Abstract
This study determined the effect of feed fermentation with Lactobacillus reuteri on growth performance and the abundance of enterotoxigenic Escherichia coli (ETEC) in weanling piglets. L. reuteri strains produce reuteran or levan, exopolysaccharides that inhibit ETEC adhesion to the mucosa, and feed fermentation was conducted under conditions supporting exopolysaccharide formation and under conditions not supporting exopolysaccharide formation. Diets were chosen to assess the impact of organic acids and the impact of viable L. reuteri bacteria. Fecal samples were taken throughout 3 weeks of feeding; at the end of the 21-day feeding period, animals were euthanized to sample the gut digesta. The feed intake was reduced in pigs fed diets containing exopolysaccharides; however, feed efficiencies did not differ among the diets. Quantification of L. reuteri by quantitative PCR (qPCR) detected the two strains used for feed fermentation throughout the intestinal tract. Quantification of E. coli and ETEC virulence factors by qPCR demonstrated that fermented diets containing reuteran significantly (P < 0.05) reduced the copy numbers of genes for E. coli and the heat-stable enterotoxin in feces compared to those achieved with the control diet. Any fermented feed significantly (P < 0.05) reduced the abundance of E. coli and the heat-stable enterotoxin in colonic digesta at 21 days; reuteran-containing diets reduced the copy numbers of the genes for E. coli and the heat-stable enterotoxin below the detection limit in samples from the ileum, the cecum, and the colon. In conclusion, feed fermentation with L. reuteri reduced the level of colonization of weaning piglets with ETEC, and feed fermentation supplied concentrations of reuteran that may specifically contribute to the effect on ETEC.
INTRODUCTION
The combined stress of weaning and movement to a different environment increases the potential for poor growth performance, nutrient malabsorption, and diseases in piglets (1–3). Diarrhea caused by enterotoxigenic Escherichia coli (ETEC) is a major disease of weaning piglets (4). ETEC establishes infection by specific fimbria mediating intestinal attachment and colonization (4–7). Following colonization, ETEC produces enterotoxins that induce watery diarrhea (8, 9). Control of ETEC infections of weanling piglets is currently achieved by antibiotics (10). The use of antibiotics in animal production, however, selects for antibiotic-resistant intestinal microbiota and favors the transfer of antibiotic resistance genes from livestock microbiota to human pathogens (7). Consequently, antimicrobial growth promoters have been banned in several jurisdictions (11), challenging the livestock industry to replace feed antibiotics without compromising animal performance or animal health.
Probiotic bacteria, prebiotics, organic acids, or antiadhesive glycans were proposed to replace feed antibiotics in pig production for improved control of ETEC (4, 6). Organic acids were shown to reduce postweaning diarrhea in pigs (12). Probiotics also decreased the incidence and severity of ETEC-caused diarrhea (13, 14). Neoglycans of porcine albumin conjugated with galacto-oligosaccharides reduced ETEC attachment in vitro (15). ETEC adhesion in vitro was also reduced by reuteran and levan, exopolysaccharides produced by Lactobacillus reuteri (16). The protective effect of reuteran was confirmed in vivo in the small intestinal segment loop perfusion model (17). Evidence for the effectiveness of probiotic cultures or antiadhesive glycans for the prevention of ETEC infection in vivo, however, remains limited. Moreover, a combination of different additives is likely required to obtain effective and economically viable alternatives for feed antibiotics (18, 19). Feed fermentation can deliver a combination of viable and probiotic lactobacilli, organic acids, and exopolysaccharides that prevent pathogen adhesion (20).
L. reuteri is a member of the commensal microbiota in swine (21, 22); it is also used industrially as a starter culture in cereal fermentations (23). L. reuteri converts maltose and glucose to lactic and acetic acids; sucrose is converted to the alternative end products organic acids, mannitol, oligosaccharides, or exopolysaccharides (24). The growth of specific strains of L. reuteri in cereal substrates also supports the formation of reutericyclin, an antibiotic with specific activity against Gram-positive pathogens (25, 26). Moreover, the exopolysaccharides reuteran and levan, which prevent adhesion of ETEC K88 fimbriae to the porcine intestinal mucosa (16, 17), are produced during the growth of L. reuteri in cereals (27, 28). However, the specific contribution of exopolysaccharide formation to the inhibition of intestinal pathogens by L. reuteri remains unknown. This study therefore aimed to determine the effect of feed fermentation with L. reuteri on the growth performance as well as the abundance of intestinal ETEC organisms in weanling piglets. Strains of L. reuteri were chosen to include the reuteran-producing strain L. reuteri TMW1.656 and the levan-producing strain L. reuteri LTH5794 (16). Fermented and chemically acidified feed served as a control to differentiate between the effects of organic acids and those of viable L. reuteri organisms. To identify the specific effects of exopolysaccharide formation, feed was fermented with addition of 10% sucrose to support reuteran or levan formation by L. reuteri or without sucrose addition to obtain the same cell counts and the same concentration of organic acids but no bacterial exopolysaccharides.
Past studies to determine the effect of feed additives used ETEC-challenged pigs (13, 29, 31). This study employed piglets that were housed and fed under conditions that are close to those used in industrial practice but that were not challenged with ETEC. This approach allowed investigation of the effect of fermentation on the diverse ETEC strains that are present in unchallenged piglets and to assess the effect of feed fermentation on animal performance, in addition to its effect on animal health.
MATERIALS AND METHODS
Microorganisms and growth condition.
L. reuteri TMW1.656 and L. reuteri LTH5794, which produce reuteran and levan, respectively, from sucrose (16), were routinely grown on modified MRS agar (32) and incubated anaerobically at 37°C for 48 h. To obtain working cultures for feed fermentation, colonies were subcultured twice in modified MRS broth. E. coli strain ECL13795 (O149, virotype STb:LT:EAST1:F4) (17) was used as a positive control to determine the specificity of primers targeting E. coli and E. coli virulence factors.
Optimization of reuteran and levan production in feed fermentations and feed fermentations.
To optimize conditions for reuteran and levan formation, white wheat and corn flour (provided by the University of Alberta Swine Research and Technology Centre [SRTC]) were mixed with an equal amount of tap water. Sucrose was added at 10 or 20% (weight sucrose/weight flour). Cells from overnight cultures of L. reuteri were washed with sterile tap water and added at cell counts of approximately 107 CFU g−1. After 24 h of fermentation at 37°C, samples were taken and the pH, cell counts, concentrations of organic acids and ethanol, and concentrations of reuteran and levan were determined as previously described (33). In brief, organic acids and ethanol were extracted from fermented feed and quantified by high-pressure liquid chromatography after separation on an Aminex HPX-87 column and detection on a refractive index detector (33). Reuteran and levan were extracted by aqueous extraction from freeze-dried samples, dialyzed against distilled water, and quantified by size exclusion chromatography (33).
For feed fermentations, a seed sourdough was prepared with ground wheat and 10% sucrose as described above and transported to the SRTC. Ground wheat was prepared with wheat of the variety Harvest HRS (2012 harvest year), which was ground through the weaned pig screen (size, 3/32). The seed sourdough was used to inoculate the first feed fermentation with a 10% inoculum. After 24 h of fermentation, 90% of the batch was used to feed the piglets; the remaining 10% was used to inoculate the subsequent batch of fermented feeds. After four fermentation cycles with 10% inoculum and 24 h of fermentation each, seed sourdoughs were prepared in the laboratory from the culture stock and used to inoculate the feed fermentations. Wheat fermented with the same strains and addition of 5% (wt/wt) glucose and 5% (wt/wt) fructose in place of sucrose served as exopolysaccharide-negative controls. A chemically acidified control was prepared with 5% (wt/wt) fructose, 5% (wt/wt) glucose, 4 parts of lactic acid (80%), and 1 part of glacial acidic acid (100%) to reach a pH of 3.8.
To verify that the strains used to inoculate fermented feed dominated the fermentation, the pH of each batch was measured after 24 h of fermentation. The cell counts in each batch of fermented feed were determined by serial dilutions and surface plating on MRS agar plates. All colonies on the modified MRS plates exhibited a uniform colony morphology that matched the colony morphology of the inoculum. In sourdoughs started with defined strains of lactobacilli, a matching and uniform colony morphology is a reliable measure for the absence of contaminants (34).
Animals and diets.
