Abstract
In broiler chickens, feed additives, including prebiotics, are widely used to improve gut health and to stimulate performance. Xylo-oligosaccharides (XOS) are hydrolytic degradation products of arabinoxylans that can be fermented by the gut microbiota. In the current study, we aimed to analyze the prebiotic properties of XOS when added to the broiler diet. Administration of XOS to chickens, in addition to a wheat-rye-based diet, significantly improved the feed conversion ratio. XOS significantly increased villus length in the ileum. It also significantly increased numbers of lactobacilli in the colon and Clostridium cluster XIVa in the ceca. Moreover, the number of gene copies encoding the key bacterial enzyme for butyrate production, butyryl-coenzyme A (butyryl-CoA):acetate CoA transferase, was significantly increased in the ceca of chickens administered XOS. In this group of chickens, at the species level, Lactobacillus crispatus and Anaerostipes butyraticus were significantly increased in abundance in the colon and cecum, respectively. In vitro fermentation of XOS revealed cross-feeding between L. crispatus and A. butyraticus. Lactate, produced by L. crispatus during XOS fermentation, was utilized by the butyrate-producing Anaerostipes species. These data show the beneficial effects of XOS on broiler performance when added to the feed, which potentially can be explained by stimulation of butyrate-producing bacteria through cross-feeding of lactate and subsequent effects of butyrate on gastrointestinal function.
INTRODUCTION
Cereal fibers are composed of carbohydrate polymers that are resistant to digestion in the small intestines of monogastric animals but are completely or partially fermented in the distal gut, and they are believed to stimulate gut health (1). The main components of the cereal fiber fraction are arabinoxylans (AX), pectins, resistant starch, cellulose, β-glucans, and lignin (2). Hydrolytic degradation of the heteropolymer AX results in a mixture of arabinose-substituted xylo-oligosaccharides (arabinoxylan-oligosaccharides) (AXOS) and nonsubstituted xylo-oligosaccharides (XOS) (3). XOS are oligomers consisting of xylose units linked through β-(1-4) linkages (4). Selective fermentation of XOS has been shown to induce changes in both the composition and activity of the gastrointestinal microbiota, improving the health and well-being of the host. This suggests that XOS could fulfill the definition of a prebiotic (5). The production of lactate and short-chain fatty acids (SCFA), including butyrate, upon fermentation of XOS, has been confirmed in several in vitro and in vivo studies (3, 6). Lactate can stimulate butyrate production due to cross-feeding between lactate-producing bacteria and lactate-utilizing butyrate-producing bacteria from Clostridium cluster XIVa (7). Butyrate has proven beneficial effects on gastrointestinal function, since it has anti-inflammatory properties, fuels epithelial cells, and increases the intestinal epithelial integrity. In addition, butyrate has been shown to improve growth performance in production animals and to change the microbiota composition and metabolic activity of the microbial ecosystem in the intestine (8, 9).
Beneficial effects of XOS have already been described in rats. In these studies, XOS was shown to significantly increase the population of bifidobacteria and lactobacilli in the cecum (10, 11). An in vitro study using swine fecal microbiota showed the highest SCFA production during fermentation of XOS (12). To our knowledge, there is not much published research on the effect of XOS on the gastrointestinal health of chickens except for the recent publication of Zhenping et al. (13), showing increased growth performance, enhanced endocrine metabolism, and improved immune function in broiler chickens after in-feed supplementation of straw-derived XOS. However, the effect of XOS on the microbiota composition in broilers has not yet been described.
In the broiler chicken, the distal ileum, the cecum, and the colon are regarded as fermentation chambers whose function is determined by the microbiota composition (14, 15). The chicken gut microbiota is dominated by species belonging to the phyla Firmicutes (up to 75%) and Bacteroidetes (between 10% and 50%) (16–22). Around 90% of the bacteria in the chicken gastrointestinal tract are unknown species, indicating that the knowledge of the intestinal microbiota of chickens is incomplete (23, 24). The majority of sequences within the Firmicutes phylum belong to the families Ruminococcaceae and Lachnospiraceae, the so-called Clostridium cluster IV and XIVa, respectively (25). Both families contain numerous members that are known to produce butyrate as a fermentation end product and are therefore linked to beneficial effects on gastrointestinal function (26, 27). Whether the abundance of these groups in the distal gut of chickens is affected by XOS is unclear.
In the current study, we analyzed the effect of XOS administration on the performance of broilers. In addition, we aimed to identify the shifts in microbiota composition induced by XOS to explain possible beneficial effects on gastrointestinal health, with emphasis on butyrate production.
MATERIALS AND METHODS
Additives/substrates.
In the in vivo study, corncob-derived XOS35 (Longlive Bio-technology, Shandong, China) was used as a feed additive. XOS35 is a mixture of 35% XOS with a degree of polymerization (DP) between 2 and 7 and 65% maltodextrin. In the in vitro fermentation study, XOS35, maltodextrin (Sigma-Aldrich, St. Louis, MO), and XOS95 (Longlive Bio-technology, Shandong, China), a mixture of 95% XOS with a DP of 2 to 7 and 5% xylose, were used. The XOS95 and maltodextrin were used to confirm that the effects of XOS35 in the in vivo trial were explained by the XOS.
Animals and diets.
A total of 192 male and 192 female 1-day-old Ross-308 broiler chickens were randomly divided in 12 pens (3 pens of female and 3 pens of male birds per treatment and 32 chickens per pen) and housed on solid floors covered with wood shavings. The light schedule was set to provide an 18-h light/6-h dark cycle. Infrared bulbs (1 per pen during the first week) together with a central heating system provided the optimal temperature. All animals were fed a wheat-rye-based diet with XOS (experimental group) or without XOS (control group), the composition of which is shown in Table 1. The experimental starter feed (fed from the first day of age until day 13) was supplemented with 0.2% XOS, and the grower feed (fed from day 14 until day 26) and the finisher feed (fed from day 27 until day 39) were supplemented with 0.5% XOS. At 13, 26, and 39 days of age, all broilers and the feed leftovers were weighed per pen to calculate the feed conversion ratio (FCR), weight gain (WG), and feed intake (FI). At 26 days of age, three chickens per pen were euthanized by an intravenous overdose of 20% sodium pentobarbital (Kela, Hoogstraten, Belgium). The complete content of cecum and colon was collected and stored at −70°C, while a part of the ileum at the level of Meckel's diverticulum was fixed in 4% formaldehyde.
TABLE 1.
