Abstract
Thermoproteales (phylum Crenarchaeota) populations are abundant in high-temperature (>70°C) environments of Yellowstone National Park (YNP) and are important in mediating the biogeochemical cycles of sulfur, arsenic, and carbon. The objectives of this study were to determine the specific physiological attributes of the isolate Pyrobaculum yellowstonensis strain WP30, which was obtained from an elemental sulfur sediment (Joseph's Coat Hot Spring [JCHS], 80°C, pH 6.1, 135 μM As) and relate this organism to geochemical processes occurring in situ. Strain WP30 is a chemoorganoheterotroph and requires elemental sulfur and/or arsenate as an electron acceptor. Growth in the presence of elemental sulfur and arsenate resulted in the formation of thioarsenates and polysulfides. The complete genome of this organism was sequenced (1.99 Mb, 58% G+C content), revealing numerous metabolic pathways for the degradation of carbohydrates, amino acids, and lipids. Multiple dimethyl sulfoxide-molybdopterin (DMSO-MPT) oxidoreductase genes, which are implicated in the reduction of sulfur and arsenic, were identified. Pathways for the de novo synthesis of nearly all required cofactors and metabolites were identified. The comparative genomics of P. yellowstonensis and the assembled metagenome sequence from JCHS showed that this organism is highly related (∼95% average nucleotide sequence identity) to in situ populations. The physiological attributes and metabolic capabilities of P. yellowstonensis provide an important foundation for developing an understanding of the distribution and function of these populations in YNP.
INTRODUCTION
Microbial communities in high-temperature (>70°C) hypoxic environments often contain abundant archaeal populations within the orders Sulfolobales, Desulfurococcales, and Thermoproteales (phylum Crenarchaeota) (1–4). Sulfolobales populations are abundant in low-pH (e.g., pH < 5) sulfidic sediments and iron oxide mats and have been shown to utilize both organic and inorganic carbon for growth (5, 6). Acidilobus-like organisms (order Desulfurococcales) are also abundant in hypoxic sulfur sediments (pH ∼3 to 6) and likely degrade complex organic constituents via fermentation (4). Members within the Thermoproteales are chemoorganoheterotrophs and/or facultative chemolithoautotrophs and currently consist of six genera: Thermofilum, Thermocladium, Caldivirga, Vulcanisaeta, Thermoproteus, and Pyrobaculum (7). Nucleotide sequences related to all six genera have been identified in metagenome data sets obtained from sulfidic geothermal systems in Yellowstone National Park (YNP); Vulcanisaeta and Caldivirga-like sequences predominate in acidic (pH 4 to 6) environments, whereas Pyrobaculum-like populations are more abundant at the pH values noted above 6 (1, 2).
The genus Pyrobaculum (8) is currently represented by eight isolates from geothermal systems in Iceland, Italy, the Philippines, Japan, and Russia. All are rod shaped, and all exhibit optimum growth conditions at temperatures ranging from 75 to 100°C and pH values ranging from 5 to 9. Pyrobaculum spp. use a variety of electron acceptors, such as elemental sulfur, nitrate, arsenate, and ferric iron (9, 10); however, Pyrobaculum aerophilum, Pyrobaculum oguniense, and Pyrobaculum calidifontis have been shown to be facultative aerobes, which grow in the presence of oxygen. A partial cytochrome aa3 terminal oxidase (type A subunit 1 heme-Cu oxidase) has been purified and characterized in P. oguniense (11, 12), and gene homologs of cytochrome aa3 have been identified in P. aerophilum (10, 13) and P. calidifontis (10). All Pyrobaculum isolates grow as chemoorganoheterotrophs on complex organic carbon (e.g., yeast extract [YE], peptone, tryptone), although Pyrobaculum neutrophilum, Pyrobaculum islandicum, P. aerophilum, and Pyrobaculum arsenaticum can also grow chemolithoautotrophically on hydrogen. These Pyrobaculum spp. fix carbon dioxide via the dicarboxylate/4-hydroxybutyrate (DC/4-HB) cycle, which contains three diagnostic genes (encoding 4-hydroxybutyryl coenzyme A [CoA] dehydratase [4-BUDH], phosphoenolpyruvate [PEP] carboxylase, and pyruvate synthase) (14–17). Enzyme activity studies using P. islandicum have also suggested that the reductive tricarboxylic acid (rTCA) cycle, which contains ATP citrate lyase and AMP-forming acetate:CoA ligase (not ATP citrate synthase), is functional in these organisms (18). Pyrobaculum-like organisms are important in many geothermal habitats in YNP, and the isolation of predominant organisms is important for characterizing relevant physiological processes occurring in situ.
Prior work in hypoxic sulfur sediments (80 to 82°C) has provided considerable information on the distribution, abundance, and potential function of Pyrobaculum-like organisms in YNP (1, 2, 19, 20). The metagenome sequence from several high-temperature sites suggests that the metabolic potential of Pyrobaculum spp. involves heterotrophy and the reduction of elemental sulfur and/or arsenic using novel dimethyl sulfoxide (DMSO)-molybdopterins (MPTs) (2). Native Pyrobaculum populations contain pathways for the degradation of polysaccharides, proteins, and lipids and are likely involved in important interactions with other members of these low-complexity communities. However, no members of the Thermoproteales from YNP have been isolated or sequenced, despite their ubiquity and importance in numerous high-temperature environments.
Pyrobaculum sp. strain WP30 (referred to here as Pyrobaculum yellowstonensis) was recently isolated (20) from the high-arsenic, sulfidic source pool (∼78 to 80°C, pH 6.1, ∼20 μM HS−/H2S, ∼135 μM As; see Fig. S1 in the supplemental material) of Joseph's Coat Hot Spring (JCHS) and was shown to grow in the presence of elemental sulfur using YE as a carbon and energy source. Similar Pyrobaculum spp. are abundant in habitats that contain copious amounts of elemental sulfur and/or dissolved sulfide over a pH range of 6 to 9 (20). Consequently, the objectives of this study were to (i) determine the specific physiological attributes of strain WP30 that relate to geochemical processes occurring in situ, (ii) measure different arsenic and sulfur species to understand the effects of strain WP30 on arsenic and sulfur cycling, and (iii) reconstruct the metabolic potential of this isolate from manual curation and annotation of the closed genome. The integrated results of microbiological, geochemical, and genomic studies provide a detailed understanding of the distribution and function of P. yellowstonensis strain WP30 populations in high-temperature environments of YNP.
