Abstract
The ability to acetylate lysine residues is conserved across organisms, and acetylation of lysine residues plays important roles in various cellular functions. Maintaining intracellular pH homeostasis is crucial for the survival of enteric bacteria in the acidic gastric tract. It has been shown that eukaryotes can stabilize the intracellular pH by histone deacetylation. However, it remains unknown whether bacteria can utilize a reversible protein acetylation system to adapt to an acidic environment. Here we demonstrate that protein acetylation/deacetylation is critical for Salmonella enterica serovar Typhimurium to survive in an acidic environment. We used RNA sequencing to analyze the transcriptome patterns under acid stress and found that the transcriptional levels of genes involved in NAD+/NADH metabolism were significantly changed, leading to an increase in the intracellular NAD+/NADH ratio. Moreover, acid stress downregulated the transcriptional level of pat, encoding acetyltransferase, and genes cyaA and crp, encoding adenylate cyclase and cyclic AMP receptor protein, respectively, which are positive regulators of pat. It was found that the acid signal alerts the tricarboxylic acid cycle to promote the consumption of acetyl coenzyme A (Ac-CoA), an acetyl group donor for the acetylation reaction. A lowered acetylation level not only was the bacterial response to acid stress but also increased the survival rate of S. Typhimurium under acid stress. The pat deletion mutant had a more stable intracellular pH, which paralleled the higher survival rate after acid treatment compared with that of both the wild-type strain and the cobB (encoding deacetylase) deletion mutant. Our data indicate that bacteria can downregulate the protein acetylation level to prevent the intracellular pH from further falling under acid stress, and this work may provide a new perspective to understand the bacterial acid resistance mechanism.
INTRODUCTION
Salmonella enterica serovar Typhimurium has long been recognized to be a foodborne pathogen. As a facultative human pathogen, it is the causative agent of nontyphoid salmonellosis and can survive at an extremely low pH. As an enteric pathogen, S. Typhimurium must survive in the strong acidic environment (pH 2) of the stomach until it can further invade the intestinal epithelium and deeper organs, including the spleen and liver (1). Therefore, the ability to sense and respond to the acidic environment is crucial for its pathogenesis. In fact, S. Typhimurium has indeed evolved a variety of elegant regulatory mechanisms to protect itself from acid stress. For example, when bacteria encounter acid stress, hydrogen ions flow into the cell and cause a decrease in the intracellular pH, which activates the expression of amino acid decarboxylases, including AdiA, CadA, and SpeF. These inducible amino acid decarboxylases are pyridoxal phosphate-containing enzymes that replace the α-carboxyl groups of their cognate amino acid substrates with a proton consumed from the cytoplasm (2). AdiA, CadA, and SpeF decarboxylate arginine, lysine, and ornithine, respectively, to produce agmatine, cadaverine, and putrescine, respectively, and at the same time, internal protons are consumed to keep the intracellular pH stable. In addition, low pH increases the accumulation of PhoP/Fur/RpoS, involved in the log-phase acid tolerance response, and RpoS/OmpR, involved in the stationary-phase acid tolerance response, all of which can control distinct sets of acid shock proteins to prevent and repair the damage to macromolecules caused by acid stress (3).
Recently, it has been reported that histone acetylation can regulate the intracellular pH (pHi) (4). When pHi decreases, histones can be deacetylated by histone deacetylases (HDACs) to produce acetate anions, and then protons and acetate anions are exported out of the cell by monocarboxylate transporters (MCTs), thus preventing pHi from further decreasing.
Protein acetylation, which used to be considered a predominant phenomenon in eukaryotes, broadly impacts bacterial physiological processes. Previous studies with S. Typhimurium and Escherichia coli identified a large number of acetylated proteins which are involved in a variety of metabolic processes (5). Proteome-wide lysine acetylation profiling of Mycobacterium tuberculosis identified 226 acetylation sites in 137 proteins participating in glycolysis/gluconeogenesis, the tricarboxylic acid (TCA) cycle, and fatty acid metabolism (6). The conserved reversible lysine acetylation in bacteria is catalyzed by the acetyl coenzyme A (Ac-CoA)-dependent, Gcn5-like protein Pat, which is an N-acetyltransferase (7), and the Sir2 homolog CobB, which is an NAD (NAD+)-dependent deacetylase (8). In S. Typhimurium, reversible lysine acetylation is a mechanism critical for the regulation of central metabolism (9).
Based on the importance of maintaining acid homeostasis in S. Typhimurium and the histone acetylation involved in pH regulation in mammalian cells, we speculate that S. Typhimurium may utilize a reversible protein acetylation system to resist acid stress. To test this hypothesis, we first studied the transcriptome of S. Typhimurium under acid stress and found that the transcription of pat was repressed. Meanwhile, the transcription levels of genes involved in NAD+/NADH transformation and Ac-CoA metabolism were significantly changed. Western blot analysis showed that the overall acetylation of intracellular proteins decreased after acid challenge. Moreover, the deletion mutant of pat had a more stable pHi, paralleling the higher survival rate, compared with that of a cobB deletion mutant under acid stress. Our data demonstrate that bacteria can better adapt themselves to an acidic environment by adjusting the global protein acetylation level.