The animal study was approved by the Animal Care and Use Committee of the University of Alberta according to the guidelines of the Canadian Council on Animal Care and was approved to be conducted at the SRTC. A total of 36 crossbred castrated male piglets were selected at weaning at 21 days of age. Each piglet was housed in an individual pen (0.5 by 1.22 m) in a temperature-controlled room (28 ± 2.5°C) during the 3-week experiment.
The piglets were randomly divided into six blocks containing six piglets each. One piglet per block was assigned to one of six experimental diets for a total of six observations per diet. Wheat or fermented wheat was included in the diets as follows: diet 1, unfermented wheat; diet 2, unfermented wheat acidified to pH 3.8 with lactic acid and acetic acid and supplemented with 5% glucose and 5% fructose; diet 3, wheat fermented with L. reuteri TMW1.656 and supplemented with 10% sucrose to support reuteran production; diet 4, wheat fermented with L. reuteri TMW1.656 and supplemented with 5% glucose and 5% fructose, which do not support reuteran formation; diet 5, wheat fermented with L. reuteri LTH5794 and supplemented with 10% sucrose to support levan production; diet 6, wheat fermented with L. reuteri LTH5794 and supplemented with 5% glucose and 5% fructose, which do not support levan formation. The diet was formulated to meet or to exceed National Research Council Canada (NRC; 2012) nutrient recommendations for 5- to 10-kg pigs (Table 1). Two phases of the diets were fed sequentially within each treatment. From day 0 to day 6, 20% fermented wheat was added to the basal diet (phase 1 diet); from days 7 to 21, the proportion of fermented wheat in the diet was increased to 50% (phase 2 diet). Titanium dioxide (TiO2) was added as an indigestible marker to each of the test diets to calculate the total tract digestibility coefficients of the nutrients. Piglets were offered free access to feed from a pen feeder and water from a nipple drinker. They were fed in mash form twice daily, at 8 a.m. and 4 p.m. At the end of the trial, piglets were fed their final meal 3 to 4 h before being euthanized to ensure that the digesta had reached each section of the small intestine. Body weight was recorded on days 0, 7, 14, and 21. Feed intake was measured each day. All of the data were used to determine average daily weight gain (ADG), average daily feed intake (ADFI), and feed efficiency (ratio of weight gain to feed intake [G/F]).
TABLE 1.
Composition of experimental diets on an as-fed basis
| Ingredient | Composition (%) |
|
|---|---|---|
| Phase 1 diet (days 0 to 6) | Phase 2 diet (days 7 to 21) | |
| Ground and fermented or unfermented wheat | 20.00 | 50.00 |
| Corn | 31.54 | 1.76 |
| Lactose | 15.00 | 10.00 |
| Soy protein concentrate | 3.00 | 2.50 |
| Herring meal | 6.00 | 2.50 |
| Brassica napus canola meal | 5.00 | |
| Wheat DD GSa | 5.00 | |
| Soybean meal | 15.00 | 15.00 |
| Canola oil | 4.00 | 3.40 |
| Other vitamin and mineral ingredients | 5.46 | 4.84 |
| Total | 100 | 100 |
DD GS, distillers dried grains with solubles.
Sample collection and preparation.
Fresh feces was collected in a plastic bag by hand grabbing of the feces from the floor of each pen on days 0, 7, 14, and 21 and stored at −20°C. After the piglets were euthanized, gut digesta were collected from the stomach, jejunum, ileum, cecum, and midcolon and placed in sterile plastic containers. Samples were stored at −20°C.
Frozen fecal and gut digesta samples were thawed and mixed aseptically with a spatula. For bacterial analysis, two 1.0- to 1.5-g subsamples were taken and stored at −80°C. Tissue samples from the jejunum and ileum (about 5 cm) were aseptically excised. The segments were opened longitudinally, and the mucosa was removed by scraping with a flame-sterilized metal spatula (35). The mucosal scrapings were stored in individual tubes at −80°C for bacterial analysis.
Genomic DNA extraction for quantitative PCR (qPCR).
Total bacterial DNA was extracted from fecal and gut digesta samples using a QIAamp DNA stool minikit (Qiagen, Inc., Valencia, CA, USA) (36) following the manufacturer's instructions. Briefly, about 120 mg (wet weight) of fecal sample was homogenized in buffer ASL (Qiagen) and heated at 95°C for 5 min to lyse the bacterial cells. After centrifugation at 20,000 × g for 1 min at room temperature (approximately 20°C), the supernatant was incubated with an InhibitEx tablet to absorb DNA-damaging compounds and PCR inhibitors (QIAamp DNA stool minikit handbook). Proteins in the lysates were removed by treatment of the samples with proteinase K and buffer AL (Qiagen) at 70°C for 10 min. Ethanol (96 to 100%) was added to the lysate to precipitate the DNA, and the mixture was applied to the QIAamp spin columns provided in the kit. The columns were washed with buffers AW1 and AW2 (Qiagen), and the DNA was eluted in buffer AE (Qiagen).
The DNA concentration was measured in a NanoDrop spectrophotometer (ND-1000; Thermo Fisher Scientific Inc., Wilmington, NC, USA), and the DNA purity was assessed by determining the ratio of the absorbance at 260 to the absorbance at 280 nm. All DNA samples had a 260 nm/280 nm absorbance ratio of >1.8.
PCR primers.
The primers used in this study and their target organisms are listed in Table 2. Primers GTFA F and GTFA R are specific for the detection of gtfA, the gene coding for reuteransucrase in L. reuteri TMW1.656. To obtain primers that specifically detect the gene coding for reutericyclin biosynthesis in L. reuteri TMW1.656, primers RC F and RC R were designed in Primer 3 software (37) to target the rtcN gene. rtcN codes for a nonribosomal peptide synthase; the gene is essential for reutericyclin biosynthesis but is essentially absent in all other lactobacilli (GenBank accession number KJ659887.1) (26). To obtain primers for the specific detection of the gene coding for levansucrase in L. reuteri LTH5794, ftfA, primers FTF F and FTF R were designed in Primer 3 software (37) to target the gene coding for a levansucrase in L. reuteri SD2112 (GenBank accession number NC_015697). The PCR amplicons that were obtained with the chromosomal DNA of L. reuteri LTH5794 and L. reuteri TMW1.656 as the template were purified from agarose gels and sequenced by Sanger sequencing (Macrogen, Rockville, MD). The sequence data were aligned by use of the ClustalW program (38) to identify sequences that are unique to ftfA of L. reuteri LTH5794. These unique sequences were used to design primers LEV F and LEV R. The Basic Local Alignment Search Tool (BLAST) was initially used to determine the specificity of the primer sequences (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). The specificity of the primers was subsequently verified in qPCRs with DNA from L. reuteri TMW1.656 or LTH5794 as the template, followed by determination of the sizes and melting temperatures of the amplicons (Fig. 1).
TABLE 2.
Primers used to profile the microorganisms in fecal and gut digesta samples
| Target | Primer | Sequence (5′–3′) | Product size (bp) | Tmb (°C) | Reference or source |
|---|---|---|---|---|---|
| Lactobacillus groupa | Lacto F | TGGAAACAGRTGCTAATACCG | 231–233 | 64 | 52 |
| Lacto R | GTCCATTGTGGAAGATTCCC | ||||
| L. reuteri | Lreu F | CAGACAATCTTTGATTGTTTAG | 303 | 64 | 53 |
| Lreu R | GCTTGTTGGTTTGGGCTCTTC | ||||
| Reuteransucrase (gtfA) | GTFA F | AATTAAACTGGTTATACTATCTC | 160 | 57 | 27 |
| GTFA R | GAGTTCATACCATCTGCAGC | ||||
| Levansucrase (ftfA) | FTF F | TATCAATGATACAAATAATGC | 1,065 | 56 | This study |
| FTF R | GCGTTTCAGGATCATTTGGT | ||||
| LEV F | GTCAATTTGATCCTTCGCC | 460 | 57 | This study | |
| LEV R | TGCAACTAAGGAAATTAAGGGC | ||||
| Nonribosomal peptide synthase (rtcN) | RC F | GGCGGAACGTTGAATATTGT | 248 | 60 | This study |
| RC R | ATTTTGGGGGAATCATAGCC | ||||
| Universal stress protein A | Ecoli F | CCGATACGCTGCCAATCAGT | 884 | 66 | 54 |
| Ecoli R | ACGCAGACCGTAGGCCAGAT | ||||
| LT | LT F | CCGTGCTGACTCTAGACCCCCA | 480 | 68 | 55 |
| LT R | CCTGCTAATCTGTAACCATCCTCTGC | ||||
| STa | STa F | ATGAAAAAGCTAATGTTGGC | 193 | 65 | 56 |
| Sta R | TACAACAAAGTTCACAGCAG | ||||
| STb | STb F | TGCCTATGCATCTACACAAT | 113 | 60 | 57 |
| STb R | CTCCAGCAGTACCATCTCTA | ||||
| K88 fimbriae | K88 F | GCACATGCCTGGATGACTGGTG | 439 | 67 | 58 |
| K88 R | CGTCCGCAGAAGTAACCCCACCT |
The Lactobacillus group includes Lactobacillus spp., Pediococcus spp., Leuconostoc spp., and Weissella spp.
