Abstract
Campylobacter jejuni is a leading cause of human foodborne gastroenteritis worldwide. The interactions between this pathogen and the intestinal microbiome within a host are of interest as endogenous intestinal microbiota mediates a form of resistance to the pathogen. This resistance, termed colonization resistance, is the ability of commensal microbiota to prevent colonization by exogenous pathogens or opportunistic commensals. Although mice normally demonstrate colonization resistance to C. jejuni, we found that mice treated with ampicillin are colonized by C. jejuni, with recovery of Campylobacter from the colon, mesenteric lymph nodes, and spleen. Furthermore, there was a significant reduction in recovery of C. jejuni from ampicillin-treated mice inoculated with a C. jejuni virulence mutant (ΔflgL strain) compared to recovery of mice inoculated with the C. jejuni wild-type strain or the C. jejuni complemented isolate (ΔflgL/flgL). Comparative analysis of the microbiota from nontreated and ampicillin-treated CBA/J mice led to the identification of a lactic acid-fermenting isolate of Enterococcus faecalis that prevented C. jejuni growth in vitro and limited C. jejuni colonization of mice. Next-generation sequencing of DNA from fecal pellets that were collected from ampicillin-treated CBA/J mice revealed a significant decrease in diversity of operational taxonomic units (OTUs) compared to that in control (nontreated) mice. Taken together, we have demonstrated that treatment of mice with ampicillin alters the intestinal microbiota and permits C. jejuni colonization. These findings provide valuable insights for researchers using mice to investigate C. jejuni colonization factors, virulence determinants, or the mechanistic basis of probiotics.
INTRODUCTION
Campylobacter jejuni is a leading cause of human gastroenteritis worldwide. C. jejuni is a Gram-negative pathogen that grows in low-oxygen (3 to 5%) environments, including the digestive tracts of animals. C. jejuni regularly colonizes commercial chicken flocks, and human disease is usually linked to the ingestion of food cross-contaminated with raw or undercooked poultry. The clinical symptoms for C. jejuni-mediated enteritis in humans include diarrhea with blood and leukocytes in the stool, leading to exsiccosis and electrolyte loss, fever, nausea, and abdominal cramps (1, 2). Individuals infected with C. jejuni may be treated with antibiotics, including erythromycin or ciprofloxacin (1, 2). Infection also increases the risk of developing Guillain-Barré syndrome, which is currently the leading cause of flaccid paralysis (3).
The human intestinal microbiota is comprised of hundreds of distinct bacterial species, bacteriophages, archaea, and fungi (4). The intestinal microbiota of healthy mammals is typically dominated by organisms from the phyla Firmicutes (Gram-positive bacteria) and Bacteroidetes (Gram-negative bacteria) (5). Collectively, the intestinal commensal microbiota provides the host with numerous physiological benefits, including vitamin synthesis, tissue integrity, digestion, fermentation of proteins and polysaccharides, bile salt metabolism, and stimulation of the immune system (6). One additional physiological benefit of the intestinal microbiota is the enhancement of host immune defenses by inhibiting growth of potentially pathogenic microorganisms (colonization resistance). Colonization resistance prevents pathogens from establishing a niche and inhibits the outgrowth of opportunistic pathogens (7).
Mice vary in their susceptibilities to C. jejuni and can be either completely resistant to colonization or only transiently infected. Mice devoid of intestinal microbiota (germfree) and mice with a defined microbiota (gnotobiotic) have been shown to be more susceptible to C. jejuni colonization than mice with normal intestinal microbiota. For example, C. jejuni effectively colonizes germfree mice and disseminates to immune tissues, including the mesenteric lymph nodes (MLN) (8–10). However, germfree mice demonstrate altered lymphoid development, resulting in an impaired immune response (11–13). There are documented instances that mice are susceptible to colonization with C. jejuni (14, 15). However, many researchers have experienced difficulty in obtaining C. jejuni colonization of mice unless the animals have been treated with an antibiotic prior to challenge to alter the intestinal microbiota (8, 10). To this end, mice treated with a five-antibiotic cocktail over the course of 6 weeks have been shown to be more susceptible to C. jejuni (16). Transplanting fecal material containing either human or mouse microbiota into these germfree mice demonstrated that mice given human microbiota were more susceptible to C. jejuni-mediated disease than mice given mouse microbiota (16). Thus, it is known that the murine intestinal microbiota impacts C. jejuni colonization as mice with limited flora are also more susceptible to C. jejuni (14). Collectively, these results suggest that the murine intestinal microbiota is comprised of microorganisms that specifically inhibit C. jejuni colonization.
In this study, we evaluated the contribution of resident microbiota in CBA/J mice. We found that all animals were colonized with C. jejuni following treatment with a single antibiotic (ampicillin), as measured by C. jejuni burden in the colon, spleen, and mesenteric lymph nodes (MLN). The intestinal microbiota of untreated and ampicillin-treated animals was examined by both culture-dependent and culture-independent methods. We recovered an isolate of Enterococcus faecalis from the murine intestine that inhibited C. jejuni growth in vitro and reduced C. jejuni colonization of mice. Additionally, deep sequencing of DNA extracted from murine fecal pellets revealed that the microbial community of the intestine influenced resistance to C. jejuni colonization, as a decrease in representatives of the Firmicutes phylum and an increase in representatives of the Bacteroidetes phylum were found in the animals treated with ampicillin. This study provides a simple method to alter murine intestinal microorganisms, thereby changing susceptibility to C. jejuni colonization of mice, and may be applicable for additional in vivo Campylobacter models.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The C. jejuni F38011 wild-type clinical strain, the ΔflgL mutant, and the ΔflgL/flgL complemented isolate were used throughout this study. The strains were cultured under microaerobic conditions (85% N2, 10% CO2, 5% O2) on Mueller-Hinton (MH) agar plates or in MH broth (Thermo Fisher Scientific, Hanover Park, IL) supplemented with 5% citrated bovine blood at 37°C and passaged to a fresh plate at least once each 48 h. Lactobacillus acidophilus NCFM and the murine-isolated E. faecalis were cultured on deMan-Rogosa-Sharpe (MRS) agar plates or in MRS broth (Thermo Fisher Scientific) under anaerobic conditions (86% N2, >13% CO2, <0.7% O2) at 37°C. Isolates recovered from intestinal samples were serially diluted and plated on MH agar plates or Luria-Bertani (LB) agar plates (Thermo Fisher Scientific) under microaerobic conditions or on MRS agar plates under anaerobic conditions at 37°C.
