Abstract
While the food-borne pathogen Listeria monocytogenes can persist in food associated environments, there are no whole-genome sequence (WGS) based methods to differentiate persistent from sporadic strains. Whole-genome sequencing of 188 isolates from a longitudinal study of L. monocytogenes in retail delis was used to (i) apply single-nucleotide polymorphism (SNP)-based phylogenetics for subtyping of L. monocytogenes, (ii) use SNP counts to differentiate persistent from repeatedly reintroduced strains, and (iii) identify genetic determinants of L. monocytogenes persistence. WGS analysis revealed three prophage regions that explained differences between three pairs of phylogenetically similar populations with pulsed-field gel electrophoresis types that differed by ≤3 bands. WGS-SNP-based phylogenetics found that putatively persistent L. monocytogenes represent SNP patterns (i) unique to a single retail deli, supporting persistence within the deli (11 clades), (ii) unique to a single state, supporting clonal spread within a state (7 clades), or (iii) spanning multiple states (5 clades). Isolates that formed one of 11 deli-specific clades differed by a median of 10 SNPs or fewer. Isolates from 12 putative persistence events had significantly fewer SNPs (median, 2 to 22 SNPs) than between isolates of the same subtype from other delis (median up to 77 SNPs), supporting persistence of the strain. In 13 events, nearly indistinguishable isolates (0 to 1 SNP) were found across multiple delis. No individual genes were enriched among persistent isolates compared to sporadic isolates. Our data show that WGS analysis improves food-borne pathogen subtyping and identification of persistent bacterial pathogens in food associated environments.
INTRODUCTION
Listeria monocytogenes is an opportunistic food-borne pathogen responsible for approximately 250 deaths per year in the United States (1). The annual costs of listeriosis, including morbidity, mortality, and lost wages, are estimated at $2.8 billion (2). A 2003 risk assessment attributed 90% of listeriosis cases in the United States to consumption of contaminated ready-to-eat (RTE) deli meats (3), and most (>60%) of U.S. listeriosis cases linked to RTE deli meats were estimated to be due contamination during retail handling and slicing (4, 5). Consequently, the retail deli environment is a focal point for listeriosis reduction efforts.
L. monocytogenes can persist in food-associated environments for months to years (6–8). Persistent strains have been linked to outbreaks of food-borne disease. For example, L. monocytogenes that was responsible for an outbreak linked to 29 cases and 4 deaths persisted in the source plant for at least 12 years (9). Therefore, the food processing industry has implemented the seek-and-destroy process to identify and eliminate point sources of persistence through enhanced environmental monitoring, sanitation, and equipment and process design (10).
One challenge for persistent pathogen control is to differentiate true persistence from repeated reintroduction of a given strain from, for example, an external supplier (11). The term “persistence” has been used in the food-borne pathogen control literature (6, 8) to describe both (i) long-term survival of pathogens in various environments, e.g., the host, a food processing facility, and a natural environment, and (ii) empirical rules to identify when a pathogen persists. Empirical rules for calling a strain persistent generally contain two requirements (i) that isolates of a persistent strain are indistinguishable by a particular molecular subtyping method and (ii) that isolates from the persistent strain are observed over a sufficiently long time period. Observation of both strains that truly persist (are continually present) and strains that are repeatedly reintroduced can produce environmental sampling results that satisfy empirical rules for persistence. To improve the empirical differentiation of persistent from sporadic strains, we have previously developed machine learning models of expert opinion (12) that incorporate information into classification such as the isolation location and the relative frequency of a subtype, but that use the until-recently gold standard technique of pulsed-field gel electrophoresis (PFGE) for L. monocytogenes subtyping.
Since whole-genome sequencing (WGS) technology is rapidly transforming epidemiology (13) and food safety (14), we surmised that WGS could improve subtyping and empirical rules to identify persistent L. monocytogenes. WGS technology has improved food-borne disease epidemiology of Salmonella (15, 16), Escherichia coli (17), and L. monocytogenes (18), including tracing a listeriosis outbreak back to a food processing facility source (19). In the present study, we utilized data from an existing longitudinal environmental study of L. monocytogenes from 30 retail delis in three U.S. states over 2 years (20) to improve the identification of persistent L. monocytogenes by PFGE results. Our goals were to (i) apply WGS-based methods, including single-nucleotide polymorphism (SNP)-based phylogenetics to improve molecular subtyping of L. monocytogenes from the retail deli environment, (ii) explore SNP differences as a quantitative metric to differentiate persistent from repeatedly reintroduced strains, and (iii) identify genetic determinants of L. monocytogenes persistence.
MATERIALS AND METHODS
Isolate selection.
A previously reported longitudinal study of L. monocytogenes in the retail deli environment (20) provided L. monocytogenes isolates for the study reported here. Briefly, this previous study consisted of phase 1, monthly longitudinal sampling of 15 retail delis for 3 months before the start of daily operations, and phase 2, monthly longitudinal sampling of 30 delis (including the previous 15) for 6 months during daily operation. In addition, there were two not-yet-published phases: phase 3, enhanced standard sanitation operating procedures, and phase 4, a monthly longitudinal sampling of the same 30 delis for 6 months during operation. All isolates were previously subtyped by AscI and ApaI PFGE. A given L. monocytogenes PFGE type was identified as putatively persistent in a given deli if an indistinguishable PFGE type was isolated on >1 phase 1 or 2 sampling. This approach identified 31 putative persistence events among 14 of 30 delis, with 19 PFGE types represented.
Details for all isolates selected for sequencing are found in Table S1 in the supplemental material and can be accessed through www.foodmicrobetracker.com (search by reference). Both persistent and sporadic (i.e., nonpersistent) isolates were sequenced. For each persistence event, one random isolate was selected from each sampling day a given PFGE type was isolated from a given deli; this yielded 139 “persistent” isolates. For all PFGE types represented among persistent isolates, 1 random “sporadic” isolate of the same PFGE type was selected from each deli where the PFGE type was found on only 1 day; this yielded 29 sporadic isolates with 11 unique PFGE types from 19 delis. Two sets of comparison isolates with PFGE types represented among persistent isolates were chosen, including (i) 7 isolates from phase 4 of the same study and (ii) 13 “other” isolates putatively persistent in a seafood plant, sausage plant, dairy farm, and another retail deli study.
Genome sequencing.
Isolates were maintained at −80°C in 15% (vol/vol) glycerol-brain heart infusion (BHI) medium in a 96-deep-well format (Nunc U96 PP2ml Deepwell Natural; Fisher Scientific, Pittsburgh, PA). Prior to DNA extraction, isolates were inoculated into 1.5 ml of BHI, incubated overnight at 37°C, and pelleted. DNA extraction (using a 96 DNeasy Blood and Tissue kit; Qiagen, Valencia, CA) proceeded according to the manufacturer's protocol with a Gram-positive pretreatment in 180 μl of lysozyme at 20 mg/ml for 30 min at 56°C and longer centrifugation times (e.g., 15 min for lysate binding) at a lower centrifugal force (3,320 relative centrifugal force). DNA was eluted in 50 μl of Tris-HCl at pH 8.0. Initial spectrophotometric quality assessment (Take3 carriage on the Synergy H1 microplate reader; BioTek Instruments, Inc., Winooski, VT) revealed an acceptable DNA concentration (median, 40 ng/μl; minimum, 14 ng/μl) and purity (A260/280 median = 1.87, minimum = 1.75; A260/230 median = 2.44, minimum = 2.00) ratios.
DNA samples were quantified with a fluorescent nucleic acid dye (Picogreen; Invitrogen, Paisley, United Kingdom), and libraries were prepared for sequencing using a Nextera XT DNA sample preparation kit and an associated Nextera XT index kit with 96 indices (Illumina, Inc., San Diego, CA). Pooled samples were sequenced with two lanes of a HiSeq 2500 rapid run with 2×151-bp paired-end sequencing. Due to low initial coverage (<2×), two isolates (FSL R8-5528 and FSL R8-7653) were resequenced with a MiSeq 2×301-bp run with a Nextera Mate pair sample prep kit.
Automated genome assembly and kSNP tree pipeline.
An in-house pipeline was scripted in UNIX Bash shell modules for preprocessing, de novo genome assembly, alignment free SNP calling, and phylogenetic analysis. Raw read quality was assessed with FastQC (v0.10.1 [http://www.bioinformatics.babraham.ac.uk/projects/fastqc/]). Illumina adapter sequences and low-quality sequence were trimmed using Trimmomatic (v0.32) (21) default settings. Trimmed, paired reads were de novo assembled using SPAdes (v3.0.0) (22) with the suggested k-mer set for prokaryotic assembly: 21, 33, 55, 77, 99, and 127 bp. Any contig <500 bp in length or with a <1× average k-mer coverage was removed. Filtered contigs were used for alignment-free SNP-based phylogenetics using kSNP (v2.0) (23) and a k-mer size of 19. Here, a diversity set of 44 additional L. monocytogenes genomes was added to the analysis (see Table S2 in the supplemental material). Local support values for the core (present in all genomes) SNP, maximum-likelihood tree were calculated using FastTree (v2.1.7) (24). Phylogenetic trees were drawn using the ape package (v3.1-1) (25) for R (v3.0.1 [http://www.r-project.org/]).
Additional phylogenetic analyses.
