Abstract
In contrast to the well-characterized and more common maleylpyruvate isomerization route of the gentisate pathway, the direct hydrolysis route occurs rarely and remains unsolved. In Pseudomonas alcaligenes NCIMB 9867, two gene clusters, xln and hbz, were previously proposed to be involved in gentisate catabolism, and HbzF was characterized as a maleylpyruvate hydrolase converting maleylpyruvate to maleate and pyruvate. However, the complete degradation pathway of gentisate through direct hydrolysis has not been characterized. In this study, we obtained from the NCIMB culture collection a Pseudomonas alcaligenes spontaneous mutant strain that lacked the xln cluster and designated the mutant strain SponMu. The hbz cluster in strain SponMu was resequenced, revealing the correct location of the stop codon for hbzI and identifying a new gene, hbzG. HbzIJ was demonstrated to be a maleate hydratase consisting of large and small subunits, stoichiometrically converting maleate to enantiomerically pure d-malate. HbzG is a glutathione-dependent maleylpyruvate isomerase, indicating the possible presence of two alternative pathways of maleylpyruvate catabolism. However, the hbzF-disrupted mutant could still grow on gentisate, while disruption of hbzG prevented this ability, indicating that the direct hydrolysis route was not a complete pathway in strain SponMu. Subsequently, a d-malate dehydrogenase gene was introduced into the hbzG-disrupted mutant, and the engineered strain was able to grow on gentisate via the direct hydrolysis route. This fills a gap in our understanding of the direct hydrolysis route of the gentisate pathway and provides an explanation for the high yield of d-malate from maleate by this d-malate dehydrogenase-deficient natural mutant.
INTRODUCTION
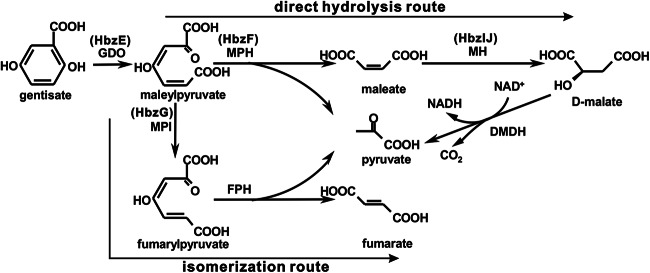
Gentisate (2,5-dihydroxybenzoate) is an important ring cleavage intermediate present in the bacterial catabolic pathway of many aromatic compounds. In the gentisate pathway, gentisate 1,2-dioxygenase catalyzes the ring cleavage oxidation of gentisate to yield maleylpyruvate, which is then further degraded by either direct hydrolysis to maleate and pyruvate (1) or isomerization to fumarylpyruvate and subsequent hydrolysis to fumarate and pyruvate (2, 3). For the isomerization route, glutathione (GSH)-, mycothiol (MSH)-, or l-cysteine-dependent maleylpyruvate isomerases have been biochemically and genetically characterized in Gram-negative (4), high-G+C-content Gram-positive (5, 6), or low-G+C-content Gram-positive (7) bacterial strains, respectively. As an alternative to the isomerization route, maleylpyruvate can also be degraded via the direct hydrolysis route, which is catalyzed by maleylpyruvate hydrolase to yield maleate and pyruvate (Fig. 1). In contrast to the well-studied isomerization route, the direct hydrolysis route has been studied only in the 2,5- and 3,5-xylenol utilizer Pseudomonas alcaligenes NCIMB 9867 (8, 9), in which m-cresol, xylenols, and 3-hydroxybenzoate are degraded via gentisate. In this strain, a maleylpyruvate hydrolase-encoding gene, hbzF, was characterized (10) and a hydratase catalyzing the transformation of maleate to d-malate was purified (11). In addition, a d-malate dehydrogenase, which catalyzes the conversion of d-malate to pyruvate and which is encoded by the dmlA gene, has been characterized for d-malate catabolism in Escherichia coli K-12 (12), which led us to speculate the complete direct hydrolysis route for gentisate biodegradation, as shown in Fig. 1.
FIG 1.
Two alternative routes for maleylpyruvate catabolism in the gentisate pathway. GDO, gentisate 1,2-dioxygenase (HbzE in strain SponMu) (19); MPH, maleylpyruvate hydrolase (HbzF in strain SponMu) (10); MH, maleate hydratase (HbzIJ in strain SponMu) (this study); DMDH, d-malate dehydrogenase; MPI, maleylpyruvate isomerase (HbzG in strain SponMu) (this study); FPH, fumarylpyruvate hydrolase.
In the archaeon Methanocaldococcus jannaschii, two genes were identified to encode large and small subunits of isopropylmalate isomerase catalyzing the isomerization between α-isopropylmalate and β-isopropylmalate (13), which is the second reaction of the biosynthetic pathway of leucine (14). This enzyme has a broad substrate specificity and also catalyzes the hydration of maleate to form d-malate (13). However, the physiological role of its hydration activity has not been elucidated.
