sample for each group; †, no ompA amplification for sequencing; ‡, for this bird a second swab was taken and identified as 13-2791_BC076.
Neg, negative.
FIG 2.
ompA-based dendrogram constructed from a global alignment of about 900 bp, including northern gannet specimens. Representative sequences from Chlamydiaceae species and various C. psittaci genotypes and their relevant strains are included. Symbols: ●, northern gannet (group 1); ○, northern gannet (group 2).
In order to ascertain the species identity of these samples, 16S and 23S rRNA genes were obtained from six samples (three samples from each group). Phylogenetic analyses resulted in an almost identical topology (Fig. 1), with all six northern gannet samples being grouped together with the C. psittaci-C. abortus cluster, but in a separate subcluster (Fig. 1).
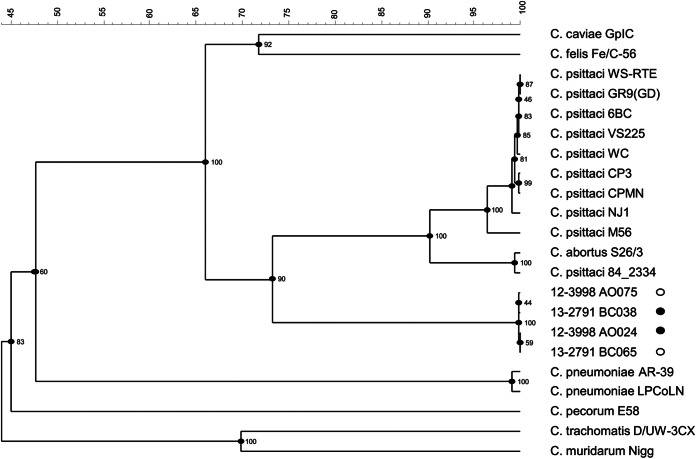
On the basis of the MLST scheme established for Chlamydiaceae, the four northern gannet samples with the highest DNA concentration were selected for sequence analysis, namely, 12-3998_AO024 and 13-2791_BC038 (group 1) and 12-3998_AO075 and 13-2791_BC065 (group 2). Although all of the seven housekeeping gene fragments were successfully amplified for all group 1 samples, new fumC primers had to be designed for group 2. MLST analysis resulted in the detection of four novel allelic profiles assigned as ST84, ST85, ST129, and ST130 on the Chlamydiales MLST website harboring novel alleles (Table 5). Among the four samples examined, variation of the MLST genes was limited in hflX, gidA, and enoA locus (Table 5). Phylogenetic analysis based on concatenated MLST gene sequences resulted in a dendrogram with an almost identical overall topology to those inferred by 16S and 23S rRNA gene sequences; the northern gannet samples were separated from the C. psittaci and C. abortus lineages forming a distinct lineage (Fig. 3). When MLST loci were analyzed separately, except for oppA and hflX loci, northern gannet samples were found closer to the C. psittaci lineage, so that they could be descendants of a common ancestor (data not shown).
FIG 3.
Phylogenetic analyses of concatenated sequences of 7 MLST housekeeping gene fragments (enoA, fumC, gatA, gidA, hemN, hlfX, and oppA) for four northern gannet specimens (12-3998_AO024, 12-3998_AO075, 13-2791_BC038, and 13-2791_BC065) and established Chlamydiaceae species. Symbols: ●, northern gannet (group 1); ○, northern gannet (group 2).
DISCUSSION
This study aimed to investigate the cloacal shedding of Chlamydiaceae in seabirds admitted to a WRC from the northeast Atlantic Ocean in order to investigate their epidemiology and distribution, as well as the resulting risk for animals in care and the medical staff. The study was conducted on birds from the Bay of Biscay (northeast Atlantic Ocean). Studies had previously been published on seabirds, either from the United States on birds found dead during an outbreak of chlamydiosis (18) or caught in the wild in the South Georgia archipelago (21), the Faroe Islands (32), the Bering Sea (26) or, more recently, in the Baltic Sea (22). Investigation of the seabirds brought into care centers is an easy way to access wild birds, without having to organize catches on site. Although the studies conducted to date in WRCs have only included a limited number of individuals from one to two seabird species (25, 34, 35), the present 2.5-year longitudinal study provided the opportunity to sample a larger panel, including several species and families.