MATERIALS AND METHODS
Culture conditions.
An overnight culture was diluted 1:100 in 5 ml of minimal E glucose (EG) medium (MgSO4, 0.098 g/liter; citric acid monohydrate, 2.0 g/liter; K2HPO4·3H2O, 13.1 g/liter; NaNH4HPO4, 2.29 g/liter; glucose, 0.4% [wt/vol], pH 7.7) (10), followed by incubation at 37°C with shaking. When the optical density at 600 nm (OD600) reached 0.4, 5 ml of culture was centrifuged at 2,500 × g for 4 min. The pellet was washed once with EG medium at pH 3.0 and then resuspended in 5 ml of EG medium at pH 3.0 (the pH was adjusted with HCl). A control culture was resuspended in 5 ml of EG medium at pH 7.7 instead of pH 3.0. The two cultures were further incubated for 15 min and harvested for RNA isolation. For measurement of the survival rate, an overnight culture was inoculated into fresh EG medium until the OD600 reached 0.4 (log phase) or 2.7 (stationary phase). After the bacteria were harvested by centrifugation, bacteria at log phase were resuspended in EG medium at pH 3.0 for an additional 1 h of incubation, and stationary-phase bacteria were resuspended in EG medium at pH 3.0 for an additional 2 h. For counting of the number of CFU, the culture was serially diluted 10-fold and plated on Luria-Bertani (LB) agar plates.
RNA isolation and RNA-seq.
RNA was isolated by using the TRIzol reagent (Invitrogen), and DNase I digestion was conducted as described previously (11).
For RNA sequencing (RNA-seq) analysis, RNA was fragmented by heating at 95°C for 10 min and annealed to biotinylated random primers. A 5′ adaptor, which contained an Illumina primer, was added to the sequence. First-strand cDNA was synthesized by reverse transcription (RT). Then, Illumina primers were used to obtain double-stranded cDNA by the PCR method. cDNA fragments of 300 to 500 bp were harvested by gel extraction and then directly amplified with a TruSeq PE cluster kit (Illumina, America). Sequencing reactions were performed on an Illumina HiSeq 2000 sequencer.
Mapping of sequenced reads and differential gene expression.
First, reads containing sequencing adaptors and reads of low quality (reads containing >5 Ns [uncertain bases]) were removed (2% to 3%) to get clean reads. Then, the sequence reads were mapped against the reference S. Typhimurium 14028S genome with Bowtie (version 2) software. The coverage rate of 80% of the genes was about 90 to 100%. The number of reads for each gene from each sample was converted to the read numbers per million reads (RPM) (12). A multivariate adaptive plot-based method was used to calculate the difference in the abundance of expression of each gene in the samples by use of a random sampling model (MARS) from the DEGseq program package (13). A false discovery rate (FDR) value of less than 0.001 indicated a significant difference.
Validation of differential expression of genes by quantitative RT-PCR.
To validate the RNA-seq data, 25 upregulated or downregulated genes were selected for quantitative RT-PCR (qPCR) analysis. The primers used are listed in Table S1 in the supplemental material. Samples were run in triplicate and amplified using the SYBR Premix Ex Taq II reagent (TaKaRa). The relative transcriptional level was determined by the 2−ΔΔCT threshold cycle (CT) method. The expression of 16S rRNA, which is relatively constant in bacteria, was used as an internal control.
Determination of NAD+ and NADH concentrations.
The intracellular concentrations of NAD+ and NADH were determined using an enzymatic cycling assay kit (BioVision). Total NAD+ and NADH concentrations were normalized to the bacterial density, and measurement of the concentrations was performed in three independent experiments.
Bacterial strain and plasmid construction.
