Abstract
The soil microbial community plays an important role in terrestrial carbon and nitrogen cycling. However, microbial responses to climate warming or cooling remain poorly understood, limiting our ability to predict the consequences of future climate changes. To address this issue, it is critical to identify microbes sensitive to climate change and key driving factors shifting microbial communities. In this study, alpine soil transplant experiments were conducted downward or upward along an elevation gradient between 3,200 and 3,800 m in the Qinghai-Tibet plateau to simulate climate warming or cooling. After a 2-year soil transplant experiment, soil bacterial communities were analyzed by pyrosequencing of 16S rRNA gene amplicons. The results showed that the transplanted soil bacterial communities became more similar to those in their destination sites and more different from those in their “home” sites. Warming led to increases in the relative abundances in Alphaproteobacteria, Gammaproteobacteria, and Actinobacteria and decreases in Acidobacteria, Betaproteobacteria, and Deltaproteobacteria, while cooling had opposite effects on bacterial communities (symmetric response). Soil temperature and plant biomass contributed significantly to shaping the bacterial community structure. Overall, climate warming or cooling shifted the soil bacterial community structure mainly through species sorting, and such a shift might correlate to important biogeochemical processes such as greenhouse gas emissions. This study provides new insights into our understanding of soil bacterial community responses to climate warming and cooling.
INTRODUCTION
Global climate change greatly affects terrestrial ecosystems, particularly in polar or alpine regions (1). However, due to the complexity of soil microbiome, how global warming affects their diversity, abundance, and structure is poorly understood (2, 3). To show the responses of microorganisms to climate changes, we must identify those microbes sensitive or acclimated to temperature and their relationships to biogeochemical processes, such as greenhouse gas emissions.
Warming is able to directly affect soil bacterial physiology (4) and indirectly affect microbes through changing plant and soil properties (5). For example, an increase in temperature may lead to a preponderance of thermally adapted microorganisms (6). Artificial warming is widely used to study the effects of warming on soil ecosystems. Integrated metagenomic and functional analyses of a long-term warming experiment in a grassland ecosystem show that one of the outcomes of climate warming is a shift of the microbial community composition, which most likely leads to the reduced temperature sensitivity of heterotrophic soil respiration (3). Also, multifactorial warming experiments show that warming and precipitation alter the microbial community composition in a constructed natural grass prairie (7). Likewise, another long-term in situ warming experiment experiences a moderate natural drought, and warming decreases the diversity and significantly alters the community composition in normal precipitation years (8). However, these artificial warming studies seldom compare the sensitivity of a microbial community along the temperature gradient and determine whether cooling exerts an opposite effect to warming on the microbial community structure.
In a mesic ecosystem, artificial warming usually causes a decrease in soil moisture content and other concurrent changes (9). These observed changes are different from those in alpine and polar regions where climate warming is often accompanied by higher soil moisture caused by glacier and permafrost melting (1). Soil transplant experiments can offer an opportunity to expose the microbial community to natural climate regimes and provide a valid field experiment platform for the study of microbial responses to climate changes (10–12). Using this method, a previous study observed the structural changes of the “home” and “away” bacterial communities on an unvegetated glacier forefield (11). In a California oak-grassland ecosystem, transplant experiments indicate that the sensitivity of soil microbial communities to climate change is dependent on historical exposure to a range of environmental conditions, such as soil temperature and moisture (10). Transplanting organic surface horizons of boreal soils into warmer regions alters the microbial community but not the temperature sensitivity of decomposition (13). Although the microbial community is generally recognized to be sensitive to disturbance (14), many previous studies using low-resolution microbial profiling methods, such as denaturing gradient gel electrophoresis, are not able to discern whether particular taxonomic groups are more or less sensitive to environmental factors (10, 11, 15).
Alpine grassland accounts for roughly 35% of the Qinghai-Tibet plateau (16), which is recognized as a region very sensitive to climate change (17). Qinghai-Tibet plateau experienced climate warming at three times the global warming rate since 1960 (18). Using an elevation gradient of an alpine ecosystem in this region as an analog of climate gradient can provide a nature model to analyze climatic change on ecosystem structure and function. This approach has been successfully used to document the effects of global change on vegetation (19, 20) and soil biogeochemical processes (21). However, little is known about the responses of soil microbial communities to climate changes in the Qinghai-Tibet plateau, although they are integral in driving biogeochemical processes such as carbon and nitrogen cycling, greenhouse gas mitigation, and ecosystem services.
In this study, we aimed to elucidate the responses of the bacterial community to climate warming or cooling in the alpine meadow ecosystem in Qinghai-Tibet plateau. We hypothesized that (i) alpine soil bacterial community structure would respond significantly to climate changes in which soil temperature and above-ground vegetation contribute significantly to shaping the bacterial community structure, (ii) climate warming and cooling would exert opposite effects on the shift of dominant bacteria, and (iii) the changes in the relative abundances of certain bacterial groups, e.g., nitrifiers, may be correlated to the flux of relevant greenhouse gases (e.g., N2O). To test these hypotheses, reciprocal soil transplant experiments were conducted along an elevation gradient in an alpine meadow ecosystem in the Qinghai-Tibet plateau to simulate climate warming or cooling. We analyzed the bacterial phylogenetic compositions after transplanting soil blocks downwards or upwards using a 16S rRNA gene-based pyrosequencing technique and assessed the relationships between the bacterial community structure and environmental factors (e.g., temperature). The results from this study generally support our hypotheses. This study provides new insights into our understanding of soil bacterial communities in response to climate warming and cooling in alpine meadow ecosystems.
MATERIALS AND METHODS
Study site description.