A number of bacterial strains have been exploited for the production of d-malate from maleate. Among these strains, Pseudomonas alcaligenes NCIMB 9867 exhibited the best production yield, and d-malate was formed with an enantiomeric purity of more than 99.97% (15). However, strain NCIMB 9867 is not able to utilize maleate as the sole carbon source, and the maleate hydratase activity was significantly induced only when the strain was grown on 3-hydroxybenzaote or gentisate (16). In this study, we report on the characterization of hbzIJ in the gentisate catabolic cluster, which encodes the two subunits of maleate hydratase but which does not have isopropylmalate isomerase activity, in strain SponMu, a spontaneous mutant of Pseudomonas alcaligenes NCIMB 9867. The stoichiometric production of enantiomerically pure d-malate from maleate was achieved by this mutant strain. However, due to the absence of the d-malate dehydrogenase gene, the direct hydrolysis route of the gentisate pathway in this strain is incomplete. The data presented herein fill a gap in our understanding of the direct hydrolysis route of the gentisate pathway and also provide an explanation for the high yield of d-malate from maleate by this d-malate dehydrogenase-deficient natural mutant.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, media, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The Escherichia coli strains were grown in lysogeny broth (LB) with 20 μg/ml tetracycline (Tc) or 50 μg/ml kanamycin (Km), as necessary. Pseudomonas alcaligenes strains were grown at 30°C in LB, minimal medium (MM) (17), or M9 medium (18) with 2.5 mM 3-hydroxybenzoate or gentisate as the sole source of carbon. The primer sequences used for PCR are available upon request.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 ΔlacU169 (ϕ80dlacZΔM15) recA1 endA1 hsdR17 thi-1 gyrA96 relA1 | Novagen |
| BL21(DE3) | F− ompT hsdS (rB− mB−) gal dcm lacY1(DE3) | Novagen |
| S17-1 | Donor strain for conjugation, Tpr Smr recA hsdR thi pro RP4-2-Tc::Mu-Km::Tn7 λpir | 25 |
| K-12 substrain W3110 | Source of d-malate dehydrogenase gene | 20 |
| P. alcaligenes | ||
| SponMu | Spontaneous mutant of P. alcaligenes NCIMB 9867 | NCIMB |
| SponMuΔhbzF | hbzF gene-disrupted mutant of SponMu (Kmr) | This study |
| SponMuΔhbzG | hbzG gene-disrupted mutant of SponMu (Kmr) | This study |
| SponMu-hbzG::dmlA | Derivative constructed from SponMuΔhbzG in which hbzG was replaced by dmlA | This study |
| Plasmids | ||
| pET28a | Expression vector, Kmr | Novagen |
| pEX18Tc | Tcr sacB+, gene replacement vector with a multiple-cloning site from pUC18 | 23 |
| pTnMod-OKm | Kmr, source of neomycin phosphotransferase II gene (nptII) | 24 |
| pET28a-hbzIJ | Kmr, pET28a derivative for expression of hbzIJ | This study |
| pET28a-hbzG | Kmr, pET28a derivative for expression of hbzG | This study |
| pET28a-dmlA | Kmr, pET28a derivative for expression of dmlA | This study |
| pHbzF::NptII | Tcr Kmr, hbzF gene-knockout vector containing DNA fragments homologous to the upstream and downstream regions of hbzF and nptII from pTnMod-Okm | This study |
| pHbzG::NptII | Tcr Kmr, hbzG gene-knockout vector containing DNA fragments homologous to the upstream and downstream regions of hbzG and nptII from pTnMod-OKm | This study |
| pHbzG::DmlA | Tcr, vector containing DNA fragments homologous to the upstream and downstream regions of hbzG and dmlA from E. coli K-12 for replacement of hbzG by dmlA | This study |
Reagents.
Gentisate sodium salt and d-malate were obtained from Sigma-Aldrich (St. Louis, MO). Maleate, l-malate, and 3-hydroxybenzoate were from Fluka (Buchs, Switzerland). Pyruvate was purchased as its sodium salt from Sinopharm Chemical Reagent Co. (Shanghai, China). Protein purification medium (Ni-Sepharose, high performance) was obtained from GE Healthcare (Uppsala, Sweden).
Construction of recombinant plasmids for overexpression of hbzIJ, hbzG, and dmlA in E. coli.
The entire hbzIJ and hbzG genes were amplified from the genomic DNA of strain SponMu using primer pairs based on the reported hbz cluster (19), and the dmlA gene was amplified from Escherichia coli K-12 (20). NdeI and HindIII sites were incorporated into the respective 5′ ends of the amplified genes described above. These fragments were cloned into pET28a in frame with the His6-coding sequence to produce pET28a-hbzIJ, pET28a-hbzG, and pET28a-dmlA, which resulted in the expression of N-terminal His6-tagged fusion proteins. The plasmids were transformed into E. coli BL21(DE3) as described previously (18).
Protein purification and determination.
E. coli cells carrying pET28a-hbzIJ, pET28a-hbzG, or pET28a-dmlA were grown in LB containing Km at 37°C to an optical density at 600 nm (OD600) of 0.6 and then induced for 4 h by addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C. The cells were harvested by centrifugation (6,000 × g, 8 min, 4°C) and suspended in binding buffer (50 mM sodium phosphate buffer containing 0.5 M sodium chloride and 5% glycerol, pH 7.4), and cell extracts were prepared by sonication in an ice water bath for 60 cycles of 5 s each with a 5-s interval between each cycle, after which the cell debris was removed by centrifugation at 13,000 × g for 1 h at 4°C. The supernatant was used for enzyme assays as well as for protein purification.
All purification procedures were carried out at 4°C. Crude cell extracts were applied to a column containing 4 ml of Ni-Sepharose high-performance affinity beads and washed with 50 ml of binding buffer. The elution buffer was the same as the binding buffer, except that it contained imidazole at a concentration of 120 mM. The eluted fractions were pooled and dialyzed with binding buffer without sodium chloride for removal of imidazole and sodium salts. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed for molecular weight determination.
Enzyme assays.
Maleate hydratase activity was measured from the decrease in the absorption at 240 nm. The reaction mixtures (final volume, 0.5 ml) contained 1 mM maleate, 45 μg of extracts of E. coli cells containing pET28a-hbzIJ, and 50 mM phosphate buffer (pH 7.4). The reference cuvette contained all of these compounds except the substrate, and the assay was initiated by addition of substrate. The molar extinction coefficient of maleate at 240 nm was taken to be 1,900 M−1 cm−1 (11). One unit of maleate hydratase activity was defined as the amount required for the decrease of 1 μmol of maleate per min at 23°C. Specific activities are expressed as units per milligram of protein, and the values are expressed as means ± standard deviations (SDs), calculated from triplicate assays. The protein concentration was determined according to the Bradford method with bovine serum albumin (BSA) as the standard. For kinetic analysis, three independent sets of experiments were performed with at least six substrate concentrations ranging from 50 to 800 μM. Michaelis-Menten kinetics were calculated by nonlinear regression analysis (OriginPro, version 9.1, software). Maleylpyruvate was prepared by oxidation of gentisate, catalyzed by NagI, from Ralstonia sp. strain U2, and maleylpyruvate isomerase activity was determined as described previously (4).
The identity of d-malate from HbzIJ-catalyzed maleate hydration was determined by enzyme assay of the purified DmlA, a d-malate dehydrogenase in E. coli K-12 (12, 21). The enzyme assay of d-malate dehydrogenase was carried out as previously described (22).
HPLC analysis.
