Abstract
Limonene, a major component of citrus peel oil, has a number of applications related to microbiology. The antimicrobial properties of limonene make it a popular disinfectant and food preservative, while its potential as a biofuel component has made it the target of renewable production efforts through microbial metabolic engineering. For both applications, an understanding of microbial sensitivity or tolerance to limonene is crucial, but the mechanism of limonene toxicity remains enigmatic. In this study, we characterized a limonene-tolerant strain of Escherichia coli and found a mutation in ahpC, encoding alkyl hydroperoxidase, which alleviated limonene toxicity. We show that the acute toxicity previously attributed to limonene is largely due to the common oxidation product limonene hydroperoxide, which forms spontaneously in aerobic environments. The mutant AhpC protein with an L-to-Q change at position 177 (AhpCL177Q) was able to alleviate this toxicity by reducing the hydroperoxide to a more benign compound. We show that the degree of limonene toxicity is a function of its oxidation level and that nonoxidized limonene has relatively little toxicity to wild-type E. coli cells. Our results have implications for both the renewable production of limonene and the applications of limonene as an antimicrobial.
INTRODUCTION
Limonene, the major component of citrus peel oil, has a variety of industrial and microbiological applications. Its antimicrobial properties make it a popular component of disinfectants and food preservatives and an environmentally friendly solvent used at the industrial scale (1–4). More recently, limonene or its hydrogenated forms have been identified to be potential jet fuel components (5–7). As such, the anticipation of a greater global demand for limonene has provided significant motivation for the renewable production of this compound from plant biomass through a microbial process (8–10), and recent efforts to optimize production have resulted in titers of over 400 mg/liter at the bench scale (11). In this context, the toxicity of limonene to the microbial host presents a major challenge, as the accumulation of toxic products limits growth and metabolic activity.
Curiously, the toxicity of limonene has been reported to be significantly higher than that of other monoterpenes or solvents with similar hydrophobicities (12, 13), suggesting that this acute toxicity is due to something other than its solvent-like properties. However, molecular-level studies of limonene toxicity in microbes are limited. While several studies in Escherichia coli have noted an impact of limonene on lipid composition (14) and suggested a role for reactive oxygen species (ROS) (15), no specific hypothesis for how limonene causes these cellular perturbations has been proposed.
In this work, we investigated the basis of limonene toxicity in the model Gram-negative bacterium E. coli. We identified a mutant with significantly enhanced tolerance to limonene and show that the majority of the toxicity in wild-type (WT) cells is due not to limonene itself but to a common oxidation product of limonene, limonene hydroperoxide.
MATERIALS AND METHODS
Strains.
The original FM003 strain was isolated under the following conditions: LB agar plates were covered with a thin layer of 100% limonene and allowed to dry. After streaking individual Keio collection strains on these plates and incubating at room temperature, colonies of spontaneous mutants appeared within 2 days. One of these colonies was chosen for further analysis as FM003.
BW25113 wild-type and ΔahpC, ΔacrA, and ΔacrB strains were obtained from the Keio collection, and the kanamycin resistance cassette was removed by FLP-FRT recombination (16). The mutant ahpC gene (ahpCL177Q, encoding an L-to-Q change at position 177) and wild-type ahpC (ahpCWT) were amplified from the genomic DNA of FM003 and BW25113, respectively, the sequences were verified by Sanger sequencing, and the genes were cloned into the pBbS2K (Tet promoter) vector (17). j5 DNA assembly design software (18) was used for all primer and construct design, and plasmids were assembled using the method described by Gibson et al. (19). A complete list of plasmids and strains used in this work can be obtained at http://public-registry.jbei.org, which also provides a mechanism for researchers to request strains (20).
Whole-genome resequencing.
DNA from both the parent stain and strain FM003 was randomly sheared into ∼400-bp fragments, and the resulting fragments were used to create Illumina libraries. These libraries were sequenced on Illumina sequencers, generating 100-bp paired-end reads. These reads were aligned to the reference genome with GenBank accession number NC_000913 using the Burrows-Wheeler alignment tool (21) and down sampled to generate an average read depth of 250 times. A total of 55 putative variants were identified using the samtools and mpileup (22) programs. Variants that failed our filtering criteria (minimum quality = 10, minimum P value for strand bias = 0.0001, minimum P value for end bias = 0.0001, single-nucleotide polymorphisms within 10 bp around a gap) were removed from the analysis, leaving 28 putative variants, 26 of which were found in both the parent strain and FM003. The genotypes of the variants in the parent strain matched the expected genotype of BW25113 ΔdmsD. Of the two variants that were unique to FM003, the only nonsynonymous one was in codon 177 of ahpC (CtG/CaG, where the lowercase nucleotides represent the variants), resulting in amino acid change L177Q. The other variant was a synonymous change in frlA at codon 123 (GAt/GAc).
Media and growth conditions.
All growth characterization experiments were performed at 30°C in EZRich medium (Teknova), modified to contain 4 g/liter glucose, 1.5 mM PO4, and 1 mM SO4. Anhydrotetracycline (aTc; 40 nM) was used for induction of strains containing Tet promoter constructs. Overnight cultures grown on LB with 50 mg/liter kanamycin were diluted 1/250 into EZRich medium (with antibiotic and inducer, if appropriate), grown to an optical density at 600 nm (OD600) of 0.3 to 0.6, and inoculated 1/200 into 96-well plates containing appropriate amounts of limonene. Growth in 96-well plates was monitored using an automated reader, shaker, and incubator (F200 or F200pro; Tecan). To reduce differences in growth due to the use of antibiotic for plasmid-containing strains, a reduced concentration of kanamycin (5 mg/liter) was used in the EZRich cultures. For experiments whose results are shown in Fig. 5, conditions were identical to those described above, except that 25-ml cultures in 250-ml nonbaffled flasks were used.