The study site was located at the Haibei Alpine Meadow Ecosystem Research Station, Chinese Academy of Sciences (HBAMERS, Qinghai, China; 37°37′N, 101°12′E), in the northeastern Qinghai-Tibet plateau. The mean elevation of the valley bottom is 3,200 m. The average precipitation is 570 mm, and the annual mean air temperature is −1.7°C (22). The soil is silty clay loam of Mat Cry-gelic Cambisols with a pH of between 7.17 and 7.97 at depths of 10 cm. Detailed soil properties at each site, e.g., total organic carbon (TOC) and total nitrogen (TN), are presented in Table S1 in the supplemental material. Four elevations of 3,200 m (37°36′42.3″N, 101°18′47.9″E), 3,400 m (37°39′55.1″N, 101°19′52.7″E), 3,600 m (37°41′46.0″N, 101°21′33.4″E), and 3,800 m (37°42′17.7″N, 101°22′09.2″E) were chosen to set up four field transplant experiment sites in May 2007 (20). The spatial distances between adjacent elevations were 6.2 km (3,200 to 3,400 m), 4.2 km (3,400 to 3,600 m), and 1.3 km (3,600 to 3,800 m), respectively. Based on the field measurement in the growing season (May to September) of 2008 and 2009, the mean soil temperatures at a 5-cm depth were 12.91 ± 1.77, 11.84 ± 1.47, 9.77 ± 1.09, and 8.86 ± 1.86°C at 3,200, 3,400, 3,600, and 3,800 m, respectively; thus, the temperature differences among the sites ranged from 0.91 to 4.05°C. The background information about the vegetation in each site was described in detail by Wang et al. (20).
Experimental design.
For the soil transplant, an intact soil block (100 cm long by 100 cm wide by 30 cm deep) was divided into four equal parts and dug out with attached vegetation. Each part was enveloped by straw ropes and immediately transferred to target sites. The blocks were reinstalled in the target sites by putting four equal parts together. Plastic film was used to isolate the soil block from surrounding soil. The soil blocks were transplanted upwards (cooling) to a higher elevation from the valley bottom at 3,200 m to 3,400 m (per our naming convention, the sample was designated 3200-3400), 3,600 m, and 3,800 m, respectively. Soil blocks from 3,800 m were also transplanted downward (warming) to 3,600, 3,400, and 3,200 m after the soils started to thaw in early May 2007. Triplicate soil blocks were transplanted from one elevation to another. Soil blocks were fully randomized throughout the study site. The setup of transplant experiment was described in detail by Wang et al. (20). Soil blocks from 3,400 m were transplanted downward to 3,200 m and upward to 3,600 m and 3,800 m. The translocation of the intact soil block caused minimal damage to the plant roots because 85% of the total root biomass is distributed above a 20-cm depth (23). Three blocks from each altitude were also removed and then reinstated at the same holes to produce “home control” blocks that had been handled as similarly as possible to those blocks moved to other elevations. Thus, there were triplicate transfers from each altitude, except for the loss of one 3,200- to 3,600-m sample.
Soil samples were collected in August 2009. Five soil cores with the depth of 0 to 10 cm were taken randomly at each plot and pooled. Soil samples were transported to the lab on ice, sieved with 2-mm mesh to remove visible grass roots and stones, stored at −20°C, and used for genomic DNA extraction and soil property measurements. Soil and vegetation property measurements, including soil temperature, soil moisture, plant biomass, soil pH, NO3−-N, NH4+-N, TOC, TN, and released CH4, released CO2, and released N2O in August 2009, were described previously (24). The greenhouse gases (CO2, N2O, and CH4) were measured by chamber per plot setting at the 5-cm depth of soil, as previously described (25).
DNA extraction, PCR amplification, and pyrosequencing.
Soil genomic DNA was extracted using a FastDNA spin kit for soil (MP Biomedical, Carlsbad, CA) according to the manufacturer's instructions. To amplify the V4-V5 hypervariable regions of 16S rRNA genes, the universal primers 515F (5′-GTGYCAGCMGCCGCGGTA-3′) and 909R (5′-CCCCGYCAATTCMTTTRAGT-3′) were used in PCR analyses (26). The PCR and other experimental procedures were described in detail by Li et al. (15). The bar-coded amplicons were pooled in an equimolar concentration for 454 pyrosequencing using a GS FLX system (454 Life Sciences, Branford, CT).
Pyrosequencing data processing.
The raw sequences were sorted based on unique sample tags, and trimmed for sequence quality using the RDP Pipeline Initial Process (http://pyro.cme.msu.edu/). A total of 170,161 high-quality and chimera-free reads with an average length of 408 bp were obtained by pyrosequencing. The processed pyrosequences were aligned using the fast, secondary-structure Aware Infernal Aligner in the RDP pyrosequencing pipeline (27). The aligned 16S rRNA gene sequences were used for a chimera check using the Uchime algorithm (28). Resampling to the same sequence depth (2,291 sequences per sample) using daisychopper.pl (http://www.festinalente.me/bioinf/downloads/daisychopper.pl) was conducted before the downstream analysis. Sequences were clustered by the complete-linkage clustering method incorporated in the RDP platform. Operational taxonomic units (OTU) were classified using a 97% nucleotide sequence similarity cutoff. Singletons were removed from the OTU table for downstream analysis. Rarefaction, Shannon index, and Chao1 estimators were calculated in RDP at the 97% sequence similarity. The phylogenetic affiliation of each 16S rRNA gene sequence was analyzed by the RDP Classifier at a confidence level of 80%. The original pyrosequencing data are available from the MG-RAST database (accession no. 4565936.3 to 4565973.3).
Statistical analysis.
Overall, structural changes of bacterial communities were evaluated by principal coordination analysis (PCoA) in Fast UniFrac (http://unifrac.colorado.edu/static/welcome.html). The statistical significance of differences between data sets was assessed by PerMANOVA using the weighted PCoA scores in PAST (http://folk.uio.no/ohammer/past/). Environmental variables providing the highest Spearman's correlation coefficients with bacterial communities were selected, and variation partitioning analysis (VPA) was performed to quantify the relative contributions of environmental variables by a previously described method (29). The Mantel test was applied to evaluate the correlations between bacterial communities with environmental variables using the Mantel procedure in the R package Vegan. Indicator species analysis at the OTU level was applied using the R package indicspecies (http://cran.r-project.org/web/packages/indicspecies/index.html), and the untransformed data of bacterial relative abundance at the OTU level were used as input data. Climate, soil, and vegetable properties were normalized and used as environmental variables. The differences in relative abundances of taxonomic units between samples were tested by one-way analysis of variance (ANOVA). The mean values, standard errors, and P values for these statistical analyses were based on twelve treatments in triplicate and one treatment in duplicate.