High-performance liquid chromatography (HPLC) analysis was carried out with an Agilent series 1200 system (Agilent Technologies, Palo Alto, CA). For the determination of pyruvate or malate, the system was equipped with an Aminex HPX-87H column (300 by 7.8 mm; Bio-Rad, Hercules, CA) running at 30°C with 5 mM H2SO4 as the eluent at a flow rate of 0.6 ml/min. Detection was by UV detection at 210 nm. For the determination of chiral isomers of d/l-malate, an Xtimate C18 column (Welch, Shanghai, China) was employed, running at 35°C with 8 mM copper(II) acetate and 16 mM l-valine (pH 4.5) as the eluent at a flow rate of 1.0 ml/min. Detection was by UV detection at 320 nm.
RT-PCR analysis.
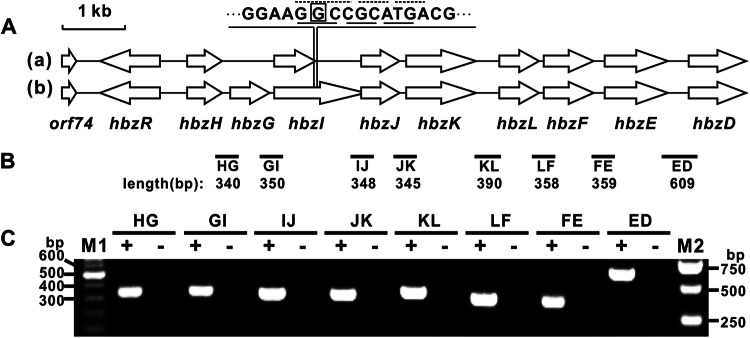
Total RNA of 3-hydroxybenzoate-induced Pseudomonas alcaligenes cells was extracted with an RNAprep pure kit for cells/bacteria (Tiangen, Beijing, China). Removal of DNA contamination and subsequent reverse transcription-PCR (RT-PCR) into cDNA were carried out using a PrimeScript reverse transcriptase reagent kit (TaKaRa, Dalian, China) in accordance with the manufacturer's instructions. The relative location of the primers used in the subsequent PCR are indicated in Fig. 2B, with the resulting cDNA being used as the template and the total RNA from which DNA was removed being the negative control.
FIG 2.
(A) Organization of the hbz gene cluster of strain SponMu compared to the formerly reported one. The arrows indicate the size and direction of transcription of each gene or ORF. The cluster organization in this study (cluster b) is modified from that of a cluster reported previously (19) (cluster a). A new ORF, designated hbzG, was identified, and the termination codon for hbzI was relocated on the basis of the resequencing results obtained in this study. The G in the box at the top was missing from the cluster in the previous report, resulting in a frameshift and early termination of the ORF. The dotted lines above the sequence indicate the false reading frames of hbzI, whereas the solid lines beneath the sequence are the corrected ones. (B) Locations of the primer sets used for RT-PCR and the lengths of the DNA fragments amplified by RT-PCR. (C) Agarose gel electrophoresis of RT-PCR products obtained from total RNA isolated from 3-hydroxybenzoate-induced SponMu cells. The primer pairs used are denoted above the lanes. Lanes +, reactions carried out with reverse transcriptase; lanes −, negative-control reactions carried out without reverse transcriptase; lanes M1 and M2, different ladder DNA markers with fragments of the indicated sizes.
Construction of hbzF- and hbzG-knockout mutant strains and replacement of hbzG by dmlA.
Fragments flanking the upstream and downstream regions of hbzF or hbzG were amplified and had product lengths of 420 to 460 bp. The primers for amplification of the upstream fragments contained EcoRI/KpnI sites at their 5′ ends, and the primers for amplification of the downstream fragments contained KpnI/XbaI sites. The two fragments were cloned into the gene replacement vector pEX18Tc (23) by three-fragment ligation, and the nptII (from pTnMod-OKm [24]) or dmlA gene cassette was subsequently inserted into the KpnI site between the two fragments, resulting in pHbzG::NptII, pHbzF::NptII, and pHbzG::DmlA, where the entire target genes were replaced by either nptII or dmlA. The constructed plasmids were transformed into the mobilizer strain E. coli S17-1 (25) before being conjugated into the recipient strain by biparental mating as described previously (26).
For hbzF- and hbzG-knockout strain selection, Km-resistant strains were screened on an LB plate containing Km, followed by the selection of double-crossover recombinants on sucrose plates (26). The resulting strains, SponMuΔhbzF and SponMuΔhbzG, were confirmed on the basis of their kanamycin resistance and by PCR analysis. Strain SponMu-hbzG::dmlA was constructed and is based on strain SponMu ΔhbzG, which is not able to grow on 3-hydroxybenzaoate or gentisate. The nptII gene of SponMuΔhbzG was replaced by dmlA through biparental mating, and strain SponMu-hbzG::dmlA was selected on an MM plate containing 3-hydroxybenzoate as the sole source of carbon, following the selection of double-crossover recombinants on sucrose plates.
Growth analysis of Pseudomonas alcaligenes strains.
Strain SponMu and its derivatives were grown in LB overnight and harvested when the OD600 reached 0.6. Washed cells were inoculated into M9 medium containing 2.5 mM gentisate or 3-hydroxybenzoate as the sole source of carbon. The growth of the strains was measured on a Bioscreen C plate reader system (Labsystems, Helsinki, Finland) by monitoring the optical density at 600 nm. The growth curves were fitted by the modified Gompertz equation, as described previously (27).
Biotransformation of maleate by permeabilized cells of Pseudomonas alcaligenes strains.
Pseudomonas alcaligenes strains were grown at 30°C in 300 ml LB containing 5 mM 3-hydroxybenzoate and harvested when the OD600 reached 0.6. The permeabilization was carried out by resuspending the cells to a final volume of 7 ml in 50 mM phosphate buffer (pH 7.6) with 0.5% (vol/vol) Triton X-100, as described previously (28). The resulting permeabilized cells were used to catalyze the transformation of maleate. The reaction system was in 50 mM phosphate buffer (pH 7.6) containing 10 μmol of maleate, 0.25 ml of permeabilized cells, and 50 μmol of NAD+, with the final volume being 9 ml. At each testing point, 0.9 ml of the reaction mixture was taken and the reaction was terminated by adding 0.1 ml of 6 M HCl. Quantitative analysis of the generated d-malate was performed by HPLC, as described above.