FIG 5.
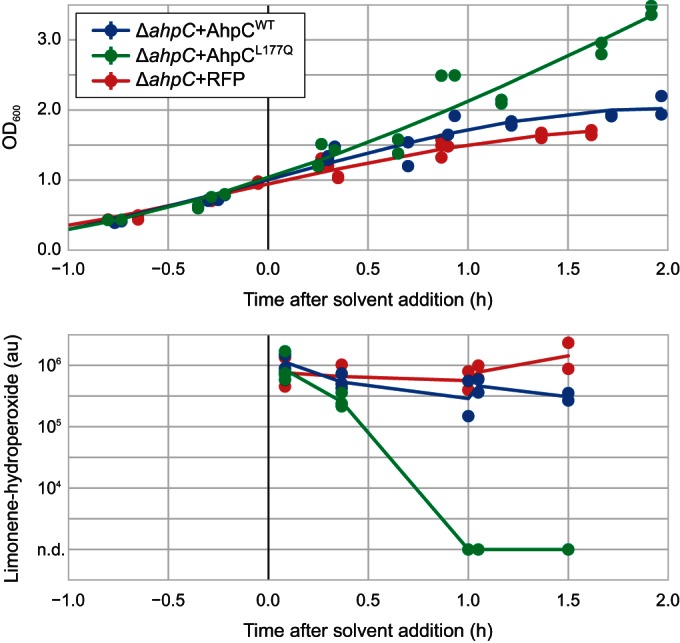
Kinetics of intracellular limonene hydroperoxide upon limonene addition to growing cultures. Partially oxidized limonene (1%, vol/vol) was added at time zero. Results for four independent biological replicates (from two different days) are shown together. Lines are spline fits. au, arbitrary units; n.d., not detected (the amount was below the detection limit).
Anaerobic growth experiments followed a protocol similar to that described above. Aerobic overnight LB cultures were diluted 1/250 into EZRich medium, sealed, sparged of air using a mixture of N2 and CO2, and grown to an OD600 of 0.3. Main cultures in the same medium were prepared in an anaerobic chamber as 1/100 dilutions of the preculture. Growth in 100-well plates was monitored in a Bioscreen C MBR reader, shaker, and incubator (Growth Curves Ab Ltd.). All media used for anaerobic growth contained 20 g/liter glucose.
Chemicals.
All chemicals were purchased from Sigma-Aldrich unless otherwise specified. S-Limonene was used for the toxicity experiments whose results are shown in Fig. 1, 3, 5, and 6, though no difference in toxicity or hydroperoxide formation was generally observed between the S and R enantiomers. The data shown in Fig. 4 are for both enantiomers from stocks purchased from several different manufacturers (Sigma-Aldrich, Acros Organics, Alfa Aesar) over several years. To anaerobically maintain nonoxidized limonene, new bottles were purchased from Sigma-Aldrich and immediately sparged with N2 gas and sealed. Liquid chromatography (LC)-time of flight (TOF) mass spectrometry (MS) and gas chromatography (GC)-MS analyses showed minimal hydroperoxide content in these samples.
FIG 1.
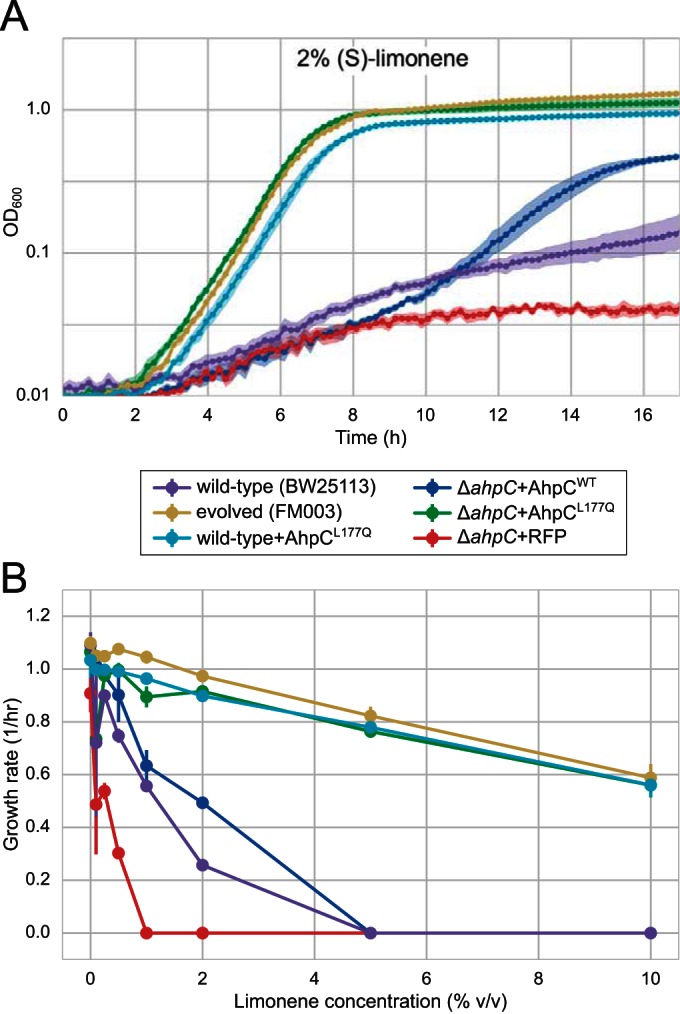
Growth of E. coli strains expressing either AhpCWT or AhpCL177Q. (A) Growth of six strains, including the spontaneous mutant FM003 as well as the reconstituted strains expressing AhpCL177Q, in defined rich medium with 2% (vol/vol) limonene. Shaded areas represent the experimental variation (standard error of the mean [SEM]) for three biological replicates. (B) Specific growth rates at different limonene concentrations. Error bars represent SEMs for three biological replicates.
