Abstract
The occurrence of Cryptosporidium oocysts in drinking source water can present a serious public health risk. To rapidly and effectively assess the source and human-infective potential of Cryptosporidium oocysts in water, sensitive detection and correct identification of oocysts to the species level (genotyping) are essential. In this study, we developed three real-time PCR genotyping assays, two targeting the small-subunit (SSU) rRNA gene (18S-LC1 and 18S-LC2 assays) and one targeting the 90-kDa heat shock protein (hsp90) gene (hsp90 assay), and evaluated the sensitivity and Cryptosporidium species detection range of these assays. Using fluorescence resonance energy transfer probes and melt curve analysis, the 18S-LC1 and hsp90 assays could differentiate common human-pathogenic species (C. parvum, C. hominis, and C. meleagridis), while the 18S-LC2 assay was able to differentiate nonpathogenic species (such as C. andersoni) from human-pathogenic ones commonly found in source water. In sensitivity evaluations, the 18S-LC2 and hsp90 genotyping assays could detect as few as 1 Cryptosporidium oocyst per sample. Thus, the 18S-LC2 and hsp90 genotyping assays might be used in environmental monitoring, whereas the 18S-LC1 genotyping assay could be useful for genotyping Cryptosporidium spp. in clinical specimens or wastewater samples.
INTRODUCTION
Waterborne Cryptosporidium spp. present a serious threat to human health because of the ubiquitous presence of Cryptosporidium spp. in water and resistance of the oocysts to environmental conditions, various disinfectants, and many treatment practices (1). As watersheds are vulnerable to contamination with both human-pathogenic and nonpathogenic species, sensitive detection of Cryptosporidium oocysts in water and correct identification of oocysts to the species/genotype level are essential for source water management and risk assessment. Traditional microscopy-based detection tools such as U.S. Environmental Protection Agency (EPA) Method 1622/1623 cannot differentiate Cryptosporidium species. Thus, genotyping tools are increasingly used in the assessment of the human-infective potential and source of Cryptosporidium oocysts in source or finished water (2–12).
Currently, numerous molecular techniques have been developed for Cryptosporidium detection and genotyping, based mostly on PCR (single-round PCR, nested PCR, real-time PCR, and multiplex PCR, etc.) followed by restriction fragment length polymorphism (RFLP) analysis, DNA sequencing, melt curve analysis, or single-strand conformation polymorphism (SSCP) analysis (13). Among these methods, the real-time PCR technique with melt curve analysis has recently evolved as a sensitive, specific, and time-saving tool for the detection and genotyping of Cryptosporidium spp. in clinical and environmental samples, relying on several gene targets, including the small-subunit (SSU) rRNA, the 70-kDa heat shock protein (hsp70), the Cryptosporidium oocyst wall protein (COWP), the 60-kDa glycoprotein (gp60), and others (14–21). The SSU rRNA gene is the most commonly used target because of its multicopy nature and the presence of both semiconserved and hypervariable regions, which are needed for the development of genus-specific genotyping tools (22).
In previous studies, real-time PCR assays were developed mostly for genus-level identification of Cryptosporidium spp. or specific detection of a single Cryptosporidium species/genotype (14, 16, 17, 20, 21, 23, 24). Combinations of multiple assays are used to genotype Cryptosporidium spp. in clinical and environmental samples. Few assays have the capability to differentiate the common human-pathogenic species C. parvum, C. hominis, and C. meleagridis simultaneously (18, 19). Even though some species/genotypes, such as C. andersoni, may not be highly infectious to humans, their presence in water may indicate the likely source of oocyst contamination. This calls for a rapid and sensitive assay for the differentiation of nonpathogenic species from human-pathogenic ones.
In the present study, we have developed three real-time PCR assays, two targeting the SSU rRNA gene and one targeting the 90-kDa hsp90 gene. These assays use fluorescence resonance energy transfer (FRET) probes and melt curve analysis for genotyping of common human-pathogenic species as well as differentiation of human-pathogenic from nonpathogenic species in water samples.
MATERIALS AND METHODS
Development of FRET probe-based real-time PCR assays.
Three real-time PCR assays were developed for use on the LightCycler 480 II machine (Roche, Indianapolis, IN), including two assays based on the SSU rRNA gene (18S-LC1 and 18S-LC2 assays) and one based on the hsp90 gene (hsp90 assay). The PCR primers and FRET probes used in these assays are shown in Table 1. The PCR mixture consisted of 1 μl of DNA, 500 nM primers, 400 nM FRET probes, 200 μM deoxyribonucleotide triphosphate (dNTP) mix (Promega, Madison, WI), 3 mM MgCl2 (Promega), 400 ng/μl of nonacetylated bovine serum albumin (BSA) (Sigma, St. Louis, MO), 1× GeneAmp PCR buffer (Applied Biosystems, Foster City, CA), and 2.5 U of GoTaq DNA polymerase (Promega) in a total volume of 50 μl.
TABLE 1.
Nucleotide sequence polymorphisms in the primer and probe regions used for three FRET probe-based real-time PCR assays among common Cryptosporidium species and genotypesa
| Assay | Species and/or genotype | Primer region, sequence | Probe region, sequence |
|---|---|---|---|
| 18S-LC1 | − | F4, 5′-GGAAGGGTTGTATTTATTAGATAAAG | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG-[fluorescein] |
