Abstract
Nucleic acid amplification technique (NAT)-based assays (referred to here as NAT assays) are increasingly used as an alternative to culture-based approaches for the detection of mycoplasma contamination of cell cultures. Assay features, like the limit of detection or quantification, vary widely between different mycoplasma NAT assays. Biological reference materials may be useful for harmonization of mycoplasma NAT assays. An international feasibility study included lyophilized preparations of four distantly related mycoplasma species (Acholeplasma laidlawii, Mycoplasma fermentans, M. orale, M. pneumoniae) at different concentrations which were analyzed by 21 laboratories using 26 NAT assays with a qualitative, semiquantitative, or quantitative design. An M. fermentans preparation was shown to decrease the interassay variation when used as a common reference material. The preparation was remanufactured and characterized in a comparability study, and its potency (in NAT-detectable units) across different NATs was determined. The World Health Organization (WHO) Expert Committee on Biological Standardization (ECBS) established this preparation to be the “1st World Health Organization international standard for mycoplasma DNA for nucleic acid amplification technique-based assays designed for generic mycoplasma detection” (WHO Tech Rep Ser 987:42, 2014) with a potency of 200,000 IU/ml. This WHO international standard is now available as a reference preparation for characterization of NAT assays, e.g., for determination of analytic sensitivity, for calibration of quantitative assays in a common unitage, and for defining regulatory requirements in the field of mycoplasma testing.
INTRODUCTION
Contamination of eukaryotic cell cultures with species from the bacterial class Mollicutes (its trivial name, mycoplasmas, is used throughout this report) may introduce changes in cell metabolism and phenotype. Mycoplasmas are small bacteria (diameter, 0.2 μm) that lack a cell wall and that may pass through bacterial filters. Mycoplasma contamination of cell cultures may affect both the scientific results of cell culture-based research and the quality of biological medicines manufactured by cell culture in the biopharmaceutical industry (1, 2). Biologics, such as vaccines, recombinant proteins, or monoclonal antibodies, are produced in bioreactors, and mycoplasma contamination significantly impacts product quality and safety, with major economic consequences. Regulations in different parts of the world require the testing of cell banks (master cell banks, working cell banks) for the presence of viable mycoplasma as well as downstream cell cultures prior to harvest (unprocessed bulk) (3–7).
Historically, international (e.g., European Union and U.S.) regulations on the evaluation of potential mycoplasma growth have been based on the inoculation of test samples in parallel on solid agar media and in liquid enrichment media favorable for slowly growing strains of mycoplasma, which are subsequently tested on solid agar. Mycoplasmas grown on solid agar usually form colonies with a typical fried egg shape. An additional mycoplasma test includes the use of so-called indicator cell lines necessary for the propagation of potential noncultivable mycoplasma strains which need living eukaryotic cells for their growth. After incubation, the indicator cells are fixed and stained with a DNA-binding fluorescent dye and analyzed under a fluorescence microscope. Here, mycoplasmas are identified by detection of their characteristic particulate or filamentous pattern on the cell surface.
Even though the culture-based detection methods described in the various pharmacopoeias are quite sensitive, as defined in product-specific test validation studies using suitable reference strains as test organisms, these tests need up to 28 days until a final result is obtained. With the development of specific and sensitive test systems based on nucleic acid amplification techniques (NATs), the question of whether NAT-based assays (referred to here as NAT assays) could replace the traditional mycoplasma detection systems arose (8–15). Since 2007, the European Pharmacopoeia (Ph. Eur.) has allowed the traditional mycoplasma detection tests to be replaced by suitably validated NAT assays. In legislation enacted elsewhere, NAT assays are accepted as alternatives to traditional tests, if they are of sufficient sensitivity and properly validated. In section 2.6.7 of the Ph. Eur., the minimal sensitivity of an NAT is defined to correspond to 10 CFU/ml for replacement of the culture-based assays or to 100 CFU/ml for replacement of the indicator cell culture method (5), whereas in the corresponding chapter (chapter 63) of the United States Pharmacopeia, the sensitivity required for a validated NAT-based assay to replace the compendial culture methods is not specified (6). Ph. Eur. section 2.6.7 also elaborates the requirements for the validation of NAT assays, including detailed recommendations for studies regarding specificity, sensitivity (limit of detection), robustness, and comparability studies. These requirements have been implemented by different NAT assays (16–19). For sensitivity, the Ph. Eur. states that “an equivalent limit of detection in terms of the number of copies of mycoplasma nucleic acid in the test sample (using suitable reference standard of mycoplasma nucleic acid)” may be chosen (5). In related fields of mandatory testing of biologicals, the Ph. Eur. defines a minimal NAT sensitivity in international units (IU) per milliliter, on the basis of the respective World Health Organization (WHO) international standards (ISs); e.g., the minimal sensitivity is 100 IU/ml for NAT assays for hepatitis C virus performed on human plasma pools used for manufacturing of plasma derivatives (20). The project to establish an IS for mycoplasma DNA detection assays was endorsed by the WHO Expert Committee on Biological Standardization (ECBS) in October 2010.
The standardization project described here was coordinated by the Paul-Ehrlich-Institut (PEI; Langen, Germany), a WHO Collaborating Centre for in vitro diagnostics (IVDs), blood and blood products, as well as vaccines. The project consisted of two phases. The first phase investigated the feasibility of standardization of generic mycoplasma detection by a wide range of current NAT assays of different designs. In the feasibility study, eight preparations of four distantly related mycoplasma species, i.e., Acholeplasma laidlawii, Mycoplasma fermentans, Mycoplasma orale, and Mycoplasma pneumoniae, were evaluated to see whether there was an improvement in reporting between laboratories and the various assays available worldwide. As a result of this feasibility study, a mycoplasma species was to be selected as the most suitable species as the candidate WHO IS. The second phase (comparability study) was to assign a value (in international units) to the lyophilized candidate reference preparation on the basis of the mean number of NAT-detectable units determined by a range of NAT assays of different generic detection designs shown to be proficient in the feasibility study.
MATERIALS AND METHODS
Mycoplasma preparations.