FIG 3.
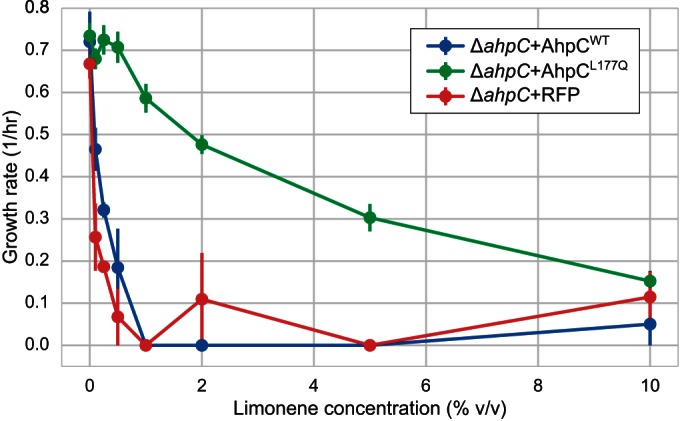
Anaerobic growth of E. coli strains expressing AhpCWT or AhpCL177Q. The specific growth rate at different limonene concentrations is shown for a ΔahpC strain expressing either of the two AhpC variations or RFP and growing anaerobically in defined rich medium. Error bars represent SEMs for three biological replicates.
FIG 6.
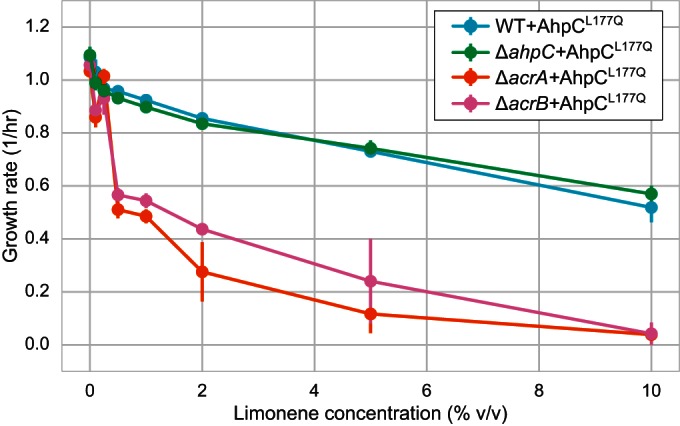
Effect of acrA and acrB gene knockouts on limonene tolerance in strains expressing AhpCL177Q. Error bars represent SEMs for three biological replicates.
FIG 4.
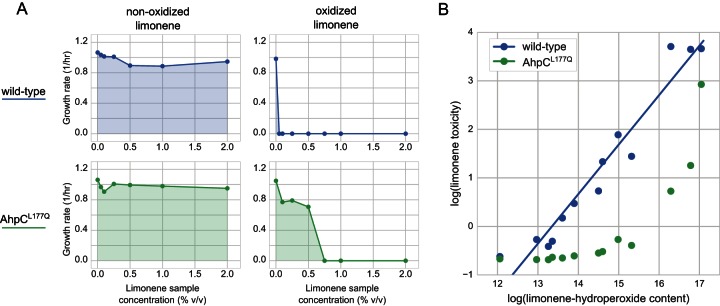
Toxicity of limonene at different degrees of oxidation. (A) Specific growth rate plotted against the limonene concentration for nonoxidized, anaerobically stored limonene (left) and fully oxidized limonene (exposed to air and light for 3 weeks) (right). (Top) Wild-type BW25113; (bottom) a ΔahpC strain expressing AhpCL177Q. (B) Limonene toxicity, defined as the inverse of the shaded areas in panel A, plotted against the limonene hydroperoxide content (LC-TOF-MS peak area, in arbitrary units). Each point represents 1 of 13 limonene bottles subjected to various degrees of oxidation. Blue points, wild-type BW25113; green points, a ΔahpC strain expressing AhpCL177Q.
Metabolite analysis.