S. Typhimurium strain 14028S was used as the parental strain in this study. A bacteriophage λ Red recombination system was employed to construct Δpat and ΔcobB mutants. A pair of primers with 50-nucleotide (nt) extensions complementary to the regions immediately adjacent to the targeted gene that was deleted and 20-nt extensions complementary to the region immediately adjacent to the antibiotic resistance gene flanked by FLP recognition target (FRT) sites was synthesized (see Table S1 in the supplemental material). Antibiotic resistance gene-carrying plasmid pKD3 (Table 1) was used as the template for PCR amplification of the knockout cassette with an Invitrogen PCR kit (Invitrogen, Carlsbad, CA). The PCR products were recovered by use of a QIAquick gel extraction kit (Qiagen, Germany) and then transformed into S. Typhimurium carrying pKD46 via electroporation by using an electroporator (ECM 630; BTX, San Diego, CA) and a 0.1-cm cuvette according to the manufacturer's instructions. Pulsed cells were rescued with 0.9 ml super optimal broth with catabolite repression (SOC) medium and incubated for 1 h at 30°C and 20 h at room temperature, before being spread onto agar plates (31). The successful deletion of cobB and pat in the mutants was verified by PCR.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| S. enterica 14028S | Wild type | Laboratory stock |
| S. enterica 14028S Δpat | Δpat mutant | This study |
| S. enterica 14028S ΔcobB | ΔcobB mutant | This study |
| Plasmids | ||
| pKD46 | Expresses bacteriophage λ Red recombinase | Laboratory stock |
| pKD3 | Source for chloramphenicol acetyltransferase cassette | Laboratory stock |
| pCP20 | Expresses FLP recombinase | Laboratory stock |
| pTrc99a-gfpmut3b | Expresses GFPmut3b | 16 |
| pQE80-YX1 | Expression vector | Laboratory stock |
| pQE80-YX1-pat | Expresses pat | This study |
pat and cobB were amplified from S. Typhimurium genomic DNA by using primer sets pat-pQE80-AfeI-F/pat-pQE80-FseI-R and (cobB-pQE80-AfeI-F/cobB-pQE80-FseI-R), respectively (see Table S1 in the supplemental material). The PCR products were purified by use of an Easy-DNA kit (Invitrogen, Carlsbad, CA). The target fragment was inserted into pQE80-YX1 using the FseI site and the AfeI site. Each cloned gene was confirmed by DNA sequencing.
Measurement of cytoplasmic pH with a GFP reporter plasmid.
GFPmut3b is a pH-sensitive green fluorescent protein (GFP) (14, 15). To detect the pHi, pTrc99a-gfpmut3b was transformed into the Δpat, ΔcobB, or wild-type (WT) strain. Standard pH curves were generated for each of these three strains. The overnight culture was diluted 1:100 in pH 7.7 EG medium supplemented with 40 μM IPTG (isopropyl-β-d-thiogalactopyranoside) and 100 μg/ml ampicillin. Cells were grown to an OD600 of 0.6, pelleted by centrifugation at 2,500 × g for 4 min, and then resuspended to the same OD600 in EG medium with a different pH. The pHs of pH 5.5, pH 6.0, pH 6.5, and pH 7.0 EG medium were adjusted with HCl, and the pH of pH 7.5 EG medium was adjusted with NaOH. To equilibrate the cytoplasmic pH with the external pH, sodium benzoate at a final concentration of 30 mM was added to the medium (16).
Measurement of the cytoplasmic pH of experimental samples was performed as described above, except that cultures were grown in pH 4.0 EG medium and sodium benzoate was omitted. Cultures were kept on ice prior to recording of the GFPmut3b excitation spectra.
Fluorescence measurement of pH reporter plasmids.
The protocol used to measure the fluorescence intensity of GFPmut3b was described previously (16). The excitation spectra of GFPmut3b were recorded using a BioTek Synergy 2 fluorescence microplate reader. Excitation was measured at 485 ± 10 nm, using an emission wavelength of 528 ± 10 nm. The excitation of three biological replicates was measured. To determine the standard curve correlating the internal pH with the fluorescence intensity, GFPmut3b fluorescence intensities at pH 5.5, pH 6.0, pH 6.5, pH 7.0, and pH 7.5 were obtained. The intensities from pH 5.5 to pH 7.5 were fitted to a linear equation, y = mx + b, where y is the GFPmut3b intensity, x is the pH value, m is the slope, and b is the y intercept. The equation was used to convert the signal intensities obtained from experimental samples to pH units. Student's t test was used for statistical analysis, and results were considered statistically significant when the P value was less than 0.05.
Western blotting.
Bacteria were grown to an OD600 of 0.4 in pH 7.7 EG medium, treated with pH 3.0 EG medium for 30 min, washed once with phosphate-buffered saline (PBS), resuspended in PBS, mixed with 5× sample buffer, and boiled for 5 min. The samples were separated on a 12% SDS-polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, and immunoblotted using a polyclonal antiacetyl antibody. The Western blot was developed using anti-rabbit IgG horseradish peroxidase-linked antibody and an enhanced chemiluminescence detection system.
RNA-seq data accession number.
The final RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo) under accession no. GSE67925.
RESULTS
Overview of transcriptome analysis under acid stress.