The type strains of four mycoplasma species representing distant phylogenetic branches within the bacterial class Mollicutes were selected for inclusion in the feasibility study panels: Mycoplasma fermentans (PG18T, NCTC 10117), Mycoplasma orale (CH19299T, NCTC 10112), Mycoplasma pneumoniae (FHT, NCTC 10119), and Acholeplasma laidlawii (PG8T, NCTC 10116). For cultivation of the mycoplasma strains, Mycosafe Friis medium (Mycosafe Diagnostics GmbH, Vienna, Austria), which had been tested and found to be negative for mycoplasma DNA, was used. The different strains were grown to a target concentration of approximately 105 CFU/ml (M. fermentans, M. orale, and M. pneumoniae) or 107 CFU/ml (A. laidlawii) and harvested during the exponential growth phase. An aliquot of each preparation was additionally diluted 1:100 using Mycosafe Friis medium as a diluent. The actual number of CFU was determined for both the neat and the 1:100-diluted preparations (harvest) and again before and after lyophilization (Table 1).
TABLE 1.
Cell titers and mycoplasma DNA concentrations in mycoplasma culture preparationsa
| Preparation and Mycoplasma species or medium | Panel member no. | Total cell titer (no. of CFU/ml) |
Mycoplasma DNA |
|||||
|---|---|---|---|---|---|---|---|---|
| No. of copies/ml |
Mean estimated no. of NAT-detectable units/ml (95% confidence interval) after lyo | |||||||
| Target | Harvest | Before lyo | After lyo | Before lyo | After lyo | |||
| Feasibility study panel | ||||||||
| Acholeplasma laidlawii | 1 | 1.0E+05 | 0.86E+05 | 0.69E+05 | 6.90E+02 | 4.77E+05 | 4.08E+05 | |
| 9 | 1.0E+07 | 0.98E+07 | 0.33E+07 | 5.45E+04 | 7.02E+07 | 4.03E+07 | 1.04E+07 (3.55E+06–3.16E+07) | |
| Mycoplasma fermentans | 2 | 1.0E+03 | 1.75E+03 | 0.57E+03 | 9.25E+01 | 1.44E+04 | 1.71E+04 | |
| 8 | 1.0E+05 | 1.67E+05 | 0.69E+05 | 0.10E+05 | 2.62E+06 | 2.23E+06 | 5.75E+05 (2.57E+05–1.31E+06) | |
| Mycoplasma orale | 3 | 1.0E+03 | 0.87E+03 | 0.45E+03 | 0.31E+03 | 1.02E+03 | 1.56E+03 | |
| 7 | 1.0E+05 | 0.84+05 | 0.46E+05 | 0.24E+05 | 1.44E+05 | 2.37E+05 | 1.35E+05 (6.45E+04–2.81E+05) | |
| Mycoplasma pneumoniae | 4 | 1.0E+03 | 1.47E+03 | 1.10E+03 | 0.19E+03 | 2.53E+03 | 5.90E+03 | |
| 6 | 1.0E+05 | 1.47E+05 | 1.15E+05 | 0.24E+05 | 1.99E+06 | 1.07E+06 | 3.01E+5 (1.44E+05–6.45E+05) | |
| Friis culture medium | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| WHO IS preparation, Mycoplasma fermentans | 1.0E+05 | 0.79E+05 | 0.75E+05 | 2.63E+03 | 1.24E+06 | 0.73E+06 | 1.82E+05 (0.81E+05–4.31E+05) | |
Mycoplasma preparations were tested at different stages by the culture method (which provided results as the number of CFU per milliliter) and by NAT (which provided results as the number of copies per milliliter or the number of NAT-detectable units per milliliter). Stages included source materials (harvest), materials obtained just before lyophilization (lyo), and materials obtained after reconstitution of the lyophilized preparations. The amount of mycoplasma DNA in materials before and after lyophilization was determined with a quantitative NAT assay reporting the number of copies per milliliter (Intego Mycoplasma). The mean number of NAT-detectable units and the corresponding 95% confidence intervals were calculated for the neat lyophilized preparations on the basis of the results of 19 different NAT assays.
For manufacture of the WHO candidate IS preparation, M. fermentans (PG18T, NCTC 10117) was grown to a target concentration of 105 CFU/ml and harvested during the exponential growth phase.
The species identity and purity of each mycoplasma culture were confirmed by 16S rRNA gene sequence analysis, which revealed nearly 100% identity with the 16S rRNA gene sequence of the respective mycoplasma type strain. These data verified that there were no other bacterial species present in the culture preparations. In addition, the identity of the strain in each culture was confirmed by randomly amplified polymorphic DNA (RAPD) PCR analysis. Furthermore, sterility tests performed under aerobic and anaerobic conditions and with both liquid and solid media indicated that no viable bacteria other than the chosen mycoplasma species were present in the reconstituted candidate WHO IS preparation, even though some manufacturing steps (e.g., aliquoting) were performed under nonaseptic conditions.
Lyophilization and residual moisture determination.
Mycoplasma preparations were stored frozen at −60°C until freeze-drying (lyophilization), which was performed by a company certified to perform the EN ISO 13485:2003 standard method. The materials were thawed, and while the materials were gently stirred, 0.5-ml volumes were dispensed into 3-ml screw-cap glass vials and immediately frozen. The coefficient of variation (CV) of filling was calculated from weight determination to be 1.2% for the feasibility panel members and 1.0% for the WHO candidate IS. Lyophilization was performed for 62 h using a Christ Epsilon 2-25 instrument with its LPC-16/NT process documentation. Before sealing of the vials, the system was flooded with dry nitrogen to replace the oxygen and moisture. The freeze-dried preparations were stored at −20°C prior to use or until delivery.
The amount of residual moisture in the freeze-dried samples was determined for a representative number of vials using an accredited method (i.e., Karl Fischer titration) according to the guidelines in the Ph. Eur. The amount of residual moisture was between 0.6% and 1.9%, in line with current guidance from the WHO (21).
Mycoplasma NAT assays.
The assays used by the participants of the feasibility or comparability study and for characterization of the materials were either quantitative (reporting e.g., the number of copies per milliliter, the number of genome copies per milliliter), semiquantitative (reporting cycle threshold [CT] values), or qualitative (reporting results as reactive or nonreactive). In the study, quantitative and semiquantitative real-time NAT assays were grouped as (semi)quantitative. The NAT assays (which were nearly exclusively based on real-time PCR) in combination with their extraction systems were comprised of 7 in-house-developed and 14 commercial systems (Table 2; for the commercial test systems, see the user guides of the manufacturers for references). The commercially available (semi)quantitative NATs were Microsart AMP Mycoplasma (Sartorius Stedim Biotech GmbH, Göttingen, Germany), Intego Mycoplasma and Venor GeM qEP (both from Minerva Biolabs, Berlin, Germany), PLEX-ID Mycoplasma detection assay (IBIS Biosciences, Abbott, Carlsbad, CA, USA), MycoTool Mycoplasma real-time PCR (Roche Diagnostics, Mannheim, Germany), and MycoSEQ Mycoplasma real-time PCR (Applied Biosystems, Foster City, CA, USA). Commercially available qualitative NATs included CytoInspect PCR/microarray (Greiner Bio-One GmbH, Frickenhausen, Germany), MilliPROBE (Merck Millipore, Billerica, MA, USA), MycoTool Mycoplasma amplification and detection kits (Roche Diagnostics GmbH, Mannheim, Germany), and Venor GeM Advance (Minerva Biolabs, Berlin, Germany).