For analysis of the oxidized forms of limonene, limonene samples were diluted with ethyl acetate and analyzed directly via LC-TOF-MS or GC-MS. LC-TOF-MS was performed on an Agilent 6210 TOF instrument using atmospheric chemical pressure ionization (APCI). Analyses were conducted at 55°C with a Kinetex XB-C18 column (length, 100 mm; internal diameter, 3 mm; particle size, 2.6 μm; Phenomenex, Inc.) using a 1200 series high-performance liquid chromatography (HPLC) system (Agilent Technologies). The mobile phase was composed of water (solvent A) and methanol (solvent B) (HPLC grade; Honeywell Burdick & Jackson, CA). Three-microliter samples were injected and separated with the following gradient: the gradient was 60% to 98% solvent B for 3.47 min, held at 98% solvent B for 7.03 min, decreased to 60% solvent B in 0.5 min, and held at 60% solvent B for a further 1.5 min. A flow rate of 0.42 ml/min from 0 to 8.67 min was used, and the flow rate was increased to 0.65 ml/min for 0.33 min and held at 0.65 ml/min for a further 3.5 min. The total analysis run time was 12.5 min. The selected mass range was 110 to 250 m/z. All other APCI-TOF MS conditions were as described previously (23). Data acquisition and processing were performed by use of an Agilent MassHunter workstation and qualitative analysis, respectively.
GC-MS was performed on an Agilent 6890 series GC equipped with an Agilent 5973 MS detector and an Agilent DB-5ms column (11). All integration and quantification were performed with software provided by the manufacturer. For compounds detected by both methods, results were quantitatively consistent. For final quantification, LC-TOF-MS data were used for limonene hydroperoxide, while GC-MS data were used for limonene, limonene oxide, carveol, and carvone.
For analysis of intracellular metabolites, 2 ml of culture was centrifuged at 14,000 × g for 1 min, the supernatant was rapidly decanted, and the cell pellet was frozen in liquid nitrogen. Pellets were extracted with 50:50 water-ethyl acetate. For extracellular metabolites, 800 μl of supernatant was extracted with 400 μl of ethyl acetate. In both cases, the organic phase was analyzed by LC-TOF-MS and GC-MS as described above.
SRA accession number.
Raw data are available from the NCBI Sequence Read Archive (SRA) under accession number SRX838687.
RESULTS
Identification and genome sequencing of a limonene-resistant mutant.
During routine screening of the Keio gene deletion collection (16) for strains with differential sensitivity to limonene, we observed the spontaneous appearance of mutants with high tolerance to limonene. One such strain, which arose from the ΔdmsD strain of the Keio collection, was named FM003 and chosen for further analysis. The phenotype of growth on LB agar plates with a limonene overlay was verified, and growth was also verified in liquid cultures with defined complex or minimal medium containing up to 5% (vol/vol) limonene. Given the robust phenotype, we sequenced the genome of FM003 to identify mutations responsible for limonene tolerance. The only nonsilent mutation identified with high confidence in FM003 compared to the sequence of the parent strain was a point mutation in the coding sequence of ahpC, resulting in amino acid change L177Q.
To determine if the mutation in ahpC resulted in a loss of function, we assayed limonene tolerance in the ΔahpC strain taken from the Keio collection. Deletion of ahpC failed to confer tolerance; in fact, the ΔahpC strain was slightly more sensitive to limonene than the wild type, suggesting that the mutation had enhanced the role of AhpC in limonene tolerance. Therefore, we cloned the ahpCL177Q allele from FM003 into a low-copy-number expression plasmid and expressed AhpCL177Q, AhpCWT, or a red fluorescent protein (RFP) control in a ΔahpC strain background to avoid heterogeneity effects. Expression of AhpCL177Q was sufficient to essentially reconstitute the limonene-tolerant phenotype of FM003, whereas expression of AhpCWT resulted in approximately wild-type levels of limonene tolerance (Fig. 1). These results strongly suggest that the phenotype of the evolved strain can be attributed to the ahpCL177Q mutation. No difference in phenotype was observed when AhpCL177Q was expressed in wild-type strains and when it was expressed in the ΔdmsD strain background in which the mutation was originally isolated (data not shown), so the extremely slight fitness advantage that remained in the FM003 strain may be attributable to the proper regulation of AhpC expression from its native locus.
Role of alkyl hydroperoxide reductase.
AhpC forms a multimeric complex with AhpF to form the enzyme alkyl hydroperoxide reductase, which is responsible for relieving peroxide stress by reducing hydroperoxides to their corresponding alcohols, using NADH as an electron donor (24) (Fig. 2A). AhpC is the peroxidase component of the enzyme, responsible for binding and reducing the peroxide substrate, while AhpF catalyzes the second step of reducing the AhpC disulfide bond and recycling AhpC. We hypothesized that AhpCL177Q may have an altered substrate specificity that allows reduction of a toxic hydroperoxide formed by oxidation of limonene (Fig. 2B), which would be consistent with the increased sensitivity of the ΔahpC strain. Further support for this hypothesis came from a previous observation that oxidation products, such as limonene hydroperoxide, are rapidly formed from limonene under aerobic conditions and are responsible for the allergenic properties of limonene in mammals (25). Additionally, previous work uncovered a separate mutation in ahpC that conferred tolerance to tetralin via hydroperoxide reduction (26). In that study, the authors had sequenced a tetralin-resistant mutant and found an ahpCG142V mutation. The mutant protein was shown to reduce tetralin hydroperoxide to 1,2,3,4-tetrahydro-1-naphthol in vitro and to alleviate the toxicity of both tetralin and tetralin hydroperoxide in vivo.
FIG 2.
(A) Reaction catalyzed by alkyl hydroperoxide reductase, formed by the enzymes AhpC and AhpF, in vivo. (B) Hypothetical mechanism of limonene hydroperoxide formation and detoxification by AhpCL177Q.