MATERIALS AND METHODS
Culturing.
The synthetic base medium contained 6.2 mM NH4Cl, 2.4 mM KH2PO4, 1.6 mM MgCl·6H2O, 1 mg liter−1 resazurin sodium salt (Sigma-Aldrich Chemical Co., Milwaukee, WI, USA), 1 ml liter−1 vitamin solution (21), and 1 ml liter−1 trace element solution (22). The medium was autoclaved after the addition of the carbon source and the electron acceptor and pH adjustment with either HCl or NaOH (pH 6). The sterile medium was aliquoted into individual serum bottles, sealed with a septum, and made anoxic by 3 cycles of vacuuming (−15 mm Hg) and/or flushing of the headspace with filtered (pore size, 0.2 μm) >99.96% N2(g) for 30 min before the addition of 0 to 70 μM cysteine to scavenge the remaining O2. All growth experiments were performed at 75°C, the optimal growth temperature of this organism (20). Growth curves were obtained in triplicate 100-ml cultures in the presence of 0.02% YE (Difco) and either 1 mM arsenate (Na2HAsO4·7H2O; Sigma-Aldrich Chemical Co.) or excess elemental sulfur (10 g liter−1; Sigma-Aldrich). The inoculum, transferred at late-log-phase growth, was also filtered (pore size, 0.2 μm) into a fourth anaerobic 100-ml serum bottle to serve as a negative control. Direct cell counting and/or the concurrent measurement of HS−/H2S or arsenate was used to monitor growth. Cell counts were performed by incubating a sample with 2× SYBR gold (Molecular Probes, Eugene, OR, USA) in the dark for at least 30 min and then enumerating the cells either with a Petroff-Hausser counting chamber (1/50-mm cell depth, improved Neubauer, 1/400-mm square ruling pattern; Hausser Scientific, Horsham, PA, USA) or by filtering on 0.4-μm-pore-size black polycarbonate track-etched filters (GE Osmonics). A Zeiss Axioskop 2+ fluorescence microscope (Carl Zeiss, Inc.) was used to confirm the cell morphology and count the cells. Dissolved HS−/H2S was measured using a modified, low-volume version of the amine-sulfuric acid method (20, 23). Cells were enumerated directly in growth experiments in which the carbon source remained YE (0.02%) and the electron acceptors were varied and included 5 mM (each) K2SO4, Na2S2O3, or KNO3 or 1 mM (each) NaSeO4·10H2O, NaSbO3 (Acros Organics, Fair Lawn, NJ, USA), or KClO4 in 20- to 25-ml culture volumes. Sulfide production rates were calculated from HS−/H2S generation curves in 20-ml cultures grown in the presence of excess elemental sulfur (5 g liter−1) and various carbon sources, including 0.02% (each) Casamino Acids (Thermo Fisher Scientific Inc., Waltham, MA, USA), tryptone, peptone, and tryptic soy broth (Sigma-Aldrich Chemical Co.). Sulfide production was not observed in the presence of elemental sulfur or 0.02% glucose, sucrose, d-ribose, fructose, d-lactose, cellobiose, starch, acetate, or citric acid. WP30 was unable to ferment 0.02% YE. Other electron acceptors, including SO42−, S2O32−, NO3−, SeO42−, SbO32−, and ClO4−, did not support the growth of strain WP30 in the presence of 0.02% YE. Attempts to grow WP30 autotrophically were not successful and were performed in 100-ml cultures in the presence of 5 mM bicarbonate plus various electron donors and acceptors, including elemental sulfur with 5% H2 (95% N2) in the headspace, elemental sulfur and 1 mM arsenate with 5% H2 in the headspace, or arsenate and HS−/H2S (1 mM each) with 100% in the N2 headspace.
Polysulfide and thioarsenate determination.
The polysulfide and arsenic-sulfur species in 50-ml cultures of strains grown in the presence of 5 mM arsenate and/or excess elemental sulfur (10 g liter−1) and controls containing filtered (pore size, 0.2 μm) inoculum were measured. Samples for arsenic speciation analysis were filtered (pore size, 0.2 μm), flash frozen [with CO2(s)], and stored at −20°C until analysis, while polysulfide samples were filtered (pore size, 0.2 μm) and immediately derivatized. Derivatization of polysulfides to form dimethylpolysulfanes was performed by adding 100 μl sample and 100 μl methyl trifluoromethanesulfonate (CF3SO3CH3; Sigma-Aldrich Chemical Co.) simultaneously to a mixture of methanol (800 μl) and 100 μl 50 mM phosphate buffer (in which the pH was adjusted to match the pH of the sample) (24). Derivatized samples were analyzed on a Merck Hitachi high-pressure liquid chromatograph (HPLC) using a reversed-phase C18 column (Waters-Spherisorb; ODS2; particle size, 5 μm; 250 by 4.6 mm) and a methanol-water eluent gradient at a flow rate of 1 ml min−1 (25). Detection was performed with an L-2420 UV-visible detector at a 230-nm wavelength. A commercially available standard (C2H6S3; Acros Organics) and fractions of a synthesized dimethylpolysulfane mixture were used for quantification (25). The abiotic monothioarsenate control (Na3AsO3S·7H2O; formula weight, 350) was synthesized as previously described (26, 27), and the elemental sulfur (10 g liter−1) and 5 mM arsenite (NaAsIIIO2; Sigma-Aldrich Chemical Co.) abiotic control was prepared with base medium and incubated at 75°C for 4 days before derivatization. Samples for arsenic speciation analysis were thawed in an anaerobic hood (95% N2, 5% H2 atmosphere) to minimize oxidation. Arsenite, arsenate, and thioarsenates were detected by anion-exchange chromatography (Dionex ICS-3000 SP, AG16/AS16 IonPac column) coupled to inductively coupled plasma mass spectrometry (XSeries2; Thermo Fisher) (AEC-ICP-MS) (28, 29).
Electron microscopy.
Cells of P. yellowstonensis were fixed in 1.5% glutaraldehyde, filtered onto 0.4-μm-pore-size filters, and then rinsed with sterile water before imaging on a Zeiss Supra 55VP field emission scanning electron microscope in low-voltage mode (Imaging and Chemical Analysis Laboratory, Montana State University, Bozeman, MT, USA). Five microliters of cell suspension was applied to 100-mesh Cu grids covered with Formvar support film sputtered with carbon (Electron Microscopy Sciences, Hatfield, PA). The cells were allowed to adhere to the grids for 1 min before the grids were blotted with a filter paper and negatively stained with a 5-μl drop of Nano-W stain (Nanoprobes, Yaphank, NY). After 30 s, the excess liquid was removed by wicking, and the sample was allowed to air dry. Samples were examined with a Tecnai T-12 transmission electron microscope (TEM) at 120 kV with a LaB6 filament. The high-resolution imaging was done at the Titan scanning/transmission electron microscope (S/TEM; 300 kV; FEI). Images were collected digitally with an Ultrascan 1000 charge-coupled device (Gatan). DigitalMicrograph software was used for imaging and image analyses of cellular features.