| R3, 5′-CTTGTCACTACCTCCCTGTATT | P6, [Red 640]-5′-GTCACATTAATTGTGATCCGTAAAG-phosphate | ||
| C. hominis/C. cuniculus | F4, 5′-GGAAGGGTTGTATTTATTAGATAAAG | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG | |
| R3, 5′-CTTGTCACTACCTCCCTGTATT | P6, 5′-GTCACATTAATTGTGATCCGTAAAG | ||
| C. meleagridis | F4, 5′-GGAAAGGTTGTATTTATTAGATAAAG | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG | |
| R3, 5′-CTTGTCACTACCTCCCTGTATT | P6, 5′-GTCACATAAATTGTGATCCGTAAAG | ||
| C. parvum | F4, 5′-GGAAGGGTTGTATTTATTAGATAAAG | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG | |
| R3, 5′-CTTGTCACTACCTCCCTGTATT | P6, 5′-GTCACATTTAATGTGATCCGTAAAG | ||
| C. canis | F4, 5′-GGAAAGGTTGTATTTATTAGATAAAG | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG | |
| R3, 5′-CTTGTCACTACCTCCCTGTATT | P6, 5′-GTCACATAAAATGTGATCCGTAAAG | ||
| C. felis | F4, 5′-GGAAAGGTCGTATTTATTAGATAAAG | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG | |
| R3, 5′-CTTGTCACTACCTCCCTGTATT | P6, 5′-GTCACAAAATAAATT-ATTGTGATCCGTAAAG | ||
| W1 | F4, 5′-GGAAGGGTTGTATTTATTAGATAAAG | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG | |
| R3, 5′-CTTGTCACTACCTCCCTGTATT | P6, 5′-GTCACATTTATTGTGATCCGTAAAG | ||
| W13 | NA | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG | |
| R3, 5′-CTTGTCACTACCTCCCTGTATT | P6, 5′-GTCACATTAAAATGTGATCCGTAAAG | ||
| W7 | F4, 5′-GGAAGGGTTGTATTTATTAGATAAAG | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG | |
| R3, 5′-CTTGTCACTACCTCCCTGTATT | P6, 5′-GTCACATCAAAAAATGTGATCCGTCAAAG | ||
| C. andersoni/C. muris | F4, 5′-GGAAGGGTTGTATTTATTAGATAAAG | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG | |
| R3, 5′-CTTGTCACTACCTCCCTGTGTC | P6, 5′-GTCGCATCAGAGATGCGATCCGTAAAG | ||
| C. ubiquitum | F4, 5′-GGAAGGGTTGTATTTATTAGATAAAG | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG | |
| R3, 5′-CTTGTCACTACCTCCCTGTATT | P6, 5′-GTCACATATAAA-TGTGATCCGTAAAG | ||
| C. bovis | F4, 5′-GGAAGGGTTGTATTTATTAGATAAAG | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG | |
| R3, 5′-CTTGTCACTACCTCCCTGTATT | P6, 5′-GTCACAT-AAT-GTGATCCGTAAAG | ||
| C. baileyi | F4, 5′-GGAAGGGTTGTATTTATTAGATAAAG | P7, 5′-AAGCTGATAGGTCAGAAACTTGAATG | |
| R3, 5′-CTTGTCACTACCTCCCTGTGTC | P6, 5′-GTCACATAAAT-GTGATCCGTAAAG | ||
| 18S-LC2 | − | F2, 5′-AAGTATAAACCCCTTTACAAGTA | P3, 5′-GAAATTTTACTTTGAGAAAATTAGAGTG-[fluorescein] |
| R2, 5′-TATTATTCCATGCTGGAGTATTC | P4 [Red 640]-5′-TTAAAGCAGGCATATGCCTTGAAT-phosphate | ||
| C. parvum/C. hominis/C. meleagridis/C. cuniculus/W1 | F2, 5′-AAGTATAAACCCCTTTACAAGTA | P3, 5′-GAAATTTTACTTTGAGAAAATTAGAGTG | |
| R2, 5′-TATTATTCCATGCTGGAGTATTC | P4, 5′-TTAAAGCAGGCATATGCCTTGAAT | ||
| C. andersoni | F2, 5′-AAGTATAAACCCCTTTACGAGTA | P3, 5′-GAAATTTTACTTTGAGAAAATTAGAGTG | |
| R2, 5′-TATTATTCCATGCTGGAGTATTC | P4, 5′-TTAAAGCAGGCAACTGCCTTGAAT | ||
| W7 | F2, 5′-AAGTATAAACCCCTTTACAAGTA | P3, 5′-GAAATTTTACTTTGAGAAAATTAGAGTG | |
| R2, 5′-TATTATTCCATGCTGTAGTATAC | P4, 5′-TTAAAGCAGGCATTAGCCTTGTAT | ||
| C. ubiquitum | F2, 5′-AAGTATAAACCCCTTTACAAGTA | P3, 5′-GAAATTTTACTTTGAGAAAATTAGAGTG | |
| R2, 5′-TATTATTCCATGCTGGAGTATTC | P4, 5′-TTAAAGCAGGCATTAGCCTTGAAT | ||
| W13 | F2, 5′-AAGTATAAACCCCTTTACAAGTA | P3, 5′-GAAATTTTACTTTGAGAAAATTAGAGTG | |
| R2, 5′-TATTATTCCATGCTGAAGTATTC | P4, 5′-TTAAAGCAGGCTATTGCCTTGAAT | ||
| C. muris | F2, 5′-AAGTATAAACCCCTTTACGAGTA | P3, 5′-GAAACTTTACTTTGAGAAAATTAGAGTG | |
| R2, 5′-TATTATTCCATGCTGGAGTATTC | P4, 5′-TTAAAGCAGGCAACTGCCTTGAAT | ||
| C. bovis | F2, 5′-AAGTATAAACCCCTTAACAAGTA | P3, 5′-GAAACTTTACTTTGAGAAAATTAGAGTG | |
| R2, 5′-TATTATTCCATGCTGGAGTATTC | P4, 5′-TTAAAGCAGGCTATTGCCTTGAAT | ||
| C. baileyi | F2, 5′-AAGTATAAACCCCTTTACAAGTA | P3, 5′-GAAACTTTACTTTGAGAAAATTAGAGTG | |
| R2, 5′-TATTATTCCATGCTGGAGTATTC | P4, 5′-TTAAAGCAGGCTATTGCCTTGAAT | ||
| C. canis | F2, 5′-GAGTATAAACCCCTTTACAAGTA | P3, 5′-GAAACTTTACTTTGAGAAAATTAGAGTG | |
| R2, 5′-TATTATTCCATGCTCTAGTATTC | P4, 5′-TTAAAGCAGGCTTTTGCCTTGAAT | ||
| C. felis | F2, 5′-AAGTATAAACCCCTTTACAAGTA | P3, 5′-GAAATTTTACTTTGAGAAAATTAGAGTG | |
| R2, 5′-TATTATTCCATGCTGGAGTATTC | P4, 5′-TTAAAGCAGGCTTTTGCCTTGAAT | ||
| hsp90 | − | F5, 5′-ATTAACTCTCTCTATTCTCAGAA | P5, 5′-CTTCTCCAATGCATCTGCAGAATT-[fluorescein] |
| R6, 5′-CATACCGATACCAGTATCAG | P6 [Red 640]-5′-AGTAACTCTCTAAGAAATACGTCT-phosphate | ||
| C. hominis | F5, 5′-ATTAACTCTCTCTATTCTCAGAA | P5, 5′-CTTCTCCAATGCATCTGCAGAATT | |
| R6, 5′-CATACCGATACCAGTATCAG | P6, 5′-AGTAACTCTCTAAGAAATACGTCT | ||
| C. meleagridis | F5, 5′-ATTAACTCTCTCTATTCTCAGAA | P5, 5′-CTTCTCCAATGCATCTGCAGAATT | |
| R6, 5′-CATACCGATACCAGTATCAG | P6, 5′-AGTAACTCTCTGAGAAATACGTCT | ||
| C. parvum | F5, 5′-ATTAACTCTCTCTATTCTCAGAA | P5, 5′-CTTCTCCAATGCATCTGCAGAATT | |
| R6, 5′-CATACCGATACCAGTATCAG | P6, 5′-AGTAACTCTCTGAGAAATACATCT | ||
| C. andersoni | F5, 5′-ATTAACTCTCTCTATTCTCAGAA | P5, 5′-CTTTTCTAGTGCATCTGCAGAGTT | |
| R6, 5′-CATACCAACACCAGTATCAG | P6, 5′-AGTAACTCTCTCAAGAAGACATCC | ||
| C. muris | F5, 5′-ATTAACTCTCTCTATTCTCAGAA | P5, 5′-CTTCTCTAGTGCATCTGCAGAGTT | |
| R6, 5′-CATACCAACACCAGTATCAG | P6, 5′-AGTAACTCTCTCAAGAAGACATCC | ||
| C. canis | F5, 5′-ATTAACTCTCTCTATTCTCAGAA | P5, 5′-CTTTTCCAATGCATCAGCAGAGTT | |
| R6, 5′-CATACCAATACCTGTATCAG | P6, 5′-AATAATTCTCTGAGAAATACATCT | ||
| C. ubiquitum | F5, 5′-ATTAACTCTCTCTATTCTCAGAA | P5, 5′-CTTCTCCAGTGCATCTGCAGAATT | |
| R6, 5′-CATACCGATACCAGTATCAG | P6, 5′-AGTAGTTCTCTAAGAAAAACATCT |
The nucleotide mismatches to the sequences of primers and probes are underlined (the underlined hyphens represent deletions). W1, deer mouse genotype III; W7, muskrat genotype I; W13, skunk genotype; −, deletions; NA, not available (the nucleotide sequence of the forward primer region was unavailable in GenBank).
