Abstract
Inanimate surfaces are regarded as key vehicles for the spread of human norovirus during outbreaks. ISO method 15216 involves the use of cotton swabs for environmental sampling from food surfaces and fomites for the detection of norovirus genogroup I (GI) and GII. We evaluated the effects of the virus drying time (1, 8, 24, or 48 h), swab material (cotton, polyester, rayon, macrofoam, or an antistatic wipe), surface (stainless steel or a toilet seat), and area of the swabbed surface (25.8 cm2 to 645.0 cm2) on the recovery of human norovirus. Macrofoam swabs produced the highest rate of recovery of norovirus from surfaces as large as 645 cm2. The rates of recovery ranged from 2.2 to 36.0% for virus seeded on stainless-steel coupons (645.0 cm2) to 1.2 to 33.6% for toilet seat surfaces (700 cm2), with detection limits of 3.5 log10 and 4.0 log10 RNA copies. We used macrofoam swabs to collect environmental samples from several case cabins and common areas of a cruise ship where passengers had reported viral gastroenteritis symptoms. Seventeen (18.5%) of 92 samples tested positive for norovirus GII, and 4 samples could be sequenced and had identical GII.1 sequences. The viral loads of the swab samples from the cabins of the sick passengers ranged from 80 to 31,217 RNA copies, compared with 16 to 113 RNA copies for swab samples from public spaces. In conclusion, our swab protocol for norovirus may be a useful tool for outbreak investigations when no clinical samples are available to confirm the etiology.
INTRODUCTION
Human noroviruses are the leading cause of epidemic and sporadic acute gastroenteritis (AGE) worldwide (1). Most outbreaks are reported in semiclosed environments, such as long-term-care facilities, hospitals, and schools (2, 3). Because the majority of infections are spread either directly, via the person-to-person route, or indirectly, through environmental surfaces or food, contaminated fomites and inanimate surfaces are regarded as important vehicles for the spread of the virus during outbreaks (4–6). In addition, the virus is easily transferred between inanimate surfaces and human skin (5, 7, 8).
Many laboratory studies have been performed to validate the efficacy of disinfectants or disinfection processes to prevent the spread of norovirus. Some of these processes have been implemented routinely in health care facilities (9–12). However, little is known about the correlation between the level of surface contamination and increased risks of norovirus infection, and this lack of understanding may affect the implementation of adequate hygiene practices. Surface-sampling methods have been used successfully to monitor the level and/or duration of environmental contamination (13). Protocols to detect norovirus on environmental surfaces and fomites in outbreak settings use swab rinse methods (7, 14, 15) or antistatic wipes (16, 17). The ISO 15216 standard protocol for the detection of norovirus and hepatitis A virus on food preparation surfaces and fomites includes the use of cotton swabs (18).
Standardized validated swab rinse protocols enable comparison of the sampling efficiencies of commercial swabs. In previous studies, different elution media and swab materials for the recovery of rotavirus, MS2, feline calicivirus (FCV), and bacteriophage P22 were evaluated (19–22). However, extrapolation of the results from these studies to a validated protocol for human norovirus is difficult, since many test variables, including the surrogate virus used for assessment of the recovery of infectious virus, the type of swab material, and the area of the swabbed surface, have not been evaluated and tested under field conditions.
In the present study, we evaluated a novel swab rinse protocol for the detection of human norovirus on inanimate surfaces using different swab materials, as well as methods for the concentration of virus from the swab eluates, and also investigated the effect of the area of the swabbed surface. The optimized sampling protocol was further field tested on samples collected from high-contact surfaces that had been contaminated by people with clinical norovirus symptoms.
MATERIALS AND METHODS
Viruses.
A norovirus GII.4-positive stool specimen obtained from a cruise ship gastroenteritis outbreak in 2010 was used in this study. A 10-to-20% stool suspension was made in phosphate-buffered saline (PBS), centrifuged (at 5,000 × g for 10 min) to remove organic particles, and further concentrated by ultrafiltration using centrifugal filter units with a molecular size cutoff of 50 kDa (Millipore, Billerica, MA). The final virus preparation was filtered through a 0.45-μm Millex-HA syringe filter (Millipore, Billerica, MA) that had been pretreated with a 0.1% (vol/vol) Tween 80 solution in order to remove bacteria and fungi. The viral RNA titer was approximately 107.5 RNA copies per ml of filtered stool based on a standard curve of GII.4 RNA transcripts (23).
Bacteriophage MS2 (ATCC 15597-B1), propagated using Escherichia coli Famp (ATCC 700891) as described previously (10), was dispensed in aliquots of 106 PFU/ml and was stored at −80°C.
Swab materials.
Four commercially available swab materials were tested in this study: cotton (Fisher Scientific, Pittsburgh, PA), polyester (BD Science, Franklin Lakes, NJ), rayon (Puritan Medical Products, Guilford, ME), and macrofoam (ITW Texwipe, Kernersville, NC). The sizes (diameter by length) of the cotton, polyester, rayon, and macrofoam swab heads were 2.11 by 12.87, 4.76 by 15.88, 4.76 by 15.88, and 19 by 26.7 mm, respectively. We also tested an antistatic wipe (Sodibox, France), which was kindly provided by Ingeborg Boxman at the Food and Consumer Product Safety Authority in the Netherlands. The Sodibox swab fabrics (320 mm by175 mm) (Raisio Diagnostic, Nieuwerkerk aan den IJssel, the Netherlands) used in this study were ready to use and were premoistened with 10 ml of Ringer's solution.