C. jejuni inoculation of mice, necropsy, and bacterial quantification.
Animal experiments were conducted according to National Institutes of Health (NIH) guidelines under supervision by the Washington State University (WSU) Animal Care and Use Committee (ASAF 04313). CBA/J mice (females, 6 to 8 weeks old) were obtained commercially from Harlan (Harlan Laboratories, Indianapolis, IN) and maintained in a specific-pathogen-free colony in microisolator cages at WSU (≤5 animals/cage). Mice were inoculated with C. jejuni (∼1010 CFU/ml in 200 μl of phosphate-buffered saline [PBS]), and control animals were given sterile PBS (uninfected) by oral gavage and observed daily for clinical signs of disease. In preliminary experiments, animals were administered 200 μl of antibiotics by oral gavage at 48 and 24 h prior to oral challenge with C. jejuni at the following concentrations: 1 mg/ml ampicillin (Fisher BioReagents, Fair Lawn, NJ), 1 mg/ml metronidazole (Acros Organics, Morris Plains, NJ), 50 μg/ml novobiocin (Sigma, St. Louis, MO), 20 μg/ml streptomycin (Sigma), and 15 μg/ml trimethoprim (Sigma). Based on the data obtained from preliminary experiments, ampicillin treatment was chosen for the remainder of the experiments. Mice were euthanized and necropsied promptly when clinical signs of disease developed or at 2, 7, and 35 days postinfection (dpi). The colon, spleen, and MLN of mice were removed aseptically and resuspended in 1 ml of MH broth. The number of C. jejuni bacteria in each organ was determined by plating serial dilutions on Campy Cefex plates. Intestinal samples were collected by excising 1-in. ileum sections. Each section was initially put in a tube containing 0.5 ml of RPMI culture medium supplemented with 1× penicillin and streptomycin (Life Technologies Corporation, Carlsbad, CA). All samples were adjusted to obtain equal weight per unit volume prior to cytokine/chemokine analysis. The second sample was placed in 10% formaldehyde for histopathological analysis. For experiments using CBA/J mice reconstituted with inhibitory bacteria, mice were treated with ampicillin at 48 h and 24 h prior to administration of inhibitory bacteria, as described above. Following ampicillin treatment, 200 μl (∼109 CFU/ml) of E. faecalis bacteria was administered to a group of mice, and L. acidophilus was administered to another group of mice by oral gavage at 24 h and 12 h prior to challenge with C. jejuni.
Cytokine/chemokine analysis.
Cytokines/chemokines were assessed in the ex vivo distal ileum sections following 24 h of incubation at 37°C with 5% CO2. Interleukin-6 (IL-6) and CXCL2/MIP-2 were assessed using a mouse IL-6 OptEIA kit (BD Biosciences, San Jose, CA) and mouse CXCL2/MIP-2 DuoSet (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions, respectively.
Gross pathology and histopathology.
Assessment of gross pathology was performed using a rubric, as described elsewhere (17). Histopathology was assessed from paraffin-embedded tissue sections (5 μm) mounted on slides and visualized by microscopy by a board-certified pathologist blinded to the samples using a Nikon Eclipse at a ×200 magnification at Washington State University's Washington Animal Disease Diagnostic Laboratory.
Isolation and sequence typing of inhibitory microbiota in CBA/J mice.
Culture-dependent microbiota analysis was performed using the bacteria recovered from the intestinal contents of mice with and without ampicillin treatment. DNA was extracted from individual colony isolates, and PCR of the 16S rRNA subunit was performed using the 27F primer (5′-AGAGTTTGATCMTGGCTCAGAACG-3′) (18) and 1435R primer (5′-CGATTACTAGCGATTCCRRCTTCA-3′) (Kelly A. Brayton, personal communication), where M is A or C and R is A or G. The primers used for sequencing included the 27F primer, 1435R primer, 533F primer (5′-GTGCCAGCMGCCGCGGTAA-3′) (19), and 519R primer (5′-GTATTACCGCGGCTGCTGG-3′) (20). Sequences were trimmed and analyzed by BLAST for identification and submitted to the National Center for Biotechnology Information (NCBI).
Multilocus sequence typing of E. faecalis was performed using primers for gdh, gyd, pstS, gki, aroE, xpt, and yqiL, as described elsewhere (21).
HPLC analysis of lactic acid production.
A chromatographic analysis method was used to determine lactic acid levels in MH broth (22, 23). The samples were analyzed via high-performance liquid chromatography (HPLC) by a standard addition method using a Hitachi L-6220 pump system with an AS-4000 auto-sampler and a PerkinElmer LC-85B UV-visible light (Vis) detector (analysis wavelength of 212 nm) controlled by Hitachi D-6000 HPLC manager software (Hitachi High Tech Americas, Inc., Schaumburg, IL). A total of 250 μl of the respective sample of growth medium was diluted with MiliQ water alone or with an addition of an appropriate aliquot of lactic acid standard to a final volume of 1.5 ml. Three samples of each growth medium were prepared for the standard addition: a diluted medium, a diluted medium with 500 mg/liter lactic acid, and a diluted medium with 1,000 mg/liter lactic acid. Each individual solution was measured in triplicate. For each analysis, 10 μl was injected onto an Acclaim Organic Acid column (5-μm particle size, 120-Å pore size, 4.0 by 120 mm) from Dionex (Thermo Scientific, Waltham, MA). An isocratic elution method was used with a mobile phase of 0.100 M Na2SO4 at a pH of 2.65 (adjusted by addition of methane sulfonic acid) and a flow rate of 0.6 ml/min at room temperature.
Determining the microbiota in CBA/J mice.
Culture-independent intestinal microbiome analysis was performed using fresh fecal pellets collected prior to and following antibiotic treatment of 15 individual CBA/J mice. The pellets were flash frozen. Genomic DNA extraction was performed using a MoBio UltraClean soil DNA isolation kit (MoBio Laboratories, Inc., Carlsbad, CA) and an MP FastPrep instrument (MP Biomedicals, Solon, OH) set at 40 s for rapid cell lysis. Each of the individually extracted DNA samples was quantified using a high-sensitivity Qubit kit (Life Technologies), and 15 ng of DNA was PCR amplified using specific bar-coded primers around the V3-V5 16S rRNA, as described previously (24). Amplified samples were sequenced using a 16S rRNA high-throughput next-generation Illumina MiSeq (Illumina, San Diego, CA) sequencing platform. A total of 42,537,848 sequence reads were generated and found suitable for further analysis. Sequence reads were aligned using the Illinois-Mayo Taxon Operations for RNA Data Set Organization (IM_TORNADO), and paired-end reads were used for determination of operational taxonomical units (OTU), as previously described (25). The OTUs were then analyzed using QIIME software for taxonomy comparison between treatment groups (26). Specific analyses included rarefaction on all samples to ensure equality of sequence number for subsequent comparisons of beta diversity (principal-component analysis [PCoA], Shannon's index, and taxonomical summary).
Statistical analysis.
All in vitro experiments consisted of at least three replicates performed on separate days to demonstrate reproducibility. Within each experiment, the samples were measured in triplicate. Results are presented as means ± standard errors of the means (SEM). Statistical analyses were performed using GraphPad Prism, version 6 (La Jolla, CA), and statistical significance was measured at a P value of ≤0.05. A nonparametric Kruskal-Wallis one-way analysis of variance (ANOVA) followed by post hoc Dunn's multiple comparisons or Tukey's test of the means was used for all in vivo assays. Results are presented as means ± SEM, and statistical significance was measured at a P value of ≤0.05. To determine differences between animals treated with ampicillin, permutational multivariate analysis of variance (PERMANOVA) or Student's t test was applied to determine statistical significance and to calculate a two-tailed P value, with a P value of ≤0.05 considered significant, using Primer, version 6, statistical software (Primer-E, Ltd., United Kingdom).