18S-LC1 real-time PCR.
The 18S-LC1 assay targeted the highly polymorphic region of the SSU rRNA gene between nucleotides 193 and 454 (GenBank accession number AF093491 for C. parvum). This ∼262-bp region had a higher GC content, and sequence polymorphism was mostly in the form of nucleotide substitutions with a few insertions or deletions, which facilitated the design of real-time PCR primers and FRET probes (Table 1). Two fluorescein-labeled probes based on nucleotides 247 to 271 and 276 to 301 (accession number AF093491) were used. The following amplification conditions were used: an initial denaturation step at 95°C for 3 min; 50 cycles of denaturation at 95°C for 2 s, annealing at 50°C for 10 s, and extension at 72°C for 15 s; melt curve analysis at 95°C for 2 s, 45°C for 30 s, and 0.1°C melt steps from 45°C to 80°C; and cooling at 40°C for 30 s.
18S-LC2 real-time PCR.
The 18S-LC2 assay targeted an ∼278-bp fragment of the hypervariable regions encompassing nucleotides 499 to 776 of the SSU rRNA gene (GenBank accession number AF093491) (Table 1). The two fluorescein-labeled probes were based on nucleotides 705 to 732 and 734 to 757 (accession number AF093491). The following amplification conditions were used: an initial denaturation step at 95°C for 3 min; 55 cycles of denaturation at 95°C for 2 s, annealing at 55°C for 15 s, and extension at 72°C for 10 s; melt curve analysis at 95°C for 2 s, 48°C for 30 s, and 0.1°C melt steps from 48°C to 95°C; and cooling at 40°C for 30 s.
hsp90 real-time PCR.
The hsp90 assay targeted an ∼195-bp region of the hsp90 gene between nucleotides 418 and 612 (GenBank accession number XM663146 for C. hominis). The sequence heterogeneity within the hsp90 gene among Cryptosporidium species/genotypes was largely in the form of nucleotide substitutions (Table 1). The two fluorescein-labeled probes were based on nucleotides 441 to 464 and 469 to 492 (accession number XM663146). The following amplification conditions were used: an initial denaturation step at 95°C for 3 min; 50 cycles of denaturation at 95°C for 2 s, annealing at 52°C for 10 s, and extension at 72°C for 15 s; melt curve analysis at 95°C for 2 s, 45°C for 30 s, and 0.1°C melt steps from 45°C to 80°C; and cooling at 40°C for 30 s.
Optimization of the magnesium concentration in real-time PCR assays.
Several concentrations (1.5, 3.0, 4.5, and 6.0 mM) of magnesium (Mg2+) were used for the optimization of the three FRET probe-based real-time PCR assays. One fecal specimen each of two Cryptosporidium species, C. parvum and C. hominis (specimens 19283 and 19310, respectively), was selected to compare the threshold cycle (CT) and melting temperature (Tm) values in the three assays at different Mg2+ concentrations. Melt curve patterns from the three assays were compared among the four Mg2+ concentrations. The optimal concentration of Mg2+ was selected for each real-time assay based on the best combination of sensitivity and melt peaks.
Evaluation of Cryptosporidium species detection range of real-time assays.
The Cryptosporidium species detection ranges of the three FRET probe-based real-time PCR assays were evaluated by using fecal specimens of nine Cryptosporidium species/genotypes commonly found in source water: C. parvum, C. hominis, C. andersoni, C. ubiquitum, C. meleagridis, C. cuniculus, deer mouse genotype III (W1), muskrat genotype I (W7), and skunk genotype (W13). Fecal specimens of C. parvum, C. hominis, and C. meleagridis were obtained from HIV patients in Peru; C. ubiquitum, deer mouse genotype III, muskrat genotype I, and skunk genotype specimens were obtained from wildlife in New York; C. andersoni specimens were obtained from cattle in China; and C. cuniculus specimens were obtained from rabbits in China. DNA was extracted from these stool specimens by using the FastDNA Spin kit for soil (MP Biomedicals, Irvine, CA) and identified to the species level by SSU rRNA-based PCR-RFLP analysis (5). Two specimens each of the nine species/genotypes were used in the evaluation, using the PCR conditions and amplification programs described above.
Evaluation of sensitivity of real-time PCR assays.
The sensitivities of the three FRET probe-based real-time PCR assays were evaluated by analyzing seeded human fecal specimens and seeded water concentrates. The C. parvum oocysts used in seeding were purified from the fecal specimens by sucrose and cesium chloride density gradients (25) and further purified and counted by flow cytometry sorting (FACSAria III; BD Biosciences, San Jose, CA) after staining with an immunofluorescent monoclonal antibody. For oocyst seeding, fecal specimens were collected from microscopy-negative children in Lima, Peru, and Cryptosporidium-negative water concentrates were collected by the filtration of at least 10 liters of lake water through an Envirochek HV filter (Pall Gelman Laboratory, Ann Arbor, MI) and elution of the filtrate according to EPA Method 1623. A total of 0.5 ml of each Cryptosporidium-negative fecal specimen or water concentrate was seeded with four levels of flow cytometry-counted oocysts (100, 50, 20, and 10 oocysts for fecal specimens and 50, 25, 10, and 5 oocysts for water samples). Five replicates at each seeding level were used in the evaluation of the sensitivity of each assay. Genomic DNA was extracted from these fecal specimens by using the FastDNA Spin kit for soil and eluted with 100 μl of distilled water. Oocysts in the seeded water samples were isolated by immunomagnetic separation (IMS) according to EPA Method 1623. DNA was extracted from these IMS-purified oocysts by using the QIAamp DNA minikit (Qiagen, Valencia, CA) and eluted with 100 μl of distilled water. In real-time PCRs, 2 μl of 100 μl DNA extracted from each fecal specimen or water sample was used as the template, equivalent to 2, 1, 0.4, and 0.2 oocysts (40, 20, 8, and 4 theoretical gene copies, respectively) per PCR for the four oocyst levels of fecal specimens and 1, 0.5, 0.2, and 0.1 oocysts (20, 10, 4, and 2 theoretical gene copies, respectively) per reaction for the four oocyst levels of water samples.
RESULTS
Development of three FRET probe-based real-time PCR assays.
