Abstract
Temporal changes in the distribution of Salmonella subtypes in livestock populations may have important impacts on human health. The first objective of this research was to determine the within-farm changes in the population of subtypes of Salmonella on Michigan dairy farms that were sampled longitudinally in 2000-2001 and again in 2009. The second objective was to determine the yearly frequency (2001 through 2012) of reported human illnesses in Michigan associated with the same subtypes. Comparable sampling techniques were used to collect fecal and environmental samples from the same 18 Michigan dairy farms in 2000-2001 and 2009. Serotypes, multilocus sequence types (STs), and pulsed-field gel electrophoresis (PFGE) banding patterns were identified for isolates from 6 farms where >1 Salmonella isolate was recovered in both 2000-2001 and 2009. The distribution of STs was significantly different between time frames (P < 0.05); only two of 31 PFGE patterns were identified in both time frames, and each was recovered from the same farm in each time frame. Previously reported within-farm decreases in the frequency of multidrug-resistant (MDR) Salmonella were due to recovery of MDR subtypes of S. enterica serotypes Senftenberg and Typhimurium in 2000-2001 and genetically distinct, pansusceptible subtypes of the same serotypes in 2009. The annual frequency of human illnesses between 2001 and 2012 with a PFGE pattern matching a bovine strain decreased for patterns recovered from dairy farms in 2000-2001 and increased for patterns recovered in 2009. These data suggest important changes in the population of Salmonella on dairy farms and in the frequency of human illnesses associated with cattle-derived subtypes.
INTRODUCTION
Globally, an estimated 93.8 million illnesses and 155,000 deaths are caused by Salmonella enterica annually (1). Livestock are important reservoirs for Salmonella, and efforts to reduce the contamination of food will likely be enhanced by controlling the pathogen on farms. While the incidence of other food-borne pathogens has decreased, the incidence of salmonellosis has remained stable, and national goals for reducing incidence have not been met (2). Cattle serve as a reservoir of Salmonella which can be transmitted to people (3), and beef and dairy products account for a notable proportion of traceable Salmonella outbreaks (4). Salmonella found on dairy farms could be transmitted through contamination and inadequate pasteurization of dairy products, contamination of carcasses and lymph nodes (cull dairy cattle), or indirect transmission through the environment. There is substantial overlap in the antimicrobial resistance (AMR) phenotypes, serotypes, and pulsed-field gel electrophoresis (PFGE) patterns of Salmonella strains recovered from dairy cattle and those that cause disease in humans (5–7). In general, a diverse population of Salmonella serotypes are recovered from cattle, but only a small number of serotypes account for the majority of illnesses. Therefore, temporal changes in the prevalence, distribution, or antimicrobial resistance profiles of Salmonella serotypes and molecular subtypes recovered on dairy farms may have important impacts on human health.
Nation-level studies suggest substantial changes in the population of Salmonella on dairy farms in the United States. Between cross-sectional studies of dairy farms in 1996, 2002, and 2007, the proportions of farms and cows that were positive for Salmonella approximately doubled (8). The same studies also demonstrated decreases in the proportion of Salmonella strains resistant to antimicrobials. Approximately 17.7% of isolates were resistant to at least one antimicrobial in 2002, compared to 3.4% of isolates in 2007 (8). Consistent with these data, our prior retro-prospective study demonstrated within-farm decreases in the proportion of multidrug-resistant (MDR) Salmonella strains between 2000-2001 and 2009 (9).
Salmonella can be found on up to 90% of dairy farms (10) and may persist within dairy herds for years (11–13), but most observational studies of Salmonella on dairy farms are cross-sectional or of limited duration, which may not be sufficient to understand long-term or within-farm population changes caused by the acquisition and/or disappearance of strains. Furthermore, previous molecular epidemiological studies of Salmonella often use diagnostic laboratory collections of isolates which provide the necessary diversity to study phylogenetic relatedness (14, 15); however, these inferences cannot be extrapolated to the population of Salmonella colonizing dairy farms. Virulence is not a property of all Salmonella strains (16), and the distribution of serotypes in livestock differs from the distribution of serotypes causing disease in humans (17), suggesting that some serotypes may be better adapted for asymptomatic colonization than for causing disease. Therefore, changes in the population of Salmonella on dairy farms may result in changes in the impact of dairy farms on human health. Likewise, the emergence or disappearance of MDR strains may change the frequency of resistant human infections. The proportion of nontyphoidal Salmonella strains recovered from humans in the United States that were resistant to at least one antimicrobial, for example, was observed to decrease between 1999 and 2010 (18).
Based on current knowledge gaps and previously reported decreases in AMR, we conducted a retro-prospective study to measure within-farm changes in the prevalence and distribution of Salmonella subtypes recovered on Michigan dairy farms that were sampled longitudinally in 2000-2001 and again in 2009. The objectives of this study were 3-fold: (i) to use a combination of subtyping techniques to describe the distribution of subtypes of Salmonella recovered on farms that participated in a 2000-2001 longitudinal study of Michigan dairy farms (10), (ii) to sample the same farms using comparable techniques and compare the relatedness of the strains from 2000-2001 to prevalent strains of Salmonella recovered in 2009 to better understand previously reported changes in AMR (9), and (iii) to describe temporal changes in the frequency of human illnesses associated with the subtypes found on dairy farms in 2000-2001 and 2009.
MATERIALS AND METHODS
Study design.
This study used a retro-prospective study design, and the data consisted of two components: retrospective data collected from Michigan dairy farms in 2000-2001 and prospective data collected 10 years later from the same farms. Retrospective data were retrieved from a 2000-2001 multicenter, longitudinal study of Salmonella shedding on randomly selected dairy farms in Michigan, New York, Wisconsin, and Minnesota (10). Stored Salmonella isolates collected in 2000-2001 were retrieved from the Center for Comparative Epidemiology (CCE) at Michigan State University (MSU). Samples from the same farms were collected in August of 2009. Approvals from the MSU Institutional Animal Care and Use Committee and the MSU Institutional Review Board were obtained prior to the initiation of this research. In 2000-2001, 31 dairy farms in Michigan that met the following criteria were selected: less than 100 miles from Michigan State University, milking greater than 30 Holstein cows, raising their own calves for replacements, and shipping milk year-round (10) For the data collected in 2009, all Michigan dairy farms that participated in 2000 were recruited to participate.
Sample collection.
Comparable sampling plans for collecting fecal and environmental samples were used in both 2000-2001 and 2009, as reported previously (9). Briefly, in 2000-2001, farms were sampled every other month, resulting in five sampling events (10). Two farms (101 and 102) were initially sampled weekly for 8 consecutive weeks in the spring/summer of 2000 and then subsequently sampled every 2 months for five consecutive visits. In July or August 2009, 16 farms were sampled once, and two farms were sampled twice. Fecal samples were collected from the rectums of dairy cattle using a single-use rectal sleeve and from calves using digital rectal retrieval. In both 2000-2001 and 2009, healthy lactating cows and “target” animals were sampled from each farm. Target animals were defined as dairy animals most likely to be shedding Salmonella, including preweaned calves, cows identified as sick by the farm management, cows within 14 days of their calving date, and cows scheduled to be culled within 14 days. The number, types, and collection of environmental samples were as previously described (9). All samples were stored in commercial bags, placed in a cooler with ice, and processed the following day.
Salmonella isolation and serotyping.
The techniques used for isolation of Salmonella for this set of isolates have been previously described (9). Briefly, isolation of Salmonella was performed in the same laboratory with highly similar protocols in 2000-2001 and 2009. Salmonella isolates harvested in 2000 were frozen in tryptic soy broth-glycerol solution at −80 C and stored in cryovials. In 2009, these were retrieved and underwent further biochemical confirmation before serotyping and antimicrobial susceptibility testing. Serotype identification was performed in 2009 for all isolates at the Diagnostic Center for Population and Animal Health (DCPAH) at MSU using the Kauffman-White scheme (19). Where a group of isolates collected from the same farm on the same day had indistinguishable PFGE banding patterns, only one isolate was selected, and the remaining isolates are reported as the same serotype.
PFGE.
For pulsed-field gel electrophoresis (PFGE), up to two randomly selected isolates from each Salmonella-positive fecal or environmental sample were selected. If more than two isolates were recovered from a single sample, then two isolates were randomly chosen to undergo PFGE. PFGE was conducted at the DCPAH using a CDC standardized protocol and the XbaI rare-cutting restriction enzyme (20). Dendrograms were constructed by applying hierarchical agglomerative clustering techniques (unweighted pair group method with arithmetic means) to similarity matrices calculated using the Dice coefficient of similarity. Band tolerance and optimization settings of 1.5% were used. If two isolates from the same sample had indistinguishable PFGE patterns, then only one of the isolates was included in the statistical analysis and summary statistics. Unique PFGE patterns identified at the DCPAH were assigned names to denote where the pattern was identified, serotype, and unique pattern number within serotype (e.g., MSU.Sty.1).