Isolates classified into genetic lineage I represented the vast majority (179/188) of isolates analyzed. Therefore, isolates from lineage I and II were analyzed separately to allow for adapting analytical methods to the large difference in isolate set sizes. The Cortex variation assembler (26) was used to detect SNPs, insertions and deletions (indels), and complex variants (e.g., phased SNPs). Primary analysis used the reference based independent analysis workflow, which includes only variants with positions present in the reference genome (lineage I reference genome J1776, GenBank accession number CP006598.1; lineage II reference genome EGDe, GenBank accession number AL591824.1 ). For secondary analysis of accessory genome SNPs, lineage I genomes were also analyzed with an independent, nonreference workflow. Nearly all complex variants were phased SNPs. They were filtered out of phylogenetic analysis because they are closely spaced (27), and their positions corresponded to regions of recombination detected using Gubbins (28).
The BEAST software package (v2.1.3) (29) was used to estimate separate phylogenies for lineage I and lineage II isolates. For lineage I isolates, a tip-dated phylogeny using isolation dates estimated dates of divergence from most recent common ancestors (MRCA) after two model selection steps. First, maximum-likelihood model selection using the default parameters in MEGA (v6) (30) identified the general time-reversible model as the best-fitting (lowest Bayesian information criterion) nucleotide substitution model. Then, model selection by path sampling and calculation of Bayes factors (31) was used to select between either a strict or relaxed lognormal clock and either a coalescent constant or Bayesian skyline population model. Model selection used ten steps of 5 to 10 million generations. The best supported model was computed in ten individual runs of 100 million generations and tracer was used to determine burn-in and verify convergence and appropriate mixing. There were insufficient lineage II isolates (n = 9) to estimate a tip-dated phylogeny, so a nondated phylogenetic tree was estimated from two individual runs of 100 million generations.
Congruence between kSNP identification of core SNPs and Cortex_var identification of high-quality SNPs was tested using Concaterpillar (v1.8) (32) default settings. Only the lineage I retail deli isolates common to both trees (171 isolates) were used. Topological congruence was tested first. Topologically congruent trees were tested for branch length congruence.
In silico MLST and sigB allelic typing.
Short Read Sequence Typer (v2, SRST2) (33) was used to determine seven gene multilocus sequence types (MLSTs; downloaded on 3 September 2014 from the Institut Pasteur [http://www.pasteur.fr/recherche/genopole/PF8/mlst/Lmono.html] [34]). Only the first 400,000 quality-trimmed sequencing reads were mapped, for an average of 30× coverage. Mismatches to existing alleles were scored as novel sequence types. The discriminatory power of MLST-, PFGE-, and SNP-based subtyping was compared by using Simpson's index of diversity (35).
Comparisons to previous subtyping methods were used to identify, and exclude, sequence data from when an incorrect isolate was likely sequenced. SRST2 was used to assign sigB allelic types to each genome sequence. If the sigB allelic type for a genome sequence did not match an isolate's sigB type previously determined by Sanger sequencing (20), then this isolate was excluded from analysis (two isolates were excluded: FSL R8-5744 and FSL R8-5866). In addition, isolate FSL N3-0993 was excluded because the genetic lineage of the sequenced isolate (lineage II) did not match the previously determined genetic lineage for the subtype determined from PCR serogrouping (the isolate was previously assigned to serotypes 1/2b, 3b, or 7 based on reference 36, a finding consistent with genetic lineage I [37]). The three isolates described above are not included in the 188 isolates that are discussed throughout the present study (whereas 191 isolates were sequenced, only 188 are reported on).
Whole-genome alignment.
Contigs from selected isolates were aligned using progressiveMauve (MAUVE, v2.3.1) (38) to identify genomic regions unique to particular PFGE types. The “−pan_genome_matrix” option of cortex_var was then used to screen all sequenced isolates for presence (>70% coverage by 33-bp k-mers) of identified genomic regions. BLASTn analysis of the three identified genomic regions found matches to prophage related genes. Therefore, these regions were submitted to PHAST (http://phast.wishartlab.com/) (39) for prophage annotation.
Gene presence or absence analyses.
To identify genes that may contribute to persistence or may be responsible for PFGE type differences, ITEP (the Integrated Toolkit for Exploration of microbial Pangenomes) (40) was used to generate a database of orthologous genes present in a representative subset of lineage I isolates. To make computation feasible, the isolate set was reduced to 121 isolates by (i) retaining only phase 1 and 2 isolates and (ii) retaining at most three isolates for each persistence event, isolates from the first time point, last time point, and a random isolate from the middle time points of isolation. The reduced set included 92 putatively persistent and 29 sporadic isolates (see Table S1 in the supplemental material). A Fisher exact test with a false discovery rate (FDR) correction was used to identify genes significantly enriched among groups of isolates while controlling the overall FDR to 5% of the significant genes.
Quantitative persistence analysis.
To quantitatively discriminate persistent strains from repeatedly reintroduced strains, we computed (i) the SNP count difference between all persistent isolates with a given SNP-based subtype in a given deli and (ii) the SNP count difference between the isolates used for the initial SNP count and all isolates with the same SNP-based subtype obtained in other delis. These measures are designated as (i) the “within-persistence-event SNP count” and (ii) the “comparison SNP count.” Aggregated SNP counts were displayed as box plots using the ggplot2 (41) package of R (42). These calculations were performed for core genome SNPs (referenced based workflow in Cortex_var), presented in the main body of the text, and accessory genome SNPs (independent workflow), presented in the supplemental material. A permutation-based statistic was used to determine whether median “within-persistence-event SNP counts” were significantly fewer than median “comparison SNP counts” using 1,000 permutations of deli labels. FDR-adjusted P values of <0.05 were considered significant. Prior to aggregation, three sets of PFGE patterns that differed by three or fewer bands were grouped together as follows: SNP group 1, grouping isolates with PFGE pattern CU-11-320, CU-8-96, and CU-40-96; SNP group 2, grouping CU-258-322, CU-258-323 and CU-259-322; and SNP group 3, grouping CU-262-391 and CU-262-79. Each of the SNP groups were treated as unique subtypes for the within and between group analyses.
Nucleotide accession numbers.
Raw sequence data and de novo assembled contigs have been deposited to the appropriate GenBank database (Sequence Read Archive [SRA] and Whole Genome Shotgun) under BioProject accession number PRJNA245909. Individual genome sequencing metrics and GenBank accessions are listed in Table S3 in the supplemental material.
RESULTS
WGS improves molecular subtyping.
A total of 188 L. monocytogenes isolates, including 175 isolates from a longitudinal study of L. monocytogenes persistence in retail delis and 13 comparison isolates from other food associated environments, were characterized by whole-genome sequencing (WGS). Median genome coverage for these isolates was 94-fold (ranging from 8.0- to 360-fold), with de novo assembly yielding a median of 26 contigs per genome (ranging from 12 to 456 contigs) and a median assembled genome size of 3.09 Mb (ranging from 2.88 to 3.14 Mb; see Table S3 in the supplemental material).
In an initial analysis, L. monocytogenes MLST data (34) extracted from the 188 genome sequences differentiated the isolates into 10 sequence types (STs), yielding a Simpson's index of discrimination (SID) of 0.74. Previously generated PFGE data differentiated these isolates into 19 PFGE types (SID = 0.91). PFGE and MLST findings showed a many-to-many relationship (Table 1). Isolates of the 10 MLST STs were differentiated into 1, 4, or 6 PFGE types for each ST. Isolates of 17 of the 19 PFGE types had only a single MLST ST for each PFGE type; the remaining two PFGE types were differentiated into a predominant MLST, as well as a second ST represented by a single isolate which differs from the majority ST in only one gene (Table 1).
TABLE 1.
Comparison of isolate PFGE type to in silico MLST
| Isolate PFGE type | No. of isolates observed with in silico MLST: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 257 | 296 | 323 | 5 | 6 | 635 | 87 | 9 | Novel | |
| CU-11-282 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| CU-11-320 | 0 | 0 | 0 | 0 | 36 | 0 | 0 | 0 | 0 | 0 |
| CU-11-326 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| CU-182-173 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 |
| CU-258-322 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 |
| CU-258-323 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 |
| CU-258-69 | 0 | 0 | 0 | 0 | 0 | 18 | 0 | 0 | 0 | 0 |
| CU-259-322 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 |
| CU-262-318 | 3 | 1a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CU-262-319 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CU-262-334 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CU-262-79 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CU-294-321 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| CU-296-330 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| CU-40-96 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 |
| CU-55-266 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CU-57-267 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 |
| CU-8-340 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 |
| CU-8-96 | 0 | 0 | 0 | 0 | 19 | 0 | 0 | 0 | 0 | 1b |
Sequence type 257 differs from sequence type 2 only in the ldh allelic type.
The novel sequence type differs from sequence type 5 only in the dat allelic type.