Cloacal shedding of Chlamydiaceae was detected here in four bird families, including the Sulidae family, never studied before. To date, Chlamydiaceae species had been detected in the Stercoraccidae, Procellaridae, Scolopacidae, Alcidae, Anatidae, Sternidae, and Laridae families (21, 22, 25, 26, 32). At the bird species level, the present study revealed the occurrence of Chlamydiaceae in six seabird species, with three of them being more represented in significant numbers (European herring gulls, northern gannets, and common murres, in rank order) and more frequently admitted into WRCs from the French coasts of the Bay of Biscay and the Channel (Union Française des Centres de Sauvegarde, unpublished data).
The prevalence of Chlamydiaceae shedders was 18.5%. Previous studies also based on molecular detection and in species with significant sample numbers (about 30 birds) reported a prevalence of 11% on average (ranging from <1 to 38% depending on the species) (21, 22, 26, 32). Regarding Laridae, Christerson et al. (26) reported an average prevalence of 17%. In the same study, a shedder prevalence of 18% was also reported from a species of the Alcidae family (pigeon Guillemot, Cepphus colomba), while in the common murres belonging to the same family we found a prevalence of 7%. In our study a higher prevalence of shedders (41%) for seabird species with a significant sample number was observed for northern gannets (Sulidae), a species for which no previous data on chlamydiosis was available. In these seabirds, a high level of Chlamydiaceae shedding (represented by the mean Cq values) was also observed. These birds were sampled after being found washed up on beaches after many days in distress on sea. An intense shedding has then probably been induced in these birds, more than in healthy ones. Indeed, Chlamydiaceae can survive in a commensal relationship in the gastrointestinal tract (53), but stress factors (in wild birds, e.g., nesting, migration, food shortages, and coinfections) can exacerbate Chlamydiaceae shedding and pathogenicity (7, 8, 10). However, the European herring gulls sampled in the present study and a priori also exposed to a distress-induced stress exhibited a lower prevalence rate and low to moderate Chlamydiaceae shedding levels. The exact reasons for the elevated Chlamydiaceae excretion observed in the northern gannets are unknown. Hypotheses to explain this finding include a higher infection rate of this species in the wild or different levels of fecal shedding, depending on the species and/or the level of distress-induced stress. In any case, the cloacal swabbing was performed <2 days after the birds were rescued, making a recent infection unlikely (54).
In northern gannets, both Chlamydiaceae prevalence and excretion level was higher during the summer than in autumn and winter taken together. Seasonal fluctuations have been already reported for pigeons (17, 55). Since the admission causes were identical from one season to another and that the same person swabbed all of the birds in the same way during the whole study period, an explanation could be that during the summer, when birds breed in colonies, physiological stress (adults foraging far to feed their offspring, immature birds leaving the nest) induce Chlamydiaceae shedding as already reported for other pathogens (56). In contrast to northern gannets, the prevalence was not significantly different from one season to another in European herring gulls, species more opportunistic and less dependent on specific food resources.
Regarding the chlamydial species determined in this study, non-C. psittaci Chlamydiaceae were detected from European herring gulls, as well as from common scoters, common murres, lesser black-backed gulls, and yellow-legged gulls. Based on the 16S rRNA and 23S rRNA sequences, these ones were shown to be two distinct descent lines within Chlamydiaceae separated from all other hitherto established Chlamydia species, with one of these lines clearly related to a “Chlamydiaceae-like” C122 specimen detected from Laridae species across the seas (26) and then later on a European herring gull specimen from Sweden (23). Detection of these atypical Chlamydiaceae in birds from different geographical origins suggest a fairly widespread.