MLST.
To define sequence types (STs), the partial gene sequences of seven housekeeping genes (thrA, purE, sucA, hisD, aroC, hemD, and dnaN) were determined according to a standard multilocus sequence typing (MLST) protocol for Salmonella (http://mlst.warwick.ac.uk/mlst/). Isolates with different PFGE patterns, collected from different farms, or collected in different time frames (2000-2001 or 2009) were selected for MLST. Primer sequences and amplification conditions were according to standardized protocols (21). Edited sequences were uploaded to the MLST website (http://mlst.warwick.ac.uk/mlst/) to identify the allelic numbers and STs for each isolate. Where a group of isolates collected from the same farm on the same day had indistinguishable PFGE banding patterns, only one isolate was selected, and the remaining isolates are reported as the same serotype and ST.
Comparison with human clinical isolates.
PFGE patterns of Salmonella strains recovered from Michigan dairy farms in this study were compared to all the PFGE patterns of the same serotypes that were identified at the Michigan Department of Community Health (MDCH) during the same time frame. For the serotypes recovered from Michigan dairy farms in 2000-2001 or 2009, raw images of PFGE patterns recovered from reported illnesses between the years 2000 and 2012 were obtained. Serotypes rarely identified at the MDCH (<100 times between 2000 and 2012) were excluded; however, serotypes with PFGE patterns that persisted between the two time frames were included in the analysis regardless of frequency. The yearly frequency (2000 to 2012) at MDCH was determined for patterns that were indistinguishable from the patterns recovered on dairy farms. Names of the PFGE patterns used at the MDCH are reported alongside the pattern names given by investigators at the DCPAH.
Statistical analysis.
Fisher's exact tests were used to compare within-farm differences in the proportions of samples positive in 2000-2001 and 2009 for samples taken from cows, calves, or the environment. To increase the validity of comparisons of prevalence between the two time periods, the summer sampling visit from 2000-2001 that most closely matched the summer sampling date from the same farm in 2009 was included in the statistical analysis. Cow or calf samples were not taken in the summer months (July, August, or September) for two herds (102 and 131), so samples from June 2001 were used instead. Descriptive statistics were generated to describe changes in the frequency of STs, serotypes, and PFGE patterns within farms between different sample types, between farms, and between years. The frequency distributions of STs between time frames were compared using a chi-square test of independence. The new information on strain characteristics is reported in light of previously identified changes in antimicrobial resistance for this set of isolates (9).
RESULTS
Prevalence of Salmonella in 2000-2001 and 2009.
In total, 18 of the 31 dairy farms that participated in the 2000-2001 study agreed to participate again in 2009. From this subset of farms in 2000-2001, a total of 5,358 samples were collected from healthy cows (2, 994), healthy calves (770), target animals (675), and the environment (919) (Table 1). Across all five sampling visits in 2000-2001, at least one isolate was recovered from 77% (14/18) of herds and 6% (264/5,358) of samples (Table 1). There was a seasonal pattern of shedding, with 2.8% (49/1,733), 5.8% (103/1,790), 10% (86/862), and 2.7% (26/973) of samples positive in the winter (January to March), spring (April to June), summer (July to September), and fall (October to December), respectively, in the 2000-2001 time frame. Two high-prevalence herds (114 and 111) accounted for 73% of the positive samples (Table 2), and the proportion of samples positive on farms 111 and 114 ranged between 13% and 62% on each visit. For the remaining 12 positive herds, the prevalence was <10%, and nine herds had fewer than three positive samples across all five visits (Table 2).
TABLE 1.
Numbers of samples from the same 18 dairy farms in 2000-2001 and 2009
| Sample type | 2000-2001 |
Summer visits |
||||
|---|---|---|---|---|---|---|
| 2000 or 2001 |
2009 |
|||||
| No. of samples | Proportion positive for Salmonella | No. of samples | Proportion positive for Salmonella | No. of samples | Proportion positive for Salmonella | |
| Cow samples | ||||||
| Healthy | 2,994 | 0.05 | 518 | 0.09 | 238 | 0.14 |
| Cull | 77 | 0.03 | 8 | 0.13 | 33 | 0.06 |
| Closeup | 176 | 0.02 | 38 | 0.03 | 73 | 0.11 |
| Fresh | 257 | 0.06 | 42 | 0.12 | 76 | 0.12 |
| Sick | 165 | 0.04 | 32 | 0.13 | 44 | 0.23 |
| Total | 3,669 | 0.05 | 638 | 0.09 | 464 | 0.13 |
| Calf samples | 770 | 0.06 | 145 | 0.08 | 163 | 0.07 |
| Environmental samples | ||||||
| Sick pen | 50 | 0 | 9 | 0 | 11 | 0.45 |
| Manure storage area | 103 | 0.14 | 18 | 0.17 | 17 | 0.29 |
| Hair coat of cull cow | 57 | 0.07 | 8 | 0 | 11 | 0 |
| Maternity pen | 85 | 0.04 | 16 | 0 | 18 | 0.17 |
| Milk filter | 99 | 0.05 | 17 | 0.06 | 16 | 0.06 |
| Waterer | 105 | 0.06 | 18 | 0.11 | 18 | 0.06 |
| Calf pen | 104 | 0.04 | 18 | 0.06 | 17 | 0.12 |
| Other | 306 | 0.03 | ||||
| Missing information | 10 | 3 | 4 | 0.25 | ||
| Total | 919 | 0.05 | 107 | 0.07 | 112 | 0.16 |
| Total | 5,358 | 0.05 | 890 | 0.09 | 739 | 0.12 |
TABLE 2.
Percentages of samples positive for Salmonella and herd sizes on 18 dairy farms sampled in the summers of 2000 or 2001 and 2009
| Farm | 2000-2001 |
2009 |
||||||
|---|---|---|---|---|---|---|---|---|
| Herd sizea | Prevalence |
Herd size | Prevalence, summer visits |
|||||
| All visits |
Summer visits |
|||||||
| % positive | No. positive/total | % positive | No. positive/total | % positive | No. positive/total | |||
| 101b | 78 | 3.6 | 19/528 | 5.4 | 2/37 | 60 | 62.9 | 22/35 |
| 102 | 113 | 0.4 | 3/696 | 2 | 1/51 | 197 | 0.0 | 0/36 |
| 108 | 77 | 0 | 0/221 | 0 | 0/38 | 145 | 0.0 | 0/38 |
| 109 | 318 | 0 | 0/302 | 0 | 0/60 | 700 | 57.4 | 27/47 |
| 111b | 108 | 44.9 | 118/263 | 64.2 | 34/53 | 116 | 6.2 | 5/81 |
| 112 | 74 | 1 | 2/194 | 2.9 | 1/34 | 83 | 0.0 | 0/38 |
| 114b | 110 | 29.3 | 76/259 | 23.9 | 11/46 | 193 | 29.8 | 14/47 |
| 118 | 191 | 0 | 0/268 | 0 | 0/57 | 158 | 0.0 | 0/46 |
| 119 | 116 | 0.4 | 1/250 | 2.2 | 1/46 | 99 | 0.0 | 0/35 |
| 121b | 473 | 0.7 | 2/267 | 1.6 | 1/63 | 818 | 20.0 | 10/50 |
| 122 | 186 | 0 | 0/293 | 0 | 0/57 | 503 | 8.5 | 4/47 |
| 125b | 257 | 5.2 | 15/289 | 25 | 14/56 | 511 | 23.9 | 11/46 |
| 126 | 305 | 0.3 | 1/301 | 0 | 0/59 | 1,067 | 0.0 | 0/46 |
| 127 | 84 | 0.5 | 1/207 | 2.5 | 1/40 | 90 | 0.0 | 0/37 |
| 128 | 151 | 0.3 | 1/288 | 1.9 | 1/54 | 170 | 0.0 | 0/42 |
| 129b | 247 | 6.9 | 22/319 | 18 | 11/61 | 456 | 2.2 | 2/91 |
| 131 | 87 | 1 | 2/201 | 0 | 0/36 | 63 | 3.2 | 1/31 |
| 132 | 90 | 0.5 | 1/212 | 2.4 | 1/42 | 76 | 2.7 | 1/37 |
| Total | 4.9 | 264/5,358 | 8.9 | 79/890 | 12.4 | 97/830 | ||
Approximate total number of lactating cows.