WGS for all 188 isolates were also used for a SNP-based phylogenetic analysis, which incorporated isolate source information to classify isolates into WGS-based SNP clades. Briefly, WGS-derived SNP data (16,097 core SNPs) were used to first generate a maximum-likelihood phylogeny of all 188 isolates plus a set of 44 reference isolates (see Fig. S1 in the supplemental material). A total of 179 and 9 of the 188 isolates classified into previously reported L. monocytogenes lineages I and II (43). A tip-dated phylogenetic analysis was conducted for the 179 lineage I isolates using 9,376 differentiating SNPs (Fig. 1) and the best-fitting Bayesian Skyline population model with relaxed lognormal clock (see Table S4 in the supplemental material). The tree using 9,376 SNPs differentiating lineage I isolates was not topologically incongruent (P value of >0.05) with the tree (see Fig. S1 in the supplemental material) from 16,097 core genome SNPs differentiating linage I and II isolates. Branch lengths did differ significantly (P < 0.001).
FIG 1.
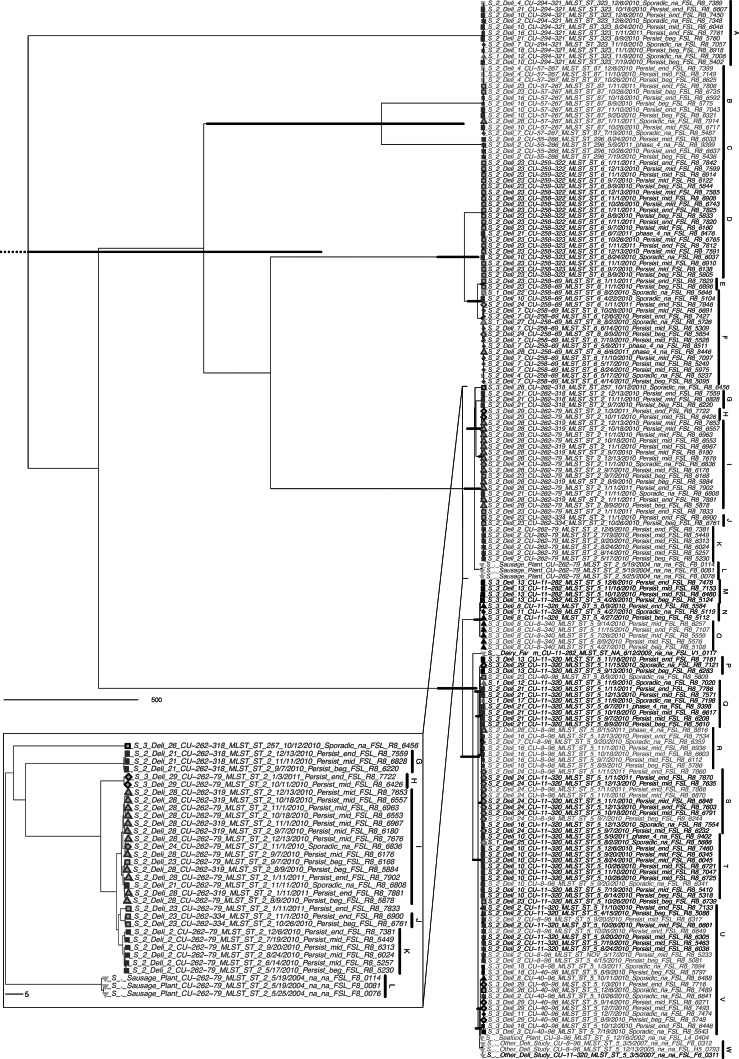
Maximum clade credibility tree from 9,376 core genome SNPs differentiating 179 lineage I isolates. Node bars indicate the 95% highest posterior density interval for the divergence time for all nodes with >90% posterior probability; the scale is in years. Tree tips are shaded by PFGE type and list isolate metadata: state number, deli number, PFGE type, MLST sequence type, isolation date, persistence status and timing, and isolate ID number. Tip symbols are shaded by state (light gray is state 1 or a comparison isolate not from the retail deli study, gray is state 2, black is state 3) and shaped by deli of isolation. The inset panel shows the higher-resolution details available in this large phylogenetic tree. Clade labels correspond to Table 4.
The Bayesian analysis revealed 23 clades among lineage I isolates (Fig. 1), including 167 persistent or sporadic isolates and 6 comparison isolates. A separate Bayesian phylogeny of lineage II isolates revealed three epidemiologically relevant clades representing four persistent isolates and five comparison isolates (Fig. 2). A clade was considered epidemiologically relevant if it (i) is well supported (>90% posterior probability; here, all had 100% support) and (ii) contains isolates predominantly from either a given deli or a given state; four clades (A, F, Q, and V) contained isolates from multiple states but were classified as specific clades since they represented a single PFGE type (described in more detail below). Three clades grouped isolates from other studies separately from isolates from retail deli project isolates with identical MLST and PFGE subtypes. These clades comprise lineage I isolates from a sausage plant (clade L, Fig. 1) and a separate retail deli study (clade W, Fig. 1) and lineage II isolates from a smoked seafood plant (clade Z, Fig. 2). In addition to the 26 clades, WGS-based SNP data identified six isolates that each represented a distinct WGS-SNP group. Based on these 32 groups, the SID for WGS-SNP-based subtyping was 0.95, compared to 0.91 and 0.75 for PFGE and MLST.
FIG 2.
Maximum clade credibility tree from 2,993 core genome SNPs differentiating nine lineage II isolates. A “●” symbol indicates a node with >90% posterior probability. Tree tips are shaded by PFGE type. Tip symbols are shaded by state (the light gray is state 1 or a comparison isolate not from the retail deli study; the dark gray is state 2) and shaped by deli of isolation. Clade labels correspond to Table 4.
Comparative genomic analyses identify prophage regions correlated with PFGE pattern differences of three or fewer bands.
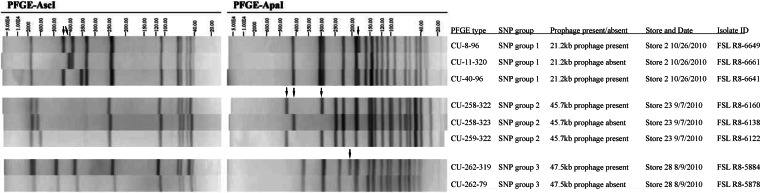
WGS-SNP-based phylogenetics (Fig. 1) revealed isolates with a given PFGE type either grouped into a monophyletic clade (e.g., clades A, B, and C) or clustered with isolates representing one or two other PFGE types (e.g., clade D). Three well-supported phylogenetic groups included multiple PFGE types: clade D (PFGE types CU-259-322, CU-258-322, and CU-258-323), clade I (PFGE types CU-262-79 and CU-262-319), and clades P to W (PFGE types CU-11-320, CU-8-96, and CU-40-96, see Fig. 1). Within each of these three monophyletic groups, isolates had PFGE patterns that differed by three or fewer bands in each of the two restriction enzyme patterns (Fig. 3).
FIG 3.
Representative PFGE patterns for three sets of PFGE types (i) grouped by WGS-SNP-based phylogenetics into well-supported clades and (ii) differed by three or fewer bands for each restriction enzyme; these sets were subsequently grouped into SNP groups. In each of the three sets, a single prophage region was identified (see Table 2) that explains the difference between the top two PFGE patterns in the set; band differences are indicated by arrows. Genomic regions that could explain the differences in the third PFGE pattern for the top two sets of patterns (CU-40-96 and CU-259-322, respectively) could not be identified.
Whole-genome alignments and subsequent read mapping (Table 2), as well as gene presence or absence analysis (see Table S4 in the supplemental material), identified 20- to 50-kb prophage regions that differentiate monophyletic PFGE types. For example, present in all six isolates with PFGE pattern CU-262-319 but absent in all isolates with PFGE pattern CU-262-79 is a 47.5-kb region encoding an intact prophage with 72 of 79 coding sequences most homologous to Listeria phage B054. Similarly, the presence of a 21.2- or 45.7-kb prophage correlated to the difference between patterns CU-8-96 and CU-11-320 or between patterns CU-258-322 and CU-258-323, respectively. Gain or loss of a prophage may be responsible for differences in PFGE patterns.
TABLE 2.