TABLE 2.
NAT assays included in feasibility study
| NAT assay use and amplification/detection method | Extraction method | Assay no. |
|---|---|---|
| NAT assays used for (semi)quantitative evaluation | ||
| In-house RT-PCR A | QIAamp viral RNA minikit | |
| In-house RT-PCR B | DNeasy blood and tissue kit | |
| In-house RT-PCR C | QiaSymphony | |
| In-house RT-PCR D | Phenol-chloroform | |
| Intego Mycoplasma | InviMag universal kit/IG | 4 |
| Intego Mycoplasma | Chemagen RSMI | 11 |
| Intego Mycoplasma | QIAamp DNA blood minikit | 15 |
| Microsart AMP Mycoplasma | InviMag universal kit/IG | 3 |
| MycoSEQ Mycoplasma real-time PCR | PrepSEQ sample preparation kit | 5, 8, 9, 10 |
| MycoSEQ Mycoplasma real-time PCR | NucliSENS easyMAG | 7 |
| MycoTool Mycoplasma real-time PCR | MagNA Pure | 1 |
| PLEX-ID Mycoplasma detection assay | Bead-beating lysis | 2 |
| Venor GeM qEP | QIAamp DNA blood minikit | 15 |
| NAT assays used for qualitative evaluation | ||
| CytoInspect PCR/microarray | CytoInspect DNA extraction kit | 18, 21, 24 |
| In-house nested PCR E | QIAamp DNA minikit | |
| In-house RT-PCR F | Phenol-chloroform | |
| In-house RT-PCR G | Silica columns | |
| MilliPROBE | Target capture (rRNA) | 26 |
| MycoTool Mycoplasma | Manual, 2-propanol | 19 |
| Venor GeM Advance | MB DNA extraction kit | 20 |
| Venor GeM Advance | QIAamp DNA blood minikit | 25 |
The potential impact of lyophilization on the detectability of mycoplasma DNA was addressed by comparative evaluation of lyophilized specimens (feasibility study panel members, candidate WHO IS) and their corresponding liquid source materials stored frozen at −80°C. Three different quantitative NAT assays (Venor GeM qEP, Intego Mycoplasma, and MycoSEQ Mycoplasma real-time PCR) were used to evaluate the potential effects of lyophilization on detection of the different mycoplasma preparations.
Stability testing.
An accelerated stability test program simulating storage/transport temperatures of −20°C, +4°C, +23°C, +37°C, and +45°C was initiated. The potential degradation of mycoplasma DNA under an elevated temperature was determined at regular intervals using two quantitative real-time NAT assays (Venor GeM qEP, Intego Mycoplasma). The stability testing has been performed for more than 3 years for the lyophilized A. laidlawii preparation (panel member 9) and more than 2 years for the WHO IS preparation; these studies are being continued.
Feasibility study design.
The study participants' names and the organizations with which they are affiliated are provided in Acknowledgments. For the feasibility study, two different panels of mycoplasma species were designed for either (semi)quantitative or qualitative NAT assays. All panel members were coded. Both panels contained the four selected mycoplasma species as common panel members. Reconstitution of panel members was performed with 0.5 ml molecular-grade, nuclease-free water; all dilutions were to be performed with the diluent representing the usual negative test matrix of the laboratory. A variety of different diluents were used by the participants, including isotonic buffers, saline, culture medium, cultured cells, cell culture supernatant, virus bulk harvest, or water. The study protocols differed between the two approaches: users of (semi)quantitative assay were asked to perform one dilution step (1:10) for the high-concentration panel members and to report numerical results, e.g., numbers of copies per milliliter or CT values. Users of qualitative assays were asked to perform endpoint dilutions and to report back positive or negative results for the respective dilutions.
For (semi)quantitative NAT assays, the feasibility study panel consisted of 9 coded members (panel members 1 to 9), representing the lyophilized mycoplasma preparations of different concentrations and a medium control (Table 1). Each participant received three identical panels for three separate test runs. Following the protocol, each participant performed three separate runs, resulting in at least three results per panel member and per 1:10 dilution, where proposed.
For qualitative NAT assays, a feasibility study panel which consisted of lyophilized preparations representing panel members 1, 2, 3, 4, and 9 was designed (Table 1). Each participant received three identical panels for three test runs. In the first test run, participants were asked to test the coded preparations neat and in log10 dilution series until test results became negative in order to determine the preliminary endpoint dilution for each panel member (the lowest concentration that tested positive). For the subsequent two test runs, 5 half-log10 dilutions around the predetermined endpoint were tested.
Comparability study design.
For comparison of the candidate WHO IS preparation with the M. fermentans preparation included in the feasibility study panel (panel member 8), the users of selected (semi)quantitative NAT assays were asked to determine the relative concentrations by testing replicates of both preparations neat and as a 1:10 dilution and to report back either the numbers of copies per milliliter or the CT values; users of qualitative NAT assays were asked to test in replicate endpoint dilution series of the two preparations, following a protocol analogous to that for the feasibility study.
Statistical methods.
Statistical analysis was performed with SAS/STAT software (version 9.3; SAS System for Windows). Estimations of the endpoint dilution and relative potency were determined using CombiStats software (version 5.0, release 2013; EDQM/Council of Europe).
(i) Relative potencies.
Evaluation of quantitative assays was performed without removing any outlying data. Assays giving CT values and those reporting numbers of copies per milliliter were evaluated separately. The potencies of samples relative to the potency of panel member 8 (M. fermentans, neat material) or panel member 2 (M. fermentans, 1:100 dilution), which were given an assigned arbitrary value of 5.00 log10 IU/ml or 3.00 log10 IU/ml, respectively, were estimated by a parallel-line assay with log-transformed data (quantitative protocol) or probit-transformed data (endpoint dilution protocol).