Ethics statement.
All animal work was performed using protocols approved by the Institutional Animal Care and Use Committee (IACUC; protocol no. 04313) at WSU. All efforts were made to raise and euthanize the animals humanely.
Sequence data accession numbers.
The 16S rRNA sequence of E. faecalis KLC3001 is available in GenBank under accession number KP942841. The 16S rRNA gene sequences used to determine the microbiota in CBA/J mice are available in the NCBI Sequence Read Archive (SRA) under accession number SRP057511.
RESULTS
Ampicillin-treated CBA/J mice are efficiently colonized with C. jejuni.
The intestinal microbiota of mice provide colonization resistance to C. jejuni. We tested whether antibiotics could alter the community of microorganisms to allow C. jejuni to colonize more efficiently. In preliminary experiments, mice were treated with a panel of antibiotics, including ampicillin, metronidazole, novobiocin, streptomycin, and trimethoprim, and tested for colonization resistance to C. jejuni. At 7 days postinfection (dpi), we were able to recover C. jejuni from mice treated only with ampicillin (data not shown). Furthermore, C. jejuni bacteria were not recovered from mice not given antibiotics.
Based on our preliminary findings, we investigated C. jejuni colonization in ampicillin-treated CBA/J mice. CBA/J mice are widely used as a general-purpose strain and have been shown to elicit an inflammatory response to enteric pathogens (27, 28). Ampicillin-treated CBA/J mice were challenged with the C. jejuni wild-type strain, a ΔflgL mutant, the ΔflgL/flgL complemented mutant or PBS (uninfected). Bacterial burden and markers of intestinal inflammation were assessed at 2, 7, and 35 dpi (Fig. 1). In C. jejuni, flgL encodes a flagellar hook-filament junction protein. We included the C. jejuni ΔflgL mutant in our analysis because it does not produce a functional flagellum. This mutant should be attenuated in mice as C. jejuni utilizes the flagellum for both motility and secretion of virulence proteins that are delivered to host epithelial cells (29, 30).
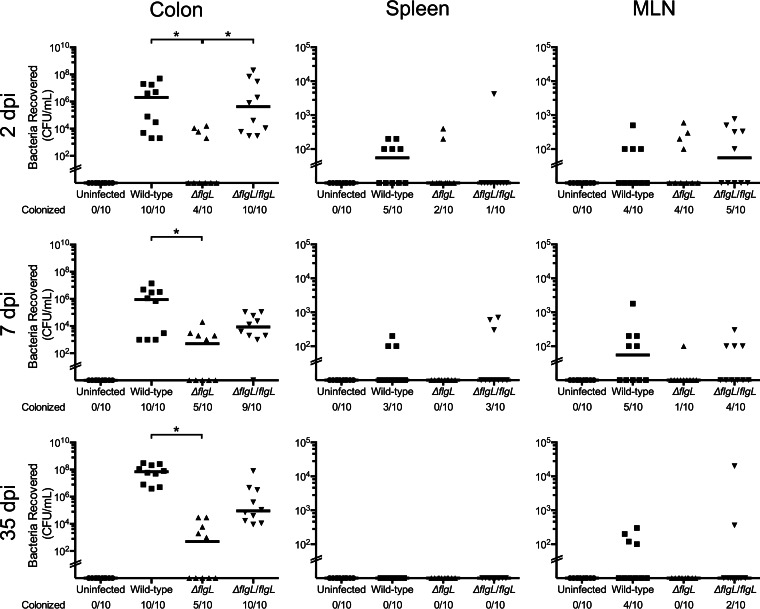
FIG 1.
C. jejuni burden in the colon, spleen, and MLN of CBA/J mice treated with ampicillin at 2, 7, and 35 dpi. CBA/J mice were treated with ampicillin and inoculated by oral gavage with a C. jejuni wild-type strain, a ΔflgL mutant, the ΔflgL/flgL isolate, or PBS (uninfected). The colon, spleen, and MLN were removed aseptically and homogenized, and serial dilutions were plated to determine CFU counts. Each data point represents the CFU/ml of C. jejuni recovered from the tissue of a single mouse. The median for each group is indicated. *, P < 0.05 for the indicated comparison (by ANOVA with Kruskal-Wallis posttest). The limit of detection was 102 CFU. MLN, mesenteric lymph nodes.
The C. jejuni wild-type strain was recovered from the colon, spleen, and MLN at all time points, with the exception that no organisms were recovered from the spleen at 35 dpi, indicating that ampicillin treatment promotes stable colonization in CBA/J mice. Although not all of the mice inoculated with the C. jejuni ΔflgL mutant completely cleared the infection, there were significantly fewer C. jejuni bacteria recovered from the colons of mice infected with the mutant than from the colons of those infected with the wild-type strain (Fig. 1). The colons of animals infected with either the C. jejuni wild-type strain or the ΔflgL/flgL complemented isolate showed signs of edema and apparent stool softening in contrast to uninfected mice or mice given the ΔflgL mutant (see Fig. S1 in the supplemental material). However, histopathological differences in ileocecocolic junction sections between animals infected with C. jejuni and uninfected controls were not obvious (see Fig. S2 in the supplemental material). Inflammatory cytokines, including the pleiotropic cytokine IL-6 and the chemotactic cytokine MIP-2 (the murine counterpart of IL-8), were assessed in ex vivo distal ileum sections to determine local inflammation within the intestinal tract. IL-6 levels were not significantly different between mice infected with the C. jejuni wild-type strain and the uninfected control at 2 dpi (2.9 ± 1.0 versus 6.7 ± 1.2 ng/g tissue), 7 dpi (9.5 ± 0.5 versus 3.3 ± 2.4 ng/g tissue), or 35 dpi (2.6 ± 1.1 versus 2.6 ± 0.9 ng/g tissue). Similarly, MIP-2 levels were not different at 2 dpi (5.3 ± 1.6 versus 8.9 ± 0.9 ng/g tissue), 7 dpi (4.6 ± 1.2 versus 4.2 ± 2.0 ng/g tissue), or 35 dpi (2.6 ± 0.7 versus 2.5 ± 0.8 ng/g tissue). Taken together, the data indicate that ampicillin-treated CBA/J mice demonstrate stable colonization with high pathogen burden and spread to extraintestinal tissues but lack the parameters of a disease model (limited histopathology and cytokine response). Based on these results, ampicillin-treated CBA/J mice provide an ideal model to investigate and identify inhibitory microbiota that normally prevents C. jejuni colonization.
Isolation of C. jejuni-inhibitory microbiota in CBA/J mice.
