Abstract
Rotavirus is the leading cause of infantile diarrhea in developing countries, where it causes a high number of deaths among infants. Two vaccines are available, being highly effective in developed countries although markedly less efficient in developing countries. As a complementary treatment to the vaccines, a Lactobacillus strain producing an anti-rotavirus antibody fragment in the gastrointestinal tract could potentially be used. In order to develop such an alternative therapy, the effectiveness of Lactobacillus rhamnosus GG to produce and display a VHH antibody fragment (referred to as anti-rotavirus protein 1 [ARP1]) on the surface was investigated. L. rhamnosus GG is one of the best-characterized probiotic bacteria and has intrinsic antirotavirus activity. Among four L. rhamnosus GG strains [GG (CMC), GG (ATCC 53103), GG (NCC 3003), and GG (UT)] originating from different sources, only GG (UT) was able to display ARP1 on the bacterial surface. The genomic analysis of strain GG (UT) showed that the genes welE and welF of the EPS cluster are inactivated, which causes a defect in exopolysaccharide (EPS) production, allowing efficient display of ARP1 on its surface. Finally, GG (UT) seemed to confer a level of protection against rotavirus-induced diarrhea similar to that of wild-type GG (NCC 3003) in a mouse pup model, indicating that the EPS may not be involved in the intrinsic antirotavirus activity. Most important, GG (EM233), a derivative of GG (UT) producing ARP1, was significantly more protective than the control strain L. casei BL23.
INTRODUCTION
Rotavirus is the leading cause of infantile diarrhea, and it is reported that approximately half a million children, most of them in developing countries, succumb to the infection every year (1). There are currently two licensed antirotavirus vaccines, RotaTeq and Rotarix, which represent a major step forward for disease control. Nevertheless, their effectiveness is markedly lower in developing countries (39.3 to 61.2% efficacy) (2–4) than in developed countries (>85% efficacy) (5–7). Thus, other alternative measures, applied either alone or in combination with the vaccines, might be of considerable value.
Nanobodies or VHHs have previously been shown to be useful for treatment of rotavirus-associated diarrhea (8, 9). Nanobodies are fragments of antibodies derived from immunoglobulins devoid of light chains that can be found in camelids (10). VHHs exhibit several properties that make them attractive as therapeutic molecules. They are the smallest molecules that efficiently bind antigens, and due to their simple conformation they are easily produced in heterologous systems (11). Finally, they are markedly more acid and heat resistant than conventional antibodies (12, 13).
An attractive approach for treatment of viral and bacterial infections is the use of lactobacilli as a vector for delivery of antibody fragments, as they resist the harsh environment of the gastrointestinal tract (GIT) and can deliver the antibodies directly to the mucosal surface. Lactobacilli are considered safe, as they are part of the mucosal microbiota of healthy individuals and some strains have been used in food fermentation processes for centuries. As a proof of concept, we have previously demonstrated that Lactobacillus casei BL23 (previously known as Lactobacillus zeae ATCC 393 pLZ15− and Lactobacillus paracasei BL23), producing the anti-rotavirus VHH ARP1 (formerly known as VHH1) (9) anchored on the cell wall, was able to confer protection against rotavirus in a mouse pup model (14). The ARP1 antibody fragment neutralizes a wide variety of rotavirus serotypes in vitro (15) and was shown to be safe and effective in reducing the severity of rotavirus-induced diarrhea in children (16).
Mucosal delivery of ARP1 by L. casei BL23 was further optimized using an apf expression cassette (17) integrated into the chromosome in order to generate a stable, food grade strain that produces ARP1 without the need for antibiotic resistance genes. The display of ARP1 on the surface of L. casei BL23 was successfully achieved using the cell wall anchor domain of the proteinase PrtP (18). This proteinase, found in lactic acid bacteria, has an LPXTG motif that mediates the covalent attachment of proteins to the peptidoglycan layer through the interaction of the carboxyl group of the glycine and an N-terminal glycine of the cross-bridges (18).
In order to develop an effective therapy against rotavirus in humans, it is of utmost importance to select suitable strains of Lactobacillus to deliver the ARP1 fragment. In our study, the strain Lactobacillus rhamnosus GG was chosen as it is a well-known probiotic strain with intrinsic antirotavirus activity (19). Furthermore, the complete genome of this strain has been sequenced (20). In the process of generating a strain of L. rhamnosus GG expressing ARP1 anchored in the cell wall, we found that only a naturally occurring variant [strain GG (UT)] was able to display ARP1 on the bacterial surface. This article describes the optimization of the expression cassette for production of anchored ARP1 in the mutant GG (UT) strain and the characterization of the mutant strain at the genomic and transcriptional levels.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Table 1 shows a list of the plasmids and strains used in this study. Escherichia coli strains were grown in Luria-Bertani (LB) broth at 37°C with 200 rpm orbital shaking or on LB-agar plates at 37°C. Lactobacillus strains were grown in MRS broth at 37°C under static conditions and on MRS agar plates at 37°C under anaerobic conditions. L. rhamnosus strain GG (Gefilus), marketed by Valio Ltd., was isolated by plating 10-fold serial dilutions of yoghurt on MRS plates that were subsequently incubated 2 days at 37°C under anaerobic conditions. Several isolated colonies with an L. rhamnosus appearance (round, white, and creamy) were picked to extract chromosomal DNA using the Qiamp DNA minikit (Qiagen). The DNA samples from the selected colonies were used as the templates in an L. rhamnosus GG-specific PCR using primers GG-I and GG-II as described previously (21). A GG-PCR-positive isolate [L. rhamnosus GG (Gefilus)] was used in this study. The following antibiotics were added for Lactobacillus transformants when required: 10 μg/ml erythromycin and 20 μg/ml chloramphenicol. In addition, 100 μg/ml ampicillin and 300 μg/ml erythromycin were added for E. coli transformants.
TABLE 1.
Strains and plasmids used in this studya
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK−, mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Invitrogen, Carlsbad, CA |
| E. coli DH11S | mcrA Δ(mrr-hsdRMS-mcrBC) Δ(lac-proAB) Δ(rec1398) deoR rpsL srl- thi-/F′proAB+ lacIqZΔM15 | Invitrogen, Carlsbad, CA |
| L. casei BL23 | Previously known as L. zeae ATCC 393 pLZ15− | 14 |
| L. rhamnosus GG (CMC) | Wild type | CMC |
| L. rhamnosus GG (ATCC 53103) | Wild type | ATCC |
| L. rhamnosus GG (NCC 3003) | Wild type | NCC |
| L. rhamnosus GG (Gefilus) | Wild type | Gefilus (Valio Ltd.) |
| L. rhamnosus GG (UT) | Natural welFE mutant | UT |
| L. rhamnosus GG (EM233) | GG (UT) with the original ARP1 cassette (anchor domain 243-aa PrtP of L. casei) integrated into the chromosome | This study |
| Plasmids | ||
| pIAV7 | Broad-range vector, Err, lacZ, pWV01 replication origin | 24 |
| pEM233 | pUC19E-borne six1, A2-int-attP six2, with expression cassette of pAF900-ARP1 | 17 |
| pEM94 | pG+host9-borne β-recombinase gene, carrying Cmr | 47 |
| pSP72 | Cloning vector | Promega |
| pAF900-ARP1 | Original ARP1 cassette (anchor domain 243-aa PrtP of L. casei) in pIAV7 | 17 |
| pLB11 | Modified ARP1 cassette (anchor domain 450-aa PrtP GG) in pIAV7 | This study |
| pLB12 | Modified ARP1 cassette (anchor domain 500-aa PrtP GG) in pIAV7 | This study |
| pLB13 | Modified ARP1 cassette (anchor domain 550-aa PrtP GG) in pIAV7 | This study |
| pLB19 | Modified ARP1 cassette (anchor domain 865-aa PrtP GG) in pIAV7 | This study |
| pLB20 | Modified ARP1 cassette (anchor domain 1,275-aa PrtP GG) in pIAV7 | This study |
| pLB31 | welFE genes in pIAV7 | This study |
CMC, Christian Medical College, Vellore, India; ATCC, American Type Culture Collection; NCC, Nestlé Culture Collection; UT, University of Tartu, Estonia.