One of six farms where isolates were selected for additional characterization.
Comparable sampling techniques were used to sample the same herds in 2009, and Salmonella was recovered from 12% (97/830) of samples and 56% (10/18) of farms (Table 1). The within-farm prevalence over all animal and environmental samples ranged from 0% to 63% and was over 50% for 2 herds, between 20% and 50% for 3 herds, and less than 10% for 13 herds. Farms with herd sizes (number of lactating cows) of >500, 100 to 499, and <100 had a prevalences of 22.0% (52/236), 5.5% (21/381), and 11.2% (24/213), respectively (Table 2). Samples with the highest proportion of positive results came from the sick pen (5/11, 45%), manure storage area (6/19, 32%), and sick adult cattle (10/49, 20%). The overall prevalence in the summer of 2009 (12%) was higher than the prevalence of Salmonella in the summer of 2000 or 2001 (Table 1).
Eleven of the 18 herds had less than 10% prevalence in both summers, including three farms where Salmonella was not recovered in either year (Table 2). For the high-prevalence herds in 2000-2001 (farms 111 and 114), farm 111 had a 6% prevalence in 2009, and farm 114 had a 30% prevalence in 2009 (Table 2). A Fisher exact test was used to compare the summer prevalence of Salmonella in cow, calf, and environmental samples between the summers of 2000 or 2001 and 2009. The within-farm cow prevalence was significantly higher (P < 0.05) in 2009 for two farms (101 and 109), and significantly lower in 2009 for farm 111. The proportion of calf samples was significantly lower in 2009 for farm 129, and the proportion of environmental samples was significantly higher for two farms (109 and 121) in 2009 than in 2000 or 2001.
To understand within-farm changes in the population of Salmonella, the serotype, ST, and PFGE pattern was identified for isolates originating from farms where at least two isolates were recovered in both sampling time frames. Six farms had >1 isolate in both time periods, and on these six farms, Salmonella was recovered from 16% (252/1,925) and 18% (64/350) of samples in 2000-2001 and 2009, respectively. There were 35 samples with >1 distinguishable PFGE pattern, which were evenly distributed across animal (11.1%, 29/262) and environmental (11.8%, 6/51) samples. A larger number of positive samples in 2009 (20.6%, 13/64) than in 2000-2001 (8.8%, 22/252) had multiple patterns. In total, 271 and 77 isolates from 2000-2001 and 2009, respectively, were included in subsequent molecular and statistical analyses.
Serotypes, sequence types, and PFGE patterns in 2000-2001 and 2009.
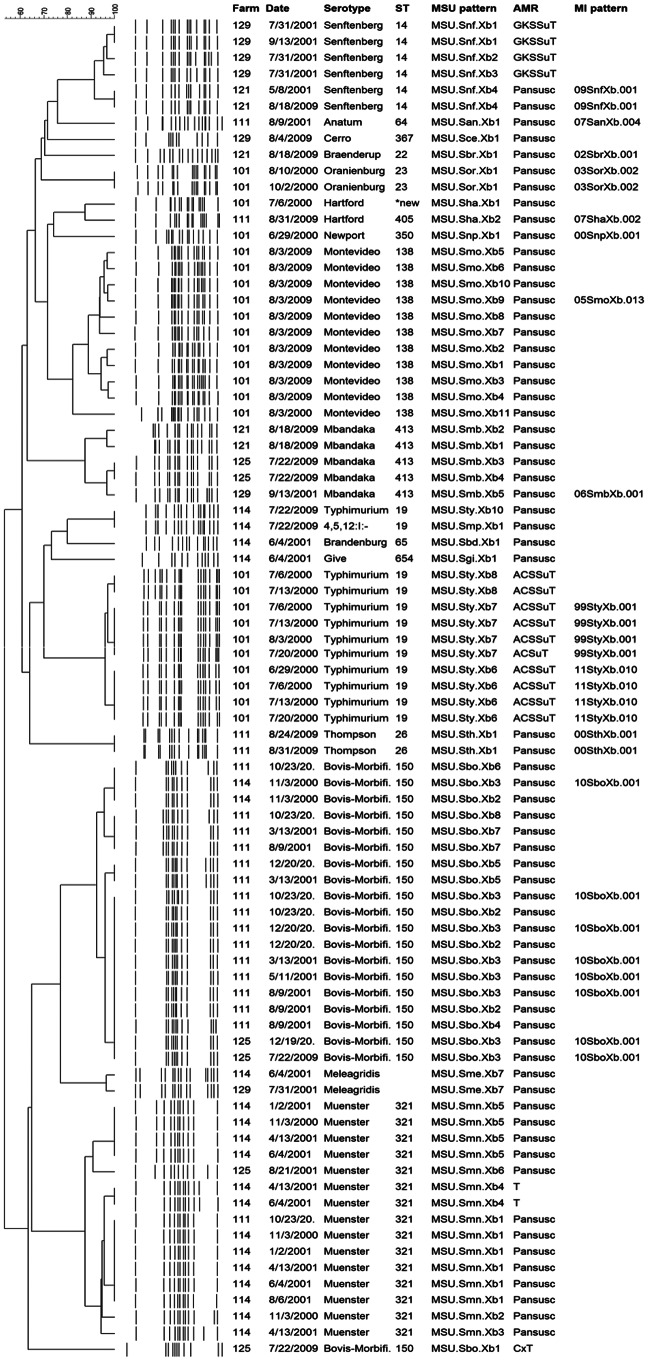
Out of 271 isolates recovered during the longitudinal sampling period from 2000 to 2001, a total of 13 serotypes, 11 STs, and 31 distinguishable PFGE patterns were identified. In 2009, 12 different serotypes, 12 sequence types, and 21 unique PFGE patterns were identified out of 77 isolates recovered from the same six herds (Fig. 1). In all but one case, there was a single ST for each serotype, regardless of the farm or year the sample was taken. A novel ST of S. enterica serotype Hartford (recovered in 2000) differed by a single base pair in the dnaN allele and had a PFGE pattern distinguishable from that for ST405 of S. Hartford recovered from in 2009 (Table 3). Two serotypes, S. enterica serotypes Typhimurium and 4:5:12:i:−, were both ST 19 and had indistinguishable PFGE patterns in 2009. The number of PFGE patterns within individual STs ranged from 1 to 11; S. enterica serotype Montevideo (ST 138) was the most diverse with the largest number of PFGE patterns.
FIG 1.
Serotypes, multilocus sequence types, and unique PFGE patterns identified on each sampling date for Michigan dairy farms sampled in 2000-2001 and 2009. ST, sequence type; MSU pattern, pattern name assigned by investigators; MI pattern, pattern name assigned by the Michigan Department of Community Health (MDCH); A, ampicillin; C, chloramphenicol; Cx, ceftriaxone; G, gentamicin; K, kanamycin; S, streptomycin; Su, sulfisoxazole; T, tetracycline.
TABLE 3.
Serotypes and sequence types of nontyphoidal salmonellae recovered on the same Michigan dairy farms sampled in 2000-2001 and 2009
| Farm | 2000-2001 |
2009 |
||||||
|---|---|---|---|---|---|---|---|---|
| Serogroup | Serotype | ST | No. of isolates | Serogroup | Serotype | ST | No. of isolates | |
| 101 | B | S. Typhimurium | 19 | 17 | C1 | S. Montevideo | 138 | 29 |
| B | S. Oranienburg | 65 | 4 | |||||
| C1 | S. Hartford | Newa | 1 | |||||
| C1 | S. Montevideo | 138 | 1 | |||||
| C2 | S. Newport | 350 | 1 | |||||
| 111 | C2 | S. Bovismorbificans | 150 | 122 | C1 | S. Thompson | 26 | 4 |
| E1 | S. Anatum | 64 | 2 | C1 | S. Hartford | 405 | 1 | |
| E1 | S. Muenster | 88 | 1 | |||||
| 114 | E1 | S. Muenster | 88 | 73 | B | S. Typhimurium | 19 | 8 |
| C1 | S. Brandenburg | 65 | 3 | B | S. 4,5,12:I:− | 19 | 8 | |
| E1 | S. Give | 654 | 3 | |||||
| C2 | S. Bovismorbificans | 150 | 2 | |||||
| E1 | S. Meleagridis | NDb | 1 | |||||
| 121 | E4 | S. Senftenberg | 14 | 1 | C1 | S. Braenderup | 22 | 4 |
| C1 | S. Mbandaka | 413 | 5 | |||||
| E4 | S. Senftenberg | 14 | 3 | |||||
| 125 | E1 | S. Muenster | 88 | 14 | C2 | S. Bovismorbificans | 150 | 10 |
| C2 | S. Bovismorbificans | 150 | 1 | C1 | S. Mbandaka | 413 | 3 | |
| 129 | E4 | S. Senftenberg | 14 | 22 | K | S. Cerro | 367 | 2 |
| C1 | S. Mbandaka | 413 | 1 | |||||
| E1 | S. Meleagridis | ND | 1 | |||||
| Total | 271 | 77 | ||||||
Novel sequence type.