Compositions and nutrient contents of the wheat-rye diets administered to chickensa
| Parameter | Starter diet | Grower diet | Finisher diet |
|---|---|---|---|
| Feedstuffs (%) | |||
| Wheat | 44.00 | 46.51 | 49.78 |
| Rye | 5.00 | 5.00 | 5.00 |
| Soybean meal (48) | 23.30 | 19.78 | 16.61 |
| Soybeans | 7.50 | 5.00 | 5.00 |
| Sunflower meal 27 | 2.50 | 2.50 | 2.50 |
| Rapeseed meal | 7.50 | 10.00 | 10.00 |
| Animal fat | 3.90 | 5.00 | 5.12 |
| Soy oil | 2.80 | 2.82 | 2.61 |
| Vitamins + trace elements (Vitamix) | 1.00 | 1.00 | 1.00 |
| CaCO3 | 0.55 | 0.56 | 0.75 |
| Di-Ca-phosphate | 0.90 | 0.62 | 0.37 |
| NaCl | 0.21 | 0.21 | 0.19 |
| Na-bicarbonate | 0.10 | 0.10 | 0.10 |
| l-Lys-HCl | 0.14 | 0.15 | 0.20 |
| dl-Methionine | 0.50 | 0.70 | 0.70 |
| l-Threonine | 0.04 | 0.03 | 0.03 |
| Phytase | 0.02 | 0.02 | 0.02 |
| Nutrient composition | |||
| Crude protein (%) | 23.00 | 21.50 | 20.50 |
| Crude fat (%) | 10.23 | 10.66 | 10.91 |
| Nonsoluble polysaccharides (%) | 13.87 | 13.98 | 13.83 |
| Metabolizable energy (MJ/kg) | 11.72 | 12.15 | 12.25 |
| d-Lysine (%) | 1.12 | 1.03 | 1.00 |
| d-Sulfur amino acids (%) | 1.10 | 0.77 | 0.75 |
| d-Threonine (%) | 0.73 | 0.67 | 0.65 |
| d-Valine (%) | 0.84 | 0.76 | 0.72 |
| Ca (%) | 0.85 | 0.80 | 0.75 |
| Available P (%) | 0.40 | 0.35 | 0.30 |
| NaCl + KCl (meq/kg) | 247 | 225 | 208 |
| Linoleic acid (18:2) (%) | 3.34 | 3.15 | 3.17 |
The starter diet was given from day 1 until day 13, the grower diet was given from day 14 until day 26, and the finisher diet was given from day 27 until day 39.
Morphological examination.
Formalin-fixed ileum segments taken at the level of Meckel's diverticulum were dehydrated in xylene, embedded in paraffin, and sectioned in 4-μm slides. The sections were deparaffinized (twice for 5 min each) in xylene, rehydrated in isopropylene (5 min), 95% alcohol (5 min), and 50% alcohol (5 min), and stained with hematoxylin and eosin. The sections were examined using light microscopy. The villus length and thickness of the tunica muscularis were measured by random measurement of 10 villi and 10 measurements of the tunica muscularis per section using a Leica DM LB2 Digital (Leica Microsystems Belgium BVBA, Diegem, Belgium) and a PC-based image analysis system (Leica Application suite V3; Leica, Diegem Belgium).
Microbiota composition. (i) DNA extraction.
DNA was extracted from cecum and colon contents using the hexadecyltrimethylammonium bromide (CTAB) method as described previously (28, 29). To 100 mg of intestinal content, 0.5 g unwashed glass beads (Sigma-Aldrich, St. Louis, MO), 0.5 ml CTAB buffer (5% [wt/vol] hexadecyltrimethylammonium bromide, 0.35 M NaCl, 120 mM K2HPO4) and 0.5 ml phenol-chloroform-isoamyl alcohol mixture (25:24:1) (Sigma-Aldrich, St. Louis, MO) were added, followed by homogenization in a 2-ml destruction tube. The samples were shaken 6 times for 30 s each using a beadbeater (MagnaLyser; Roche, Basel, Switzerland) at 6,000 rpm with 30 s between shakings. After centrifugation (10 min, 8000 rpm), 300 μl of the supernatant was transferred to a new tube. The rest of the tube content was reextracted with 250 μl CTAB buffer and again homogenized with a beadbeater. The samples were centrifuged for 10 min at 8,000 rpm, and 300 μl supernatant was added to the first 300 μl supernatant. The phenol was removed by adding an equal volume of chloroform-isoamyl alcohol (24:1) (Sigma-Aldrich, St. Louis, MO) and performing a short spin. The aqueous phase was transferred to a new tube. The nucleic acids were precipitated with two volumes of polyethylene glycol (PEG) 6000 solution (30% [wt/vol] PEB, 1.6 M NaCl) for 2 h at room temperature. After centrifugation (20 min, 13,000 rpm), the pellet was rinsed with 1 ml of ice-cold 70% (vol/vol) ethanol. The pellet was dried and resuspended in 100 μl RNA-free water (VWR, Leuven, Belgium).
(ii) Quantitative PCR (qPCR) for total bacteria and the butyryl-CoA:acetate-CoA transferase gene.
The numbers of total bacteria and butyryl-coenzyme A (butyryl-CoA):acetate-CoA transferase genes were quantified in 3 samples per pen (18 samples per treatment). To determine the number of total bacteria, primers Uni 331F (5′-TCCTACGGGAGGCAGCAGT-3′) and Uni 797R (5′-GGACTAACCAGGGTATCTAATCCTGTT-3′) were used (30). Amplification and detection were performed using the CFX384 Bio-Rad detection system (Bio-Rad, Nazareth-Eke, Belgium). Each reaction was done in triplicate in a 12-μl total reaction mixture using 2× SensiMix SYBR No-ROX mix (Bioline, Kampenhout, Belgium), a 0.5 μM final primer concentration, and 2 μl of DNA (50 ng/μl). The amplification program consisted of 1 cycle at 95°C for 10 min followed by 40 cycles of 1 min at 94°C, 1 min at 53°C, and 2 min at 60°C. The fluorescent products were detected at the last step of each cycle. A melting curve analysis was done after amplification and was obtained by slow heating from 60°C to 95°C at a rate of 0.5°C/5 s to confirm the specificity of the reaction.
To quantify the number of gene copies encoding the butyryl-CoA:acetate-CoA transferase enzyme, primers BCoATscrF (5′-GCIGAICATTTCACITGGAAYWS-3′) and BCoATscrR (5′-CCTGCCTTTGCAATRTCIACRA ANGC-3′) were used (31). Each reaction was done in triplicate in a 12-μl total reaction mixture using 2× SensiMix SYBR No-ROX mix (Bioline, Kampenhout, Belgium), a 2.5 μM final primer concentration and 2 μl of DNA (50 ng/μl). The amplification program consisted of 1 cycle at 95°C for 10 min followed by 40 cycles of 30 s at 95°C, 30 s at 53°C, and 30 s at 72°C.
(iii) 16S rRNA gene sequencing to identify microbiota composition.
Fecal samples derived from one animal per pen (6 per treatment) were used for 16S rRNA gene sequencing. For each sample, 16S rRNA gene PCR libraries were generated with the primers E9-29 and E514-430 (32), targeting hypervariable regions V1 to V3. The oligonucleotide design included 454 Life Sciences's A or B sequencing titanium adapters (Roche Diagnostics, Vilvoorde, Belgium) and multiplex identifiers (MIDs) fused to the 5′ end of each primer. The amplification mix contained 5 U of FastStart high-fidelity polymerase (Roche Diagnostics, Vilvoorde, Belgium), 1× enzyme reaction buffer, 200 μM deoxynucleoside triphosphates (dNTPs) (Eurogentec, Liège, Belgium), 0.2 μM each primer, and 100 ng of genomic DNA in a volume of 100 μl. Thermocycling conditions consisted of a denaturation at 94°C for 15 min followed by 25 cycles at 94°C for 40 s, 56°C for 40 s, and 72°C for 1 min and a final elongation step of 7 min at 72°C. These amplifications were performed on an Ep Master System gradient apparatus (Eppendorf, Hamburg, Germany). Electrophoresis of the PCR products was done on a 1% agarose gel, and the DNA fragments were plugged out and purified using the SV PCR purification kit (Promega Benelux, Leiden, The Netherlands). The quality and quantity of the products were assessed with a Picogreen double-stranded DNA (dsDNA) quantitation assay (Isogen, St-Pieters-Leeuw, Belgium). All libraries were run in the same titanium pyrosequencing reaction using Roche MIDs. All amplicons were sequenced using the Roche GS-Junior Genome Sequencer instrument (Roche, Vilvoorde, Belgium), and the sequence number of each sample is normalized to 2,323 reads.