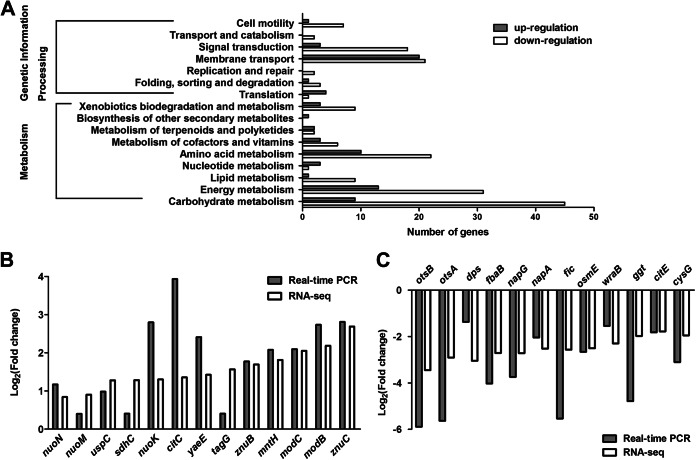
When bacteria encounter stressful conditions, they must make a prompt and global response to survive. In order to have an overall and comprehensive understanding about changes to the transcriptome under acid stress, we used RNA-seq analysis to determine the effect of acid treatment on the levels of gene transcription in S. Typhimurium. Sequencing using the Illumina HiSeq 2000 sequencer yielded 10,384,420 reads for the control group (pH 7.7) and 10,236,644 reads for the acid (pH 3.0)-treated group. The log2 ratios of the RPM values were used to identify differentially expressed genes. Consequently, 659 genes that showed a difference in expression of at least 2-fold (log2 ratios of the RPM values, ≥1 or ≤−1) between the control group and the acid (pH 3.0)-treated group were defined to be differentially expressed (see Table S2 in the supplemental material). Compared with the gene expression in the control group, 136 genes were upregulated and 523 genes were downregulated under acid stress. Since 12.1% (659/5,424) of the S. Typhimurium genes were significantly affected, we conclude that acid challenge has a crucial impact on the S. Typhimurium transcriptome. The Kyoto Encyclopedia of Genes and Genomes (KEGG) was used to classify genes into different categories. Among the 659 genes, 253 genes could be classified into categories (Fig. 1A). The cluster for carbohydrate metabolism was the largest group (54/253, 21.3%), followed by the clusters for energy metabolism (44/253, 17.4%) and membrane transport (41/253, 16.2%). It seems that S. Typhimurium alters carbohydrate and energy metabolism when it encounters a low-pH stress. We validated the RNA-seq data by qPCR. Twenty-five of the selected genes had comparable levels of expression by qPCR and RNA-seq (Fig. 1B and C).
FIG 1.
Transcriptome of Salmonella Typhimurium under acid stress. (A) Genes that were significantly differentially expressed under acid stress in Salmonella Typhimurium were classified using KEGG. (B and C) Comparison of transcriptional levels of 25 selected genes by qPCR and RNA-seq. (B) Upregulated genes; (C) downregulated genes.
In this study, the expression of the genes for several enzymes for the catabolism of carbohydrate and amino acids was shown to be pH dependent, which is consistent with previous reports (17, 18). For example, genes involved in processes of glycolysis and the TCA cycle, such as acnB (aconitate hydratase 2/2-methylisocitrate dehydratase) and sdhABC (succinate dehydrogenase subunit), respectively, were shown to be elevated in the acid treatment group. Maltose ABC transporter-coding genes malG and malF were reported to be repressed by acid stress (19), and our RNA-seq data showed the same expression pattern.
It has been known that the genes for membrane-bound systems for proton and electron transport are upregulated by acid shock (17). In our RNA-seq analysis, the genes for NADH dehydrogenase (nuoABCEFGHIJKLMN), which exports H+, were upregulated. The sdhABC (succinate dehydrogenase) genes, which provide electrons for proton export, were upregulated in response to the acid signal as well. It was reported that the genes for the lysine/arginine/ornithine decarboxylase system could be induced upon acid treatment (20). However, this phenomenon was not observed in this study. Stancik et al. also reported that the expression of the genes for this decarboxylase system did not change under an acidic environment in aerobic cultures (18). This may be because the highly aerobic culture conditions repressed the expression of the genes for these systems.
The expression of the genes for many envelope and periplasmic components has been identified to be pH dependent (18, 21). For example, hdeB is an acid chaperone and could be induced under acidic conditions (21). It is known that metal ion solubility increases at low pH, which would lead to the induction of the fep gene cluster (which encodes the iron-enterobactin transport system), znuABC (which encodes the zinc transport system), sitABCD (which encodes the manganese/iron transport system) and promote the uptake of metal ions (18). Our results show these genes are upregulated at pH 3.0 and confirm that extracellular pH can affect the membrane components.
Changes to metabolism related to acetylation.