Construction of expression cassettes.
For production of cell wall-anchored ARP1, the pAF900 plasmid containing the promoter, signal peptide, C-terminal domain, and terminator from the apf gene of Lactobacillus crispatus M247 was used (17) (Fig. 1A). The ARP1 gene was fused with the sequence encoding an E-tag for detection, followed by the coding region of the last 243 amino acids (aa) of the C-terminal part of the proteinase PrtP from L. casei BL23 for cell wall anchoring. This cassette was integrated into the chromosome of L. rhamnosus strain GG (UT) as previously described (22). Briefly, the strain was transformed with the integrative plasmid pEM233 (17) as previously described (23). This plasmid, besides the mentioned cassette, harbors the phage A2 integrase gene (A2-int gene) and an A2-attP site. The A2 integrase mediates the insertion of the DNA vector by recombination of the A2-attP site with an attB site present in the genome. For removal of the DNA of plasmid origin, the strain was subsequently transformed with the temperature-sensitive plasmid pEM94 carrying the β-recombinase gene that catalyzes the recombination between two six sites that flank the ARP1 expression cassette. After curing the plasmid pEM94 at 37°C, the resulting strain was named GG (EM233) (Table 1). The stability of the integration system was tested in our previous study (17), where we determined that the integrated gene is stable after 50 generations.
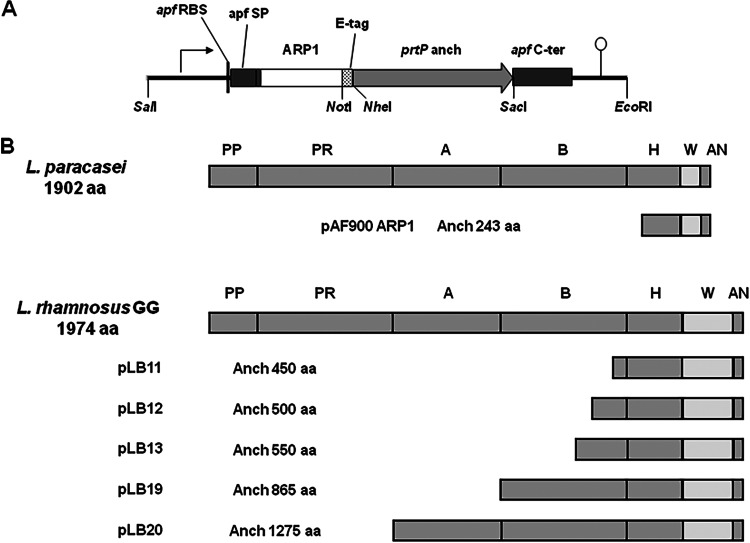
FIG 1.
(A) Original expression cassette for production of ARP1 anchored on the cell wall by 243 amino acids of the C-terminal part of PrtP from L. casei BL23 (prtP anch). The cassette contains the following elements of the apf gene from L. crispatus M247 (GenBank accession number AF492458): promoter (arrow), ribosomal binding site (apf RBS), signal peptide (apf SP), apf C-terminal part (apf C-ter), and terminator (lollipop). The ARP1 coding gene is fused with an E-tag sequence for detection. (B) Domains and different anchor regions of PrtP from L. casei BL23 and L. rhamnosus GG used for the construction of the different cassettes. PP, pre-pro domain; PR, proteinase domain; A, A-domain; B, B-domain; H, helical domain; W, cell wall domain; AN, anchor domain.
In order to optimize the display of ARP1 on the surface of L. rhamnosus GG cells, the anchoring domain was replaced by increasing lengths of the C-terminal part of PrtP from L. rhamnosus GG (ATCC 53103) (Fig. 1B). Briefly, the apf expression cassette from the plasmid pAF900-ARP1 (17) was excised using the restriction enzymes SalI and EcoRI and cloned into pSP72 (Promega) digested with the same restriction enzymes. The different parts of the prtP gene were amplified with the corresponding pair of primers (see Table S1 in the supplemental material) using L. rhamnosus GG (ATCC 53103) genomic DNA as a template. The anchor domain in the expression cassette was replaced with the different amplicons using NheI and SacI or NotI and SacI depending on the occurrence of NheI and NotI restriction sites on the amplicons. Finally, the series of modified expression cassettes with different lengths of anchor domains were cloned into pIAV7 (24) using the restriction enzymes SalI and EcoRI. The resulting constructs (Table 1) were introduced into L. rhamnosus GG by electroporation as previously described (25, 26).
Western blot analysis.
The expression of ARP1 in transformed lactobacilli was determined using Western blotting. The transformants were grown in MRS containing erythromycin (10 μg/ml) until an optical density at 600 nm (OD600) of 0.8 was reached. Cells were collected by centrifugation (5,000 × g, 15 min at 4°C), washed once with phosphate-buffered saline (PBS), and boiled in Laemmli buffer (Bio-Rad, Hemel-Hampstead, United Kingdom). Cell extracts were centrifuged for 5 min at 12,000 × g, and the supernatants were run on a 12% SDS-polyacrylamide gel at 170 V. Proteins were subsequently transferred onto nitrocellulose membranes (Hybond-ECL; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). The membranes were blocked with PBS with 0.05% (vol/vol) Tween 20 plus 5% (wt/vol) milk powder and successively incubated with monoclonal mouse anti-E-tag antibodies (Phadia AB, Uppsala, Sweden) and horseradish peroxidase (HRP)-labeled goat anti-mouse antibodies (Dako A/S, Glostrup, Denmark). The signal was detected by chemiluminescence using the ECL Plus Western blotting detection system (GE Healthcare).
Fluid-based assay and flow cytometry.
For detection of ARP1 on the cell surface, a fluid-based assay was performed. Briefly, bacterial cells from exponential-phase cultures were incubated with mouse monoclonal anti-E-tag antibodies (1 μg/ml; Phadia AB, Uppsala, Sweden) and alkaline phosphatase (AP)-conjugated rabbit anti-mouse antibodies (diluted 1:1,000; Dako A/S, Glostrup, Denmark) with PBS washes in between. Cells were resuspended in diethanolamine buffer (1 M, pH 10) and transferred to wells of a microtiter plate. p-Nitrophenyl phosphate (pNPP) was used as a substrate, and the optical density was read at 405 nm.
When a more accurate determination of the ARP1 display was needed, flow cytometry was performed. In this case, the bacterial cells were sequentially incubated with monoclonal mouse anti-E-tag antibodies (5 μg/ml; Phadia AB, Uppsala, Sweden) and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibodies (dilution, 1:200; Jackson ImmunoResearch Laboratories). Cells were washed before fixation with 0.5% paraformaldehyde. The samples were analyzed using a fluorescence-activated cell sorter (FACS) instrument (FACSCalibur; Becton Dickinson). The binding of rhesus rotavirus (RRV) particles to anchored ARP1 on bacteria was also determined by flow cytometry. For this assay, the cells were sequentially incubated with RRV (107 to 108 focus-forming units [FFU]/ml), polyclonal rabbit anti-RRV antibodies K230 (dilution, 1:200; a gift from Lennart Svensson, University of Linköping) and phycoerythrin (PE)-conjugated donkey anti-rabbit antibodies (dilution, 1:200; Jackson ImmunoResearch) with washes in between.