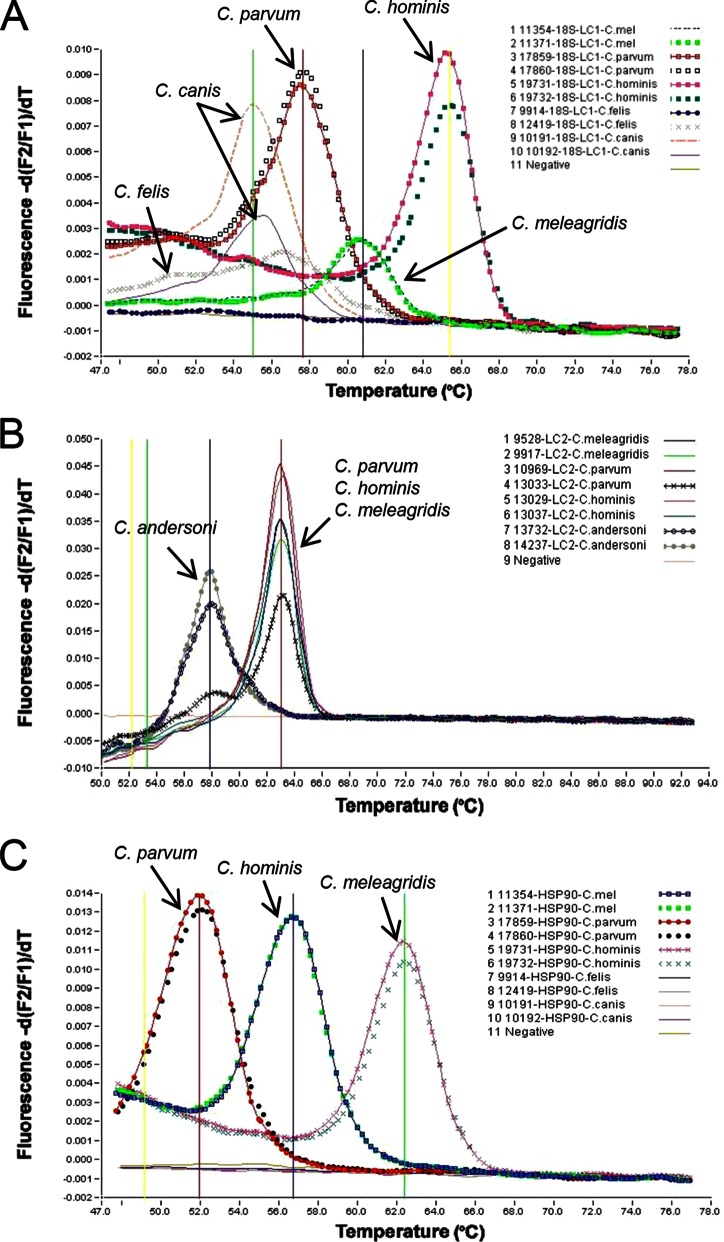
The genotyping capacity of the 18S-LC1 real-time PCR assay was evaluated by using two DNA preparations from each of the five common Cryptosporidium spp. in humans. The PCR amplified the DNAs of all species tested. Melt curve analysis was able to differentiate all five species, C. hominis, C. parvum, C, meleagridis, C. canis, and C. felis (Fig. 1A). For the 18S-LC2 assay, the genotyping capacity was evaluated by using two DNA preparations each of the three most common Cryptosporidium spp. (C. parvum, C. hominis, and C. meleagridis) in humans and the most common Cryptosporidium sp. (C. andersoni) in water. The 18S-LC2 assay amplified the DNAs of all species. Melt curve analysis was able to differentiate C. andersoni from common human-pathogenic species (Fig. 1B). Similarly, the genotyping capacity of the hsp90 assay was evaluated by using two DNA preparations each of the five common Cryptosporidium spp. in humans. The assay amplified the DNAs of the three most common human-pathogenic species, C. parvum, C. hominis, and C. meleagridis, which could be differentiated from each other by melt curve analysis (Fig. 1C).
FIG 1.
Development of three FRET probe-based real-time PCR genotyping assays. (A) Differentiation of five human-pathogenic Cryptosporidium species (C. hominis, C. parvum, C, meleagridis, C. canis, and C. felis) in clinical specimens by 18S-LC1-based melt curve analysis. (B) Differentiation of the most common Cryptosporidium species (C. andersoni) in water from the three most common Cryptosporidium species (C. parvum, C. hominis, and C. meleagridis) in humans by 18S-LC2-based melt curve analysis. (C) Differentiation of three major human-pathogenic Cryptosporidium species (C. hominis, C. parvum, and C. meleagridis) by hsp90-based melt curve analysis. The melt peaks for each Cryptosporidium species are labeled.
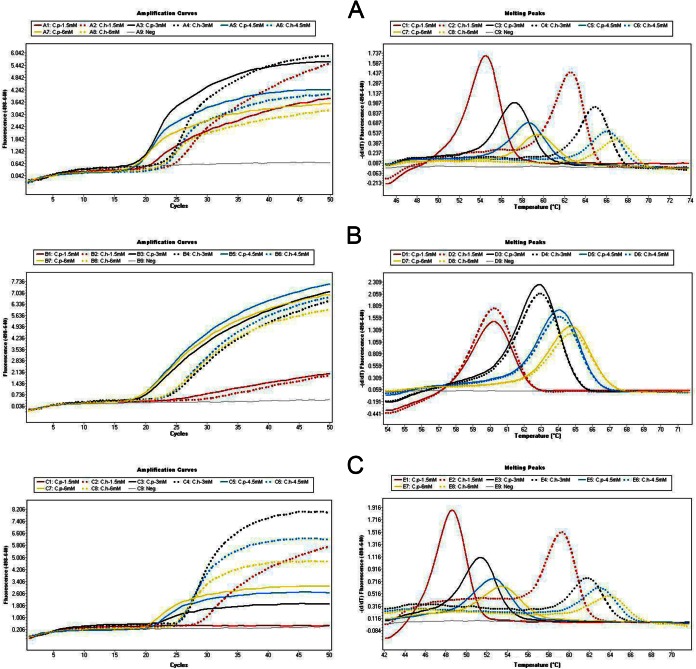
Optimization of the Mg2+ concentration in three real-time PCR assays.
The Mg2+ concentration was optimized in the three FRET probe-based real-time PCR assays. For all three assays, the use of higher Mg2+ concentrations generated lower CT values and higher Tm values for both C. parvum and C. hominis (Table 2). The melt peaks were lower when higher Mg2+ concentrations were used for PCR (Fig. 2). Based on the best combination of sensitivity and melt curve patterns (Fig. 2A to C), the optimal concentration of Mg2+ was selected to be 3.0 mM Mg2+ for all three PCR assays.
TABLE 2.
Comparison of relative sensitivities of three FRET probe-based real-time PCR assays using various Mg2+ concentrations
| Assay | Specimen | Species | Mg2+ concn (mM) | CT | Tm (°C) |
|---|---|---|---|---|---|
| 18S-LC1 | 19283 | C. parvum | 1.5 | 19.65 | 54.27 |
| 3.0 | 18.80 | 57.29 | |||
| 4.5 | 18.02 | 58.65 | |||
| 6.0 | 17.61 | 59.67 | |||
| 19310 | C. hominis | 1.5 | 24.36 | 62.39 | |
| 3.0 | 23.12 | 64.99 | |||
| 4.5 | 22.54 | 66.22 | |||
| 6.0 | 21.82 | 67.08 | |||
| 18S-LC2 | 19283 | C. parvum | 1.5 | 23.41 | 60.12 |
| 3.0 | 19.41 | 62.81 | |||
| 4.5 | 19.13 | 64.02 | |||
| 6.0 | 18.96 | 64.71 | |||
| 19310 | C. hominis | 1.5 | 29.52 | 60.16 | |
| 3.0 | 23.99 | 62.81 | |||
| 4.5 | 23.54 | 64.02 | |||
| 6.0 | 23.39 | 64.71 | |||
| hsp90 | 19283 | C. parvum | 1.5 | 28.30 | 48.50 |
| 3.0 | 21.21 | 51.36 | |||
| 4.5 | 20.22 | 52.55 | |||
| 6.0 | 19.62 | 53.55 | |||
| 19310 | C. hominis | 1.5 | 28.51 | 59.17 | |
| 3.0 | 25.74 | 62.09 | |||
| 4.5 | 24.94 | 63.25 | |||
| 6.0 | 24.27 | 64.24 |
FIG 2.