Stainless-steel and toilet seat coupons.
Stainless steel coupons (5.1 by 5.1 cm or 25.4 by 25.4 cm) were cut from a sheet of S-180 grade T-304 stainless steel (Phoenix Metals Company, Norcross, GA). Each coupon was pretreated with 0.1% Tween 80, rinsed first in sterile distilled water and then in 70% ethanol, air dried, wrapped in aluminum foil, and autoclaved for 15 min at 121°C prior to use. Church white wood round toilet seats were purchased from Lowe's, and the upper surfaces of the toilet seats were precleaned with 0.1% Tween 80, rinsed first in sterile distilled water and then in 70% ethanol, and air dried prior to use.
Viral RNA extraction, purification, and concentration.
Viral RNA was extracted from 50 μl of the swab eluent by using the MagMAX-96 viral RNA isolation kit (Life Technologies Corporation, Carlsbad, CA), and the KingFisher purification instrument (Fisher Scientific, Pittsburgh, PA), as described previously (24). Also, two kinds of spin columns were consecutively used to purify and concentrate viral RNA from swab eluents. Specifically, 1 ml of swab eluent was lysed with 1 ml of a guanidinium-based lysis buffer as described previously (25) using Midi columns (Omega Biotek, Norcross, GA), and nucleic acid was further concentrated by using RNA Clean & Concentrator-5 spin columns (Zymo Research Corporation, Irvine, CA) according to the manufacturer's instructions. All oligonucleotide primers and probes used in this study were obtained from Life Technologies Corporation (Carlsbad, CA).
Detection of norovirus by RT-qPCR.
Reverse transcription–TaqMan real-time PCR (RT-qPCR) assays for the detection of genogroup I (GI) and GII human norovirus were carried out on an ABI 7500 platform (Life Technologies Corporation, Carlsbad, CA) (26, 27). Coliphage MS2 was included as an external extraction control prior to RNA extraction. Samples with a threshold cycle (CT) value of ≥30 for MS2 were retested undiluted and 1/10 diluted. Standard curves of GI.4 RNA and GII.7 RNA transcripts were included in each run (23).
Optimizing the surface-sampling methodology. (i) Reference test condition.
A 50-μl aliquot of a pooled viral suspension was seeded onto a 25.8-cm2 stainless-steel surface and was then dried for 1 h under ambient conditions (16 to 22°C; relative humidity, 45 to 60%). Prior to sampling, each swab was dipped into a tube containing 2.5 ml of swab elution buffer (PBS containing 0.02% Tween 80 [PBST]) and was then pressed against the side of the tube to squeeze out excess liquid. The entire surface area of the stainless-steel coupon was swabbed three times by 1 stroke of the swab in the horizontal direction, 1 stroke in the vertical direction, and 1 stroke in the diagonal direction. The swab was then dipped back into the elution buffer in a 15-ml tube, mixed by vortexing for at least 10 s, and then pressed against the side of the tube to elute the PBST. This eluent was then stored at −70°C until testing. To determine the maximum amount of virus that could be recovered, the virus inoculum was seeded onto the swabs or onto stainless-steel surfaces and was eluted immediately.
(ii) Effect of virus desiccation.
To measure the effect of virus desiccation on virus recovery, 50 μl seeding inoculum was seeded on 25.8-cm2 stainless-steel coupons and was dried for 0, 1, 8, 24, or 48 h. The coupons were then swabbed and were processed as described above.
(iii) Effect of the area of the swabbed surface.
To examine the influence of the surface area on viral recovery, 50 μl seeding inoculum was seeded onto stainless-steel coupons of varying sizes (5.1 by 5.1 cm, 7.6 by 7.6 cm, 10.2 by 10.2 cm, 12.7 by 12.7 cm) and was dried for 1 h at room temperature. The coupons were then swabbed and were processed as described above.
(iv) Comparing a macrofoam swab with an antistatic wipe for the recovery of GII.4 norovirus from large surface areas.
Stainless coupons with surface areas of 161.3 cm2 and 645 cm2 were each contaminated with 500 μl of a clarified GII.4 stool suspension (105.6 RNA copies per ml) and were air dried for approximately 48 h under ambient conditions (16 to 22°C; relative humidity, 45 to 60%). GII.4 norovirus was recovered with either a macrofoam swab or an antistatic wipe.
Evaluation of a macrofoam-based environmental swab protocol.
The level of virus recovery and the detection limit of the macrofoam-based swab rinse protocol were evaluated by seeding stainless-steel coupons (645 cm2) and toilet seat coupons (approximately 670 cm2) with 2-fold or 3-fold serial dilutions of a clarified GII.4 norovirus-positive stool suspension. The dilutions were prepared in a norovirus-negative stool suspension in order to keep the amount of stool matrix identical in the different dilutions. After drying of the inoculum for 48 h at room temperature, the coupons were sampled with a macrofoam swab prewetted with 2.5 ml of PBST, and the virus was concentrated and extracted from 1 ml of PBST eluate as described above.