Genome sequencing.
The genomes of strains GG (UT), GG (ATCC 53103), and GG (Gefilus) were sequenced using a hybrid assembly approach. For each strain, a mate-pair library with an insert size of 3 kb was constructed using the SOLiD Mate-Pair Library construction kit and sequenced on a SOLiD v3 sequencer with two 50-bp read lengths according to the manufacturer's instructions (Life Technologies, Carlsbad, CA, USA). In addition, 200-bp fragment libraries for Ion Torrent were constructed and sequenced on an IonPGM sequencer according to the manufacturer's instructions (Life Technologies, Carlsbad, CA, USA). IonTorrent reads were assembled using Mira v3.4.1. Scaffolding of Mira contigs together with the SOLiD mate-pair data was done using Hybrid Assembly Pipeline with SOLiD reads (HAPS; v0.1.296) and in-house scripts. The scaffolds were assembled into a draft genome using the genome reference strain (GenBank accession number NC_013198) as a backbone. Remaining gaps were manually filled from Sanger sequences.
The final genome sequences for GG (UT), GG (ATCC 53103), and GG (Gefilus) were compared to each other to identify potential mutations. The genetic variations found were confirmed by PCR amplification and Sanger sequencing.
Transcriptome analysis.
The transcriptomes of strains GG (UT), GG (ATCC 53103), and GG (Gefilus) were analyzed and compared. For hybridization with the microarrays, high-quality RNA from four independent replicates of exponential-phase cultures was extracted as follows. One milliliter of bacterial culture (OD600, between 0.7 and 0.8) was mixed by vortexing for 5 s with 3 ml of RNAprotect Bacteria Reagent (Qiagen). After 5 min of incubation at room temperature (RT), the tubes were centrifuged at 5,000 × g at RT for 10 min, and the supernatant was discarded. Cells were resuspended in 100 μl of RNase-free water and transferred immediately to a screw-cap tube containing 1 ml of TRI reagent (Sigma-Aldrich, St. Louis, MO, USA) and 0.5 g of zirconia beads. Samples were homogenized by beating 3 times for 45 s with 1 min of incubation on ice between runs and subsequently incubated for 5 min at RT. One hundred microliters of 1-bromo-3-chloropropane (Sigma) was added to each sample, and after vortexing for 30 s, the samples were incubated for 10 min at RT. The aqueous phase of each sample was recovered after centrifuging at 12,000 × g at 4°C for 5 min and treated with 1 volume of chloroform. Following another centrifugation step, 0.5 volume of absolute ethanol was added to the isolated aqueous phase, and each sample was transferred to an RNeasy minikit column (Qiagen). Purification of the total RNA was then performed according to the manufacturer's instructions. The quantity and quality of the RNA were assessed with a 2100 Bioanalyzer (Agilent Technologies).
The design of the expression array used is described elsewhere (27). The microarray hybridization process and data analysis were performed by the Bioinformatics and Expression analysis core facility (BEA) at Karolinska Institutet. One hundred nanograms of each RNA sample was labeled with Cy3 and hybridized on the array according to the protocol from Agilent Technologies for “Agilent one-color microarray-based exon analysis.” Images from scanning were analyzed using the Agilent Feature Extraction software version 10.7.3.1. Generated text files with background-adjusted signals were imported into the Partek Genomics Suite software version 6.6 and subjected to quantile normalization. Processed signals were exported to Excel, and expression levels in different strains were compared using two-tailed Student's t test. A 3-fold change in gene expression and a P value of <0.05 were used to identify genes showing potential differential expression between the samples.
Based on the genome and transcriptome analysis of strains GG (UT), GG (ATCC 53103), and GG (Gefilus), 16 open reading frames (ORFs) that had shown differential expression with at least one probe were selected for further analysis by real-time PCR. For this analysis, the RNA was extracted as described above with addition of an on-column DNase digestion step (Qiagen). A first-strand cDNA synthesis kit (GE Healthcare) was used to generate cDNA from total RNA amplifying with pd(N)6 random hexamer primers according to the manufacturer's instructions. The expressions of the identified open reading frames were assessed by SYBR green quantitative PCR (qPCR) with a KAPA SYBR FAST universal qPCR kit (KAPA Biosystems, Boston, MA, USA) on an ABI Prism 7500 sequence detection system (Applied Biosystems, Foster City, CA, USA). Dilutions of cDNA were run in 20-μl reaction mixtures containing 1 μl cDNA and 8 pmol of each set of gene-specific primers. PCR conditions were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. The data were analyzed using the ABI 7500 v2.0.4 software (Applied Biosystems). Transcript levels were calculated based on a standard curve derived from dilutions of pooled cDNA from replicates of all strains. The primers designed for the detection and quantification of the transcripts from the respective ORFs are listed in Table S2 in the supplemental material.
Complementation of welFE mutations.
To complement the mutations in the EPS cluster of strain GG (UT), a fragment of DNA containing the genes welF-welE was amplified by PCR from GG (ATCC 53103) genomic DNA (see Table S1 in the supplemental material for the primers used) and cloned into pIAV7 using the restriction enzymes SalI and EcoRI, resulting in the plasmid pLB31 (Table 1). This plasmid was introduced by electroporation into the cells of strain GG (EM233) producing surface-anchored ARP1.
TEM.
Cells from Lactobacillus cultures grown in MRS (OD600, 0.8) were fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer containing 0.1 M sucrose and 3 mM CaCl2, pH 7.4, at RT and fixed overnight at 4°C. After fixation, the cells were rinsed in 0.1 M phosphate buffer (pH 7.4) and centrifuged (1,700 × g). The pellets were then postfixed in 2% osmium tetroxide in 0.1 M phosphate buffer (pH 7.4) at 4°C for 2 h, dehydrated in ethanol followed by acetone, and embedded in LX-112 (Ladd, Burlington, VT, USA). Sections were contrasted with uranyl acetate followed by lead citrate and examined in a Leo 906 transmission electron microscope (TEM; Oberkochen, Germany) at 80 kV. Digital images were captured using a Morada digital camera (Soft Imaging System, GmbH, Münster, Germany).
Mouse pup model of rotavirus infection.
Pregnant BALB/c mice were purchased from Charles River Laboratories, Germany. Four-day-old pups were used for the study. The duration of the experiment was 6 days. Lactobacillus strains were administered orally once daily in a volume of 10 μl containing approximately 108 CFU from day −1 until day 3. Pups were infected on day 0 with 1.3 × 106 FFU of RRV. The occurrence and severity of diarrhea were assessed daily until day 4 using the following score system: watery diarrhea and loose stool were given the scores 2 and 1, respectively. No stool or normal stool was given a score of 0. Prevalence was compared among groups in a day-wise manner using Fisher's exact test. Severity (sum of scores during the experiment for each pup) and duration (total days with diarrhea) were analyzed with Kruskal-Wallis and Dunn tests. Two-tailed P values of <0.05 were considered statistically significant. All the animal experiments were approved by the local ethical committee of the Karolinska Institutet at Karolinska University Hospital (Huddinge).
Nucleotide sequence accession numbers.
The sequence of the EPS cluster from strain GG (UT) was deposited in the NCBI database under accession number KP969453. The sequences with the rest of the genetic variations of strains GG (UT) and GG (ATCC 53103) were deposited under accession numbers KT026036, KT026037, KT026038, KT026039, KT026040, and KT026041, corresponding to the positions of the reference sequences 57657, 240673, 264278, 656561, 1241467, and 1994717, respectively (see Table S3 in the supplemental material).