Based on results obtained using specific qPCR systems and ompA sequence analysis, two variants among northern gannet samples being incA-qPCR C. psittaci-positive were detected, leading to the definition of two distinct groups of birds. However, complementary MLST, 16S, and 23S rRNA gene analysis showed that all northern gannet samples are related to the well-known cluster of C. psittaci and C. abortus species, but in a separate subcluster. Examination of these samples with the DNA microarray for Chlamydiaceae (57) revealed no reaction with species-specific probes, thus suggesting that these samples do not belong to any of the established chlamydial species (data not shown). For C. psittaci detection, both ompA- and incA-based qPCR systems were used in the present study. However, for the incA-based PCR system cross-reactions with C. abortus strains have already been identified (unpublished data); meanwhile, ompA is known to be one of the most polymorphic genes in the family Chlamydiaceae and a hot spot for mutations and interstrain recombinations (58, 59). Hence, the use of these two qPCR systems could lead to misidentifications regarding the C. psittaci and/or C. abortus species. As previously reported, despite obvious pathogenicity differences, analysis of gene sequences shows that C. psittaci comprises an unresolved cluster of strains, from which C. abortus is differently evolving. The position of the ompA variants detected in northern gannet specimens should be clarified by more comprehensive studies based on the whole-genome sequencing analysis of strains which have to be isolated beforehand. These two ompA variants were detected in northern gannet specimens at each sampling times. It could be hypothesized that these two groups of birds came from two distinct breeding colonies and that ompA variants could be used as markers of their origin. Thus, taking into account that the northern gannet is a species breeding in colonies with seasonal and age distinct patterns of distributions (60–63), adults collected in the summer in our study harbored one or another of the two ompA variants and came then probably from mixed groups of birds whose breeding colonies were located in the Channel or on the south coasts of Great Britain or Ireland (Celtic Sea) (62). To confirm the potential of the chlamydial typing for the colony determination and then for ecological studies of the northern gannet species, analysis of Chlamydiaceae species harbored by birds from different breeding colonies across Europe would be interesting.
In conclusion, this study confirms the diversity of Chlamydiaceae occurring in birds, as well as the large number of bird species harboring Chlamydiaceae without clinical signs. It is important that veterinarians, medical practitioners, and persons with professional or leisure activities involving contact with wild birds should be aware of the potential exposure to Chlamydiaceae, especially in the case of ecological disasters, such as oil spills, where many seabirds are housed together in close contact with rehabilitation teams. The use of sensitive and broad-range molecular tools is a prerequisite for the detection of all Chlamydiaceae, including the novel distinct descent lines observed in seabirds that we observed here. Preliminary typing results from northern gannet specimens suggest more diversity within the cluster constituting from the closely related C. psittaci and C. abortus species. As previously recommended by Van Van Loock et al. (64), the characterization of C. psittaci and C. abortus strains should not be done based on a single gene and a single method.
Supplementary Material
ACKNOWLEDGMENTS
This study used samples collected from birds admitted to the CVFSE, which is funded by Nantes Métropole, the Conseil Général de Loire-Atlantique, the Conseil Régional des Pays de la Loire, the Direction Régional de l'Environnement, de l'Aménagement et du Logement Pays de la Loire, and private partners—especially Total S.A., Total Exploration Production, Total Raffinage Chimie, and Total Raffinage France.