We hypothesized that limonene hydroperoxide may form quickly in vivo due to the presence of the reactive oxygen species (ROS) formed during aerobic respiration (27). In this case, we would expect limonene to be significantly less toxic during anaerobic growth. Surprisingly, we found no decrease in limonene toxicity under anaerobic conditions and still observed a significant increase in tolerance provided by expression of AhpCL177Q (Fig. 3). This result suggested that if the toxic species was indeed limonene hydroperoxide, it was formed before addition of limonene to the E. coli culture.
Direct measurement of limonene oxidation products.
To determine directly if limonene hydroperoxide was present as a contaminant of the limonene used in our toxicity assays, we developed a method to quantify oxidized forms of limonene using liquid chromatography and time of flight mass spectrometry (LC-TOF-MS). Since pure standards for limonene hydroperoxide were not commercially available, we oxidized limonene by leaving it exposed to air for 3 weeks with gentle stirring, a procedure previously shown to lead to the formation of significant quantities of limonene hydroperoxide (25). The oxidized limonene was analyzed by LC-TOF-MS and showed a greater than 10-fold increase in the signal corresponding to the exact mass of limonene hydroperoxide. While multiple chromatographic peaks were observed, we did not attempt to distinguish isomers such as limonene-1-hydroperoxide and limonene-2-hydroperoxide or cis and trans forms. Several other likely oxidation products of limonene, such as limonene-1,2-oxide and carvone, were also observed, while approximately 50% of the sample remained in the form of pure limonene.
We also assayed the toxicity of the oxidized limonene sample enriched in limonene hydroperoxide to wild-type and AhpCL177Q-expressing E. coli strains. The oxidized limonene sample was found to be significantly more toxic, completely inhibiting the growth of wild-type E. coli even at 0.1% (vol/vol). AhpCL177Q-expressing strains were able to tolerate up to 0.5% (vol/vol) oxidized limonene, with only minor defects in the growth rate being detected (Fig. 4A). Since these results further corroborated the hypothesis that limonene toxicity is caused by the presence of the limonene hydroperoxide contaminant, we created a library of limonene samples oxidized to various degrees and assayed their hydroperoxide content and toxicity. The library consisted of 11 different limonene bottles from several manufacturers which had been subject to various levels of routine laboratory use, as well as 2 bottles that were sparged with nitrogen gas immediately upon receipt from the vendor and stored anaerobically. We observed a remarkable correlation between toxicity and hydroperoxide content (Fig. 4B), with the anaerobically stored limonene showing minimal toxicity to wild-type E. coli.
As mentioned above, mass spectrometry analysis showed several other oxidation products of limonene present in the oxidized samples. Using a combination of LC-TOF-MS and gas chromatography-mass spectrometry (GC-MS) analysis, we performed absolute quantification of carvone, carveol, and limonene-1,2-oxide, for which commercial standards were available. Carvone was determined to make up at most 5% of the sample, while carveol and limonene-1,2-oxide accounted for less than 1% (vol/vol). As mentioned above, about 50% of the sample was in the form of pure limonene, suggesting that another large fraction is accounted for by polymers of the oxidation products, which would be consistent with previous reports from a similar oxidation procedure (25).
To determine if carvone, carveol, or limonene-1,2,-oxide could be the toxic component of the oxidized limonene, we assayed their toxicity to both wild-type and AhpCL177Q-expressing E. coli strains. The pure compounds were dissolved in nonoxidized (anaerobically stored) limonene at various concentrations, and the mixture was added to the medium at 1% (vol/vol). Limonene with up to 20% carvone or limonene-1,2-oxide or with up to 5% carveol had no effect on the growth rate of either strain (see Fig. S1 in the supplemental material). These results offer further evidence that the predominant toxic species is limonene hydroperoxide.
To examine the role of AhpCL177Q in alleviating toxicity, we quantified intracellular limonene hydroperoxide levels in cells grown in the presence of limonene. Exponential-phase cells expressing AhpCWT, AhpCL177Q, or RFP were treated with 1% (vol/vol) partially oxidized limonene (i.e., a limonene sample imposing an intermediate level of toxicity), and cell samples were taken at several time points after limonene addition. LC-TOF-MS analysis of intracellular metabolites showed that significant quantities of intracellular limonene hydroperoxide appeared directly after limonene addition in all three strains (Fig. 5). In the strains expressing RFP or AhpCWT, growth was dramatically impacted and the levels of intracellular limonene hydroperoxide remained high. In contrast, the strain expressing AhpCL177Q quickly resumed growth and the level of limonene hydroperoxide dropped below the detection limit within 1 h, strongly supporting the hypothesis that AhpCL177Q reduces the toxic hydroperoxide to a less toxic compound.
On the basis of the known function of the AhpCF complex, the reduction product is likely to be carveol or the isomer p-mentha-2,8-dien-1-ol (Fig. 2). We observed a very slight accumulation of carveol and its isomers in the AhpCL177Q strain (see Fig. S2 in the supplemental material). However, the data were appreciably noisy due to the fact that these alcohols were already present in the oxidized limonene sample. Thus, our data are consistent with the proposed function of AhpCL177Q but do not allow a definitive statement about the final detoxification product.
Role of AcrAB efflux pump.
