Abstract
Biocontrol agents isolated outside Africa have performed inconsistently under field conditions in Africa. The development of indigenous phytobeneficial microbial strains that suit local environments may help enhance competitiveness with in situ microorganisms and effectiveness at suppressing local pathogen strains. We isolated bacteria from the rhizosphere of maize growing in southwestern Nigeria and assessed them for growth-promoting characteristics. The best isolates were characterized using 16S rRNA genes and were further evaluated in the greenhouse on maize seedlings. Four isolates (EBS8, IGBR11, EPR2, and ADS14) were outstanding in in vitro assays of antagonistic activity against a local strain of Fusarium verticillioides, phosphate solubilization efficiency, chitinase enzyme activity, and indole-3-acetic acid production. Inoculation of maize seeds with these isolates resulted in ≥95% maize seed germination and significantly enhanced radicle and plumule length. In the greenhouse, maize seedling height, stem girth, number of leaves, leaf area, shoot mass (dry matter), and nutrient contents were significantly enhanced. The bioprotectant and phytobeneficial effects were strongest and most consistent for isolate EBS8, which was identified as a Bacillus strain by 16S rRNA gene analysis. As a bacterial strain that exhibits multiple growth-promoting characteristics and is adapted to local conditions, EBS8 should be considered for the development of indigenous biological fertilizer treatments.
INTRODUCTION
Maize was traditionally grown as a subsistence crop on small plots in home gardens in West and Central Africa, but it has recently expanded into a local “cash crop,” especially in southwestern Nigeria, where >30% of cropland is devoted to maize production (1). However, maize productivity is often low in Nigeria due to a combination of agrobiological (2), climatic (3, 4), and technological (5) factors coupled with high postharvest losses (6). In addition to continued soil nutrient limitation (5), diseases such as downy mildew, rust, leaf blight, leaf spot, maize streak virus, and root, stalk, collar, and ear rots (7, 8) are among the major factors that limit maize production in southwestern Nigeria. Thus, national and international bodies have raised a global call to promote maize production through the management of biological communities associated with agroecosystems, which is an environmentally friendly and cost-effective strategy (4). In this regard, the use of plant growth-promoting rhizobacteria (PGPR) can play an important role in developing sustainable systems of crop production (9). PGPR can play a pivotal role in crop production by means of siderophore production, antagonism to soilborne root pathogens, phosphate solubilization, and dinitrogen fixation (10). PGPR are also known to alter root morphology and enhance the uptake of essential nutrients by plants (10).
Despite the potential benefits of using rhizobacteria to enhance crop protection (11), they remain largely untapped in the effort to improve maize production in Africa. In addition, biocontrol agents isolated outside the region and imported to Africa have performed inconsistently under field conditions (12). The development of indigenous biocontrol strains that suit local environments may help enhance competitiveness with in situ microorganisms and effectiveness at suppressing local pathogen strains (13). Similarly, Howell (14) suggested that biocontrol agents should be isolated from the soil locality where they are expected to function in disease control. In addition, too few phytobeneficial indigenous rhizobacterial isolates been identified using molecular methods in Nigeria. Molecular characterization of isolates using 16S rRNA genes is necessary to gain insight into the taxonomy of beneficial rhizobacteria and the properties of related strains prior to field application. This study was therefore undertaken to assess the phytobeneficial effects and biotechnological properties of indigenous bacterial isolates with a view to the improvement of local maize production. Isolates were initially screened in vitro for potential plant growth-promoting characteristics, and then select isolates were tested for their effects on maize seed germination and seedling growth.
MATERIALS AND METHODS
Collection of soil samples and bacterial isolation.
Soil was collected from several fields where maize had been grown for 5 to 10 years, cutting across different ecological zones in southwestern Nigeria (Guinea savannah, derived savannah, lowland rainforest, freshwater swampy forest, mangrove forest/coastal vegetation). In each field, rhizosphere soils adhering to maize roots at a depth of 5 to 15 cm were collected in five locations and were mixed together to form a composite soil sample. A serial-dilution–pour plate technique was used to isolate bacteria (15) on nutrient agar (NA; Oxoid Chemicals, Loughborough, United Kingdom). Inoculated petri plates were incubated at 25 ± 2°C for 24 h. Isolates differing in morphological appearance on NA were selected and were streaked onto new plates until pure cultures were obtained. Pure cultures of bacterial isolates were maintained on NA slants and were stored at 4°C.
Prior to measurement of the growth-promoting characteristics of isolates, preliminary identification was carried out using biochemical tests. We performed Gram staining and tests for catalase, starch hydrolysis, casein hydrolysis, growth in 4% NaCl, gelatin hydrolysis, and sugar fermentation by following standard protocols (16).
Growth-promoting characteristics of isolates.
The following in vitro plant growth-promoting characteristics were measured: pathogen-antagonistic bioassay, phosphate solubilization, chitinase enzyme activity, and indole-3-acetic acid (IAA) production.
An in vitro bioassay (17) evaluated the antagonistic potential of the bacterial isolates against Fusarium verticillioides, which was obtained from the Institute of Agricultural Research and Training (IAR&T) Plant Pathology Unit, Moore Plantation, Ibadan, Nigeria. Nutrient yeast broth agar (NYBA) (8.0 g nutrient broth, 2.0 g yeast extract, 2.0 g K2HPO4, 2.0 g KH2PO4, 2.5 g glucose, and 15.0 g agar per liter of water, adjusted to pH 6.7) plates were inoculated with two 5-mm agar plugs of F. verticillioides at opposite sides of each plate. After 24 h of incubation, an agar plug of one bacterial isolate was placed upside down at the center of each plate so that the bacterial culture was in direct contact with the agar plate. Five replicate plates were inoculated with each bacterial isolate and were incubated at 28 ± 2°C for 7 days. Results were scored after 7 days for antagonist-pathogen interaction. The diameter of the zone of inhibition of fungal growth around the point of bacterial inoculation was measured in millimeters and was compared to that on plates with no bacteria.
The phosphate solubilization abilities of the isolates were tested as described by Sharma et al. (18). Five grams of CaHPO4 was used as the source of phosphate in agar plates that also contained 2.5 g of glucose, 1 g of MgSO4·7H2O, and 20 g of agar per liter (pH 6.8). Each isolate was spot-inoculated onto five replicate plates and was incubated at 28 ± 2°C for 48 h. The halo zone (medium clearing) surrounding the colonies was measured, and the phosphate solubilization efficiency (PSE), expressed as a percentage, was calculated as (solubilization diameter × 100)/(growth diameter).