The 16S rRNA gene sequence reads were processed with the MOTHUR package (33). The quality of all sequence reads was denoised using the Pyronoise algorithm implemented in MOTHUR and filtered with the following criteria: minimal length of 425 bp, an exact match to the barcode, and 1 mismatch allowed to the proximal primer. The sequences were evaluated for the presence of chimeric amplifications using Uchime (34). The resulting read sets were compared to a reference data set of aligned sequences of the corresponding region derived from the SILVA database 1.15 of full-length rRNA gene sequences (http://www.arb-silva.de/) implemented in MOTHUR (35). The final reads were clustered into operational taxonomic units (OTUs) using the nearest-neighbor algorithm, using MOTHUR with a 0.03 distance unit cutoff. At the OTU level of analysis (OTU definition level for a 0.02 distance matrix), a total of 3,052 OTUs were created. A taxonomic identity was attributed to each OTU by comparison with the SILVA database (80% homogeneity cutoff). As a secondary analysis, all unique sequences for each OTU were compared to the SILVA data set 1.15 using the BLASTN algorithm (36). For each OTU, a consensus detailed taxonomic identification was given based upon the identity (less than 1% mismatch with the aligned sequence) and the metadata associated with the best hit (validated bacterial species or not).
In vitro fermentation. (i) Bacterial strains, growth, and coculture studies.
The butyrate-producing strain Anaerostipes butyraticus LMG 24724T and the lactate-producing strain Lactobacillus crispatus LMG 9479T were purchased from the LMG culture collection. A. butyraticus and L. crispatus were grown in M2GSC (37) and Man-Rogosa-Sharpe (MRS) medium, respectively, in an anaerobic chamber (Ruskinn Technology, Bridgend, United Kingdom) with 84% N2, 8% H2, and 8% CO2 at 37°C.
The in vitro fermentation study was conducted using a nutrient-poor medium described by Moura et al. (38) with minor modifications (0.85 g/liter Casitone, 0.15 g/liter enzymatic digest of soybean, 0.25 g/liter NaCl, 0.125 g/liter K2HPO4, 5.0g/liter Bacto peptone, 5.0 g/liter yeast nitrogen base, and 0.5g/liter resazurin); after autoclaving, 1 mg/ml cysteine-HCl, 1% (vol/vol) of salt solution A (100.0 g/liter NH4Cl, 10.0 g/liter MgCl2 · 6H2O, 10.0 g/liter CaCl2 · 2H2O), 1% (vol/vol) trace solution (0.025 g/liter MnCl2 · 4H2O, 0.02 g/liter FeSO4 · 7H2O, 0.025 g/liter ZnCl2, 0.025 g/liter CuCl2 · 2H2O, 0.05 g/liter CoCl2 · 6H2O, 0.05 g/liter SeO2, 0.25 g/liter NiCl2 · 6H2O, 0.25 g/liter Na2MoO4 · 2H2O, 0.314 g/liter NaVO3, 0.25 g/liter H3BO3 dissolved in 0.02 M HCl), and 1.2% (vol/vol) vitamin/phosphate solution (0.0204 g/liter biotin, 0.0205 g/liter folic acid, 0.164 g/liter Ca d-pentothenate, 0.164 g/liter nicotinamide, 0.164 g/liter riboflavin, 0.164 g/liter thiamine HCl, 0.164 g/liter pyridoxine HCl, 0.201 g/liter para-aminobenzoid acid, and 0.0205 g/liter cyanocobalamin dissolved in 54.7 g/liter KH2PO4, filter sterilized) containing a mixture of SCFAs (final concentrations: acetate, 31 mM; propionate, 9 mM; isobutyrate, isovalerate, and valerate, 1 mM each) were added. A 5% stock solution of XOS35, maltodextrin, and XOS95 was prepared in the nutrient-poor medium, filter sterilized (0.2 μm), and diluted in the nutrient-poor medium to a final concentration of 0.5% (vol/vol). Unsupplemented nutrient-poor medium was used as control (blank). The final pH of the medium was adjusted to 6.5 ± 0.1. The media were preincubated in an anaerobic cabinet until anaerobiosis, as indicated by the colorless state of resazurin in the medium. A. butyraticus and L. crispatus, precultured in M2GSC and MRS broth, respectively, at 37°C under anaerobic conditions for 24 ± 1 h without shaking, were diluted 100-fold in the supplemented and nonsupplemented nutrient-poor medium. The coculture of A. butyraticus and L. crispatus was prepared using equal portions of the inoculum (1/200 each) from the 2 pure cultures. After 24 h of anaerobic incubation at 37°C, bacterial growth was monitored by measuring the optical density (OD) at 650 nm. After measuring the pH, the cultures were centrifuged at 14,000 rpm for 10 min at room temperature. The supernatants were stored at −20°C until lactate and butyrate concentrations were determined using high-performance liquid chromatography (HPLC) analysis. The in vitro fermentation assay was done twice in triplicate.
(ii) Determination of butyrate and lactate concentrations.
dl-Lactate and butyrate were quantified using HPLC with UV detection, as described by De Baere et al. (39). The supernatant was acidified using concentrated hydrochloric acid and extracted with diethyl ether for 20 min. The upper ether phase was transferred to another extraction tube and extracted again for 20 min with sodium hydroxide. The aqueous phase was transferred to an autosampler vial, and concentrated hydrochloric acid was added. An aliquot was injected on the HPLC-UV instrument. The HPLC instrument consisted of a P1000X type quaternary gradient pump, an AS3000 type autosampler, a UV1000 type UV detector, and an SN4000 type system controller, all from ThermoFisher Scientific (Breda, The Netherlands). Chromatographic separation was achieved using a hypersilGold aQ column (150 by 4.6 mm; particle size, 3 μm; ThermoFisher Scientific). Gradient elution (80/20) was performed using NaH2PO4 in HPLC-grade water and HPLC-grade acetonitrile as mobile phases A and B, respectively. The detector was set at a wavelength of 210 nm. The Chromquest software (ThermoFisher Scientific) was used for data processing.
Statistical analysis.
The comparison of the performance data was performed with an independent-sample t test (SPSS 22.0). The qPCR and morphology data were analyzed by means of a linear mixed-effects model with pen included as random effect (S-Plus). The differences were considered statistically significant at a P value of ≤0.05 and were considered a tendency at a P value of ≤0.1. Statistical differences in relative abundance in bacterial population between groups were assessed by using the nonparametric Kruskal-Wallis H test with the Benjamin-Hochberg false-discovery rate screen and the Tukey-Kramer post hoc test. Moreover, differences in specific bacterial population relative abundances based on 16S rRNA gene profiling were analyzed with a nonparametric Mann-Whitney test using a two-tailed P value calculation. GraphPad Prism software version 5 was used to perform the statistical analysis for the in vitro fermentation. All quantitative parameters (pH, OD, and SCFA concentrations) were compared using the Kruskal-Wallis test. The Dunn post hoc test was applied for multicomparisons of these variables if there was a significant difference with the Kruskal-Wallis test.
Nucleotide sequence accession number.
The raw sequences from this work have been deposited in the NCBI BioProject database (accession number PRJNA277118).
RESULTS
Broiler performance after supplementation of broiler feed with XOS35.