ND, not determined.
Within-farm distribution and relatedness of Salmonella subtypes.
In 2000-2001, subtypes of Salmonella frequently persisted across multiple bimonthly sampling visits through the longitudinal sampling period in 2000-2001. There were 11 different PFGE patterns that were repeatedly recovered on >1 sampling visit from the same farm. The number of days between the first and last recovery of a subtype ranged between 21 and 290. Two herds (111 and 114) had a persistently high prevalence of Salmonella. On farm 111, one S. enterica serotype Bovismorbificans (MSU.Sbo.Xb3) pattern represented 84% (106/126) of the recovered isolates. Similarly, on farm 114, 12 unique PFGE patterns representing three serotypes were recovered, but a single pattern of S. enterica serotype Muenster (MSU.Smn.Xb1) represented 78% (65/83) of the isolates. Other subtypes in the 2000-2001 time frame were recovered from <10% of samples across all visits. In 2009, three herds (101, 114, and 125) with a >20% prevalence were infected with Salmonella serotypes Montevideo, Typhimurium, and Bovismorbificans (Table 3). A single PFGE pattern made up 28% (8/29), 100% (16/16), and 90% (9/10) of the isolates recovered from each of the three farms (101, 114, and 125), respectively.
Indistinguishable subtypes were distributed across different production classes and multiple environments within the same farm, suggesting frequent transmission and widespread environmental contamination. For each sampling visit, comparable proportions of cow, calf, and environmental samples were positive for each subtype (Table 4). MDR subtypes showed the largest difference in the within-farm prevalence across cow and calf samples. For instance, a higher proportion of calf samples (47%, 15/32) than of cow samples (4%, 3/80) on farm 129 were positive for the MDR subtype of S. enterica serotype Senftenberg (MSU.Snf.Xb1). Similarly, a higher proportion of calf samples (60%, 3/5) than of cow samples (4%, 1/28) were positive for any MDR pattern of Salmonella Typhimurium (MSU.Sty.Xb6) within the same sampling visit (Table 4).
TABLE 4.
Percentages of cow, calf, and environmental samples positive for a PFGE pattern on Michigan dairy farmsa
| Farm | Serotype | PFGE pattern | AMR pattern | Date (mo/day/yr) | All samples |
Cow samples |
Calf samples |
Environmental samples |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % positive | No. positive/total | % positive | No. positive/total | % positive | No. positive/total | % positive | No. positive/total | |||||
| 101 | S. Montevideo | MSU.Smo.Xb5 | 8/3/2009 | 19 | 8/42 | 28 | 8/29 | 0 | 0/6 | 0 | 0/7 | |
| 101 | S. Montevideo | MSU.Smo.Xb9 | 8/3/2009 | 17 | 7/42 | 10 | 3/29 | 0 | 0/6 | 57 | 4/7 | |
| 101 | S. Typhimurium | MSU.Sty.Xb6 | ACSSuT | 7/20/2000 | 10 | 4/41 | 4 | 1/28 | 60 | 3/5 | 0 | 0/8 |
| 111 | S. Bovismorbificans | MSU.Sbo.Xb3 | 10/23/2000 | 54 | 30/56 | 50 | 22/44 | 50 | 2/4 | 75 | 6/8 | |
| 111 | S. Bovismorbificans | MSU.Sbo.Xb3 | 12/20/2000 | 36 | 18/50 | 39 | 14/36 | 50 | 3/6 | 13 | 1/8 | |
| 111 | S. Bovismorbificans | MSU.Sbo.Xb3 | 3/13/2001 | 26 | 15/58 | 18 | 7/38 | 30 | 3/10 | 50 | 5/10 | |
| 111 | S. Bovismorbificans | MSU.Sbo.Xb3 | 5/11/2001 | 29 | 14/48 | 26 | 9/34 | 33 | 2/6 | 38 | 3/8 | |
| 111 | S. Bovismorbificans | MSU.Sbo.Xb3 | 8/9/2001 | 48 | 29/60 | 59 | 24/41 | 38 | 3/8 | 18 | 2/11 | |
| 114 | S. Muenster | MSU.Smn.Xb1 | 11/3/2000 | 48 | 24/50 | 47 | 17/36 | 67 | 4/6 | 38 | 3/8 | |
| 114 | S. Muenster | MSU.Smn.Xb1 | 1/2/2001 | 12 | 6/52 | 3 | 1/35 | 44 | 4/9 | 13 | 1/8 | |
| 114 | S. Muenster | MSU.Smn.Xb1 | 4/13/2001 | 18 | 9/51 | 11 | 4/36 | 50 | 3/6 | 22 | 2/9 | |
| 114 | S. Muenster | MSU.Smn.Xb1 | 6/4/2001 | 23 | 15/64 | 25 | 11/44 | 0 | 0/8 | 33 | 4/12 | |
| 114 | S. Muenster | MSU.Smn.Xb1 | 8/6/2001 | 22 | 11/49 | 24 | 9/37 | 0 | 0/4 | 25 | 2/8 | |
| 114 | S. 4,5,12:I:- | MSU.Smp.Xb1 | 7/22/2009 | 16 | 8/49 | 18 | 6/34 | 22 | 2/9 | 0 | 0/6 | |
| 114 | S. Typhimurium | MSU.Sty.Xb10 | 7/22/2009 | 16 | 8/49 | 15 | 5/34 | 22 | 2/9 | 17 | 1/6 | |
| 125 | S. Bovismorbificans | MSU.Sbo.Xb3 | 7/22/2009 | 19 | 9/48 | 22 | 6/27 | 20 | 3/15 | 0 | 0/6 | |
| 125 | S. Muenster | MSU.Smn.Xb6 | 8/21/2001 | 24 | 14/59 | 33 | 13/40 | 10 | 1/10 | 0 | 0/9 | |
| 129 | S. Senftenberg | MSU.Snf.Xb1 | GKSSuT | 7/31/2001 | 15 | 10/66 | 5 | 2/40 | 41 | 7/17 | 11 | 1/9 |
| 129 | S. Senftenberg | MSU.Snf.Xb1 | GKSSuT | 9/13/2001 | 16 | 10/64 | 3 | 1/40 | 53 | 8/15 | 11 | 1/9 |
Shown where the total proportion of positive samples was higher than 10%.
Between-farm distribution and relatedness of Salmonella subtypes.
The same serotype was recovered from multiple farms within the 2000-2001 (S. enterica serotypes Bovismorbificans, Meleagridis, Senftenberg, and Muenster) and 2009 (S. enterica serotype Mbandaka) time frames. PFGE patterns differed across farms, with the exception of two PFGE patterns (MSU.Sbo.Xb3, MSU.Smn.Xb1) representing Salmonella serotypes Bovismorbificans and Muenster, respectively, which were each recovered on farms 111 and 114 in the 2000-2001 sampling time frame. On farm 111 the MSU.Sbo.Xb3 pattern was recovered from over 25% of samples on each of five visits, while on farm 114 the same pattern was recovered from only a single sample. Conversely, a single sample on farm 111 was positive for the MSU.Smn.Xb1 subtype, while 20.6% (65/250) of samples from farm 114 were positive for this pattern. While important differences between the strains exist, the identification of the same strains on two different farms at very different prevalence levels suggests important differences between farm environments.
Relatedness of Salmonella subtypes between 2000-2001 and 2009.