Each isolate was screened for chitinase production according to the method of El-Mehalawy et al. (19). Colloidal chitin agar (CCA) was prepared by a method modified from that of Lima et al. (20). The medium contained 1.0 g bacteriological peptone, 0.3 g urea, 1.4 g (NH4)2SO4, 0.3 g MgSO4·7H2O, 0.3 g CaCl2·6H2O, 1.0 g glucose, 15.0 g colloidal chitin prepared from crab shell chitin (21), 1 ml trace element solution (1 mM Fe2+, Zn2+, and Co2+), and 20.0 g agar (Oxoid Chemicals) per liter of water, adjusted to pH 6.0. Each isolate was inoculated on five replicate CCA plates and was incubated at 28 ± 2°C for 48 h, when zones of chitin clearing were seen around colonies, indicating chitinase activity. The chitinase enzyme activity (CEA) of each isolate, expressed as a percentage, was calculated as (clear zone diameter × 100)/(growth diameter).
The potential of each isolate to produce IAA was measured according to the work of Khakipour et al. (22). Isolates were grown in triplicate in tryptophan nutrient broth (5 g tryptophan per liter of nutrient broth; Oxoid Chemicals) and were incubated with shaking for 48 h at 28 ± 2°C. Visually turbid cultures were centrifuged at 4°C for 10 min at 14,462 × g. Then 2 ml of the supernatant was mixed with 4 ml of Salkowsky reagent (50 ml 35% perchloric acid, 1 ml 0.5 M FeCl3 solution), and the mixture was kept in the dark. After 20 min, light absorption (540 nm) by this mixture was measured using a spectrophotometer. Light absorption was compared to a standard curve in order to determine IAA production by each isolate in milligrams per liter.
Ribosomal sequencing and phylogeny of isolates.
Each bacterium was grown in nutrient agar broth culture at 25 ± 2°C for 24 h and was then centrifuged at 4,722 × g for 5 min. The pellet was washed with phosphate-buffered saline (PBS) twice and was then resuspended in 200 μl of PBS for DNA extraction. DNA was extracted by using the ZR Fungal/Bacterial DNA MiniPrep kit (Zymo Research, Irvine, CA, USA) according to the manufacturer's instructions. The 16S rRNA gene was amplified using universal primers for bacteria (23). PCR was carried out in 25-μl volumes containing 1.5 mM MgCl2, 0.125 U/μl Taq polymerase, 2.5 mM each deoxynucleoside triphosphate (dNTP), 0.25 μM forward primer (8f) (5′-AGAGTTTGATCCTGGCTCAG-3′), 0.25 μM reverse primer (1392R) (5′-ACGGGCGGTGTGTAC-3′), and ∼0.1 ng template DNA. PCR was performed under the following conditions: 10 min at 95°C, followed by 30 cycles of 45 s at 95°C, 30 s at 54°C, and 90 s at 72°C, followed by a final extension at 72°C for 5 min. The PCR-amplified samples were loaded onto a 1.5% agarose gel and were run at 220 V for 45 min. Gels were stained with ethidium bromide for 15 min and were visualized with a UV transilluminator.
PCR products were purified using the UltraClean PCR Clean-Up kit (Mo Bio Laboratories) according to the manufacturer's instructions. The quantity of DNA was then determined using a PicoGreen DNA assay (Invitrogen). DNA standards were prepared using Lambda DNA (Promega, Madison, WI). Fluorescence was quantified using a microplate reader (BioTek Instruments, Winooski, VT, USA). DNA was then sequenced at the Plant-Microbe Genomics Facility at Ohio State University. The sequences were taxonomically classified using the Na¿ve Bayesian Classifier implemented by the Ribosomal Database Project (RDP) (24). Ribosomal sequences were edited using Bioedit, version 7.0.5 (25). Reference sequences representing nearest neighbors were obtained from the RDP and were included in phylogenetic analysis. Evolutionary distances were computed using the maximum composite likelihood method (26). A phylogenetic tree was constructed using MEGA 5 software (27). Evolutionary history was inferred using the neighbor-joining method (28). The tree topologies were evaluated by bootstrap analysis (29) based on 500 resamplings.
Seed germination bioassay.
The maize variety most commonly grown in southwestern Nigeria (SUWAN-1-Y) was used for laboratory and greenhouse experiments. Maize seeds were surface sterilized with 0.5% NaOCl for 2 min, followed by 30 s in 70% ethanol and two rinses in distilled water, followed by air drying (30). Bacterial inocula were prepared by incubating bacterial cultures for 24 h and were diluted with sterile distilled water to give a concentration of approximately 106 cells ml−1 (106 CFU ml−1) adjusted with a hemocytometer. Seeds were inoculated with an isolate by immersion in a suspension of bacteria containing 106 CFU ml−1 for 30 min and were then dried in a laminar flow cabinet for 1 to 2 h. The effect of each bacterial isolate on maize seed germination was then measured using blotter techniques (31). This experiment was carried out in five replicated petri plates, and the result was compared with that for control seeds treated with water instead of a bacterial isolate. Ten seeds inoculated with each bacterium were placed in 9-cm-diameter petri dishes lined with sterilized moistened filter paper and were incubated for 7 days at 28 ± 2°C. Germinated seeds were counted at day 7. The average radicle and plumule lengths for each petri dish were also recorded for calculation of the vigor index (32). The vigor index was calculated as (mean of plumule + radicle lengths) × germination rate.
Bioprotectant effect of isolates in the presence of F. verticillioides.