Tm, melting temperature.
FIG 1.
Validation of specificity of primers for strain-specific PCR detection of L. reuteri TMW1.656 and LTH5794. Specific amplification was verified by determination of the uniform and matching melting temperatures (Tms) in qPCRs and by determination of the sizes of the amplicons.
Primers were synthesized by Integrated DNA Technologies Inc., diluted to a final concentration of 10 μg of primer per μl with autoclaved Milli-Q water upon receipt, and stored at −20°C.
Quantification of bacteria and bacterial metabolites or toxins by qPCR.
qPCR was performed on a 7500 Fast real-time PCR system (Applied Biosystems, Foster City, CA, USA). The total reaction volume of 20 μl contained 10 μl of QuantiFast SYBR green master mix (Applied Biosystems), 2 μl (10 μM) of primers, and 1 μl of template DNA from fecal or gut digesta samples. Each reaction was run in duplicate in a MicroAmp Fast Optical 96-well reaction plate sealed with MicroAmp optical adhesive film (Applied Biosystems). The concentration of template DNA was about 100 mg liter−1. PCR amplicons were purified by using QIAquick PCR purification kit (250), according to the manufacturer's instructions.
Standard curves were generated using serial 10-fold dilutions of the purified PCR amplicons, which were amplified by PCRs with the same primers (Table 2) and genomic DNA from L. reuteri strains or pig feces. The initial concentration of the purified PCR amplicons was determined by use of a NanoDrop spectrophotometer. Amplification conditions generally involved 1 cycle at 95°C for 5 min for initial denaturation, 40 cycles of denaturation at 95°C for 30 s, annealing with optimal annealing temperatures (Table 2) for 30 s, and extension at 72°C for 30 s. In the melting curve stage, the reaction conditions included 1 cycle at 95°C for 15 s, 1 cycle at 60°C for 1 min, and a stepwise increase of the temperature from 55 to 95°C (at 10 s per 0.5°C). To ensure correct amplification results, the melting curves were checked to verify that the PCR amplicons yielded a single melting peak.
Statistical analysis.
Data were analyzed using Statistical Analysis System (SAS; version 9.3) software (SAS Institute, 2012). Data for fecal bacteria and growth performance were analyzed according to a randomized complete block design with repeated measurement using the mixed procedure (Proc MIXED). The model included diet, period, and the interaction of diet and period as fixed effects. The blocks were considered random effects, and the individual animals were considered experimental units. Data for the microbiota of the gut digesta were subjected to analysis of variance as a randomized complete block design using the general linear model procedure (Proc GLM). Treatment comparisons were determined by contrast. The experimental unit was the piglet, and differences with a P value of <0.05 with a Bonferroni adjustment for multiple comparisons (SAS version 9.3) were considered to be statistically significant. The Kolmogorov-Smirnoff test (39) was used to test for the normality of all variables. The results of growth performance analyses are presented as means, while data from analyses of the bacteria are presented as least-squares means with standard errors.
RESULTS
Production of reuteran and levan in feed fermentations.
Reuteran and levan formation in feed fermentations was evaluated with wheat and corn. The in situ production of reuteran or levan by L. reuteri TMW1.656 in corn fermentation was low compared to that in wheat fermentation with the same sucrose addition (data not shown). Therefore, ground wheat was used for feed fermentations.
The levels of metabolite formation after fermentation by L. reuteri in wheat are shown in Table 3. The highest reuteran yield from L. reuteri TMW1.656 was obtained in fermentations with addition of 10% sucrose (Table 3). Addition of 10% sucrose provided fermented feed with lower acetate concentrations than addition of 20% sucrose (Table 3). Because acetic acid may reduce the palatability of fermented feed, wheat flour with addition of 10% sucrose was used to obtain fermented feed.
TABLE 3.
Concentration of metabolites in feed after 24 h of fermentation with L. reuteria
| L. reuteri strain (exopolysaccharide produced) | Amt of sucrose added (%) | Metabolite concn (mmol/kg feed) |
Reuteran or levan concn (g/kg feed)b | ||
|---|---|---|---|---|---|
| Lactate | Acetate | Ethanol | |||
| TMW1.656 (reuteran) | 10 | 87.1 ± 5.8 | 51.0 ± 4.2 | 36.2 ± 1.8 | 5.6 ± 1.0 |
| 20 | 79.4 ± 0.1 | 71.3 ± 1.3 | 2.5 ± 0.0 | 3.7 ± 1.1 | |
| LTH5794 (levan) | 10 | 80.7 ± 4.6 | 57.3 ± 3.7 | 27.31 ± 4.9 | 3.2 ± 0.6 |
| 20 | 70.7 ± 3.3 | 67.1 ± 3.4 | 3.41 ± 4.8 | 3.7 ± 0.6 | |
Wheat flour with addition of 10 or 20% sucrose was used as the substrate. The pH of unfermented wheat after inoculation was 6.1, and the pH after 24 h of fermentation was 3.7 for all fermentations.
The six diets used in the present study were chosen to assess the impacts of organic acids (control versus chemically acidified feed), of viable L. reuteri (unfermented controls versus four fermented diets), and of reuteran and levan (fermented diets supplemented or not supplemented with sucrose to support reuteran and levan formation).
Growth performance of pigs and animal health.
Data indicating the growth performance in the 21-day trial are presented in Table 4. Pigs fed diets containing reuteran or levan displayed a reduced average daily feed intake and a reduced average daily weight gain compared with those for pigs fed unfermented diets. The average daily feed intake was lower in pigs fed fermented diets than pigs fed unfermented diets. However, feed efficiency did not differ among the diets. All pigs remained healthy throughout the experimental period and did not develop diarrhea.
TABLE 4.
Growth performance of pigs fed supplemental fermented diet for 21 days
| Parameter | AFDI (g DMd/day) | ADG (g/day) | G/F ratio |
|---|---|---|---|
| Dietary treatmenta | |||
| CTRL | 305 | 255 | 0.76 |
| ACID | 295 | 264 | 0.84 |
| TMW1.656 | |||
| REU+ | 268 | 241 | 0.86 |
| REU− | 283 | 240 | 0.79 |
| LTH5794 | |||
| LEV+ | 266 | 210 | 0.74 |
| LEV− | 265 | 240 | 0.84 |
| SEM | 19 | 19 | 0.05 |
| P valueb | |||
| Acids | 0.659 | 0.712 | 0.231 |
| Exopolysaccharides | 0.644 | 0.403 | 0.628 |
| L. reuteri + reuteran or levan | 0.029c | 0.046 | 0.894 |
| L. reuteri | 0.085 | 0.24 | 0.725 |
| Fermentation | 0.025 | 0.067 | 0.899 |
Diets were supplemented with organic acid and fermented sourdough. Pigs were fed a phase 1 diet for the first 7 days, followed by a phase 2 diet from days 7 to 21. Abbreviations for the diets are as follows: CTRL, control; ACID, chemically acidified feed; REU+ and REU−, feed fermented with L. reuteri TMW1.656 with addition of sucrose to support reuteran formation (REU+) or with addition of glucose and fructose (REU−); LEV+ and LEV−, feed fermented with L. reuteri LTH5794 with addition of sucrose to support levan formation (LEV+) or with addition of glucose and fructose (LEV−).
For acid effects, P values are for the control diet versus the chemically acidified diet; for reuteran or levan effects, P values are for sucrose-supplemented diets REU+ and LEV+ versus REU− and LEV−; for L. reuteri + reuteran or levan, P values are for reuteran- or levan-containing fermented diets (REU+ and LEV+) versus unfermented diets (CTRL and ACID); for L. reuteri, P values are for fermented diets without exopolysaccharides (REU− and LEV−) versus unfermented diets (CTRL and ACID); for fermentation, P values are for all fermented diets (REU+, REU−, LEV+, and LEV−) versus unfermented diets (CRTL and ACID).