RESULTS
Changes in the overall bacterial community structure after reciprocal soil transplant.
After the reciprocal soil transplant, the principal coordination analysis (PCoA) of the overall composition and structure of soil bacterial communities revealed that most transplanted samples were well separated from their “home” controls using a weighted UniFrac method (Fig. 1). When we used unweighted (the presence or absence of an OTU) method, it provided a similar result to the weighted analysis (data not shown) but lower variations explained by the first two axes (PCo1-axis 7.6% and PCo2-axis 4.9%). This indicated that major differences between samples were in the relative abundances of the OTU rather than in the absence or presence of specific taxa.
FIG 1.
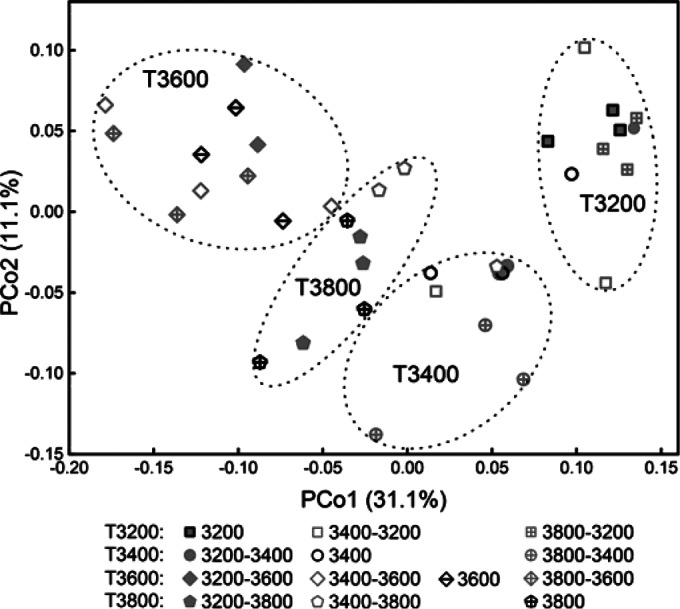
PCoA score plot of transplant samples (gray-edge symbols) and their controls at “home” sites (black-edge symbols) based on weighted UniFrac metrics. Values on PCoA axes indicate the percentages of total variation explained by each axis. Samples were named by their elevations or transplanted to other elevations, e.g., sample 3200 means that it was collected in a “home control” block at the elevation of 3,200 m; sample 3200-3400 means that soil was collected from a plot transplanted from 3,200 m to 3,400 m. T3200 means the group samples collected at the elevation of 3,200 m from both transplanted and “home control” blocks. The relative abundances of OTU were used as input in the analysis.
The weighted PCoA pattern (PCo1 versus PCo2) showed that most of the samples collected at the same elevation formed a cluster regardless of which elevations where they were originally located (Fig. 1). For example, bacterial communities in soil transplanted from 3,200, 3,400, and 3,800 m to 3,600 m tended to cluster together. The PerMANOVA test based on the Bray-Curtis distance measures showed that the bacterial community structure was significantly (P < 0.02) different among these clusters grouped by objective elevations (see Table S2 in the supplemental material). It was further confirmed that the change in bacterial community structure was significantly correlated with soil temperature, plant biomass, and NO3−-N (P < 0.01) in the Mantel test (Table 1).
TABLE 1.
Spearman's correlation of environmental variables with bacterial community structure as determined by the Mantel testa
| Variable | Spearman correlation coefficient (r) | P |
|---|---|---|
| Soil temp | 0.567 | 0.0001 |
| Plant biomass | 0.331 | 0.0001 |
| NO3−-N | 0.057 | 0.186 |
| NH4+-N | –0.069 | 0.856 |
| TOC | 0.169 | 0.002 |
| TN | 0.093 | 0.026 |
| Moisture | 0.109 | 0.025 |
| Soil pH | –0.017 | 0.594 |
Permutations, 9,999 (using the relative abundances of OTU as input in analysis).
Bacterial richness and diversity were highest in the samples originally from 3,800 m based on the number of OTU, Chao1 richness, Shannon's diversity, and evenness indices (see Table S3 and Fig. S1 in the supplemental material). After soil was transplanted from 3,800 m downward to other elevations, the Shannon's diversity and evenness indices were significantly decreased compared to those in their “home” controls (ANOVA, P < 0.05). However, the diversity indices in all other transplanted or mock samples were not significantly different.
Changes of dominant bacterial groups.