Genome sequencing, assembly, and annotation.
Subsamples of approximately 8.9 × 109 total cells were harvested from two 600-ml WP30 cultures grown to 4.5 × 107 cells ml−1 in the presence of elemental sulfur (3.33 g liter−1) and As(V) (5 mM arsenate). Cells were pelleted by centrifugation (16,000 × g for 30 min at 4°C), flash frozen in liquid N2, and stored at −80°C until extraction. DNA was extracted with an MP FastDNA spin kit for soil (QBioGene, Solon, OH, USA), according to the manufacturer's directions, yielding ∼50 μg of long DNA fragments (>15 kb). Approximately 3 μg of DNA was sufficient to generate two complete runs on a 454 GS junior titanium sequencer at Montana State University (454 Life Sciences, Branford, CT, USA). Emulsion PCR and rapid library preparation were performed according to the manufacturer's protocols. The coassembly of the two sequencing runs with a 95% nucleotide sequence identity overlap (Newbler Assembler software, v.2.5p1) resulted in 1.993 Mb on 5 contigs at 47× coverage (see Table S1 in the supplemental material). Gap sequences were verified by Sanger sequencing (Plant-Microbe Genomic Facility, Ohio State University) of the PCR amplicons generated with primer pairs designed on the flanking contigs (see Table S2 in the supplemental material). Automated annotation pipelines, including the Integrated Microbial Genomes Expert Review (IMG-ER) system (30, 31) and the Rapid Annotations using Subsystems Technology (RAST) server (32), were initially used to identify and annotate open reading frames, followed by the manual curation of genes (e.g., frameshifts, rRNA introns) and metabolic pathways (see Table S3 in the supplemental material). tRNAs were identified with the tRNAscan-SE server (v.1.3.1) (33, 34).
Phylogenetic analysis and comparative genomics.
All sequence alignments (16S rRNA genes, 58 single-copy proteins, DMSO-molybdopterin catalytic proteins) were generated using MUSCLE (35) and/or ClustalW software, manually edited, and then used to construct phylogenetic trees by the maximum likelihood and/or neighbor-joining method (bootstrap values were determined with either 100 or 1,000 resamplings) in MEGA (v.5.0) software (36). The 58 single-copy proteins (see Table S4 in the supplemental material) shared across the Archaea were concatenated prior to phylogenetic analysis (4, 37).
Orthologous proteins from the genomes of P. yellowstonensis strain WP30, P. arsenaticum DSM 13514, P. islandicum DSM 4184, P. calidifontis JCM 11548, P. aerophilum strain IM2, P. neutrophilum V24Sta, P. oguniense TE7, and Pyrobaculum sp. strain 1860 were identified by use of the InParanoid algorithm (release 7) (38). The genome sequence of strain WP30 was compared to other Pyrobaculum genome sequences by use of NCBI-BLAST algorithms, IMG-ER tools, the Artemis comparison tool (ACT) (39), and the MAUVE (v.2) program (40, 41).
Nucleotide sequence accession number.
The NCBI GenBank accession number for the complete P. yellowstonensis strain WP30 genome is available under registered BioProject record number PRJNA258558. The IMG-ER project identification number (ID) for this genome is 8081.
RESULTS
Growth characteristics.
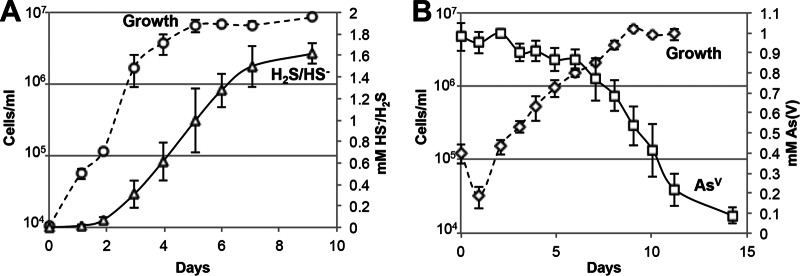
The maximum growth rate of P. yellowstonensis (strain WP30) in the presence of elemental sulfur and yeast extract (YE) was 0.14 ± 0.04 generations h−1 (doubling time, 7.3 ± 2.1 h) and corresponded to a sulfide production rate of 14.1 ± 5.2 μM h−1 (Fig. 1A). Variable rates of sulfide production were observed when P. yellowstonensis strain WP30 was grown in the presence of elemental sulfur and different carbon and energy sources, including Casamino Acids (20.9 ± 11.6 μM h−1), tryptone (20.2 ± 3.1 μM h−1), peptone (16.6 ± 0.9 μM h−1), and tryptic soy broth (6.5 ± 1.2 μM h−1), and these resulted in final sulfide concentrations ranging from 1.4 to 1.6 mM. Growth in the presence of YE and 1 mM arsenate was 3.5 times slower than that in the presence of elemental sulfur, resulting in a rate of 0.04 ± 0.02 generations h−1 (doubling time, 27.3 ± 15.5 h) and a maximum arsenate reduction rate of 7.1 ± 2.0 μM h−1 (Fig. 1B). Growth was 1.5 times faster in the presence of YE, elemental sulfur, and 5 mM arsenate (0.21 ± 0.04 generations h−1; doubling time, 4.8 ± 0.8 h) than in the presence of elemental sulfur and resulted in an estimated arsenate reduction rate of ∼44 μM h−1 (Fig. 2).
FIG 1.
Growth of Pyrobaculum yellowstonensis strain WP30 (75°C) in the presence of yeast extract and elemental sulfur (A) or arsenate (B).
FIG 2.