Prophage regions associated with isolate PFGE typesa
| Isolates with PFGE pattern (AscI-ApaI) | No. of isolates with a: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 21.2-kb prophage with unknown homologyb |
45.7-kb prophage with homology to LP_030_3c |
47.5-kb prophage with homology to B054d |
|||||||
| No. absent | No. present | %RP | No. absent | No. present | %RP | No. absent | No. present | %RP | |
| CU-8-340 | 0 | 5 | 96 (96–96) | 5 | 0 | <35 | 5 | 0 | <35 |
| CU-40-96 | 0 | 12 | 97 (91–98) | 12 | 0 | <35 | 12 | 0 | <35 |
| CU-8-96 | 0 | 20 | 97 (94–100) | 20 | 0 | <35 | 20 | 0 | <35 |
| CU-11-320 | 36 | 0 | <35 | 36 | 0 | <35 | 36 | 0 | <35 |
| CU-259-322 | 5 | 0 | <35 | 0 | 5 | 100 (100–100) | 5 | 0 | <35 |
| CU-258-322 | 7 | 0 | <35 | 0 | 7 | 100 (99–100) | 7 | 0 | <35 |
| CU-258-323 | 8 | 0 | <35 | 8 | 0 | <35 | 8 | 0 | <35 |
| CU-258-69 | 18 | 0 | <35 | 18 | 0 | <35 | 0 | 18 | 85 (83–97) |
| CU-262-318 | 4 | 0 | <35 | 4 | 0 | <35 | 0 | 4 | 75 (72–83) |
| CU-262-334 | 2 | 0 | <35 | 2 | 0 | <35 | 0 | 2 | 97 (97–97) |
| CU-262-319 | 6 | 0 | <35 | 6 | 0 | <35 | 0 | 6 | 100 (100–100) |
| CU-262-79 | 21 | 0 | <35 | 21 | 0 | <35 | 21 | 0 | <35 |
| CU-11-282 | 5 | 0 | <35 | 5 | 0 | <35 | 5 | 0 | <35 |
| CU-11-326 | 3 | 0 | <35 | 3 | 0 | <35 | 3 | 0 | <35 |
| CU-294-321 | 11 | 0 | <35 | 11 | 0 | <35 | 11 | 0 | <35 |
| CU-55-266 | 4 | 0 | <35 | 4 | 0 | <35 | 4 | 0 | <35 |
| CU-57-267 | 12 | 0 | <35 | 12 | 0 | <35 | 12 | 0 | <35 |
The numbers absent or present were determined by calculating the percentage of 33-bp k-mers in prophage present in each isolate's genome. Prophage was considered present if >70% of the k-mers were present. All calls of absent had <35% of the prophage k-mers. Italicized data indicate PFGE types grouped into SNP groups: SNP group 1 (PFGE patterns CU-11-320, CU-8-96, and CU-40-96), SNP group 2 (CU-258-322, CU-258-323, and CU-259-322), and SNP group 3 (CU-262-319 and CU-262-79). Boldface data indicate pairs of PFGE types which differ by ≤3 bands (Fig. 3) and isolates that differ by prophage presence. %RP, percent region present. %RP values are reported as medians; the corresponding range is indicated in parentheses.
That is, a prophage identified in 23-kb contig 38 from the FSL R8-5523 genome. PHAST (www.phast.wishartlab.com/) annotated an intact, 21.2-kb prophage with 26 coding sequences (CDS). No single reference phage showed homology to the majority of the CDS.
That is, a prophage identified in 101-kb contig 9 from the FSL R8-7842 genome. PHAST annotated an intact, 45.7-kb prophage with 76 coding sequences. Of those CDS, 43 were homologous to Listeria phage LP_030_3 (the sequence was reported previously [45]).
That is, a prophage identified in 169-kb contig 8 from the FSL R8-5884 genome. PHAST annotated an intact, 47.5-kb prophage with 79 coding sequences. Of those CDS, 72 were homologous to Listeria phage B054 (the sequence was reported previously [44]).
We could not identify a genetic element that could explain the two band differences between PFGE patterns CU-259-322 and CU-258-322 or between CU-40-96 and CU-8-96, including by screening for eight previously described L. monocytogenes plasmids (see Table S6 in the supplemental material). For subsequent analyses, we grouped isolates that represented monophyletic PFGE patterns into SNP group 1 (PFGE patterns CU-11-320, CU-8-96, and CU-40-96), SNP group 2 (CU-258-322, CU-258-323, and CU-259-322), and SNP group 3 (CU-262-319 and CU-262-79).
WGS-based phylogenies suggest distinct transmission and distribution patterns of L. monocytogenes clades with recent common ancestors of <10 years.
Initial phylogenetic analysis (detailed above) identified 26 clades, three of which were exclusively composed of isolates from sources other than retail deli study (Table 3) and are not further discussed. All 23 of the remaining clades represented isolates of either (i) a single PFGE type (12 clades; e.g., clades A, B, and C, Fig. 1) or (ii) a single SNP group composed of 2 to 3 PFGE types (11 clades; e.g., clades D, I, S, Fig. 1); WGS differentiated distinct clades within a given SNP group (e.g., SNP group 1 was differentiated into 8 clades). Classifying clades based on geographic origin of the isolates (Table 4) revealed (i) deli-specific clades (all isolates from a single deli), (ii) state-specific clades (all isolates from delis located in a single state), and (iii) multistate clades (isolates from delis in multiple states).
TABLE 3.
Epidemiologically relevant phylogenetic clades of isolates from studies other than the retail deli study reported by Simmons et al. (20)
| Clade ID | PFGE type or SNP group subtype represented within cladea | 95% HPD for date of divergence from MRCA (yr)b | Isolate source (reference) | No. of median pairwise SNP differences among isolates of the clade (no. of isolates within the clade) |
|---|---|---|---|---|
| L | SNP group 3 (CU-262-79) | 7.3–10.5 | Sausage plant (62) | 2 (3) |
| W | SNP group 1 (CU-11-320 and CU-8-96) | 5.7–9.2 | Other retail deli study (63) | 5 (3) |
| Z | CU-182-173 | NEc | Smoked seafood plant (55, 64, 65) | 24 (5) |
A SNP group subtype is comprised of two or three individual PFGE types (indicated in parentheses where applicable). See the text for additional details.
HPD, highest posterior density. MRCA, most recent common ancestor.
NE, not estimated (the MRCA was not estimated for genetic lineage II isolates due to insufficient isolates to allow for adequate estimation by tip dating).
TABLE 4.
Epidemiologically relevant phylogenetic clades from the retail deli study
| Phylogeographic clade classificationa | Clade ID | PFGE type or SNP group subtype represented within cladeb | 95% HPD for date of clade divergence from MRCA (yr)c | Deli ID of isolates putatively: |
Median SNP count differences among isolates of the same PFGE type or SNP group: |
|||
|---|---|---|---|---|---|---|---|---|
| Persistent | Sporadic | Within a putative persistence event (no. of isolates within deli) | Among isolates from a given deli and all other delis (no. of isolates in other delis) | FDR-corrected P value for significantly fewer median SNP differencesf | ||||
| Deli specific | M | CU-11-282 | 1.3–4.3 | 13 | NAd | 3 (4) | NA | NA |
| J | CU-262-334 | 0.8–2.6 | 23 | NA | 2 (2) | NA | NA | |
| C | CU-55-266 | 1.2–4.3 | 2 | NA | 3 (4) | NA | NA | |
| O | CU-8-340 | 1.8–7.0 | 8 | NA | 10.5 (5) | NA | NA | |
| E | CU-258-69 | 1.2–8.4 | 23 | NA | 13 (2) | 77 (16) | 0.025 | |
| G | CU-262-318 | 1.0–3.5 | 21 | NA | 2 (3) | 102 (1) | 0.357 | |
| U | SNP group 1 (CU-11-320 and CU-8-96) | 2.2–7.0 | 2 | NA | 9 (11) | 24 (53) | 0.023 | |
| K | SNP group 3 (CU-262-79) | 1.4–3.7 | 2 | NA | 2 (10) | 10 (18) | <0.001 | |
| H | SNP group 3 (CU-262-79) | 0.9–3.8 | 29 | NA | 4 (2) | 25 (22) | 0.008 | |
| X | CU-296-330 | NEe | 23 | NA | 4 (2) | NA | NA | |
| Y | CU-182-173 | NE | 19 | NA | 1 (2) | NA | NA | |
| State specific | N | CU-11-326 | 1.3–3.1 | 8 | 11 | 2 (2) | 1 (1) | 1.000 |
| B | CU-57-267 | 1.6–5.6 | 10 | 7, 28 | 3 (3) | 5 (9) | 0.159 | |
| 16 | 0 (2) | 3.5 (10) | 0.071 | |||||
| 23 | 3 (2) | 4 (10) | 0.433 | |||||
| 4 | 5 (3) | 5 (9) | 0.784 | |||||
| P | SNP group 1 (CU-11-320) | 1.5–7.7 | 13 | 29 | 12 (2) | 41 (62) | 0.082 | |
| R | SNP group 1 (CU-8-96) | 2.6–10.8 | 16 | 7 | 22 (6) | 51 (58) | <0.001 | |
| S | SNP group 1 (CU-11-320 and CU-8-96) | 1.5–5.1 | 24 | 16 | 4 (10) | 18 (54) | 0.007 | |
| D | SNP group 2 (CU-258-323, CU-258-322, and CU-259-322) | 1.9–6.6 | 23 | 10, 21 | 7 (18) | 9 (2) | 0.159 | |
| I | SNP group 3 (CU-262-79 and CU-262-319) | 1.6–4.4 | 23 | 21, 24 | 10 (2) | 8.5 (22) | 0.784 | |
| 28 | 4 (12) | 9 (12) | <0.001 | |||||
| Multistate | F | CU-258-69 | 2.2–7.8 | 24 | 4, 10, 22, 27, 28 | 10 (2) | 8 (16) | 0.808 |
| 7 | 5 (9) | 8 (9) | 0.025 | |||||
| A | CU-294-321 | 1.4–4.3 | 10 | 2, 4, 7, 12 | 2 (3) | 2 (8) | 0.808 | |
| 16 | 2 (2) | 2 (9) | 0.808 | |||||
| 21 | 3 (2) | 3 (9) | 0.959 | |||||
| T | SNP group 1 (CU-11-320 and CU-8-96) | 2.0–6.8 | 10 | 25 | 6 (10) | 20 (54) | 0.004 | |
| V | SNP group 1 (CU-40-96) | 1.8–7.0 | 18 | 2, 3, 8, 11, 26 | 6 (3) | 46 (61) | 0.008 | |
| 29 | 3 (3) | 43 (59) | <0.001 | |||||
| Q | SNP group 1 (CU-11-320) | 1.4–5.7 | 21 | 12, 17 | 4 (6) | 22 (58) | 0.004 | |
Deli-specific clade, all isolates in a clade were isolated from a single deli; state-specific clade, all isolates in a given clade were isolated from the same states; multistate clade, all isolates in a given clade were isolated from multiple delis across multiple states.