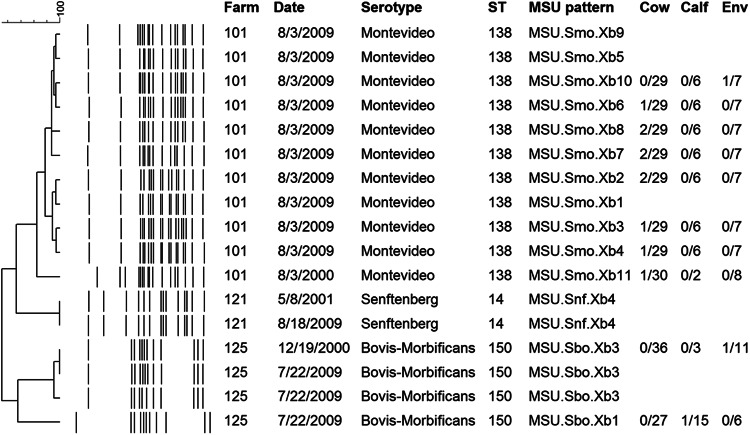
Of the six farms positive in both years, three had the same serotype recovered at both time points. Two PFGE patterns (MSU.Sbo.Xb3 and MSU.Snf.Xb4), representing Salmonella serotypes Bovismorbificans and Senftenberg, were recovered in both 2000 and 2009, and each was recovered within the same farm in both time frames (Fig. 2), suggesting long-term persistence of the strains within the same farm. The two subtypes, MSU.Sbo.Xb3 and MSU.Snf.Xb4, were recovered from single samples in 2001 on farms 125 (1/288) and 121 (1/267) and subsequently recovered from 6% (3/50) and 20% (9/46) of samples on the same farms in 2009, respectively. Three other serotypes, Salmonella serotypes Typhimurium, Hartford, and Mbandaka, were recovered in both time frames but from different herds. On each farm, strains had distinguishable banding patterns (Fig. 1).
FIG 2.
Pulsed-field gel electrophoresis patterns and dendrogram of serotypes recovered from the same farms in both the 2000-2001 and 2009 time frames. On two farms, indistinguishable patterns were recovered within the same farm in both time frames (farm 125–S. enterica serotype Bovis-Morbificans and farm 121–S. enterica serotype Senftenberg) Positive/total samples for each subtype on each sampling date for adult cows, preweaned calves, and environmental swabs are shown.
The overall distribution of sequence types was significantly different between time periods (P < 0.05). Differences in strains between the two time points reflected a higher prevalence of strains of serogroup C1 and a lower prevalence of serogroups E1 and E4. Serogroups represent important antigenic properties of strains, and changes in populations of serogroups may indicate important changes in the ecology of Salmonella. Strains of the same serogroups may occupy the same niche. For instance, expansion in the population of Salmonella enterica serotype Enteritidis may have filled an ecological void vacated by Salmonella enterica serotypes Pullorum and Gallinarum following eradication campaigns (22). In this study, the predominant ST of Salmonella within each farm was different between years for all six farms (Table 3). Strains of serogroup C1 were the predominant serotype for three of the six farms in 2009 (Table 3), compared to none in 2000-2001. Only 2.5% (7/273) of the recovered isolates in 2000-2001 were strains of serogroup C1, compared to 60% of the isolates in 2009. This change in the population of serogroups may indicate a change in the environment on dairy farms.
Population changes associated with changes in AMR.
A detailed analysis of within-farm changes in the phenotypic antimicrobial resistance (AMR) of Salmonella was previously reported (9). This report is intended to detail the genotypic changes in the population of Salmonella associated with the previously observed changes in phenotype. Briefly, although the prevalence of Salmonella was higher in 2009 than in 2000-2001, the frequency of MDR subtypes was lower in 2009 than in 2000-2001. The proportions of isolates resistant to at least one antimicrobial were 16% (43/271) and 1.3% (1/77) in 2000-2001 and 2009, respectively (Table 5). The proportion of resistant isolates recovered in the summer of 2009 (1%) was lower relative than those recovered in the summers of 2001 (27%) and 2000 (84%). MDR subtypes of Salmonella Typhimurium with the ACSSuT phenotype (resistant to ampicillin, chloramphenicol, streptomycin, sulfisoxazole, and tetracycline) and Salmonella Senftenberg with the GKSSuT phenotype (resistant to gentamicin, kanamycin, streptomycin, sulfisoxazole, and tetracycline) were recovered in 2000-2001 from farms 101 and 129, respectively. In 2009, only pansusceptible strains of Salmonella Typhimurium and Salmonella Senftenberg (farms 114 and 121, respectively) were recovered (Fig. 3 and 4). The apparently persistent subtypes of Salmonella Bovismorbificans (MSU.Sbo.Xb3) were mostly pansusceptible in both 2000-2001 (105/107 isolates) and 2009 (9/9 isolates). In 2009, however, a distinct strain of Salmonella Bovismorbificans resistant to ceftriaxone and tetracycline was recovered from the same farm. Other serotypes recovered in both years (Salmonella Hartford, Salmonella Mbandaka, and Salmonella Montevideo) were susceptible to all tested antimicrobial in both time periods (Fig. 1).
TABLE 5.
Frequency of antimicrobial resistance profiles of Salmonella serotypes that were recovered in both 2000-2001 and 2009
| Serotype | 2000-2001 |
2009 |
||
|---|---|---|---|---|
| Profilea | No. of isolates | Profile | No. of isolates | |
| S. Bovismorbificans | Susceptible | 123 | Susceptible | 9 |
| S | 1 | CxT | 1 | |
| Su | 1 | |||
| S. Hartford | Susceptible | 1 | Susceptible | 1 |
| S. Montevideo | Susceptible | 1 | Susceptible | 29 |
| S. Mbandaka | Susceptible | 1 | Susceptible | 8 |
| S. Typhimurium | ACSSuT | 15 | Susceptible | 8 |
| ACSSu | 1 | |||
| ACSuT | 1 | |||
| S. Senftenberg | Susceptible | 1 | Susceptible | 3 |
| GKSSuT | 13 | |||
| GSSuT | 4 | |||
| GSuT | 2 | |||
| SuT | 2 | |||
| GKSuT | 1 | |||
| Total | 168 | 59 | ||
A, ampicillin; C, chloramphenicol; Cx, ceftriaxone; G, gentamicin; K, kanamycin; S, streptomycin; Su, sulfisoxazole; T, tetracycline.
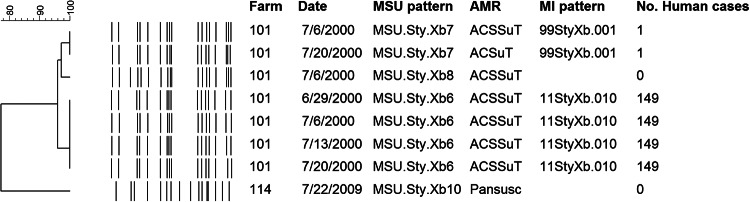
FIG 3.
Unique PFGE patterns of Salmonella Typhimurium (sequence type 19) recovered at each sampling date from Michigan dairy farms in 2000 and 2009. Salmonella Typhimurium strains recovered in 2000 that were multidrug resistant and associated with human illnesses had a distinct PFGE pattern compared to the pansusceptible strain recovered in 2009, which was not associated with any reported human illnesses in Michigan between 2000 and 2012.
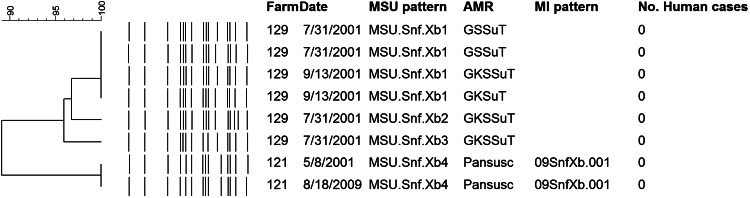
FIG 4.
Unique PFGE patterns of Salmonella Senftenberg (sequence type 14) recovered from Michigan dairy farms in 2001 and 2009. The Salmonella Senftenberg strains recovered in 2001 were MDR but were not associated with human illnesses in Michigan. Pansusceptible isolates of Salmonella Senftenberg from 2001 were indistinguishable from a Salmonella Senftenberg isolate recovered from the same farm in 2009.
Changes in the frequency of human illnesses caused by dairy farm Salmonella subtypes.