Peroral ampicillin treatment permitted C. jejuni to colonize CBA/J mice, likely by reducing key bacterial communities that contributed to the colonization resistance to C. jejuni. Therefore, experiments were performed to identify the bacteria that inhibit C. jejuni colonization. We collected the colonic fecal content from five mice either treated or untreated with ampicillin and serially diluted and plated the intestinal contents on three separate culture media (MHB or LB under microaerobic conditions and MRS under anaerobic conditions). We intentionally sought to recover lactic acid bacteria to demonstrate proof of principal (recovery of an ampicillin-sensitive bacterium that might inhibit C. jejuni colonization of mice) as we have previously documented that specific Lactobacillus strains inhibit C. jejuni colonization of animals (31). Thirteen visually distinct colonies were chosen for analysis (see Table S1 in the supplemental material). The viability of C. jejuni was tested by coculture with each of the individual isolates, as described elsewhere (31). Five intestinal isolates limited C. jejuni growth in vitro during coculture, and their identification and classification were determined by sequencing PCR-amplified regions of the 16S rRNA. DNA typing revealed that the three inhibitory isolates (MEK1, MEK2, and MEK3) were identified as Enterococcus faecalis (see Table S2 in the supplemental material), while MEK4 matched sequences of Rothia species and MEK5 matched Staphylococcus epidermidis. Although Rothia sp. and S. epidermidis have been isolated from the upper gastrointestinal tract, neither is considered to be a major constituent in the intestinal microbiota (32). However, E. faecalis is a major constituent of the human and murine intestinal tract. Each recovered isolate of E. faecalis was determined to be sequence type 55 (ST55), as determined by multilocus sequence typing (MLST) analysis (see Table S3 in the supplemental material). Given that the three E. faecalis isolates were indistinguishable from one another, we chose to further characterize only MEK1 (redesignated E. faecalis KLC3001). E. faecalis KLC3001 was susceptible (≥17-mm diameter) to ampicillin (10-μg susceptibility disk) in a Kirby-Bauer disk diffusion assay, with an average zone of inhibition of 33.3 ± 5.2 mm. Considering that MEK1 was sensitive to ampicillin and would likely be reduced in ampicillin-treated mice, we assessed this strain further for anti-Campylobacter activity using both in vitro and in vivo assays.
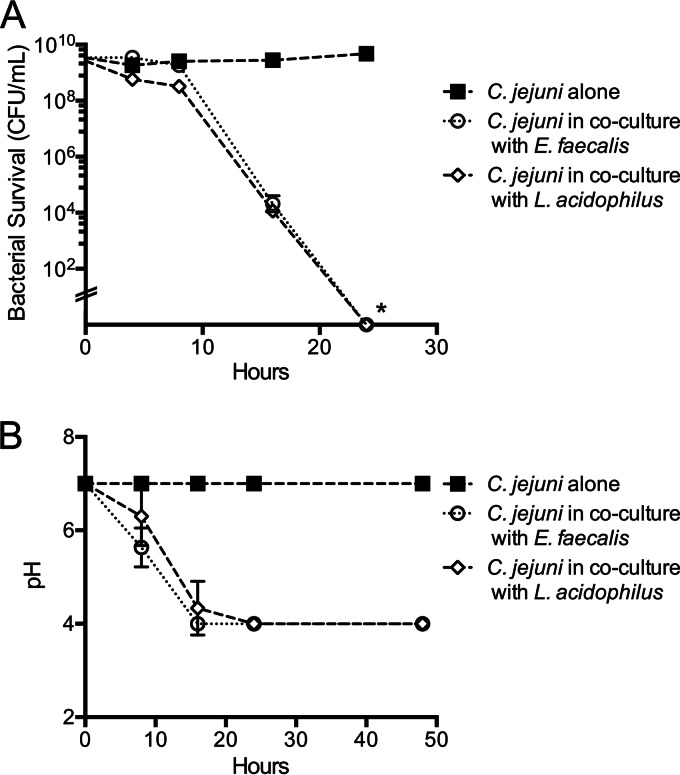
The ability of E. faecalis to mediate resistance to C. jejuni during in vitro growth assays was assessed by inoculating C. jejuni into pasteurized milk either in monoculture or coculture with E. faecalis and monitoring growth for 24 h. The reason for using pasteurized milk for this assay is because both C. jejuni and lactic acid bacteria are metabolically active in this medium (31). We also included Lactobacillus acidophilus in this assay as this bacterium has previously been shown to inhibit C. jejuni growth in vitro through production of organic acids (31). While C. jejuni was able to survive in monoculture for 24 h, the growth of C. jejuni in coculture with E. faecalis or L. acidophilus was significantly inhibited. More specifically, no viable C. jejuni bacteria were recovered at 24 h, with a detection limit of 102 CFU/ml (Fig. 2A). C. jejuni viability correlated with acidification of the medium and with the production of lactic acid by the homofermentative E. faecalis KLC3001 and L. acidophilus NCFM strains (Fig. 2B). Lactic acid production in supernatants harvested from overnight broth cultures for E. faecalis KLC3001 (210 mM ± 30 mM) and L. acidophilus NCFM (180 mM ± 25 mM) was elevated compared to the level from broth alone but not significantly different, as judged by HPLC analysis. In summary, these experiments demonstrated that E. faecalis KLC3001 inhibits the growth of the C. jejuni F38011 strain in vitro.
FIG 2.
E. faecalis inhibits C. jejuni growth during in vitro coculture. (A) C. jejuni strain F3011 was inoculated into pasteurized milk samples either in monoculture (C. jejuni alone) or in coculture with E. faecalis KLC3001 and L. acidophilus NCFM (control) and incubated for 24 h. The average number of CFU ± standard deviation is shown for each time point for at least three independent replicates. *, P < 0.05 for the comparison between results for C. jejuni alone and in coculture with either E. faecalis or L. acidophilus at 24 h (by one-way ANOVA). (B) Concurrently, the pH of each milk sample was assessed during the experiment shown in panel A.
Murine-isolated Enterococcus faecalis inhibits C. jejuni colonization of CBA/J mice.
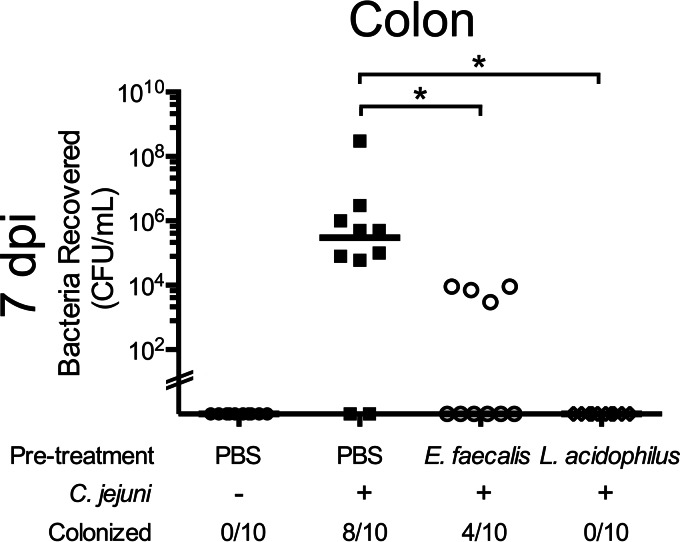
To test the hypothesis that specific bacteria mediate colonization resistance, we assessed whether the E. faecalis KLC3001 isolate could inhibit C. jejuni colonization of mice. We developed a colonization resistance model in which ampicillin-treated CBA/J mice were given oral inoculations with E. faecalis prior to infection with C. jejuni. L. acidophilus was used as a test control, and PBS-treated (uninfected) mice were used to demonstrate C. jejuni colonization. At 7 dpi, Campylobacter burden in the colon was assessed. The results indicated that there was a significant reduction in C. jejuni bacteria recovered from the colons of animals inoculated with E. faecalis prior to C. jejuni challenge compared to levels in mice given PBS (Fig. 3). Additionally, mice inoculated with L. acidophilus exhibited colonization resistance to C. jejuni. These results indicate that the E. faecalis KLC3001 strain limits C. jejuni colonization of mice.