We are grateful to the public and members of environmental protection associations for live bird collection and to the staff of the CVFSE/Oniris, especially Caroline Ladan, for the bird sampling. We also acknowledge the useful feedback from the reviewers of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00778-15.
REFERENCES
- 1.Schachter J. 1999. Infection and disease epidemiology, p 139–169. In Stephens RS. (ed), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, DC. [Google Scholar]
- 2.Sachse K, Laroucau K. 2014. Avian chlamydiosis: two more bacterial players discovered. Vet J 200:347–348. doi: 10.1016/j.tvjl.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 3.Sachse K, Bavoil PM, Kaltenboeck B, Stephens RS, Kuo CC, Rosselló-Móra R, Horn M. 2015. Emendation of the family Chlamydiaceae: proposal of a single genus, Chlamydia, to include all currently recognized species. Syst Appl Microbiol doi: 10.1016/j.syapm.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Kaleta EF, Taday EM. 2003. Avian hosts range of Chlamydophila spp. based on isolation, antigen detection, and serology. Avian Pathol 32:435–462. doi: 10.1080/03079450310001593613. [DOI] [PubMed] [Google Scholar]
- 5.Sachse K, Laroucau K, Vorimore F, Magnino S, Feige J, Müller W, Kube S, Hotzel H, Schubert E, Slickers P, Ehricht R. 2009. DNA microarray-based genotyping of Chlamydophila psittaci strains from culture and clinical samples. Vet Microbiol 135:22–30. doi: 10.1016/j.vetmic.2008.09.041. [DOI] [PubMed] [Google Scholar]
- 6.Madani SA, Peighambari SM. 2013. PCR-based diagnosis, molecular characterization and detection of atypical strains of avian Chlamydia psittaci in companion and wild birds. Avian Pathol 42:38–44. doi: 10.1080/03079457.2012.757288. [DOI] [PubMed] [Google Scholar]
- 7.Harkinezhad T, Geens T, Vanrompay D. 2009. Chlamydophila psittaci infections in birds: a review with emphasis on zoonotic consequences. Vet Microbiol 135:68–77. doi: 10.1016/j.vetmic.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 8.Speck S, Duff JP. 2012. Chlamydiaceae infections. Psittacosis/ornithosis, p 337–342. In Gavier-Widen D, Duff JP, Meredith A (ed), Infectious diseases of wild mammals and birds in Europe. Section 2: bacterial infections, vol 26 Blackwell Publishing, New York, NY. [Google Scholar]
- 9.Vanrompay D, Karkinezhad T, van de Walle M, Beeckman D, von Droogenbroeck C, Verminnen K, Leten R, Martel A, Cauwerts K. 2007. Chlamydophila psittaci transmission from pet birds to humans. Emerg Infect Dis 13:1108–1110. doi: 10.3201/eid1307.070074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersen AA, Franson JC. 2007. Avian chlamydiosis, p 303–316. In Thomas NJ, Hunter DB, Atkinson CT (ed), Infectious diseases of wild birds. Section 2: bacterial and fungal diseases, vol 15 Blackwell Publishing, New York, NY. [Google Scholar]
- 11.Beeckman DS, Vanrompay DC. 2009. Zoonotic Chlamydophila psittaci infections from a clinical perspective. Clin Microbiol Infect 15:11–17. doi: 10.1111/j.1469-0691.2008.02669.x. [DOI] [PubMed] [Google Scholar]
- 12.Smith KA, Bradley KK, Stobierski MG, Tengelsen LA. 2005. Compendium of measures to control Chlamydophila psittaci (formerly Chlamydia psittaci) infection among humans (psittacosis) and pet birds. J Am Vet Med Assoc 226:532–539. doi: 10.2460/javma.2005.226.532. [DOI] [PubMed] [Google Scholar]
- 13.Magnino S, Haag-Wackernagel D, Geigenfein I, Helmecke S, Dovc A, Prukner-Radovcic E, Residbegović E, Ilieski V, Laroucau K, Donati M, Martinov S, Kaleta EF. 2009. Chlamydial infections in feral pigeons in Europe: review of data and focus on public health implications. Vet Microbiol 135:54–67. doi: 10.1016/j.vetmic.2008.09.045. [DOI] [PubMed] [Google Scholar]