P values of less than 0.05 are indicated in boldface.
DM, dry matter.
Genus-, species-, and strain-specific detection of lactobacilli in fecal samples.
qPCR with strain-specific primers was employed to determine whether the strains employed for feed fermentation remained present throughout the gastrointestinal transit. Organisms of the Lactobacillus group and L. reuteri were quantified to determine whether the dietary L. reuteri bacteria influenced the abundance of autochthonous lactobacilli and L. reuteri. The genes coding for reuteransucrase and levansucrase in L. reuteri TMW1.656 and LTH5794, gtfA and ftfA, respectively, were used as strain-specific markers. The copy numbers of the gene representing the Lactobacillus group remained stable or increased slightly during the study period (Table 5). The copy numbers of the genes representing L. reuteri increased after 7 days of feeding in all groups; with the exception of piglets fed chemically acidified diets, the fecal copy numbers of the genes representing L. reuteri decreased again at day 21 (Table 5). In animals fed chemically acidified diets, L. reuteri numbers were maintained at the same high level from day 7 to day 21 (Table 5). At days 14 and 21, the L. reuteri numbers in piglets that were fed diets fermented with L. reuteri TMW1.656 were lower than those in the group fed chemically acidified diets.
TABLE 5.
Gene copy numbers for the Lactobacillus group, L. reuteri, gtfA, rtcN, and ftfA obtained on days 0, 7, 14, and 21a
| Organism or gene and time (day) | Log(gene copy no./g) for the following dietb: |
|||||
|---|---|---|---|---|---|---|
| Control | Chemically acidified | TMW1.656 with sucrose | TMW1.656 with Glu + Fru | LTH5794 with sucrose | LTH5794 with Glu + Fru | |
| Lactobacillus group | ||||||
| 0 | 9.13 ± 0.17A,X | 8.93 ± 0.17A,X | 8.86 ± 0.21A,X | 8.92 ± 0.29A,X | 9.4 ± 0.19A,X | 8.69 ± 0.19A,B,X |
| 7 | 9.35 ± 0.17A,B,C,X | 9.69 ± 0.17A,Y | 9.02 ± 0.17C,X | 9.51 ± 0.17A,B,X | 9.32 ± 0.17A,B,X | 9.1 ± 0.17B,C,X,Y |
| 14 | 8.99 ± 0.17A,X | 9.3 ± 0.17A,B,X,Y | 9.32 ± 0.17A,B,X | 9.48 ± 0.17B,X | 9.05 ± 0.17A,B,X | 9.06 ± 0.17A,B,X,Y |
| 21 | 9.02 ± 0.17A,X | 9.35 ± 0.17A,X,Y | 9.23 ± 0.17A,X | 9.21 ± 0.17A,X | 9.22 ± 0.17A,X | 9.17 ± 0.17A,Y |
| L. reuteri | ||||||
| 0 | 7.85 ± 0.28A,X | 7.47 ± 0.28A,B,X | 6.87 ± 0.33B,X | 7.44 ± 0.44A,B,X | 8.14 ± 0.3A,X | 7.48 ± 0.3A,B,X |
| 7 | 8.68 ± 0.27A,Y,Z | 8.93 ± 0.27A,Y | 8.25 ± 0.27A,Y | 8.42 ± 0.27A,Y | 8.81 ± 0.27A,Y | 8.84 ± 0.27A,Y |
| 14 | 8.98 ± 0.19A,Z | 8.94 ± 0.19A,Y | 8.17 ± 0.19B,Y | 8.39 ± 0.19B,X,Y | 8.59 ± 0.19A,B,X,Y | 8.7 ± 0.19A,B,Y |
| 21 | 8.38 ± 0.16A,B,X,Y | 8.83 ± 0.16B,Y | 7.44 ± 0.16C,X | 8.05 ± 0.16A,X,Y | 8.03 ± 0.16A,X | 8.13 ± 0.16A,X |
| Reuteransucrase (gtfA) | ||||||
| 0 | <5 | <5 | <5 | <5 | <5 | <5 |
| 7 | <5 | 5.65 ± 0.26A | 5.93 ± 0.26A | 6.41 ± 0.26B | <5 | <5 |
| 14 | <5 | 5.36 ± 0.26A | 6.42 ± 0.26B | 6.45 ± 0.26B | <5 | <5 |
| 21 | <5 | 5.65 ± 0.26A | 5.95 ± 0.26A | 6.8 ± 0.26B | <5 | <5 |
| Levansucrase (ftfA) | ||||||
| 0 | 6.67 ± 0.3 | <6 | <6 | <6 | <6 | <6 |
| 7 | 6.08 ± 0.17A | <6 | <6 | <6 | 8.36 ± 0.17B | 8.23 ± 0.17B |
| 14 | 6.9 ± 0.13A | 6.72 ± 0.13A | 6.79 ± 0.13A | 6.69 ± 0.13A | 7.82 ± 0.13B | 8.26 ± 0.13C |
| 21 | 6.22 ± 0.2A | <6 | <6 | <6 | 8.09 ± 0.2B | 8.38 ± 0.2B |
| Nonribosomal peptide synthase (rtcN) | ||||||
| 0 | <6 | <6 | <6 | <6 | <6 | <6 |
| 7 | <6 | 6.46 ± 0.29 | 6.81 ± 0.29 | 7.17 ± 0.29 | <6 | <6 |
| 14 | <6 | <6 | 6.57 ± 0.34 | 6.92 ± 0.34 | <6 | <6 |
| 21 | <6 | <6 | 6.24 ± 0.25 | 6.86 ± 0.25 | <6 | <6 |
gtfA is a marker for L. reuteri TMW1.656 in feces, rtcN is a second marker for L. reuteri TMW1.656 in feces, and ftfA is a marker for L. reuteri LTH5794 in feces. Data are presented as least-squares means (n = 36) ± standard errors of the means. Superscripts A, B, and C denote significant differences (P < 0.05) between diets at each time point (comparison across rows); superscripts X, Y, and Z denote significant differences (P < 0.05) within a diet over time (comparison across columns). Values that do not share a superscript are significantly different. The detection limit for gtfA was 5 log10 gene copies/g of feces (wet weight); the detection limit for the Lactobacillus group, L. reuteri, rtcN, and ftfA was 6 log10 gene copies/g of feces (wet weight).
For TMW1.656 and LTH5794, the diets consisted of feed fermented with TMW1.656 and LTH5794, respectively, and supplemented with the indicated sugar.
Before the treatments (day 0), gtfA or rtcN was not detected in any sample (Table 5), while ftfA was detected only in pigs assigned to the control group (Table 5). The gene for reuteransucrase and rtcN were detected at days 7, 14, and 21 in pigs receiving feed fermented with L. reuteri TMW1.656 and at lower copy numbers in pigs fed chemically acidified diets (Table 5). The copy numbers of the ftfA gene of L. reuteri LTH5794 were 2 log units higher than the copy numbers of the gene in the feces of pigs fed other diets (Table 5); however, the copy numbers of ftfA were above the detection limit in piglets fed the unfermented control diet at all times and in piglets of the other groups at day 14 (Table 5).
Genus-, species-, and strain-specific detection of lactobacilli in gut samples.
Lactobacilli were quantified in gut samples at day 21. The copy numbers of the gene representing the Lactobacillus group were high in all intestinal compartments except the ileum, where the numbers were 1 to 2 log(gene copies g−1) lower (data not shown). The copy numbers of the gene representing L. reuteri in the stomach, the ileum, and the cecum were higher for piglets fed fermented diets than piglets fed chemically acidified diets (data not shown); however, the copy numbers of the gene representing L. reuteri in the colons of animals fed L. reuteri TMW1.656 were reduced (data not shown).