FIG 3.
E. faecalis prevents C. jejuni colonization of CBA/J mice. CBA/J mice were treated with ampicillin, followed by oral gavage with PBS, E. faecalis KLC3001, and L. acidophilus NCFM, and then challenged with either PBS (uninfected) or C. jejuni after 48 h. At 7 dpi, the mice were euthanized, the colons were removed aseptically and homogenized, and serial dilutions were plated to determine CFU counts. Each data point represents the CFU/ml of C. jejuni recovered from the colon of a single mouse. The median for each group is indicated. *, P < 0.05 for the indicated comparison (by ANOVA with Kruskal-Wallis posttest).
Ampicillin treatment alters the intestinal microbiota in CBA/J mice.
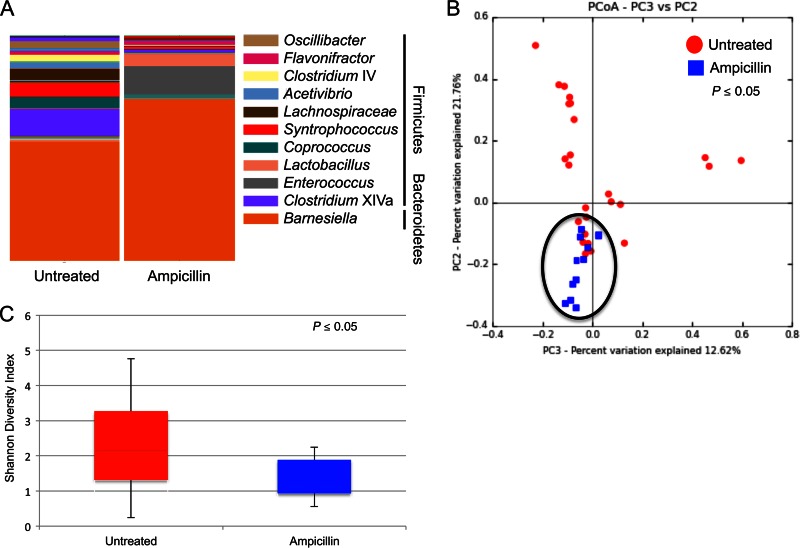
Considering the complexity of the intestinal microbiota and the limitations of culture-dependent analysis, we also assessed the culture-independent diversity of the intestinal bacterial community in mice treated or untreated with ampicillin. Fresh fecal pellets were collected from CBA/J mice treated or untreated with ampicillin, DNA was extracted, and the V3-V5 16S rRNA was amplified and sequenced using an Illumina MiSeq sequencing platform. Sequence reads were aligned and clustered into operational taxonomic units (OTUs), and differences between taxonomies were compared for each treatment group (Fig. 4A). The results indicated that ampicillin treatment of mice decreased the overall taxonomical complexity of the intestinal microbiota, with an observed increase in representatives of the Bacteroidetes (compare 52.8% of OTUs in untreated animals with 72.1% in ampicillin-treated animals), namely, Barnesiella sp. (71.9%). Furthermore, we observed a decrease in representatives of the Firmicutes in ampicillin-treated animals compared to levels in untreated mice (compare 47.2% in untreated animals with 25.9% in ampicillin-treated animals). Complete taxon identifications for all OTUs identified are listed in Table S4 in the supplemental material. In addition to taxonomic structure analysis, we assessed the OTUs common to all animals within a treatment group (i.e., the core microbiome) (see Table S5 in the supplemental material). This analysis revealed two genera present in the intestines of all untreated and ampicillin-treated CBA/J mice. This analysis also revealed seven genera present in all of the untreated mice and three genera present only in the ampicillin-treated mice. Microbial community structures were clearly impacted by ampicillin treatment, as judged by principal coordinate analysis (Fig. 4B), and this change was likely due to a significant decrease in overall diversity among community structures following ampicillin treatment (Fig. 4C). The intestinal microbial composition of animals was more similar between mice that received ampicillin treatment than between control mice (untreated). There was no significant alpha diversity (within-community diversity). These results indicate that ampicillin treatment reduces the complexity of the intestinal microbiota, including shifting the total community toward a single Bacteroidetes genus, and this shift corresponded to the observed disruption of the murine colonization resistance to C. jejuni. Ampicillin treatment changes the composition of the murine intestinal microbiota, resulting in increased C. jejuni colonization susceptibility, and enables dissemination beyond the intestine.
FIG 4.
Taxonomy summary and microbial diversity of OTUs from fecal samples of CBA/J mice with and without peroral ampicillin treatment. DNA extracted from fecal samples was used for 16S rRNA PCR amplification, sequenced, and clustered into OTUs. The taxonomy summary (A), principal coordinate analysis (B), and Shannon diversity index (C) were analyzed using QIIME software to determine differences in microbial community structures. The sequences from at least 15 animals per treatment are shown in each analysis. The taxonomy summary in panel A represents OTUs with greater than 1% total representation; OTUs with less than 1% representation are listed in Table S4 in the supplemental material. Statistical difference (P ≤ 0.05) in panel C was determined by PERMANOVA.
DISCUSSION
The intestinal tract of animals is colonized by a large number of commensal and symbiotic microorganisms, collectively known as microbiota, with levels in the colon exceeding 1011 bacterial cells per gram of fecal content (33). Colonization resistance, or the ability of the commensal microbiota to prevent colonization by exogenous pathogens or opportunistic commensals, is an important part of the host defense (34). Intestinal microbiota provide colonization resistance to pathogens in multiple ways, including competition for niches by preventing pathogens from attaching to their target sites, depleting essential nutrients for pathogen viability, producing bacteriocins or other metabolites that inhibit pathogen function, producing organic acids that alter intraluminal pH levels to acidic conditions unfavorable to pathogens, or utilizing the limited oxygen available in the gut contributing to the anaerobic and capnophilic environment (35–38). Furthermore, the commensal microbiota provides immune-mediated colonization resistance to pathogens by stimulating the development of immune cell populations involved in innate and adaptive immune processes as well as the production of antimicrobial and proinflammatory factors (39, 40). The goal of this study was to develop a simple and reproducible method that allows C. jejuni to colonize mice.