- 14.Sachse K, Laroucau K, Riege K, Wehner S, Dilcher M, Creasy HH, Weidmann M, Myers G, Vorimore F, Vicari N, Magnino S, Liebler-Tenorio E, Ruettger A, Bavoil PM, Hufert FT, Rosselló-Móra R, Marz M. 2014. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Syst Appl Microbiol 37:79–88. doi: 10.1016/j.syapm.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Vorimore F, Hsia R, Huot-Creasy H, Bastian S, Deruyter L, Passet A, Sachse K, Bavoil P, Myers G, Laroucau K. 2013. Isolation of a new chlamydia species from the feral sacred ibis (Threskiornis aethiopicus): Chlamydia ibidis. PLoS One 8:e74823. doi: 10.1371/journal.pone.0074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pantchev A, Sting R, Bauerfeind R, Tyczka J, Sachse K. 2010. Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comp Immunol Microbiol Infect Dis 33:473–484. doi: 10.1016/j.cimid.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Sachse K, Kuehlewind S, Ruettger A, Schubert E, Rohde G. 2012. More than classical Chlamydia psittaci in urban pigeons. Vet Microbiol 157:476–480. doi: 10.1016/j.vetmic.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Franson JC, Pearson JE. 1995. Probable epizootic chlamydiosis in wild California (Larus californicus) and ring-billed (Larus delawarensis) gulls in North Dakota. J Wildl Dis 31:424–427. doi: 10.7589/0090-3558-31.3.424. [DOI] [PubMed] [Google Scholar]
- 19.Olsen B, Persson K, Broholm KA. 1998. PCR detection of Chlamydia psittaci in faecal samples from passerine birds in Sweden. Epidemiol Infect 121:481–483. doi: 10.1017/S0950268898001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schettler E, Fickel J, Hotzel H, Sachse K, Streich WJ, Wittstatt U, Frölich K. 2003. Newcastle disease virus and Chlamydia psittaci in free-living raptors from eastern Germany. J Wildl Dis 39:57–63. doi: 10.7589/0090-3558-39.1.57. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann B, Rahman R, Bergström S, Bonnedahl J, Olsen B. 2000. Chlamydophila abortus in a brown skua (Catharacta Antarctica lonnbergi) from a Subantarctic Island. Appl Environ Microbiol 66:3654–3656. doi: 10.1128/AEM.66.8.3654-3656.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blomqvist M, Christerson L, Waldenström J, Herrmann B, Olsen B. 2012. Chlamydia psittaci in Swedish wetland birds: a risk to zoonotic infection? Avian Dis 56:737–740. doi: 10.1637/10105-022812-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 23.Blomqvist M, Christerson L, Waldenström J, Lindberg P, Helander B, Gunnarsson G, Herrmann B, Olsen B. 2012. Chlamydia psittaci in birds of prey, Sweden. Infect Ecol Epidemiol 2:8435. doi: 10.3402/iee.v2i0.8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beckmann KM, Borel N, Pocknell AM, Dagleish MP, Sachse K, John SK, Pospischil A, Cunningham AA, Lawson B. 2014. Chlamydiosis in British Garden Birds (2005-2011): retrospective diagnosis and Chlamydia psittaci genotype determination. Eco Health 11:544–563. doi: 10.1007/s10393-014-0951-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalmar ID, Dicxk V, Dossche L, Vanrompay D. 2014. Zoonotic infection with Chlamydia psittaci at an avian refuge centre. Vet J 199:300–302. doi: 10.1016/j.tvjl.2013.10.034. [DOI] [PubMed] [Google Scholar]
- 26.Christerson L, Blomqvist M, Grannas K, Thollesson M, Laroucau K, Waldenström J, Eliasson I, Olsen B, Herrmann B. 2010. A novel Chlamydiaceae-like bacterium found in fecal specimens from sea birds from the Bering Sea. Environ Microbiol Rep 2:605–610. doi: 10.1111/j.1758-2229.2010.00174.x. [DOI] [PubMed] [Google Scholar]
- 27.Raso TF, Teixeira RH, Carrasco AO, Araújo JP Jr, Pinto AA. 2013. Chlamydophila psittaci infections in hyacinth macaws (Anodorhynchus hyacinthinus) confiscated in Brazil. J Zoo Wildl Med 44:169–172. doi: 10.1638/1042-7260-44.1.169. [DOI] [PubMed] [Google Scholar]
- 28.Ortega N, Apaza D, Gonzalez F, Salinas J, Caro MR. 2012. Occurrence of Chlamydiaceae in nonsymptomatic free-living raptors in Spain. Eur J Wildl Res 58:351–355. doi: 10.1007/s10344-011-0538-6. [DOI] [Google Scholar]
- 29.Pennycott TW, Dagleish MP, Wood AM, Garcia C. 2009. Chlamydophila psittaci in wild birds in the UK. Vet Rec 164:157–158. doi: 10.1136/vr.164.5.157. [DOI] [PubMed] [Google Scholar]
- 30.Colvile KM, Lawson B, Pocknell AM, Dagleish MP, John SK, Cunningham AA. 2012. Chlamydiosis in British songbirds. Vet Rec 171:177. doi: 10.1136/vr.100506. [DOI] [PubMed] [Google Scholar]