RESULTS
Display of ARP1 on the cell surface by L. rhamnosus GG (UT).
L. rhamnosus GG (NCC 3003) was initially transformed with plasmid pAF900-ARP1, and the production and display of ARP1 on the cell surface were monitored by Western blotting and a fluid-based assay, respectively. The transformed strain GG (NCC 3003) pAF900-ARP1 was able to produce amounts of ARP1 similar to those produced by the L. casei pAF900-ARP1 strain, but the antibody fragment was not displayed on its surface (see Fig. S1 in the supplemental material). To investigate whether the inability of surface display of ARP1 by GG (NCC 3003) was a specific problem of this strain, L. rhamnosus strains GG (CMC), GG (ATCC 53103), and GG (UT) with different origins were also transformed with plasmid pAF900-ARP1. All the strains efficiently produced the fragment ARP1 as shown in the Western blot, yet only GG (UT) was able to display the fragment of antibody on its surface (see Fig. S1 in the supplemental material). In the Western blot analysis, it was also observed that the recombinant protein showed higher molecular mass than expected and there was some degree of degradation.
Optimization of the anchoring system.
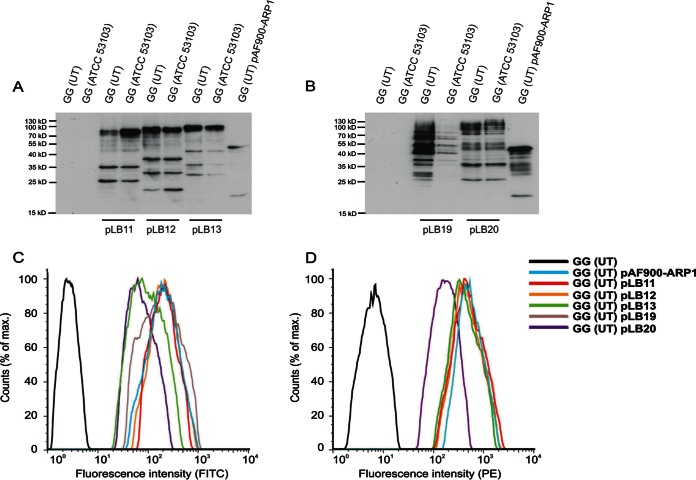
In an attempt to achieve a correct display of ARP1 in the strain GG, a series of new cassettes with different lengths of the C-terminal part (from 450 to 1,275 amino acids) of PrtP from GG (ATCC 53103) were constructed (Fig. 1B). The 1,275-amino-acid PrtP fragment was the longest possible without including the proteinase domain. Strains GG (ATCC 53103) and GG (UT), transformed with the plasmids containing the different cassettes (Table 1), were tested for production and display of ARP1 by Western blotting and in a fluid-based assay, respectively. Similar amounts of ARP1 were detected in the cell extracts from the different transformants from the two strains (Fig. 2A and B), but only the GG (UT) transformants were able to display the recombinant antibody with all expression constructs (see Fig. S2 in the supplemental material). As previously observed with the construct pAF900-ARP1, the recombinant proteins of different lengths showed higher molecular masses than the theoretical ones. Moreover, the proteins with the longer PrtP fragments corresponding to the constructs pLB19 and pLB20 showed increased degradation (Fig. 2B).
FIG 2.
Production, display, and functionality of ARP1 in the different constructs generated in this study. (A and B) Production of ARP1 determined in culture cell extracts by Western blotting using monoclonal mouse anti-E-tag antibodies and HRP-labeled goat anti-mouse antibodies as primary and secondary antibodies, respectively. The signal was detected by chemiluminescence using the ECL Plus Western blotting detection system (GE Healthcare). The theoretical molecular masses of the different ARP1 constructs are 39.7 kDa (pAF900-ARP1), 59.7 kDa (pLB11), 64.9 kDa (pLB12), 70.2 kDa (pLB13), 104 kDa (pLB19), and 148.7 kDa (pLB20). (C and D) Flow cytometric analyses to study the display of ARP1 (C) and binding to rotavirus (D) in GG (UT) using different anchor domains. For the analysis of histogram C, samples were incubated with a monoclonal mouse anti-E-tag antibodies and FITC-conjugated goat anti-mouse antibodies. For the analysis of histogram D, samples were incubated with RRV particles, rabbit anti-RRV K230, and PE-conjugated donkey anti-rabbit antibodies.
Given that strain GG (UT) was able to display ARP1 on its surface, the level of display and functionality of the antibody fragment was analyzed by flow cytometry in the different GG (UT) transformants to determine which anchor domain was optimal for production and display of ARP1. No significant differences were found in the levels of display of ARP1 fused with different anchor domains except for the 550- and 1,275-amino-acid anchor domains that presented lower levels (Fig. 2C). However, the functionality of ARP1, determined as binding to RRV particles, was similar for all the constructs except for the longest C-terminal part (1,275 amino acids), which showed a reduced capacity of binding to RRV (Fig. 2D).
Genome and transcriptome analysis comparisons.
The genomes of strains GG (UT), GG (ATCC 53103), and GG (Gefilus) were sequenced and analyzed to detect potential mutations and/or variations that could be involved in the capacity of displaying ARP1 on the cell surface. From the total of 8 genetic variations found, including insertions, deletions, and amino acid changes, 5 were unique to GG (ATCC 53103) and 3 were unique to GG (UT) (see Table S3 in the supplemental material).
In strain GG (UT), the gene kdgK, which encodes a 2-dehydro-3-deoxygluconokinase that is involved in pectin and galacturonate catabolism, is disrupted by a DNA fragment containing LGG_00143, which codes for a hypothetical protein.
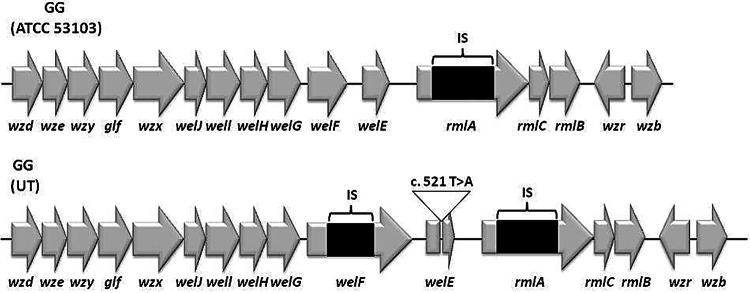
The other two mutations unique to strain GG (UT) were located within the exopolysaccharide (EPS) biosynthesis cluster. The sequence of the whole EPS cluster of GG (UT) was confirmed by PCR amplification and sequencing of the obtained amplicons. The first mutation (reference position 2095506) is a nucleotide change (T to A) that leads to a stop codon within the welE gene. This gene encodes the priming glycosyltransferase in the EPS biosynthesis cluster of L. rhamnosus, while the second mutation (reference position 2096942) is an insertion containing two transposase-encoding genes within the welF gene in GG (UT), which is located immediately upstream of welE (Fig. 3). The welF gene encodes one of the unique glycosyltransferases that add the remaining sugars to EPS.
FIG 3.
EPS cluster organization in strains GG (ATCC 53103) and GG (UT). The cluster organization of GG (ATCC 53103) is taken from the genome sequence determined in this study, and it is identical to the cluster organization of GG (Gefilus) and the one described previously (33). The change of T to A in nucleotide 521 of the welE gene of GG (UT) is indicated as c.521 T>A. IS, insertion sequence.
In spite of the genetic variations found, the transcriptome profiles of the strains GG (UT), GG (ATCC 53103), and GG (Gefilus) determined with expression arrays and real-time PCR were similar (see Table S4 in the supplemental material; also data not shown).