Field testing of macrofoam swabs.
Since the level of norovirus recovery obtained with the macrofoam swab was better than that with the other swab materials, we sampled environmental surfaces on a cruise ship on which a number of suspected norovirus cases had been reported (Table 1). A total of 24 swab samples were collected from different environmental surfaces in cabins occupied by passengers who had reported norovirus symptoms. The surfaces included toilet seat, faucet, door handle, and telephone surfaces, and importantly, the cabins were not cleaned until the swabs had been collected. In addition, 68 swab samples from frequently touched surfaces of common areas on the ship (e.g., table top, ice cream dispenser, and table condiment container) were collected. Swab samples were kept at 4°C for <72 h prior to shipping on dry ice to the laboratory, where they were processed and tested for norovirus (see Fig. 5).
TABLE 1.
Norovirus-positive environmental swab samples on a cruise ship with reported clinical cases of viral gastroenteritis
| Sample areaa | Sample point description | Avg CT value (no. of positive assay results/assays performed) | Genotype | No. of norovirus RNA copies per sampled areac |
|---|---|---|---|---|
| Atrium | Handrail | 34.3 (1/2) | GII | 16 |
| Cabin A | Toilet seat | 31.4 (2/2) | GII.1b | 31,217 |
| Cabin A | Hand sink faucet | 37.5 (1/2) | GII | 491 |
| Cabin A | Door handle | 35.0 (2/2) | GII | 2,675 |
| Cabin A | Remote control | 38.6 (2/2) | GII.1b | 233 |
| Cabin B | Toilet seat | 33.5 (2/2) | GII.1b | 986 |
| Lido | Dispenser handle of ice cream machine | 34.2 (2/2) | GII | 16 |
| Lido | Table condiments | 35.2 (1/2) | GII | 15 |
| Lido | Table top | 35.3 (1/2) | GII | 14 |
| Pizzeria | Counter surface | 35.7 (1/2) | GII | 14 |
| Main galley | Touch-screen video game machine | 37.1 (1/2) | GII | 64 |
| Vending machine | Touchable surface | 38.8 (1/2) | GII | 18 |
| Crew lounge | Keyboard and computer mouse | 36.8 (1/2) | GII | 80 |
| Cabin C | Faucet and door handle | 31.6 (2/2) | GII.1b | 26,458 |
| Cabin C | Telephone | 36.4 (2/2) | GII | 1,035 |
| Cabin C | Keyboard | 33.0 (2/2) | GII | 1,317 |
| Medical center | Clipboard | 36.0 (2/2) | GII | 113 |
Cabins A, B, and C had been occupied by individuals who had been clinically ill with viral gastroenteritis symptoms.
Four of the 17 GII-positive swab samples could be genotyped.
RNA copies were calculated based on a standard curve of GII.7 RNA transcripts.
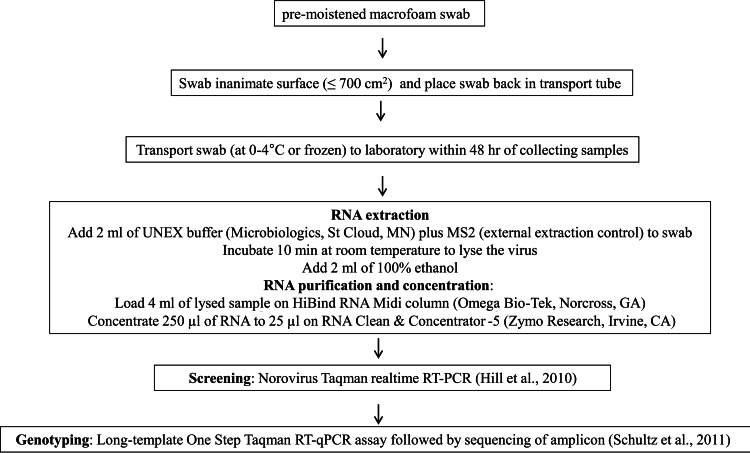
FIG 5.
Flow chart of final protocol for the sampling of norovirus on environmental surfaces. Screening was conducted by the method of Hill et al. (26), and viruses were genotyped by the method of Schultz et al. (28).
Confirmation of norovirus-positive swab samples by sequencing.
Nucleic acids from swab samples positive for norovirus by RT-qPCR-were reamplified using a long-template TaqMan assay (L-RT-qPCR) as described previously (28). L-RT-qPCR products of an appropriate size (378 bp) were separated by electrophoresis on 2% agarose gels, gel purified, and cycle sequenced using BigDye chemistry. Samples were analyzed and genotyped by local BLAST searches against GI and GII norovirus reference sequences at the CDC.
Data analysis.