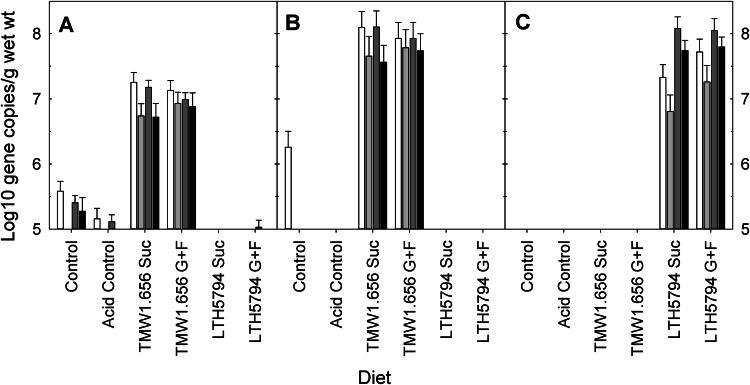
The copy numbers of the gtfA gene of L. reuteri TMW1.656 were high in all samples from piglets that were fed L. reuteri TMW1.656, and the copy numbers of the gtfA gene were low or the gene was absent in samples from other piglets (Fig. 2A). Matching results were obtained with primers targeting rtcN, the second strain-specific marker for L. reuteri TMW1.656 (Fig. 2B). ftfA of L. reuteri LTH5794 was detected only in pigs that were fed diets fermented with this strain (Fig. 2C).
FIG 2.
Copy numbers of the genes gtfA as a marker for L. reuteri TMW1.656 (A), rtcN as a second marker for L. reuteri TMW1.656 (B), and ftfA as a marker for L. reuteri LTH5794 (C) in digesta obtained from the stomach (white bars), ileum (light gray bars), cecum (dark gray), or colon (black). Data are presented as least-squares means (n = 36), with their standard errors being represented by vertical bars. The copy numbers of the gtfA gene were significantly (P < 0.05) higher in samples from animals fed L. reuteri TMW1.656 than in samples from all other animals. The copy numbers of the rtcN gene were significantly (P < 0.05) higher in samples from animals fed L. reuteri TMW1.656 than in samples from all other animals. The detection limits for the Lactobacillus group, L. reuteri, and levansucrase were 6 log10 gene copies/g of digesta (wet weight). Suc, sucrose; G, glucose; F, fructose.
Detection of E. coli and ETEC virulence factors in fecal samples.
E. coli and ETEC were quantified by qPCR to determine the effect of feed fermentation and reuteran or levan production by L. reuteri on the numbers of intestinal E. coli bacteria (Table 6). E. coli and ETEC were detected in the feces of all pigs at all times (Table 6). The copy numbers of genes representing E. coli decreased over time in animals that were fed fermented diets but not in animals that were fed the control diet or the chemically acidified diet (Table 6). At day 21, the gene copy numbers of genes representing E. coli were the lowest in animals in animals receiving fermented diets containing reuteran (Table 6). The levels of the genes for virulence factors of ETEC, heat-stable enterotoxin b (STb), heat-labile enterotoxin (LT), and K88 fimbriae, peaked at day 7 or day 14 in all animals and decreased at day 21. The copy numbers of the gene for STb were lower in animals receiving fermented diets containing reuteran than in the control animals or animals that were fed the chemically acidified diet (Table 6). Moreover, samples from animals receiving reuteran were the only samples where the copy numbers of the gene for LT were below the detection limit (Table 6). The copy numbers of the gene for K88 fimbriae were below the detection limit in all animals except animals in the control group or the group receiving feed fermented with L. reuteri LTH5794 but without sucrose (Table 6).
TABLE 6.
Gene copy number for E. coli, STb, LT, K88 fimbriae, and STa in feces obtained on days 0, 7, 14, and 21a
| Bacterium or bacterial toxin and time (day) | Log(gene copy no./g) for the following dietb: |
|||||
|---|---|---|---|---|---|---|
| Control | Acidified control | TMW1.656 with sucrose | TMW1.656 with Glu + Fru | LTH5794 with sucrose | LTH5794 with Glu + Fru | |
| E. coli | ||||||
| 0 | 6.66 ± 0.45A,X | 6.7 ± 0.45A,X | 6.91 ± 0.55A,X,Y | 6.74 ± 0.78A,X | 7.72 ± 0.49A,X | 7.09 ± 0.49A,X |
| 7 | 6.86 ± 0.45A,B,X | 6.61 ± 0.45A,X | 7.98 ± 0.45B,X | 6.61 ± 0.45A,X | 7.74 ± 0.45A,B,X | 7.59 ± 0.45A,B,X |
| 14 | 7.13 ± 0.45A,X | 6.95 ± 0.45A,X | 6.17 ± 0.45A,Y | 6.51 ± 0.45A,X | 7.29 ± 0.45A,X | 7.02 ± 0.45A,X |
| 21 | 6.42 ± 0.45A,X | 5.96 ± 0.45A,B,X | 4.77 ± 0.45B,Z | 5.36 ± 0.45A,B,X | 5.22 ± 0.45A,B,Y | 5.21 ± 0.45A,B,Y |
| STb | ||||||
| 0 | <4 | <4 | <4 | <4 | <4 | <4 |
| 7 | 5.07 ± 0.82A,X | 5.83 ± 0.82A,X | 7.84 ± 0.82B,X | 6.1 ± 0.82A,B,X | 6.78 ± 0.82A,B,X,Y | 6.79 ± 0.82A,B,X,Y |
| 14 | 7.76 ± 0.76A,Y | 7.7 ± 0.76A,B,X | 6.51 ± 0.76A,X | 6.29 ± 0.76A,X | 8.05 ± 0.76A,Y | 7.03 ± 0.76A,Y |
| 21 | 6.94 ± 0.69A,X | 6.51 ± 0.69A,B,X | 4.27 ± 0.69C,Y | 5.04 ± 0.69A,B,C,X | 4.91 ± 0.69B,C,X | 4.69 ± 0.69B,C,X |
| LT | ||||||
| 0 | <4 | 4.29 ± 0.26X | 4.24 ± 0.33X | <4 | 4.23 ± 0.29X | 4.84 ± 0.29X |
| 7 | 6.16 ± 0.73X | 5.73 ± 0.73Y | 6.56 ± 0.73Y | 6.22 ± 0.73X | 6.19 ± 0.73Y | 5.78 ± 0.73X |
| 14 | 7.29 ± 0.75X | 7.19 ± 0.75Y | 6.11 ± 0.75Y | 6.68 ± 0.75X | 6.91 ± 0.75Y | 6.34 ± 0.75X |
| 21 | 4.97 ± 0.33Y | 4.1 ± 0.33X | <4 | 4.08 ± 0.33Y | 4.44 ± 0.33X | 4.85 ± 0.33X |
| K88 fimbriae | ||||||
| 0 | <4 | <4 | <4 | <4 | <4 | <4 |
| 7 | 5.43 ± 0.74X,Y | 4.86 ± 0.74X | 5.82 ± 0.74Y | 5.59 ± 0.74X | 5.63 ± 0.74X | 5.15 ± 0.74X |
| 14 | 6.53 ± 0.82X | 6.39 ± 0.82X | 5.29 ± 0.82X | 5.9 ± 0.82X | 6.15 ± 0.82X | 5.31 ± 0.82X |
| 21 | 4.32 ± 0.42Y | <4 | <4 | <4 | <4 | 4.15 ± 0.42X |
| STa | ||||||
| 0 | <4 | <4 | <4 | <4 | <4 | 4.11 ± 0.16 |
| 7 | <4 | <4 | <4 | <4 | <4 | <4 |
| 14 | <4 | <4 | <4 | 4.13 ± 0.16 | <4 | <4 |
| 21 | <4 | <4 | <4 | <4 | <4 | <4 |
Data are presented as least-squares means (n = 36) ± standard errors of the means. Superscripts A, B, and C denote significant differences (P < 0.05) between diets at each time point (comparison across rows); superscripts X, Y, and Z denote significant differences (P < 0.05) within a diet over time (comparison across columns). Values that do not share a superscript are significantly different. The detection limit for E. coli, STb, LT, K88 fimbriae, and STa was 4 log10 gene copies/g of feces (wet weight).
For TMW1.656 and LTH5794, the diets consisted of feed fermented with TMW1.656 and LTH5794, respectively, and supplemented with the indicated sugar.
Detection of genes coding for E. coli and ETEC virulence factors in gut samples.