Previous work showed that even minimal levels of E. coli tolerance to limonene are completely dependent on the presence of a functional efflux pump, such as the E. coli AcrAB-TolC complex (13). AcrAB-TolC is a broad-specificity efflux pump and has crucial roles in tolerance to a wide variety of other organic solvents as well as other toxins, such as antibiotics (28). To determine if limonene efflux by AcrAB-TolC was still required in cells expressing AhpCL177Q, we expressed AhpCL177Q in strains with deletions of either acrA or acrB.
While the ΔacrA and ΔacrB strains showed almost no fitness defect in medium without limonene, growth was significantly impaired by limonene (as described above, partially oxidized limonene with an intermediate level of toxicity was used). Expression of AhpCL177Q conferred no additional tolerance to limonene in these strains (Fig. 6). This suggests that, consistent with prior knowledge, intracellular limonene is efficiently secreted by the AcrAB-TolC efflux pump, relieving the toxicity of limonene itself. However, an entirely orthogonal stress is imposed by the oxidized form, limonene hydroperoxide, and this toxicity is relieved only by the mutant alkyl hydroperoxidase AhpCL177Q.
DISCUSSION
The toxicity of limonene to E. coli has been somewhat enigmatic. The working assumption for the toxicity of organic solvents is that they work largely by disrupting the cell membrane, which in turn interferes with a variety of processes, such as respiration, transport, and maintenance of ion gradients (28). Under this model, the toxicity is largely a function of the compound's hydrophobicity, and several studies have shown the inverse correlation between toxicity and the log octanol-water partition coefficient (POW), a measure of hydrophobicity (29). However, limonene is significantly more toxic than other solvents in the same log POW range and very closely related compounds, like pinene (12, 13). In this work, we show that this is most likely due to the presence of the limonene oxidation product limonene hydroperoxide as a contaminant of limonene.
A key finding in our study is that limonene hydroperoxide is likely to be present in most laboratory limonene stocks, as we observed that significant hydroperoxide formation and associated toxicity appeared within several weeks of routine handling of limonene bottles. Special care, such as storage under anaerobic conditions, appears to be required for limonene if it is used for toxicity studies. Ideally, chemical manufacturers should also be encouraged to store and ship limonene sparged with an inert gas. With the recent interest in the mechanism of limonene toxicity in microbes (14, 15, 30, 31), it is critical to ensure that the toxicity attributed to limonene is not in fact due to limonene hydroperoxide. Our findings suggest that nonoxidized limonene has effects similar to those of other solvents in the same log POW range. These solvents have relatively little effect on wild-type E. coli, although they are highly toxic to cells with an impaired efflux capability, such as the cells of ΔacrA or ΔacrB strains (13). The AcrAB-TolC efflux pump is particularly critical for compounds with a log POW in the range of 3.9 to 5.5 (29); thus, it is not surprising that limonene, with a log POW of approximately 4.1, is efficiently effluxed by AcrAB-TolC.
The AhpCL177Q protein, discovered in a strain of E. coli that evolved on medium containing limonene and, presumably, limonene hydroperoxide, appears to be highly efficient at reducing limonene hydroperoxide to a less toxic product. Given the increased sensitivity of the ΔahpC mutant, wild-type AhpC likely has some minimal activity against limonene hydroperoxide, but the activity is greatly enhanced by the L177Q mutation. Analysis of the protein structure could illuminate the effect of the mutation, and the structure of E. coli AhpC was recently solved by X-ray crystallography to a 3.3-Å resolution (32). Unfortunately, the C-terminal tail, which includes the L177 residue, could not be resolved in the structure. However, L177 is likely to be located close to the active site at C166, and the structure is consistent with the C-terminal tail being involved in substrate recognition. An alternative, though unlikely, scenario is that the L177Q mutation generally increases AhpC activity against a variety of substrates. Further in vitro studies with AhpCL177Q and other mutant versions of AhpC would be necessary to resolve these questions.
We were mildly surprised to find that AhpCL177Q was equally efficient in a wild-type and a ΔahpC strain background (Fig. 1B), since AhpC forms a decamer in its functional form (32, 33) and the mutation was originally isolated in a strain with only the single ahpCL177Q allele. This strongly suggests that the heteromer is functional, which would be consistent with the fact that each decamer has 10 independent substrate-accessible active sites. Alternatively, very small quantities of homomeric AhpCL177Q may be sufficient for activity, which would also be consistent with our findings that even very low (leaky) expression of AhpCL177Q is sufficient for tolerance and no increase in tolerance is associated with higher levels of expression (see Fig. S3 in the supplemental material).
Although the oxidation of limonene to limonene hydroperoxide was observed half a century ago (34, 35), there is a limited body of literature on the biological properties of limonene hydroperoxide. A large fraction of this literature focuses on the allergenic properties of limonene, which were shown to be due to limonene hydroperoxide (25, 36–38). This direct parallel to bacterial toxicity suggests a general cellular damage mechanism for limonene hydroperoxide. In general, hydroperoxides, like other reactive oxygen species, cause oxidative damage to a variety of cellular macromolecules, such as DNA, proteins, and lipids (39). Lipid peroxidation could be particularly relevant in the case of limonene hydroperoxide, as the hydrophobic limonene moiety is likely to insert into the membrane.