An isolate-pathogen interaction study was conducted in sterilized sandy-loam soil using the pathogenic fungus F. verticillioides (33, 34). Topsoil was collected from the Moore Plantation, IAR&T, Ibadan, Nigeria, and was sieved through 2-mm mesh to remove plant roots and debris. The experiment was set up in a greenhouse with plastic pots (15 cm in diameter) containing 2.5 kg of hot-steam-sterilized soil. Treatments were replicated five times in a completely randomized design. A conidial suspension of phytopathogenic F. verticillioides was prepared and was adjusted to 1 × 106 spores/ml using a hemocytometer. Four milliliters of F. verticillioides inoculum was mixed with 50 ml sterile water and was used to directly inoculate the soil (35), after which pots were immediately covered with black polyethylene bags for 48 h. To prepare maize seeds, presterilized SUWAN-1-Y seeds were coated with gum arabic (Sigma-Aldrich, Seelze, Germany) as an adhesive and were mixed with each bacterial cell suspension (1 × 106 cells/ml) until uniformly coated, while a mixture of maize and water was used as the control. Three inoculated maize seeds were planted per pot at a depth of 2 cm and were immediately covered with soil. Soil was held at 43.5% of water-holding capacity. Watering and weeding were done throughout the experiment. After 21 days, disease symptoms were assessed according to the following indices. Disease incidence (DI) was calculated as the percentage of maize seedlings in a pot that showed visible signs of infection (36). Disease severity (DS) was scored qualitatively based on the observable symptoms in the most diseased plant in each pot, with slight modifications (37, 38). The scores were as follows: 0, apparently healthy seedling; 1, 1 leaf infected; 2, 2 to 3 leaves infected/traces of stem rot; 3, all leaves infected/stunted growth/stem rot; and 4, damping off/wilting/seedling death. The disease reduction percentage (DRP) was calculated using the following formula (39): DRP = 100 × [1 − (DI of treatment/DI of controls)].
Phytobeneficial effects of isolates on maize seedling health and growth.
Unsterilized sandy-loam soil was used in a greenhouse experiment similar to that described above to determine the effects of bacterial isolates on maize growth in the presence of naturally occurring pathogens and other soil microbes (35). Maize seeds (SUWAN-1-Y) were surface sterilized and were inoculated with each bacterial isolate as described above. Three inoculated maize seeds were planted per pot in 2.5 kg unsterilized soil. Control pots were planted with seeds that received no bacterial inoculum. Treatments were replicated five times and were completely randomized. Pots were maintained at 34.5% of soil water-holding capacity. Watering and weeding were done throughout the experimental period. Disease expression (DE) was calculated as the percentage of maize seedlings in each pot showing visible signs of infection. The average maize seedling height (in centimeters), stem girth (in centimeters), number of leaves, leaf area (in square centimeters), and shoot mass (g dry matter) were also determined for each pot. After the experiment, maize seedling shoots were collected, dried at 70°C to a constant weight, and then ground using Wiley ED-5 milling equipment to enable passage through a 2-mm sieve. Nutrient analysis was performed on seedling tissue that was pooled across individuals in each pot at the Analytical Laboratory of the International Institute of Tropical Agriculture, Ibadan, Nigeria, where 2 g of ground dried samples was digested in a hot sulfuric acid solution using black selenium powder as a catalyst (40). The digested solutions were read colorimetrically in a Technicon AutoAnalyzer II instrument (41) for the simultaneous determination of nitrogen and phosphorus contents. The potassium content was determined by flame photometry (42).
Statistical analyses.
Experimental treatments were compared using SAS software, version 9.1 (SAS Institute, Cary, NC, USA) (43). Nineteen bacterial isolate treatments were evaluated for in vitro growth-promoting characteristics. Dependent variables in laboratory bioassay experiments were subjected to analysis of variance (ANOVA), followed by post hoc pairwise comparisons using the Student-Newman-Keuls multiple-range test. The four best-performing isolates were then used in the experiments examining effects on maize seed germination and seedling growth in the greenhouse. The effects of isolates on maize were also compared by ANOVA and the Student-Newman-Keuls test.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the NCBI GenBank database under accession numbers KP792804 to KP792815.
RESULTS
Growth-promoting characteristics of isolates.
Nineteen bacterial isolates obtained from rhizosphere soil samples were selected based on their cultural and morphological differences (data not shown). All 19 isolates displayed some antagonistic activity against F. verticillioides on NYBA in vitro (Table 1). The isolates with the greatest antagonistic activity were EBS8 (diameter of zone of inhibition, 10.6 mm), EPR2 (7.5 mm), IGBR11 (7.4 mm), and ADS14 (7.6 mm). Except for EPR3, all isolates showed clear halos of phosphate and chitin solubilization around points of inoculation (Table 1). EBS8 had the highest phosphate solubilization efficiency (69.5%), followed by ADS14 (62.4%). Similarly, EPR2 had the highest chitinase enzyme activity (69.6%), followed by ADS14 (45.6%). The largest amount of IAA was produced by ADS14 (10.1 mg/liter), followed by IGBR11 (7.2 mg/liter).
TABLE 1.
Growth-promoting characteristics of isolatesa
| Isolateb | Antagonistic effect (diam of zone of inhibition [mm])c | Phosphate solubilization efficiency (%) | Chitinase enzyme activity (%) | IAA production (mg/liter) |
|---|---|---|---|---|
| ABS6 | 5.3 ± 0.1 d | 55.7 ± 0.0 e | 13.7 ± 0.5 h | 2.0 ± 0.0 h |
| ADS14* | 7.6 ± 0.1 b | 62.3 ± 0.1 b | 45.6 ± 0.1 b | 10.1 ± 0.0 a |
| AKR5 | 2.0 ± 0.0 e | 38.3 ± 0.2 j | 1.4 ± 0.0 j | 2.2 ± 0.1 h |
| AT-IKS | 1.1 ± 0.0 f | 22.9 ± 0.8 m | 1.7 ± 0.0 j | 7.4 ± 0.2 b |
| AT-ILR | 2.2 ± 0.0 e | 37.4 ± 0.3 j | 2.1 ± 0.0 j | 4.8 ± 0.0 f |
| AT-SKR | 1.1 ± 0.0 f | 29.5 ± 0.3 l | 1.2 ± 0.1 j | 1.6 ± 0.2 i |
| EBS8* | 10.6 ± 0.0 a | 69.5 ± 0.4 a | 42.2 ± 0.2 c | 5.5 ± 0.2 e |
| EPR2* | 7.5 ± 0.3 b | 61.9 ± 0.0 b | 69.6 ± 0.0 a | 6.3 ± 0.1 d |
| EPR3 | 1.6 ± 0.5 ef | 0.0 ± 0.0 n | 0.0 ± 0.0 j | 1.1 ± 0.1 i |
| EPR4 | 5.2 ± 0.0 d | 60.4 ± 0.4 c | 26.7 ± 0.2 f | 6.1 ± 0.0 d |
| EPR7 | 2.1 ± 0.0 e | 31.1 ± 0.0 k | 13.8 ± 0.8 h | 5.4 ± 0.2 e |
| IBS8 | 5.1 ± 0.0 d | 52.4 ± 0.2 f | 35.1 ± 2.9 d | 4.7 ± 0.0 f |
| IGBR11* | 7.4 ± 0.3 b | 62.0 ± 0.0 b | 41.4 ± 0.4 c | 7.2 ± 0.0 b |
| IGGR11 | 6.2 ± 0.0 c | 57.6 ± 0.2 d | 20.3 ± 0.0 g | 6.4 ± 0.0 cd |
| ILS13 | 6.7 ± 0.2 c | 44.9 ± 0.7 g | 20.6 ± 0.1 g | 6.8 ± 0.0 bc |
| IPR1 | 5.1 ± 0.0 d | 41.8 ± 0.8 hi | 11.1 ± 0.0 i | 6.5 ± 0.0 cd |
| OSR7 | 5.3 ± 0.2 d | 40.7 ± 0.4 i | 31.8 ± 0.0 e | 2.6 ± 0.2 h |
| TDS9 | 5.3 ± 0.2 d | 42.3 ± 0.1 h | 14.7 ± 0.0 h | 5.8 ± 0.0 de |
| UNS9 | 5.4 ± 0.2 d | 55.2 ± 0.4 e | 21.3 ± 0.1 g | 3.6 ± 0.4 g |
| None | 0.0 ± 0.0 g | 0.0 ± 0.0 n | 0.0 ± 0.0 j | 0.0 ± 0.0 j |
Three isolates were used for testing IAA production and five for the other assays. Values are means ± standard deviations. Values followed by different letters within a column indicate significant differences according to the Student-Newman-Keuls multiple-range test (α = 0.05).