RESULTS
Description of phenotype and genotype of spontaneous mutant strain SponMu.
Previously, two gene clusters, xln (GenBank accession no. AF173167) (29) and hbz (GenBank accession no. DQ394580) (19), were reported to be involved in gentisate catabolism in strain NCIMB 9867, which was identified to be an m-cresol and 2,5- and 3,5-xylenol utilizer (30). Additionally, it was reported that strain NCIMB 9867 could spontaneously mutate to lose its ability to utilize m-cresol or 2,5- or 3,5-xylenol but still retain the ability to grow on 3-hydroxybenzoate and gentisate (9). In this study, the revived culture obtained from NCIMB exhibited the same growth phenotype as the mutant mentioned above, and so did the culture of the 2nd purchase. In this spontaneous mutant, the lack of the xln cluster was confirmed by PCR, suggesting the possible involvement of this cluster in the catabolic steps in the degradation of m-cresol and xylenols. This mutant strain is referred to as SponMu in this study.
Sequence reanalysis of the hbz cluster.
In the previously reported hbz cluster of P. alcaligenes NCIMB 9867 (19), two genes were identified to be involved in gentisate metabolism. hbzE encodes a strictly inducible gentisate 1,2-dioxygenase (19), and hbzF encodes a maleylpyruvate hydrolase which catalyzes the direct hydrolysis of maleylpyruvate without prior isomerization (10). Here, an additional open reading frame (ORF), designated hbzG, was identified between hbzH and hbzI, as shown in Fig. 2A. Besides, reanalysis of this cluster found that the previously annotated hbzI was unexpectedly terminated, resulting in the formation of a polypeptide shorter than its homologs. Resequencing of this region indicated that hbzI was actually 1,443 bp in length (and encoded a polypeptide of 480 amino acids) rather than the previously reported 681 bp. This difference was apparently due to a sequencing error in which a guanine (G) at position 4292 was missed (Fig. 2A). During the transcription study of this cluster, correctly sized amplified products were detected by RT-PCR, as shown in Fig. 2B and C. This indicates that hbzHGIJKLFED are cotranscribed in a single polycistronic mRNA.
Among the newly identified ORFs, HbzG exhibits 46% identity with NagL from Ralstonia sp. strain U2, which is a typical GSH-dependent maleylpyruvate isomerase catalyzing the isomerization of maleylpyruvate to fumarylpyruvate (4). On the other hand, tandemly located hbzI and hbzJ genes are deduced to encode two polypeptides of 51.1 and 24.4 kDa, respectively, and share significant identity (up to 64%) with the large and small subunits of 3-isopropylmalate dehydratase (or isopropylmalate isomerase; EC 4.2.1.33). Coincidentally, their sizes are similar to those of the pair of large and small subunits (57 and 24 kDa) of previously purified maleate hydratase (sequence unknown) from Pseudomonas alcaligenes NCIMB 9867 (11).
HbzIJ catalyzed the hydration of maleate.
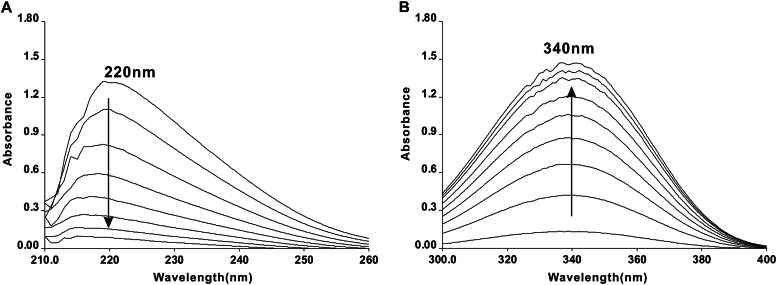
Maleate is the product of direct hydrolysis of maleylpyruvate, which is catalyzed by HbzF in strain NCIMB 9867 (10). However, the catabolic steps of maleate via the direct hydrolysis route of gentisate still remain unresolved. A previous study revealed that the absorption of the saturated hydroxyacid (malate) was negligible compared with the absorption of the unsaturated acid (maleate) (11). Herein, hydratase activity was measured by monitoring the decrease in the absorption of the substrate maleate. As shown in Fig. 3A, the spectrophotometric changes that occurred during the reaction indicated that the cell extract of E. coli BL21(DE3) carrying pET28a-hbzIJ was able to catalyze the transformation of maleate. The specific activity was 2.12 ± 0.05 U/mg, and the Km value for maleate was 0.707 ± 0.068 mM. However, no transformation occurred when fumarate, the cis-trans isomer of maleate, was used as the substrate. Additionally, no activity against maleate was detected during the same procedure performed with cell extracts of E. coli BL21(DE3) harboring pET28a with no insert. Nevertheless, HbzIJ exhibited no activity to catalyze the isomerization of α-isopropylmalate, determined as described previously (14).
FIG 3.
(A) Spectrophotometric changes during the transformation of maleate catalyzed by cell extracts of E. coli BL21(DE3) carrying pET28a-hbzIJ. Before the reaction started, 45 μg of cell extract was added to both the sample and the reference cuvettes containing 50 mM phosphate buffer (pH 7.4). The reaction was initiated by addition of 400 μM maleate. The spectra were recorded every minute after the addition of cell extracts. (B) Subsequent changes after the addition of 50 μg of purified d-malate dehydrogenase (DmlA), 1 mM DTT, 0.4 mM Mn2+, and 1 mM NAD+ into the sample and reference cuvettes after the reaction described for panel A. The formation of NADH (maximum λ, 340 nm) indicates the presence of d-malate in the product from HbzIJ-catalyzed maleate hydration.
d-Malate is the product of maleate hydration catalyzed by HbzIJ.
As an intermediate of the tricarboxylic acid (TCA) cycle, l-malate exists in all living cells. However, as its chiral isomer, d-malate is generally considered the unnatural form of malate. The genetic determinant of d-malate metabolism had remained unclear until recent years. d-Malate dehydrogenase (DmlA) from E. coli K-12 is a d-isomer-specific enzyme and catalyzes the dehydrogenation (decarboxylating) of d-malate to pyruvate and CO2, accompanied by the formation of NADH from NAD+ (12, 21). Thus, purified DmlA (as shown in Fig. 4B) was applied for the detection of d-malate in the products of HbzIJ-catalyzed maleate transformation. When DmlA and NAD+ were added to the products, the generation of NADH (maximum λ, 340 nm) was observed, as shown in Fig. 3B, indicating the presence of d-malate. Meanwhile, pyruvate was found by HPLC analysis to be the product from d-malate dehydrogenation. This result not only confirmed the formation of d-malate from maleate hydration but also suggested that d-malate dehydrogenase may catalyze the last step of the direct hydrolysis route of the gentisate pathway.