(ii) Absolute potencies.
(a) Quantitative assays. Evaluation of the results reported by quantitative assays was restricted to dilutions in the range where the assays produced comparable data (linear range). For comparison of laboratories, the replicate results of each laboratory, corrected for the dilution factor, were combined as the arithmetic mean of the log10 number of copies per milliliter. Furthermore, these estimates were combined to obtain an overall estimation for each sample.
(b) Qualitative assays (endpoint dilution procedure). The results from the independent runs were pooled to give a series of the number positive out of the number tested at each dilution. The pooled results of the single assays were evaluated by probit analysis to estimate the concentration at which 63% of the samples tested were positive (i.e., the dilution at which, on average, a single copy per sample tested could be expected, assuming a Poisson distribution). The calculated endpoint was used to give estimates expressed in NAT-detectable units per milliliter after correction for an equivalent volume of the test sample. This estimation was performed by a parallel-line assay on probit-transformed data using CombiStats software (version 4.0, release 2008; EDQM/Council of Europe). The correction included the volume extracted, the volume eluted, and the fraction used for amplification.
RESULTS
Characterization of mycoplasma preparations.
The different mycoplasma preparations for the feasibility study were characterized at different stages to determine if the performance of the preparations was affected by the processing steps: after harvest, after freeze-thawing prior to lyophilization, and after reconstitution of the lyophilized material. Table 1 summarizes the results obtained. The freeze-thaw step prior to lyophilization reduced mycoplasma viability by a factor of up to 3. The postlyophilization viability of the different mycoplasma preparations of the feasibility study panels was determined for the lyophilized specimens and compared with the mycoplasma viability of the respective liquid source materials (for which an additional freeze-thaw cycle simulating the lyophilization procedure was included). Mycoplasma viability was reduced by lyophilization by a factor of approximately 0.2 log10 (M. orale), 1 log10 (M. fermentans, M. pneumoniae), and 2 log10 (A. laidlawii). The potential effect of lyophilization on mycoplasma nucleic acid integrity and/or detectability was addressed by a comparative investigation of the lyophilized materials and their liquid source materials by the NAT assays. In contrast to viability, DNA detection was equivalent between liquid and lyophilized materials (Table 1).
The stability of the analyte mycoplasma DNA in two different lyophilized mycoplasma preparations (panel member 9, A laidlawii; WHO IS, M. fermentans) was investigated over a time period of up to 3 years at different storage temperatures, using two different quantitative NATs. Over this period, the analyte mycoplasma DNA was observed to have a high degree of stability, with no indication of degradation at storage temperatures of between −20°C (the recommended storage condition) and +45°C (data not shown).
Feasibility study.
Interest in voluntary participation in the feasibility study was received from numerous organizations representing governmental authorities, biopharmaceutical manufacturers, IVD manufacturers, and contract testing providers (see Acknowledgments). There was no preselection of laboratories or NAT assays; all laboratories that expressed a willingness to participate were included in the study. Panels of lyophilized mycoplasma preparations were dispatched at ambient temperature to 21 participating organizations for evaluation in 26 (semi)quantitative or qualitative mycoplasma NAT assays.
The amplification/detection systems and extraction procedures used by the participants are listed in Table 2. The assay numbers representing commercially available tests are indicated with the agreement of the manufacturers. Two of the assays (PLEX-ID Mycoplasma detection assay, CytoInspect PCR/microarray) were able to differentiate mycoplasma species. Both assays correctly identified the four mycoplasma species of the different panel members.
All assays in this evaluation targeted mycoplasma DNA, with the exception of two assays targeting mycoplasma RNA. NAT assays for mycoplasma DNA detection were numbered 1 to 25 [assays 1 to 16] were (semi)quantitative assays; (assays 17 to 25 were qualitative assays), which do not reflect the order in Acknowledgments or Table 2. The results reported back for one of the RNA detection assays were incomplete and did not allow statistical evaluation; therefore, results obtained by this assay are not included further in this study report. The data obtained with the remaining RNA detection assay (assay 26, MilliPROBE) were not included in the relative and absolute potency calculations for assays detecting mycoplasma DNA.
There were no false-positive results obtained with the negative specimen (panel member 5) included in the panel for (semi)quantitative assays, with the exception of one in-house reverse transcription-PCR (RT-PCR) assay (assay 13), which reported high CT values for this panel member.
All assays were able to detect the four mycoplasma species included in the panels, with the exception of a (semi)quantitative in-house assay (assay 6) designed for detection of M. pneumoniae exclusively, which did not cross-react with the other species, and two qualitative assays which failed to amplify either M. orale (assay 20) or A. laidlawii and M. pneumoniae (assay 22). Results from these assays were excluded from the statistical evaluations for the respective species.
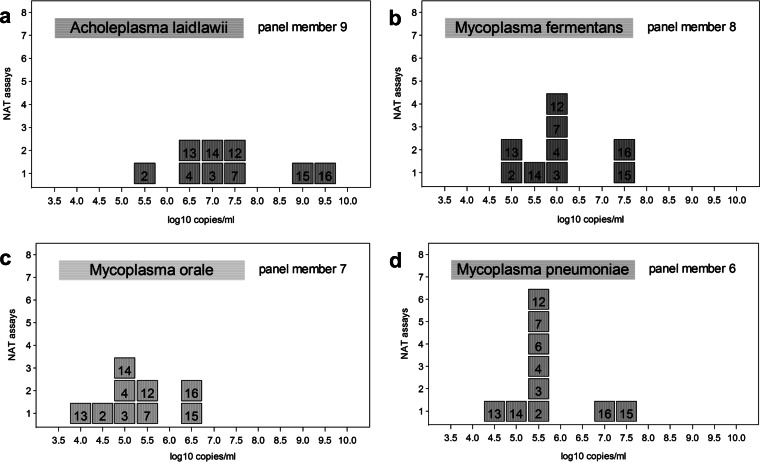
The results reported by quantitative assays for the same panel members (e.g., in numbers of copies per milliliter) differed between assays by a factor of up to 5 log10. The distribution of concentrations reported for neat panel members of the four mycoplasma species are shown in Fig. 1.
FIG 1.