Isolates followed by an asterisk were chosen for further characterization.
In vitro antagonistic effect against F. verticillioides.
Molecular characterization of isolates.
Twelve bacterial isolates were selected for 16S ribosomal sequencing based on their growth-promoting characteristics. The RDP Naïve Bayesian Classifier assigned sequences to the following genera: Myroides (isolates AT-IKS, EPR4, IPR1, and TDS9), Enterobacter (IGBR11, OSR7), Bacillus (EBS8, ILS13), Lysinibacillus (EPR2), Citrobacter (ADS14), Stenotrophomonas (UNS9), and unclassified Pseudomonadaceae (ABS6). All sequences except ABS6 were classified to genus level with a confidence of >80%.
The phylogenetic positions of the four isolates with the greatest growth-promoting characteristics (EBS8, IGBR11, EPR2, and ADS14) were further investigated by the construction of a phylogenetic tree using representative sequences obtained from RDP. After construction of the phylogenetic tree, the four isolates were separated into four distinct clusters based on their genera (Fig. 1). The bootstrap support of the relationships between EBS8, EPR2, ADS14, and IGBR11 and their nearest neighbors was 94%, 99%, 98%, and 86%, respectively (Fig. 1).
FIG 1.
Phylogenetic relationship of EBS8, EPR2, ADS14, and IGBR11, based on 16S rRNA genes and inferred using the neighbor-joining method. Type strains used for comparison are given. Numbers above each node are bootstrap confidence levels (expressed as percentages) generated from 500 bootstrap trees.
Maize seed germination.
Of the 19 isolates evaluated, four (EBS8, EPR2, IGBR11, and ADS14) exhibited consistent growth-promoting characteristics and were chosen for further evaluation in association with maize. In a seed germination assay, each of these bacterial isolates enhanced maize seed germination (Table 2). These bacteria also significantly enhanced both radicle and plumule length (P, <0.05, except for the effect of IGBR11 on plumule length), with ADS14 and EBS8 inducing the largest effects. The best vigor index was obtained for seeds inoculated with isolate EBS8 (Table 2).
TABLE 2.
Effects of isolates on maize seed germination and vigor indexa
| Treatment | % Germination | Radicle length (cm) | Plumule length (cm) | Vigor index |
|---|---|---|---|---|
| Control | 72.5 (3.5) b | 2.7 (0.1) d | 1.2 (0.0) d | 326.4 (0.1) e |
| Isolate | ||||
| EBS8 | 95.0 (0.0) a | 8.7 (0.4) a | 2.8 (0.3) b | 1,037.7 (5.0) a |
| EPR2 | 95.0 (0.0) a | 3.9 (0.0) c | 1.8 (0.1) c | 498.1 (0.5) c |
| IGBR11 | 95.0 (0.0) a | 5.9 (0.2) b | 1.4 (0.1) d | 643.8 (0.2) b |
| ADS14 | 90.0 (0.0) a | 8.3 (0.3) a | 3.4 (0.1) a | 403.0 (0.6) d |
Results are mean values (standard deviations) for five replicates. Values followed by different letters within a column indicate significant differences according to the Student-Newman-Keuls multiple-range test (α = 0.05).
Suppression of diseases in maize due to F. verticillioides.
Control seedlings exposed to F. verticillioides but not inoculated with any bacteria expressed diseases such as leaf curl, leaf wilting, leaf blight, and stem rot and were often dead at day 21 (Table 3). In contrast, isolates EBS8 and IGBR11 showed outstanding antifungal effects and complete reduction of disease symptoms (Table 3). Isolate EPR2 also significantly reduced disease severity in maize seedlings (29% disease reduction relative to the controls), with seedlings showing traces of only leaf curl. Inoculation with isolate ADS14 resulted in 19% disease reduction, with seedlings showing traces of both leaf curl and stem rot.
TABLE 3.
Disease assessment in maize seedlings inoculated with different bacterial isolates and then germinated in soil exposed to F. verticillioidesa
| Treatment | Disease incidence (%) | Disease severity score | Disease reduction (%) |
|---|---|---|---|
| Control | 100.0 (0.0) a | 4.0 (0.0) a | 0.0 (0.0) c |
| Isolate | |||
| EBS8 | 0.0 (0.0) c | 0.0 (0.0) c | 100.0 (0.0) a |
| EPR2 | 30.0 (4.1) b | 1.5 (0.7) b | 29.0 (4.1) b |
| IGBR11 | 0.0 (0.0) c | 0.0 (0.0) c | 100.0 (0.0) a |
| ADS14 | 20.1 (0.1) bc | 1.0 (0.0) bc | 19.0 (0.0) b |
Results are mean values (standard deviations) for five replicates. Values followed by different letters within a column indicate significant differences according to the Student-Newman-Keuls multiple-range test (α = 0.05).