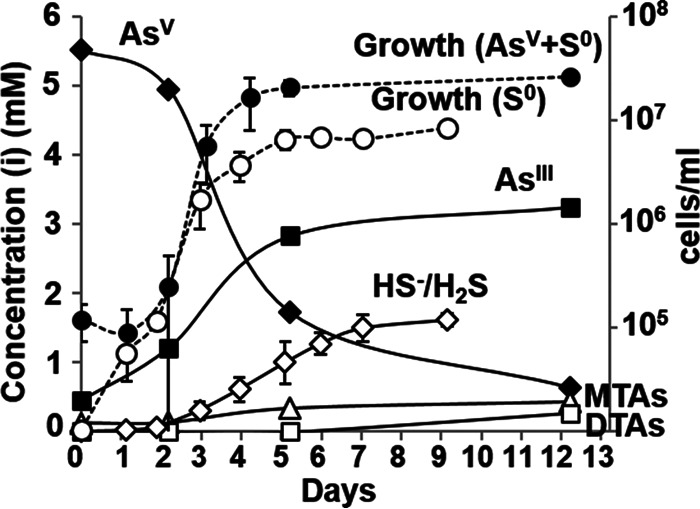
Concentrations (in millimolar) of arsenite (■), arsenate (⬥), monothioarsenate (Δ), dithioarsenate (□), and sulfide (◇) during growth (number of cells per milliliter) of Pyrobaculum yellowstonensis strain WP30 on elemental sulfur (open circles) or elemental sulfur and arsenate (closed circles). MTAs, total monothioarsenate; DTAs, total dithioarsenate.
The production of sulfide and arsenite during growth in the presence of elemental sulfur and arsenate resulted in significant changes in the distribution of sulfur and arsenic species (Fig. 2). Arsenite was the dominant arsenic species (∼70%) detected after cells entered stationary phase. Monothioarsenate concentrations increased ∼2.5-fold during log-phase growth and reached a final concentration (at 12 days) of 0.44 mM. Conversely, dithioarsenate was detected (0.27 mM) only at 12 days. In sterile controls, arsenate remained the primary As species; the concentrations of mono- and dithioarsenate were below the detection limit (data not shown). Similar distributions of polysulfide species were detected in strain WP30 cultures grown in the presence of elemental sulfur and 5 mM arsenate, in anoxic sterile controls of elemental sulfur and 5 mM arsenite, and in anoxic 1 mM monothioarsenate solutions (see Table S5 in the supplemental material). Polysulfides were not detected either in sterile elemental sulfur and arsenate controls or in strain WP30 cultures grown exclusively in the presence of elemental sulfur. These results show that the formation of polysulfide species is dependent on the presence of arsenite reacting with elemental sulfur as well as the concentration of monothioarsenate (42, 43).
Cell morphology.
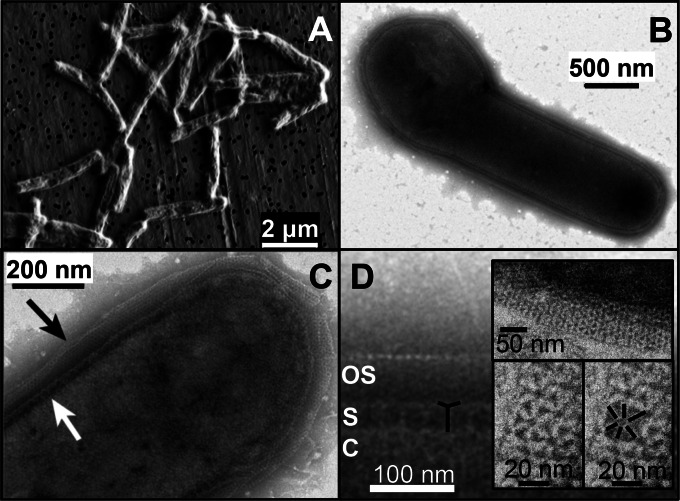
P. yellowstonensis strain WP30 is rod shaped (2 to 15 μm by 0.5 μm), although diploid pairs and club-shaped and budding cells were observed (Fig. 3A and B). Transmission electron micrographs (TEMs) (Fig. 3B to D) revealed a distinct S layer and an external sheath comprised of individual subunits. The S layer of P. yellowstonensis consists of regularly spaced (p6 symmetry) cytoplasmic membrane-anchored pillars, which support radially extended filiform structures (Fig. 3C and D), and is very similar to the S layers characterized in Thermoproteus tenax, P. islandicum, and Pyrobaculum organotrophum (44, 45). The pillar length averaged 40 ± 10 nm and was spaced from adjacent pillars by 32 ± 6 nm. The outer sheath (Fig. 3B to D) consists of a single layer of regularly spaced (13 ± 2 nm) small subunits (6 to 7 nm). This sheath forms an ordered crystalline fabric that envelops the cell at 52 ± 11 nm from the S layer, although this distance can decrease significantly toward the poles of the cells (Fig. 3B). A similar outer sheath was previously identified in P. organotrophum (44). Other surface appendages, including pili and flagella, were not observed in strain WP30, which is consistent with the absence of motility genes.
FIG 3.
Cell morphology of Pyrobaculum yellowstonensis strain WP30. (A to D) Scanning electron micrograph (SEM) of strain WP30 grown in the presence of elemental sulfur and arsenate (A) and transmission electron micrographs (TEMs) of strain WP30 grown in the presence of elemental sulfur (B to D). Black and white arrows in panel C correspond to the outer sheath and primary S layer, respectively. In panel D, the outer sheath (OS), S layer (S), and cytoplasm (C) are labeled, while the basic subunit of the S layer is emphasized in black. (Insets) Structure (p6 symmetry) of the primary S layer at high resolution.
P. yellowstonensis strain WP30 genome.
The closed genome (1,993,257 bp; G+C content, 58.3%) of P. yellowstonensis (strain WP30) contains a single set of rRNA genes, 8 clustered regularly interspaced short palindromic repeat (CRISPR)/Cas regions, and ∼2,270 protein-coding genes (Table 1). All 20 tRNA synthetase genes and only 28 tRNA genes were identified (see Table S3 in the supplemental material). The genes for tRNAs for histidine, asparagine, and the aromatic amino acids (tryptophan, tyrosine, phenylalanine) were not identified; however, the anticodons of 4 tRNA genes were undetermined (see Table S6 in the supplemental material). Introns were detected in 10 tRNA genes, which is a universal attribute of tRNA genes in the genus Pyrobaculum (46).
TABLE 1.