The numbers of the quantitative mycoplasma NAT assays are indicated in boxes, and each box represents a different assay (Table 2). The results reported by the assays for the different mycoplasma species, A laidlawii (a), M. fermentans (b), M. orale (c), and M. pneumoniae (d) (contained in panel members 9, 8, 7, and 6, respectively), are represented on a log10 scale. Despite the high variation of the reported results between the assays, individual quantitative NAT assays exhibited similar relative quantities across the different mycoplasma species; e.g., assays 15 and 16 consistently reported the highest concentrations of mycoplasma DNA.
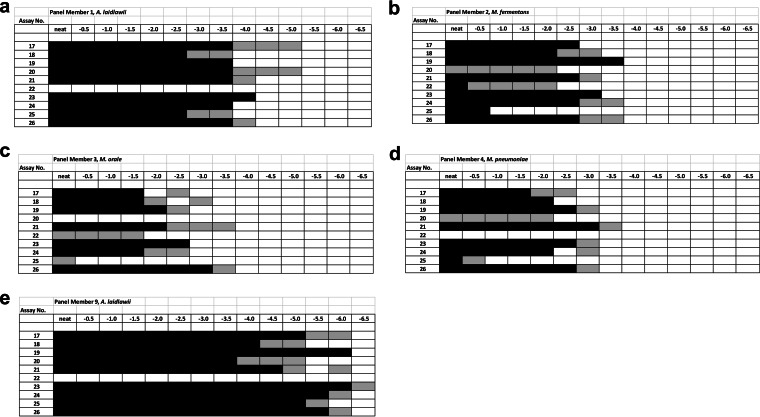
The results reported for qualitative assays showed a difference in sensitivity of up to 2 log10 between assays of the diluted samples for most assays and species. Figure 2 illustrates the reactivity of individual qualitative assays (assay numbers 17 to 26) with replicate testing of the respective dilution series, providing for each assay either consistent positive or negative results (black or white boxes) or inconsistent results (gray boxes) at the detection limits of the assays due to the Poisson distribution of the analyte.
FIG 2.
The numbers of the qualitative NAT assays (assays 17 to 26; see also Table 2) are indicated on the left, and the dilution steps performed for the different mycoplasma preparations are given on the top. Serial half-log dilutions (neat to −6.5 log10) of the mycoplasma species A. laidlawii (low concentration) (a), M. fermentans (b), M. orale (c), M. pneumoniae (d), and A. laidlawii (high concentration) (e) (contained in panel members 1, 2, 3, 4, and 9, respectively) were tested as replicates. Results obtained on testing of replicates are indicated as black boxes (consistently positive), as white boxes (consistently negative), or as gray boxes (positive or negative, probably due to the Poisson distribution of the analyte). Several qualitative NAT assays showed a similar relative dilutional sensitivity across the different mycoplasma species; e.g., assays 19, 21, and 26 consistently exhibited the highest dilutional sensitivities.
Statistical evaluation of feasibility study results. (i) (Semi)quantitative NAT assays.
The design of the panel members combined with the proposed test protocol revealed results for three different (by log10) concentrations (high, medium, low) for each of the four mycoplasma species included in the panel. The panel itself included two preparations with a 100-fold difference (high, low), with the high concentration also being diluted 1:10 by the participant to the medium concentration.
The relative potency (relative to the potency of a reference preparation assigned an arbitrary potency) for the remaining panel members was determined by the respective NAT assays. For this calculation, either the numbers of copies per milliliter (reported by quantitative assays) or the CT values reported by semiquantitative real-time NATs were taken. For the statistical evaluation, geometric mean values obtained for three individual panels in three test runs were used. Overall, there was a high level of reproducibility between different test runs of an assay (intralaboratory variability), with only a few outlier results being excluded.
Estimation of relative potencies was done by means of a parallel-line model with the validity preconditions of (i) linearity within a dilution series for a mycoplasma species and (ii) parallelism between the results obtained for different mycoplasma species. The linearity is necessary for a potency calculation covering at least three concentration levels per species, and the parallelism (similar slopes) confirms that the amplification efficiency of an NAT assay system is equivalent for the different mycoplasma species. If both preconditions are fulfilled for two different mycoplasma species, the relative potency between these species reported by the specific NAT assay may be calculated. For the vast majority of assays and mycoplasma preparations, these preconditions were fulfilled.
(ii) Qualitative NAT assays.
The data obtained by an NAT assay in three different dilution series were pooled to give the number positive out of the number tested at each dilution. As for the (semi)quantitative assays, the potencies of individual panel members relative to those of the preparations included in the panel used as a reference, e.g., M. fermentans, were estimated.
Harmonization of mycoplasma NATs.
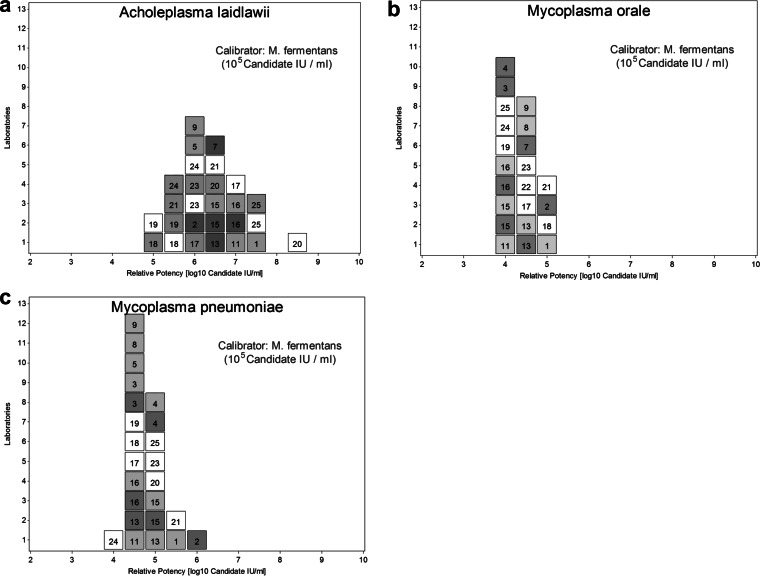
In the relative potency calculation, a mycoplasma preparation may be defined as a common calibrator to which an arbitrary unitage (e.g., an arbitrary number of IU per milliliter) is assigned, and the relative potency may be calculated in IU per milliliter for the other mycoplasma preparations. The overall distribution of results, expressed in IU per milliliter, obtained by the different assays reveals if the use of a common calibrator helps to harmonize the results compared to those of an analysis where no common calibrator is used, e.g., numbers of copies per milliliter. Harmonization is reflected by a reduction of interassay or interlaboratory variation. Respective calculations were performed using A. laidlawii, M. orale, or M. fermentans as candidate calibrators. With A. laidlawii (panel member 9), a harmonization of the NAT assay results compared to the distribution of the numbers of copies per milliliter reported by the assays was obtained. However, the A. laidlawii preparations were not consistently detected by all assays, and different concentration levels were nonlinear for some assays. Furthermore, compared to the results obtained for M. fermentans, the results between the runs of some assays appeared to be less consistent for A. laidlawii. M. orale was missed or underestimated by some assays, and M. pneumoniae, as an agent with a higher biological hazard classification, would cause logistical shipment problems if it was chosen as an international reference preparation. In contrast, if the M. fermentans preparation (panel member 8, assigned an arbitrary concentration of 105 candidate IU mycoplasma DNA/ml) was used as a common calibrator, the harmonization in candidate units reported for the other panel members was striking, especially for M. orale or M. pneumoniae, although it was slightly less striking for A. laidlawii (Fig. 3). Therefore, M. fermentans was selected as the most suitable candidate species for a WHO IS.