Phytobeneficial effects of isolates.
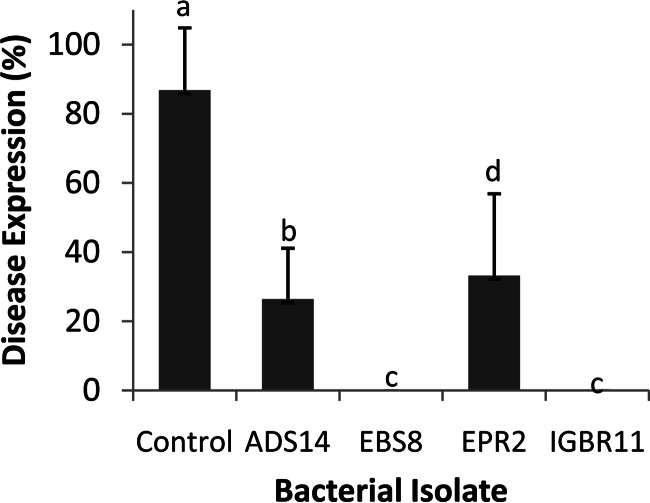
In the greenhouse experiment with unsterilized soil, the level of disease expression in control maize seedlings that received no bacterial inoculum was 90% (Fig. 2). All isolates provided good protection from disease relative to the controls, but only seedlings inoculated with isolate EBS8 or IGBR11 had 0% disease expression, with no evidence of physical disease symptoms. Maize seedlings inoculated with bacterial isolates had significant increases in shoot mass (dry matter), height, stem girth, and number of leaves over those of the control (P = 0.05) (Table 4). Isolate EBS8 produced a shoot mass (dry matter) nearly four times that of the control and double that obtained with isolate IGBR11, although IGBR11-inoculated seedlings had significantly higher leaf area (Table 4). In addition, isolates EPR2 and ADS14 enhanced maize seedling shoot mass (dry matter) 3-fold over that of the control. All bacterial isolates enhanced nitrogen content in maize seedling shoots (Table 4); EBS8 inoculation resulted in the highest N concentration (3.9 ppm), followed by ADS14 (3.5 ppm). EBS8 was also the only isolate that significantly increased both phosphorus and potassium contents in maize seedling shoots (Table 4).
FIG 2.
Disease expression (expressed as a percentage) of maize seedlings inoculated with different bacterial isolates and planted in unsterilized soil. Mean values for five replicates are shown. Error bars, standard deviations. Different letters above the bars indicate significant differences according to the Student-Newman-Keuls multiple-range test (α = 0.05).
TABLE 4.
Phytobeneficial effects of bacterial isolates on maize seedling growtha
| Treatment | Height (cm) | Stem girth (cm) | No. of leaves | Leaf area (cm2) | Shoot mass (g dry matter) | Nutrient content (ppm) |
||
|---|---|---|---|---|---|---|---|---|
| N | P | K | ||||||
| Control | 23.7 (0.4) a | 0.4 (0.3) b | 7.0 (0.0) b | 225.2 (24.7) c | 1.8 (0.0) d | 1.4 (0.1) d | 0.8 (0.0) b | 2.3 (0.3) d |
| Isolate | ||||||||
| EBS8 | 29.0 (2.2) a | 1.4 (0.1) a | 8.0 (0.0) a | 242.0 (1.1) bc | 7.1 (0.6) a | 3.9 (0.0) a | 1.1 (0.0) a | 4.5 (0.5) b |
| EPR2 | 28.7 (2.1) a | 1.5 (0.0) a | 8.0 (0.3) a | 284.7 (4.6) b | 4.7 (0.1) b | 1.8 (0.1) c | 0.6 (0.1) c | 6.4 (0.2) a |
| IGBR11 | 24.6 (0.1) a | 1.1 (0.0) a | 8.0 (0.3) a | 337.4 (0.7) a | 3.5 (0.1) c | 1.8 (0.1) c | 0.4 (0.1) d | 1.7 (0.1) d |
| ADS14 | 27.8 (3.0) a | 1.8 (0.3) a | 8.0 (0.0) a | 276.5 (14.5) b | 4.0 (0.1) c | 3.5 (0.1) b | 0.4 (0.0) d | 3.6 (0.3) c |
Results are means (standard deviations) for five replicates. Values followed by different letters within a column indicate significant differences according to the Student-Newman-Keuls multiple-range test (α = 0.05).
DISCUSSION
In this study, we have isolated and characterized rhizosphere bacterial strains related to the genera Bacillus (isolate EBS8), Citrobacter (ADS14), Enterobacter (IGBR11), and Lysinibacillus (EPR2) that show great promise both for the control of maize diseases caused by F. verticillioides and for the improvement of maize nutrition. Laditi et al. (44) have shown that indigenous Nigerian soil microbial isolates performed better at enhancing maize shoot mass (dry matter) than imported commercial isolates marketed as bioinoculants, which proved ineffective. Thus, although our isolates need further testing under field conditions, we have confidence in their transferability to the field, because the strains were isolated from the same environment in which they are intended to be used (southwestern Nigeria).
A great deal of effort has gone into the identification of effective biocontrol agents with multiple plant growth-promoting traits (10, 45). We have taken an integrated approach, similar to that of Ahmad et al. (13), testing isolates for a variety of plant growth-promoting characteristics. This provides insight into the functional differences between isolates and is necessary for careful selection of beneficial indigenous isolates. Some of our isolated rhizobacteria exhibited more than one plant growth-promoting trait, which is expected to be advantageous for seedling growth under multiple types of adverse conditions (10).
All four of the isolates on which we focused demonstrated antifungal potential in both in vitro and soil-based assays. Remarkably, isolates EBS8 and IGBR11 completely suppressed disease symptoms caused by a native soil community or by inoculated F. verticillioides. The biocontrol activity of these isolates may result from their ability to produce the largest amounts of chitinase, or other undetermined lytic enzymes, among the isolates tested (46, 47). Their effectiveness may also be related to the fact that they were isolated from the rhizosphere of the host plant that they were intended to protect (14), and thus, they had previous exposure to indigenous pathogens and competitor rhizosphere bacteria. Interestingly, the high in vitro antifungal activity and high chitinase activity produced by isolates EPR2 and ADS14 was not as indicative of biocontrol activity in soil assays as it was for isolates EBS8 and IGBR11. It is known that tests based on in vitro mycelial growth inhibition and root colonization do not always correlate with biocontrol efficacy under natural conditions (46, 48). The production of antifungal metabolites is subject to complex regulation by an array of environmental factors, and these metabolites may not be equally expressed under in vitro and natural soil conditions (49, 50).