Comparison of relative sensitivities and melt curve patterns of three FRET probe-based real-time PCR assays using 1.5 mM (red), 3.0 mM (black), 4.5 mM (blue), and 6.0 mM (yellow) final concentrations of Mg2+. Solid lines, C. parvum 19283; dotted lines C. hominis 19310. (A) 18S-LC1 assay; (B) 18S-LC2 assay; (C) hsp90 assay.
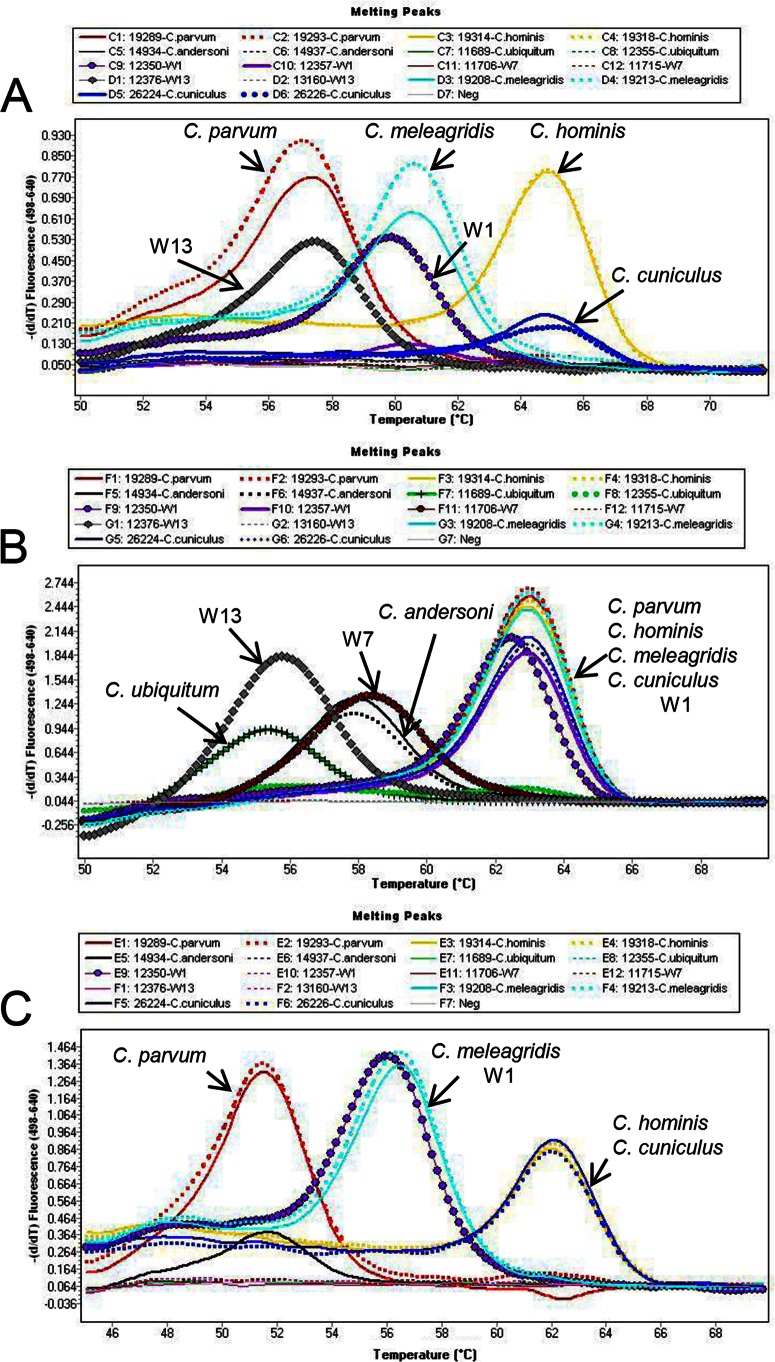
Cryptosporidium species detection ranges of the three real-time PCR assays.
The abilities of the three real-time PCR assays to detect and differentiate nine Cryptosporidium species/genotypes commonly found in source water were compared by using two DNA preparations of each species/genotype. These species and genotypes were differentiated by melt curve analysis. By using the optimized 18S-LC1 real-time PCR assay (i.e., 3 mM Mg2+), DNA extracts of six species/genotypes commonly found in source water were amplified (Table 3). Melt curve analysis was able to differentiate C. parvum/W13, C. meleagridis, W1, and C. hominis/C. cuniculus from each other (paired organisms have similar melting temperatures and thus cannot be differentiated) (Table 3 and Fig. 3A). For 18S-LC2, DNA extracts of all nine species/genotypes evaluated were amplified by this assay (Table 3). Melt curve analysis was able to differentiate C. ubiquitum/W13, C. andersoni/W7, and C. parvum/C. hominis/C. meleagridis/W1/C. cuniculus (Table 3 and Fig. 3B). For the hsp90 PCR assay, DNA extracts of five species/genotypes were amplified, and melt curve analysis differentiated C. parvum, C. meleagridis/W1, and C. hominis/C. cuniculus (Table 3 and Fig. 3C).
TABLE 3.
Differentiation of Cryptosporidium species/genotypes commonly found in source water by using three FRET probe-based real-time PCR assaysa
| Specimen | Species or genotype | 18S-LC1 assay |
18S-LC2 assay |
hsp90 assay |
|||
|---|---|---|---|---|---|---|---|
| CT | Tm (°C) | CT | Tm (°C) | CT | Tm (°C) | ||
| 19289 | C. parvum | 25.04 | 57.48 | 25.44 | 62.93 | 27.03 | 51.58 |
| 19293 | C. parvum | 23.01 | 57.22 | 23.81 | 62.92 | 25.55 | 51.52 |
| 19314 | C. hominis | 16.98 | 65.08 | 17.88 | 62.92 | 19.13 | 62.28 |
| 19318 | C. hominis | 21.33 | 65.09 | 21.23 | 62.93 | 22.66 | 62.28 |
| 14934 | C. andersoni | − | − | 29.83 | 57.77 | 37.14b | 51.54b |
| 14937 | C. andersoni | − | − | 34.08 | 57.77 | − | − |
| 11689 | C. ubiquitum | − | − | 27.57 | 55.27 | − | − |
| 12355 | C. ubiquitum | − | − | 35.42 | 56.28 | − | − |
| 12350 | W1 | 26.65 | 60.01 | 28.36 | 62.36 | 30.27 | 56.27 |
| 12357 | W1 | − | − | 35.76 | 62.92 | − | − |
| 11706 | W7 | − | − | 37.38 | 58.28 | − | − |
| 11715 | W7 | − | − | − | − | − | − |
| 12376 | W13 | 28.86 | 57.43 | 31.70 | 55.70 | − | − |
| 13160 | W13 | − | − | − | − | − | − |
| 19208 | C. meleagridis | 27.44 | 60.81 | 28.87 | 62.92 | 30.30 | 56.74 |
| 19213 | C. meleagridis | 23.46 | 60.78 | 24.30 | 62.92 | 25.35 | 56.69 |
| 26224 | C. cuniculus | 30.23 | 65.13 | 33.54 | 62.92 | 35.49 | 62.35 |
| 26226 | C. cuniculus | 30.73 | 64.81 | 34.49 | 62.89 | 37.59 | 62.35 |
−, negative in PCR amplification; W1, deer mouse genotype III; W7, muskrat genotype I; W13, skunk genotype.