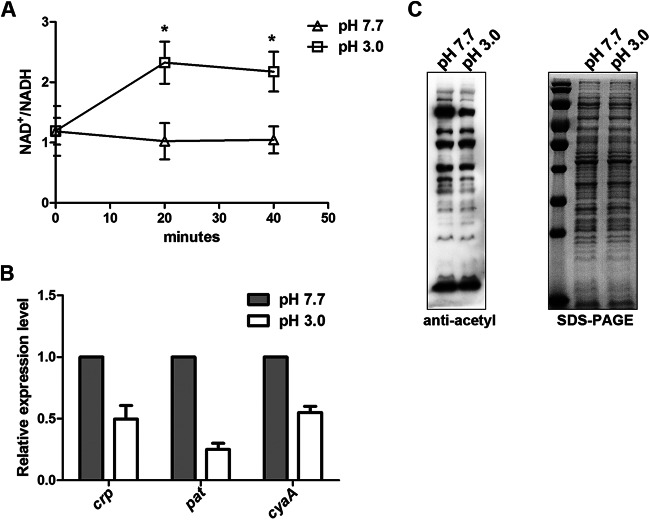
By RNA-seq analysis, we found that 44 genes related to energy metabolism were differentially expressed after acid stimulation (see Table S3 in the supplemental material). Most of these differentially expressed genes were downregulated, suggesting that cellular energy metabolism was inhibited under acid stress. The nuo gene cluster, encoding NADH dehydrogenase subunits, which catalyze the conversion of NADH and quinone into NAD+ and quinol, respectively, was upregulated. nirBCD, encoding nitrite reductase and the nitrite transporter, which catalyzes the conversion of NAD+ into NADH, were downregulated. These changes may collectively affect the concentration of NAD+ and NADH in cells, leading to an increased ratio of NAD+ to NADH. To confirm these findings, we measured the intracellular NAD+ and NADH concentrations of S. Typhimurium under acid stress. Normally, the ratio of NAD+ to NADH is about 1, but after acid stimulation for 20 min, it increased and was maintained at about 2 thereafter (Fig. 2A). Since CobB is an NAD+-dependent deacetylase, the activity of CobB may be higher under acid stress than at pH 7.7, leading to the reduction of protein acetylation.
FIG 2.
Changes in metabolism related to acetylation. (A) Intracellular NAD+/NADH ratios of WT S. Typhimurium maintained in EG medium at pH 3.0 or pH 7.7 at the indicated time points. The value for each time point represents the mean from three independent experiments. Significant differences in the NAD+/NADH ratio between pH 3.0 and pH 7.7 were observed after 20 min or 40 min of treatment (P = 0.047 for both time points). (B) Transcriptional levels of crp, pat, and cyaA. S. Typhimurium cultures were grown to an OD600 of 0.4. After incubation in medium at pH 3.0 or pH 7.7 for 15 min, RNA was extracted and then analyzed by RT-qPCR. (C) Acetylation spectra in medium at pH 3.0 or pH 7.7. Bacteria were grown to an OD600 of 0.4 in EG medium at pH 7.7 and switched to EG medium at pH 3.0 or pH 7.7 for another 30 min. Cell lysates were separated by SDS-PAGE and analyzed by antiacetyllysine Western blotting.
The RNA-seq data showed that under acidic conditions, the transcriptional level of pat (STM14_3248) decreased 1.61-fold. In addition, the cyaA and crp genes (encoding cyclic AMP [cAMP] synthetase and cAMP receptor protein [CRP], respectively) were downregulated 1.85-fold and 1.49-fold, respectively, under acid stress. Since the cAMP-CRP complex can bind to the pat promoter and positively regulate the transcription of pat (22), the additive effect of cyaA and crp downregulation could further reduce the expression of pat. We confirmed the results by qPCR. The transcription levels of all three genes, cyaA, crp, and pat, were lower in an acidic environment (Fig. 2B).
A total of 170 genes related to metabolism were differentially expressed, and these accounted for the largest category by KEGG clustering (Fig. 1A). These included 54 genes involved in carbohydrate metabolism. sdhABC and acnB, encoding succinate dehydrogenase subunits and aconitate hydratase 2/2-methylisocitrate dehydratase, respectively, which promote progression of the TCA cycle, were upregulated. While the frdABC and citDEF genes, encoding, respectively, fumarate reductase subunits and the citrate lyase subunit, which inhibit the TCA cycle, were downregulated. Altogether, these results suggest that acid stress promotes progression of the TCA cycle and consumes acetyl-CoA. As acetyl-CoA is the acetyl group donor, its consumption may reduce the level of acetylation.
Considering the fact that the cells had a higher NAD+/NADH ratio and repressed pat transcription after acid treatment, we predict that the intracellular acetylation level may decrease under acid stress. To test this idea, we treated cells with media of different pHs for 30 min and performed Western blotting with whole-cell lysates. As expected, the overall protein acetylation level in the group treated with the pH 3.0 medium was lower than that in the control group treated with the pH 7.7 medium (Fig. 2C). This result indicates that acid stress can globally downregulate the acetylation level.
The acetylation level affects survival and pH maintenance under acid stress.