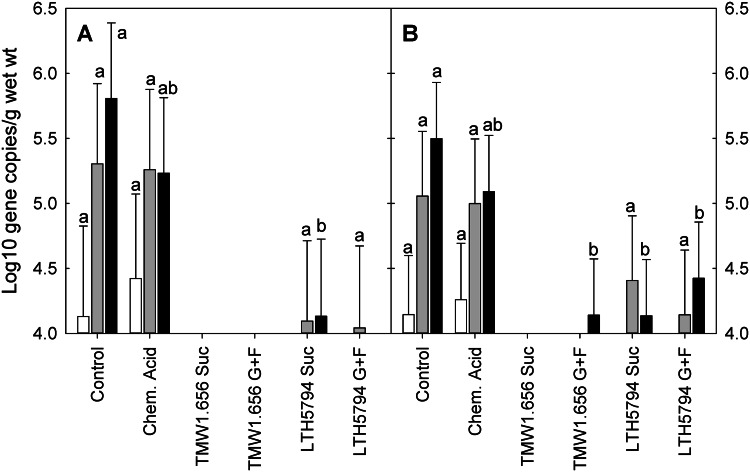
Genes coding for E. coli and STb were detected in the gut digesta of pigs fed unfermented diets; in contrast, the copy numbers of the genes for E. coli and STb were below the detection limit for all samples from pigs fed a reuteran-containing diet fermented with L. reuteri TMW1.656 (Fig. 3). In animals fed L. reuteri LTH5794, the gene copy numbers for E. coli and ETEC were below the detection limit in ileal samples; the gene copy numbers in the colon were lower than those in samples from animals fed unfermented diets (Fig. 3). Other virulence factors of ETEC, including LT, heat-stable enterotoxin a (STa), and K88 fimbriae, were not detected in any of the samples from gut digesta (ileum, cecum, and colon). Neither E. coli nor its virulence factors were detected in samples of mucosal scrapings from the jejunum.
FIG 3.
Gene copy numbers for E. coli (A) and its STb virulence factor (B) in the gut digesta of pigs fed control feed (white bars), chemically acidified feed (light gray bars), feed fermented with L. reuteri TMW1.656 with addition of sucrose to support reuteran formation (black bars), feed fermented with L. reuteri TMW1.656 with addition of glucose and fructose (white bars with slashes), feed fermented with L. reuteri LTH5794 with addition of sucrose to support levan formation (light gray bars with slashes), or feed fermented with L. reuteri LTH5794 with addition of glucose and fructose (dark gray bars with slashes). Data are presented as least-squares means (n = 36), with their standard errors being represented by vertical bars. a, b, and c denote significant differences (P < 0.05) between diets at each site. Values not having the same letter are significantly different. The detection limit was 4 log10 gene copies/g of digesta (wet weight). Chem. Acid., chemically acidified; Suc, sucrose; G, glucose; F, fructose.
DISCUSSION
ETEC causes diarrhea in newborn and weaned pigs, resulting in serious morbidity and mortality and major financial losses in the swine industry (6, 40). The present study demonstrated that feed fermentation with two exopolysaccharide-producing strains of L. reuteri reduced the abundance of ETEC in weanling piglets. Beneficial effects of feed fermentation related predominantly to the ingestion of viable cells of L. reuteri. In addition, the presence of reuteran produced during feed fermentation further reduced the numbers of ETEC bacteria.
Fermented feeds may benefit gut health and improve the growth performance of pigs (20, 41). In previous studies, the average daily weight gain and the average daily feed intake of pigs fed fermented liquid diets were reduced compared to those of pigs receiving unfermented feed; however, feed efficiency did not differ (41, 42). The present study confirmed the reduced feed intake and unchanged feed efficiency in pigs receiving fermented feed. Remarkably, this effect was less pronounced in the groups that received fermented feed without reuteran or levan. The formation of reuteran and levan from sucrose reduces the sweet taste of the feed because sucrose is converted to oligosaccharides and polysaccharides that do not taste sweet (24, 43). In fermented or unfermented control feeds supplemented with glucose and fructose, the levels of acidity are comparable but the concentrations of monosaccharides and mannitol are higher. Pigs thus might prefer a balance of sweet and sour tastes. This finding suggests the possibility that feed intake may be increased through the formulation of feeds with the aim of improving their taste.
Organic acids promote the growth of weaning pigs due to their antimicrobial properties and because they lower the gastric pH (12, 44); however, the present study did not reveal positive effects of organic acids on growth performance or inhibition of E. coli. The abundance of lactobacilli and L. reuteri increased in animals fed chemically acidified diets. Chemically acidified feed was supplemented with glucose and fructose so that the sugar levels matched those in the fermented diets and thus provided more fermentable substrates to autochthonous lactobacilli in the stomach (45).
qPCR is widely used to detect bacteria in fecal and gut content samples (46). Because the viable L. reuteri bacteria that are present in the feed or the upper intestinal tract remain viable throughout gastrointestinal transit (21, 28, 47), quantification of DNA by qPCR indicates the presence of viable cells. The primers were designed to target strain-specific sequences in L. reuteri TMW1.656 and L. reuteri LTH5794, and the melting temperature of amplicons was routinely checked to verify specific amplification. Cross-contamination was essentially absent in intestinal tissue samples. However, low levels of gtfA and ftfA were detected in fecal samples from pigs that were not fed the corresponding strains. This may reflect the cross contamination of fecal samples after defecation. Despite this limitation, the qPCR methodology differentiated between feed-fermenting L. reuteri strains and autochthonous strains of lactobacilli (21, 23). Both strains persisted in the guts of the piglets, were excreted with feces in high abundance, and accounted for 1% to >10% of the total L. reuteri bacteria. L. reuteri TMW1.656 and LTH5794 are rodent lineage strains that differ physiologically and phylogenetically from porcine strains of L. reuteri (21, 23). L. reuteri LTH5794 had no apparent influence on autochthonous lactobacilli, in keeping with the findings of previous studies feeding probiotic lactobacilli (47, 48). Feed fermented with L. reuteri TMW1.656, however, decreased the abundance of autochthonous L. reuteri bacteria. L. reuteri TMW1.656 produces reutericyclin (25), a low-molecular-weight antimicrobial compound inhibiting Gram-positive bacteria, including L. reuteri (49), that may account for the reduced numbers of L. reuteri bacteria.
Oral administration of lactic acid bacteria to swine reduced the amounts of fecal Enterobacteriaceae and coliform bacteria (47); oral administration of Lactobacillus amylovorus also reduced the level of colonization of weaning piglets with ETEC in a challenge study (36). The present study demonstrated that feed fermentations reduce the abundance of intestinal E. coli, including ETEC, compared to that achieved with both the unfermented control and the acidified unfermented control. Although all piglets remained healthy throughout the study, a reduced number of ETEC bacteria indicates a reduced risk of ETEC-induced diarrhea. This health benefit justifies the designation of the two strains as probiotic strains (50).
The effect of feed fermentation with L. reuteri on the abundance of fecal E. coli or fecal levels of ETEC virulence factors was dependent on the strain and on the presence of reuteran or levan. The reuteran-containing fermented diet was the only diet that significantly reduced the fecal levels of STb compared to those in both unfermented control diets; this diet was also the only diet reducing the levels of fecal LT to levels below the detection limit at week 3 and reducing the levels of E. coli and STb to levels below the detection limit in all intestinal tissue samples (Table 6 and Fig. 3). Exopolysaccharides produced by lactic acid bacteria reduce the adherence of pathogenic bacteria, such as E. coli, to the intestinal mucosa (51). Specifically, the reuteran produced by L. reuteri TMW1.656 reduces the adhesion of ETEC K88. In the present study, the reuteran-positive groups had lower copy numbers of genes for E. coli and the ETEC virulence factors than groups fed diets containing L. reuteri but not reuteran or levan, which may reflect the antiadhesive properties of reuteran in vitro (16) and in an in vivo model (17). Together with the findings presented in those prior reports, this study suggests that reuteran reduces the level of ETEC adhesion to the intestinal mucosa in vivo. However, the levels of E. coli and ETEC bacteria in animals receiving L. reuteri TMW1.565-fermented diets with or without reuteran were not significantly different. This may relate to the low levels of reuteran that may be formed from the sucrose that is present in wheat flour (43).
In conclusion, feed fermentation with L. reuteri reduced the level of colonization of weaning piglets with ETEC bacteria and additionally supplied reuteran in concentrations that may specifically contribute to the prevention of ETEC adhesion to the intestinal mucosa. The study thus constitutes a step toward understanding the metabolic activities that confer probiotic properties to lactic acid bacteria.