FIG 3.
The distribution of candidate numbers of IU per milliliter was calculated for (semi)quantitative assays (assays 1 to 16; dark gray squares, calculations based on the reported numbers of copies per milliliter; light gray squares, calculations based on CT values) and for qualitative assays (assays 17 to 25). The assay number (Table 2) is included in each box. All calculations were based on M. fermentans preparations with an arbitrary concentration of 105 candidate IU/ml (panel member 8) or 103 candidate IU/ml (panel member 2). The harmonization of the assays by a common calibrator is obvious by the closer distribution of the test results in IU per milliliter, e.g., compared to the distribution of numbers of copies per milliliter reported by quantitative assays (Fig. 1). Calculations were performed for the preparations containing A. laidlawii (a), M. orale (b), or M. pneumoniae (c). For A. laidlawii, calculations for qualitative assays (assays 17 to 25) were performed both for the neat concentration (panel member 9), represented as gray boxes, and for the 1:100 dilution (panel member 1), represented as white boxes.
Comparability study.
A new lyophilized M. fermentans preparation was manufactured using a protocol identical to that used for preparation of the feasibility study panels. The M. fermentans concentration of the candidate WHO IS was targeted to be in the range of that of panel member 8. The measured mycoplasma DNA concentration and the number of CFU per milliliter both in the source material used for the candidate WHO IS and in the lyophilized materials were slightly lower than those in the respective materials of panel member 8 (Table 1).
Eleven different NAT assays that were shown to be proficient in the feasibility study in regard to the consistent detection of distantly related mycoplasma species were used to compare the two lyophilized preparations. In total, the two materials were compared by four quantitative assays reporting results in numbers of copies per milliliter, by two semiquantitative assays generating CT values, and by five qualitative assays in regard to potency differences. The results obtained by this set of assays are summarized in Table 3. The mycoplasma DNA content in the candidate WHO IS was determined to be slightly lower than that in panel member 8; the weighted mean difference factor was 0.316.
TABLE 3.
Comparability study results: potency of proposed WHO IS relative to that of panel member 8 (the reference)a
| Assay | Assay type | Potency of WHO IS candidate relative to that of panel member 8 | 95% confidence interval | No. of copies/ml reported for WHO IS candidate |
|---|---|---|---|---|
| In-house RT-PCR A | Quantitative | 0.288 | 0.163–0.477 | 7.4 × 105 |
| In-house RT-PCR B | Quantitative | 0.272 | 0.110–0.535 | 5.2 × 105 |
| Microsart AMP Mycoplasma | Quantitative | 0.282 | 0.204–0.382 | 2.7 × 105 |
| Intego Mycoplasma | Quantitative | 0.309 | 0.223–0.421 | 3.1 × 105 |
| MycoTool Mycoplasma real-time PCR | Semiquantitative | 0.270 | 0.202–0.358 | |
| MycoSEQ Mycoplasma real-time PCR | Semiquantitative | 0.392 | 0.274–0.549 | |
| In-house RT-PCR F | Qualitative | 0.150 | 0.000–4.251 | |
| In-house RT-PCR G | Qualitative | 0.750 | 0.356–1.582 | |
| CytoInspect PCR/microarray | Qualitative | 0.603 | 0.062–5.707 | |
| Venor GeM Advance | Qualitative | 0.747 | 0.204–2.658 | |
| MycoTool Mycoplasma amplification and detection kits | Qualitative | 0.383 | 0.140–1.049 | |
| Combinedb | 0.316 | 0.277–0.360 |
Relative potencies were estimated by means of a parallel line model (quantitative data) and probit analysis (qualitative data; the Spearman-Kaerber method was used instead of the probit method in cases where the slope of the curves could not be estimated). The overall potency estimator is based on a weighted combination of results. No outliers were removed from the combination due to relatively homogeneous results.
Weighted combination estimator.
Proposed unitage for the WHO IS.
Analytes in complex biological materials are often not traceable to the International System of Units (SI), e.g., moles or grams; therefore, the respective WHO ISs are assigned an arbitrary unitage, the international unit (IU). For NAT ISs developed for infectious diseases, the IU has generally been aligned as closely as possible to the nucleic acid copy number (e.g., the genome copy number) or to the NAT-detectable unit. Therefore, the mean numbers of copies reported by different quantitative assays for the feasibility study panel members were combined with the corresponding mean number of NAT-detectable units calculated from the different qualitative assays' endpoint dilution results. For this calculation, the results of the feasibility study were used. The mean potencies and 95% confidence intervals for the neat materials of the feasibility study are shown in Table 1. The mean value of the M. fermentans concentration in panel member 8 was 5.75 log10 NAT-detectable units/ml (575,439 NAT-detectable units/ml). The weighted mean difference between the candidate WHO IS and panel member 8 was a factor of 0.316, as determined in the comparability study (Table 3), resulting in 181,838 NAT-detectable units/ml for the candidate WHO IS. Therefore, it was proposed that a unitage of 2 × 105 IU/ml, a value close to the number of NAT-detectable units determined, be assigned to the candidate WHO IS.