Metabolic engineering of microbes for production of fuels and commodity chemicals has been challenging, and the toxicity of the final product is thought to be a major impediment to increasing production titers (40). In the specific case of microbial production of limonene, our findings are very encouraging, as they suggest that the limonene produced inside the cell will not be highly toxic, as long as it is efficiently secreted. Consistent with this, expression of AhpCL177Q did not confer any significant improvement in limonene production in 24- to 72-h batch cultures of engineered E. coli (see Fig. S4 in the supplemental material). However, industrial fermentations can last several weeks or more, and we observed significant limonene oxidation to limonene hydroperoxide on this time scale. While a scaled-up production process would be tailored toward preventing significant product oxidation, even small quantities of the hydroperoxide could be toxic, and the use of strains not subject to this toxicity would be preferred. Since strains expressing AhpCL177Q showed no obvious defects in growth or metabolism, they are excellent candidates for use in microbial limonene production processes at the industrial scale. Furthermore, the ahpCL177Q mutation appeared after only a few days of growth under the stress condition, and it is possible that even more tolerant strains could be obtained by continued laboratory evolution.
Supplementary Material
ACKNOWLEDGMENTS
This work, conducted by the Joint BioEnergy Institute, was supported by the Office of Science, Office of Biological and Environmental Research, of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. F.M. was supported by Total New Energies USA, Inc., Emeryville, CA, as part of a joint project between the Joint BioEnergy Institute and Total New Energies USA, Inc.
We thank Heather Szmidt-Middleton for help with initial strain archiving and testing and Margaret Brown and Tristan de Rond for other technical assistance and helpful discussions.
J.D.K. has financial interests in Amyris and Lygos.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01102-15.
REFERENCES
- 1.Bomgardner M. 2011. Orange crush. Chem Eng News Arch 89:21. doi: 10.1021/cen-v089n014.p021. [DOI] [Google Scholar]
- 2.Ciriminna R, Lomeli-Rodriguez M, Carà PD, Lopez-Sanchez JA, Pagliaro M. 2014. Limonene: a versatile chemical of the bioeconomy. Chem Commun (Camb) 50:15288–15296. doi: 10.1039/C4CC06147K. [DOI] [PubMed] [Google Scholar]
- 3.Dellutri J. November 1986. Mixture of citric oil, vinegar and water. US patent 4, 620, 937 A.
- 4.Settanni L, Palazzolo E, Guarrasi V, Aleo A, Mammina C, Moschetti G, Germanà MA. 2012. Inhibition of foodborne pathogen bacteria by essential oils extracted from citrus fruits cultivated in Sicily. Food Control 26:326–330. doi: 10.1016/j.foodcont.2012.01.050. [DOI] [Google Scholar]
- 5.Chuck CJ, Donnelly J. 2014. The compatibility of potential bioderived fuels with jet A-1 aviation kerosene. Appl Energy 118:83–91. doi: 10.1016/j.apenergy.2013.12.019. [DOI] [Google Scholar]
- 6.Meylemans HA, Quintana RL, Harvey BG. 2012. Efficient conversion of pure and mixed terpene feedstocks to high density fuels. Fuel 97:560–568. doi: 10.1016/j.fuel.2012.01.062. [DOI] [Google Scholar]
- 7.Renninger NS, Ryder JA, Fisher KJ. May 2011. Jet fuel compositions and methods of making and using same. US patent 7,935,156 B2.
- 8.Carter OA, Peters RJ, Croteau R. 2003. Monoterpene biosynthesis pathway construction in Escherichia coli. Phytochemistry 64:425–433. doi: 10.1016/S0031-9422(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 9.Behrendorff JB, Vickers CE, Chrysanthopoulos P, Nielsen LK. 2013. 2,2-Diphenyl-1-picrylhydrazyl as a screening tool for recombinant monoterpene biosynthesis. Microb Cell Fact 12:76. doi: 10.1186/1475-2859-12-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halfmann C, Gu L, Zhou R. 2014. Engineering cyanobacteria for the production of a cyclic hydrocarbon fuel from CO2 and H2O. Green Chem 16:3175–3185. doi: 10.1039/C3GC42591F. [DOI] [Google Scholar]
- 11.Alonso-Gutierrez J, Chan R, Batth TS, Adams PD, Keasling JD, Petzold CJ, Lee TS. 2013. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab Eng 19:33–41. doi: 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Brennan TCR, Turner CD, Krömer JO, Nielsen LK. 2012. Alleviating monoterpene toxicity using a two-phase extractive fermentation for the bioproduction of jet fuel mixtures in Saccharomyces cerevisiae. Biotechnol Bioeng 109:2513–2522. doi: 10.1002/bit.24536. [DOI] [PubMed] [Google Scholar]
- 13.Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A. 2011. Engineering microbial biofuel tolerance and export using efflux pumps. Mol Syst Biol 7:487. doi: 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Pasqua R, Hoskins N, Betts G, Mauriello G. 2006. Changes in membrane fatty acids composition of microbial cells induced by addiction of thymol, carvacrol, limonene, cinnamaldehyde, and eugenol in the growing media. J Agric Food Chem 54:2745–2749. doi: 10.1021/jf052722l. [DOI] [PubMed] [Google Scholar]