Genome properties of Pyrobaculum yellowstonensis strain WP30 compared to other Pyrobaculum spp.a
| Genome | Size (Mb) | G+C content (%) | ANI (%) | No. of genes | No. of CDSs | No. of tRNAs | No. of CRISPRs |
|---|---|---|---|---|---|---|---|
| P. yellowstonensis WP30 | 1.99b | 58.3 | 100 | 2,304 | 2,269 | 28 | 8 |
| P. neutrophilum | 1.77 | 59.9 | 73.5 | 2,053 | 2,006 | 44 | 10 |
| P. calidifontis | 2.01 | 57.2 | 71.3 | 2,213 | 2,184 | 24 | 5 |
| Pyrobaculum sp. strain 1860 | 2.47 | 57.0 | 72.8 | 2,888 | 2,824 | 31 | 8 |
| P. arsenaticum | 2.12 | 55.1 | 71.3 | 2,410 | 2,363 | 43 | 6 |
| P. oguniensec | 2.44 | 55.1 | 71.2 | 3,014 | 2,869 | 48 | 5 |
| P. aerophilum | 2.22 | 51.4 | 70.6 | 2,625 | 2,575 | 47 | 5 |
| P. islandicum | 1.83 | 49.6 | 70.5 | 2,063 | 2,014 | 45 | 6 |
ANI, average nucleotide sequence identity (by BLAST analysis) to the nucleotide sequence of strain WP30; CDSs, coding sequences; CRISPRs, clustered regularly interspaced short palindromic repeats.
Specifically, 1,993,257 bp.
Excludes an extrachromosomal element (0.02 Mb, 50.6% G+C content, 35 coding sequences).
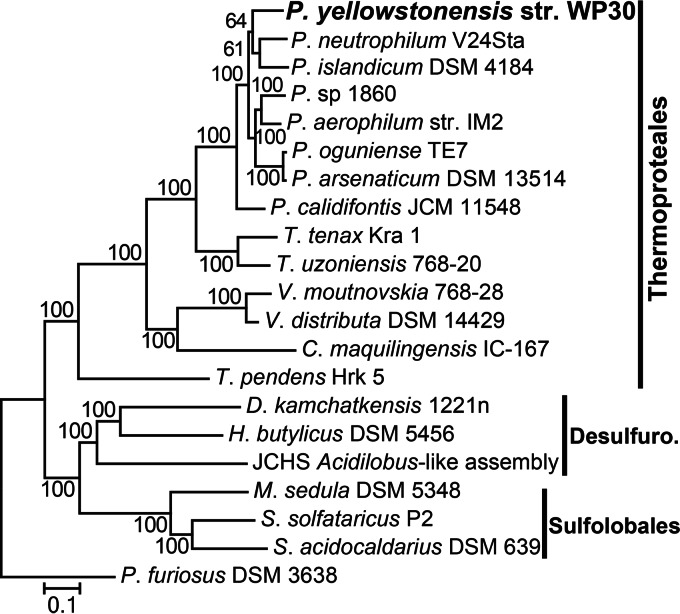
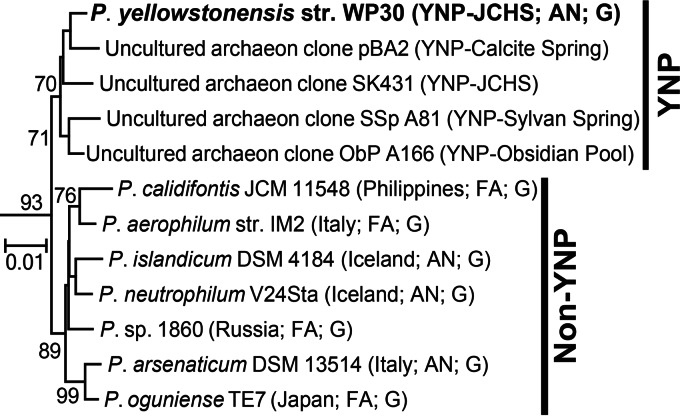
Phylogenetic analysis of 58 aligned and concatenated single-copy proteins shared in Archaea revealed that P. yellowstonensis (strain WP30) is most closely related to the strictly anaerobic isolates P. islandicum and P. neutrophilum V24Sta, both of which were obtained from Iceland (Fig. 4) (8, 47). Phylogenetic analysis using the 16S rRNA gene also confirmed the placement of strain WP30 in the genus Pyrobaculum (order Thermoproteales). The 16S rRNA gene of this isolate is highly related (>99% nucleotide sequence identity) to 16S rRNA gene sequences observed in numerous hot springs in YNP which form a group distinct from the 16S rRNA gene sequences of Pyrobaculum populations that are not from YNP (Fig. 5). Analysis of genome relatedness indices confirmed that strain WP30 is a new Pyrobaculum species (48). Specifically, average nucleotide sequence identities (ANIs) to the nucleotide sequence of other Pyrobaculum genomes ranged from 70.5 to 73.5% (ANIblast; Table 1) and 82.4 to 88.6% (ANImummer) (species demarcation was classified at 95 to 96% identity) (49). Values from genome BLAST distance phylogeny (GBDP) analysis ranged from 0.23 to 0.26 for identity to other Pyrobaculum genomes with a threshold for species demarcation of 0.26 (50). Moreover, the average identity of the nucleotide sequences of 40 marker genes to those of the marker genes of P. neutrophilum was 78.9%, which is considerably lower than the 96.5% species threshold (51).
FIG 4.
Phylogenetic analysis of Pyrobaculum yellowstonensis strain WP30. Phylogenetic analysis of 58 concatenated, single-copy proteins identified in Archaea. The tree was constructed using maximum likelihood analysis; bootstrap values were determined by resampling 100 replicate trees. The scale bar represents the number of substitutions per 100 positions. T. uzoniensis, Thermoproteus uzoniensis; T. pendens, Thermofilum pendens. Other genus names are abbreviated as follows: D., Desulfurella; H., Hyperthermus; M., Metallosphaera; S., Sulfolobus; and P., Pyrococcus. Desulfuro., Desulfurococcales.
FIG 5.
Phylogenetic tree (neighbor joining) of Pyrobaculum 16S rRNA genes. Bootstrap values (values of ≥70% are shown) were determined by resampling 1,000 replicate trees. The isolation location and other characteristics (FA, facultative anaerobe; AN, anaerobe; G, publically available genome) are indicated. Bootstrap values were determined by resampling 100 replicate trees. The scale bar represents the number of substitutions per 100 positions.
rRNA gene introns.