Bereswill and colleagues recently published an article that shows that mice treated with a quintuple-antibiotic cocktail develop clinical signs of C. jejuni disease (16). We first attempted to reproduce this model, with the idea of possibly refining the methods to identify a simpler method of altering the intestinal microbiota in a manner to permit C. jejuni colonization (16). However, no animals were colonized by C. jejuni (data not shown). Our results may be attributed to differences in the C. jejuni strain used or to variations in the human intestinal microbiota used to reconstitute the animals. In support of this notion, human susceptibility to infection by Campylobacter is associated with the species composition of the human intestinal microbiome (41). We then performed studies using the streptomycin ad libitum model, which has been used to investigate Salmonella pathogenesis (42). However, the animals in this model had cleared C. jejuni by 2 dpi, demonstrating that Campylobacter differs from Salmonella in this model (data not shown). Based on these findings, we initiated studies to determine whether the treatment of mice with various antibiotics would permit C. jejuni colonization. We treated mice with one of five different antibiotics (ampicillin, metronidazole, novobiocin, streptomycin, and trimethoprim). Most of these antibiotics were chosen because they have a limited bacterial spectrum of activity; the idea was to not completely disrupt the intestinal microbiota. In our preliminary experiments, ampicillin treatment of mice resulted in the greatest change (increase) in C. jejuni colonization. More specifically, we found that the treatment of CBA/J mice with ampicillin resulted in C. jejuni colonization with recovery of the pathogen from extraintestinal tissues (Fig. 1). These results are in agreement with the findings of Stahl et al., demonstrating that SIGIRR (single immunoglobulin IL-1R-related molecule)-deficient mice treated with a single antibiotic (vancomycin) can be colonized with C. jejuni (43). We also found that a functional flagellum was necessary for in vivo colonization. More specifically, we found that a ΔflgL mutant shows a significant reduction in its ability to colonize mice compared to the ability of a C. jejuni wild-type isolate. This finding is consistent with work performed with Salmonella enterica serovar Typhimurium, whereby flagellar defects were found to impair the fitness of a pathogen due to its inability to utilize the nutrients released in the inflamed intestine (44). In summary, our results demonstrate that the administration of ampicillin prior to infection is sufficient to permit C. jejuni colonization of all treated mice.
To demonstrate that ampicillin treatment of mice permits C. jejuni colonization by altering the intestinal microbiota, we performed a proof-of-concept experiment. We purposely biased our selection of microbes that might inhibit C. jejuni growth by plating the intestinal contents from mice treated with ampicillin on MRS plates (MRS medium supports the growth of lactic acid bacteria, including enterococci and lactobacilli). The E. faecalis KLC3001 isolate recovered from mice was indeed found to be sensitive to ampicillin and inhibited C. jejuni growth in vitro (Fig. 2). We also found that E. faecalis KLC3001 contributes to the in vivo colonization resistance to C. jejuni. More specifically, inoculation of ampicillin-treated mice with E. faecalis KLC3001 showed reduced C. jejuni colonization levels compared to levels in C. jejuni-inoculated mice not administered E. faecalis (Fig. 3). In comparison to the E. faecalis-inoculated mice, the bacteria recovered from the control mice at 7 dpi showed more diversity in colony morphology and fewer Enterococcus-like colonies (data not shown). Of additional interest is that the inhibitory effects of E. faecalis and L. acidophilus on C. jejuni in vitro were indistinguishable from one another, but L. acidophilus provided greater resistance to C. jejuni colonization of mice.
Given that ampicillin treatment of mice permits C. jejuni colonization by altering the intestinal microbiota, we wanted to identify the changes in the intestinal microbiota following treatment with ampicillin using deep sequencing. The principal coordinate analysis of total microbiota indicated that the microbial communities in the intestines were more similar in animals treated with ampicillin than in animals not receiving treatment (Fig. 4). Of note, the relative abundances of the two phyla (Bacteroidetes and Firmicutes) found in both ampicillin-treated and untreated CBA/J mice were significantly impacted by ampicillin treatment. Specifically, we observed that the treatment of mice with ampicillin resulted in a significant increase in representatives of the Bacteroidetes (genus Barnesiella) and a decrease in representatives of the Firmicutes (genus Clostridium XIVa). Bacteroidetes and Firmicutes are the two most prominent phyla that comprise the mouse intestinal microbiota, as well as the human intestinal microbiota, and a shift in the ratio of these phyla has been associated with many disease conditions (45–47). The influence of Barnesiella in C. jejuni colonization of mice is not known. Nevertheless, it is clear that the microbiota can influence pathogen clearance or susceptibility. For example, infant mice with elevated Escherichia coli levels are more susceptible to C. jejuni (48). The decrease in Clostridium XIVa bacteria in ampicillin-treated animals is also of interest given the findings of Atarashi and colleagues (49). The investigators reported that colonization of gnotobiotic mice with mouse-derived clostridial XIVa bacteria enhances anti-inflammatory signaling by directing the expansion of lamina propria and systemic regulatory T cells associated with secretion of the anti-inflammatory protein IL-10. Considering that C. jejuni colonizes animals with increased inflammation, including mice with an altered genetic background (IL-10−/−), the decrease in Clostridium XIVa bacteria may contribute to colonization by facilitating a favorable intestinal environment (49). Taking these results together, the overall complexity of the intestinal environment was reduced in ampicillin-treated animals.
Based on the recovery of the E. faecalis KLC3001 isolate from mice treated with ampicillin, the observation of an increase in Enterococcus taxa following ampicillin treatment by deep sequencing was enigmatic. However, isolates of E. faecalis vary considerably in their phenotypic properties, including resistance to antibiotics. Moreover, E. faecalis strains have been used as probiotics to balance the intestinal microbiota or used in dairy products (50, 51), whereas other strains are opportunistic pathogens associated with nosocomial infections (52). Consistent with our work to recover ampicillin-sensitive bacteria that could demonstrate anti-Campylobacter activity, Robyn and coworkers found that E. faecalis (strain MB 5229) has anti-Campylobacter activity in vitro. However, the anti-Campylobacter activity demonstrated by this strain was ineffective in broiler chickens (53). Our results indicate that E. faecalis KLC3001 has anti-Campylobacter activity in vivo. Based on the striking differences in the intestinal microbiota from the untreated and ampicillin-treated mice, as judged by deep-sequence analysis, there are undoubtedly additional culturable and nonculturable microorganisms that influence Campylobacter colonization.