C. andersoni specimen 14934 appears to have a low level of C. parvum coinfection.
FIG 3.
Differentiation of Cryptosporidium species/genotypes commonly found in source water (C. parvum, C. hominis, C. andersoni, C. ubiquitum, deer mouse genotype III [W1], muskrat genotype I [W7], skunk genotype [W13], C. meleagridis, and C. cuniculus) by using melt curve analysis in the three FRET probe-based real-time PCR assays. (A) 18S-LC1 assay; (B) 18S-LC2 assay; (C) hsp90 assay. The melt peaks for each Cryptosporidium species are labeled.
Sensitivity of three real-time PCR assays for analysis of fecal specimens.
We used Cryptosporidium-negative human fecal specimens seeded with five levels of low numbers of C. parvum oocysts to evaluate the sensitivities of the three real-time assays. A summary of the results is shown in Table 4. For 18S-LC1, four seeded specimens (one at the level of 2 oocysts/PCR and three at the level of 1 oocyst/PCR) could be amplified with relatively low CT values, and the C. parvum oocysts seeded could be differentiated from those of C. hominis (positive-control DNA extracted from a C. hominis-positive fecal specimen) by melt curve analysis, although the melt peaks were low (data not shown). For 18S-LC2, 14 C. parvum-seeded specimens could be amplified (5 at the level of 2 oocysts/PCR, 5 at the level of 1 oocyst/PCR, 3 at the level of 0.4 oocysts/PCR, and 1 at the level of 0.2 oocysts/PCR). In melt curve analysis, all specimens of C. parvum showed the expected Tm value and melt peak (data not shown). For hsp90, seven seeded specimens (one at the level of 2 oocysts/PCR, five at the level of 1 oocyst/PCR, and one at the level of 0.2 oocysts/PCR) could be amplified, and the C. parvum oocysts present could be differentiated from those of C. hominis by melt curve analysis (data not shown). Thus, 18S-LC2 had a higher sensitivity than did hsp90 and 18S-LC1 in detecting low levels of Cryptosporidium oocysts in clinical specimens.
TABLE 4.
Sensitivity evaluations of three FRET probe-based real-time PCR assays using fecal specimens and water sample concentrates seeded with various levels of low numbers of C. parvum oocystsa
| Specimen type | Level | No. of oocysts/PCR | 18S-LC1 assay |
18S-LC2 assay |
hsp90 assay |
|||
|---|---|---|---|---|---|---|---|---|
| Avg CT ± SD (no. of positive specimens/total no. of specimens) | Avg Tm (°C) ± SD (no. of positive specimens/total no. of specimens) | Avg CT ± SD (no. of positive specimens/total no. of specimens) | Avg Tm (°C) ± SD (no. of positive specimens/total no. of specimens) | Avg CT ± SD (no. of positive specimens/total no. of specimens) | Avg Tm (°C) ± SD (no. of positive specimens/total no. of specimens) | |||
| Feces | I | 2 | 28.76 (1/5) | 57.31 ± 0.20 (4/5) | 35.35 ± 1.59 (5/5) | 62.69 ± 0.09 (5/5) | 36.06 (1/5) | 51.13 ± 0.17 (4/5) |
| II | 1 | 29.23 ± 0.49 (3/5) | 57.19 ± 0.06 (3/5) | 35.13 ± 1.38 (5/5) | 62.76 ± 0.06 (5/5) | 34.19 ± 2.70 (5/5) | 51.27 ± 0.18 (5/5) | |
| III | 0.4 | − | 58.07 (1/5) | 37.31 ± 1.54 (3/5) | 62.50 ± 0.34 (3/5) | − | − | |
| IV | 0.2 | − | − | 37.13 (1/5) | 62.75 (1/5) | 36.66 (1/5) | 51.35 (1/5) | |
| Water | V | 1 | − | 57.19 (1/5) | 35.18 ± 0.74 (5/5) | 62.83 ± 0.04 (5/5) | 37.11 ± 1.17 (4/5) | 51.44 ± 0.02 (4/5) |
| VI | 0.5 | 28.14 (1/5) | 56.92 (1/5) | 34.99 ± 1.87 (4/5) | 62.93 ± 0.05 (4/5) | 38.35 ± 1.06 (3/5) | 51.53 ± 0.12 (3/5) | |
| VII | 0.2 | − | − | 37.42 ± 1.76 (5/5) | 62.86 ± 0.03 (5/5) | 38.93 (1/5) | 51.67 (1/5) | |
| VIII | 0.1 | − | − | 36.81 ± 0.92 (3/5) | 62.80 ± 0.04 (3/5) | 38.04 (1/5) | 51.46 (1/5) | |
−, not detected.
Sensitivities of the three real-time PCR assays for analysis of water samples.
In evaluations with water sample concentrates seeded with low numbers of C. parvum oocysts, only one seeded sample (one at the level of 0.5 oocysts/PCR) could be amplified by 18S-LC1 PCR, and the C. parvum oocysts present could be differentiated from those of C. hominis (positive-control DNA extracted from a C. hominis-positive fecal specimen) by melt curve analysis (Table 4). For 18S-LC2, DNA extracts from 17 seeded samples were amplified (5 at the level of 1 oocyst/PCR, 4 at the level of 0.5 oocysts/PCR, 5 at the level of 0.2 oocysts/PCR, and 3 at the level of 0.1 oocysts/PCR). In melt curve analysis, all samples of C. parvum showed the expected melt peaks and Tm values, which were similar to those of C. hominis (positive control; the assay was meant to differentiate the human-pathogenic species C. parvum, C. hominis, and C. meleagridis from C. andersoni). For hsp90, DNA extracts of nine seeded samples (four at the level of 1 oocyst/PCR, three at the level of 0.5 oocysts/PCR, one at the level of 0.2 oocysts/PCR, and one at the level of 0.1 oocysts/PCR) could be amplified, and the C. parvum oocysts present could be differentiated from those of C. hominis (positive control) by melt curve analysis. Thus, 18S-LC2 and hsp90 had higher sensitivities than did 18S-LC1 in detecting low levels of Cryptosporidium oocysts in water.