ACKNOWLEDGMENTS
We are grateful to Xiaoyan Chen, Kim Williams, Yalu Yan, and Cindy Jing Zhao for support with the animal experiments. Yan Yang is grateful to Michael E. Stiles for advice and support during manuscript preparation.
The Alberta Livestock and Meat Agency is acknowledged for funding (grant no. 2010R012R and 2013R002R). Michael G. Gänzle acknowledges the Canada Research Chair program for funding.
REFERENCES
- 1.Kohler EM, Moon H. 1984. Enteric colibacillosis of newborn pigs. Purdue University Cooperative Extension Service publication PIH-30. Purdue University, West Lafayette, IN. [Google Scholar]
- 2.Barnett KL, Kornegay ET, Risley CR, Lindemann MD, Schurig GG. 1989. Characterization of creep feed consumption and its subsequent effects on immune-response, scouring index and performance of weanling pigs. J Anim Sci 67:2698–2708. [DOI] [PubMed] [Google Scholar]
- 3.Hedemann MS, Jensen BB. 2004. Variations in enzyme activity in stomach and pancreatic tissue and digesta in piglets around weaning. Arch Anim Nutr 58:47–59. doi: 10.1080/00039420310001656677. [DOI] [PubMed] [Google Scholar]
- 4.Fairbrother JM, Nadeau É, Gyles CL. 2005. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim Health Res Rev 6:17–39. doi: 10.1079/AHR2005105. [DOI] [PubMed] [Google Scholar]
- 5.Parry SH, Rooke DM. 1985. Adhesins and colonization factors of Escherichia coli, p 79–155. In Sussman M. (ed), The virulence of Escherichia coli: reviews and methods. Special publications of the Society for General Microbiology, vol 13 Academic Press, London, United Kingdom. [Google Scholar]
- 6.Nagy B, Fekete PZ. 2005. Enterotoxigenic Escherichia coli in veterinary medicine. Int J Med Microbiol 295:443–454. doi: 10.1016/j.ijmm.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Docic M, Bilkei G. 2003. Differences in antibiotic resistance in Escherichia coli, isolated from east-European swine herds with or without prophylactic use of antibiotics. J Vet Med B Infect Dis Vet Public Health 50:27–30. doi: 10.1046/j.1439-0450.2003.00609.x. [DOI] [PubMed] [Google Scholar]
- 8.Gyles CL. 1994. Escherichia coli enterotoxins, p 337 In Gyles CL. (ed), Escherichia coli in domestic animals and humans. CAB International, Wallingford, Oxon, United Kingdom. [Google Scholar]
- 9.Blanco M, Blanco JE, Gonzalez EA, Mora A, Jansen W, Gomes TAT, Zerbini LF, Yano T, DeCastro AFP, Blanco J. 1997. Genes coding for enterotoxins and verotoxins in porcine Escherichia coli strains belonging to different O:K:H serotypes: relationship with toxic phenotypes. J Clin Microbiol 35:2958–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton MD. 2000. Antibiotic use in animal feed and its impact on human health. Nutr Res Rev 13:279–299. doi: 10.1079/095442200108729106. [DOI] [PubMed] [Google Scholar]
- 11.Casewell M, Friis C, Marco E, McMullin P, Phillips I. 2003. The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J Antimicrob Chemother 52:159–161. doi: 10.1093/jac/dkg313. [DOI] [PubMed] [Google Scholar]
- 12.Tsiloyiannis VK, Kyriakis SC, Vlemmas J, Sarris K. 2001. The effect of organic acids on the control of porcine post-weaning diarrhoea. Res Vet Sci 70:287–293. doi: 10.1053/rvsc.2001.0476. [DOI] [PubMed] [Google Scholar]
- 13.Kyriakis SC, Tsiloyiannis VK, Vlemmas J, Sarris K, Tsinas AC, Alexopoulos C, Jansegers L. 1999. The effect of probiotic LSP 122 on the control of post-weaning diarrhoea syndrome of piglets. Res Vet Sci 67:223–228. doi: 10.1053/rvsc.1999.0308. [DOI] [PubMed] [Google Scholar]
- 14.Daudelin JF, Lessard M, Beaudoin F, Nadeau E, Bissonnette N, Boutin Y, Brousseau JP, Lauzon K, Fairbrother JM. 2011. Administration of probiotics influences F4 (K88)-positive enterotoxigenic Escherichia coli attachment and intestinal cytokine expression in weaned pigs. Vet Res 42:69. doi: 10.1186/1297-9716-42-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarabia-Sainz HM, Armenta-Ruiz C, Sarabia-Sainz JAI, Guzman-Partida AM, Ledesma-Osuna AI, Vazquez-Moreno L, Montfort GRC. 2013. Adhesion of enterotoxigenic Escherichia coli strains to neoglycans synthesised with prebiotic galactooligosaccharides. Food Chem 141:2727–2734. doi: 10.1016/j.foodchem.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Gänzle MG, Schwab C. 2010. Exopolysaccharide synthesized by Lactobacillus reuteri decreases the ability of enterotoxigenic Escherichia coli to bind to porcine erythrocytes. Appl Environ Microbiol 76:4863–4866. doi: 10.1128/AEM.03137-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen XY, Woodward AW, Zijlstra RT, Gänzle MG. 2014. Exopolysaccharides synthesized by Lactobacillus reuteri protect against enterotoxigenic Escherichia coli in piglets. Appl Environ Microbiol 80:5752–5760. doi: 10.1128/AEM.01782-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pluske JR. 2013. Feed- and feed additives-related aspects of gut health and development in weanling pigs. J Anim Sci Biotechnol 4:1. doi: 10.1186/2049-1891-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Lange CFM, Pluske J, Gong J, Nyachoti CM. 2010. Strategic use of feed ingredients and feed additives to stimulate gut health and development in young pigs. Livestock Sci 134:124–134. doi: 10.1016/j.livsci.2010.06.117. [DOI] [Google Scholar]
- 20.van Winsen RL, Urlings BAP, Lipman LJA, Snijders JMA, Keuzenkamp D, Verheijden JHM, van Knapen F. 2001. Effect of fermented feed on the microbial population of the gastrointestinal tracts of pigs. Appl Environ Microbiol 67:3071–3076. doi: 10.1128/AEM.67.7.3071-3076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frese SA, MacKenzie DA, Peterson DA, Schmaltz R, Fangman T, Zhou Y, Zhang CM, Benson AK, Cody LA, Mulholland F, Juge N, Walter J. 2013. Molecular characterization of host-specific biofilm formation in a vertebrate gut symbiont. PLoS Genet 9:e1004057. doi: 10.1371/journal.pgen.1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leser TD, Amenuvor JZ, Jensen TK, Lindecrona RH, Boye M, Møller K. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl Environ Microbiol 68:673–690. doi: 10.1128/AEM.68.2.673-690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su MSW, Oh PL, Walter J, Gänzle MG. 2012. Intestinal origin of sourdough Lactobacillus reuteri isolates as revealed by phylogenetic, genetic, and physiological analysis. Appl Environ Microbiol 78:6777–6780. doi: 10.1128/AEM.01678-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gänzle MG, Vermeulen N, Vogel RF. 2007. Carbohydrate, peptide and lipid metabolism of lactobacilli in sourdough. Food Microbiol 24:128–138. doi: 10.1016/j.fm.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Gänzle MG, Vogel RF. 2003. Contribution of reutericyclin production to the stable persistence of Lactobacillus reuteri in an industrial sourdough fermentation. Int J Food Microbiol 80:31–45. doi: 10.1016/S0168-1605(02)00146-0. [DOI] [PubMed] [Google Scholar]
- 26.Lin XB, Lohans CT, Duar R, Zheng J, Vederas JC, Walter J, Gänzle MG. 2015. Genetic determinants of reutericyclin biosynthesis in Lactobacillus reuteri. Appl Environ Microbiol 81:2032–2041. doi: 10.1128/AEM.03691-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gänzle MG, Schwab C. 2009. Ecology of exopolysaccharide formation by lactic acid bacteria: sucrose utilization, stress tolerance, and biofilm formation, p 263–278. In Ullrich M. (ed), Bacterial polysaccharides: current innovations and future trends. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 28.Schwab C, Gänzle GM. 2005. Exopolysaccharide production by intestinal lactobacilli, p 83–96. In Tannock GW. (ed), Probiotics and prebiotics: scientific aspects. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 29.Jansman AJM, Wagenaars CMF, van der Meulen J. 2010. Response of weaned piglets to a challenge with enterotoxigenic Escherichia coli (ETEC) when fed diets with pea or pea fractions. Livestock Sci 133:229–231. doi: 10.1016/j.livsci.2010.06.072. [DOI] [Google Scholar]
- 30.Reference deleted.