Maize inoculated with the four isolates had higher germination rates than the control, a result similar to findings for sorghum (51) and pearl millet (52). Although suppression of seed pathogens could be involved in this improvement in seed germination, these findings may also be due to the synthesis of hormones such as IAA by the isolates in this study (53). IAA can trigger the activity of specific enzymes that promote early germination and increased plumule and radicle length (54), and seed inoculation with IAA-producing rhizobacteria has been shown to enhance early seedling establishment (55). Particular isolates may also have been involved in the production and metabolism of auxin, which is responsible for cellular elongation (56), or cytokinin, which stimulates cellular division (57).
The significant enhancement in maize growth parameters (height, girth, and leaf number) could result from biological activity of the isolates such as antagonizing plant pathogens, synthesizing phytohormones, and increasing the availability and uptake of nutrients (58). In Nigeria, P deficiency is a serious threat to maize growth (59). One of the secondary objectives of this study was to evaluate indigenous soil bacteria for their potential in helping to reduce P deficiencies in Nigerian maize fields through inoculants capable of mineral P solubilization. This was found to be a common trait: all the bacterial isolates tested were able to solubilize CaHPO4, which indicates that bacterial isolates will likely be useful components in the sustainable production of maize in Nigeria (60).
In comparison to other isolates, EBS8 exhibited not only a strong bioprotectant effect but also good physiological growth-promoting characteristics on maize seedlings. Bacillus species have been reported previously in the rhizosphere of maize (61) and have been shown to act as bioprotectants and plant growth-promoting bacteria (10). Isolate EBS8 was effective at enhancing nitrogen content in maize seedlings, as reported previously for Bacillus megaterium on wheat (62). Apart from nitrogen, maize seedling shoots inoculated with isolate EBS8 had the highest phosphorus contents, whereas maize inoculated with isolate IGBR11 or ADS14 had a phosphorus content even lower than that of the control. This is an indication that isolates IGBR11 and ADS14 lack the ability to solubilize phosphorus in an unsterilized soil. The high levels of phosphate solubilization by isolate EBS8 are similar to the outstanding performance of Bacillus strain BPR7 reported by Kumar et al. (63).
Our findings confirm that rhizosphere bacteria have the potential to exhibit multiple growth-promoting traits that can directly influence seedling establishment and growth (10, 63). The rhizosphere of maize grown in Nigeria has proven to be a promising environment in which to search for bacteria that can be developed into indigenous phytobeneficial bacteria of potential use in the management and sustainability of food crops. Further work with these strains is now needed in field trials, under different local environmental conditions, with additional methods of inoculation (such as incorporation into the soil), and with the full participation of farmers. Exploration of indigenous phytobeneficial bacteria may benefit the sustainability of food crops in diverse countries.
ACKNOWLEDGMENTS
We thank the International Institute of Tropical Agriculture, Nigeria, Cereals Unit of the Institute of Agricultural Research & Training (IAR&T) for technical support. The West African Research Association of Boston University, Boston, MA, USA, provided a grant for partial support of this work. This study was also supported by grants from the U.S. National Science Foundation (DEB-0918240 and DEB-0918878) and the U.S. Department of Energy (DE-SC000433).
We also thank Suhana Chattopadhyay and Matthew Gacura for assistance with molecular and bioinformatics methods.
REFERENCES
- 1.Alabi RA, Onolemhemhen PO. 2001. Relative economic advantage of maize-soybean mixed cropping. Niger J Agric Bus Rural Dev 2:13–21. [Google Scholar]
- 2.IITA. 2009. CGIAR research program MAIZE. http://www.iita.org/projects-asset;jsessionid=1B6ED4A781EEE57FA3D351254FDC17C2?p_p_id=101_INSTANCE_9Kn9&p_p_lifecycle=0&p_p_state=normal&p_p_mode=view&p_p_col_id=column-1&p_p_col_pos=2&p_p_col_count=4&_101_INSTANCE_9Kn9_struts_action=%2Fasset_publisher%2Fview_content&_101_INSTANCE_9Kn9_urlTitle=cgiar-research-program-maize&_101_INSTANCE_9Kn9_type=content&redirect=%2Fprojects.
- 3.Agbola T, Ojeleye D. 2007. Climate change and food crop production in Ibadan, Nigeria. Afr Crop Sci Conf Proc 8:1423–1433. [Google Scholar]
- 4.CIMMYT. 2010. DT maize will greatly profit African farmers. CIMMYT E-News 7:2. [Google Scholar]
- 5.Iken JE, Amusa NA. 2004. Maize research and production in Nigeria. Afr J Biotechnol 3:302–307. doi: 10.5897/AJB2004.000-2056. [DOI] [Google Scholar]
- 6.Agoda S, Atanda S, Usanga OE, Ikotun I, Isong IU. 2011. Post-harvest food losses reduction in maize production in Nigeria. Afr J Agric Res 6:4833–4839. [Google Scholar]
- 7.White JF, Bacon CW. 2000. Microbial endophytes. Marcel Dekker, New York, NY. [Google Scholar]
- 8.Andrés-Ares JL, Alonso-Ferro RC, Campo-Ramírez L, Moreno-González J. 2004. Fusarium graminearum Schwabe, a maize root and stalk rot pathogen isolated from lodged plants in northwest Spain. Span J Agric Res 2:249–252. doi: 10.5424/sjar/2004022-82. [DOI] [Google Scholar]
- 9.Shoebitz M, Ribaudo CM, Pardo MA, Cantore ML, Ciampi L, Cura JA. 2009. Plant growth promoting properties of a strain of Enterobacter ludwigii isolated from Lolium perenne rhizosphere. Soil Biol Biochem 41:1768–1774. doi: 10.1016/j.soilbio.2007.12.031. [DOI] [Google Scholar]
- 10.Hayat R, Ali S, Amara U, Khalid R, Ahmed I. 2010. Soil beneficial bacteria and their role in plant growth promotion: a review. Ann Microbiol 60:579–598. doi: 10.1007/s13213-010-0117-1. [DOI] [Google Scholar]
- 11.Talubnak C, Soytong K. 2010. Biological control of vanilla anthracnose using Emericella nidulans. J Agric Technol 6:47–55. [Google Scholar]
- 12.Debananda SN, Suchitra S, Salam N. 2009. Screening of actinomycete isolates from niche habitats in Manipur for antibiotic activity. Am J Biochem Biotechnol 5:221–225. doi: 10.3844/ajbbsp.2009.221.225. [DOI] [Google Scholar]
- 13.Ahmad F, Ahmad I, Khan MS. 2008. Screening of free-living rhizobacteria for their multiple plant growth promoting activities. Microbiol Res 163:173–181. doi: 10.1016/j.micres.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Howell CR. 2003. Mechanisms employed by Trichoderma spp. in the biological control of plant diseases: the history and evolution of current concepts. Plant Dis 87:4–10. doi: 10.1094/PDIS.2003.87.1.4. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds J. 30 September 2005. Serial dilution protocols. ASM MicrobeLibrary. http://www.microbelibrary.org/library/laboratory-test/2884-serial-dilution-protocols.