Complementation of the mutant GG (UT).
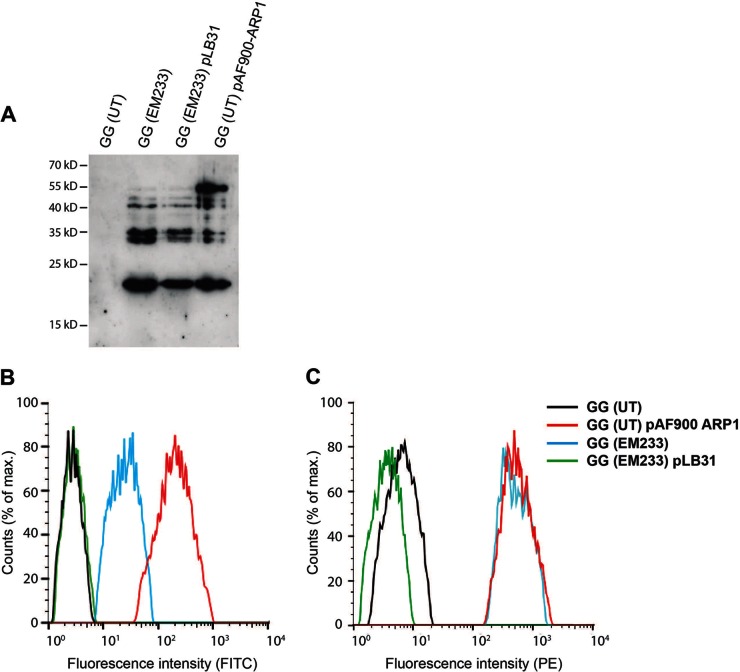
To confirm the involvement of the mutations of the genes welF and welE in the ability to display ARP1 on the cell surface of GG, complementation studies using the intact genes with their own promoters and terminators were performed. The strain GG (EM233), which harbors a chromosomal insertion of the ARP1 cassette fused with the original anchor domain (243 aa of PrtP from L. casei BL23) was transformed with the plasmid pLB31 (carrying the welF and welE genes).
The production of ARP1 in GG (EM233) was not affected by the complementation plasmid, pLB31, as observed by Western blotting (Fig. 4A). However, ARP1 was not detected on the cell surface of strain GG (EM233) pLB31 (carrying the welF and welE genes) using an anti-E-tag antibody in flow cytometry (Fig. 4B). In accordance with this, the binding capacity of cell-anchored ARP1 to RRV particles tested by flow cytometry was also impaired in the complemented strain (Fig. 4C).
FIG 4.
Complementation of GG (UT) with the genes welF and welE. (A) Production of ARP1 determined in culture cell extracts by Western blotting. (B and C) Flow cytometric analyses to study the display (B) and binding (C) of ARP1 in GG (EM233) and complemented strain. Strains GG (UT) and GG (UT) pAF900-ARP1 were included as negative and positive control, respectively. For the analysis of histogram B, samples were incubated with a monoclonal mouse anti-E-tag antibodies and FITC-conjugated anti-mouse antibodies. For the analysis of histogram C, samples were incubated with RRV particles, rabbit anti-RRV K230, and PE-conjugated donkey anti-rabbit antibodies.
To determine whether an EPS layer on the cell surface of GG (ATCC 53103) masked ARP1, the cell walls of strains GG (ATCC 53103), GG (UT), and GG (UT) pLB31 were analyzed by TEM. An EPS layer was detected on the cell surface of strain GG (ATCC 53103) and the complemented strain GG (UT) pLB31 but was not present in the original GG (UT) strain (Fig. 5). Thus, the absence of this EPS layer in GG (UT) might permit better exposure of ARP1 on the bacterial surface.
FIG 5.
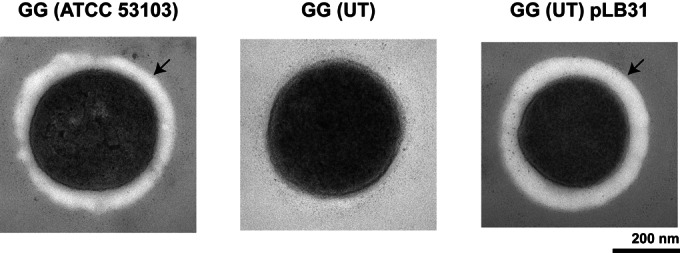
Transmission electron microscopy analysis of the cell walls of strains GG (ATCC 53103), GG (UT), and GG (UT) pLB31. The arrow points to an EPS layer on the surface of the bacterium.
Protection against RRV infection in vivo.
The EPS-deficient strain GG (UT) was tested in a mouse pup model to determine whether this strain retains its protective capacity against rotavirus infection. Strain GG (EM233) was also tested to investigate whether the production of ARP1 can enhance the protective capacity of GG. Groups of mice treated with strains GG (NCC 3003) and L. casei BL23 were included as positive and negative controls, respectively. The groups of mice treated with the original strain GG (NCC 3003) and the mutant strain GG (UT) presented similar values of the infection parameters (Table 2). Although not significant, the clinical parameters of the group treated with strain GG (EM233) were reduced in comparison to the two groups treated with the nontransformed GG strains. The differences in the prevalence (peak of infection at day 3), duration, and severity between the GG (EM233) group and the L. casei group were statistically significant (P < 0.05).
TABLE 2.
Prevalence, duration, and severity of rotavirus-induced diarrhea in the mouse pup model
| Organism | No. of animals | Prevalence of diarrhea (max %) | Mean duration ± SE (day) | Mean severity score ± SE |
|---|---|---|---|---|
| L. casei BL23 | 8 | 75 | 1.625 ± 0.420 | 2.625 ± 0.730 |
| L. rhamnosus GG (NCC 3003) | 9 | 33.33 | 0.556 ± 0.242 | 1.000 ± 0.408 |
| L. rhamnosus GG (UT) | 8 | 37.5 | 0.750 ± 0.250 | 1.125 ± 0.398 |
| L. rhamnosus GG (EM233) | 9 | 11.11a | 0.222 ± 0.222b | 0.333 ± 0.333b |
P < 0.05 versus L. casei group by Fisher's test.
P < 0.05 versus L. casei group by Kruskal-Wallis and Dunn's test.
DISCUSSION
In order to develop an oral therapy against rotavirus-induced diarrhea, the well-known probiotic strain L. rhamnosus GG was chosen to produce ARP1 using the apf expression cassette. The probiotic properties of this strain have been described in previous articles and related to their ability to strengthen the intestinal barrier and to trigger innate and adaptive immune responses (28–30). Most importantly for our purpose, L. rhamnosus GG has intrinsic antirotavirus activity (19). Another appealing characteristic of L. rhamnosus GG is that it has been shown to transiently colonize the small intestine (31), so that a genetically modified strain can potentially produce ARP1 in situ when the therapy is needed and be naturally eliminated after this period. When testing the ARP1 display in the L. rhamnosus GG strains of different origins, the antibody fragment was detected only on the surface of GG (UT). This strain was obtained from the bacterial collection of the University of Tartu (Estonia). A cassette with 450 amino acids of the C-terminal part of PrtP was the shortest anchoring domain among the ones tested mediating optimal display and functionality of ARP1 on the cell surface of GG (UT). When analyzed by Western blotting, all the fusion proteins of ARP1 with the different PrtP anchoring domains presented some degree of degradation and higher molecular masses than the theoretical ones. This could be due to conformational complexity, isopeptide bond formation, or even glycosylation. It has been described that L. rhamnosus GG secretes O-glycosylated proteins (32). Given these results, the strain GG (UT), which was able to display ARP1 on its surface and the cassette with 450 amino acids of the C-terminal part of PrtP, was selected to further develop therapeutically effective lactobacilli.