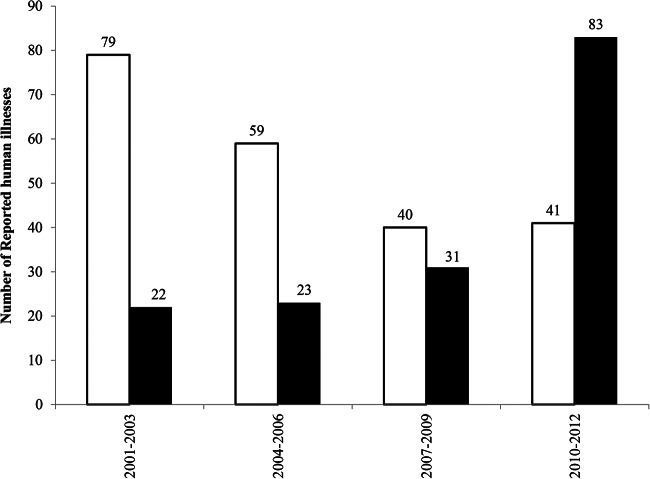
A total of 11,326 isolates from human cases of salmonellosis were submitted to the MDCH for serotyping between 2001 and 2012. Forty-two percent (4,705/11,326) of the human cases were associated with one of the 16 serotypes that were also found on Michigan dairy farms in either 2000-2001 or 2009. Eleven of the 16 serotypes were chosen for further analysis, including 9 serotypes that were identified at the MDCH >100 times between 2001 and 2012 and two serotypes (Salmonella Bovismorbificans and Salmonella Senftenberg) that were recovered from the same farm in 2000-2001 and 2009. These 11 serotypes accounted for 95% (4,445/4,705) of the human cases associated with the 16 serotypes detected on dairy farms in either time period. In total, 48 PFGE patterns representing 11 serotypes recovered on Michigan dairy farms in 2000-2001 or 2009 were compared to 716 PFGE patterns of the same serotypes recovered from human illnesses in Michigan between the years 2000 and 2012. Of the 48 PFGE patterns, 12 (25%) matched at least one pattern representing strains associated with human infections. Two PFGE patterns of Salmonella serotype Typhimurium and one pattern representing each of the remaining 10 serotypes matched a PFGE pattern recovered from humans (Table 6). Most subtypes found on the dairy farms were cattle specific, with no match to strains associated with human illness. Specifically, only 387/11,326 (3.4%) of salmonellosis cases reported between 2001 and 2012 were associated with a PFGE pattern that matched a pattern found on Michigan dairy farms in either time period. Nonetheless, 38% (29/77) and 49% (132/271) of isolates recovered on dairy farms for the 11 serotypes analyzed had PFGE patterns that matched at least one human pattern. There were also opposing temporal changes in the yearly frequency of human illnesses associated with PFGE patterns recovered from dairy farms in either 2000-2001 or 2009. The yearly frequency of human illnesses associated with PFGE patterns that were recovered from Michigan dairy farms only in 2000-2001 decreased between 2001 and 2012, primarily due to decreases in the frequency of the MSU.Sty.Xb6 pattern of Salmonella Typhimurium (Fig. 5; Table 6). Similarly, the total frequency of reported human illnesses caused by PFGE patterns that were recovered from Michigan dairy farms only in 2009 increased between 2000 and 2012. Primarily, the frequency of a PFGE pattern of S. enterica serotype Thompson (MSU.Sth.Xb1) increased over that time frame, with smaller increases for patterns of S. enterica serotypes Braenderup (MSU.Sbr.Xb1) and Hartford (MSU.Sha.Xb2). The PFGE patterns of Salmonella Senftenberg and Salmonella Bovismorbificans that persisted within farms over the 10-year period were associated with only six human illnesses. These data demonstrate that most subtypes of Salmonella found on dairy farms were infrequent causes of human illness; however, there were opposing temporal changes in the yearly frequency of reported human illness between 2001 and 2012 for subtypes of Salmonella Typhimurium and Salmonella Thompson corresponding to the year of recovery on Michigan dairy farms.
TABLE 6.
Frequency of identification of PFGE patterns at the Michigan Department of Community Health that were indistinguishable from a PFGE pattern recovered from Michigan dairy farms in 2000-2001, 2009, or both time frames
| Time frame(s) | Serotype | MSU pattern | MDCH Pattern | PulseNet XbaI pattern | Identification during 3-yr submission period at MDCH |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2001–2003 | 2004–2006 | 2007–2009 | 2010–2012 | |||||||||
| Frequency of matching pattern | Total for serotype | Frequency of matching pattern | Total for serotype | Frequency of matching pattern | Total for serotype | Frequency of matching pattern | Total for serotype | |||||
| 2000-2001 | S. Typhimurium | MSU.Sty.Xb6 | 99StyXb.001 | JPXX01.0003 | 70 | 504 | 43 | 528 | 21 | 524 | 15 | 285 |
| S. Oranienburg | MSU.Sor.Xb1 | 03SorXb.002 | JJXX01.0020 | 4 | 63 | 14 | 77 | 12 | 54 | 14 | 50 | |
| S. Newport | MSU.Snp.Xb1 | 00SnpXb.001 | JJPX01.0119 | 5 | 209 | 1 | 189 | 5 | 154 | 6 | 183 | |
| S. Mbandaka | MSU.Smb.Xb5 | 06SmbXb.001 | 0 | 10 | 1 | 28 | 0 | 23 | 0 | 29 | ||
| S. Anatum | MSU.San.Xb1 | 07SanXb.004 | 0 | 66 | 0 | 24 | 1 | 33 | 0 | 14 | ||
| S. Typhimurium | MSU.Sty.Xb7 | 11StyXb.010 | JPXX01.1757 | 0 | 504 | 0 | 528 | 0 | 524 | 1 | 285 | |
| Total | 79 | 1,373 | 59 | 1,390 | 40 | 1,328 | 41 | 786 | ||||
| 2009 | S. Thompson | MSU.Sth.Xb1 | 00SthXb.001 | JP6X01.0001 | 18 | 66 | 19 | 43 | 20 | 69 | 65 | 94 |
| S. Braenderup | MSU.Sbr.Xb1 | 02SbrXb.001 | JBPX01.0002 | 4 | 34 | 2 | 47 | 9 | 37 | 11 | 48 | |
| S. Hartford | MSU.Sha.Xb2 | 07ShaXb.002 | 0 | 23 | 0 | 25 | 1 | 38 | 7 | 56 | ||
| S. Montevideo | 05SmoXb.013 | 05SmoXb.013 | 0 | 29 | 2 | 63 | 1 | 30 | 0 | 58 | ||
| Total | 22 | 152 | 23 | 178 | 31 | 174 | 83 | 195 | ||||
| 2000-2001 and 2009 | S. Senftenberg | 09SnfXb.001 | 09SnfXb.001 | 0 | 6 | 0 | 4 | 1 | 10 | 0 | 3 | |
| S. Bovismorbificans | 10SboXb.001 | 10SboXb.001 | 0 | 11 | 0 | 12 | 0 | 6 | 5 | 7 | ||
| Total | 0 | 17 | 0 | 16 | 1 | 16 | 5 | 10 | ||||
FIG 5.
Frequency of reported human illnesses at the Michigan Department of Community Health for PFGE patterns recovered from Michigan dairy farms in either 2000-2001 (white bars) or 2009 (black bars).
DISCUSSION
This study utilized a unique approach to enable long-term temporal comparisons of the genetic relatedness and distribution of Salmonella subtypes recovered from the same Michigan dairy farms in 2000-2001 and 2009. The increasing prevalence of Salmonella and decreasing AMR over time observed in this set of farms are consistent with nation-level data (8). The emergence and/or disappearance of novel strains can cause temporal shifts in the population of Salmonella on farms, which could result in changes in the distribution of subtypes associated with human infections. Given the ability of Salmonella to persist within farms for long periods of time, longitudinal studies with greater temporal separation are important to discern population changes over time and determine the importance of cattle-derived genotypes in human infections.
Within-farm distribution of Salmonella subtypes.
These results demonstrate clonal dominance and persistence of specific Salmonella subtypes between bimonthly visits in 2000-2001, widespread environmental dissemination within dairy farms, and likely transmission between different production classes of animals. Some of the subtypes seemed to have the same propensity to colonize both cows and calves. For instance, on farm 111, a single PFGE pattern of Salmonella Bovismorbificans was recovered on 5 consecutive sampling visits and in relatively equal proportions of cow samples (39%, 76/193) and calf samples (38%, 13/34) (Table 4). Similarly, 21% (42/199) and 33% (11/33) of cow and calf samples, respectively, were positive for the same subtype of Salmonella Muenster on farm 114. MDR subtypes of Salmonella, in contrast, may be more likely to colonize the immature gastrointestinal tract of calves. An MDR subtype of Salmonella Senftenberg (MSU.Snf.Xb1) was recovered from a higher proportion of calves (47%, 15/32) than cows (4%, 3/80) on the same farm. Similarly, a higher proportion of calf samples (60%, 3/5) than of cow samples (4%, 1/28) were positive for any MDR pattern of Salmonella Typhimurium (MSU.Sty.Xb6) on the same farm and within the same sampling visit.
Relatedness of Salmonella subtypes between 2000-2001 and 2009.