FIG 4.
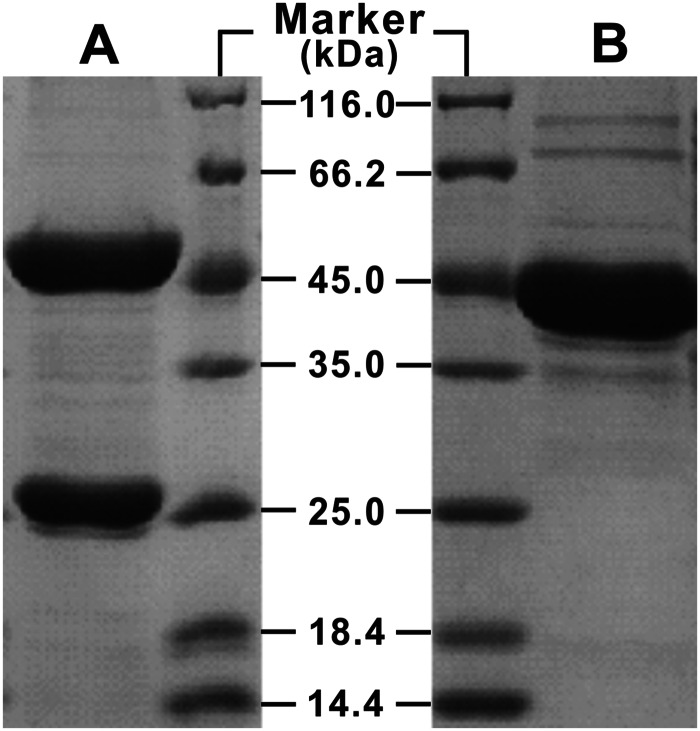
SDS-PAGE of purified His6-HbzIJ and His6-DmlA. (A) Purified His6-HbzIJ (3.4 μg); (B) purified His6-DmlA (3.9 μg). Molecular mass standards (in kilodaltons) are indicated in the lanes labeled Marker.
Quantitative analysis of d-malate in the HbzIJ-catalyzed hydration of maleate was carried out by use of d-malic acid and l-malic acid assay kits (Megazyme, Wicklow, Ireland) and HPLC. One milliliter of a reaction mixture containing 5 mM maleate and 90 μg of cell extracts of E. coli BL21(DE3)(pET28a-hbzIJ) was incubated at 30°C for 30 min. The reaction product was diluted 20 times to meet the detection limit of the kits. Five replicate tests with the d-malic acid assay kit indicated that 5.015 ± 0.016 mM d-malate was formed during the hydration of 5 mM maleate catalyzed by HbzIJ, and no l-malate was detected by the l-malic acid assay kit. HPLC analysis distinguished these two chiral isomers, with retention times of 8.7 min for l-malate and 12.9 min for d-malate being determined. A standard curve of the d-malate concentration was generated from a series of concentrations ranging from 0.2 to 1.2 mM d-malate. When 1 mM maleate was transformed by HbzIJ, the concentration of d-malate in the product was determined to be 0.993 mM with reference to the standard curve. Thus, both the enzyme assay and HPLC analysis revealed that HbzIJ catalyzes the stoichiometric conversion of maleate to enantiomerically pure d-malate.
Purification of HbzIJ.
HbzIJ belongs to the aconitase superfamily (NCBI Conserved Domain Database accession no. cl00285). Proteins in this superfamily usually require a 4Fe-4S iron-sulfur cluster in their active form. The constructed plasmid pET28a-hbzIJ was able to express N-terminus-tagged fusion protein His6-HbzI and HbzJ with no tag, and both His6-HbzI and HbzJ were collected in the elution fractions, as shown in the SDS-polyacrylamide gel in Fig. 4A. However, the purified pale brown apoenzyme lost its activity, probably because of the oxidation of the 4Fe-4S cluster. It has been reported that certain apoforms of 4Fe-4S cluster-containing proteins can be activated in vitro by the addition of S2− and Fe2+ (31). In this study, 1.1 mg of purified HbzIJ was mixed with 3 mM dithiothreitol (DTT), 1.5 mM Fe(NH4)2(SO4)2, and Na2S in a final volume of 7 ml. The mixture was sealed and placed in an ice water bath for 2 h. Its hydration activity of 0.655 ± 0.036 U/mg was then restored.
Alternative routes of the gentisate pathway in strain SponMu.
No bacterial strain has been reported to have both the isomerization and hydrolysis routes of the gentisate pathway. In addition to identification of the maleylpyruvate hydrolase gene (hbzF) in strain NCIMB 9867 (10), HbzG expressed from E. coli BL21(DE3) harboring pET28a-hbzG was demonstrated in this study to be a typical GSH-dependent maleylpyruvate isomerase with a specific activity of 0.026 U/mg, calculated as previously described (32). Biotransformation of maleylpyruvate was carried out to investigate whether both enzymes were simultaneously functional in strain SponMu. When 40 μM maleylpyruvate was transformed by the cell extracts of strain SponMu, both maleate and fumarate were detected by HPLC at respective concentrations of 8.02 μM and 10.74 μM, which indicated that maleylpyruvate could be transformed simultaneously via both hydrolysis and isomerization in this strain, as illustrated in Fig. 1.
To investigate the physiological role of maleylpyruvate hydrolase (HbzF) and maleylpyruvate isomerase (HbzG) in the gentisate catabolism of this strain, hbzF- and hbzG-disrupted strains of SponMu, designated strain SponMuΔhbzF and strain SponMuΔhbzG, respectively, were constructed. During measurement of their growth, as shown in Fig. 5, strain SponMuΔhbzG was no longer able to grow in M9 medium with gentisate as the sole carbon source, while strain SponMuΔhbzF retained this ability. Meanwhile, maleylpyruvate hydrolase was still detectable in cell extracts of strain SponMuΔhbzG with a specific activity of 0.121 ± 0.019 U/mg, suggesting that the disruption of hbzG did not affect the expression of maleylpyruvate hydrolase HbzF. This result indicates that the maleylpyruvate isomerization route is a complete pathway for gentisate catabolism in strain SponMu, whereas the direct hydrolysis route is not.