The level of virus recovery (expressed as a percentage) was calculated by dividing the total RNA copies detected from the swab by the total RNA copies from the initial inoculum and multiplying by 100. Data from independent variables (elution medium type, size of surface, drying time, assay method, and swab material type) were analyzed by n-way analysis of variance (ANOVA) (29). Tukey's post hoc test was used to determine the effects of independent variables on viral recovery by using PASW Statistics, version 18 (IBM SPSS Inc., New York, NY). Additionally, the Mann-Whitney test was used to determine differences between the levels of norovirus recovered in the cabins occupied by passengers with AGE and the levels of norovirus detected in common areas. Significance was concluded if the P value was <0.05. Data from at least 5 replicates were included for each data point.
RESULTS
Comparison of rates of virus recovery from different swab materials.
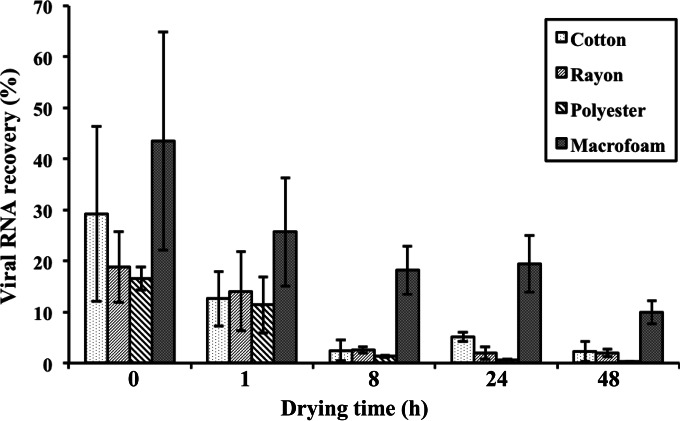
The rates of recovery of GII.4 norovirus from swabs that had been directly inoculated with the virus were higher than 60% for all four swab materials tested. The rate of recovery of GII.4 virus from 26-cm2 stainless-steel coupons without drying was highest for macrofoam swabs (43.5% ± 21.4%), followed by cotton (29.2% ± 17.1%), rayon (18.8% ± 6.9%), and polyester (16.6% ± 2.3%) swabs (Fig. 1). After the virus was dried for 1 h at room temperature, the rate of virus recovery using the macrofoam swab was reduced to 25.7% ± 10.6%, which was still higher than those with the cotton (12.6% ± 5.4%), rayon (14.0% ± 7.7%), and polyester (12.5% ± 9.4%) swabs (Fig. 1).
FIG 1.
Effects of drying time of GII.4 norovirus on stainless-steel surfaces on virus recovery from different swab materials. Results are averages for at least four replicates.
Effect of virus desiccation on virus recovery.
For macrofoam swabs, the rates of virus recovery ranged from 18.2% to 25.7% when the drying time of the inoculum was ≤24 h, whereas after 48 h of drying, the rate of virus recovery was reduced significantly, to 10.0% ± 2.3% (P < 0.05) (Fig. 1). In contrast, the rates of virus recovery using the three fiber-tipped swabs (cotton, rayon, and polyester swabs) were reduced to ≤2.5% after 8 h of drying. Overall, the level of virus recovery with macrofoam swabs was the highest for each variable tested, and at 48 h of drying time, this level was >4.4-fold higher than that with any of the three fiber-tipped swabs.
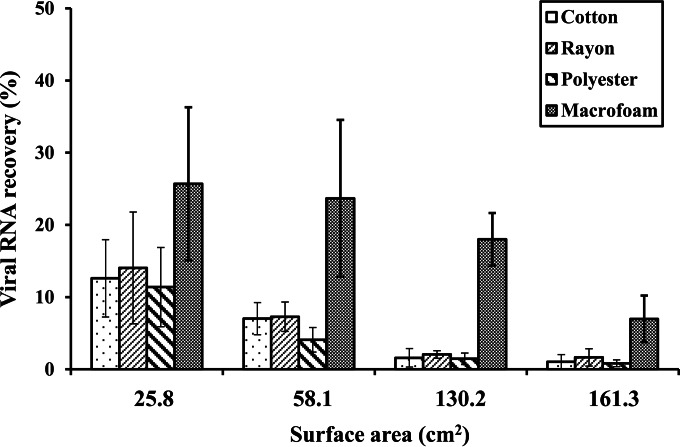
Effect of surface area on virus recovery.
Stainless-steel coupons of varying sizes (25.8 cm2, 58.1 cm2, 130.2 cm2, and 161.3 cm2) were sampled in order to determine the effect of the area of the swabbed surface on virus recovery (Fig. 2). An n-way ANOVA of our data showed that both the swab type and the area of the swabbed surface were significant factors for virus recovery (P < 0.001). When macrofoam swabs were used, the rate of virus recovery was ≥18.0% ± 3.6% for a surface area as large as 130.2 cm2 but decreased to 7.0% ± 3.2% for 161.3-cm2 coupons. In contrast, when fiber-tipped swabs were used on ≥130.2-cm2 coupons, the rate of virus recovery by use of any of the three fiber-tipped swabs was ≤2%.
FIG 2.
Recovery of GII.4 norovirus from stainless-steel coupons of different sizes by use of four different swab materials. Results are averages for at least four replicates.