Several intron sequences were identified within the 23S and 16S rRNA genes of P. yellowstonensis (Table 2). The presence of rRNA gene introns is a distinguishing characteristic of Pyrobaculum spp. and, more broadly, the entire order Thermoproteales (52, 53, 53–56). Two of the three 16S rRNA gene introns (PyWP30.S919 and PyWP30.S1391) (57) encode a homing endonuclease that contains at least one LAGL-IDADG DNA binding motif. The LAGL-IDADG family of homing endonucleases (Pfam14528) is divided into two groups that possess either one or two copies of this motif (58). Intron PyWP30.S1093 is short (24 bp) and forms a transcribed hairpin structure (data not shown).
TABLE 2.
Intron sequences identified in the 16S and 23S rRNA genes of Pyrobaculum yellowstonensis strain WP30
| Introna | Coordinates |
HP or CDSb | Sizec |
No. of LAGLIDADG motif copiesd | ||
|---|---|---|---|---|---|---|
| Gene | Genome | nt | aa | |||
| PyWP30.S916 | 886–1633 | 723533–724280 | CDS | 748 | 234 | 2 |
| PyWP30.S1090 | 1798–1821 | 724448–724471 | HP | 24 | ||
| PyWP30.S1389 | 2128–2790 | 724778–725440 | CDS | 663 | 208 | 1 |
| PyWP30.L1914 | 2028–2690 | 727669–728331 | CDS | 663 | 192 | 1 |
| PyWP30.L1953 | 2730–3343 | 728371–728984 | CDS | 614 | 126 | 1 |
The nomenclature was described previously (57); e.g., S916 is small subunit 16S rRNA position 916 (Escherichia coli numbering), and L1914 is large subunit 23S rRNA position 1914 (E. coli numbering).
HP, hairpin forming; CDS, coding sequence.
nt, number of nucleotides; aa, number of amino acids.
Pfam14528 domains.
Two of the introns in strain WP30 (PyWP30.S919 and PyWP30.S1391) are located in highly conserved regions of the 16S rRNA gene that impact (in silico) the annealing of universal primers Ab906F, U926R, Ab927R, and Ab934R or U1406R (see Fig. S2 in the supplemental material). Two 23S rRNA homing endonuclease-encoding introns (PyWP30.L1914 [663 nucleotides {nt}] and PyWP30.L1953 [614 nt]) were also identified in strain WP30 and were separated by 41 nt (Table 2). Both deduced protein sequences contained one LAGL-IDADG motif, but they showed no similarity to each other and <55% amino acid identity to other homing endonuclease sequences. Predicted secondary structures of all five transcribed introns (59) include a bulge-helix-bulge motif at the insertion locus, which is universally recognized by the tRNA splicing endoribonuclease responsible for intron excision during the formation of a functional RNA (60–63).
Metabolic reconstruction.
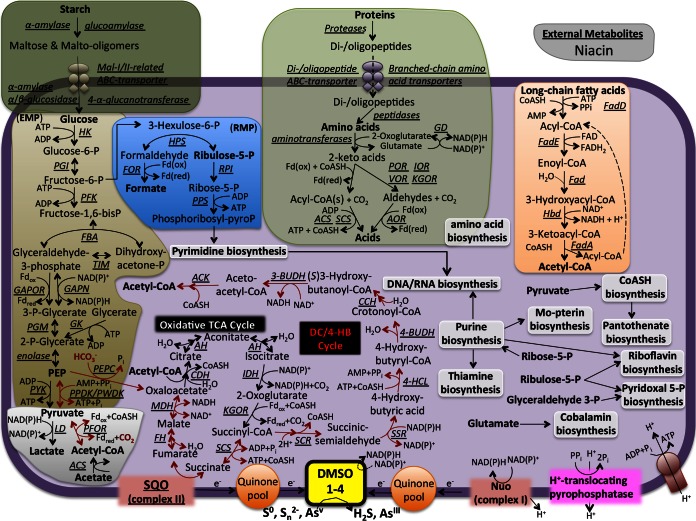
The biochemical pathways identified in the genome of P. yellowstonensis are consistent with the observed growth phenotype of this organism. Specifically, this organism is a chemoorganoheterotroph that derives cell biomass and reducing power from the oxidation of organic compounds (e.g., carbohydrates, proteins, and lipids) coupled with the reduction of elemental sulfur and/or arsenate (Fig. 6; see also Table S3 in the supplemental material). A complete maltose ABC transporter and a putative glucose/arabinose transporter were identified in P. yellowstonensis (glcS [PyWP30_02245], glcT [PyWP30_02244], and glcU [PyWP30_02243], similar to genes identified in Sulfolobus solfataricus [64]); however; the glcV gene (ATP binding subunit) is not present in P. yellowstonensis (similar to the findings for several other Pyrobaculum genomes). Acetyl-CoA generated from glycolysis (the Embden-Meyerhof-Parnas pathway), the catabolism of amino acids, and/or the β-oxidation of lipids can be oxidized in the TCA cycle. WP30 contains two sugar kinases that are unrelated to the pyrophosphate-dependent phosphofructokinase (PFK) characterized in Thermoproteus tenax (65). Instead, one of these WP30 genes (PyWP30_00078) is 37% identical (amino acid sequence identity) to the ATP-dependent PFKB characterized in Aeropyrum pernix (66) and may function similarly.
FIG 6.