- 31.McKenzie R, Darsley M, Thomas N, Randall R, Carpenter C, Forbes E, Finucane M, Sack RB, Hall E, Bourgeois AL. 2008. A double-blind, placebo-controlled trial to evaluate the efficacy of PTL-003, an attenuated enterotoxigenic E. coli (ETEC) vaccine strain, in protecting against challenge with virulent ETEC. Vaccine 26:4731–4739. doi: 10.1016/j.vaccine.2008.06.064. [DOI] [PubMed] [Google Scholar]
- 32.Meroth CB, Walter J, Hertel C, Brandt MJ, Hammes WP. 2003. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol 69:475–482. doi: 10.1128/AEM.69.1.475-482.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galle S, Schwab C, Arendt E, Gänzle M. 2010. Exopolysaccharide forming Weissella strains as starter cultures for sorghum and wheat sourdoughs. J Agric Food Chem 58:5834–5841. doi: 10.1021/jf1002683. [DOI] [PubMed] [Google Scholar]
- 34.Lin XB, Gänzle MG. 2014. Quantitative high-resolution melting PCR analysis for monitoring of fermentation microbiota in sourdough. Int J Food Microbiol 186:42–48. doi: 10.1016/j.ijfoodmicro.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 35.Said HM, Redha R, Nylander W. 1989. Biotin transport in the human intestine: inhibition by anticonvulsant drugs. Am J Clin Nutr 49:127–131. [DOI] [PubMed] [Google Scholar]
- 36.Konstantinov SR, Smidt H, Akkermans AD, Casini L, Trevisi P, Mazzoni M, De Filippi S, Bosi P, de Vos WM. 2008. Feeding of Lactobacillus sobrius reduces Escherichia coli F4 levels in the gut and promotes growth of infected piglets. FEMS Microbiol Ecol 66:599–607. doi: 10.1111/j.1574-6941.2008.00517.x. [DOI] [PubMed] [Google Scholar]
- 37.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. [DOI] [PubMed] [Google Scholar]
- 38.Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young IT. 1977. Proof without prejudice—use of Kolmogorov-Smirnov test for analysis of histograms from flow systems and other sources. J Histochem Cytochem 25:935–941. doi: 10.1177/25.7.894009. [DOI] [PubMed] [Google Scholar]
- 40.Melkebeek V, Goddeeris BM, Cox E. 2013. ETEC vaccination in pigs. Vet Immunol Immunopathol 152:37–42. doi: 10.1016/j.vetimm.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 41.Canibe N, Jensen BB. 2012. Fermented liquid feed—microbial and nutritional aspects and impact on enteric diseases in pigs. Anim Feed Sci Technol 173:17–40. doi: 10.1016/j.anifeedsci.2011.12.021. [DOI] [Google Scholar]
- 42.Amezcua MDR, Friendship R, Dewey C, Weese JS, de Lange C, Reid G. 2007. Effects on growth performance, feed efficiency, and health of weanling pigs fed fermented liquid whey inoculated with lactic acid bacteria that inhibit Escherichia coli in vitro. J Swine Health Prod 15:320–329. [Google Scholar]
- 43.Korakli M, Rossmann A, Gänzle MG, Vogel RF. 2001. Sucrose metabolism and exopolysaccharide production in wheat and rye sourdoughs by Lactobacillus sanfranciscensis. J Agric Food Chem 49:5194–5200. doi: 10.1021/jf0102517. [DOI] [PubMed] [Google Scholar]
- 44.Heo JM, Opapeju FO, Pluske JR, Kim JC, Hampson DJ, Nyachoti CM. 2013. Gastrointestinal health and function in weaned pigs: a review of feeding strategies to control post-weaning diarrhoea without using in-feed antimicrobial compounds. J Anim Physiol Anim Nutr 97:207–237. doi: 10.1111/j.1439-0396.2012.01284.x. [DOI] [PubMed] [Google Scholar]
- 45.Schwab C, Tveit AT, Schleper C, Urich T. 2014. Gene expression of lactobacilli in murine forestomach biofilms. Microb Biotechnol 7:347–359. doi: 10.1111/1751-7915.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filion Me. 2012. Quantitative real-time PCR in applied microbiology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 47.De Angelis M, Siragusa S, Caputo L, Ragni A, Burzigotti R, Gobbetti M. 2007. Survival and persistence of Lactobacillus plantarum 4.1 and Lactobacillus reuteri 3S7 in the gastrointestinal tract of pigs. Vet Microbiol 123:133–144. doi: 10.1016/j.vetmic.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 48.du Toit M, Franz CMAP, Dicks LMT, Schillinger U, Haberer P, Warlies B, Ahrens F, Holzapfel WH. 1998. Characterisation and selection of probiotic lactobacilli for a preliminary minipig feeding trial and their effect on serum cholesterol levels, faeces pH and faeces moisture content. Int J Food Microbiol 40:93–104. doi: 10.1016/S0168-1605(98)00024-5. [DOI] [PubMed] [Google Scholar]
- 49.Gänzle MG, Höltzel A, Walter J, Jung G, Hammes WP. 2000. Characterization of reutericyclin produced by Lactobacillus reuteri LTH2584. Appl Environ Microbiol 66:4325–4333. doi: 10.1128/AEM.66.10.4325-4333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rijkers GT, Bengmark S, Enck P, Haller D, Herz U, Kalliomaki M, Kudo S, Lenoir-Wijnkoop I, Mercenier A, Myllyluoma E, Rabot S, Rafter J, Szajewska H, Watzl B, Wells J, Wolvers D, Antoine JM. 2010. Guidance for substantiating the evidence for beneficial effects of probiotics: current status and recommendations for future research. J Nutr 140:671S–676S. doi: 10.3945/jn.109.113779. [DOI] [PubMed] [Google Scholar]
- 51.Ruas-Madiedo P, Gueimonde M, Margolles A, Reyes-Gavilan CGDL, Salminen S. 2006. Exopolysaccharides produced by probiotic strains modify the adhesion of probiotics and enteropathogens to human intestinal mucus. J Food Prot 69:2011–2015. [DOI] [PubMed] [Google Scholar]
- 52.Byun R, Nadkarni MA, Chhour KL, Martin FE, Jacques NA, Hunter N. 2004. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J Clin Microbiol 42:3128–3136. doi: 10.1128/JCM.42.7.3128-3136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song YL, Kato N, Liu CX, Matsumiya Y, Kato H, Watanabe K. 2000. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol Lett 187:167–173. doi: 10.1016/S0378-1097(00)00196-8. [DOI] [PubMed] [Google Scholar]
- 54.Chen J, Griffiths MW. 1998. PCR differentiation of Escherichia coli from other Gram-negative bacteria using primers derived from the nucleotide sequences flanking the gene encoding the universal stress protein. Lett Appl Microbiol 27:369–371. doi: 10.1046/j.1472-765X.1998.00445.x. [DOI] [PubMed] [Google Scholar]
- 55.Kotlowski R, Bernstein CN, Sepehri S, Krause DO. 2007. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 56:669–675. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han W, Liu B, Cao B, Beutin L, Kruger U, Liu H, Li Y, Liu Y, Feng L, Wang L. 2007. DNA microarray-based identification of serogroups and virulence gene patterns of Escherichia coli isolates associated with porcine postweaning diarrhea and edema disease. Appl Environ Microbiol 73:4082–4088. doi: 10.1128/AEM.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Casey TA, Bosworth BT. 2009. Design and evaluation of a multiplex polymerase chain reaction assay for the simultaneous identification of genes for nine different virulence factors associated with Escherichia coli that cause diarrhea and edema disease in swine. J Vet Diagn Invest 21:25–30. doi: 10.1177/104063870902100104. [DOI] [PubMed] [Google Scholar]
- 58.Setia A, Bhandari SK, House JD, Nyachoti CM, Krause DO. 2009. Development and in vitro evaluation of an Escherichia coli probiotic able to inhibit the growth of pathogenic Escherichia coli K88. J Anim Sci 87:2005–2012. doi: 10.2527/jas.2008-1400. [DOI] [PubMed] [Google Scholar]