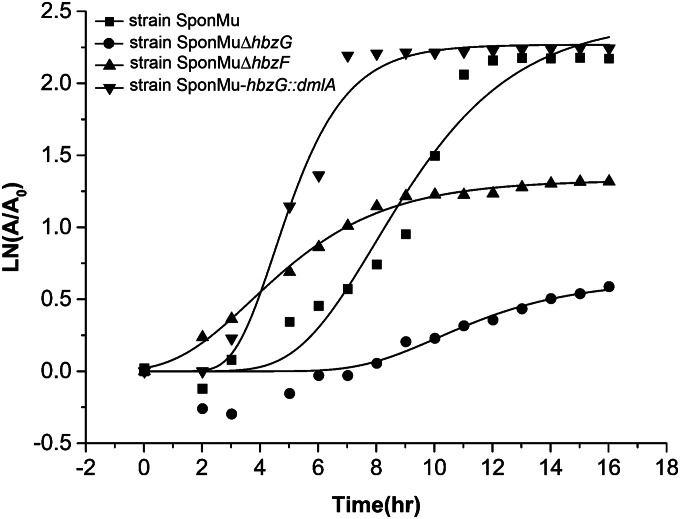
FIG 5.
Growth curves of P. alcaligenes SponMu and its mutant derivatives in liquid M9 medium containing 2.5 mM gentisate as the sole source of carbon. Growth was monitored by reading the optical density at 600 nm every 1 h over a period of 16 h on a Bioscreen C system. The growth curves were fitted by the modified Gompertz equation (27) with OriginPro (version 9.1) software, and all points represent the mean values from triplicate trials. LN, natural logarithm; A, OD600 value of the cells; A0, initial OD600 value of the cells. The maximum specific growth rates (μmax) for strains SponMu, SponMuΔhbzG, SponMuΔhbzF, and SponMu-hbzG::dmlA were 0.33, 0.09, 0.19, and 0.60, respectively.
The introduction of a d-malate dehydrogenase gene confers to strain SponMu ΔhbzG the ability to grow on gentisate.
On the basis of the observations presented above, it is rational to speculate that the d-malate dehydrogenase catalyzing the dehydrogenation of d-malate to pyruvate, which is likely the final step of the direct hydrolysis route of the gentisate pathway before entering the TCA cycle, is not present in strain SponMu, as shown in Fig. 1. A newly constructed strain, SponMu-hbzG::dmlA, in which hbzG (encoding maleylpyruvate isomerase) was replaced by dmlA (encoding d-malate dehydrogenase from E. coli K-12), was able to grow on gentisate, as shown in Fig. 5. This result confirmed our hypothesis that the absence of the d-malate dehydrogenase in this strain resulted in its inability to utilize gentisate via the direct hydrolysis route.
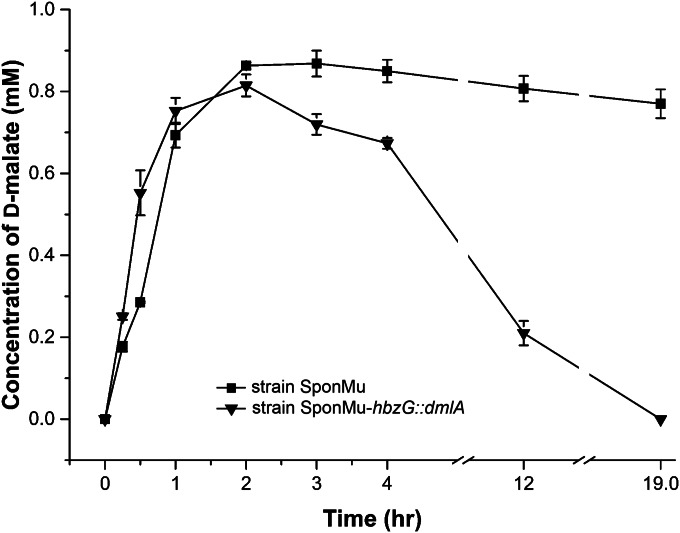
d-Malate accumulation from maleate in strains SponMu and SponMu-hbzG::dmlA.
The permeabilized cells of strains SponMu and SponMu-hbzG::dmlA were employed as catalytic units to produce d-malate from maleate as described previously (28). As shown in Fig. 6, in both strains the maximum production of d-malate was achieved during the initial period (about 2 h). Subsequently, d-malate was gradually assimilated by strain SponMu-hbzG::dmlA, and no d-malate was detected at 19 h. However, no significant decrease in the amount of d-malate was observed in strain SponMu during the same period. This biochemical evidence clearly indicates that the introduction of dmlA into strain SponMu prevents this strain from accumulating d-malate.
FIG 6.
d-Malate accumulation from maleate in strains SponMu and SponMu-hbzG::dmlA. The transformation of 1 mM maleate was catalyzed by permeabilized cells of strains SponMu and SponMu-hbzG::dmlA. The reaction conditions and sample management are described in Materials and Methods. Triple replicates were applied for each reaction. Data are presented as means ± SDs. The concentration of d-malate was monitored by HPLC over a period of 19 h.