DISCUSSION
Several scientific publications indicate high interlaboratory and interassay variability for the determination of mycoplasma contamination. This refers to the measurement of living mycoplasma cells by biological assays and to the determination of mycoplasma nucleic acids by NAT assays (22, 23). The high variability between NAT assays was confirmed in the studies performed in this project and reemphasizes the need for standardization of these assays. The collaborative study for the establishment of a WHO IS for mycoplasma DNA consisted of two parts. In the first phase, the feasibility study, it could be shown that most of the assays designed for generic detection are able to consistently detect the different mycoplasma species provided in the panel. In the study, only one assay that was designed for the specific detection of M. pneumoniae (assay 6) did not cross-react with the other mycoplasma species. The majority of assays of generic design detected the four mycoplasma species contained in the panel. Only a few in-house-developed qualitative assays failed to detect one or more of the mycoplasma species or showed different sensitivities for different species. In this initial study, the need for an international reference preparation was also confirmed by the high variation of quantitative assay results for the same preparations, best explained by a current lack of standardization of assays.
We performed different calculations assuming that individual panel members would be used as a common calibrator. Some degree of harmonization between assays would have been achieved with any panel member; however, the M. fermentans preparation appeared to be the most suitable candidate. The effect of harmonization of assays by use of the M. fermentans preparation as a common calibrator is obvious when the potencies of the other mycoplasma preparations are expressed relative to the potency of the M. fermentans preparation.
Due to the complex composition of biological materials, the target analyte, e.g., the nucleic acid of a virus or an organism, has to be extracted and purified prior to analysis. This kind of analyte is often not traceable to SI units, e.g., grams or moles. WHO ISs are instead often assigned in IU per milliliter to have a common value for the content of the analyte. In the field of NAT assays, the numbers of IU per milliliter assigned to the WHO ISs have been in a range broadly similar to the numbers of copies per milliliter reported by quantitative assays and the number of PCR-detectable units per milliliter calculated from qualitative assay results obtained by replicate limiting dilutions. In the comparability study, the weighted mean difference factor between panel member 8 and the corresponding candidate WHO IS was calculated using a number of assays of different types (quantitative, semiquantitative, qualitative) and designs. On the basis of this differential factor and the absolute mean potency of panel member 8 in the feasibility study, it was proposed to assign a concentration of 2 × 105 IU/ml to the candidate WHO IS for mycoplasma DNA. This value is in the same range as the numbers of copies per milliliter both reported by different quantitative NAT assays (Table 3) and confirmed by an NAT-independent method for measuring genomic mycoplasma DNA in panel member 8 after staining with PicoGreen, a fluorochrome selectively binding to double-stranded DNA (S. Czurda, data not shown).
The unitage associated with the candidate WHO IS is for mycoplasma DNA and not RNA. First, only one assay (assay 26) in the feasibility study delivered complete results based on RNA detection. The results obtained for assay 26 (MilliPROBE) were reported for information purposes only and were not included in the statistical evaluation of the DNA detection assays. Second, mycoplasma RNA as an analyte representing mycoplasma contamination is potentially very different from the analyte mycoplasma genomic DNA. The composition and levels of different mycoplasma RNAs depend on the metabolic status of the mycoplasma cell (e.g., cultivation conditions), and test results may depend on the specific target RNAs selected by different assays. Furthermore, there are strong indications that even in lyophilized specimens, mycoplasma RNA is less stable under accelerated and stress conditions than mycoplasma DNA (T. Hämmerle, personal communication). In conclusion, assays targeting (potentially different) mycoplasma RNA may be much more difficult to standardize by use of a reference preparation of the current design.
The feasibility study revealed that one mycoplasma species may be representative of another, with even distantly related mycoplasma species being much more consistently detected after harmonization of NAT assays designed for generic mycoplasma detection. As a result of these studies, the candidate WHO IS chosen is a culture-based preparation of Mycoplasma fermentans which has been freeze-dried for long-term stability.
Based on the data summarized here, the WHO ECBS established on 25 October 2013 the lyophilized M. fermentans preparation as the “1st World Health Organization international standard for mycoplasma DNA for nucleic acid amplification technique-based assays designed for generic mycoplasma detection” (24) with an assigned unitage of 2 × 105 IU/ml. This new WHO IS is now available from the WHO Collaborating Centre at PEI (www.pei.de).
ACKNOWLEDGMENTS
We thank the members of the PDA Mycoplasma Task Force for advice, especially Barbara Potts, Sven Deutschmann, Thomas Hämmerle, and Laurent Mallet.
The members of the Mycoplasma Collaborative Study Group who participated in the feasibility study and/or the comparability study (in alphabetical order of institutions) are as follows: Thomas Hämmerle, Baxter AG, Orth/Donau, Austria; Cynthia Martino, Bionique Testing Laboratories, Saranac Lake, NY, USA; Michael Hantman, Charles River Biopharmaceutical Services, Malvern, PA, USA; Alena Dabrazhynetskaya and Vladimir Chizhikov, FDA/CBER/LMD, Kensington, MD, USA; Walter Rudorfer and Joerg Stappert, Greiner Bio-One, Frickenhausen, Germany; Vicki Chalker, Health Protection Agency, London, United Kingdom; Rangarajan Sampath, IBIS Biosciences, Abbott, Carlsbad, CA, USA; Dietmar Mayer, IDT Biologika, Dessau, Germany; Francesca Bonci, Kedrion Biopharmaceuticals, Castelvecchio Pascoli, Italy; Susan Brand-Hoefs, Merck, Oss, Netherlands; Fabrizio Lecce, Merck Serono, Ivrea, Italy; Freek Blanken and Nigel Stapleton, Microsafe Laboratories, Leiden, Netherlands; Matthias Hornschuh and Dirk Vollenbroich, Minerva Biolabs, Berlin, Germany; Stefan Czurda, Ursula Ulrych, and Renate Rosengarten, Mycosafe Diagnostics GmbH, Vienna, Austria; Yuko Sasaki, National Institute of Infectious Diseases, Tokyo, Japan; Alexandra Priessner and Michael Molitor, Novartis Vaccines and Diagnostics, Marburg, Germany; Claudia König and Oliver Karo, Paul-Ehrlich-Institut, Langen, Germany; Holger Kavermann and Sven Deutschmann, Roche Diagnostics, Penzberg, Germany; Eric Abachin and Laurent Mallet, Sanofi Pasteur, Paris, France; Andreas Lindauer, Synlab MVZ, Weiden, Germany; Brian Mondeja Rodriguez, Tropical Medicine Institute, Havana, Cuba.