- 31.Telfer BL, Moberley SA, Hort KP, Branley JM, Dwyer DE, Muscatello DJ, Correll PK, England J, McAnulty JM. 2005. Probable psittacosis outbreak linked to wild birds. Emerg Infect Dis J 11:391–397. doi: 10.3201/eid1103.040601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrmann B, Persson H, Jensen JK, Joensen H, Klint M, Olsen B. 2006. Chlamydophila psittaci in Fulmars, the Faroe Islands. Emerg Infect Dis J 12:330–332. doi: 10.3201/eid1202.050404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rehn M, Ringberg H, Runehagen A, Herrmann B, Olsen B, Petersson AC, Hjertqvist M, Kühlmann-Berenzon S, Wallensten A. 2013. Unusual increase of psittacosis in southern Sweden linked to wild bird exposure, January to April 2013. Euro Surveill 18:20478. [PubMed] [Google Scholar]
- 34.Vlahovic K, Matica B, Bata I, Pavlak M, Pavicic Z, Popovic M, Nejedli S, Dovč A. 2004. Campylobacter, salmonella, and chlamydia in free-living birds of Croatia. Eur J Wildl Res 50:127–132. doi: 10.1007/s10344-004-0052-1. [DOI] [Google Scholar]
- 35.Sharples E, Baines SJ. 2009. Prevalence of Chlamydophila psittaci-positive cloacal PCR tests in wild avian casualties in the UK. Vet Rec 164:16–17. doi: 10.1136/vr.164.1.16. [DOI] [PubMed] [Google Scholar]
- 36.Grogan A, Kelly A. 2013. A review of RSPCA research into wildlife rehabilitation. Vet Rec 172:211. doi: 10.1136/vr.101139. [DOI] [PubMed] [Google Scholar]
- 37.Rouffaer LO, Haesebrouck F, Martel A. 2014. Extended-spectrum B-lactamanase-producing Enterobacteriaceae isolated from feces of Falconidae, Accipitridae, and Laridae in bird rescue centers in Belgium. J Wildl Dis 50:957–960. doi: 10.7589/2013-08-208. [DOI] [PubMed] [Google Scholar]
- 38.Carter HR. 2003. Oil and California's seabirds: an overview. Mar Ornithol 31:1–7. [Google Scholar]
- 39.Balseiro A, Espi A, Marquez V, Perez V, Ferreras MC, Garcia Marin JF, Prieto JM. 2005. Pathological features in marine birds affected by the Prestige's oil spill in the North of Spain. J Wildl Dis 41:371–378. doi: 10.7589/0090-3558-41.2.371. [DOI] [PubMed] [Google Scholar]
- 40.Empower. 2014. Incidents. European management programme for oiled wildlife and other marine wildlife emergency responses. http://www.oiledwildlife.eu/background_information/incidents. [Google Scholar]
- 41.Cedre. 2014. Spill cartography. Centre of Documentation, Research, and Experimentation on Accidental Water Pollution, Brest, France: http://www.cedre.fr/en/spill/spill-cartography.php. [Google Scholar]
- 42.Ehricht R, Slickers P, Goellner S, Hotzel H, Sachse K. 2006. Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Mol Cell Probes 20:60–63. doi: 10.1016/j.mcp.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Ménard A, Clerc M, Subtil A, Mégraud F, Bébéar C, de Barbeyrac B. 2006. Development of a real-time PCR for the detection of Chlamydia psittaci. J Med Microbiol 55:471–473. doi: 10.1099/jmm.0.46335-0. [DOI] [PubMed] [Google Scholar]
- 44.Zocevic A, Vorimore F, Vicari N, Gasparini J, Jacquin L, Sachse K, Magnino S, Laroucau K. 2013. A real-time PCR assay for the detection of atypical strains of Chlamydiaceae from pigeons. PLoS One 8:e58741. doi: 10.1371/journal.pone.0058741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pannekoek Y, Morelli G, Kusecek B, Morré SA, Ossewaarde JM, Langerak AA, van der Ende A. 2008. Multi locus sequence typing of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis. BMC Microbiol 8:42. doi: 10.1186/1471-2180-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pannekoek Y, Dickx V, Beeckman DS, Jolley KA, Keijzers WC, Vretou E, Maiden MC, Vanrompay D, van der Ende A. 2010. Multilocus sequence typing of Chlamydia reveals an association between Chlamydia psittaci genotypes and host species. PLoS One 5:e14179. doi: 10.1371/journal.pone.0014179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pudjiatmoko Fukushi H, Ochiai Y, Yamaguchi T, Hirai K. 1997. Phylogenetic analysis of the genus Chlamydia based on 16S rRNA gene sequences. Int J Syst Bacteriol 47:425–431. doi: 10.1099/00207713-47-2-425. [DOI] [PubMed] [Google Scholar]
- 48.Thomas V, Casson N, Greub G. 2006. Criblamydia sequanensis, a new intracellular Chlamydiales isolated from Seine river water using amoebal coculture. Environ Microbiol 8:2125–2135. doi: 10.1111/j.1462-2920.2006.01094.x. [DOI] [PubMed] [Google Scholar]