To investigate the origin of the differences in the surface ARP1 display, genomic analysis was carried out on strains GG (UT), GG (ATCC 53103), and GG (Gefilus). The result of this analysis indicate that GG (UT) and GG (ATCC 53103) appeared to be derived from GG (Gefilus), given the genetic variations among these three strains. All three presented the gene rmlA, which codes for a glucose-1-phosphate thymidylyl transferase and forms part of the EPS cluster, disrupted by an insertion (IS) element (ISLrh2). Nevertheless, this disruption is most probably not relevant since an intact rmlACBD operon is found at another locus of the genome (20, 33). GG (UT) presented three unique genetic variations that are likely generated after several passages in vitro. Propagation of strains isolated from the intestinal gut, including L. rhamnosus GG, in the absence of selective pressure probably results in metabolic simplification and gene mutations. One of these variations that are unique for GG (UT) includes an insertion within the gene kdgK, which encodes α-2-dehydro-3-deoxygluconokinase. The α-2-dehydro-3-deoxygluconokinase is involved in pectin and galacturonate catabolism (34, 35), and the disruption might thus cause GG (UT) to be unable to use galacturonate as a carbon source. Galacturonate can be found in the intestine, coming from the diet and as part of glycoproteins and glycolipids of the intestinal mucus (36), but it is unclear whether this mutation could affect the behavior and persistence of GG (UT) in the GIT.
It has been shown that L. rhamnosus GG possesses a thick layer of EPS composed of two types of polysaccharides: the first, most abundant, being galactose-rich polysaccharides and the second, shorter glucose-rich polysaccharides (37). The galactose-rich polysaccharides are composed of hexasaccharide repeated units that, besides d-galactose, contain l-rhamnose and N-acetyl-d-glucosamine residues (38). The exact biochemical composition of the shorter glucose-rich polysaccharides is unknown (37). The genes that control the production of the galactose-rich polysaccharides are localized in the EPS gene cluster that contains 17 putative ORFs, of which 16 putatively encode proteins involved in the biosynthesis of bacterial polysaccharides and 1 (orf1) encodes a putative transposase (33). The occurrence of spontaneous mutations that lead to altered EPS production has been reported before in strains of Lactobacillus (39, 40). In the present study, two naturally occurring genetic variations were found in the welE and welF genes of the EPS gene cluster of GG (UT), which abolished the production of cell-bound EPS as shown in electron microscopy. The knockout of the welE gene was previously shown to prevent the production of long galactose-rich EPS molecules and increase the adherence of GG to mucus from human intestine and epithelial cells due to better exposure of the adhesins on the bacterial surface (33, 41). Furthermore, it has been shown that the mucus-specific surface adhesin of L. rhamnosus GG was detected by immunogold only in a welE knockout mutant that was deficient in EPS production (42). Similarly, the exopolysaccharides might mask the ARP1 on the surface of strains GG (NCC 3003), GG (ATCC 53103), and GG (CMC). In accordance with this hypothesis, plasmid complementation of welE and welF restored the production of exopolysaccharide in strain GG (UT) as observed by electron microscopy and prevented the detection of ARP1 and binding of rotavirus on the surface of engineered lactobacilli GG (EM233) as shown by flow cytometry. Recently, we successfully expressed the IgG binding domains of protein G on the surface of L. rhamnosus GG (UT), confirming that this strain is suitable for cell surface display (43). Moreover, supporting that the lack of EPS is important for a correct display of ARP1, L. casei BL23, which does not produce long galactose-rich EPS (44), was shown to efficiently expose this molecule on its surface (17).
The role of EPS in connection to the probiotic properties of GG is not completely clear. The EPS of L. rhamnosus GG was reported to protect the bacterium against innate immune factors such as complement and cathelicidins (41) and to be an important factor for survival of the bacteria inside the GIT. On the other hand, the EPS negatively affected the adhesive capacity of the microorganism to intestinal mucus by masking fimbriae and adhesins (33). Furthermore, the expression of genes within the EPS cluster of GG was downregulated in the presence of bile (27) (a possible indicator of gut entrance), which suggests that EPS production could be regulated depending on the needs for protection against immune attacks versus optimal adherence.
We previously showed that treatment with GG (NCC 3003) reduced the prevalence, duration, and severity of rotavirus-induced diarrhea in a mouse pup model whereas L. casei BL23 was not protective (45). In the present study, GG (UT) seemed to confer a level of protection similar to that of GG (NCC 3003), suggesting that the EPS of GG might not be involved in the protective capacity of the strain in the mouse model. It is also important to highlight that strain GG (EM233) producing ARP1 significantly reduced the clinical parameters compared to the control strain. In our previous study using GG, we observed that when used in the animal model, GG (NCC 3003) in combination with anti-rotavirus hyperimmune bovine colostrum, the infection parameters were reduced in comparison with those observed with use of GG (NCC 3003) alone (45). Using the recombinant GG strain expressing the anti-rotavirus antibody fragment as a therapy would, however, be easier to produce and more cost-effective than a mix of purified antibodies and probiotics.
Taken together, the results suggest that a modified GG (UT) strain expressing ARP1 anchored on the cell wall could form the base for an oral therapy against rotavirus. In view of the finding that some L. rhamnosus GG isolates present genomic deletions that include the genes spaCBA (46), it is worth noting that this is not the case for strain GG (UT). The genes spaCBA code for pilin subunits that are believed to be involved in human mucus binding and persistence in the GIT. However, it should be taken into account that the lack of EPS and the disruption of the gene kdgK may affect survival and persistence in the GIT. This should be tested in a future safety trial with human volunteers.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Bioinformatics and Expression Analysis core facility (BEA), Karolinska Institutet, Huddinge, Sweden, for the microarray hybridization process and data analysis, Kjell Hultenby from the electron microcopy unit (Emil), Karolinska University Hospital, Huddinge, Sweden, for the electron microscopy pictures, Kari Johansen, Department of Microbiology, Tumor and Cell Biology, Karolinska Institute, Stockholm, Sweden, for the preparations of RRV, Lennart Svensson, Department of Clinical and Experimental Medicine, University of Linköping, for providing the polyclonal rabbit anti-RRV antibodies K230, Leon Frenken, Unilever Research & Development, Vlaardingen BV, Netherlands, for providing the ARP1-encoding gene, Marika Mikelsaar, Department of Microbiology, University of Tartu, Tartu, Estonia, for providing strain GG (UT), and Gagandeep Kang, Department of Gastrointestinal Sciences, Christian Medical College (CMC) Vellore, India, for providing strain GG (CMC).
This work was supported by the European Union (EU)-funded project LACTOBODY(202162).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00945-15.