DISCUSSION
In addition to the well-characterized isomerization route of the gentisate pathway, a direct hydrolysis route of maleylpyruvate was also reported to exist in some Pseudomonas strains (1, 8, 33). Recently, a novel hbzF gene was identified to encode maleylpyruvate hydrolase in Pseudomonas alcaligenes NCIMB 9867, catalyzing the hydrolysis of maleylpyruvate to form maleate and pyruvate (10). However, maleate is not an intermediate of the central metabolism pathway in bacteria. Thus, whether maleate is metabolized in the direct hydrolysis route of the gentisate pathway is a key question to be answered. In some maleate-assimilating strains, maleate could be converted into fumarate by maleate isomerase (34, 35), and this reaction was also the last step of the nicotinic acid pathway in Pseudomonas putida KT2440 (36) and the nicotine pathway in Pseudomonas putida S16 (37), while in strain NCIMB 9867 it was reported that a maleate hydratase catalyzed the metabolism of maleate generated from gentisate, resulting in the formation of d-malate (1). Maleate hydratase was initially reported to exist in corn kernels (38) or mammalian kidneys (39). This enzyme was also once purified from Pseudomonas alcaligenes NCIMB 9867 (11) and Arthrobacter sp. strain MCI2612 (40), exhibiting large and small subunits on SDS-PAGE. Notably, an enzyme with a broad specificity from the archaeon Methanocaldococcus jannaschii, consisting of large and small subunits (MJ0499 and MJ1277), was reported to catalyze both isopropylmalate isomerization and maleate hydration. Their encoding genes were located separately from each other in the genome (13). However, in this study, we demonstrate that tandemly located hbzIJ encodes maleate hydratase, which catalyzes maleate hydration to stoichiometrically form enantiomerically pure d-malate but which has no isopropylmalate isomerase activity. As an unnatural form, enantiomerically pure d-malate may potentially be applied as a precursor in the synthesis of a wide variety of pharmaceutical and chemical compounds. The identification of the maleate hydratase gene in a gentisate catabolic cluster not only enhances our knowledge of the direct hydrolysis route of the gentisate pathway but also enables the construction of an engineered bacterial strain with an optimized pathway for the production of d-malate from 3-hydroxybenzoate through the gentisate pathway or directly from maleate hydration. It was observed that purified HbzIJ lost the ability to catalyze maleate hydration, but its activity could be restored after reconstitution of the 4Fe-4S cluster. However, the purified maleate hydratase, which consisted of two subunits (57 and 24 kDa) from native strain NCIMB 9867, was reported to require no cofactor for full activity (11). It was not known whether this discrepancy was caused by its expression in E. coli instead of Pseudomonas alcaligenes or the difference in the purification procedure. Since the previous study did not report the sequence information for the polypeptide of this purified enzyme, it cannot be confirmed that HbzIJ was actually the previously purified maleate hydratase.
Spontaneous mutant strain SponMu, which lacks the xln cluster, could not grow in m-cresol or 2,5- or 3,5-xylenol. However, the remaining hbz cluster enabled us to study the role of the isomerization route and direct hydrolysis route of the gentisate pathway in this strain. Disruption of maleylpyruvate isomerase-encoding gene hbzG in strain SponMu resulted in the loss of the ability to grow in 3-hydroxybenzoate or gentisate, and the introduction of the d-malate dehydrogenase-encoding gene in the derivative restored its growth ability. In this case, we conclude that, in strain SponMu, gentisate is still metabolized via the isomerization route and the direct hydrolysis route plays no role in gentisate assimilation because of the lack of d-malate dehydrogenase. It was also reported that strain NCIMB 9867 was unable to grow on MM with maleate as the sole source of carbon, possibly due to a lack of its transporter (15). Here, we demonstrate that the lack of d-malate dehydrogenase in the strain could be the decisive reason for this.
It was reported that when permeabilized NCIMB 9867 cells were incubated with 15 mM maleate under anaerobic conditions, maleate was degraded within 1 h with the stoichiometric formation of d-malate, and d-malate consumption was not observed in 5 h (15). In this study, we have provided an adequate explanation for this interesting phenomenon by monitoring the difference in the accumulation and assimilation of d-malate between strain SponMu and its derivative into which the d-malate dehydrogenase gene dmlA was introduced. The deficiency in d-malate assimilation in strain SponMu coincidently enables the remarkable production of d-malate from maleate without further degradation.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (31400028 and 31271333) and the China Postdoctoral Science Foundation (2014M551403).
REFERENCES
- 1.Hopper DJ, Chapman PJ, Dagley S. 1968. Enzymic formation of d-malate. Biochem J 110:798–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lack L. 1959. The enzymic oxidation of gentisic acid. Biochim Biophys Acta 34:117–123. doi: 10.1016/0006-3002(59)90239-2. [DOI] [PubMed] [Google Scholar]
- 3.Lack L. 1961. Enzymic cis-trans isomerization of maleylpyruvic acid. J Biol Chem 236:2835–2840. [PubMed] [Google Scholar]
- 4.Zhou NY, Fuenmayor SL, Williams PA. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J Bacteriol 183:700–708. doi: 10.1128/JB.183.2.700-708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng J, Che YS, Milse J, Yin YJ, Liu L, Ruckert C, Shen XH, Qi SW, Kalinowski J, Liu SJ. 2006. The gene ncgl2918 encodes a novel maleylpyruvate isomerase that needs mycothiol as cofactor and links mycothiol biosynthesis and gentisate assimilation in Corynebacterium glutamicum. J Biol Chem 281:10778–10785. doi: 10.1074/jbc.M513192200. [DOI] [PubMed] [Google Scholar]
- 6.Liu TT, Xu Y, Liu H, Luo S, Yin YJ, Liu SJ, Zhou NY. 2011. Functional characterization of a gene cluster involved in gentisate catabolism in Rhodococcus sp. strain NCIMB 12038. Appl Microbiol Biotechnol 90:671–678. doi: 10.1007/s00253-010-3033-1. [DOI] [PubMed] [Google Scholar]