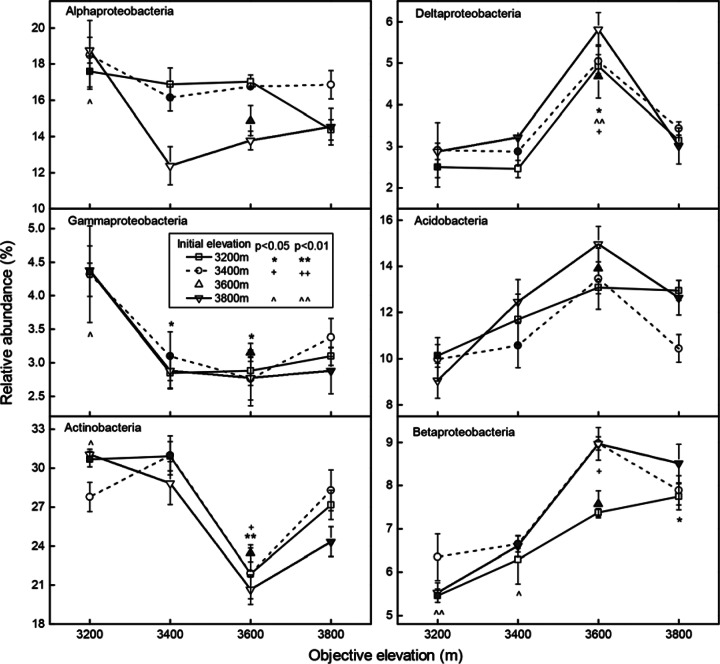
The relative abundances of some dominant phyla significantly changed after the reciprocal soil transplant. When soil was transplanted from 3,200 m upward (cooling condition), the relative abundances of Gammaproteobacteria and Actinobacteria significantly decreased, whereas the Betaproteobacteria significantly increased (ANOVA, P < 0.05) (Fig. 2; see also Table S4 in the supplemental material). However, Deltaproteobacteria only increased significantly at 3,600 m. When soil was transplanted from 3,800 m downward (warming condition), the relative abundances of Gammaproteobacteria, Alphaproteobacteria, and Actinobacteria increased significantly at 3,200 m. Betaproteobacteria decreased significantly (P < 0.05) at 3,200 and 3,400 m, whereas Deltaproteobacteria were more abundant at 3,600 m. Generally speaking, no matter which elevation the samples originated from or in the “home” control soil, Actinobacteria and Gammaproteobacteria were more abundant in lower elevations (3,200 m and 3,400 m) than those in higher elevations (3,600 m and 3,800 m); Betaproteobacteria and Verrucomicrobia increased at 3,600 and 3,800 m; Acidobacteria, Deltaproteobacteria, and Nitrospirae were most abundant at 3,600 m (see Table S4 in the supplemental material). Warming and cooling exerted opposite influences on the relative abundances of dominant groups, including Actinobacteria, Betaproteobacteria, Deltaproteobacteria, Gammaproteobacteria, and Acidobacteria (Fig. 2). The relative abundances of these taxa significantly (P < 0.01) correlated to temperature (Table 2).
FIG 2.
Changes in the dominant bacterial groups (i.e., the percentage compared to the total number of reads) at the phylum or class level along the elevational gradient. Significant differences between the transplant samples (open symbols) and their controls at “home” sites (filled symbols) were calculated by ANOVA. Means ± the standard errors are shown in the figures.
TABLE 2.
Spearman correlation coefficients between the relative abundances of dominant taxa and environmental variables
| Dominant taxon | Spearman correlation coefficient (r)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| Soil temp | Plant biomass | NO3−-N | NH4+-N | TOC | TN | Moisture | Soil pH | |
| Actinobacteria | 0.496** | 0.641** | –0.360* | –0.090 | 0.321* | 0.086 | –0.198 | 0.050 |
| Order Actinomycetales | 0.593** | 0.621** | –0.317 | –0.074 | 0.282 | 0.047 | –0.245 | 0.050 |
| Alphaproteobacteria | 0.377* | 0.151 | –0.174 | 0.057 | 0.496** | 0.501** | –0.279 | 0.064 |
| Order Rhizobiales | 0.562** | 0.469** | –0.190 | 0.017 | 0.527** | 0.381* | –0.163 | 0.223 |
| Betaproteobacteria | –0.747** | –0.653** | 0.249 | –0.068 | –0.403* | –0.080 | 0.179 | –0.258 |
| Order Burkholderiales | –0.354* | –0.057 | 0.062 | 0.058 | –0.336* | –0.379* | 0.320 | –0.267 |
| Deltaproteobacteria | –0.347* | –0.691** | 0.384* | –0.005 | –0.306 | 0.089 | 0.213 | –0.044 |
| Order Myxococcales | –0.126 | –0.569** | 0.418** | –0.104 | –0.254 | 0.108 | 0.177 | –0.023 |
| Gammaproteobacteria | 0.409* | 0.289 | –0.275 | –0.033 | 0.160 | 0.187 | –0.140 | –0.219 |
| Order Xanthomonadales | 0.613** | 0.503** | –0.391* | –0.095 | 0.385* | 0.281 | –0.381* | 0.046 |
| Acidobacteria | –0.481** | –0.475** | 0.400* | 0.148 | –0.386* | –0.378* | 0.201 | 0.062 |
| Class Acidobacteria Gp4 | –0.644** | –0.618** | 0.307 | 0.119 | –0.281 | –0.090 | 0.080 | –0.033 |
Significance: *, P < 0.05; **, P < 0.01.
Among the most abundant genera, the relative abundance of Gp4, Gp7 (Acidobacteria), and Spartobacteria genera incertae sedis (Verrucomicrobia) was high at high elevations regardless of the sample's initial elevation, and genera Gp4, Gp7, Nitrospira were most abundant at 3,600 m. Genera Gp6, Pseudonocardia, and Blastococcus decreased at high elevations. The Solirubrobacter (Actinobacteria) and Nocardioides (Actinobacteria) were the lowest at 3,600 m (P < 0.05; see Table S4 in the supplemental material).
In total, 1,003 OTU were significantly (P < 0.05) associated with habitats at different elevations according to indicator species analysis (see Table S5 and S6 in the supplemental material). A total of 511 indicator OTU were associated with only one of four sites; more OTU were associated with 3,200 m and 3,600 m, and fewer were associated with 3,800 m. The microbial groups associated with 3,200 m contained a higher number of OTU affiliated to Actinobacteria (e.g., Streptomyces and Pseudonocardia) and Alphaproteobacteria (e.g., Geminicoccus, Microvirga, Mesorhizobium, and Pedomicrobium), whereas those associated with 3,800 m contained a higher number of OTU affiliated with Acidobacteria (e.g., Gp1, Gp2, Gp4, and Gp7), and Deltaproteobacteria (e.g., Anaeromyxobacter and Cystobacter). In addition, 332 indicator OTU were associated with two sites. They were mainly divided into two groups. The first group, G1100 (low-elevation group, mainly associated with 3,200 m and 3,400 m), contained more OTU affiliated with Actinobacteria and Alphaproteobacteria, whereas the second group, G0011 (high-elevation group, mainly associated with 3,600 m and 3,800 m), contained a higher number of OTU affiliated with Acidobacteria and Verrucomicrobia. Indicator species analysis confirmed that OTU associated with 3,200 m and 3,400 m were quite different from those associated with 3,600 m and 3,800 m. This agreed with the changes in the relative abundances of dominant phyla after the soil transplant along the elevation gradient.
Correlations between the bacterial communities and environmental variables.