A SNP group subtype is comprised of two or three individual PFGE types (indicated in parentheses where applicable), See the text for details.
HPD, highest posterior density; MCRA, most recent common ancestor.
NA, not applicable. A deli-specific clade cannot contain any isolates from other delis.
NE, not estimated (due to insufficient lineage II isolates for tip-dated phylogenetics).
Significantly different values are indicated in boldface.
Isolates in a “deli-specific” clade are found only in a given deli and not in other delis, providing initial support for persistence in a given deli. Importantly, WGS-SNP-based clades further discriminated isolates of a given PFGE type. For example, isolates with PFGE type CU-262-79 were putatively persistent in four delis (2, 23, 28, and 29) and a sausage plant but were differentiated into two deli-specific clades (H and K), one clade from the sausage plant (clade L), and one state-specific clade (I, Fig. 1, inset panel).
Classification into a “state-specific” clade indicates that isolates in a clade are found across multiple delis in the same state but not in other states. Often, isolates within a given clade represented a strain that was putatively persistent in one or multiple delis and also isolated sporadically from other delis in a given state. For example, the PFGE type CU-57-267 (clade B, Table 4) was putatively persistent (isolated two or three times) in four distinct delis (4, 10, 16, and 23) and isolated sporadically in delis 7 and 28 but never isolated from delis in states other than the one common to these six delis. These findings suggest that certain L. monocytogenes strains may be present in a given state, or supply chain, and dispersed into multiple delis, with subsequent establishment of persistence in some.
Classification into a “multistate” clade indicates that isolates found in a clade were found across multiple states. Isolates within a given clade represented a strain that was putatively persistent in one or multiple delis in a single state and also isolated sporadically from one or more delis in a different state. For example, the PFGE type CU-294-321 (clade A, Table 4 and Fig. 1) was putatively persistent in three delis (10, 16, and 21) from state 2, as well as sporadically isolated from three delis in the same state but also isolated from deli 12, which was located in a different state (state 1).
Bayesian phylogenetic analysis using the isolation dates estimated each of 23 relevant clades to have diverged from individual most recent common ancestors (MRCA) less than 11 years ago (Tables 3 and 4). For deli-specific clades, MRCA estimates support that isolates from a given deli likely emerged within the plausible lifetime of a deli, a finding consistent with persistence in the deli. Even the divergence times of clades with statewide or multistate dispersion are relatively recent, e.g., 1.4 to 4.3 years in the past for clade A CU-294-321 isolates (Table 4). These recent divergence times for multistate clades suggest a possible point source that allows for broad distribution of an L. monocytogenes strain across different delis (e.g., a national supplier).
SNP counts show that some putatively persistent strains of L. monocytogenes are more closely related to each other than other isolates of the same subtype, suggesting that they represent persistent strains.
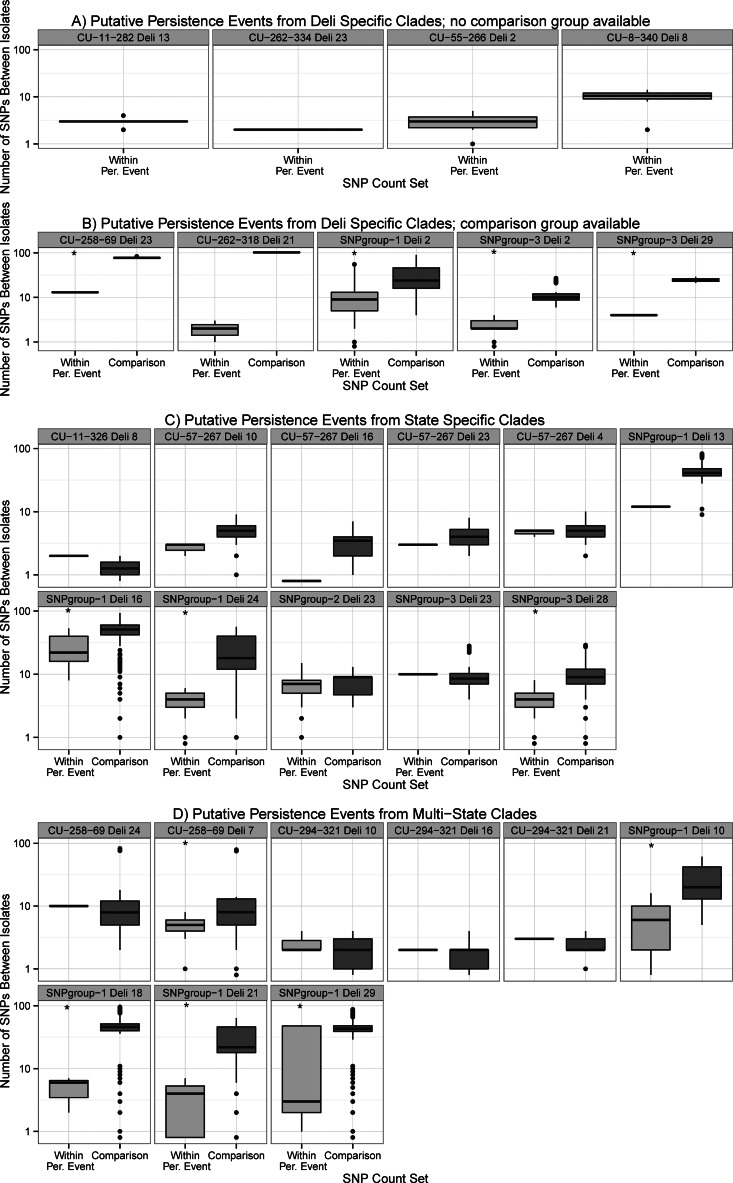
We reasoned that quantitative evidence, based on WGS-SNP data, could further support persistence in a given deli. Briefly, one may assume pathogen persistence within a particular facility is caused by a single introduction of a common ancestor of the persistent isolates with subsequent replication in a facility. Under that assumption, SNP counts within a population of persistent isolates (“within persistent event SNP count”) should be significantly lower than SNP counts between any persistent isolate and any other isolate with the same subtype from a different deli (“comparison SNP count”). The strain is (i) likely persistent when the median within-persistence-event SNP count is significantly smaller than the comparison SNP count and (ii) repeatedly reintroduced from some common source when no fewer SNP differences are observed. These SNP counts are summarized in Fig. 4 using a permutation-based statistic to determine significant differences (P values in Table 4). Overall, 12 of the 25 testable putative persistence events showed significantly fewer median within-persistence-event SNP counts (range, 2 to 22 SNPs) than median comparison SNP counts (range, 8 to 77 SNPs), supporting that these isolates represent persistent strains.
FIG 4.
SNP count box plots to identify persistent strains of L. monocytogenes using 9,376 core genome SNPs among lineage I isolates. (A) Within-persistence-event SNP counts for isolates that form a deli-specific clade where no comparison isolates of the same subtype are available. (B) Within-persistence-event and comparison SNP counts for isolates that form a deli-specific clade and comparison isolates. (C) SNP counts for putative persistence events for isolates that form a state-specific clade. (D) SNP counts for putative persistence events for isolates that form a multistate clade. To accommodate a log-scale y axis, counts of zero SNP differences are plotted just below the 1 SNP difference y axis minimum. An asterisk (*) indicates significantly fewer median SNP count differences for the within-persistence-event group than for the comparison group (FDR-adjusted P value of <0.05; Table 4). For each box, the solid line is the median SNP difference count, the box height is the inner quartile range (IQR), whiskers extend to the most extreme value within 1.5 × IQR of the box, and outliers are plotted as points.
Among the 11 deli-specific clades, there were six putative persistence events wherein the putatively persistent strain had a subtype that did not appear among isolates from any other deli (Fig. 4A), and five where the subtype did appear in other delis (Fig. 4B). Median within-persistence-event SNP counts ranged from 1 to 13 SNPs (Fig. 4 and Table 4), and pairs of isolates from a single deli often showed only one, or even zero, SNP differences (Fig. 4, e.g., SNP group 1, deli 2). Among the five deli-specific clades with a comparison group, four showed significantly fewer within-persistence-event SNP counts (e.g., clade K, median 2, and 10 SNPs for the “within” and “comparison” groups, respectively). The large, but insignificant, median SNP count difference for clade G, subtype CU-262-318 (2 and 102 SNPs, within and comparison, respectively), was caused by the four total isolates giving insufficient power to show significance.
Among the seven state-specific clades, there were 11 putative persistence events; for three of these events the within-persistence-event SNP counts were significantly fewer than the comparison SNP counts (Table 4 and Fig. 4C). For example, within clade I, the 12 SNP group 3 isolates from deli 28 likely represented a persistent strain (significant SNP count difference), whereas the two isolates from deli 23 showed no evidence for persistence (nonsignificant difference). Among the five multistate clades, five of the nine individual putative persistence events showed statistical evidence for persistence (Table 4 and Fig. 4D).