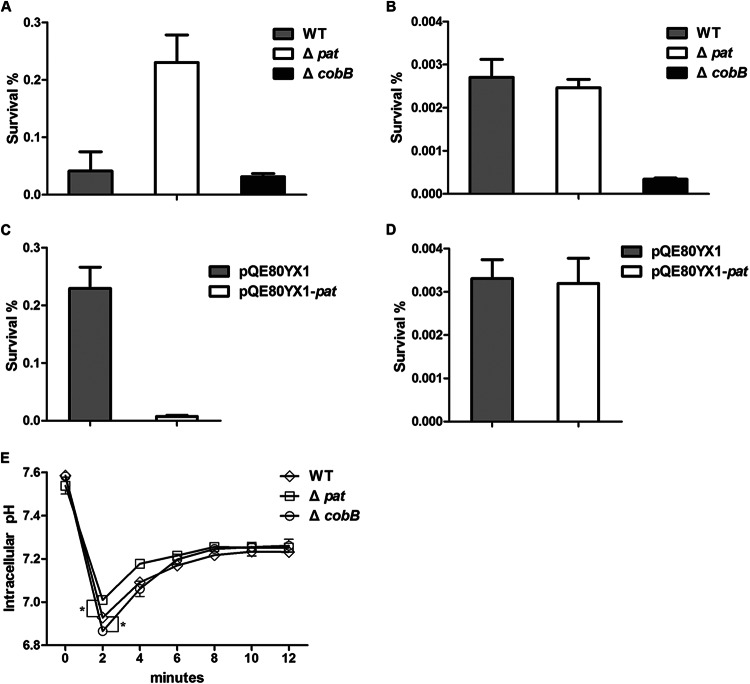
From the results of RNA-seq analysis presented above, we found that the acid signal may decrease the acetylation level through the regulation of pat transcription and the NAD+/NADH ratio of the cell. To explore whether a lower level of acetylation is beneficial for bacteria to fight against acid stress, we constructed S. Typhimurium mutants in which pat or cobB was deleted and compared their abilities to withstand acid challenge with the ability of the WT strain to withstand acid challenge at both log phase and stationary phase. The overnight culture was inoculated into fresh EG medium until the OD600 reached 0.4 (log phase). After 1 h of incubation at pH 3.0, the survival rate of the Δpat mutant (2.30%) was significantly higher than that of the WT (0.41%) or the ΔcobB mutant (0.31%) (Fig. 3A). It has been shown that S. Typhimurium has an increased resistance to acid stress in stationary phase compared with that in log phase (10). Therefore, we collected the bacteria at stationary phase and extended the acid treatment to 2 h to maximize the survival difference between strains. We found that the survival rate of the ΔcobB mutant (0.0034%) was significantly lower than that of the WT (0.027%) or the Δpat mutant (0.024%) (Fig. 3B). These results suggest that lowering of the acetylation level of cellular proteins is critical for bacterial survival under acidic conditions. To further confirm the role of pat and cobB under acid stress, we complemented their expression in the corresponding knockout strains. Overexpression of pat with pQE80YX1-pat in the Δpat strain decreased the survival rate from 2.29% to 0.074% at log phase (Fig. 3C) but not at stationary phase (Fig. 3D). The complementation of cobB expression with pQE80YX1-cobB in the ΔcobB strain led to cell aggregation for some unknown reasons, preventing us from further pursuing study of its phenotype. Taken together, our current results indicate that protein acetylation status, maintained by the balance between the Pat acetyltransferase and the CobB deacetylase, is critical for the resistance to acid stress in S. Typhimurium.
FIG 3.
Survival and cytoplasmic pH of different strains in acidic medium. (A and B) Survival rates of the WT, the Δpat mutant, or the ΔcobB mutant in EG medium at pH 3.0 at log phase (A) and at stationary phase (B). (C and D) Survival rates of the Δpat mutant carrying plasmid pQE80YX1-pat, which overexpresses pat, or the vector control at log phase (C) or stationary phase (D). (E) The cytoplasmic pH in the WT, Δpat mutant, or ΔcobB mutant determined by green fluorescent protein fluorimetry. Cells were resuspended in medium at pH 4.0, and the GFP excitation spectra were recorded without addition of benzoate. Fluorescence measurements were converted to pH units using a standard curve. The data for each time point represent the mean value and standard error from three independent experiments.
Intracellular pH is affected by protein acetylation level.
Under acid stress, bacteria have evolved systems of pH homeostasis to stabilize their pHi values (3). To explore why the pat and cobB mutants had different survival rates under acidic conditions, we used a pH-sensitive GFP reporter plasmid to determine the change in pHi after acid treatment. Basically, the fluorescence emission of GFP corresponds to a certain pH (23). By equilibrating the pH of the bacteria to the external pH using sodium benzoate, we first generated the standard curve correlating pHi with fluorescence intensity in the Δpat, ΔcobB, and WT strains. To measure the pHi of these strains after acid treatment, the cultures were centrifuged and resuspended in EG medium at pH 4.0. After the cells were shifted to medium at pH 4.0, the pHi decreased dramatically in all strains (Fig. 3E). This result was consistent with the previous finding that an instantaneous H+ influx leads to a decrease in the pHi (24). However, the pHi values of all three strains were significantly different. Based on the standard curves, we calculated the pHi of log-phase bacteria (OD600 = 0.4) after 2 min of treatment in medium at pH 4.0. The calculation showed that the pHi of the Δpat mutant was 7.01 ± 0.001 but that the pHi values of the ΔcobB mutant and WT were 6.86 ± 0.008 and 6.93 ± 0.015, respectively. This finding indicates that after acid challenge the Δpat mutant has a more stable pHi than the ΔcobB and WT strains and that the ΔcobB strain has the most drastic change of pHi. In other words, the acetylation/deacetylation system is involved in maintaining pH homeostasis in S. Typhimurium.