To evaluate the effect of XOS on broiler performance, the body weight and feed intake were measured, and FCR and growth were calculated. When considering the starter and grower periods together (day 0 to day 26), the FCR was significantly (P = 0.003) more favorable for chickens fed the XOS-supplemented diet than for chickens fed the control diet (Table 2). For the whole trial period (day 0 to 39), the FCR was also significantly improved (lower) for the group receiving the XOS-supplemented diet (P = 0.04). The average body weight at the different time points was nonsignificantly higher for chickens fed the diet supplemented with 0.5% XOS than for the chickens given the nonsupplemented diet. These results, together with the significantly improved FCR, show a biologically relevant improved performance for chickens given the XOS-supplemented diet.
TABLE 2.
Effect of XOS supplementation on growth performance of chickensa
| Interval (days) | XOS | FCR | BW (g) | FI (g/day) | WG (g/day) |
|---|---|---|---|---|---|
| 0–26 | − | 1.50 ± 0.01 | 1,364 ± 15.39 | 67.3 ± 2.3 | 44.8 ± 1.50 |
| + | 1.45 ± 0.01 | 1,421 ± 16.97 | 69.4 ± 2.04 | 47.8 ± 1.72 | |
| P value | 0.003** | 0.30 | 0.50 | 0.19 | |
| 0–39 | − | 1.66 ± 0.01 | 2,401 ± 60.01 | 100.0 ± 2.75 | 60.4 ± 1.53 |
| + | 1.63 ± 0.01 | 2,446 ± 57.26 | 100.4 ± 2.43 | 61.6 ± 1.46 | |
| P value | 0.04* | 0.60 | 0.93 | 0.60 |
The feed conversion ratio (FCR), body weight (BW), feed intake (FI), and weight gain (WG) were measured at three time intervals for animals fed a wheat-rye-based diet without or with 0.2% XOS (days 1 to 13) and 0.5% XOS (days 14 to 26 and 27 to 39). Values are the means for 6 pens with 32 chickens ± standard errors of the mean. Statistical analysis was done with SPSS. An independent-sample t test was used to determine statistical differences between groups receiving nonsupplemented and XOS-supplemented diets. P values of less than 0.05 (*) and 0.001 (**) were considered significant.
Intestinal morphology.
Supplementation of 0.5% XOS to the broiler feed significantly (P = 0.04) increased the villus length in the ileum (Table 3). The tunica muscularis was shown (P = 0.38) to be thicker in the group fed the XOS-supplemented diet (Table 3).
TABLE 3.
Effects of XOS supplementation on the intestinal morphology of chickens on day 26a
| Parameter | Value |
P value | |
|---|---|---|---|
| Without 0.5% XOS | With 0.5% XOS | ||
| Length of villi (μm) | 1,059 ± 40.00 | 1,228 ± 59.79 | 0.04 |
| Thickness of tunica muscularis (μm) | 167.0 ± 11.01 | 178.9 ± 6.32 | 0.38 |
The data are means and standard errors of the means of measurements taken at day 26 from ileal sections of animals fed a wheat-rye-based diet without or with 0.5 % XOS (n = 18). The length and the thickness were measured for 10 randomly selected villi and 10 different places for the tunica muscularis, using a PC-based analysis system. Statistical analysis was done with S-Plus using a linear mixed-effects model with pen as random factor. P values of less than 0.05 were considered significant.
Microbiota composition as determined by qPCR and 16S rRNA gene sequencing.
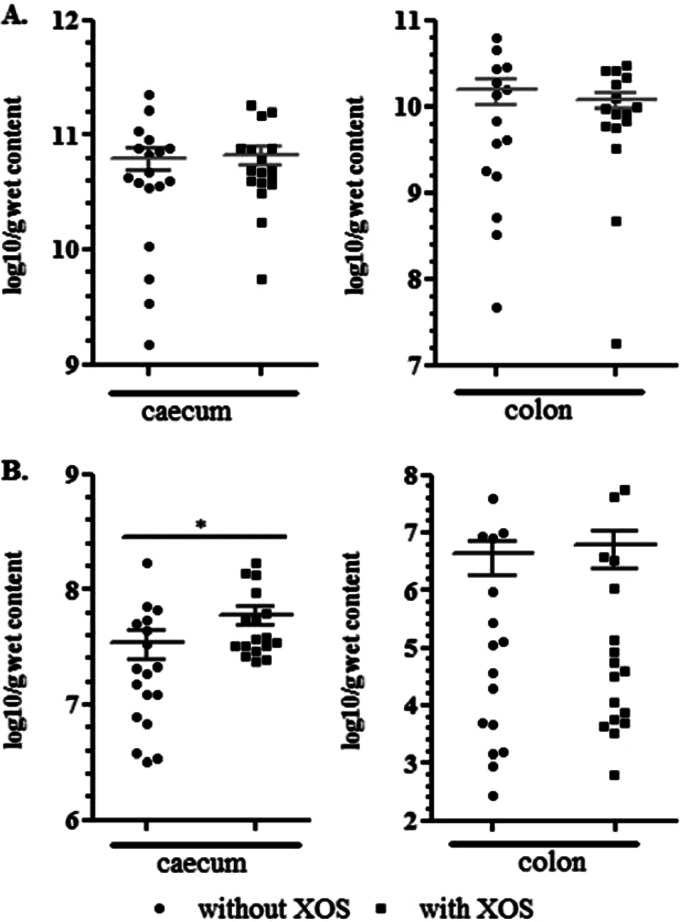
There was no difference in the number of total bacteria between the XOS-supplemented and nonsupplemented groups in both the cecum and the colon (Fig. 1A). The number of gene copies encoding the butyryl-CoA:acetate-CoA transferase was significantly (P = 0.02) higher in the ceca of the chickens that received 0.5% XOS (Fig. 1B).
FIG 1.
Numbers of total bacteria (A) and butyryl-CoA:acetate-CoA transferase gene copies (B), expressed as log10 copy number of the gene per gram of wet content in the cecal and colonic contents of 26-day-old chickens fed a wheat-rye-based diet either supplemented or not with 0.5% XOS (18 chickens for each treatment). Statistical analysis was done with S-Plus using a linear mixed-effects model with pen as a random factor to determine statistical differences between groups of animals fed a wheat-rye-based diet without and with XOS. *, P ≤ 0.05.