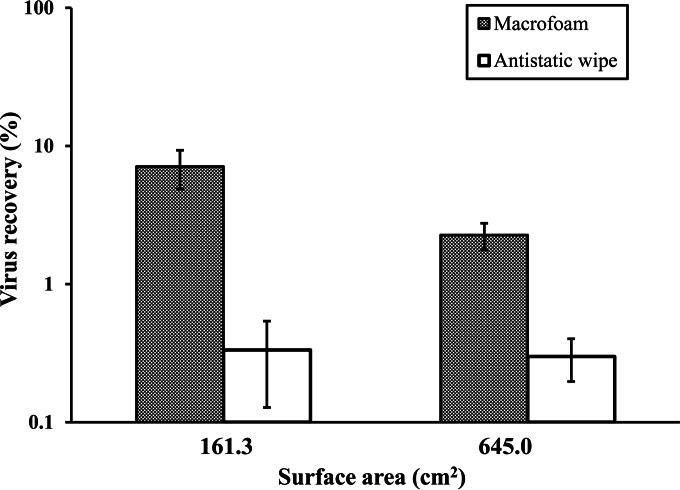
Comparison of macrofoam swabs with antistatic wipes for virus recovery from large surface areas.
The rates of virus recovery with a macrofoam swab from stainless-steel coupons of 161.3 and 645 cm2 were 7.08% ± 2.21% and 2.3% ± 0.5%, respectively (P < 0.001) (Fig. 3), whereas with antistatic wipes, the rates of recovery were 0.33% ± 0.21% and 0.30% ± 0.10%, respectively (P = 0.123).
FIG 3.
Comparison of a macrofoam swab with an antistatic wipe for the recovery of GII.4 norovirus from large sampling areas (161.3 or 645 cm2). The level of virus recovery (expressed as a percentage) was calculated by dividing the total RNA copies in the eluates (2.5 ml for macrofoam and 15 ml for antistatic wipe) by the total RNA copies in the initial inoculum (500 µl) and multiplying by 100. Results are averages for at least four replicates.
Comparison of rates of virus recovery from stainless-steel and toilet seat surfaces.
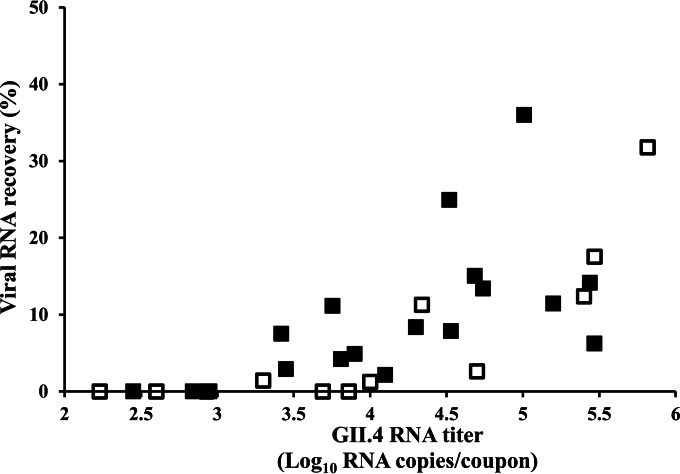
The rates of virus recovery from stainless-steel and toilet seat coupons with surface areas of 645 cm2 and 700 cm2 ranged from 2.2% to 36.0% and from 1.2% to 33.6%, respectively (Fig. 4). Macrofoam swabs were able to detect norovirus from stainless-steel and toilet seat coupons at seeding titers of ≥3.4 log10 and 4 log10 RNA copies, respectively.
FIG 4.
Characterization of sampling performance of the macroform swab-based sampling methodology. Stool sample suspensions (500 μl) with varying norovirus titers were seeded onto stainless-steel coupons (■) and toilet seats (□), dried for 48 h, and sampled with macrofoam swabs. GII.4 RNA was extracted, purified, and concentrated as described in Materials and Methods. Data were obtained from at least two independent experiments. Standard curves for GII.7 RNA transcripts were used to convert CT values into RNA copies. Results are averages for at least four replicates.
Sampling using macrofoam swabs to detect norovirus on a cruise ship.
We field tested the macrofoam swab on different environmental surfaces on a cruise ship with reported cases of suspected norovirus gastroenteritis. Norovirus GII was detected in 17 (18%) of the 92 swab samples (Table 1). Eight (33%) of the 24 swab samples collected from surfaces in cabins where passengers showed norovirus symptoms and 9 (15%) of the 68 samples collected from common areas tested positive. The viral loads for the positive samples ranged widely, from 16 to 31,217 RNA copies. The median viral load recovered from cabins with suspected clinical norovirus cases was 3.6 log10 RNA copies (range, 2.37 log10 to 4.49 log10 RNA copies), significantly higher than that recovered from common areas (1.20 log10 RNA copies; range, 1.2 log10 to 2.1 log10 RNA copies) (P < 0.001). Four of 17 positive swab samples could be genotyped and had identical GII.1 sequences.