REFERENCES
- 1.Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD. 2012. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis 12:136–141. doi: 10.1016/S1473-3099(11)70253-5. [DOI] [PubMed] [Google Scholar]
- 2.Armah GE, Sow SO, Breiman RF, Dallas MJ, Tapia MD, Feikin DR, Binka FN, Steele AD, Laserson KF, Ansah NA, Levine MM, Lewis K, Coia ML, Attah-Poku M, Ojwando J, Rivers SB, Victor JC, Nyambane G, Hodgson A, Schodel F, Ciarlet M, Neuzil KM. 2010. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet 376:606–614. doi: 10.1016/S0140-6736(10)60889-6. [DOI] [PubMed] [Google Scholar]
- 3.Madhi SA, Cunliffe NA, Steele D, Witte D, Kirsten M, Louw C, Ngwira B, Victor JC, Gillard PH, Cheuvart BB, Han HH, Neuzil KM. 2010. Effect of human rotavirus vaccine on severe diarrhea in African infants. N Engl J Med 362:289–298. doi: 10.1056/NEJMoa0904797. [DOI] [PubMed] [Google Scholar]
- 4.Zaman K, Dang DA, Victor JC, Shin S, Yunus M, Dallas MJ, Podder G, Vu DT, Le TP, Luby SP, Le HT, Coia ML, Lewis K, Rivers SB, Sack DA, Schodel F, Steele AD, Neuzil KM, Ciarlet M. 2010. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in Asia: a randomised, double-blind, placebo-controlled trial. Lancet 376:615–623. doi: 10.1016/S0140-6736(10)60755-6. [DOI] [PubMed] [Google Scholar]
- 5.Glass RI, Parashar UD. 2006. The promise of new rotavirus vaccines. N Engl J Med 354:75–77. doi: 10.1056/NEJMe058285. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz-Palacios GM, Pérez-Schael I, Velázquez FR, Abate H, Breuer T, Clemens SC, Cheuvart B, Espinoza F, Gillard P, Innis BL, Cervantes Y, Linhares AC, López P, Macías-Parra M, Ortega-Barría E, Richardson V, Rivera-Medina DM, Rivera L, Salinas B, Pavía-Ruz N, Salmerón J, Rüttimann R, Tinoco JC, Rubio P, Nuñez E, Guerrero ML, Yarzábal JP, Damaso S, Tornieporth N, Sáez-Llorens X, Vergara RF, Vesikari T, Bouckenooghe A, Clemens R, De Vos B, O'Ryan M. 2006. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med 354:11–22. doi: 10.1056/NEJMoa052434. [DOI] [PubMed] [Google Scholar]
- 7.Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, Rodriguez Z, Dallas MJ, Heyse JF, Goveia MG, Black SB, Shinefield HR, Christie CDC, Ylitalo S, Itzler RF, Coia ML, Onorato MT, Adeyi BA, Marshall GS, Gothefors L, Campens D, Karvonen A, Watt JP, O'Brien KL, DiNubile MJ, Clark HF, Boslego JW, Offit PA, Heaton PM. 2006. Safety and efficacy of a pentavalent human–bovine (WC3) reassortant rotavirus vaccine. N Engl J Med 354:23–33. doi: 10.1056/NEJMoa052664. [DOI] [PubMed] [Google Scholar]
- 8.Garaicoechea L, Olichon A, Marcoppido G, Wigdorovitz A, Mozgovoj M, Saif L, Surrey T, Parreno V. 2008. Llama-derived single-chain antibody fragments directed to rotavirus VP6 protein possess broad neutralizing activity in vitro and confer protection against diarrhea in mice. J Virol 82:9753–9764. doi: 10.1128/JVI.00436-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Vaart JM, Pant N, Wolvers D, Bezemer S, Hermans PW, Bellamy K, Sarker SA, van der Logt CP, Svensson L, Verrips CT, Hammarstrom L, van Klinken BJ. 2006. Reduction in morbidity of rotavirus induced diarrhoea in mice by yeast produced monovalent llama-derived antibody fragments. Vaccine 24:4130–4137. doi: 10.1016/j.vaccine.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 10.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. 1993. Naturally occurring antibodies devoid of light chains. Nature 363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 11.Frenken LG, van der Linden RH, Hermans PW, Bos JW, Ruuls RC, de Geus B, Verrips CT. 2000. Isolation of antigen specific llama VHH antibody fragments and their high level secretion by Saccharomyces cerevisiae. J Biotechnol 78:11–21. doi: 10.1016/S0168-1656(99)00228-X. [DOI] [PubMed] [Google Scholar]
- 12.Dolk E, van Vliet C, Perez JM, Vriend G, Darbon H, Ferrat G, Cambillau C, Frenken LG, Verrips T. 2005. Induced refolding of a temperature denatured llama heavy-chain antibody fragment by its antigen. Proteins 59:555–564. doi: 10.1002/prot.20378. [DOI] [PubMed] [Google Scholar]
- 13.van der Linden RH, Frenken LG, de Geus B, Harmsen MM, Ruuls RC, Stok W, de Ron L, Wilson S, Davis P, Verrips CT. 1999. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim Biophys Acta 1431:37–46. doi: 10.1016/S0167-4838(99)00030-8. [DOI] [PubMed] [Google Scholar]
- 14.Pant N, Hultberg A, Zhao Y, Svensson L, Pan-Hammarstrom Q, Johansen K, Pouwels PH, Ruggeri FM, Hermans P, Frenken L, Boren T, Marcotte H, Hammarstrom L. 2006. Lactobacilli expressing variable domain of llama heavy-chain antibody fragments (lactobodies) confer protection against rotavirus-induced diarrhea. J Infect Dis 194:1580–1588. doi: 10.1086/508747. [DOI] [PubMed] [Google Scholar]
- 15.Aladin F, Einerhand AW, Bouma J, Bezemer S, Hermans P, Wolvers D, Bellamy K, Frenken LG, Gray J, Iturriza-Gomara M. 2012. In vitro neutralisation of rotavirus infection by two broadly specific recombinant monovalent llama-derived antibody fragments. PLoS One 7:e32949. doi: 10.1371/journal.pone.0032949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarker SA, Jakel M, Sultana S, Alam NH, Bardhan PK, Chisti MJ, Salam MA, Theis W, Hammarstrom L, Frenken LG. 2013. Anti-rotavirus protein reduces stool output in infants with diarrhea: a randomized placebo-controlled trial. Gastroenterology 145:740–748.e748. doi: 10.1053/j.gastro.2013.06.053. [DOI] [PubMed] [Google Scholar]
- 17.Martin MC, Pant N, Ladero V, Gunaydin G, Andersen KK, Alvarez B, Martinez N, Alvarez MA, Hammarstrom L, Marcotte H. 2011. Integrative expression system for delivery of antibody fragments by lactobacilli. Appl Environ Microbiol 77:2174–2179. doi: 10.1128/AEM.02690-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siezen RJ. 1999. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie Van Leeuwenhoek 76:139–155. doi: 10.1023/A:1002036906922. [DOI] [PubMed] [Google Scholar]
- 19.Guandalini S. 2011. Probiotics for prevention and treatment of diarrhea. J Clin Gastroenterol 45(Suppl):S149–S153. doi: 10.1097/MCG.0b013e3182257e98. [DOI] [PubMed] [Google Scholar]
- 20.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, De Keersmaecker SC, Vanderleyden J, Hamalainen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjarvi T, Auvinen P, de Vos WM. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci U S A 106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brandt K, Alatossava T. 2003. Specific identification of certain probiotic Lactobacillus rhamnosus strains with PCR primers based on phage-related sequences. Int J Food Microbiol 84:189–196. doi: 10.1016/S0168-1605(02)00419-1. [DOI] [PubMed] [Google Scholar]
- 22.Martin MC, Alonso JC, Suarez JE, Alvarez MA. 2000. Generation of food-grade recombinant lactic acid bacterium strains by site-specific recombination. Appl Environ Microbiol 66:2599–2604. doi: 10.1128/AEM.66.6.2599-2604.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Keersmaecker SC, Braeken K, Verhoeven TL, Perea Velez M, Lebeer S, Vanderleyden J, Hols P. 2006. Flow cytometric testing of green fluorescent protein-tagged Lactobacillus rhamnosus GG for response to defensins. Appl Environ Microbiol 72:4923–4930. doi: 10.1128/AEM.02605-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Arellano I, Zuniga M, Perez-Martinez G. 2001. Construction of compatible wide-host-range shuttle vectors for lactic acid bacteria and Escherichia coli. Plasmid 46:106–116. doi: 10.1006/plas.2001.1531. [DOI] [PubMed] [Google Scholar]