Genome sequence-derived metabolic reconstruction of Pyrobaculum yellowstonensis strain WP30. HK, ATP-dependent hexokinase; PGI, phosphoglucose isomerase; PFK, ATP-dependent phosphofructokinase; FBA, fructose-bisphosphate aldolase/phosphatase; TIM, triosephosphate isomerase; GAPOR, glyceraldehyde-3-phosphate ferredoxin oxidoreductase; GAPN, nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase [phosphoglycerate kinase (PGK) and NAD(P)-dependent phosphorylating GAP dehydrogenase are also present but are not shown]; PGM, phosphoglycerate mutase; GK, glycerate kinase; PYK, pyruvate kinase; PPDK, pyruvate, phosphate dikinase; PWDK, pyruvate-water dikinase; PEPC, phosphoenolpyruvate carboxylase; Fdox, oxidized ferredoxin; Fdred, reduced ferredoxin; LD, lactate dehydrogenase; PFOR, pyruvate:ferredoxin oxidoreductase; ACS, AMP-forming acetyl-CoA synthetase; CDH, citrate synthase; AH, aconitase; IDH, NADP+-dependent isocitrate dehydrogenase; KGOR, 2-oxoglutarate:ferredoxin oxidoreductase; SCS, succinyl-CoA synthetase; SQO, succinate:quinone oxidoreductase; FH, fumarase (class 1); MDH, malic dehydrogenase enzyme; SCR, succinyl-CoA reductase (NADPH); SSR, succinic semialdehyde reductase (NADPH); 4-HCl, 4-hydroxybutyrate-CoA ligase (AMP forming); 4-BUDH, 4-hydroxybutyryl-CoA dehydratase; CCH, crotonyl-CoA hydratase; 3-BUDH, (S)-3-hydroxybutyryl-CoA dehydrogenase (NAD+); ACK, acetoacetyl-CoA β-ketothiolase; HPS, 3-hexulose-6-phosphate synthase; FOR, formaldehyde:ferredoxin oxidoreductase; RPI, ribose-5-phosphate isomerase; PPS, phosphoribosyl pyrophosphate synthase; CoASH, coenzyme A; GD, glutamate dehydrogenase; POR, pyruvate:ferredoxin oxidoreductase; IOR, indolepyruvate:ferredoxin oxidoreductase; VOR, 2-ketoisovalerate:ferredoxin oxidoreductase; KGOR, 2-oxoglutarate:ferredoxin oxidoreductase; AOR, aldehyde:ferredoxin oxidoreductase; FadD, acyl-CoA ligase (acyl-CoA synthetase); FadE, acyl-CoA dehydrogenase; Fad, enoyl-CoA hydratase; Hbd, 3-hydroxyacyl-CoA dehydrogenase; FadA, 3-ketoacyl-CoA thiolase; RMP, ribulose monophosphate pathway.
All genes implicated in the dicarboxylate/4-hydroxybutyrate (DC/4-HB) pathway for the fixation of carbon dioxide were also identified in strain WP30. The DC/4-HB cycle has been shown to fix CO2 in facultative chemolithoautotrophic Pyrobaculum spp. that use H2 as an electron donor (see Table S2 in the supplemental material) (14–17, 67). Both phosphoenolpyruvate carboxylase (PEPC) and pyruvate:ferrodoxin oxidoreductase (PFOR), which are responsible for CO2 fixation in the DC/4-HB pathway, were identified. However, repeated attempts to grow strain WP30 exclusively on CO2 (HCO3−) were unsuccessful. No hydrogenases (Ni-Fe, Fe-Fe, nonmetal) were identified in strain WP30, and H2 did not support growth. Facultative lithoautotrophic Pyrobaculum spp. contain homologs of the type I Ni-Fe hydrogenases, although the molecular mechanism of H2 oxidation in the order Thermoproteales has not been elucidated (14, 68). The presence of complete (or nearly complete) pathways for the de novo biosynthesis of numerous cofactors and metabolites (including purines; pyrimidines; vitamins B1, B2, B5, B6, and B12; and the molybdopterin cofactor) suggests that strain WP30 is prototrophic with respect to these metabolites. However, genes for the biosynthesis of niacin (vitamin B3; required by NAD+/NADH) were not identified, which suggests that this organism requires exogenous sources of this common cofactor. The biosynthesis pathways for phenylalanine and proline were incomplete. The pathways for tyrosine, lysine, methionine, serine, and histidine were essentially complete, although 1 to 2 genes may not have been annotated. The pathways for the remaining 13 amino acids were all complete.
P. yellowstonensis strain WP30 lacks heme-Cu oxidases (subunit 1) and all other components of characterized aerobic terminal oxidase complexes (69, 70). The absence of genes necessary for aerobic respiration is consistent with the observed growth on other electron acceptors (elemental sulfur and/or arsenate). However, this organism contains multiple copies of cydA, which is similar to the gene for subunit A of cytochrome bd ubiquinol oxidases (71, 72). Unlike the bacterial CydAB complex, the gene architecture in strain WP30 (and other archaea) consists of two copies of cydAA′ genes: subunit A of the encoded peptide is similar to that in CydAB; however, subunit A′ is a smaller, single-heme-containing protein. The deduced proteins, CydA1′ and CydA2′, are not homologous to CydB, which suggests that these complexes do not perform the same oxygen reduction reaction as CydAB. Although the functions of CydA1A1′ and CydA2A2′ are unknown at this time, numerous members of the Crenarchaeota, including native Acidilobus-like populations (4) and the majority of Pyrobaculum spp., contain homologous genes. Aerobic Pyrobaculum spp. do not contain cyd genes but have heme-Cu oxidase complexes (10–12). Genes involved in the reduction of oxygen radicals, including superoxide dismutase, peroxiredoxin, and thioredoxin, were identified in P. yellowstonensis strain WP30 (see Table S3 in the supplemental material), indicating tolerance toward reactive oxygen species.
Four novel dimethyl sulfoxide (DMSO)-molybdopterin (MPT) family proteins were identified in strain WP30 (Fig. 7). Phylogenetic analysis of the deduced DMSO-MPT proteins (catalytic subunit) showed that these entries form distinct clades, which are closely related to characterized sulfur reductases (e.g., SreA or PsrA; clades 1 and 2), tetrathionate or arsenate reductases (e.g., TtrA or ArrA; clade 3), or DmsA/BisC/TorA proteins (clade 4). Clades 1, 2, and 3 are defined almost exclusively by proteins contributed by Pyrobaculum spp. Clade 3 Pyrobaculum proteins are closely related to arsenate reductases (ArrA) and/or tetrathionate reductases (TtrA), which have been studied in other archaea and bacteria (73, 74). Clade 4 DMSO-MPT proteins are found across the domain Archaea (Fig. 6), and although they remain uncharacterized, they are distantly related to other bacterial entries responsible for DMSO (DmsA), trimethylamine N-oxide (TorA), or biotin sulfoxide (BisC) reduction. Other genes encoding complete electron transport pathways were also identified, including an NADH:quinone oxidoreductase (Nuo; complex I), a succinate:quinone oxidoreductase (SQO; complex II), and a complete V-type ATP synthase (Fig. 5; see also Table S3 in the supplemental material).
FIG 7.