DISCUSSION
We developed and evaluated a new protocol for the sampling of environmental surfaces for human norovirus (Fig. 5). Of the 4 swab materials tested, including fiber-tipped swabs made from polyester, rayon, or cotton, macrofoam-based swabs demonstrated the highest rate of recovery of virus seeded and dried on stainless-steel surfaces. The macrofoam swabs had at least 10-times-higher levels of virus recovery from large surface areas than antistatic wipes, which are widely used for field sampling (13). The superior performance of macrofoam-based swabs compared to other swab materials has also been reported for vegetative bacteria and their spores (30, 31).
When viruses are dried on surfaces, their desiccation has a significant negative effect on sampling efficiency. We did not detect differences in rates of virus recovery among the different swab materials when viruses were directly seeded, confirming that these swab materials were equally effective in releasing the absorbed viruses. Our findings indicate that the type of swab material and the area of the sampled surface are important factors for the detection of noroviruses from environmental surfaces. We found that the addition of Tween 80 to PBS enhanced the level of virus recovery, in agreement with the ability of a surfactant to increase the water content of the target surface and to facilitate the solubilization of cells or proteins from surfaces (32).
To detect norovirus contamination on environmental surfaces in outbreak settings, sampling of large surface areas is highly preferable, since high-contact surfaces, such as doorknobs and computer keyboards, are frequently implicated in the transmission of enteric viruses (14, 15, 33–35). The geometry of these frequently touched objects is irregular, and they are typically larger than 130 cm2, a size that exceeds the capacity of most fiber-tipped swabs. Antistatic wipes have been used successfully for the detection of norovirus on large surface areas in field settings (16, 17, 36). We confirmed that antistatic wipes consistently recover viruses from large surface areas but that macrofoam swabs show a higher rate of virus recovery from surfaces as large as 625 cm2.
To maximize the level of recovery of norovirus from swabs, several steps for the efficient elution and concentration of noroviruses were incorporated into our new swab protocol, including concentration of viral RNA using spin columns (Fig. 5). In addition, we found that efficient virus recovery from swab samples required transportation and storage at refrigeration temperatures.
We tested our swab protocol on a cruise ship on which several passengers had reported norovirus-like symptoms. Swab samples from all three case cabins tested positive for norovirus, with virus titers significantly higher than those of samples collected from common areas on the ship. Flushing of the toilet has been suggested as a possible risk factor contributing to environmental contamination because of the potential aerosols generated (37). Furthermore, the positive findings on items such as telephones, keyboards, and door handles support the idea that contaminated hands act as a key vehicle for the spread of norovirus in the cabins.
Our study has several limitations. Since no stool samples were collected from the sick passengers on the ship, we were not able to confirm the finding of the GII.1-positive swab samples. Given the virus detection limit of the macrofoam-based swab method, negative results should not necessarily be interpreted as the absence of viral contamination. Additionally, the efficiency of norovirus recovery from hard surfaces other than stainless steel and toilet seats may be lower and requires further evaluation.
In general, the norovirus loads on frequently touched surfaces in public areas were lower than those in cabins whose occupants had gastroenteritis, likely because of regular cleaning practices. However, because of the low infectious dose of noroviruses (38, 39), low-level norovirus contamination on surfaces in common areas presents a potential health risk. Surfaces that are frequently touched by bare hands, such as condiment containers and dispenser handles in restaurants, may facilitate the spread of noroviruses among passengers. Our data support previous data showing that contact with an infected partner and the use of contaminated toilets are risk factors associated with norovirus infections (40). The fact that we found identical GII.1 sequences in the three cabins sampled strongly suggests that this strain was the etiologic agent of the viral gastroenteritis clusters on this voyage and supports previous reports that environmental sampling may serve as an effective norovirus outbreak investigational tool (14–16, 35). Cotton swabs are currently recommended by the ISO/TS 15216 standard protocol for the sampling of food preparation surfaces (18). Our results show that the use of premoistened macrofoam swabs leads to an improved virus recovery level and therefore should be considered for the detection of norovirus on inanimate surfaces.
ACKNOWLEDGMENTS
We thank Ingeborg Boxman for providing the antistatic wipes and Captain Jaret Ames and Amy Freeland for their continued support.
The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC). Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the CDC or the U.S. Department of Health and Human Services.