- 16.Harrigan WF, McCance ME. 1996. Laboratory methods in microbiology. Academic Press, New York, NY. [Google Scholar]
- 17.Tomas MJE, Simone PMD, Congergado F, Suarez FG. 1980. Methods to assess antagonism of soil microorganisms towards fungal spore germination. Soil Biol Biochem 12:197–198. doi: 10.1016/0038-0717(80)90060-7. [DOI] [Google Scholar]
- 18.Sharma K, Dak G, Agrawal A, Bhatnagar M, Sharma R. 2007. Effect of phosphate solubilizing bacteria on the germination of seeds and seedling growth. J Herb Med Toxicol 1:61–63. [Google Scholar]
- 19.El-Mehalawy AA, Hassanein NM, Khater HM, Karam El-Din EA, Yousef YA. 2004. Influence of maize root colonization by the rhizosphere actinomycetes and yeast fungi on plant growth and on the biological control of late wilt disease. Int J Agric Biol 6:599–605. [Google Scholar]
- 20.Lima LHC, Ulhoa CJ, Fernandes AP, Felix CR. 1997. Purification of a chitinase from Trichoderma spp. and its action on Sclerotium rolfsii and Rhizoctonia solani cell walls. J Gen Appl Microbiol 43:31–37. doi: 10.2323/jgam.43.31. [DOI] [PubMed] [Google Scholar]
- 21.Hsu SC, Lockwood IL. 1975. Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl Microbiol 29:422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khakipour N, Khavazi K, Mojallali H, Pazira E, Asadirahmani H. 2008. Production of auxin hormone by fluorescent pseudomonads. Am Eurasian J Agric Environ Sci 4:687–692. [Google Scholar]
- 23.Blackwood CB, Oaks A, Buyer JS. 2005. Phylum- and class-specific PCR primers for general microbial community analysis. Appl Environ Microbiol 71:6193–6198. doi: 10.1128/AEM.71.10.6193-6198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Xiao M, Geng X, Liu J, Chen J. 2007. Horizontal transfer of genetic determinants for degradation of phenol between the bacteria living in plant and its rhizosphere. Appl Microbiol Biotechnol 77:733–739. doi: 10.1007/s00253-007-1187-2. [DOI] [PubMed] [Google Scholar]
- 25.Brown JW. 1999. The ribonuclease P database. Nucleic Acids Res 27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 30.Zinniel DK, Lambrecht P, Harris NB, Zhengyu F, Kuczmarski D, Higley P, Ishimaru CA, Arunakumari A, Barletta RG, Vidaver AK. 2002. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl Environ Microbiol 68:2198–2208. doi: 10.1128/AEM.68.5.2198-2208.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ISTA. 1999. International rules for seed testing. Seed Sci Technol 27(Suppl):333. [Google Scholar]
- 32.Zucconi F, Pera A, Forte M, De Bertoldi M. 1981. Evaluating toxicity of immature compost. Biocycle 2:54–57. [Google Scholar]
- 33.Fredricks DN, Relman DA. 1996. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin Microbiol Rev 9:18–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baniasadi F, Shahidi Bonjar GH, Baghizadeh A, Nik AK, Jorjandi M, Aghighi S, Farokhi PR. 2009. Biological control of Sclerotinia sclerotiorum, causal agent of sunflower head and stem rot disease, by use of soil borne actinomycetes isolates. Am J Agric Biol Sci 4:146–151. doi: 10.3844/ajabssp.2009.146.151. [DOI] [Google Scholar]
- 35.Ros M, Hernandez MT, Garcia C, Bernal A, Pascual JA. 2005. Biopesticide effect of green compost against Fusarium wilt on melon plants. J Appl Microbiol 98:845–854. doi: 10.1111/j.1365-2672.2004.02508.x. [DOI] [PubMed] [Google Scholar]
- 36.Michel VV, Wang JF, Midmore DY, Hartman GL. 1997. Effect of intercropping and soil amendment with urea and calcium oxide on the incidence of bacterial wilt of tomato and survival of soil-borne Pseudomonas solanacearum in Taiwan. Plant Pathol 46:600–610. doi: 10.1046/j.1365-3059.1997.d01-45.x. [DOI] [Google Scholar]
- 37.Soonthompoct P, Trevathan LE, Gonzalez MS, Tomaso-Peterson M. 2001. Fungal occurrence, disease incidence and severity, and yield of maize symptomatic for seedling disease in Mississippi. Mycopathologia 150:39–46. doi: 10.1023/A:1011032801808. [DOI] [PubMed] [Google Scholar]
- 38.Gwary DM, Saleh B, Gwary SD. 2006. Evaluation of new pearl millet lines to Maiduguri pathotype of Sclerospora graminicola. Int J Agric Biol 8:597–601. [Google Scholar]
- 39.Cao Y, Zhang Z, Ling N, Yuan Y. 2011. Bacillus subtilis SQR 9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol Fertil Soils 47:495–506. doi: 10.1007/s00374-011-0556-2. [DOI] [Google Scholar]
- 40.Novozamsky I, Houba VJG, van Eck R, van Vark W. 1983. A novel digestion technique for multi-element plant analysis. Commun Soil Sci Plant Anal 14:239–248. doi: 10.1080/00103628309367359. [DOI] [Google Scholar]
- 41.IITA. 1989. Automated and semi-automated methods for soil and plant analysis. Manual series no. 7. IITA, Ibadan, Nigeria. [Google Scholar]
- 42.Okalebo JR, Gathua KW, Woomer PL. 1993. Laboratory method of soil and plant analysis: a working manual. Tropical Soil Biology and Fertility Programme (TSBF), Nairobi, Kenya. [Google Scholar]