DISCUSSION
Identification of Cryptosporidium to the species/genotype level is especially challenging for environmental samples because of the usual presence of very low numbers of oocysts and high concentrations of PCR inhibitors and nontarget organisms. However, it is essential for the assessment of public health importance and the identification of contamination sources of the oocysts. In this study, we developed three FRET probe-based real-time PCR genotyping assays for rapid tracking of Cryptosporidium contamination and conducted a side-by-side comparison of sensitivities and Cryptosporidium species detection ranges of these assays.
The SSU rRNA gene is the most common target used for Cryptosporidium detection and genotyping in environmental samples, because it has 20 copies per oocyst, thus increasing the probability of detecting small numbers of oocysts commonly found in environmental samples. In addition, it has regions with semiconserved and hypervariable nucleotide sequences, facilitating the design of primers and probes for the development of genus-specific genotyping assays. Previously, Limor et al. (19) developed an SSU rRNA gene-based real-time PCR genotyping assay, using FRET probes for real-time detection of Cryptosporidium and melt curve analysis for genotyping of five major human-pathogenic species in clinical specimens. However, the assay showed a detection limit of 5 oocysts per PCR, which makes it inapplicable in the analysis of environmental samples (19). In this study, to increase the sensitivity of detection, we reduced the size of the amplicon from ∼830 bp to ∼262 bp and developed the 18S-LC1 real-time PCR assay for differentiation of the five major human-pathogenic Cryptosporidium species. The primers and probes were chosen based on multiple-sequence alignments of the SSU rRNA genes of various Cryptosporidium species/genotypes and other apicomplexan parasites. The sequences of LC Red-labeled hybridization probes are identical to that of C. hominis/C. cuniculus, but it has 1 base mismatch to C. meleagridis/deer mouse genotype III (W1) and 2 base mismatches to C. parvum/skunk genotype (W13). In melt curve analysis, C. hominis/C. cuniculus had the highest melt temperature, followed by C. meleagridis/W1 and C. parvum/W13. Because of more sequence divergence from the 18S-LC1 primers and probes, gastric Cryptosporidium species, such as C. andersoni and C. muris, cannot be detected by this assay. Thus, the 18S-LC1 assay is most useful in the analysis of clinical specimens from humans.
To assess the source and public health significance of Cryptosporidium oocysts in drinking source water and watersheds, it is necessary to develop methods capable of identifying the Cryptosporidium species and genotypes commonly found in source water, such as C. parvum, C. hominis, C. andersoni, and C. ubiquitum, and distinguishing between human-pathogenic and nonpathogenic species/genotypes. Thus, we developed another SSU rRNA gene-based real-time PCR assay, 18S-LC2, for the detection of major Cryptosporidium species/genotypes in water. Nine common Cryptosporidium species/genotypes can be detected by this real-time assay. Moreover, C. andersoni, the dominant non-human-pathogenic species in water samples (2, 3, 8, 11), can be differentiated from the three major human-pathogenic species (C. parvum, C. hominis, and C. meleagridis) by melt curve analysis. Other species/genotypes not highly infectious to humans, such as muskrat genotype I (W7) and skunk genotype (W13), can also be differentiated from the major human-pathogenic species by this assay. Because C. parvum, C. hominis, C. meleagridis, C. cuniculus, and deer mouse genotype III (W1) have identical sequences in the regions of the two SSU rRNA probes in 18S-LC2 (Table 1), samples of these species/genotypes showed similar melting temperatures in melt curve analyses.
In addition to genotyping tools based on the SSU rRNA gene, there is a need for a secondary and confirmatory genotyping tool targeting other genetic loci, especially when the three major human-pathogenic species, C. parvum, C. hominis, and C. meleagridis, are detected. In this study, we have developed a real-time PCR genotyping assay targeting the hsp90 gene. The primers and probes were designed based on the alignment of hsp90 sequences of various Cryptosporidium species/genotypes (Table 1). Like the 18S-LC1 assay, the hsp90 assay also has a preference for detecting the major human-pathogenic species because of the primer and probe design strategy. The probe sequence is identical to that of C. hominis/C. cuniculus but has 1 base mismatch to the C. meleagridis/deer mouse genotype III (W1) sequence and 2 base mismatches to the C. parvum sequence, allowing the differentiation of these species/genotypes by melt curve analysis. In future studies, to determine if the melting temperatures identified for each species are consistent in each assay, data from more isolates per Cryptosporidium species are needed for statistical analysis.
The three real-time PCR genotyping assays have different detection limits. For the detection of low levels of flow cytometry-sorted C. parvum oocysts seeded in fecal specimens or water concentrates, the 18S-LC2 and hsp90 assays could detect a single oocyst, indicating that it is possible for these two assays to detect low levels of oocysts in water samples. A recent study comparing the sensitivities and specificities of 10 TaqMan-based real-time PCR assays showed that 8 of the assays could reliably detect 10 flow cytometry-sorted oocysts in reagent water or an environmental matrix (26). Therefore, the 18S-LC2 and hsp90 real-time PCR genotyping assays developed in this work likely have higher sensitivities in detecting Cryptosporidium oocysts in water and thus can be potentially used as rapid genotyping tools for environmental monitoring. The 18S-LC1 genotyping assay developed in this study may be not applicable for environmental samples because of its lower sensitivity. It is more applicable for genotyping human-pathogenic species in clinical specimens or wastewater samples, which usually contain high numbers of Cryptosporidium oocysts.
In summary, the three real-time PCR assays developed in this study can genotype common human-pathogenic species as well as differentiate human-pathogenic from nonpathogenic species in water. The 18S-LC2 and hsp90 assays have the capacity to detect low levels of Cryptosporidium oocysts and thus can be potentially combined for genotyping of Cryptosporidium oocysts in the environment, by initial differentiation of human-pathogenic species from others using the 18S-LC2 assay and further identification of human-pathogenic species using the hsp90 assay. Nevertheless, field use of this approach requires further evaluations and validations by using different sample matrices (source water, storm runoff, or finished water) and testing schemes (water concentrates, IMS pellets, or Method 1623 slides). Hopefully, it can become part of a systematic assessment of the source and public health significance of Cryptosporidium oocysts in drinking source water and watersheds.
ACKNOWLEDGMENTS
This study was supported in part by project RFP4179 from the Water Research Foundation and the Water Special Project (2014ZX07104006) from the Ministry of Science and Technology, China.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. The United States Environmental Protection Agency through its Office of Research and Development collaborated in the research described here. It has been subjected to the Agency's administrative review and approved for publication.