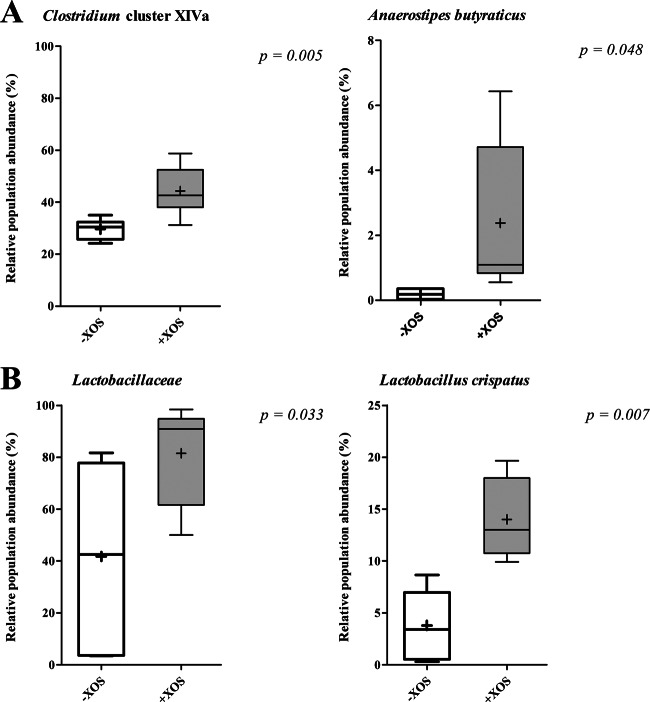
Significant changes were observed in the abundance of specific 16S rRNA gene sequences in cecum and colon samples at different taxonomic levels (Fig. 2; Table 4). Although 35 bacterial families were detected in the cecal microbiota, only the abundance of Clostridium cluster XIVa was shown to be significantly (P = 0.005) increased in animals fed an XOS-supplemented diet compared to animals fed a control diet (44.29% versus 29.65%) (Table 4). Forty-one bacterial families were detected in the colon microbiota, of which Lactobacillaceae was shown to be significantly (P = 0.033) higher in animals fed an XOS-supplemented diet than in animals fed a control diet (81.54% versus 41.73%). In the cecal samples a total of 834 species were detected, of which 16 were shown to be significantly different between the XOS-treated and control animals. A significant higher abundance was observed for Anaerostipes butyraticus, a butyrate-producing species classified within Clostridium cluster XIVa (from 0.4% to 2.5%; P = 0.048) (Fig. 2). Seven hundred twenty-one species were detected in the colon samples, of which 11 were shown to be significantly different in animals receiving dietary XOS compared to those receiving the control diet. XOS supplementation resulted in a significant increase of Lactobacillus crispatus (from 4% to 15%; P = 0.007) (Fig. 2) in the colon.
FIG 2.
Box plots showing mean relative sequence abundances of the Clostridium cluster XIVa and Anaerostipes butyraticus in the ceca (A) and of the Lactobacillaceae and Lactobacillus crispatus in the colons (B) of 26-day-old chickens fed without or with XOS-supplemented feed (6 chickens for each treatment). The plus represents the mean value, and the whiskers are the median, the minimum and maximum values, and the first and third quartiles.
TABLE 4.
Clostridium cluster XIVa and cluster IV members, identified in the ceca of chickens at day 26, for which the relative proportion was significantly different between the XOS-supplemented and unsupplemented groupsa
| Cluster and sequence | Abundance |
Highest 16S rRNA gene sequence similarity |
|||
|---|---|---|---|---|---|
| Without 0.5% XOS | With 0.5% XOS | P value | Type strain of validly named species (% 16S rRNA gene sequence similarity) | Accession no. | |
| Clostridium cluster XIVa | 29.64 | 44.29 | 0.004 | ||
| Blautia_RL199 | 0.04 | 0.15 | 0.04 | Blautia faecis (95.96) | DQ793371 |
| Lachnospiraceae_cc142 | 1.27 | 5.47 | 0.04 | Blautia schinkii (93.93) | DQ057372 |
| Lachnospiraceae_ic1296 | 0.34 | 0.79 | 0.03 | Blautia producta (92.33) | DQ057459 |
| Lachnospiraceae_GRC80 | 0.68 | 0.27 | 0.02 | Eubacterium contortum (94.27) | DQ673545 |
| Lachnospiraceae_B5-F3 | 1.19 | 7.28 | 0.01 | Blautia producta (93.77) | EF025241 |
| Lachnospiraceae_TS29 | 0.22 | 0.92 | 0.04 | Eubacterium hallii (95.71) | FJ367509 |
| Clostridium cluster IV | 29.12 | 29.31 | NS | ||
| Ruminococcaceae_BY13 | 0.69 | 0.05 | 0.04 | Pseudoflavonifactor capillosus (96.23) | DQ342336 |
| Ruminococcaceae_CFT19C1 | 0.00 | 0.08 | 0.04 | Clostridium alkalicellulosi (84.4) | DQ455843 |
| Ruminococcaceae_CFT212F12 | 0.05 | 0.16 | 0.01 | Oscillibacter valericigenes (95.73) | DQ456381 |
| Ruminococcaceae_RL246 | 0.19 | 0.78 | 0.02 | Clostridium alkalicellulosi (85.44) | DQ793581 |
| Ruminococcaceae_TS1 | 1.78 | 0.29 | 0.03 | Clostridium aldrichii (85.66) | FJ365262 |
| Ruminococcaceae_ELU0008 | 0.04 | 0.30 | 0.01 | Subdoligranulum variabile (92.69) | HQ740050 |
The results are based on the sequencing data for 6 chickens per treatment. NS, not significant.
In vitro fermentation.
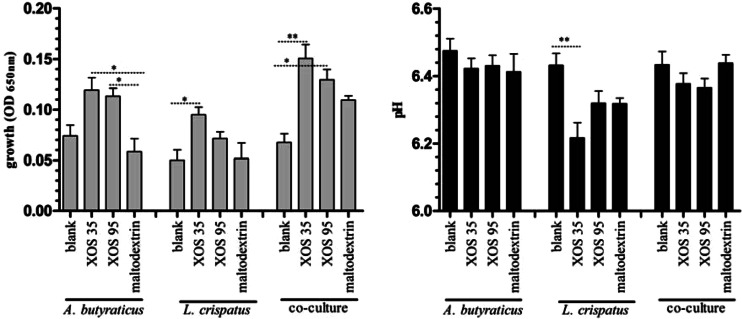
To investigate cross-feeding between L. crispatus and A. butyraticus in the presence of XOS, an in vitro fermentation assay was carried out. XOS95 and maltodextrin were used to confirm the effect of XOS in the in vivo trial. Only the monoculture of L. crispatus resulted in a small pH drop when XOS35 was added to the medium (6.4 ± 0.04 versus 6.2 ± 0.04) (Fig. 3). A. butyraticus showed a significantly increased (P = 0.007) proliferation when XOS35 and XOS95 were added to the medium compared to maltodextrin (Fig. 3). The proliferation of L. crispatus increased significantly when XOS35 was added to the medium. The proliferation of the strains in the coculture was higher when XOS35 or XOS95 was added than with the nonsupplemented medium (Fig. 3). Supplementation of maltodextrin to the medium did not cause any changes.
FIG 3.
pH values and optical densities (650 nm) after 24 h of in vitro fermentation of different substrates by A. butyraticus, L. crispatus, and both in coculture. All the in vitro fermentation experiments were done twice in triplicate. Statistical analysis was done with GraphPad Prism 5, using a Kruskal-Wallis test with a Dunn post hoc test. P values of ≤0.05 (*) and ≤0.01 (**) were considered significant.
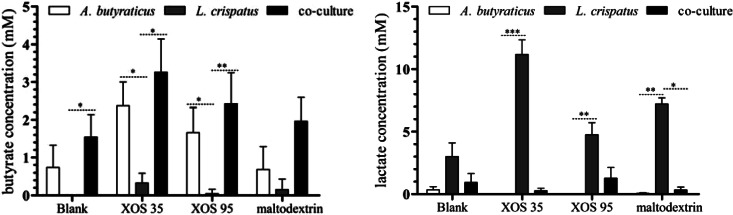
The concentrations of the fermentation acids butyrate and dl-lactate in all monocultures and cocultures were determined after incubation (Fig. 4). It was found that A. butyraticus was able to produce butyrate, while L. crispatus produced high concentrations of lactate. The concentration of butyrate or lactate produced by L. crispatus and A. butyraticus, respectively, was below the cutoff values (1 mM and 0.5 mM, respectively) as determined during optimization of the HPLC method (39). XOS35 and XOS95 significantly stimulated lactate production by L. crispatus compared with A. butyraticus, which was not able to produce lactate. In the coculture, lactate concentrations were very low, even when XOS35 or XOS95 was added to the medium, while the butyrate concentration was higher than the concentrations in the monoculture of A. butyraticus with XOS35 (3.3 ± 0.8 versus 2.3 ± 0.6) (Fig. 4), but the difference was nonsignificant. A similar observation was made for XOS95 (2.4 ± 0.8 versus 1.6 ± 0.6).