DISCUSSION
RNA-seq-based transcriptome analysis has been successfully applied in S. Typhimurium and other bacteria. For example, transcriptomic analyses by RNA-seq of S. Typhimurium under 22 distinct infection-relevant environmental conditions have been conducted (25). However, there have been no reports of transcriptome analysis of S. Typhimurium in acidic minimal medium, which mimics the physiological environment that S. Typhimurium usually encounters in vivo (10). In this study, we applied RNA-seq to analyze the global transcriptional profiles in minimal medium under acid stress. Genes involved in metabolism, some of which can convert NADH to NAD+, were significantly affected. As a result, the ratio of NAD+ to NADH increased after acid challenge (Fig. 2A), and this increase may lead to the activation of NAD+-dependent CobB. Simultaneously, the RNA-seq data showed a decreased level of pat transcription and an enhanced TCA cycle, leading to the consumption of Ac-CoA. Collectively, the overall protein acetylation level was decreased under acid stress (Fig. 2C). Our data clearly demonstrate that protein acetylation spectra are changed by an acidic stimulus.
Our results showed that the survival rate of the Δpat mutant after acid treatment was the highest among the strains tested, while that of the ΔcobB mutant was the lowest at log phase (Fig. 3A). The survival rate of the Δpat mutant was significantly increased compared with that of the WT in log phase but not in stationary phase. This may be due to the variation in the level of pat mRNA in different phases. Our previous study showed that pat is more actively transcribed in log phase than other phases (9). Therefore, pat exerted its effects on bacterial survival rate only in log phase and not in stationary phase. When we overexpressed pat, the acid resistance phenotype of the Δpat strain was impaired (Fig. 3C). Interestingly, we observed the difference in survival rates between the ΔcobB mutant and the WT in stationary phase but not in log phase. It was shown that the ratio of NAD+/NADH increases in stationary phase (26), which increases the activity of CobB. Therefore, the cobB deletion mutant showed a decreased survival rate in stationary phase compared with that of the WT strain. The data presented above suggest that a lower acetylation level is beneficial for bacteria to overcome acid stress. Protein acetylation/deacetylation is not a passive bacterial response to acid challenge. Instead, the bacteria actively downregulate their overall acetylation level to cope with the acid stress.
The ratios of NAD+/NADH and Ac-CoA/CoA are indicators of cellular energy and carbon status, respectively. Meanwhile, the activities of Pat and CobB can be regulated by the concentration of Ac-CoA and NAD+, respectively. Under acid stress, bacteria have to coordinate different metabolic pathways to allow themselves to overcome difficult environmental conditions. Acid stimulation may affect global protein acetylation through the regulation of metabolic flux. In addition, it is known that acetylation can regulate many metabolic enzymes, including acetyl-CoA synthetase (Acs) (7), glyceraldehyde-3-phosphate dehydrogenase (GapA), isocitrate lyase (AceA), and isocitrate dehydrogenase kinase/phosphatase (AceK) (9). We speculate that acetylation uses Ac-CoA and NAD+, two molecules that are directly involved in metabolic pathways, as the substrates, providing unparalleled advantages in sensing cellular pHi and thus allowing the bacteria to resist acid stress.
We found that the pat and cobB deletion mutants have different pHi values when encountering acid stress. The underlying mechanism is still elusive. Several amino acid decarboxylases could be induced and catalyze the decarboxylation reaction, in which extra internal protons would be consumed (27). Although we did not detect the transcriptional activation of lysine/arginine/ornithine decarboxylase in our RNA-seq analysis, our previous mass spectrometry data showed that the inducible lysine decarboxylase CadA was acetylated (9). We speculate that acetylation could regulate the activity of decarboxylase to affect the efflux of H+ and result in the change of the pHi.
Weinert et al. showed that acetyl phosphate (AcP) is a critical factor for lysine acetylation in E. coli (28). AcP is also an intermediate molecule of primary metabolism and involved in protein phosphorylation (29, 30). Therefore, it is highly possible that the AcP synthesis pathway could be involved in the acid stress response through both protein acetylation and metabolic modulation in S. Typhimurium.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the State Key Development Programs for Basic Research of China (973 Program no. 2015CB554203), the National Natural Science Foundation of China (no. 31270173, no. 31070114), and the Program for Professor of Special Appointment (Eastern Scholar) at the Shanghai Institutions of Higher Learning.
We are grateful to Rasika Harshey of the University of Texas at Austin for plasmid pTrc99a-gfpmut3b and to Jaemin Lee for help in measuring the pHi.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01009-15.