In general, TOC, moisture, soil CO2 and N2O fluxes, plant biomass, and plant coverage significantly increased as elevations decreased (P < 0.01; see Table S1 in the supplemental material). These variables showed significant and positive correlations with temperature (see Table S7 in the supplemental material), which implicated that warming might increase both C and N sequestration and certain greenhouse gas emissions. However, most soil properties (NO3−-N, NH4+-N, TN, TOC, and soil pH) and the number of plant species were more significantly correlated to their initial elevations, which indicated that these variables were relatively stable after a 2-year transplant. Soil CH4 flux was not significantly (P > 0.05) changed with elevation.
VPA was performed to quantify the relative contributions of different environmental variables to changes in the bacterial community structure by the “varpart” procedure. Soil temperature, plant biomass, and soil properties explained 31.4, 18.0, and 19.4% of the observed variations, respectively, leaving 57.0% of the variation unexplained (ANOVA, P = 0.001, Fig. 3). Plant biomass alone only explained 0.1% of the variation, and most of its contribution was shared with temperature (17.8%) and soil properties (8.0%). Soil temperature alone explained 13.6% of the variation, which was highest as a single factor. Two soil property parameters passed the permutation test (P < 0.05), including NO3−-N and TOC, explaining 6.8% (P = 0.002) and 4.2% (P = 0.046) of the total variation, respectively.
FIG 3.
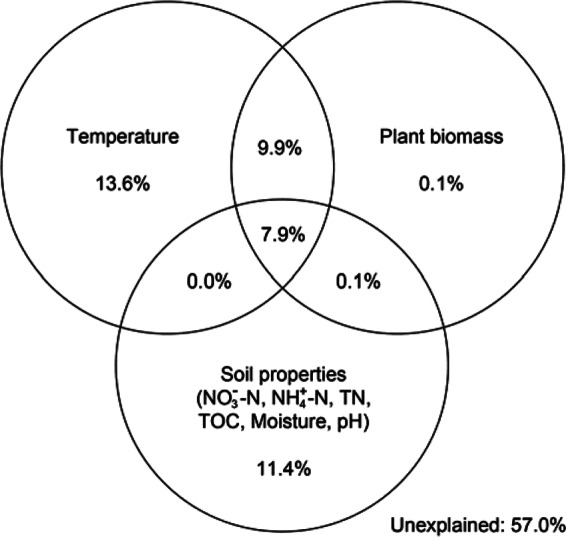
Variation partition analysis of the effects of soil temperature, plant biomass, and soil properties on bacterial community structure. Percentages are the variation of the bacterial community structure explained by the three sets of environmental factors. The relative abundances of OTU were used as input in the analysis.
The relative abundances of 47 different bacterial genera were significantly (P < 0.01) correlated with soil temperature or plant biomass (see Table S8 in the supplemental material), and 22 of them were significantly correlated with both soil temperature and plant biomass, such as Acidobacteria Gp4, Gp6, and Nitrospira. However, only four genera were exclusively and significantly correlated with soil temperature, including Pseudonocardia, Pirellula, Chitinophaga, and Sphaerobacter. This indicated that indirect impacts of temperature on bacterial communities through plants were more common than direct temperature impacts. NO3−-N was significantly (P < 0.01) correlated with 20 genera. However, only two genera, Solirubrobacter and Adhaeribacter, were significantly (P < 0.05) correlated with NH4+-N.
The relative abundances of nine genera significantly (P < 0.05) correlated with the N2O flux, in which genera Pedomicrobium and Rubrobacter showed the highest positive correlation coefficients, while Gp1 showed a negative correlation (P < 0.01) (see Table S8 in the supplemental material). The CO2 flux was significantly correlated with 33 genera at P < 0.05 and 17 genera at P < 0.01. However, the CH4 flux was only significantly correlated with three genera at P < 0.05 and two genera at P < 0.01.
DISCUSSION
Low temperature is a key factor in limiting the primary production of the alpine ecosystem in the Qinghai-Tibet plateau, which is a sensitive model ecosystem used to investigate the consequences of climate warming. Using an elevation gradient as an analog of climate change and soil transplant experiments, this model showed that soil bacterial communities originating from different elevations became more similar when they were transplanted to the same elevation and more distinct from the controls in their “home” sites. The changes of the overall community structure were found to be significant after a 2-year transplant. Meanwhile, significant changes in the relative abundances of some dominant phyla after soil transplant were also observed. Temperature was identified as the primary factor in influencing bacterial community structure. Our transplant experiment was conducted in a short geographic distance scale within soil temperature changes from 0.91 to 4.05°C in the growing season, simulating possible climate changes in the alpine ecosystem.
Soil transplant caused changes in multiple factors that influenced the bacterial community structure. Apparently, the changes in bacterial community structure were strongly influenced by soil temperature and plant biomass, which was evident by VPA and Mantel analysis (Table 1 and Fig. 3). A previous study indicates that warming directly affects plant growth and leads to shifts in plant species in this alpine ecosystem (21), which may alter the quality and quantity of soil C input and influence soil microbial community structure (30, 31). In the present study, the warming effect on aboveground vegetation was apparent as indicated by the increase of vegetation biomass, and coverage. Thus, the changes in organic C input from plants is likely an important mechanism for bacterial community responses to climate warming in the alpine meadow ecosystem. Overall, these results support our first hypothesis that alpine soil bacterial community would response significantly to climate changes, in which soil temperature and aboveground vegetation contribute significantly to shaping the bacterial community structure. We found that microbial community was more similar between 3,800 m and 3,400 m than between 3,600 m and 3,400 m (Fig. 1). The temperature is often higher at 3,800 m than at 3,600 m in May and June due to a temperature inversion effect in this region. The dynamic temperature pattern and its associated changes in plant and soil properties, e.g., moisture, may partially explain the phenomenon.