FIG 4.
Butyrate and dl-lactate concentrations after 24 h of in vitro fermentation of different substrates by A. butyraticus, L. crispatus and both in coculture. The in vitro fermentation experiments were done twice in triplicate. Statistical analysis was done with GraphPad Prism 5, using a Kruskal-Wallis test followed by a Dunn post hoc test. P values of ≤0.05 (*), ≤0.01 (**), and < 0.001 (***) were considered significant.
DISCUSSION
It is generally accepted that shifts in the intestinal microbiota composition may be the result of dietary changes, such as the addition of cereal fibers (22, 40, 41). In the current study, we demonstrated that administration of XOS to broiler feed altered the microbiota composition in the gut, with butyrate-producing bacteria and lactobacilli being more abundant in the cecum and colon, respectively.
In the chicken gut, lactobacilli are one of the predominant genera (42). These bacteria have the ability to adhere to the mucosal layers and epithelium, promoting colonization (43, 44). Through interaction with the intestinal epithelial cells, lactobacilli can cause immunomodulation and offer protection to the intestinal barrier by antagonistic activities against pathogens (44–46). In addition, the probiotic use of lactobacilli has been shown to beneficially affect performance in broilers. Broilers fed diets containing a mixture of 12 Lactobacillus strains or a single Lactobacillus acidophilus strain had a better weight gain and a better FCR (47, 48). Lactobacilli are known to ferment carbohydrates into lactic acid as major end product, which may lower the pH of the intestinal environment, resulting in the inhibition of growth of acid-sensitive pathogenic bacteria. However, this pH effect may be rather limited, as lactic acid is absorbed from the intestine or used as a substrate for lactate-utilizing bacteria, such as representatives of the genera Eubacterium, Anaerostipes, Veillonella, and Megasphaera (49, 50).
In the present study, in addition to the significantly higher abundance of lactobacilli in the colon, we found an increased number of butyryl-CoA:acetate-CoA transferase gene copies in the ceca of chickens that received an XOS-supplemented diet. Butyryl-CoA:acetate-CoA transferase is a key enzyme in the major pathway for bacterial butyrate production in the gut (7). Hippe et al. showed that this enzyme is a suitable marker for the butyrate-producing capacity of the intestinal microbiota which mainly belong to Clostridium cluster IV and XIVa (51, 52). We observed a significant increase of members from both clusters in the ceca of chickens that were administered XOS.
The increased abundance of both lactobacilli and butyrate-producing bacteria can partly be explained by cross-feeding mechanisms. Bacteria related to Eubacterium hallii and Anaerostipes caccae, both members of Clostridium cluster XIVa, are able to convert acetate and lactate into butyrate (7, 53). This metabolic cross-feeding between lactate-producing and lactate-utilizing bacteria may help to stabilize the luminal pH and may be a factor in the butyrogenic effect of certain dietary substrates (54). Our in vivo study showed a significant increase of the lactate-producing species Lactobacillus crispatus in the colon and the lactate-utilizing butyrate-producing species Anaerostipes butyraticus in the cecum. The lactic acid produced by L. crispatus in the colon may reach the cecum and become available for A. butyraticus due to antiperistalsis (55, 56). The in vitro fermentation assay showed that reference strains of both species metabolized XOS, resulting in production of high concentrations of lactic acid by L. crispatus, which were thought to be consumed by the butyrate-producing bacterium A. butyraticus. Most likely many other strains also can carry out a similar cross-feeding reaction in order to generate high butyrate levels in the chicken hindgut.
Production of butyrate most probably plays a role in the beneficial effects on gut morphology and growth performance observed in the current study. In poultry, butyrate enhances nonspecific intestinal defense mechanisms against pathogens that can affect performance, such as Clostridium perfringens, by stimulating the mucin glycoprotein expression in intestinal epithelial cells (57–59). Butyrate is a major energy source for the colonocytes and exerts anti-inflammatory activities by several mechanisms (60). One of these mechanisms is the suppression of nuclear factor kappa B (NF-κB), which regulates the expression of proinflammatory cytokines (61). Butyrate has also been shown to interfere with signaling by interferon gamma (IFN-γ) through its inhibitory effect on the activation of signal transducer and activator of transcription 1 (STAT1) (62). Butyrate also upregulates the expression of peroxisome proliferator-activated receptor gamma (PPAR-γ), a transcription factor that belongs to the nuclear hormone receptor family. PPAR-γ inhibits the expression of inflammatory cytokines and directs the differentiation of immune cells toward anti-inflammatory phenotypes (63–65).
We observed longer villi in the ileums of chickens that were fed an XOS-supplemented diet than in those of chickens fed a control diet. This effect on the small intestinal morphology may at least partly be due to butyrate production by Clostridium cluster IV and XIVa species in the hindgut, through its effect on the expression of glucagon-like peptide 2 (GLP-2). Butyrate indeed appears to be a strong stimulator of GLP-2 production. This hormone is secreted by entero-endocrine L cells and acts indirectly through multiple downstream mediators (66). Its receptor (GLP-2R) is localized on distinct subpopulations of gut endocrine cells in the stomach, small intestine, and colon but also on subepithelial myofibroblasts (67, 68). Hu et al. showed a beneficial effect of intravenous GLP-2 injection in broilers on growth performance, intestinal morphology, villus height, and crypt cell proliferation (69).
In conclusion, XOS, supplemented to the broiler diet, improved broiler performance by improving the feed conversion ratio. Administration of XOS resulted in an increased abundance of butyrate-producing bacteria in the cecum and of lactobacilli in the colon at day 26 of age. It is hypothesized that microbial cross-feeding, in which lactic acid produced by the lactobacilli is consumed by butyrate-producing bacteria in the cecum, stimulates gut heath and consequently performance through the beneficial effects of butyrate. Whether this cross-feeding also occurs in the complex gut ecosystem needs to be clarified in further in vivo work.
ACKNOWLEDGMENTS
We are grateful to Christian Puttevils and Delphine Ameye for their skillful technical assistance with the morphological examination. We acknowledge the Ph.D. students from the Department of Pathology, Bacteriology, and Avian Diseases who assisted in the sampling for the in vivo trial.
We are grateful to the PDV animal feed of The Netherlands government for financial support of this work.