The effects of antibiotics on intestinal microbiota are profound. Although our results demonstrate that ampicillin treatment is associated with an increase in C. jejuni colonization, the exact mechanism is unknown. The direct and indirect effects of antibiotic treatment on the intestinal community structure can be long lasting and results in an alteration in the total number of bacteria as well as a change in the composition and balance of specific bacteria in the community (54, 55). It is well known that a shift in community structure following antibiotic treatment can alter the function of the community, including disease susceptibility and nutrient acquisition (56). For example, antibiotic treatment has been shown to promote Clostridium difficile colonization (57, 58). In contrast, the administration of a cocktail of six microorganisms to mice has been shown to provide C. difficile colonization resistance and to inhibit stable residence (58). Reeves and colleagues also found that mice precolonized with a murine Lachnospiraceae isolate had significantly decreased C. difficile colonization compared to mice colonized with E. coli (57). In addition to altering the intestinal community, antibiotic treatment may cause bacterial lysis and the release of carbon sources as well as an increase in the level of bile acid that could be critical for pathogen colonization (59). It is likely that if two organisms are trying to occupy the same niche, the organism that most efficiently competes for the limiting nutrient will be successful (59). Moreover, if an organism already occupies a niche, an invading organism will be at a disadvantage to compete for nutrients. Furthermore, antibiotic treatment disrupts this relationship, resulting in an alteration in the ability of the endogenous community to limit colonization of a pathogen.
Our results demonstrate that ampicillin alters the intestinal microbiota, thereby allowing C. jejuni to colonize the intestinal tract. Additionally, we show that ampicillin-treated mice given E. faecalis have restored colonization resistance to C. jejuni. It is likely that there are other bacterial species involved in the murine colonization resistance to C. jejuni, as evidenced by the efficacy of L. acidophilus to prevent C. jejuni colonization. A potential next step is to look at different C. jejuni strains and their susceptibility or resistance to colonization. Genetically modified mice may also benefit from ampicillin treatment, as IL-10−/− mice cleared of microbiota with a multiantibiotic cocktail demonstrated increased C. jejuni-mediated disease leading to death (60). In conclusion, we believe our findings will allow researchers to better explore C. jejuni-host interactions, whether the focus is probiotic inhibition or virulence assessment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Galen J. Gorence and Jan E. Luft for animal care and assistance at Washington State University. We thank Christopher Gourley, Mark Nissen, Jason Neal-McKinney, and Nicholas Negretti for technical assistance. We thank Kelly Brayton for providing the sequences of all primers, including primer 1435R, used for 16S rRNA gene amplification.
This work was supported by funds awarded by the NIH to M.E.K. (R56 AI088518-01A1). J.L.O. was supported, in part, by funds awarded by the NIH T32 Training Program in Infectious Diseases and Microbial Immunology (5 T32 AI 7025-33). D.R.S. and T.P.E. were supported, in part, by funds awarded by the National Institute of General Medical Sciences ([NIGMS] T32GM083864 and T32GM008336, respectively). A.G.B.-F. and B.A.W. were supported with discretionary funds from B.A.W.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the NIGMS.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00281-15.
REFERENCES
- 1.Blaser M. 1990. Campylobacter species, p 1649–1658. In Mandell GL, Douglas RG, Bennett JE (ed), Principles and practice of infectious diseases, 3rd ed Churchill Livingstone, New York, NY. [Google Scholar]
- 2.Allos BM. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis 32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 3.Buzby JC, Allos BM, Roberts T. 1997. The economic burden of Campylobacter-associated Guillain-Barre syndrome. J Infect Dis 176(Suppl 2):S192–S197. doi: 10.1086/513785. [DOI] [PubMed] [Google Scholar]
- 4.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. 2011. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol 19:349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Blaut M., Clavel T. 2007. Metabolic diversity of the intestinal microbiota: implications for health and disease. J Nutr 137:751S–755S. [DOI] [PubMed] [Google Scholar]
- 7.Lawley TD, Walker AW. 2013. Intestinal colonization resistance. Immunology 138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jesudason MV, Hentges DJ, Pongpech P. 1989. Colonization of mice by Campylobacter jejuni. Infect Immun 57:2279–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Youssef M, Corthier G, Goossens H, Tancrede C, Henry-Amar M, Andremont A. 1987. Comparative translocation of enteropathogenic Campylobacter spp. and Escherichia coli from the intestinal tract of gnotobiotic mice. Infect Immun 55:1019–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee A, O'Rourke JL, Barrington PJ, Trust TJ. 1986. Mucus colonization as a determinant of pathogenicity in intestinal infection by Campylobacter jejuni: a mouse cecal model. Infect Immun 51:536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savidge TC, Smith MW, James PS, Aldred P. 1991. Salmonella-induced M-cell formation in germ-free mouse Peyer's patch tissue. Am J Pathol 139:177–184. [PMC free article] [PubMed] [Google Scholar]
- 12.Shroff KE, Cebra JJ. 1995. Development of mucosal humoral immune responses in germ-free (GF) mice. Adv Exp Med Biol 371A:441–446. [DOI] [PubMed] [Google Scholar]
- 13.Szeri I, Anderlik P, Banos Z, Radnai B. 1976. Decreased cellular immune response of germ-free mice. Acta Microbiol Acad Sci Hung 23:231–234. [PubMed] [Google Scholar]
- 14.Chang C, Miller JF. 2006. Campylobacter jejuni colonization of mice with limited enteric flora. Infect Immun 74:5261–5271. doi: 10.1128/IAI.01094-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaser MJ, Hopkins JA, Berka RM, Vasil ML, Wang WL. 1983. Identification and characterization of Campylobacter jejuni outer membrane proteins. Infect Immun 42:276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bereswill S, Fischer A, Plickert R, Haag LM, Otto B, Kuhl AA, Dasti JI, Zautner AE, Munoz M, Loddenkemper C, Gross U, Gobel UB, Heimesaat MM. 2011. Novel murine infection models provide deep insights into the “menage a trois” of Campylobacter jejuni, microbiota and host innate immunity. PLoS One 6:e20953. doi: 10.1371/journal.pone.0020953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gourley CR, Konkel ME. Campylobacter jejuni molecular mechanisms and animal models of colonization and disease. In Singh SK. (ed), Emerging and re-emerging human infections: genome to infectome, in press. Wiley-Blackwell, Oxford, United Kingdom. [Google Scholar]
- 18.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Hoboken, NJ. [Google Scholar]
- 19.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner S, Pryer KM, Miao VP, Palmer JD. 1999. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol 46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Garbajosa P, Bonten MJ, Robinson DA, Top J, Nallapareddy SR, Torres C, Coque TM, Canton R, Baquero F, Murray BE, del Campo R, Willems RJ. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J Clin Microbiol 44:2220–2228. doi: 10.1128/JCM.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubey UK, Mistry VV. 1996. Effect of bifidogenic factors on growth characteristics of bifidobacteria in infant formulas. J Dairy Sci 79:1156–1163. doi: 10.3168/jds.S0022-0302(96)76469-X. [DOI] [PubMed] [Google Scholar]