Shifts in the bacterial community structure were further evidenced by the changes in phylogenetic composition from phyla to OTU levels (Fig. 2; see also Table S4 in the supplemental material). Some bacterial groups sensitive to warming or cooling were identified. The relative abundances of many dominant bacterial groups shifted oppositively when the soil blocks were transplant upward and downward, and these bacterial groups showed symmetric responses to warming and cooling. The relative abundances of Actinobacteria (dominant order Actinomycetales), Alphaproteobacteria (dominant order Rhizobiales), and Gammaproteobacteria (dominant order Xanthomonadales) were positively correlated with soil temperature, while Betaproteobacteria (dominant order Burkholderiales) and Acidobacteria (dominant class Acidobacteria Gp4) were negatively correlated to soil temperature (Table 2). These bacterial groups were also found to react to soil transfer in a glacier forefield (32). The correlation of plant biomass with the relative abundances of dominant bacterial communities was similar to that of soil temperature. Consequently, there was a good correlation between local composition and local abiotic conditions, even after climate changes had occurred. These results fully support our second hypothesis that warming and cooling exert opposite effects on the shift of bacterial community. It implicates that a bacterial community could rebound to its original state after short-term warming or cooling events.
Three important mechanisms are possibly applied by microbial communities in response to temperature changes (4): acclimation, genotypic change within a species (evolution), and species sorting. Acclimation usually refers to physiological adjustments of individuals in response to a single environmental factor (6). Acclimation only induces minor shifts in the temperature response of a bacterium (33) or a community; thus, the structure of a community would be relatively stable when temperature changes in a narrow range (0.91 to 4.05°C in the present study) if acclimation was the main mechanism. The genotypic change within a species usually requires extreme environmental changes and a long time (4). Therefore, dramatic genotypic changes in a community were not expected to be dominant mechanisms. The significant differences were likely due to the changes in relative abundances of dominant taxa with temperature. This does not support that changes in community composition are mainly caused by rapid evolutionary adaptation through horizontal gene transfer since it could allow sensitive microorganisms to adapt to new environmental conditions and quickly return the community to its original composition (14). Our results also indicated that different species might respond to global change at different rates and directions, resulting in an abundance increase or decrease of certain taxa. The intraspecific variability in the taxon abundance may provide the potential for a community to respond rapidly and reversibly to warming events through plastic adjustment (species sorting). Although several mechanisms may play interactive roles in structuring bacterial communities, our data partially support the notion that species sorting is an important mechanism for a bacterial community to respond to climate changes in this alpine ecosystem, in which species genetically better adapted to a certain temperature range may outcompete other less-well-adapted species (4).
It is still challenging to understand the relationships among microbial processes, responses to climate changes, and phylogenetic positions among taxa (14). The reciprocal transplant approach is able to simultaneously test the effects of both community composition and environment on ecosystem functioning (34). Although the warming-associated increase in plant biomass resulted in an increase in soil C input, increases of soil C and N loss through CO2 and N2O effluxes were also observed, either in transplant blocks or in mock-transplant blocks. This might be due to warming increasing the rates of organic degradation and heterotrophic respiration. It was possibly explained in that the bacterial community had already shifted, and its activity became more similar to that of the destination community at least after a soil transplant for 2 years. Under such a shift, local environmental factors played important roles in determining gas flux. Further, the close relationship between the relative abundances of many heterotrophic bacteria and CO2 efflux revealed here indicated that microbial heterotrophic respiration was likely important to regulate soil CO2 efflux for this grassland and that the compositional changes of a bacterial community might influence the greenhouse gas fluxes, even though the complex mechanisms behind the relationship were not fully understood.
Since N2O emission is mainly a microbially regulated process, it is reasonable to speculate that the N2O flux would be highly correlated to changes in some specific genera. Previous studies indicated that nitrification might be an important mechanism influencing alpine grassland N2O emission (24). In the present study, we did not observe any significant correlations between well-known autotrophic nitrification bacteria with N2O flux, such as Nitrospira. The relative abundance of the slow-growing organism Nitrospira was high when the temperature decreased, which was opposite to the change in N2O flux with temperature. However, we noted that some bacterial genera distributed in multiple taxa were positively correlated with N2O flux, e.g., the genera Pedomicrobium and Rubrobacter (see Table S8 in the supplemental material). Most of these genera were heterotrophic and became more abundant when temperatures increased, possibly indicating the importance of heterotrophs to nitrification in this ecosystem, and yet we have no direct evidence to show their roles in N2O flux. In a soil transplanting study across a climate gradient, Balser and Firestone demonstrate that nitrification potential and N2O flux are likely driven by Gram-negative bacteria (characterized by PLFA data) (35). However, broad-based microbial indices such as biomass or lipid composition may not be sensitive enough to show significant correlations with the soil process rate. Although we do not know the physiological roles of the aforementioned uncultured microbes on N2O production, our data provided evidence that taxa in the new community function at least at different rates when combined at the community level compared to their original ones, and the differences in process rates can be attributed partially to compositional differences and not simply to physiological responses of the original community under new environmental conditions (14).
In contrast to climate warming, the decreases in aboveground plant biomass, total organic C and N content, and soil CO2 and N2O effluxes under cooling were documented here and in other ecosystems (36), suggesting that cooling reduced C sequestration by decreasing soil C input and repressing microbial activity. The balancing mechanisms of the warming or cooling effects on terrestrial C and N sequestration in Qinghai-Tibet grasslands may occur between the increase/decrease of plant C and N inputs to soil and the increase or decrease in microbial activities. This also suggests that the adjustment of biogeochemical processes may be a possible mechanism to stabilize soil C reservoirs under climate changes and keep the soil C and N stocks relatively stable in a certain time scale. This provided a partial explanation for the observation that topsoil C stocks in Qinghai-Tibet grasslands were remarkably stable (37) in the whole plateau surveyed from 1998 to 2004, even though the plateau experienced three times the average global warming rate since 1960 (18). The significant changes in bacterial community structure and greenhouse gas effluxes under warming and cooling that we observed further support that an adjustment of biogeochemical processes may accompany the shift of community structure and its associated physiological traits.