L. monocytogenes nearly indistinguishable by SNP count can be isolated from multiple delis across states.
SNP counts can characterize isolates of L. monocytogenes that do not appear to persist in particular delis. For example, for isolates found within multistate clade A, PFGE type CU-294-321, the median of two to three SNP count differences for putatively persistent isolates is identical to the median of two to three SNP count differences between comparison isolates from other delis, including one isolate from a separate state (Table 4). For all three delis where the strain was putatively persistent, there was at least one isolate of the same PFGE type from another deli that differed by 1 or even 0 SNPs (Fig. 4). Strikingly, the single isolate from a different state (FSL R8-7006, from state 1) was found to have 0 to 1 SNP differences from six isolates from five delis in state 2 (0 SNP differences from FSL R8-6046, deli 10; 1 SNP difference from FSL R8-5402, FSL R8-7348, FSL R8-7057, FSL R8-6918, and FSL R8-6607, delis 10, 2, 7, 16, and 21, respectively). Overall, in 13 of 25 putative persistence events a comparison isolate had 0 to 1 SNP difference from the putatively persistent strain (Fig. 4), including for persistence events with statistical significance (e.g., some comparison group isolates show 0 to 1 SNP differences from SNP group 1 isolates from delis 18 and 29).
Importantly, including accessory genome SNPs (see Fig. S2 in the supplemental material, using 14,846 SNPs, see Fig. S3 in the supplemental material for a phylogenetic tree) does not change the result that the nearly indistinguishable L. monocytogenes can be isolated from multiple delis, although the absolute numbers shift toward slightly more SNP differences. For example, the single clade A isolate from state 1 showed 0 to 1 and 1 to 3 SNP differences to six isolates from delis in state 2 across the core and accessory genome, respectively. L. monocytogenes with nearly, or even identical, SNP profiles, thus, can be isolated from multiple delis across states.
Gene presence or absence analysis fails to find correlates to a persistence phenotype.
Initial gene enrichment analyses confirmed that isolates with PFGE types with ≤3 band differences that grouped into one of three WGS-SNP groups differed by a number of genes (see Table S4 in the supplemental material). Gene enrichment analysis failed, though, to find any genes that were enriched among isolates initially (putatively) identified as either persistent (n = 92) or sporadic (n = 29; see Table S4 in the supplemental material), or when comparing isolates with statistical evidence for persistence (n = 38) to sporadic isolates of the same subtypes (n = 16; see Table S4 in the supplemental material). Separate gene enrichment analyses compared persistent and comparison isolates separately for all nine instances with significant evidence for persistence and at least three persistent isolates. In no comparison were any genes significantly enriched among persistent isolates (see Table S4 in the supplemental material). In one case, 10 genes were significantly enriched among nonpersistent isolates (SNP group 1, deli 16), but these genes were not present in a conserved region, nor were they annotated with functions related to establishing persistence or survival in a harsh environment (see Table S7 in the supplemental material). Taken together, these data suggest there are no robust gene presence or absence patterns linked to the persistent strains found here.
DISCUSSION
WGS-based subtyping has improved subtyping of bacterial pathogens in a number of contexts, including surveillance of nosocomial and food-borne pathogens (15–18, 46, 47). In addition to disease surveillance, traditional molecular subtyping methods have improved our understanding of transmission, sources, and reservoirs of food-borne pathogens along the food chain, including in farms, processing plants, and retail establishments (37). Here, we specifically used WGS-based subtyping of isolates obtained from a previous longitudinal study of L. monocytogenes in retail deli environments to obtain insights into L. monocytogenes transmission. In particular, previous PFGE studies of these isolates (20) revealed a number of findings that complicated interpretation of L. monocytogenes transmission, including (i) isolation of identical PFGE types across a number of retail delis (e.g., one PFGE type was found in eleven retail delis in three U.S. states) and (ii) isolation over time of closely related PFGE types (i.e., types that differed by ≤3 bands) in a given retail deli. Overall, WGS-based subtyping of these isolates (i) showed that WGS allows for practically relevant improved subtyping over existing molecular subtyping methods and (ii) yielded evidence for both persistence and repeat introduction from external sources in deli establishments. Our findings, including the presence of L. monocytogenes with highly similar WGS-SNP profiles and most recent common ancestors of <10 years in different retails delis, provide insights into the transmission and genomic diversity of L. monocytogenes, which will be essential for food safety applications of WGS-based subtyping.
WGS improves molecular subtyping over PFGE and MLST subtyping methods.
Here, WGS improved discriminatory power over both PFGE and MLST and sometimes differentiated isolates with identical PFGE types obtained from different delis into WGS-SNP-based clades that were only found in a given deli. These findings are consistent with the improved discrimination by WGS of outbreak-associated Salmonella (15, 16), E. coli (17), and L. monocytogenes (18). WGS also overcame three cases of overdiscrimination by PFGE due to prophage differences. This overdiscrimination may occur when L. monocytogenes isolates that share a recent common ancestor show PFGE patterns that differ by a distance (three or fewer bands [48]) that can be caused by a single genetic event such as prophage (18, 49) or plasmid (50) gains or losses. Here, we found that three sets of PFGE types that differed by three or fewer bands differed in the presence of an intact prophage regions; these findings clarified that a single L. monocytogenes clone presented by two PFGE types can persist in a deli. For example, we previously reported reisolation of two PFGE types (CU-11-320 and CU-8-96) in deli 2 (20); WGS showed that these two PFGE types represent one clone with a MRCA of <7 years.
Unlike existing molecular subtyping methods, WGS-based subtyping generates primary genome sequence data useful for secondary analyses, including in silico phage detection (51), plasmid detection (52), antibiotic resistance profile prediction and virulence gene detection (33), and serovar prediction (53). WGS data from the present study suggest that it is unlikely there are individual genes responsible for the persistence of L. monocytogenes in the retail deli environment. This finding is consistent with the observation that researchers working with ad hoc definitions of persistence and generally small sample sizes have previously failed to find robust genetic determinants of bacterial persistence in food-associated environments (6, 8).
L. monocytogenes contamination at retail represents a complicated scenario with a mixture of persistence and repeated introduction from external sources.
A key goal of the present study was to apply WGS-based subtyping to better understand the sources and transmission of L. monocytogenes in retail delis, building on a previous PFGE-based study of L. monocytogenes isolates obtained during a longitudinal study of 30 retail delis in three states (20). A specific goal was to use WGS-based subtyping to differentiate persistent L. monocytogenes contamination in delis from contamination that likely represents repeated introduction from an external source. The WGS analysis included multiple steps, including (i) SNP-based identification of relevant phylogenetic clades and (ii) quantitative comparison of the number of SNPs differentiating isolates within a deli to the number of SNPs differentiating isolates between delis.
The SNP difference approach is similar to that used by Leekitcharoenphon et al. (16), wherein the range of SNP differences among isolates within a Salmonella Typhimurium outbreak was 2 to 30 SNPs, whereas SNP differences between outbreak isolates and background ranged from 15 to 334 SNPs. SNP difference analysis has also been used in community epidemiology to identify that Staphylococcus aureus isolates collected from patients within the same New York City households were more closely related, with a median of 3 SNP differences, than isolates collected from different households in the community, with a median of 104 SNP differences (46). The SNP difference counts here were similar; isolates of the same subtype within a deli had a median of 22 SNP differences or fewer compared to a median of up to 102 SNP differences between delis.
Our previous PFGE-based study reported a number of cases where the PFGE evidence for persistence was ambiguous. For example, three PFGE types (CU-11-320, CU-262-79, and CU-8-96) were repeatedly isolated in multiple delis (four, three, and two delis, respectively [20]). In a number of cases, the WGS-SNP data reported here resolved isolates with a PFGE type into a number of well-supported clades that (i) included only isolates from a single deli and (ii) likely diverged from a most recent common ancestor less than 10 years ago. These findings are consistent with the introduction of a L. monocytogenes strain into the deli from an outside source, followed by persistence and short-term diversification within the deli. Interestingly L. monocytogenes persistence over 10 to 15 years, in food processing environments, has previously been reported (9, 54, 55).
In the cases reported here, persistence is also supported by quantitative analysis of SNP difference data; isolates within “deli-specific” clades showed very limited SNP differences (median of 2 to 11 SNPs, with instances of 0 or only 1 SNP difference). In 12 cases, isolates of deli-specific clades had significantly fewer median SNP differences between themselves compared to the median SNP differences between themselves and isolates of the same subtype from other delis. The permutation-based statistical test of significant SNP difference, developed here, provides a quantitative assessment of persistence that can be used in future studies.
We also identified well supported clades that were classified here as “state-specific” or “multistate.” In many of these cases, there was evidence for persistence of the clone representing one of these given clades in one deli, along with sporadic isolation in one or more other delis. For example, nine isolates from deli 7 with the PFGE pattern CU-258-69 showed statistically significant SNP difference evidence for persistence, and we also sporadically found five closely related isolates from delis in two states. Possible explanations for these findings include (i) a clone persisting within a given deli but being transferred to another deli or (ii) a clone being introduced into multiple delis from an external source but only establishing persistence in one (or some) delis. Alternatively, persistence may go undetected in some delis due to imperfect sampling. Scenario i is supported by a previous study that showed transfer of a persistent L. monocytogenes strain with dicing equipment transferred from one processing plant to another (56). Scenario ii is supported by a previously reported multistate U.S. outbreak of listeriosis linked to turkey deli meat (54) and a widespread outbreak of listeriosis in Quebec, Canada, linked to pasteurized cheese (57). In the former report, L. monocytogenes identical, as determined by PFGE, to the outbreak strain were isolated from three delis and subsequently the source of contamination was traced back to persistent L. monocytogenes contamination (over up to 12 years) at a deli meat processor that supplied each of these delis (54). In the latter report, L. monocytogenes identical, as determined by PFGE, to the outbreak strain were isolated from environmental or product samples from 22 retailers, and the source of contamination was subsequently traced back to a single upstream producer (57).