With the exception of two apparently persistent subtypes, the overall population of Salmonella recovered in 2009 was substantially different from the population recovered from the same set of farms in 2000-2001. Population changes may be a result of introduction of new strains into the farm environment, within-farm diversification of the strain resulting in higher fitness, or changes in the microbial environment that result in differential fitness advantages between Salmonella strains. In this study, herds were sampled once or twice in the summer of 2009 and were sampled at 5 bimonthly visits in 2000-2001. A single subtype of Salmonella typically predominated within farms between sampling visits in 2000-2001, suggesting that a single sampling visit is likely to capture a large proportion of the predominant strains on the farms in 2009. However, the majority (19/21) of the prevalent subtypes recovered in 2009 were not recovered in the more intensive sampling of the same set of herds in 2000-2001. For the two subtypes recovered in both time frames, each was recovered within the same farm at both time points. Because the more intensive sampling in 2000-2001 could be expected to capture most of the diversity in the Salmonella population, these results suggest that changes in the population of Salmonella were primarily due to introduction of Salmonella strains between the time periods.
Changes in herd demographics and management practices may result in changes in the microbial environment that have important impacts on the population of Salmonella. The prevalence of Salmonella has consistently been associated with herd size (8, 23, 24), and expansion of herds could be expected to result in overall increases in the prevalence of Salmonella. Considering summer visits only, the prevalence of Salmonella was higher in 2009 (12%) than in 2000-2001 (9%) (Table 2), and the proportion of resistant isolates recovered in the summer of 2009 (1%) was lower than those recovered in the summers of 2001 (27%) and 2000 (84%). Dairy farming management practices changed substantially between 2000 and 2009, and these practices may influence the ecology of Salmonella by selecting for specific strains. Various management practices have been previously associated with a higher prevalence of Salmonella (10, 23, 24) For instance, increases in the frequency of the use of anionic salts or vaccination for Salmonella may have differential impacts on strain types. Larger studies with more farms would be necessary to pinpoint the mechanism for changes in the prevalence and population of Salmonella on dairy farms.
This study provides additional evidence of long-term within-farm persistence of Salmonella strains over a 10-year period. Only two PFGE patterns were recovered in both years, and each pattern was recovered within the same herd. All 13 isolates representing these two patterns were pansusceptible, and only 1 human illness in Michigan between 2000 and 2012 was associated with an indistinguishable PFGE pattern for either of the apparently persistent strains. Long-term persistence within farms has previously been documented for Salmonella serotypes Cerro, Typhimurium, and Newport (11–13) and within previously clinically ill dairy cattle animals for up to a year (25). A recent study showed in greater temporal detail a within-farm serotype shift that occurred gradually over 2 years (12). At the farm level, long-term persistence could be caused more by a combination of environmental persistence, temporary chain infections, and extended excretions from individual animals (11). In this study, subtypes of Salmonella that apparently persisted within farms between the two time frames were pansusceptible and rarely associated with the reported cases of salmonellosis. The durations of shedding for isolates causing clinical disease in cattle were shown to be similar for different serotypes (25); however, it is possible that pansusceptible Salmonella strains that infrequently cause clinical illness may be more apt to colonize a large percentage of animals and persist for longer periods of time.
Population changes associated with changes in antimicrobial resistance of Salmonella.
Changes in AMR for the isolates in the study have previously been reported (9). This study identifies the changes in genotypes responsible for the associated changes in AMR. In this subset of farms, MDR isolates of Salmonella Typhimurium and Salmonella Senftenberg were recovered in 2000-2001, and only pansusceptible strains of the same serotypes were recovered in 2009 (Fig. 3 and 4). The MDR Salmonella Typhimurium PFGE pattern recovered in 2000 was consistent with the resistance phenotype of Salmonella Typhimurium DT104 and was a frequent, yet declining, cause of reported cases of human salmonellosis in Michigan. By comparison, the pansusceptible strain of Salmonella Typhimurium recovered in 2009 was not identified as a reported cause of human illness in Michigan. Recovery of genotypically distinct (with distinguishable PFGE patterns) MDR and pansusceptible strains of Salmonella Senftenberg and Salmonella Typhimurium may represent clonal displacement between time frames; however, larger studies would be necessary to determine if the same phenomenon explains decreases in nation-level estimates of the frequency of AMR of Salmonella recovered from dairy cattle. Region- or nation-level prevalence estimates of AMR in Salmonella can change through the emergence and/or disappearance of clonal subtypes, and clear temporal patterns of clonal displacement have previously been documented for nontyphoidal Salmonella (26, 27). Decreases in resistance for some antimicrobials were demonstrated for Salmonella from cattle in the Northeast between 2004 and 2011, in part due to the dissemination of pansusceptible subtypes (28); however, within-serotype increases in resistance were shown for Salmonella serotype Newport. Isolates from the environment or from cattle with subclinical shedding were excluded from that study despite being more commonly susceptible to antimicrobials and a less frequent causes of illness. Strains of the same serotypes may occupy the same ecological niche, and less-pathogenic and/or less-resistant strains may nonetheless play an important role in temporal changes in the population of Salmonella. Consequently, more research is necessary to understand how less-pathogenic Salmonella strains impact the overall ecology of the Salmonella subpopulation.
Changes in the frequency of reported human illnesses associated with the same subtypes.
Humans may be exposed to dairy farm-associated subtypes of Salmonella through consumption of inadequately pasteurized or unpasteurized milk, contamination of beef through fecal contamination of carcasses or lymph node contamination, and less-direct transmission routes, including environmental contamination and contamination of fruits and vegetables through environmental contamination. In this study, the majority of the PFGE patterns recovered from the farms were uncommon causes of human salmonellosis, in agreement with recent research (7). Many of the pansusceptible serotypes recovered are uncommonly recovered from clinical illnesses. These data are in agreement with data from the 2010 NARMS report, where 53/61, 8/8, and 4/4 isolates of Salmonella Montevideo, Salmonella Mbandaka, and Salmonella Senftenberg isolates from cattle were susceptible to all antimicrobials tested (29). Similarly, 57/60 Salmonella Montevideo isolates recovered from clinically ill humans were susceptible to all tested antimicrobials (29). However, this study suggests that temporal changes in the population of Salmonella on these dairy farms correlate with changes in the frequency of reported human illnesses associated with same PFGE patterns. Subtypes recovered on dairy farms in 2009, including patterns of Salmonella Thompson, Salmonella Braenderup, and Salmonella Hartford, were numerically more frequent causes of human illness in the later time frames (2007 to 2009 and 2010 to 2012) (Table 6). Likewise, the frequency of human illnesses with PFGE patterns indistinguishable from those of the Salmonella Typhimurium strain (MSU.Sty.Xb7) (recovered in 2000) decreased substantially through the time frame from 2001 to 2012. The resistance phenotype, ST, and PFGE banding pattern of the MDR Salmonella Typhimurium strain recovered in 2000 from farm 101 are indistinguishable from those of the globally disseminated Salmonella Typhimurium DT104 strain (30). The changes observed in this study are consistent with previous work that has shown a decline in the prevalence of Salmonella Typhimurium DT104 (8, 31). The recovery of indistinguishable PFGE patterns from humans and dairy farms does not demonstrate that the dairy farms are specific sources of illness but demonstrates that these Salmonella subtypes may have been circulating within and between these populations during the same time frame. More-discriminatory molecular techniques or PFGE with two enzymes would be particularly useful to make comparisons between strains from bovine and human populations; however, two-enzyme PFGE patterns were not available for most of the retrospective human clinical isolates, and more-discriminatory comparisons between human and bovine isolates were not possible.
Conclusions.
This study provides insights into changes in the distribution, genetic relatedness, and AMR of Salmonella subtypes recovered from the same Michigan dairy farms in 2000-2001 and 2009. During the longitudinal sampling in 2000-2001, Salmonella strains persisted within farms over a bimonthly sampling visits and were broadly distributed within farms across adult lactating cattle and calves. However, Salmonella isolates recovered from the same set of farms in 2009 most frequently had a different serotype, sequence type, and PFGE pattern. Only 2 of 21 recovered PFGE patterns in 2009 had previously been recovered in the more-intensive sampling period from 2000 to 2001. The two PFGE patterns that were recovered in both time periods were recovered within the same farm, suggesting long-term persistence. Additionally, a lower frequency of AMR in 2009 than in 2000-2001 was attributable to the recovery of MDR strains in 2000-2001 and genetically distinct, pansusceptible strains of the same serotype in 2009. Finally, the PFGE patterns recovered in 2000-2001 and 2009 were more frequently associated with human illnesses in their respective time frames, suggesting that changes in the population of Salmonella on dairy farms may have important implications for public health. These data support the conclusion that the 2009 population of Salmonella on dairy farms was distinct, more prevalent, and less antimicrobial resistant than the population of Salmonella recovered from the same subset of farms in 2000-2001. Further understanding of the drivers of population changes may lead to positive interventions for reducing the prevalence and AMR of Salmonella on dairy farms.