In summary, global mean temperature had increased by approximately 0.85°C between 1880 and 2012, and surface temperatures across the globe are predicted to rise an additional 0.3 to 4.8°C by the end of 21st century (38). Our soil transplant experiments in a short elevation gradient with narrow temperature difference from 0.91 to 4.05°C in the Qinghai-Tibet alpine ecosystem detected bacterial communities that were sensitive to climate changes. Further, our findings suggest that the soil microbiome is likely determined by the combined direct and indirect effects of temperature change. Temperature and plant biomass are likely primary factors influencing the responses of bacterial community structure to climate change in this alpine ecosystem. Climate warming or cooling shifts the soil bacterial community structure in an opposite direction mainly through species sorting (adjustment). This implies that the bacterial community structure and function are resilient within a range of temperature fluctuation under a certain time frame. A strong correlation between the relative abundance of specific bacterial taxa and the N2O flux possibly indicates a certain linkage between biogeochemical processes and the shifts in bacterial community structure likely through species adjustment and adaptation to climate changes at community levels.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grants 31300447 and 31400458), the Strategic Priority Research Program of the Chinese Academy of Sciences (XBD15010201 [to S.W.] and XDB15010300 [to X.L.]), the Information Program of the Chinese Academy of Sciences (XXH12504-3-18), and a postdoctoral program for J.A. (2013M542297). Q.L. was supported by a grant to W.L.'s laboratory for conducting part of the research experiments.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00557-15.
REFERENCES
- 1.Walther G-R, Post E, Convey P, Menzel A, Parmesan C, Beebee TJ, Fromentin J-M, Hoegh-Guldberg O, Bairlein F. 2002. Ecological responses to recent climate change. Nature 416:389–395. doi: 10.1038/416389a. [DOI] [PubMed] [Google Scholar]
- 2.Bardgett RD, Freeman C, Ostle NJ. 2008. Microbial contributions to climate change through carbon cycle feedbacks. ISME J 2:805–814. doi: 10.1038/ismej.2008.58. [DOI] [PubMed] [Google Scholar]
- 3.Zhou J, Xue K, Xie J, Deng Y, Wu L, Cheng X, Fei S, Deng S, He Z, Van Nostrand JD, Luo Y. 2012. Microbial mediation of carbon-cycle feedbacks to climate warming. Nat Climate Change 2:106–110. [Google Scholar]
- 4.Bárcenas-Moreno G, Gómez-Brandón M, Rousk J, Bååth E. 2009. Adaptation of soil microbial communities to temperature: comparison of fungi and bacteria in a laboratory experiment. Global Change Biol 15:2950–2957. doi: 10.1111/j.1365-2486.2009.01882.x. [DOI] [Google Scholar]
- 5.Shaver GR, Canadell J, Chapin FS, Gurevitch J, Harte J, Henry G, Ineson P, Jonasson S, Melillo J, Pitelka L, Rustad L. 2000. Global warming and terrestrial ecosystems: a conceptual framework for analysis. Bioscience 50:871–882. doi: 10.1641/0006-3568(2000)050[0871:GWATEA]2.0.CO;2. [DOI] [Google Scholar]
- 6.Bradford MA, Davies CA, Frey SD, Maddox TR, Melillo JM, Mohan JE, Reynolds JF, Treseder KK, Wallenstein MD. 2008. Thermal adaptation of soil microbial respiration to elevated temperature. Ecol Lett 11:1316–1327. doi: 10.1111/j.1461-0248.2008.01251.x. [DOI] [PubMed] [Google Scholar]
- 7.Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW. 2010. Soil microbial community responses to multiple experimental climate change drivers. Appl Environ Microbiol 76:999–1007. doi: 10.1128/AEM.02874-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheik CS, Beasley WH, Elshahed MS, Zhou X, Luo Y, Krumholz LR. 2011. Effect of warming and drought on grassland microbial communities. ISME J 5:1692–1700. doi: 10.1038/ismej.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knapp AK, Beier C, Briske DD, Classen AT, Luo Y, Reichstein M, Smith MD, Smith SD, Bell JE, Fay PA, Heisler JL, Leavitt SW, Sherry R, Smith B, Weng E. 2008. Consequences of more extreme precipitation regimes for terrestrial ecosystems. Bioscience 58:811–821. doi: 10.1641/B580908. [DOI] [Google Scholar]
- 10.Waldrop MP, Firestone MK. 2006. Response of microbial community composition and function to soil climate change. Microb Ecol 52:716–724. doi: 10.1007/s00248-006-9103-3. [DOI] [PubMed] [Google Scholar]
- 11.Lazzaro A, Gauer A, Zeyer J. 2011. Field-scale transplantation experiment to investigate structures of soil bacterial communities at pioneering sites. Appl Environ Microbiol 77:8241–8248. doi: 10.1128/AEM.05778-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djukic I, Zehetner F, Watzinger A, Horacek M, Gerzabek MH. 2013. In situ carbon turnover dynamics and the role of soil microorganisms therein: a climate warming study in an Alpine ecosystem. FEMS Microbiol Ecol 83:112–124. doi: 10.1111/j.1574-6941.2012.01449.x. [DOI] [PubMed] [Google Scholar]
- 13.Vanhala P, Karhu K, Tuomi M, Björklöf K, Fritze H, Hyvärinen H, Liski J. 2011. Transplantation of organic surface horizons of boreal soils into warmer regions alters microbiology but not the temperature sensitivity of decomposition. Global Change Biol 17:538–550. doi: 10.1111/j.1365-2486.2009.02154.x. [DOI] [Google Scholar]
- 14.Allison SD, Martiny JBH. 2008. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A 105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Rui J, Mao Y, Yannarell A, Mackie R. 2014. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol Biochem 68:392–401. doi: 10.1016/j.soilbio.2013.10.017. [DOI] [Google Scholar]