REFERENCES
- 1.McCleary BV. 2003. Dietary fibre analysis. Proc Nutr Soc 62:3–9. doi: 10.1079/PNS2002204. [DOI] [PubMed] [Google Scholar]
- 2.James SL, Muir JG, Curtis SL, Gibson PR. 2003. Dietary fibre: a roughage guide. Intern Med J 33:291–296. doi: 10.1046/j.1445-5994.2003.00404.x. [DOI] [PubMed] [Google Scholar]
- 3.Broekaert WF, Courtin CM, Verbeke K, Van de Wiele T, Verstraete W, Delcour JA. 2011. Prebiotic and other health-related effects of cereal-derived arabinoxylans, arabinoxylan-oligosaccharides, and xylooligosaccharides. Crit Rev Food Sci Nutr 51:178–194. doi: 10.1080/10408390903044768. [DOI] [PubMed] [Google Scholar]
- 4.Aachary AA, Prapulla SG. 2008. Corncob-induced endo-1,4-beta-d-xylanase of Aspergillus oryzae MTCC 5154: production and characterization of xylobiose from glucuronoxylan. J Agric Food Chem 56:3981–3988. doi: 10.1021/jf073430i. [DOI] [PubMed] [Google Scholar]
- 5.Gibson GR, Probert HM, Loo JV, Rastall RA, Roberfroid MB. 2004. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev 17:259–275. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- 6.Scott KP, Martin JC, Duncan SH, Flint HJ. 2014. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol Ecol 87:30–40. doi: 10.1111/1574-6941.12186. [DOI] [PubMed] [Google Scholar]
- 7.Duncan SH, Louis P, Flint HJ. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilloteau P, Martin L, Eeckhaut V, Ducatelle R, Zabielski R, Van Immerseel F. 2010. From the gut to the peripheral tissues: the multiple effects of butyrate. Nutr Res Rev 23:366–384. doi: 10.1017/S0954422410000247. [DOI] [PubMed] [Google Scholar]
- 9.Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, Calignano A. 2011. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol 17:1519–1528. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu CK, Liao JW, Chung YC, Hsieh CP, Chan YC. 2004. Xylooligosaccharides and fructooligosaccharides affect the intestinal microbiota and precancerous colonic lesion development in rats. J Nutr 134:1523–1528. [DOI] [PubMed] [Google Scholar]
- 11.Gobinath D, Madhu AN, Prashant G, Srinivasan K, Prapulla SG. 2010. Beneficial effect of xylo-oligosaccharides and fructo-oligosaccharides in streptozotocin-induced diabetic rats. Br J Nutr 104:40–47. doi: 10.1017/S0007114510000243. [DOI] [PubMed] [Google Scholar]
- 12.Smiricky-Tjardes MR, Flickinger EA, Grieshop CM, Bauer LL, Murphy MR, Fahey GC Jr. 2003. In vitro fermentation characteristics of selected oligosaccharides by swine fecal microflora. J Anim Sci 81:2505–2514. [DOI] [PubMed] [Google Scholar]
- 13.Zhenping S, Wenting L, Ruikui Y, Jia L, Honghong L, Wei S, Zhongmie W, Jingpan L, Zhe S, Yuling Q. 2013. Effect of a straw-derived xylooligosaccharide on broiler growth performance, endocrine metabolism, and immune response. Can J Vet Res 77:105–109. [PMC free article] [PubMed] [Google Scholar]
- 14.Sekelja M, Rud I, Knutsen SH, Denstadli V, Westereng B, Naes T, Rudi K. 2012. Abrupt temporal fluctuations in the chicken fecal microbiota are explained by its gastrointestinal origin. Appl Environ Microbiol 78:2941–2948. doi: 10.1128/AEM.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meimandipour A, Shuhaimi M, Hair-Bejo M, Azhar K, Kabeir BM, Rasti B, Yazid AM. 2009. In vitro fermentation of broiler cecal content: the role of lactobacilli and pH value on the composition of microbiota and end products fermentation. Lett Appl Microbiol 49:415–420. doi: 10.1111/j.1472-765X.2009.02674.x. [DOI] [PubMed] [Google Scholar]
- 16.Dumonceaux TJ, Hill JE, Hemmingsen SM, Van Kessel AG. 2006. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Appl Environ Microbiol 72:2815–2823. doi: 10.1128/AEM.72.4.2815-2823.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong J, Forster RJ, Yu H, Chambers JR, Sabour PM, Wheatcroft R, Chen S. 2002. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiology Lett 208:1–7. doi: 10.1111/j.1574-6968.2002.tb11051.x. [DOI] [PubMed] [Google Scholar]
- 18.Lepage P, Leclerc MC, Joossens M, Mondot S, Blottiere HM, Raes J, Ehrlich D, Dore J. 2013. A metagenomic insight into our gut's microbiome. Gut 62:146–158. doi: 10.1136/gutjnl-2011-301805. [DOI] [PubMed] [Google Scholar]
- 19.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Meta HITC, Bork P, Ehrlich SD, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torok VA, Ophel-Keller K, Loo M, Hughes RJ. 2008. Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Appl Environ Microbiol 74:783–791. doi: 10.1128/AEM.01384-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu J, Idris U, Harmon B, Hofacre C, Maurer JJ, Lee MD. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl Environ Microbiol 69:6816–6824. doi: 10.1128/AEM.69.11.6816-6824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knarreborg A, Simon MA, Engberg RM, Jensen BB, Tannock GW. 2002. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl Environ Microbiol 68:5918–5924. doi: 10.1128/AEM.68.12.5918-5924.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apajalahti J, Kettunen A, Graham H. 2004. Characteristics of the gastrointestinal microbial communities, with special reference to the chicken. World Poultry Sci J 60:223–232. doi: 10.1079/WPS20040017. [DOI] [Google Scholar]
- 24.Bjerrum L, Engberg RM, Leser TD, Jensen BB, Finster K, Pedersen K. 2006. Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and culture-based techniques. Poult Sc 85:1151–1164. doi: 10.1093/ps/85.7.1151. [DOI] [PubMed] [Google Scholar]
- 25.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 26.Duncan SH, Louis P, Flint HJ. 2007. Cultivable bacterial diversity from the human colon. Lett Appl Microbiol 44:343–350. doi: 10.1111/j.1472-765X.2007.02129.x. [DOI] [PubMed] [Google Scholar]
- 27.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 28.Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66:5488–5491. doi: 10.1128/AEM.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowalchuk GA, Stienstra AW, Heilig GH, Stephen JR, Woldendorp JW. 2000. Molecular analysis of ammonia-oxidising bacteria in soil of successional grasslands of the Drentsche A (The Netherlands). FEMS Microbiol Ecol 31:207–215. doi: 10.1111/j.1574-6941.2000.tb00685.x. [DOI] [PubMed] [Google Scholar]
- 30.Hopkins MJ, Macfarlane GT, Furrie E, Fite A, Macfarlane S. 2005. Characterisation of intestinal bacteria in infant stools using real-time PCR and Northern hybridisation analyses. FEMS Microbiol Ecol 54:77–85. doi: 10.1016/j.femsec.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Louis P, Flint HJ. 2007. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol 73:2009–2012. doi: 10.1128/AEM.02561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brosius J, Dull TJ, Sleeter DD, Noller HF. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol 148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 33.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol 66:1654–1661. doi: 10.1128/AEM.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moura P, Barata R, Carvalheiro F, Girio F, Loureiro-Dias MC, Esteves MP. 2007. In vitro fermentation of xylo-oligosaccharides from corn cobs autohydrolysis by Bifidobacterium and Lactobacillus strains. Food Sci Technol 40:963–972. [Google Scholar]