- 49.Everett KD, Bush RM, Andersen AA. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol 49:415–440. [DOI] [PubMed] [Google Scholar]
- 50.Denamur J, Sayada C, Souriau A, Orfila J, Rodolakis A, Elion J. 1991. Restriction pattern of the major outer-membrane protein gene provides evidence for a homogeneous invasive group among ruminant isolates of Chlamydia psittaci. J Gen Microbiol 137:2525–2530. doi: 10.1099/00221287-137-11-2525. [DOI] [PubMed] [Google Scholar]
- 51.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.BirdLife International. 2014. The data zone: species. IUCN red list for birds. Bird Life International, Cambridge, United Kingdom: http://www.birdlife.org/datazone/home. [Google Scholar]
- 53.Pospischil A, Borel N, Chowdhury EH, Guscetti F. 2009. Aberrant chlamydial developmental forms in the gastrointestinal tract of pigs spontaneously and experimentally infected with Chlamydia suis. Vet Microbiol 135:147–156. doi: 10.1016/j.vetmic.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 54.Vanrompay D, Ducatelle R, Haesebrouck F. 1995. Chlamydia psittaci infections: a review with emphasis on avian chlamydiosis. Vet Microbiol 45:93–119. doi: 10.1016/0378-1135(95)00033-7. [DOI] [PubMed] [Google Scholar]
- 55.Trávnicek M, Cisláková L, Deptuła W, Stosik M, Bhide MR. 2002. Wild pigeons and pheasants: a source of Chlamydophila psittaci for humans and animals. Ann Agric Environ Med 9:253–255. [PubMed] [Google Scholar]
- 56.Broman T, Palmgren H, Bergström S, Sellin M, Waldenström J, Danielsson-Tham ML, Olsen B. 2002. Campylobacter jejuni in black-headed gulls (Larus ridibindus): prevalence, genotypes, and influence on C. jejuni epidemiology. J Clin Microbiol 40:4594–4602. doi: 10.1128/JCM.40.12.4594-4602.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borel N, Kempf E, Hotzel H, Schubert E, Torgerson P, Slickers P, Ehricht R, Tasara T, Pospischil A, Sachse K. 2008. Direct identification of chlamydiae from clinical samples using a DNA microarray assay: a validation study. Mol Cell Probes 22:55–64. doi: 10.1016/j.mcp.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 58.Gomes JP, Bruno WJ, Nunes A, Santos N, Florindo C, Borrego MJ, Dean D. 2007. Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hot spots. Genome Res 17:50–60. doi: 10.1101/gr.5674706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brunelle BW, Sensabaugh GF. 2012. Nucleotide and phylogenetic analyses of the Chlamydia trachomatis ompA gene indicates it is a hot spot for mutation. BMC Res Notes 5:53. doi: 10.1186/1756-0500-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carboneras C. 1992. Order Pelecaniformes family Sulidae (gannets and boobies), p 696 In Del Hoyo J, Elliot A, Sargatal J (ed), Handbook of the birds of the world: ostrich to ducks, vol 1 Lynx Edicions, Barcelona, Spain. [Google Scholar]
- 61.Fort J, Pettex E, Tremblay Y, Lorentsen SH, Garthe S, Votier S, Pons JB, Siorat F, Furness RW, Grecian WJ, Bearhop S, Montevecchi WA, Grémillet D. 2012. Meta-population evidence of oriented chain migration in northern gannets (Morus bassanus). Front Ecol Environ 10:237–242. doi: 10.1890/110194. [DOI] [Google Scholar]
- 62.Wakefield ED, Bodey TW, Bearhop S, Blackburn J, Colhoun K, Davies R, Dwyer RG, Green JA, Grémillet D, Jackson AL, Jessopp MJ, Kane A, Langston RH, Lescroël A, Murray S, Le Nuz SM, Patrick SC, Péron C, Soanes LM, Wanless S, Votier S, Hamer KC. 2013. Space partitioning without territoriality in gannets. Science 341:68–70. doi: 10.1126/science.1236077. [DOI] [PubMed] [Google Scholar]
- 63.Pettex E, Bonadonna F, Enstipp MR, Siorat F, Grémillet D. 2010. Northern gannets anticipate the spatio-temporal occurrence of their prey. J Exp Biol 213:2365–2371. doi: 10.1242/jeb.042267. [DOI] [PubMed] [Google Scholar]
- 64.Van Loock M, Vanrompay D, Herrmann B, Vander Stappen J, Volckaert G, Goddeeris BM, Everett KD. 2003. Missing links in the divergence of Chlamydophila abortus from Chlamydophila psittaci. Int J Syst Evol Microbiol 53:761–770. doi: 10.1099/ijs.0.02329-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.