Our data also provide evidence for repeat introduction of a given L. monocytogenes strain from an external source to multiple retail establishments. Specifically, we identified a number of state-specific and multistate clades that were well supported, showed an MRCA < 10 years, and contained closely related isolates from a considerable number of different retail establishments. For example, all 11 isolates with PFGE pattern CU-294-321 from seven separate delis, across two states and with repeat isolation in three delis, formed a singled, well-supported clade with an MRCA of <5 years, and all isolates differed by <5 SNPs. These cases suggest persistence of a strain representing a given clade at an external site, which serves as a source of introduction into multiple delis. A strain could persist at a supplier facility and be introduced to multiple delis through product shipments, as suggested in a 2008-2009 listeriosis outbreak from pasteurized Mexican-style cheese (58), and consistent with the studies cited above (54, 57). Alternatively, a given strain with a recent MRCA could be a recently emerged, evolutionary successful strain found across multiple source environments. For example, a New York State bovine-associated clade of Salmonella Cerro has been estimated to have emerged from an MRCA in 1998 and has since shown clonal expansion with unique genomic deletions (59), in addition to being widespread among dairy herds (60). Importantly, for state-specific and multistate clades, the WGS and source data available here typically do not provide sufficient information to determine whether reisolation of a given clade in a given deli represents either (i) multiple reintroduction events or (ii) actual persistence in a given retail deli environment. Well-designed sampling plans, involving sample collection after cleaning and sanitation and before introduction of potential fomites, will remain essential for differentiating reintroduction and persistence.
Rapid clonal spread of L. monocytogenes with limited genomic diversification suggests that the use of WGS-based subtyping in the traceback of food-borne disease outbreaks and cases still requires strong epidemiological data.
Although our study showed the utility of WGS-based subtyping to understand L. monocytogenes transmission in food-associated environments, the utility of these methods will increase as large WGS data sets of pathogen isolates, including sample-associated metadata, become available. Comparison of WGS data to surveillance databases has found multiple L. monocytogenes epidemic clones and outbreak subtypes associated with a 2011 outbreak of listeriosis related to cantaloupe and linked these subtypes to other outbreak-associated isolates worldwide (49). For environmental pathogen control, comparison to large WGS data sets on L. monocytogenes isolates from different sources may allow for determination whether a given state-specific or multistate clade with a recent MRCA represents a broadly distributed epidemic clone (with introduction into delis from different sources) or a point-source-specific clone (with introduction into delis from a specific source, e.g., a national distributor). As public databases, such the NCBI GenBank and the curated subproject GenomeTrakr for food-borne pathogen sequences (http://www.fda.gov/Food/FoodScienceResearch/WholeGenomeSequencingProgramWGS/default.htm), continue to grow, the ability to classify nationally dispersed L. monocytogenes strains will improve.
While our study provides further evidence that whole-genome sequence analysis improves upon existing molecular subtyping methods for food-borne pathogens, it also revealed some important challenges when relying on WGS data to identify outbreak sources. Our data specifically show that L. monocytogenes with identical, or nearly identical, genome sequences (0 to 1 SNPs different) and MRCA of <10 years can be obtained from different deli environments in different states. This clearly indicates that detection in a food-associated environment of a L. monocytogenes isolate that shows a high level of WGS similarity to isolates from human patients does not necessarily establish a causal link between the food source and human illness. Our findings here are also consistent data from WGS of human and food isolates from a 1988 listeriosis case and a 2000 outbreak linked to the same processing facility and a strain that persisted in that facility over at least 12 years. The 2000 L. monocytogenes food isolate differed from the 1988 human case isolate by only one synonymous SNP (9). When limited SNPs difference are observed between isolates, one should also note that these SNPs may arise during laboratory passage, since the two subcultures of the year 2000 L. monocytogenes human isolate sequenced differed by a single SNP (9). WGS does not necessarily allow us to overcome all challenges associated with use of existing molecular subtyping methods. Specifically, our data suggest that, similar to findings of identical PFGE types in two processing plants, both linked to a human listeriosis outbreak (61), L. monocytogenes isolates with “identical” WGS data can be found in multiple food associated environments. Hence, WGS-based subtyping methods are not a substitute for strong context knowledge and epidemiological data but instead an improved tool for system-level approaches to tracking and controlling the sources of food-borne disease.
Supplementary Material
ACKNOWLEDGMENTS
We want to thank the Cornell Biotechnology Resource Center for helping to design the high-throughput sample preparation sequencing workflow, as well as for carrying out library preparation and sequencing.
This study was supported by a USDA-NIFA predoctoral fellowship grant (2013-67011-21117).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01049-15.
REFERENCES
- 1.Scallan E, Hoekstra R, Angulo F, Tauxe R, Widdowson M-A, Row S, Jones J, Griffin P. 2011. Foodborne illness acquired in the united states-major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ERS/USDA. 2014. Cost estimates of foodborne illnesses. Economic Research Service/US Department of Agriculture, Washington, DC: http://ers.usda.gov/data-products/cost-estimates-of-foodborne-illnesses.aspx. [Google Scholar]
- 3.CFSAN/FSIS. 2003. Quantitative assessment of relative risk to public health from food-borne Listeria monocytogenes among selected categories of ready-to-eat foods. Center for Food Safety and Applied Nutrition/Food Safety and Inspection Service, Washington, DC: http://www.fda.gov/food/foodscienceresearch/risksafetyassessment/ucm183966.htm. [Google Scholar]
- 4.Pradhan AK, Ivanek R, Grohn YT, Bukowski R, Geornaras I, Sofos JN, Wiedmann M. 2010. Quantitative risk assessment of listeriosis-associated deaths due to Listeria monocytogenes contamination of deli meats originating from manufacture and retail. J Food Prot 73:620–630. [DOI] [PubMed] [Google Scholar]
- 5.Endrikat S, Gallagher D, Pouillot R, Hicks Quesenberry H, LaBarre D, Schroeder CM, Kause J. 2010. A comparative risk assessment for Listeria monocytogenes in prepackaged versus retail-sliced deli meat. J Food Prot 73:612–619. [DOI] [PubMed] [Google Scholar]
- 6.Carpentier B, Cerf O. 2011. Persistence of Listeria monocytogenes in food industry equipment and premises. Int J Food Microbiol 145:1–8. doi: 10.1016/j.ijfoodmicro.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Tompkin RB. 2002. Control of Listeria monocytogenes in the food-processing environment. J Food Prot 65:709–725. [DOI] [PubMed] [Google Scholar]
- 8.Ferreira V, Wiedmann M, Teixeira P, Stasiewicz MJ. 2014. Listeria monocytogenes persistence in food associated environments: epidemiology, strain characteristics, and implications for public health. J Food Prot 77:150–170. doi: 10.4315/0362-028X.JFP-13-150. [DOI] [PubMed] [Google Scholar]
- 9.Orsi RH, Borowsky ML, Lauer P, Young SK, Nusbaum C, Galagan JE, Birren BW, Ivy RA, Sun Q, Graves LM, Swaminathan B, Wiedmann M. 2008. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics 9:539. doi: 10.1186/1471-2164-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malley TJ, Butts J, Wiedmann M. 2015. Seek and destroy process: Listeria monocytogenes process controls in the ready-to-eat meat and poultry industry. J Food Prot 78:436–445. doi: 10.4315/0362-028X.JFP-13-507. [DOI] [PubMed] [Google Scholar]
- 11.Malley TJ, Stasiewicz MJ, Grohn YT, Roof S, Warchocki S, Nightingale K, Wiedmann M. 2013. Implementation of statistical tools to support identification and management of persistent Listeria monocytogenes contamination in smoked fish processing plants. J Food Prot 76:796–811. doi: 10.4315/0362-028X.JFP-12-236. [DOI] [PubMed] [Google Scholar]
- 12.Vangay P, Steingrimsson J, Wiedmann M, Stasiewicz MJ. 2014. Classification of Listeria monocytogenes persistence in retail delicatessen environments using expert elicitation and machine learning. Risk Anal 34:1830–1845. doi: 10.1111/risa.12218. [DOI] [PubMed] [Google Scholar]