REFERENCES
- 1.Glass RI, Parashar UD, Estes MK. 2009. Norovirus gastroenteritis. N Engl J Med 361:1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payne DC, Vinjé J, Szilagyi PG, Edwards KM, Staat MA, Weinberg GA, Hall CB, Chappell J, Bernstein DI, Curns AT, Wikswo M, Shirley SH, Hall AJ, Lopman B, Parashar UD. 2013. Norovirus and medically attended gastroenteritis in U.S. children. N Engl J Med 368:1121–1130. doi: 10.1056/NEJMsa1206589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vega E, Barclay L, Gregoricus N, Shirley SH, Lee D, Vinjé J. 2014. Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013. J Clin Microbiol 52:147–155. doi: 10.1128/JCM.02680-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isakbaeva ET, Widdowson MA, Beard RS, Bulens SN, Mullins J, Monroe SS, Bresee J, Sassano P, Cramer EH, Glass RI. 2005. Norovirus transmission on cruise ship. Emerg Infect Dis 11:154–158. doi: 10.3201/eid1101.040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopman BA, Gastañaduy P, Park GW, Hall AJ, Parashar UD, Vinjé J. 2012. Environmental transmission of norovirus gastroenteritis. Curr Opin Virol 2:96–102. doi: 10.1016/j.coviro.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Malek M, Barzilay E, Kramer A, Camp B, Jaykus LA, Escudero-Abarca B, Derrick G, White P, Gerba C, Higgins C, Vinjé J, Glass R, Lynch M, Widdowson MA. 2009. Outbreak of norovirus infection among river rafters associated with packaged delicatessen meat, Grand Canyon, 2005. Clin Infect Dis 48:31–37. doi: 10.1086/594118. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2008. Norovirus outbreak in an elementary school—District of Columbia, February 2007. MMWR Morb Mortal Wkly Rep 56:1340–1343. [PubMed] [Google Scholar]
- 8.Cheesbrough JS, Barkess-Jones L, Brown DW. 1997. Possible prolonged environmental survival of small round structured viruses. J Hosp Infect 35:325–326. doi: 10.1016/S0195-6701(97)90230-9. [DOI] [PubMed] [Google Scholar]
- 9.Park GW, Boston DM, Kase JA, Sampson MN, Sobsey MD. 2007. Evaluation of liquid- and fog-based application of Sterilox hypochlorous acid solution for surface inactivation of human norovirus. Appl Environ Microbiol 73:4463–4468. doi: 10.1128/AEM.02839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park GW, Sobsey MD. 2011. Simultaneous comparison of murine norovirus, feline calicivirus, coliphage MS2, and GII.4 norovirus to evaluate the efficacy of sodium hypochlorite against human norovirus on a fecally soiled stainless steel surface. Foodborne Pathog Dis 8:1005–1010. doi: 10.1089/fpd.2010.0782. [DOI] [PubMed] [Google Scholar]
- 11.CDC. 2011. Updated norovirus outbreak management and disease prevention guidelines. MMWR Recomm Rep 60(RR-3):1–18. [PubMed] [Google Scholar]
- 12.MacCannell T, Umscheid CA, Agarwal RK, Lee I, Kuntz G, Stevenson KB, Healthcare Infection Control Practices Advisory Committee. 2011. Guideline for the prevention and control of norovirus gastroenteritis outbreaks in healthcare settings. Infect Control Hosp Epidemiol 32:939–969. doi: 10.1086/662025. [DOI] [PubMed] [Google Scholar]
- 13.Isenberg HD. 2007. Microbiological assay of environmental and medical-device surfaces, p 485–496. In Garcia LS. (ed), Clinical microbiology procedures handbook, 2nd ed American Society for Microbiology, Washington, DC. [Google Scholar]
- 14.Gallimore CI, Taylor C, Gennery AR, Cant AJ, Galloway A, Iturriza-Gomara M, Gray JJ. 2006. Environmental monitoring for gastroenteric viruses in a pediatric primary immunodeficiency unit. J Clin Microbiol 44:395–399. doi: 10.1128/JCM.44.2.395-399.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bright KR, Boone SA, Gerba CP. 2010. Occurrence of bacteria and viruses on elementary classroom surfaces and the potential role of classroom hygiene in the spread of infectious diseases. J Sch Nurs 26:33–41. doi: 10.1177/1059840509354383. [DOI] [PubMed] [Google Scholar]
- 16.Boxman IL, Dijkman R, te Loeke NA, Hagele G, Tilburg JJ, Vennema H, Koopmans M. 2009. Environmental swabs as a tool in norovirus outbreak investigation, including outbreaks on cruise ships. J Food Prot 72:111–119. [DOI] [PubMed] [Google Scholar]
- 17.Boxman IL, Verhoef L, Dijkman R, Hagele G, Te Loeke NA, Koopmans M. 2011. Year-round prevalence of norovirus in the environment of catering companies without a recently reported outbreak of gastroenteritis. Appl Environ Microbiol 77:2968–2974. doi: 10.1128/AEM.02354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International Organization for Standardization (ISO). 15 March 2013. Microbiology of food and animal feed—horizontal method for determination of hepatitis A virus and norovirus in food using real-time RT-PCR. ISO/TS 15216-1:2013. ISO, Geneva, Switzerland. [Google Scholar]
- 19.Julian TR, Tamayo FJ, Leckie JO, Boehm AB. 2011. Comparison of surface sampling methods for virus recovery from fomites. Appl Environ Microbiol 77:6918–6925. doi: 10.1128/AEM.05709-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taku A, Gulati BR, Allwood PB, Palazzi K, Hedberg CW, Goyal SM. 2002. Concentration and detection of caliciviruses from food contact surfaces. J Food Prot 65:999–1004. [DOI] [PubMed] [Google Scholar]