- 16.Cao G, Tang Y, Mo W, Wang Y, Li Y, Zhao X. 2004. Grazing intensity alters soil respiration in an alpine meadow on the Tibetan plateau. Soil Biol Biochem 36:237–243. doi: 10.1016/j.soilbio.2003.09.010. [DOI] [Google Scholar]
- 17.Wang SP, Luo CY, Xu GP, Chao ZG, Lin XW, Hu YG, Zhang ZH, Duan JC, Chang XF, Su AL, Li YN, Zhao XQ, Du MY, Tang YH, Kimball B. 2010. Effect of warming and grazing on litter mass loss and temperature sensitivity of litter and dung mass loss on the Tibetan plateau. Global Change Biol 16:1606–1617. doi: 10.1111/j.1365-2486.2009.02026.x. [DOI] [Google Scholar]
- 18.Li CQ, Tang MC. 1988. The climate change of Qinghai-Xizang plateau and its neighborhood in the recent 30 years. Plateau Meteorol 1:332–341. [Google Scholar]
- 19.Klein JA, Harte J, Zhao X-Q. 2004. Experimental warming causes large and rapid species loss, dampened by simulated grazing, on the Tibetan plateau. Ecol Lett 7:1170–1179. doi: 10.1111/j.1461-0248.2004.00677.x. [DOI] [Google Scholar]
- 20.Wang S, Meng F, Duan J, Wang Y, Cui X, Piao S, Niu H, Xu G, Luo C, Zhang Z, Zhu X, Shen M, Li Y, Du M, Tang Y, Zhao X, Ciais P, Kimball B, Peñuelas J, Janssens I, Cui S, Zhao L, Zhang F. 2014. Asymmetric sensitivity of first flowering date to warming and cooling in alpine plants. Ecology 95:3387–3398. doi: 10.1890/13-2235.1. [DOI] [Google Scholar]
- 21.Wang S, Duan J, Xu G, Wang Y, Zhang Z, Rui Y, Luo C, Xu B, Zhu X, Chang X, Cui X, Niu H, Zhao X, Wang W. 2012. Effects of warming and grazing on soil N availability, species composition, and ANPP in an alpine meadow. Ecology 93:2365–2376. doi: 10.1890/11-1408.1. [DOI] [PubMed] [Google Scholar]
- 22.Zhao L, Li YN, Xu SX, Zhou HK, Gu S, Yu GR, Zhao XQ. 2006. Diurnal, seasonal and annual variation in net ecosystem CO2 exchange of an alpine shrubland on Qinghai-Tibetan plateau. Global Change Biol 12:1940–1953. doi: 10.1111/j.1365-2486.2006.01197.x. [DOI] [Google Scholar]
- 23.Li X, Zhang X, Wu J, Shen Z, Zhang Y, Xu X, Fan Y, Zhao Y, Yan W. 2011. Root biomass distribution in alpine ecosystems of the northern Tibetan plateau. Environ Earth Sci 64:1911–1919. doi: 10.1007/s12665-011-1004-1. [DOI] [Google Scholar]
- 24.Yang Y, Wu L, Lin Q, Yuan M, Xu D, Yu H, Hu Y, Duan J, Li X, He Z, Xue K, van Nostrand J, Wang S, Zhou J. 2013. Responses of the functional structure of soil microbial community to livestock grazing in the Tibetan alpine grassland. Global Change Biol 19:637–648. doi: 10.1111/gcb.12065. [DOI] [PubMed] [Google Scholar]
- 25.Lin X, Wang S, Ma X, Xu G, Luo C, Li Y, Jiang G, Xie Z. 2009. Fluxes of CO2, CH4, and N2O in an alpine meadow affected by yak excreta on the Qinghai-Tibetan plateau during summer grazing periods. Soil Biol Biochem 41:718–725. doi: 10.1016/j.soilbio.2009.01.007. [DOI] [Google Scholar]
- 26.Tamaki H, Wright CL, Li X, Lin Q, Hwang C, Wang S, Thimmapuram J, Kamagata Y, Liu W-T. 2011. Analysis of 16S rRNA amplicon sequencing options on the Roche/454 next-generation titanium sequencing platform. PLoS One 6:e25263. doi: 10.1371/journal.pone.0025263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao M, Rui J, Li J, Dai Y, Bai Y, Heděnec P, Wang J, Zhang S, Pei K, Liu C, Wang Y, Zhili H, Frouz J, Li X. 2014. Rate-specific responses of prokaryotic diversity and structure to nitrogen deposition in the Leymus Chinensis steppe. Soil Biol Biochem 79:81–90. doi: 10.1016/j.soilbio.2014.09.009. [DOI] [Google Scholar]
- 30.Goldfarb KC, Karaoz U, Hanson CA, Santee CA, Bradford MA, Treseder KK, Wallenstein MD, Brodie EL. 2011. Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Front Microbiol 2:94. doi: 10.3389/fmicb.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 32.Zumsteg A, Bååth E, Stierli B, Zeyer J, Frey B. 2013. Bacterial and fungal community responses to reciprocal soil transfer along a temperature and soil moisture gradient in a glacier forefield. Soil Biol Biochem 61:121–132. doi: 10.1016/j.soilbio.2013.02.017. [DOI] [Google Scholar]
- 33.Leroi AM, Bennett AF, Lenski RE. 1994. Temperature acclimation and competitive fitness: an experimental test of the beneficial acclimation assumption. Proc Natl Acad Sci U S A 91:1917–1921. doi: 10.1073/pnas.91.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed HE, Martiny JBH. 2007. Testing the functional significance of microbial composition in natural communities. FEMS Microbiol Ecol 62:161–170. doi: 10.1111/j.1574-6941.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 35.Balser TC, Firestone MK. 2005. Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry 73:395–415. doi: 10.1007/s10533-004-0372-y. [DOI] [Google Scholar]
- 36.Dukes JS, Chiariello NR, Cleland EE, Moore LA, Shaw MR, Thayer S, Tobeck T, Mooney HA, Field CB. 2005. Responses of grassland production to single and multiple global environmental changes. PLoS Biol 3:e319. doi: 10.1371/journal.pbio.0030319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Fang J, Smith P, Tang Y, Chen A, Ji C, Hu H, Rao S, Tan K, He J. 2009. Changes in topsoil carbon stock in the Tibetan grasslands between the 1980s and 2004. Global Change Biol 15:2723–2729. doi: 10.1111/j.1365-2486.2009.01924.x. [DOI] [Google Scholar]
- 38.IPCC. 2014. Fifth assessment report—climate change 2014: impacts, adaptation, and vulnerability. IPCC, Geneva, Switzerland: http://www.ipcc.ch/report/ar5/wg2/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.