REFERENCES
- 1.Bouzid M, Steverding D, Tyler KM. 2008. Detection and surveillance of waterborne protozoan parasites. Curr Opin Biotechnol 19:302–306. doi: 10.1016/j.copbio.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Ruecker NJ, Braithwaite SL, Topp E, Edge T, Lapen DR, Wilkes G, Robertson W, Medeiros D, Sensen CW, Neumann NF. 2007. Tracking host sources of Cryptosporidium spp. in raw water for improved health risk assessment. Appl Environ Microbiol 73:3945–3957. doi: 10.1128/AEM.02788-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruecker NJ, Matsune JC, Wilkes G, Lapen DR, Topp E, Edge TA, Sensen CW, Xiao L, Neumann NF. 2012. Molecular and phylogenetic approaches for assessing sources of Cryptosporidium contamination in water. Water Res 46:5135–5150. doi: 10.1016/j.watres.2012.06.045. [DOI] [PubMed] [Google Scholar]
- 4.Jellison KL, Lynch AE, Ziemann JM. 2009. Source tracking identifies deer and geese as vectors of human-infectious Cryptosporidium genotypes in an urban/suburban watershed. Environ Sci Technol 43:4267–4272. doi: 10.1021/es900081m. [DOI] [PubMed] [Google Scholar]
- 5.Xiao L, Alderisio K, Limor J, Royer M, Lal AA. 2000. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl Environ Microbiol 66:5492–5498. doi: 10.1128/AEM.66.12.5492-5498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkes G, Ruecker NJ, Neumann NF, Gannon VP, Jokinen C, Sunohara M, Topp E, Pintar KD, Edge TA, Lapen DR. 2013. Spatiotemporal analysis of Cryptosporidium species/genotypes and relationships with other zoonotic pathogens in surface water from mixed-use watersheds. Appl Environ Microbiol 79:434–448. doi: 10.1128/AEM.01924-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kothavade RJ. 2012. Potential molecular tools for assessing the public health risk associated with waterborne Cryptosporidium oocysts. J Med Microbiol 61:1039–1051. doi: 10.1099/jmm.0.043158-0. [DOI] [PubMed] [Google Scholar]
- 8.Yang W, Chen P, Villegas EN, Landy RB, Kanetsky C, Cama V, Dearen T, Schultz CL, Orndorff KG, Prelewicz GJ, Brown MH, Young KR, Xiao L. 2008. Cryptosporidium source tracking in the Potomac River watershed. Appl Environ Microbiol 74:6495–6504. doi: 10.1128/AEM.01345-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruecker NJ, Matsune JC, Lapen DR, Topp E, Edge TA, Neumann NF. 2013. The detection of Cryptosporidium and the resolution of mixtures of species and genotypes from water. Infect Genet Evol 15:3–9. doi: 10.1016/j.meegid.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Ruecker NJ, Hoffman RM, Chalmers RM, Neumann NF. 2011. Detection and resolution of Cryptosporidium species and species mixtures by genus-specific nested PCR-restriction fragment length polymorphism analysis, direct sequencing, and cloning. Appl Environ Microbiol 77:3998–4007. doi: 10.1128/AEM.02706-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Zhao X, Chen J, Jin W, Zhou X, Li N, Wang L, Xiao L. 2011. Occurrence, source, and human infection potential of Cryptosporidium and Giardia spp. in source and tap water in Shanghai, China. Appl Environ Microbiol 77:3609–3616. doi: 10.1128/AEM.00146-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang J, Alderisio KA, Xiao L. 2005. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol 71:4446–4454. doi: 10.1128/AEM.71.8.4446-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fayer R, Xiao L. 2008. Cryptosporidium and cryptosporidiosis, 2nd ed CRC Press, Boca Raton, FL. [Google Scholar]
- 14.da Silva DC, Paiva PR, Nakamura AA, Homem CG, Souza MS, Grego KF, Meireles MV. 2014. The detection of Cryptosporidium serpentis in snake fecal samples by real-time PCR. Vet Parasitol 204:134–138. doi: 10.1016/j.vetpar.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Alonso JL, Amoros I, Guy RA. 2014. Quantification of viable Giardia cysts and Cryptosporidium oocysts in wastewater using propidium monoazide quantitative real-time PCR. Parasitol Res 113:2671–2678. doi: 10.1007/s00436-014-3922-9. [DOI] [PubMed] [Google Scholar]
- 16.Burnet JB, Ogorzaly L, Tissier A, Penny C, Cauchie HM. 2013. Novel quantitative TaqMan real-time PCR assays for detection of Cryptosporidium at the genus level and genotyping of major human and cattle-infecting species. J Appl Microbiol 114:1211–1222. doi: 10.1111/jam.12103. [DOI] [PubMed] [Google Scholar]
- 17.Stroup SE, Roy S, McHele J, Maro V, Ntabaguzi S, Siddique A, Kang G, Guerrant RL, Kirkpatrick BD, Fayer R, Herbein J, Ward H, Haque R, Houpt ER. 2006. Real-time PCR detection and speciation of Cryptosporidium infection using Scorpion probes. J Med Microbiol 55:1217–1222. doi: 10.1099/jmm.0.46678-0. [DOI] [PubMed] [Google Scholar]
- 18.Tanriverdi S, Tanyeli A, Baslamisli F, Koksal F, Kilinc Y, Feng X, Batzer G, Tzipori S, Widmer G. 2002. Detection and genotyping of oocysts of Cryptosporidium parvum by real-time PCR and melting curve analysis. J Clin Microbiol 40:3237–3244. doi: 10.1128/JCM.40.9.3237-3244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Limor JR, Lal AA, Xiao L. 2002. Detection and differentiation of Cryptosporidium parasites that are pathogenic for humans by real-time PCR. J Clin Microbiol 40:2335–2338. doi: 10.1128/JCM.40.7.2335-2338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hadfield SJ, Chalmers RM. 2012. Detection and characterization of Cryptosporidium cuniculus by real-time PCR. Parasitol Res 111:1385–1390. doi: 10.1007/s00436-012-2874-1. [DOI] [PubMed] [Google Scholar]
- 21.Jothikumar N, da Silva AJ, Moura I, Qvarnstrom Y, Hill VR. 2008. Detection and differentiation of Cryptosporidium hominis and Cryptosporidium parvum by dual TaqMan assays. J Med Microbiol 57:1099–1105. doi: 10.1099/jmm.0.2008/001461-0. [DOI] [PubMed] [Google Scholar]
- 22.Jiang J, Xiao L. 2003. An evaluation of molecular diagnostic tools for the detection and differentiation of human-pathogenic Cryptosporidium spp. J Eukaryot Microbiol 50(Suppl):542–547. doi: 10.1111/j.1550-7408.2003.tb00623.x. [DOI] [PubMed] [Google Scholar]
- 23.Amar CF, Dear PH, McLauchlin J. 2004. Detection and identification by real time PCR/RFLP analyses of Cryptosporidium species from human faeces. Lett Appl Microbiol 38:217–222. doi: 10.1111/j.1472-765X.2004.01473.x. [DOI] [PubMed] [Google Scholar]
- 24.Higgins JA, Fayer R, Trout JM, Xiao L, Lal AA, Kerby S, Jenkins MC. 2001. Real-time PCR for the detection of Cryptosporidium parvum. J Microbiol Methods 47:323–337. doi: 10.1016/S0167-7012(01)00339-6. [DOI] [PubMed] [Google Scholar]
- 25.Arrowood MJ, Donaldson K. 1996. Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. J Eukaryot Microbiol 43:89S. doi: 10.1111/j.1550-7408.1996.tb05015.x. [DOI] [PubMed] [Google Scholar]
- 26.Staggs SE, Beckman EM, Keely SP, Mackwan R, Ware MW, Moyer AP, Ferretti JA, Sayed A, Xiao L, Villegas EN. 2013. The applicability of TaqMan-based quantitative real-time PCR assays for detecting and enumerating Cryptosporidium spp. oocysts in the environment. PLoS One 8:e66562. doi: 10.1371/journal.pone.0066562. [DOI] [PMC free article] [PubMed] [Google Scholar]