- 21.Scherer K, Ellerbroek L, Schulenburg J, Johne R, Klein G. 2009. Application of a swab sampling method for the detection of norovirus and rotavirus on artificially contaminated food and environmental surfaces. Food Environ Virol 1:42–49. doi: 10.1007/s12560-008-9007-0. [DOI] [Google Scholar]
- 22.Herzog AB, Pandey AK, Reyes-Gastelum D, Gerba CP, Rose JB, Hashsham SA. 2012. Evaluation of sample recovery efficiency for bacteriophage P22 on fomites. Appl Environ Microbiol 78:7915–7922. doi: 10.1128/AEM.01370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentry J, Vinjé J, Guadagnoli D, Lipp EK. 2009. Norovirus distribution within an estuarine environment. Appl Environ Microbiol 75:5474–5480. doi: 10.1128/AEM.00111-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park GW, Barclay L, Macinga D, Charbonneau D, Pettigrew CA, Vinjé J. 2010. Comparative efficacy of seven hand sanitizers against murine norovirus, feline calicivirus, and GII.4 norovirus. J Food Prot 73:2232–2238. [DOI] [PubMed] [Google Scholar]
- 25.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. 1990. Rapid and simple method for purification of nucleic acids. J Clin Microbiol 28:495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill VR, Mull B, Jothikumar N, Ferdinand K, Vinjé J. 2010. Detection of GI and GII noroviruses in ground water using ultrafiltration and TaqMan real-time RT-PCR. Food Environ Virol 2:218–224. doi: 10.1007/s12560-010-9049-y. [DOI] [Google Scholar]
- 27.Rolfe KJ, Parmar S, Mururi D, Wreghitt TG, Jalal H, Zhang H, Curran MD. 2007. An internally controlled, one-step, real-time RT-PCR assay for norovirus detection and genogrouping. J Clin Virol 39:318–321. doi: 10.1016/j.jcv.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Schultz AC, Vega E, Dalsgaard A, Christensen LS, Norrung B, Hoorfar J, Vinjé J. 2011. Development and evaluation of novel one-step TaqMan realtime RT-PCR assays for the detection and direct genotyping of genogroup I and II noroviruses. J Clin Virol 50:230–234. doi: 10.1016/j.jcv.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D'Agostino SR, Sullivan L, Beiser A. 2006. Introductory applied biostatistics. Thomson Brooks/Cole, Belmont, CA. [Google Scholar]
- 30.Gilbert SE, Rose LJ, Howard M, Bradley MD, Shah S, Silvestri E, Schaefer FW III, Noble-Wang J. 2014. Evaluation of swabs and transport media for the recovery of Yersinia pestis. J Microbiol Methods 96:35–41. doi: 10.1016/j.mimet.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 31.Rose L, Jensen B, Peterson A, Banerjee SN, Arduino MJ. 2004. Swab materials and Bacillus anthracis spore recovery from nonporous surfaces. Emerg Infect Dis 10:1023–1029. doi: 10.3201/eid1006.030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore G, Griffith C. 2007. Problems associated with traditional hygiene swabbing: the need for in-house standardization. J Appl Microbiol 103:1090–1103. doi: 10.1111/j.1365-2672.2007.03330.x. [DOI] [PubMed] [Google Scholar]
- 33.Huslage K, Rutala WA, Sickbert-Bennett E, Weber DJ. 2010. A quantitative approach to defining “high-touch” surfaces in hospitals. Infect Control Hosp Epidemiol 31:850–853. doi: 10.1086/655016. [DOI] [PubMed] [Google Scholar]
- 34.Wadl M, Scherer K, Nielsen S, Diedrich S, Ellerbroek L, Frank C, Gatzer R, Hoehne M, Johne R, Klein G, Koch J, Schulenburg J, Thielbein U, Stark K, Bernard H. 2010. Food-borne norovirus-outbreak at a military base, Germany, 2009. BMC Infect Dis 10:30. doi: 10.1186/1471-2334-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu HM, Fornek M, Schwab KJ, Chapin AR, Gibson K, Schwab E, Spencer C, Henning K. 2005. A norovirus outbreak at a long-term-care facility: the role of environmental surface contamination. Infect Control Hosp Epidemiol 26:802–810. doi: 10.1086/502497. [DOI] [PubMed] [Google Scholar]
- 36.Boxman I, Dijkman R, Verhoef L, Maat A, van Dijk G, Vennema H, Koopmans M. 2009. Norovirus on swabs taken from hands illustrate route of transmission: a case study. J Food Prot 72:1753–1755. [DOI] [PubMed] [Google Scholar]
- 37.Barker J, Jones MV. 2005. The potential spread of infection caused by aerosol contamination of surfaces after flushing a domestic toilet. J Appl Microbiol 99:339–347. doi: 10.1111/j.1365-2672.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- 38.Teunis PFM, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, Le Pendu J, Calderon RL. 2008. Norwalk virus: how infectious is it? J Med Virol 80:1468–1476. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- 39.Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Ramani S, Hill H, Ferreira J, Graham DY. 2014. Determination of the 50% human infectious dose for Norwalk virus. J Infect Dis 209:1016–1022. doi: 10.1093/infdis/jit620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chimonas MA, Vaughan GH, Andre Z, Ames JT, Tarling GA, Beard S, Widdowson MA, Cramer E. 2008. Passenger behaviors associated with norovirus infection on board a cruise ship—Alaska, May to June 2004. J Travel Med 15:177–183. doi: 10.1111/j.1708-8305.2008.00200.x. [DOI] [PubMed] [Google Scholar]