ACKNOWLEDGMENT
We acknowledge the funding support of the Harvey J. Fiege fund for Genetic Research.
REFERENCES
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 2.CDC. 2009. Preliminary FoodNet Data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2008. MMWR Morb Mortal Wkly Rep 58:333–337. [PubMed] [Google Scholar]
- 3.Fey PD, Safranek TJ, Rupp ME, Dunne EF, Ribot E, Iwen PC, Bradford PA, Angulo FJ, Hinrichs SH. 2000. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N Engl J Med 342:1242–1249. doi: 10.1056/NEJM200004273421703. [DOI] [PubMed] [Google Scholar]
- 4.Lynch M, Painter J, Woodruff R. 2006. Surveillance for foodborne disease outbreaks: United States, 1998–2002. MMWR Morb Mortal Wkly Rep 55:1–34. [PubMed] [Google Scholar]
- 5.Soyer Y, Alcaine SD, Schoonmaker-Bopp DJ, Root TP, Warnick LD, McDonough PL, Dumas NB, Gröhn YT, Wiedmann M. 2010. Pulsed-field gel electrophoresis diversity of human and bovine clinical Salmonella isolates. Foodborne Pathog Dis 7:707–717. doi: 10.1089/fpd.2009.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcaine SD, Soyer Y, Warnick LD, Su W-L, Sukhnanand S, Richards J, Fortes ED, McDonough P, Root TP, Dumas NB, Gröhn Y, Wiedmann M. 2006. Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl Environ Microbiol 72:7575–7585. doi: 10.1128/AEM.01174-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Rivera LD, Wright EM, Siler JD, Elton M, Cummings KJ, Warnick LD, Wiedmann M. 2014. Subtype analysis of Salmonella isolated from subclinically infected dairy cattle and dairy farm environments reveals the presence of both human- and bovine-associated subtypes. Vet Microbiol 170:307–316. doi: 10.1016/j.vetmic.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.USDA. 2011. Salmonella, Listeria, and Campylobacter on U.S. dairy operations, 1996–2007. USDA, Fort Collins, CO. [Google Scholar]
- 9.Habing GG, Lo Y-J, Kaneene JB. 2012. Changes in the antimicrobial resistance profiles of Salmonella isolated from the same Michigan dairy farms in 2000 and 2009. Food Res Int 45:919–924. doi: 10.1016/j.foodres.2011.02.054. [DOI] [Google Scholar]
- 10.Fossler CP, Wells SJ, Kaneene JB, Ruegg PL, Warnick LD, Bender JB, Godden SM, Halbert LW, Campbell AM, Zwald AM. 2004. Prevalence of Salmonella spp. on conventional and organic dairy farms. J Am Vet Med Assoc 225:567–573. doi: 10.2460/javma.2004.225.567. [DOI] [PubMed] [Google Scholar]
- 11.Cobbold R, Rice D, Davis M, Besser T, Hancock D. 2006. Long-term persistence of multi-drug-resistant Salmonella enterica serovar Newport in two dairy herds. J Am Vet Med Assoc 228:585–591. doi: 10.2460/javma.228.4.585. [DOI] [PubMed] [Google Scholar]
- 12.Van Kessel JAS, Karns JS, Wolfgang DR, Hovingh E, Schukken YH. 2012. Dynamics of Salmonella serotype shifts in an endemically infected dairy herd. Foodborne Pathog Dis 9:319–324. doi: 10.1089/fpd.2011.1054. [DOI] [PubMed] [Google Scholar]
- 13.Vanselow BA, Hum S, Hornitzky MA, Eamens GJ, Quinn K. 2007. Salmonella Typhimurium persistence in a Hunter Valley dairy herd. Aust Vet J 85:446–450. doi: 10.1111/j.1751-0813.2007.00224.x. [DOI] [PubMed] [Google Scholar]
- 14.Adaska JM, Silva AJ, Berge ACB, Sischo WM. 2006. Genetic and phenotypic variability among Salmonella enterica serovar Typhimurium isolates from California dairy cattle and humans. Appl Environ Microbiol 72:6632–6637. doi: 10.1128/AEM.01038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao S, McDermott PF, White DG, Qaiyumi S, Friedman SL, Abbott JW, Glenn A, Ayers SL, Post KW, Fales WH, Wilson RB, Reggiardo C, Walker RD. 2007. Characterization of multidrug resistant Salmonella recovered from diseased animals. Vet Microbiol 123:122–132. doi: 10.1016/j.vetmic.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Gebreyes W, Thakur S, Dorr P, Tadesse DA, Post K, Wolf L. 2009. Occurrence of spvA virulence gene and clinical significance for multidrug-resistant Salmonella strains. J Clin Microbiol 47:777–780. doi: 10.1128/JCM.01660-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarwari R, Magder LS, Levine P, McNamara AM, Knower S, Armstrong GL, Etzel R, Hollingsworth J, Morris JG. 2001. Serotype distribution of Salmonella isolates from food animals after slaughter differs from that of isolates found in humans. J Infect Dis 183:1295–1299. doi: 10.1086/319671. [DOI] [PubMed] [Google Scholar]
- 18.FDA. 2011. National Antimicrobial Resistance Monitoring System—Enteric Bacteria (NARMS): 2009 executive report. Food and Drug Administration, Rockville, MD. [Google Scholar]
- 19.Kauffman F. 1975. Classification of bacteria: a realistic scheme with special reference to the classification of Salmonella- and Escherichia-species. Munksgaard, Copenhagen, Denmark. [Google Scholar]
- 20.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 21.Achtman M, Wain J, Weill F-X, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A, Dougan G, Harrison LH, Brisse S. 2012. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog 8:e1002776. doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bäumler AJ, Hargis BM, Tsolis RM. 2000. Tracing the origins of Salmonella outbreaks. Nature 287:50–52. [DOI] [PubMed] [Google Scholar]
- 23.Habing GG, Lombard J, Kopral CA, Dargatz D, Kaneene JB. 2012. Farm-level associations with the shedding of Salmonella and antimicrobial-resistant Salmonella in U.S. dairy cattle. Foodborne Pathog Dis 9:815–821. doi: 10.1089/fpd.2012.1149. [DOI] [PubMed] [Google Scholar]
- 24.Fossler CP, Wells SJ, Kaneene JB, Ruegg PL, Warnick LD, Bender JB, Eberly LE, Godden SM, Halbert LW. 2005. Herd-level factors associated with isolation of Salmonella in a multi-state study of conventional and organic dairy farms. I. Salmonella shedding in cows. Prev Vet Med 70:257–277. [DOI] [PubMed] [Google Scholar]
- 25.Cummings KJ, Warnick LD, Alexander KA, Cripps CJ, Gröhn YT, James KL, McDonough PL, Reed KE. 2009. The duration of fecal Salmonella shedding following clinical disease among dairy cattle in the northeastern USA. Prev Vet Med 92:134–139. doi: 10.1016/j.prevetmed.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butaye P, Michael GB, Schwarz S, Barrett TJ, Brisabois A, White DG. 2006. The clonal spread of multidrug-resistant non-typhi Salmonella serotypes. Microbes Infect 8:1891–1897. doi: 10.1016/j.micinf.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 27.Davis M, Hancock D, Besser T. 2002. Multiresistant clones of Salmonella enterica: the importance of dissemination. J Lab Clin Med 140:135–141. doi: 10.1067/mlc.2002.126411. [DOI] [PubMed] [Google Scholar]
- 28.Cummings KJ, Perkins G, Khatibzadeh SM, Warnick LD, Altier C. 2013. Antimicrobial resistance trends among Salmonella isolates obtained from dairy cattle in the northeastern United States, 2004–2011. Foodborne Pathog Dis 10:353–361. doi: 10.1089/fpd.2012.1285. [DOI] [PubMed] [Google Scholar]
- 29.FDA. 2012. National Antimicrobial Resistance Monitoring System—Enteric Bacteria (NARMS): 2010 executive report. Food and Drug Administration, Rockville, MD. [Google Scholar]
- 30.Lan R, Reeves PR, Octavia S. 2009. Population structure, origins and evolution of major Salmonella enterica clones. Infect Genet Evol 9:996–1005. doi: 10.1016/j.meegid.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Threlfall EJ, Fisher IST, Berghold C, Gerner-Smidt P, Tschäpe H, Cormican M, Luzzi I, Schnieder F, Wannet W, Machado J, Edwards G. 2003. Antimicrobial drug resistance in isolates of Salmonella enterica from cases of salmonellosis in humans in Europe in 2000: results of international multi-centre surveillance. Euro Surveill 8(2):pii=400 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=400. [DOI] [PubMed] [Google Scholar]