- 23.van Der Wielen PW, Biesterveld S, Notermans S, Hofstra H, Urlings BA, van Knapen F. 2000. Role of volatile fatty acids in development of the cecal microflora in broiler chickens during growth. Appl Environ Microbiol 66:2536–2540. doi: 10.1128/AEM.66.6.2536-2540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walther-Antonio MR, Jeraldo P, Berg Miller ME, Yeoman CJ, Nelson KE, Wilson BA, White BA, Chia N, Creedon DJ. 2014. Pregnancy's stronghold on the vaginal microbiome. PLoS One 9:e98514. doi: 10.1371/journal.pone.0098514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schachtschneider KM, Yeoman CJ, Isaacson RE, White BA, Schook LB, Pieters M. 2013. Modulation of systemic immune responses through commensal gastrointestinal microbiota. PLoS One 8:e53969. doi: 10.1371/journal.pone.0053969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reingold L, Rahal K, Schmiedlin-Ren P, Rittershaus AC, Bender D, Owens SR, Adler J, Zimmermann EM. 2013. Development of a peptidoglycan-polysaccharide murine model of Crohn's disease: effect of genetic background. Inflamm Bowel Dis 19:1238–1244. doi: 10.1097/MIB.0b013e31828132b4. [DOI] [PubMed] [Google Scholar]
- 28.Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, Baumler AJ. 2012. Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3(3):e00143-12. doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neal-McKinney JM, Konkel ME. 2012. The Campylobacter jejuni CiaC virulence protein is secreted from the flagellum and delivered to the cytosol of host cells. Front Cell Infect Microbiol 2:31. doi 10.3389/fcimb.2012.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samuelson DR, Eucker TP, Bell JA, Dybas L, Mansfield LS, Konkel ME. 2013. The Campylobacter jejuni CiaD effector protein activates MAP kinase signaling pathways and is required for the development of disease. Cell Commun Signal 11:79. doi: 10.1186/1478-811X-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neal-McKinney JM, Lu X, Duong T, Larson CL, Call DR, Shah DH, Konkel ME. 2012. Production of organic acids by probiotic lactobacilli can be used to reduce pathogen load in poultry. PLoS One 7:e43928. doi: 10.1371/journal.pone.0043928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zamakhchari M, Wei G, Dewhirst F, Lee J, Schuppan D, Oppenheim FG, Helmerhorst EJ. 2011. Identification of Rothia bacteria as gluten-degrading natural colonizers of the upper gastro-intestinal tract. PLoS One 6:e24455. doi: 10.1371/journal.pone.0024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu J, Gordon JI. 2003. Honor thy symbionts. Proc Natl Acad Sci U S A 100:10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thijm HA, van der Waaij D. 1979. The effect of three frequently applied antibiotics on the colonization resistance of the digestive tract of mice. J Hyg 82:397–405. doi: 10.1017/S0022172400053924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalliomaki MA, Walker WA. 2005. Physiologic and pathologic interactions of bacteria with gastrointestinal epithelium. Gastroenterol Clin North Am 34:383–399, vii. doi: 10.1016/j.gtc.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, Hinton JC, Van Immerseel F. 2006. Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl Environ Microbiol 72:946–949. doi: 10.1128/AEM.72.1.946-949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cherrington CA, Hinton M, Pearson GR, Chopra I. 1991. Short-chain organic acids at pH 5.0 kill Escherichia coli and Salmonella spp. without causing membrane perturbation. J Appl Bacteriol 70:161–165. doi: 10.1111/j.1365-2672.1991.tb04442.x. [DOI] [PubMed] [Google Scholar]
- 38.Marteyn B, Scorza FB, Sansonetti PJ, Tang C. 2011. Breathing life into pathogens: the influence of oxygen on bacterial virulence and host responses in the gastrointestinal tract. Cell Microbiol 13:171–176. doi: 10.1111/j.1462-5822.2010.01549.x. [DOI] [PubMed] [Google Scholar]
- 39.Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, Schwab SR, Littman DR. 2013. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature 494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dicksved J, Ellstrom P, Engstrand L, Rautelin H. 2014. Susceptibility to Campylobacter infection is associated with the species composition of the human fecal microbiota. mBio 5(5):e01212-14. doi 10.1128/mBio.01212-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stahl M, Ries J, Vermeulen J, Yang H, Sham HP, Crowley SM, Badayeva Y, Turvey SE, Gaynor EC, Li X, Vallance BA. 2014. A novel mouse model of Campylobacter jejuni gastroenteritis reveals key pro-inflammatory and tissue protective roles for Toll-like receptor signaling during infection. PLoS Pathog 10:e1004264. doi: 10.1371/journal.ppat.1004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stecher B, Barthel M, Schlumberger MC, Haberli L, Rabsch W, Kremer M, Hardt WD. 2008. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol 10:1166–1180. doi 10.1111/j.1462-5822.2008.01118.x. [DOI] [PubMed] [Google Scholar]
- 45.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, Beaugerie L, Cosnes J, Corthier G, Marteau P, Dore J. 2009. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 47.Hansen J, Gulati A, Sartor RB. 2010. The role of mucosal immunity and host genetics in defining intestinal commensal bacteria. Curr Opin Gastroenterol 26:564–571. doi: 10.1097/MOG.0b013e32833f1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haag LM, Fischer A, Otto B, Plickert R, Kuhl AA, Gobel UB, Bereswill S, Heimesaat MM. 2012. Intestinal microbiota shifts towards elevated commensal Escherichia coli loads abrogate colonization resistance against Campylobacter jejuni in mice. PLoS One 7:e35988. doi: 10.1371/journal.pone.0035988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holzapfel WH, Haberer P, Geisen R, Bjorkroth J, Schillinger U. 2001. Taxonomy and important features of probiotic microorganisms in food and nutrition. Am J Clin Nutr 73:365S–373S. [DOI] [PubMed] [Google Scholar]
- 51.Giraffa G, Sisto F. 1997. Susceptibility to vancomycin of enterococci isolated from dairy products. Lett Appl Microbiol 25:335–338. doi: 10.1046/j.1472-765X.1997.00241.x. [DOI] [PubMed] [Google Scholar]
- 52.Christoffersen TE, Jensen H, Kleiveland CR, Dorum G, Jacobsen M, Lea T. 2012. In vitro comparison of commensal, probiotic and pathogenic strains of Enterococcus faecalis. Br J Nutr 108:2043–2053. doi: 10.1017/S0007114512000220. [DOI] [PubMed] [Google Scholar]
- 53.Robyn J, Rasschaert G, Hermans D, Pasmans F, Heyndrickx M. 2013. In vivo broiler experiments to assess anti-Campylobacter jejuni activity of a live Enterococcus faecalis strain. Poult Sci 92:265–271. doi: 10.3382/ps.2012-02712. [DOI] [PubMed] [Google Scholar]
- 54.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun 77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson CJ, Bohannan BJ, Young VB. 2010. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev 74:453–476. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. 2012. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun 80:3786–3794. doi: 10.1128/IAI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, Deakin LJ, Pickard DJ, Duncan SH, Flint HJ, Clark TG, Parkhill J, Dougan G. 2012. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Britton RA, Young VB. 2014. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology 146:1547–1553. doi: 10.1053/j.gastro.2014.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haag LM, Fischer A, Otto B, Plickert R, Kuhl AA, Gobel UB, Bereswill S, Heimesaat MM. 2012. Campylobacter jejuni induces acute enterocolitis in gnotobiotic IL-10−/− mice via Toll-like receptor-2 and -4 signaling. PLoS One 7:e40761. doi: 10.1371/journal.pone.0040761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.