- 25.Kruger C, Hu Y, Pan Q, Marcotte H, Hultberg A, Delwar D, van Dalen PJ, Pouwels PH, Leer RJ, Kelly CG, van Dollenweerd C, Ma JK, Hammarstrom L. 2002. In situ delivery of passive immunity by lactobacilli producing single-chain antibodies. Nat Biotechnol 20:702–706. doi: 10.1038/nbt0702-702. [DOI] [PubMed] [Google Scholar]
- 26.Marcotte H, Koll-Klais P, Hultberg A, Zhao Y, Gmur R, Mandar R, Mikelsaar M, Hammarstrom L. 2006. Expression of single-chain antibody against RgpA protease of Porphyromonas gingivalis in Lactobacillus. J Appl Microbiol 100:256–263. doi: 10.1111/j.1365-2672.2005.02786.x. [DOI] [PubMed] [Google Scholar]
- 27.Koskenniemi K, Laakso K, Koponen J, Kankainen M, Greco D, Auvinen P, Savijoki K, Nyman TA, Surakka A, Salusjarvi T, de Vos WM, Tynkkynen S, Kalkkinen N, Varmanen P. 2011. Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol Cell Proteomics 10:M110.002741. doi: 10.1074/mcp.M110.002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harata G, He F, Hiruta N, Kawase M, Kubota A, Hiramatsu M, Yausi H. 2010. Intranasal administration of Lactobacillus rhamnosus GG protects mice from H1N1 influenza virus infection by regulating respiratory immune responses. Lett Appl Microbiol 50:597–602. doi: 10.1111/j.1472-765X.2010.02844.x. [DOI] [PubMed] [Google Scholar]
- 29.Kaila M, Isolauri E, Saxelin M, Arvilommi H, Vesikari T. 1995. Viable versus inactivated Lactobacillus strain GG in acute rotavirus diarrhoea. Arch Dis Child 72:51–53. doi: 10.1136/adc.72.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lebeer S, Claes I, Tytgat HL, Verhoeven TL, Marien E, von Ossowski I, Reunanen J, Palva A, Vos WM, Keersmaecker SC, Vanderleyden J. 2012. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol 78:185–193. doi: 10.1128/AEM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alander M, Satokari R, Korpela R, Saxelin M, Vilpponen-Salmela T, Mattila-Sandholm T, von Wright A. 1999. Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl Environ Microbiol 65:351–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lebeer S, Claes IJ, Balog CI, Schoofs G, Verhoeven TL, Nys K, von Ossowski I, de Vos WM, Tytgat HL, Agostinis P, Palva A, Van Damme EJ, Deelder AM, De Keersmaecker SC, Wuhrer M, Vanderleyden J. 2012. The major secreted protein MspI/p75 is O-glycosylated in Lactobacillus rhamnosus GG. Microb Cell Fact 11:15. doi: 10.1186/1475-2859-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebeer S, Verhoeven TL, Francius G, Schoofs G, Lambrichts I, Dufrene Y, Vanderleyden J, De Keersmaecker SC. 2009. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl Environ Microbiol 75:3554–3563. doi: 10.1128/AEM.02919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hugouvieux-Cotte-Pattat N, Nasser W, Robert-Baudouy J. 1994. Molecular characterization of the Erwinia chrysanthemi kdgK gene involved in pectin degradation. J Bacteriol 176:2386–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pouyssegur J, Stoeber F. 1974. Genetic control of the 2-keto-3-deoxy-d-gluconate metabolism in Escherichia coli K-12: kdg regulon. J Bacteriol 117:641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peekhaus N, Conway T. 1998. What's for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J Bacteriol 180:3495–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francius G, Lebeer S, Alsteens D, Wildling L, Gruber HJ, Hols P, De Keersmaecker S, Vanderleyden J, Dufrene YF. 2008. Detection, localization, and conformational analysis of single polysaccharide molecules on live bacteria. ACS Nano 2:1921–1929. doi: 10.1021/nn800341b. [DOI] [PubMed] [Google Scholar]
- 38.Landersjo C, Yang Z, Huttunen E, Widmalm G. 2002. Structural studies of the exopolysaccharide produced by Lactobacillus rhamnosus strain GG (ATCC 53103). Biomacromolecules 3:880–884. doi: 10.1021/bm020040q. [DOI] [PubMed] [Google Scholar]
- 39.Van Geel-Schutten GH, Faber EJ, Smit E, Bonting K, Smith MR, Ten Brink B, Kamerling JP, Vliegenthart JF, Dijkhuizen L. 1999. Biochemical and structural characterization of the glucan and fructan exopolysaccharides synthesized by the Lactobacillus reuteri wild-type strain and by mutant strains. Appl Environ Microbiol 65:3008–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peant B, LaPointe G, Gilbert C, Atlan D, Ward P, Roy D. 2005. Comparative analysis of the exopolysaccharide biosynthesis gene clusters from four strains of Lactobacillus rhamnosus. Microbiology 151:1839–1851. doi: 10.1099/mic.0.27852-0. [DOI] [PubMed] [Google Scholar]
- 41.Lebeer S, Claes IJ, Verhoeven TL, Vanderleyden J, De Keersmaecker SC. 2011. Exopolysaccharides of Lactobacillus rhamnosus GG form a protective shield against innate immune factors in the intestine. Microb Biotechnol 4:368–374. doi: 10.1111/j.1751-7915.2010.00199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Ossowski I, Reunanen J, Satokari R, Vesterlund S, Kankainen M, Huhtinen H, Tynkkynen S, Salminen S, de Vos WM, Palva A. 2010. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl Environ Microbiol 76:2049–2057. doi: 10.1128/AEM.01958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gunaydin G, Zhang R, Hammarstrom L, Marcotte H. 2014. Engineered Lactobacillus rhamnosus GG expressing IgG-binding domains of protein G: capture of hyperimmune bovine colostrum antibodies and protection against diarrhea in a mouse pup rotavirus infection model. Vaccine 32:470–477. doi: 10.1016/j.vaccine.2013.11.057. [DOI] [PubMed] [Google Scholar]
- 44.Maze A, Boel G, Zuniga M, Bourand A, Loux V, Yebra MJ, Monedero V, Correia K, Jacques N, Beaufils S, Poncet S, Joyet P, Milohanic E, Casaregola S, Auffray Y, Perez-Martinez G, Gibrat JF, Zagorec M, Francke C, Hartke A, Deutscher J. 2010. Complete genome sequence of the probiotic Lactobacillus casei strain BL23. J Bacteriol 192:2647–2648. doi: 10.1128/JB.00076-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pant N, Marcotte H, Brussow H, Svensson L, Hammarstrom L. 2007. Effective prophylaxis against rotavirus diarrhea using a combination of Lactobacillus rhamnosus GG and antibodies. BMC Microbiol 7:86. doi: 10.1186/1471-2180-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sybesma W, Molenaar D, van IJcken W, Venema K, Kort R. 2013. Genome instability in Lactobacillus rhamnosus GG. Appl Environ Microbiol 79:2233–2239. doi: 10.1128/AEM.03566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin MC, Fernandez M, Martin-Alonso JM, Parra F, Boga JA, Alvarez MA. 2004. Nisin-controlled expression of Norwalk virus VP60 protein in Lactobacillus casei. FEMS Microbiol Lett 237:385–391. doi: 10.1111/j.1574-6968.2004.tb09721.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.