- 13.Kao RR, Haydon DT, Lycett SJ, Murcia PR. 2014. Supersize me: how whole-genome sequencing and big data are transforming epidemiology. Trends Microbiol 22:282–291. doi: 10.1016/j.tim.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergholz TM, Moreno Switt AI, Wiedmann M. 2014. Omics approaches in food safety: fulfilling the promise? Trends Microbiol 22:275–281. doi: 10.1016/j.tim.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.den Bakker HC, Allard MW, Bopp D, Brown EW, Fontana J, Iqbal Z, Kinney A, Limberger R, Musser KA, Shudt M, Strain E, Wiedmann M, Wolfgang WJ. 2014. Rapid whole-genome sequencing for surveillance of Salmonella enterica serovar Enteritidis. Emerg Infect Dis 20:1306–1314. doi: 10.3201/eid2008.131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. 2014. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One 9:e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, Larios O, Allen V, Lee B, Nadon C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. doi: 10.1186/1471-2164-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.US Food and Drug Administration. 2014. Whole genome sequencing helps FDA identify dangerous bacteria. US Food and Drug Administration, Bethesda, MD: http://www.fda.gov/ForConsumers/ConsumerUpdates/ucm397287.htm. [Google Scholar]
- 20.Simmons C, Stasiewicz MJ, Wright E, Warchocki S, Roof S, Kause JR, Bauer N, Ibrahim S, Wiedmann M, Oliver HF. 2014. Listeria monocytogenes and Listeria spp. contamination patterns in retail delicatessen establishments in three U.S. states. J Food Prot 77:1929–1939. doi: 10.4315/0362-028X.JFP-14-183. [DOI] [PubMed] [Google Scholar]
- 21.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. 2012. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner SN, Hall BG. 2013. When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8:e81760. doi: 10.1371/journal.pone.0081760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 26.Iqbal Z, Turner I, McVean G. 2013. High-throughput microbial population genomics using the Cortex variation assembler. Bioinformatics 29:275–276. doi: 10.1093/bioinformatics/bts673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G. 2012. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat Genet 44:226–232. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15:13pgs. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leigh JW, Susko E, Baumgartner M, Roger AJ. 2008. Testing congruence in phylogenomic analysis. Syst Biol 57:104–115. doi: 10.1080/10635150801910436. [DOI] [PubMed] [Google Scholar]
- 33.Inouye M, Dashnow H, Raven L, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salcedo C, Arreaza L, Alcala B, de la Fuente L, Vazquez JA. 2003. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J Clin Microbiol 41:757–762. doi: 10.1128/JCM.41.2.757-762.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nightingale K. 2010. Listeria monocytogenes: knowledge gained through DNA sequence-based subtyping, implications, and future considerations. J AOAC Int 93:1275–1286. [PubMed] [Google Scholar]
- 38.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benedict MN, Henriksen JR, Metcalf WW, Whitaker RJ, Price ND. 2014. ITEP: an integrated toolkit for exploration of microbial pan-genomes. BMC Genomics 15:8. doi: 10.1186/1471-2164-15-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 42.Core Team R. 2014. R: a language and environment for statistical computing. The R Project for Statistical Computing, Los Angeles, CA: http://www.R-project.org. [Google Scholar]
- 43.Orsi RH, Bakker HC, Wiedmann M. 2011. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 301:79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Dorscht J, Klumpp J, Bielmann R, Schmelcher M, Born Y, Zimmer M, Calendar R, Loessner MJ. 2009. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J Bacteriol 191:7206–7215. doi: 10.1128/JB.01041-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denes T, Vongkamjan K, Ackermann HW, Moreno Switt AI, Wiedmann M, den Bakker HC. 2014. Comparative genomic and morphological analyses of Listeria phages isolated from farm environments. Appl Environ Microbiol 80:4616–4625. doi: 10.1128/AEM.00720-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uhlemann AC, Dordel J, Knox JR, Raven KE, Parkhill J, Holden MT, Peacock SJ, Lowy FD. 2014. Molecular tracing of the emergence, diversification, and transmission of Staphylococcus aureus sequence type 8 in a New York community. Proc Natl Acad Sci U S A 111:6738–6743. doi: 10.1073/pnas.1401006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris SR, Cartwright EJ, Torok ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of methicillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lomonaco S, Verghese B, Gerner-Smidt P, Tarr C, Gladney L, Joseph L, Katz L, Turnsek M, Frace M, Chen Y, Brown E, Meinersmann R, Berrang M, Knabel S. 2013. Novel epidemic clones of Listeria monocytogenes, United States, 2011. Emerg Infect Dis 19:147–150. doi: 10.3201/eid1901.121167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ying C, Robin MS, Sophia K. 2008. Genomic divisions/lineages, epidemic clones, and population structure, p 337–357. In Handbook of Listeria monocytogenes. CRC Press, Inc, Boca Raton, FL. [Google Scholar]
- 51.Akhter S, Aziz RK, Edwards RA. 2012. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res 40:e126. doi: 10.1093/nar/gks406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ranieri ML, Shi C, Moreno Switt AI, den Bakker HC, Wiedmann M. 2013. Comparison of typing methods with a new procedure based on sequence characterization for Salmonella serovar prediction. J Clin Microbiol 51:1786–1797. doi: 10.1128/JCM.03201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olsen SJ, Patrick M, Hunter SB, Reddy V, Kornstein L, MacKenzie WR, lane K, Bidol S, Stoltman GA, Frye DM, Lee I, Hurd S, Jones TF, LaPorte TN, Dewitt W, Graves L, Wiedmann M, Schoonmaker-Bopp DJ, Huang AJ, Vincent C, Bugenhagen A, Corby J, Carloni ER, Holcomb ME, Woron RF, Zansky SM, Dowdle G, Smith F, Ahrabi-Fard S, Ong AR, Tucker N, Hynes NA, Mead P. 2005. Multistate outbreak of Listeria monocytogenes infection linked to delicatessen turkey meat. Clin Infect Dis 40:962–967. doi: 10.1086/428575. [DOI] [PubMed] [Google Scholar]
- 55.Vongkamjan K, Roof S, Stasiewicz MJ, Wiedmann M. 2013. Persistent Listeria monocytogenes subtypes isolated from a smoked fish processing facility included both phage susceptible and resistant isolates. Food Microbiol 35:38–48. doi: 10.1016/j.fm.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 56.Lundén JM, Autio TJ, Korkeala HJ. 2002. Transfer of persistent Listeria monocytogenes contamination between food-processing plants associated with a dicing machine. J Food Prot 65:1129–1133. [DOI] [PubMed] [Google Scholar]
- 57.Gaulin C, Ramsay D, Bekal S. 2012. Widespread listeriosis outbreak attributable to pasteurized cheese, which led to extensive cross-contamination affecting cheese retailers, Quebec, Canada, 2008. J Food Prot 75:71–78. doi: 10.4315/0362-028X.JFP-11-236. [DOI] [PubMed] [Google Scholar]
- 58.Jackson KA, Biggerstaff M, Tobin-D'Angelo M, Sweat D, Klos R, Nosari J, Garrison O, Boothe E, Saathoff-Huber L, Hainstock L, Fagan RP. 2011. Multistate outbreak of Listeria monocytogenes associated with Mexican-style cheese made from pasteurized milk among pregnant, Hispanic women. J Food Prot 74:949–953. doi: 10.4315/0362-028X.JFP-10-536. [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez-Rivera LD, Moreno Switt AI, Degoricija L, Fang R, Cummings CA, Furtado MR, Wiedmann M, den Bakker HC. 2014. Genomic characterization of Salmonella Cerro ST367, an emerging Salmonella subtype in cattle in the United States. BMC Genomics 15:427. doi: 10.1186/1471-2164-15-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cummings KJ, Warnick LD, Elton M, Rodriguez-Rivera LD, Siler JD, Wright EM, Grohn YT, Wiedmann M. 2010. Salmonella enterica serotype Cerro among dairy cattle in New York: an emerging pathogen? Foodborne Pathog Dis 7:659–665. doi: 10.1089/fpd.2009.0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gottlieb SL, Newbern EC, Griffin PM, Graves LM, Hoekstra RM, Baker NL, Hunter SB, Holt KG, Ramsey F, Head M, Levine P, Johnson G, Schoonmaker-Bopp D, Reddy V, Kornstein L, Gerwel M, Nsubuga J, Edwards L, Stonecipher S, Hurd S, Austin D, Jefferson MA, Young SD, Hise K, Chernak ED, Sobel J. 2006. Multistate outbreak of listeriosis linked to turkey deli meat and subsequent changes in US regulatory policy. Clin Infect Dis 42:29–36. doi: 10.1086/498113. [DOI] [PubMed] [Google Scholar]
- 62.Ferreira V, Barbosa J, Stasiewicz M, Vongkamjan K, Moreno Switt A, Hogg T, Gibbs P, Teixeira P, Wiedmann M. 2011. Diverse geno- and phenotypes of persistent Listeria monocytogenes isolates from fermented meat sausage production facilities in Portugal. Appl Environ Microbiol 77:2701–2715. doi: 10.1128/AEM.02553-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sauders BD, Sanchez MD, Rice DH, Corby J, Stich S, Fortes ED, Roof SE, Wiedmann M. 2009. Prevalence and molecular diversity of Listeria monocytogenes in retail establishments. J Food Prot 72:2337–2349. [DOI] [PubMed] [Google Scholar]
- 64.Thimothe J, Nightingale KK, Gall K, Scott VN, Wiedmann M. 2004. Tracking of Listeria monocytogenes in smoked fish processing plants. J Food Prot 67:328–341. [DOI] [PubMed] [Google Scholar]
- 65.Lappi VR, Thimothe J, Nightingale KK, Gall K, Scott VN, Wiedmann M. 2004. Longitudinal studies on Listeria in smoked fish plants: impact of intervention strategies on contamination patterns. J Food Prot 67:2500–2514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.