- 15.Chueca B, Pagán R, García-Gonzalo D. 2014. Differential mechanism of Escherichia coli inactivation by (+)-limonene as a function of cell physiological state and drug's concentration. PLoS One 9:e94072. doi: 10.1371/journal.pone.0094072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. 2011. BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J Biol Eng 5:12. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hillson NJ, Rosengarten RD, Keasling JD. 2012. j5 DNA assembly design automation software. ACS Synth Biol 1:14–21. doi: 10.1021/sb2000116. [DOI] [PubMed] [Google Scholar]
- 19.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 20.Ham TS, Dmytriv Z, Plahar H, Chen J, Hillson NJ, Keasling JD. 2012. Design, implementation and practice of JBEI-ICE: an open source biological part registry platform and tools. Nucleic Acids Res 40:e141. doi: 10.1093/nar/gks531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goh E-B, Baidoo EEK, Keasling JD, Beller HR. 2012. Engineering of bacterial methyl ketone synthesis for biofuels. Appl Environ Microbiol 78:70–80. doi: 10.1128/AEM.06785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poole LB. 2005. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys 433:240–254. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Karlberg A-T, Shao LP, Nilsson U, Gäfvert E, Nilsson JLG. 1994. Hydroperoxides in oxidized d-limonene identified as potent contact allergens. Arch Dermatol Res 286:97–103. doi: 10.1007/BF00370734. [DOI] [PubMed] [Google Scholar]
- 26.Ferrante AA, Augliera J, Lewis K, Klibanov AM. 1995. Cloning of an organic solvent-resistance gene in Escherichia coli: the unexpected role of alkylhydroperoxide reductase. Proc Natl Acad Sci U S A 92:7617–7621. doi: 10.1073/pnas.92.17.7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Storz G, Imlay JA. 1999. Oxidative stress. Curr Opin Microbiol 2:188–194. doi: 10.1016/S1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 28.Ramos JL, Duque E, Gallegos M-T, Godoy P, Ramos-González MI, Rojas A, Terán W, Segura A. 2002. Mechanisms of solvent tolerance in Gram-negative bacteria. Annu Rev Microbiol 56:743–768. doi: 10.1146/annurev.micro.56.012302.161038. [DOI] [PubMed] [Google Scholar]
- 29.Tsukagoshi N, Aono R. 2000. Entry into and release of solvents by Escherichia coli in an organic-aqueous two-liquid-phase system and substrate specificity of the AcrAB-TolC solvent-extruding pump. J Bacteriol 182:4803–4810. doi: 10.1128/JB.182.17.4803-4810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brennan TCR, Krömer JO, Nielsen LK. 2013. Physiological and transcriptional responses of Saccharomyces cerevisiae to d-limonene show changes to the cell wall but not to the plasma membrane. Appl Environ Microbiol 79:3590–3600. doi: 10.1128/AEM.00463-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Zhu Y, Du G, Zhou J, Chen J. 2013. Exogenous ergosterol protects Saccharomyces cerevisiae from d-limonene stress. J Appl Microbiol 114:482–491. doi: 10.1111/jam.12046. [DOI] [PubMed] [Google Scholar]
- 32.Dip PV, Kamariah N, Subramanian Manimekalai MS, Nartey W, Balakrishna AM, Eisenhaber F, Eisenhaber B, Grüber G. 2014. Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli. Acta Crystallogr D Biol Crystallogr 70:2848–2862. doi: 10.1107/S1399004714019233. [DOI] [PubMed] [Google Scholar]
- 33.Parsonage D, Youngblood DS, Sarma GN, Wood ZA, Karplus PA, Poole LB. 2005. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry 44:10583–10592. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schenck GO, Gollnick K, Buchwald G, Schroeter S, Ohloff G. 1964. Zur chemischen und sterischen Selektivität der photosensibilisierten O2-Übertragung auf (+)-Limonen und (+)-Carvomenthen. Justus Liebigs Ann Chem 674:93–117. doi: 10.1002/jlac.19646740111. [DOI] [Google Scholar]
- 35.Clark BC Jr, Jones BB, Iacobucci GA. 1981. Characterization of the hydroperoxides derived from singlet oxygen oxidation of (+)-limonene. Tetrahedron 37(Suppl 1):405–409. doi: 10.1016/0040-4020(81)85078-8. [DOI] [Google Scholar]
- 36.Christensson JB, Johansson S, Hagvall L, Jonsson C, Börje A, Karlberg A-T. 2008. Limonene hydroperoxide analogues differ in allergenic activity. Contact Dermatitis 59:344–352. doi: 10.1111/j.1600-0536.2008.01442.x. [DOI] [PubMed] [Google Scholar]
- 37.Johansson S, Giménez-Arnau E, Grøtli M, Karlberg A-T, Börje A. 2008. Carbon- and oxygen-centered radicals are equally important haptens of allylic hydroperoxides in allergic contact dermatitis. Chem Res Toxicol 21:1536–1547. doi: 10.1021/tx800104c. [DOI] [PubMed] [Google Scholar]
- 38.Rudbäck J, Ramzy A, Karlberg A-T, Nilsson U. 2014. Determination of allergenic hydroperoxides in essential oils using gas chromatography with electron ionization mass spectrometry: gas chromatography. J Sep Sci 37:982–989. doi: 10.1002/jssc.201300843. [DOI] [PubMed] [Google Scholar]
- 39.Ames BN. 1983. Dietary carcinogens and anticarcinogens. Oxygen radicals and degenerative diseases. Science 221:1256–1264. [DOI] [PubMed] [Google Scholar]
- 40.Fischer CR, Klein-Marcuschamer D, Stephanopoulos G. 2008. Selection and optimization of microbial hosts for biofuels production. Metab Eng 10:295–304. doi: 10.1016/j.ymben.2008.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.