- 43.SAS (Statistical Analysis System). 2009. SAS/STAT guide for personal computers, version 9.2. SAS Institute Incorporated, Cary, NC. [Google Scholar]
- 44.Laditi MA, Nwoke OC, Jemo M, Abaidoo RC, Ogunjobi AA. 2012. Evaluation of microbial inoculants as biofertilizers for the improvement of growth and yield of soybean and maize crops in savanna soils. Afr J Agric Res 7:405–413. doi: 10.5897/AJAR11.904. [DOI] [Google Scholar]
- 45.Weller DM, Thomashow LS. 1993. Use of rhizobacteria for biocontrol. Curr Opin Biotechnol 4:306–311. doi: 10.1016/0958-1669(93)90100-B. [DOI] [Google Scholar]
- 46.Nihorimbere V, Ongena M, Cawoy H, Brostaux B, Kakana P, Jourdan E, Thonart P. 2010. Beneficial effects of Bacillus subtilis on field-grown tomato in Burundi: reduction of local Fusarium disease and growth promotion. Afr J Microbiol Res 4:1135–1142. [Google Scholar]
- 47.Fernando WGD, Nakkeeran S, Zhang Y, Savchuk S. 2007. Biological control of Sclerotinia sclerotiorum (Lib.) de Bary by Pseudomonas and Bacillus species on canola petals. Crop Prot 26:100–107. doi: 10.1016/j.cropro.2006.04.007. [DOI] [Google Scholar]
- 48.Williams GE, Asher MJC. 1996. Selection of rhizobacteria for the control of Pythium ultimum and Aphanomyces cochlioides on sugar-beet seedlings. Crop Prot 15:479–486. doi: 10.1016/0261-2194(96)00014-2. [DOI] [Google Scholar]
- 49.Duffy BK, Défago G. 1999. Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl Environ Microbiol 65:2429–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inam-ul-Haq M, Javed M, Ahmad R, Rehman A. 2003. Evaluation of different strains of Pseudomonas fluorescens for the biocontrol of Fusarium wilt of chickpea. Pak J Plant Pathol 2:65–74. doi: 10.3923/ppj.2003.65.74. [DOI] [Google Scholar]
- 51.Raju NS, Niranjana SR, Janardhana GR, Prakash HS, Shetty HS, Mathur SB. 1999. Improvement of seed quality and field emergence of Fusarium moniliforme infected sorghum seeds using biological agents. J Sci Food Agric 79:206–212. doi:. [DOI] [Google Scholar]
- 52.Niranjan SR, Deepak SA, Basavaraju P, Shetty HS, Reddy MS, Kloepper JW. 2003. Comparative performance of formulations of plant growth promoting rhizobacteria in growth promotion and suppression of downy mildew in pearl millet. Crop Prot 22:579–588. doi: 10.1016/S0261-2194(02)00222-3. [DOI] [Google Scholar]
- 53.Ng LC, Sariah M, Sariam O, Radziah O, Zainal Abidin MA. 2012. Rice seed bacterization for promoting germination and seedling growth under aerobic cultivation system. Aust J Crop Sci 6:170–175. [Google Scholar]
- 54.Kaufman PB, Wu LL, Brock TG, Kim K. 1995. Hormones and the orientation of growth, p 547–570. In Davies PJ. (ed), Plant hormones: physiology, biochemistry, and molecular biology. Kluwer Academic, Dordrecht, Netherlands. [Google Scholar]
- 55.Khalid A, Arshad M, Zahir ZA. 2004. Screening plant growth-promoting rhizobacteria for improving growth and yield of wheat. J Appl Microbiol 96:473–480. doi: 10.1046/j.1365-2672.2003.02161.x. [DOI] [PubMed] [Google Scholar]
- 56.Bharathi R, Vivekananthan R, Harish S, Ramanathan A, Samiyappan R. 2004. Rhizobacteria-based bio-formulations for the management of fruit rot infection in chillies. Crop Prot 23:835–843. doi: 10.1016/j.cropro.2004.01.007. [DOI] [Google Scholar]
- 57.Anzala FJ. 2006. Contrôle de la vitesse de germination chez le maïs (Zea mays L.): etude de la voie de biosynthèse des acides aminésissus de l'aspartate et recherche de QTLs. Ph.D. thesis University of Angers, Angers, France. [Google Scholar]
- 58.Shaukat K, Affrasayab S, Hasnain S. 2006. Growth responses of Triticum aestivum to plant growth promoting rhizobacteria used as a biofertilizer. Res J Microbiol 1:330–338. [Google Scholar]
- 59.Ayodele OJ, Omotoso SO. 2008. Nutrient management for maize production in soils of the savannah zone of south-western Nigeria. Int J Soil Sci 3:20–27. doi: 10.3923/ijss.2008.20.27. [DOI] [Google Scholar]
- 60.Zaidi A, Khan MS, Ahmad M, Oves M. 2009. Plant growth promotion by phosphate solubilizing bacteria. Acta Microbiol Immunol Hung 56:263–284. doi: 10.1556/AMicr.56.2009.3.6. [DOI] [PubMed] [Google Scholar]
- 61.Gao Z, Zhuang J, Chen J, Liu X, Tang S. 2004. Population of endophytic bacteria in maize roots and its dynamic analysis. Ying Yong Sheng Tai Xue Bao 15:1344–1348. (In Chinese.) [PubMed] [Google Scholar]
- 62.El-Komy HMA. 2005. Coimmobilization of Azospirillum lipoferum and Bacillus megaterium for successful phosphorus and nitrogen nutrition of wheat plants. Food Technol Biotechnol 43:270–277. [Google Scholar]
- 63.Kumar P, Dubey RC, Maheshwari DK. 2012. Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol Res 167:493–499. doi: 10.1016/j.micres.2012.05.002. [DOI] [PubMed] [Google Scholar]