REFERENCES
- 1.Armstrong SE, Mariano JA, Lundin DJ. 2010. The scope of mycoplasma contamination within the biopharmaceutical industry. Biologicals 38:211–213. doi: 10.1016/j.biologicals.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Laborde S, Degrave A, Lehmann D, Jouette S, Rofel C, Muller T, Hertzog N, Rook M, Ribault S. 2010. Detection of Mollicutes in bioreactor samples by real-time transcription-mediated amplification. Lett Appl Microbiol 50:633–638. doi: 10.1111/j.1472-765X.2010.02846.x. [DOI] [PubMed] [Google Scholar]
- 3.FDA. 2008. Guidance for FDA reviewers and sponsors: content and review of chemistry, manufacturing and control (CMC) information for human gene therapy investigational new drug applications (INDs). FDA, Rockville, MD: http://www.fda.gov/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/CellularandGeneTherapy/ucm072587.htm#ProductTesting. [Google Scholar]
- 4.FDA. 2010. Characterization and qualification of cell substrates and other biological materials used in the production of viral vaccines for infectious disease indications. FDA, Rockville, MD: http://www.fda.gov/downloads/biologicsbloodvaccines/guidancecomplianceregulatoryinformation/guidances/vaccines/ucm202439.pdf. [Google Scholar]
- 5.Council of Europe. 2015. 2.6.7. Mycoplasmas, p 178–183. European Pharmacopoeia 8.0. Council of Europe, Strasbourg, France. [Google Scholar]
- 6.United States Pharmacopeial Convention. 2010. Mycoplasma tests. USP 38-NF 28. United States Pharmacopeial Convention, Rockville, MD. [Google Scholar]
- 7.Knezevic I, Stacey G, Petricciani J, Sheets R. 2010. Evaluation of cell substrates for the production of biologicals: revision of WHO recommendations. Report of the WHO Study Group on Cell Substrates for the Production of Biologicals, 22-23 April 2009, Bethesda, USA Biologicals 38:162–169. doi: 10.1016/j.biologicals.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Janetzko K, Rink G, Hecker A, Bieback K, Klüter H, Bugert P. 2014. A single-tube real-time PCR assay for Mycoplasma detection as a routine quality control of cell therapeutics. Transfus Med Hemother 41:83–89. doi: 10.1159/000357096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence B, Bashiri H, Dehghani H. 2010. Cross comparison of rapid mycoplasma detection platforms. Biologicals 38:218–223. doi: 10.1016/j.biologicals.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Peredeltchouk M, David SA, Bhattacharya B, Volokhov DV, Chizhikov V. 2011. Detection of mycoplasma contamination in cell substrates using reverse transcription-PCR assays. J Appl Microbiol 110:54–60. doi: 10.1111/j.1365-2672.2010.04853.x. [DOI] [PubMed] [Google Scholar]
- 11.Pinheiro de Oliveira TF, Fonseca AA, Camargos MF, de Oliveira AM, Pinto Cottorello AC, Souza Ados R, de Almeida IG, Heinemann MB. 2013. Detection of contaminants in cell cultures, sera and trypsin. Biologicals 41:407–414. doi: 10.1016/j.biologicals.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Shahhosseiny MH, Hosseiny Z, HR Khoramkhorshid HR, Azari S, Shokrgozar MA. 2010. Rapid and sensitive detection of Mollicutes in cell culture by polymerase chain reaction. J Basic Microbiol 50:171–178. doi: 10.1002/jobm.200800174. [DOI] [PubMed] [Google Scholar]
- 13.Störmer M, Vollmer T, Henrich B, Kleesiek K, Dreier J. 2009. Broad-range real-time PCR assay for the rapid identification of cell-line contaminants and clinically important mollicute species. Int J Med Microbiol 299:291–300. doi: 10.1016/j.ijmm.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Volokhov DV, Graham LJ, Brorson KA, Chizhikov VE. 2011. Mycoplasma testing of cell substrates and biologics: review of alternative non-microbiological techniques. Mol Cell Probes 25:69–77. doi: 10.1016/j.mcp.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Eldering JA, Felten C, Veilleux CA, Potts BJ. 2004. Development of a PCR method for mycoplasma testing of Chinese hamster ovary cell cultures used in the manufacture of recombinant therapeutic proteins. Biologicals 32:183–193. doi: 10.1016/j.biologicals.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Deutschmann SM, Kavermann H, Knack Y. 2010. Validation of a NAT-based Mycoplasma assay according European Pharmacopoiea. Biologicals 38:238–248. doi: 10.1016/j.biologicals.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Vanni I, Ugolotti E, Raso A, Di Marco E, Melioli G, Biassoni R. 2012. Development and validation of a multiplex quantitative polymerase chain reaction assay for the detection of Mollicutes impurities in human cells, cultured under good manufacturing practice conditions, and following European Pharmacopoeia requirements and the International Conference on Harmonization guidelines. Cytotherapy 14:752–766. doi: 10.3109/14653249.2012.671517. [DOI] [PubMed] [Google Scholar]
- 18.Zhi Y, Mayhew A, Seng N, Takle GB. 2010. Validation of a PCR method for the detection of mycoplasmas according to European Pharmacopoeia section 2.6.7. Biologicals 38:232–237. doi: 10.1016/j.biologicals.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Asarnow D, Warford A, Fernandez L, Hom J, Sandhu G, Candichoy Z, Luna G, Goldman M, Rarich R. 2010. Validation and international regulatory experience for a mycoplasma touchdown PCR assay. Biologicals 38:224–231. doi: 10.1016/j.biologicals.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Council of Europe. 2015. Human plasma for fractionation, p 1746–1747. European Pharmacopoeia 8.0. Council of Europe, Strasbourg, France. [Google Scholar]
- 21.World Health Organization. 2006. Recommendations for the preparation, characterization and establishment of international and other biological reference standards (revised 2004), p 73–131. WHO Technical Report Series 932. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 22.Milne K, Daas A. 2006. Establishment of European Pharmacopoeia Mycoplasma reference strains. Pharmeuropa Bio 2006:57–72. [PubMed] [Google Scholar]
- 23.Dabrazhynetskaya A, Volokhov DV, Lin TL, Beck B, Gupta RK, Chizhikov V. 2013. Collaborative study report: evaluation of the ATCC experimental mycoplasma reference strains panel prepared for comparison of NAT-based and conventional mycoplasma detection methods. Biologicals 41:377–383. doi: 10.1016/j.biologicals.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 24.WHO Expert Committee on Biological Standardization. 2014. Sixty-fourth report. WHO Tech Rep Ser 987:42. [Google Scholar]