REFERENCES
- 1.Drecktrah D, Knodler LA, Ireland R, Steele-Mortimer O. 2006. The mechanism of Salmonella entry determines the vacuolar environment and intracellular gene expression. Traffic 7:39–51. doi: 10.1111/j.1600-0854.2005.00360.x. [DOI] [PubMed] [Google Scholar]
- 2.Ramos-Morales F. 2012. Acidic pH: enemy or ally for enteric bacteria? Virulence 3:103–106. doi: 10.4161/viru.19382. [DOI] [PubMed] [Google Scholar]
- 3.Bearson S, Bearson B, Foster JW. 1997. Acid stress responses in enterobacteria. FEMS Microbiol Lett 147:173–180. doi: 10.1111/j.1574-6968.1997.tb10238.x. [DOI] [PubMed] [Google Scholar]
- 4.McBrian MA, Behbahan IS, Ferrari R, Su T, Huang TW, Li K, Hong CS, Christofk HR, Vogelauer M, Seligson DB, Kurdistani SK. 2013. Histone acetylation regulates intracellular pH. Mol Cell 49:310–321. doi: 10.1016/j.molcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu BJ, Kim JA, Moon JH, Ryu SE, Pan JG. 2008. The diversity of lysine-acetylated proteins in Escherichia coli. J Microbiol Biotechnol 18:1529–1536. [PubMed] [Google Scholar]
- 6.Liu F, Yang M, Wang X, Yang S, Gu J, Zhou J, Zhang XE, Deng J, Ge F. 2014. Acetylome analysis reveals diverse functions of lysine acetylation in Mycobacterium tuberculosis. Mol Cell Proteomics 13:3352–3366. doi: 10.1074/mcp.M114.041962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starai VJ, Escalante-Semerena JC. 2004. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J Mol Biol 340:1005–1012. doi: 10.1016/j.jmb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP. 2010. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee IS, Slonczewski JL, Foster JW. 1994. A low-pH-inducible, stationary-phase acid tolerance response in Salmonella typhimurium. J Bacteriol 176:1422–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J, Hinton JC. 2012. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Feng Z, Wang X, Zhang X. 2010. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 14.Kitko RD, Cleeton RL, Armentrout EI, Lee GE, Noguchi K, Berkmen MB, Jones BD, Slonczewski JL. 2009. Cytoplasmic acidification and the benzoate transcriptome in Bacillus subtilis. PLoS One 4:e8255. doi: 10.1371/journal.pone.0008255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilks JC, Slonczewski JL. 2007. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J Bacteriol 189:5601–5607. doi: 10.1128/JB.00615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Harshey RM. 2012. Loss of FlhE in the flagellar type III secretion system allows proton influx into Salmonella and Escherichia coli. Mol Microbiol 84:550–565. doi: 10.1111/j.1365-2958.2012.08043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol 187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J Bacteriol 184:4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chagneau C, Heyde M, Alonso S, Portalier R, Laloi P. 2001. External-pH-dependent expression of the maltose regulon and ompF gene in Escherichia coli is affected by the level of glycerol kinase, encoded by glpK. J Bacteriol 183:5675–5683. doi: 10.1128/JB.183.19.5675-5683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Ordonez A, Begley M, Prieto M, Messens W, Lopez M, Bernardo A, Hill C. 2011. Salmonella spp. survival strategies within the host gastrointestinal tract. Microbiology 157:3268–3281. doi: 10.1099/mic.0.050351-0. [DOI] [PubMed] [Google Scholar]
- 21.Gajiwala KS, Burley SK. 2000. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J Mol Biol 295:605–612. doi: 10.1006/jmbi.1999.3347. [DOI] [PubMed] [Google Scholar]
- 22.Castano-Cerezo S, Bernal V, Blanco-Catala J, Iborra JL, Canovas M. 2011. cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol Microbiol 82:1110–1128. doi: 10.1111/j.1365-2958.2011.07873.x. [DOI] [PubMed] [Google Scholar]
- 23.Miesenbock G, De Angelis DA, Rothman JE. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 24.Shabala L, Budde B, Ross T, Siegumfeldt H, Jakobsen M, McMeekin T. 2002. Responses of Listeria monocytogenes to acid stress and glucose availability revealed by a novel combination of fluorescence microscopy and microelectrode ion-selective techniques. Appl Environ Microbiol 68:1794–1802. doi: 10.1128/AEM.68.4.1794-1802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz JP, Passonneau JV, Johnson GS, Pastan I. 1974. The effect of growth conditions on NAD+ and NADH concentrations and the NAD+:NADH ratio in normal and transformed fibroblasts. J Biol Chem 249:4138–4143. [PubMed] [Google Scholar]
- 27.Viala JP, Meresse S, Pocachard B, Guilhon AA, Aussel L, Barras F. 2011. Sensing and adaptation to low pH mediated by inducible amino acid decarboxylases in Salmonella. PLoS One 6:e22397. doi: 10.1371/journal.pone.0022397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinert BT, Iesmantavicius V, Wagner SA, Scholz C, Gummesson B, Beli P, Nystrom T, Choudhary C. 2013. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell 51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. 2007. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol 189:5574–5581. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verdin E, Ott M. 2013. Acetylphosphate: a novel link between lysine acetylation and intermediary metabolism in bacteria. Mol Cell 51:132–134. doi: 10.1016/j.molcel.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmid DNA. J Mol Biol 166:557–580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.