- 39.De Baere S, Eeckhaut V, Steppe M, De Maesschalck C, De Backer P, Van Immerseel F, Croubels S. 2013. Development of a HPLC-UV method for the quantitative determination of four short-chain fatty acids and lactic acid produced by intestinal bacteria during in vitro fermentation. J Pharm Biomed Anal 80:107–115. doi: 10.1016/j.jpba.2013.02.032. [DOI] [PubMed] [Google Scholar]
- 40.Shakouri MD, Kermanshahi H, Mohsenzadeh M. 2006. Effect of different non starch polysaccharides in semi purified diets on performance and intestinal microflora of young broiler chickens. Int J Poult Sci 5 6:557–561. [Google Scholar]
- 41.Torok VA, Hughes RJ, Mikkelsen LL, Perez-Maldonado R, Balding K, MacAlpine R, Percy NJ, Ophel-Keller K. 2011. Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Appl Environ Microbiol 77:5868–5878. doi: 10.1128/AEM.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei S, Morrison M, Yu Z. 2013. Bacterial census of poultry intestinal microbiome. Poult Sci 92:671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- 43.Kravtsov EG, Yermolayev AV, Anokhina IV, Yashina NV, Chesnokova VL, Dalin MV. 2008. Adhesion characteristics of Lactobacillus is a criterion of the probiotic choice. Bull Exp Biol Med 145:232–234. doi: 10.1007/s10517-008-0058-x. [DOI] [PubMed] [Google Scholar]
- 44.Sengupta R, Altermann E, Anderson RC, McNabb WC, Moughan PJ, Roy NC. 2013. The role of cell surface architecture of lactobacilli in host-microbe interactions in the gastrointestinal tract. Mediat Inflamm 2013:237921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Servin AL. 2004. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev 28:405–440. doi: 10.1016/j.femsre.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 46.Rinttila T, Apajalathi J. 2013. Intestinal microbiota and metabolites—implications for broiler chicken health and performance. J Appl Poult Res 22:647–658. doi: 10.3382/japr.2013-00742. [DOI] [Google Scholar]
- 47.Jin LZ, Ho YW, Abdullah N, Jalaludin S. 1998. Growth performance, intestinal microbial populations, and serum cholesterol of broilers fed diets containing Lactobacillus cultures. Poult Sci 77:1259–1265. doi: 10.1093/ps/77.9.1259. [DOI] [PubMed] [Google Scholar]
- 48.Jin LZ, Ho YW, Abdullah N, Jalaludin S. 2000. Digestive and bacterial enzyme activities in broilers fed diets supplemented with Lactobacillus cultures. Poult Sci 79:886–891. doi: 10.1093/ps/79.6.886. [DOI] [PubMed] [Google Scholar]
- 49.Belenguer A, Duncan SH, Holtrop G, Anderson SE, Lobley GE, Flint HJ. 2007. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl Environ Microbiol 73:6526–6533. doi: 10.1128/AEM.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harmsen HJM, Raangs GC, He T, Degener JE, Welling GW. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl Environ Microbiol 68:2982–2990. doi: 10.1128/AEM.68.6.2982-2990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hippe B, Zwielehner J, Liszt K, Lassl C, Unger F, Haslberger AG. 2011. Quantification of butyryl CoA:acetate CoA-transferase genes reveals different butyrate production capacity in individuals according to diet and age. FEMS Microbiol Lett 316:130–135. doi: 10.1111/j.1574-6968.2010.02197.x. [DOI] [PubMed] [Google Scholar]
- 52.Louis P, Flint HJ. 2009. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 53.Sato T, Matsumoto K, Okumura T, Yokoi W, Naito E, Yoshida Y, Nomoto K, Ito M, Sawada H. 2008. Isolation of lactate-utilizing butyrate-producing bacteria from human feces and in vivo administration of Anaerostipes caccae strain L2 and galacto-oligosaccharides in a rat model. FEMS Microbiol Ecol 66:528–536. doi: 10.1111/j.1574-6941.2008.00528.x. [DOI] [PubMed] [Google Scholar]
- 54.Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hodgkiss JP. 1984. Peristalsis and antiperistalsis in the chicken caecum are myogenic. Q J Exp Physiol 69:161–170. doi: 10.1113/expphysiol.1984.sp002777. [DOI] [PubMed] [Google Scholar]
- 56.Janssen PW, Lentle RG, Hulls C, Ravindran V, Amerah AM. 2009. Spatiotemporal mapping of the motility of the isolated chicken caecum. J Comp Physiol B Biochem Syst Environ Physiol 179:593–604. doi: 10.1007/s00360-009-0342-8. [DOI] [PubMed] [Google Scholar]
- 57.Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, Hinton JC, Van Immerseel F. 2006. Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl Environ Microbiol 72:946–949. doi: 10.1128/AEM.72.1.946-949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Timbermont L, Lanckriet A, Dewulf J, Nollet N, Schwarzer K, Haesebrouck F, Ducatelle R, Van Immerseel F. 2010. Control of Clostridium perfringens-induced necrotic enteritis in broilers by target-released butyric acid, fatty acids and essential oils. Avian Pathol 39:117–121. doi: 10.1080/03079451003610586. [DOI] [PubMed] [Google Scholar]
- 59.Willemsen LEM, Koetsier MA, van Deventer SJH, van Tol EAF. 2003. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E-1 and E-2 production by intestinal myofibroblasts. Gut 52:1442–1447. doi: 10.1136/gut.52.10.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. 2008. The role of butyrate on colonic function. Aliment Pharmacol Ther 27:104–119. [DOI] [PubMed] [Google Scholar]
- 61.Inan MS, Rasoulpour RJ, Yin L, Hubbard AK, Rosenberg DW, Giardina C. 2000. The luminal short-chain fatty acid butyrate modulates NF-kappa B activity in a human colonic epithelial cell line. Gastroenterology 118:724–734. doi: 10.1016/S0016-5085(00)70142-9. [DOI] [PubMed] [Google Scholar]
- 62.Klampfer L, Huang J, Sasazuki T, Shirasawa S, Augenlicht L. 2003. Inhibition of interferon gamma signaling by the short chain fatty acid butyrate. Mol Cancer Res 1:855–862. [PubMed] [Google Scholar]
- 63.Wächtershäuser A, Loitsch SM, Stein J. 2000. PPAR-γ is selectively upregulated in Caco-2 cells by butyrate. Biochem Bioph Res Commun 272:380–385. doi: 10.1006/bbrc.2000.2793. [DOI] [PubMed] [Google Scholar]
- 64.Schwab M, Reynders V, Loitsch S, Steinhilber D, Stein J, Schroder O. 2007. Involvement of different nuclear hormone receptors in butyrate-mediated inhibition of inducible NF kappa B signalling. Mol Immunol 44:3625–3632. doi: 10.1016/j.molimm.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 65.Martin H. 2010. Role of PPAR-gamma in inflammation. Prospects for therapeutic intervention by food components. Mutat Res 690:57–63. [DOI] [PubMed] [Google Scholar]
- 66.Dube PE, Brubaker PL. 2007. Frontiers in glucagon-like peptide-2: multiple actions, multiple mediators. Am J Physiol Endocrinol Metab 293:E460–E465. doi: 10.1152/ajpendo.00149.2007. [DOI] [PubMed] [Google Scholar]
- 67.Drucker DJ. 2001. The glucagon-like peptides. Endocrinology 142:521–527. doi: 10.1210/endo.142.2.7983. [DOI] [PubMed] [Google Scholar]
- 68.de Heuvel E, Wallace L, Sharkey KA, Sigalet DL. 2012. Glucagon-like peptide 2 induces vasoactive intestinal polypeptide expression in enteric neurons via phophatidylinositol 3-kinase-gamma signaling. Am J Physiol Endocrinol Metab 303:E994–E1005. doi: 10.1152/ajpendo.00291.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu XF, Guo YM, Huang BY, Bun S, Zhang LB, Li JH, Liu D, Long FY, Yang X, Jiao P. 2010. The effect of glucagon-like peptide 2 injection on performance, small intestinal morphology, and nutrient transporter expression of stressed broiler chickens. Poult Sci 89:1967–1974. doi: 10.3382/ps.2009-00547. [DOI] [PubMed] [Google Scholar]
- 70.Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, Gordon JI. 2010. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem 285:22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]