- 7.Liu T-T, Zhou N-Y. 2012. Novel l-cysteine-dependent maleylpyruvate isomerase in the gentisate pathway of Paenibacillus sp. strain NyZ101. J Bacteriol 194:3987–3994. doi: 10.1128/JB.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayly RC, Chapman PJ, Dagley S, Diberardino D. 1980. Purification and some properties of maleylpyruvate hydrolase and fumarylpyruvate hydrolase from Pseudomonas alcaligenes. J Bacteriol 143:70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poh CL, Bayly RC. 1980. Evidence for isofunctional enzymes used in m-cresol and 2,5-xylenol degradation via the gentisate pathway in Pseudomonas alcaligenes. J Bacteriol 143:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu K, Liu TT, Zhou NY. 2013. HbzF catalyzes direct hydrolysis of maleylpyruvate in the gentisate pathway of Pseudomonas alcaligenes NCIMB 9867. Appl Environ Microbiol 79:1044–1047. doi: 10.1128/AEM.02931-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Werf MJ, van den Tweel WJJ, Hartmans S. 1993. Purification and characterization of maleate hydratase from Pseudomonas pseudoalcaligenes. Appl Environ Microbiol 59:2823–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reed JL, Patel TR, Chen KH, Joyce AR, Applebee MK, Herring CD, Bui OT, Knight EM, Fong SS, Palsson BO. 2006. Systems approach to refining genome annotation. Proc Natl Acad Sci U S A 103:17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drevland RM, Waheed A, Graham DE. 2007. Enzymology and evolution of the pyruvate pathway to 2-oxobutyrate in Methanocaldococcus jannaschii. J Bacteriol 189:4391–4400. doi: 10.1128/JB.00166-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross SR, Burns RO, Umbarger HE. 1963. The biosynthesis of leucine. II. The enzymic isomerization of β-carboxy-β-hydroxyisocaproate and α-hydroxy-β-carboxyisocaproate. Biochemistry 2:1046–1052. [DOI] [PubMed] [Google Scholar]
- 15.van der Werf M, van den Tweel WJJ, Hartmans S. 1992. Screening for microorganisms producing d-malate from maleate. Appl Environ Microbiol 58:2854–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Werf MJ, Huybers P, van den Tweel WJJ, Hartmans S. 1997. Induction of maleate hydratase in Pseudomonas pseudoalcaligenes. World J Microbiol Biotechnol 13:279–282. doi: 10.1023/A:1018526822535. [DOI] [Google Scholar]
- 17.Liu H, Wang SJ, Zhou NY. 2005. A new isolate of Pseudomonas stutzeri that degrades 2-chloronitrobenzene. Biotechnol Lett 27:275–278. doi: 10.1007/s10529-004-8293-3. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 19.Yeo CC, Tan CL, Gao XL, Zhao B, Poh CL. 2007. Characterization of hbzE-encoded gentisate 1,2-dioxygenase from Pseudomonas alcaligenes NCIMB 9867. Res Microbiol 158:608–616. doi: 10.1016/j.resmic.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Bachmann BJ. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev 36:525–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukas H, Reimann J, Kim OB, Grimpo J, Unden G. 2010. Regulation of aerobic and anaerobic d-malate metabolism of Escherichia coli by the LysR-type regulator DmlR (yeaT). J Bacteriol 192:2503–2511. doi: 10.1128/JB.01665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tipton PA, Peisach J. 1990. Characterization of the multiple catalytic activities of tartrate dehydrogenase. Biochemistry 29:1749–1756. doi: 10.1021/bi00459a013. [DOI] [PubMed] [Google Scholar]
- 23.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 24.Dennis JJ, Zylstra GJ. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of Gram-negative bacterial genomes. Appl Environ Microbiol 64:2710–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 26.Schweizer HP. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol 6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 27.Zwietering MH, Jongenburger I, Rombouts FM, van't Riet K. 1990. Modeling of the bacterial growth curve. Appl Environ Microbiol 56:1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werf MJ, Hartmans S, Tweel WJJ. 1995. Permeabilization and lysis of Pseudomonas pseudoalcaligenes cells by Triton X-100 for efficient production of d-malate. Appl Microbiol Biotechnol 43:590–594. doi: 10.1007/s002530050456. [DOI] [Google Scholar]
- 29.Yeo CC, Wong MVM, Feng YM, Song KP, Poh CL. 2003. Molecular characterization of an inducible gentisate 1,2-dioxygenase gene, xlnE, from Pseudomonas alcaligenes NCIMB 9867. Gene 312:239–248. doi: 10.1016/S0378-1119(03)00619-X. [DOI] [PubMed] [Google Scholar]
- 30.Hopper DJ, Chapman PJ. 1971. Gentisic acid and its 3-methyl-substituted and 4-methyl-substituted homologues as intermediates in bacterial degradation of meta-cresol, 3,5-xylenol and 2,5-xylenol. Biochem J 122:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malkin R, Rabinowi JC. 1966. Reconstitution of clostridial ferredoxin. Biochem Biophys Res Commun 23:822–827. doi: 10.1016/0006-291X(66)90561-4. [DOI] [PubMed] [Google Scholar]
- 32.Hagedorn SR, Bradley G, Chapman PJ. 1985. Glutathione-independent isomerization of maleylpyruvate by Bacillus megaterium and other Gram-positive bacteria. J Bacteriol 163:640–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain RK. 1996. The molecular cloning of genes specifying some enzymes of the 3,5-xylenol degradative pathway. Appl Microbiol Biotechnol 45:502–508. doi: 10.1007/BF00578462. [DOI] [Google Scholar]
- 34.Hatakeyama K, Asai Y, Uchida Y, Kobayashi M, Terasawa M, Yukawa H. 1997. Gene cloning and characterization of maleate cis-trans isomerase from Alcaligenes faecalis. Biochem Biophys Res Commun 239:74–79. doi: 10.1006/bbrc.1997.7430. [DOI] [PubMed] [Google Scholar]
- 35.Fisch F, Fleites CM, Delenne M, Baudendistel N, Hauer B, Turkenburg JP, Hart S, Bruce NC, Grogan G. 2010. A covalent succinylcysteine-like intermediate in the enzyme-catalyzed transformation of maleate to fumarate by maleate isomerase. J Am Chem Soc 132:11455–11457. doi: 10.1021/ja1053576. [DOI] [PubMed] [Google Scholar]
- 36.Jimenez JI, Canales A, Jimenez-Barbero J, Ginalski K, Rychlewski L, Garcia JL, Diaz E. 2008. Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc Natl Acad Sci U S A 105:11329–11334. doi: 10.1073/pnas.0802273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang H, Yao Y, Wang L, Yu H, Ren Y, Wu G, Xu P. 2012. Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci Rep 2:377. doi: 10.1038/srep00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sacks W, Jensen CO. 1951. Malease, a hydrase from corn kernels. J Biol Chem 192:231–236. [PubMed] [Google Scholar]
- 39.Englard S, Britten JS, Listowsky I. 1967. Stereochemical course of the maleate hydratase reaction. J Biol Chem 242:2255–2259. [PubMed] [Google Scholar]
- 40.Ueda M, Asano Y, Yamada H. 1993. Purification and characterization of maleate hydratase from Arthrobacter sp. strain MCI2612. Biosci Biotechnol Biochem 57:1545–1548. doi: 10.1271/bbb.57.1545. [DOI] [Google Scholar]