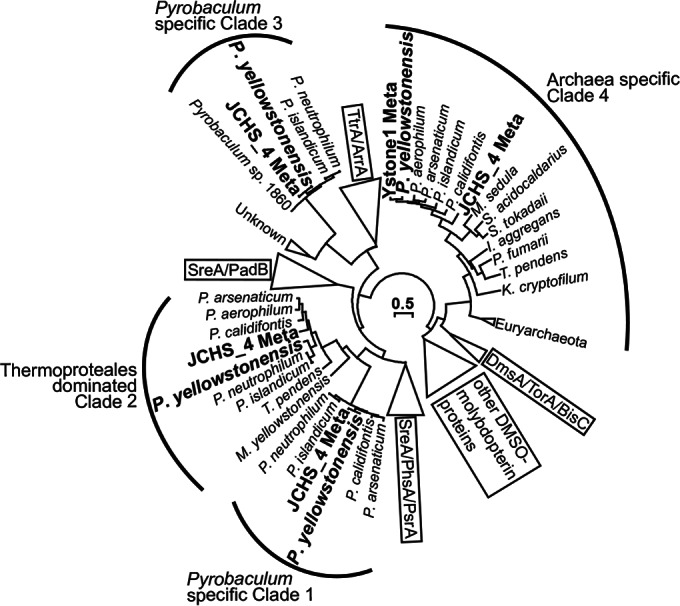
Phylogenetic tree of the DMSO-molybdopterin family of proteins. The four protein sequences identified in the Pyrobaculum yellowstonensis strain WP30 genome and sequences identified in two Joseph's Coat Hot Spring metagenome data sets (Ystone1 and JCHS_4) are shown in bold. The unrooted tree was constructed using maximum likelihood analysis, and bootstrap values were calculated by 100 resamplings. ArrA, arsenate reductase; BisC, biotin sulfoxide reductase; DmsA, dimethyl sulfoxide reductase; PadB, phenylacetyl-CoA dehydratase; PsrA/PhsA, polysulfide/polythionate reductase; SreA, elemental sulfur reductase; TorA, trimethylamine N-oxide reductase; TtrA, tetrathionate reductase; S. tokodaii, Sulfolobus tokodaii; I. aggregans, Ignisphaera aggregans; P. fumarii, Pyrolobus fumarii; K. cryptofilum, Korarchaeum cryptofilum; M. yellowstonensis; Metallosphaera yellowstonensis.
Importance of P. yellowstonensis strain WP30 in situ.
The comparative genomics of strain WP30 and two metagenomes from Joseph's Coat Hot Springs, YNP (Ystone1 and JCHS_4 [1, 2]) showed that P. yellowstonensis-like sequences are abundant in circumneutral (pH ∼6) sulfur sediments and represented ∼15 to 75% of the microbial community in two independent samples. Pyrobaculum-like sequence assemblies from JCHS (2) were compared to the complete genome of strain WP30 as well as a closed genome from P. neutrophilum (formerly Thermoproteus neutrophilus [75]). Strain WP30 recruited more metagenome sequences (n = 1,209) at a higher amino acid sequence identity (≥70%) than all other available Pyrobaculum genomes (e.g., ∼1,141 for P. neutrophilum). In fact, genes highly related to the three novel DSMO-MPT proteins (clades 1 to 3; Fig. 7) of P. yellowstonensis were observed in the JCHS populations, a finding which also suggests that the deduced proteins are important for energy conservation in sulfur sediment environments.
DISCUSSION
Pyrobaculum yellowstonensis strain WP30 was isolated from a hypoxic, reduced sulfur, and high-arsenic environment in Yellowstone National Park (JCHS; see Fig. S1 and Table S7 in the supplemental material) and shares many characteristics with other Pyrobaculum spp., including the use of organic substrates as carbon and energy sources and the presence of introns in vital tRNA and rRNA genes. Strain WP30 requires components in yeast extract (or amino acids) as a carbon and energy source and elemental sulfur and/or arsenate as an electron acceptor. Unique attributes of strain WP30 include a lower temperature optimum (i.e., 75°C), a cell-enveloping sheath (Fig. 3B to D), and a propensity for sulfur and arsenate reduction. The complete genome contains genes responsible for the catabolism of amino acids and sugars and for respiratory complexes (i.e., dimethyl sulfoxide oxidoreductases) implicated in the reduction of elemental sulfur and arsenate.
Genome sequencing of strain WP30 revealed numerous rRNA and tRNA intron sequences, including three introns identified in highly conserved regions of the 16S rRNA gene. The presence of introns at these loci reduces the frequency of annealing of many universal primers that are often used in environmental 16S rRNA gene sequencing surveys. Population abundance estimates from these surveys should be interpreted with caution, specifically, when they are obtained from high-temperature habitats where Pyrobaculum-like organisms are likely to be found. Random environmental DNA sequencing (metagenomics) confirmed the presence of additional Pyrobaculum-like 16S rRNA intron sequences in JCHS, indicating that these mobile genetic elements are abundant and diverse in Pyrobaculum-like populations (59).
The novel DMSO-MPT oxidoreductases identified in strain WP30 and in JCHS de novo assemblies of the same population type are most closely related to elemental sulfur reductases, polysulfide reductases, arsenate reductases, and tetrathionate reductases. These DMSO-MPT complexes are the only terminal reductases available to strain WP30 and in situ populations of P. yellowstonensis and are thus critical to native Pyrobaculum populations for respiration on sulfur and arsenic species, particularly in JCHS, which contains elemental sulfur and high concentrations of arsenate (∼20 μM) and thiosulfate (∼900 μM) (20). Consequently, native Pyrobaculum populations in JCHS do not appear to require oxygen and contribute to the maintenance of anoxic conditions through the regeneration of arsenite and sulfide.
Strain WP30 synthesizes all of its own cofactors and metabolites (except niacin) and could provide a source of these constituents for other auxotrophic community members. For example, the in situ JCHS Acidilobus-like populations (Desulfurococcales) cannot synthesize numerous cofactors and metabolites, including cobalamin, purines, and certain amino acids (4), which could be obtained directly or indirectly from Pyrobaculum spp. The reduction of elemental sulfur and arsenate by strain WP30 results in higher concentrations of reduced species (e.g., sulfide, polysulfides, arsenite, and thioarsenates), which participate in other thermodynamically favorable reactions (Fig. 8). Sulfide and arsenite can serve as electron donors for other community members, including other archaea (e.g., Sulfolobales) and/or bacteria (e.g., Aquificales). Polysulfides and thioarsenates formed as secondary products of sulfur and arsenate respiration may act as electron donors or acceptors for other members of the microbial community, particularly in high-pH systems where these compounds are thermodynamically stable (28, 42, 43, 76–80). Thioarsenate concentrations of 6 μM (primarily as dithioarsenate) have been measured in JCHS (20), and the formation of these species was shown to be promoted during sulfur and arsenate respiration by P. yellowstonensis under culture conditions. The chemistry and cycling of thioarsenates are extremely complex, as has been shown in prior studies, and a potential linkage with microbial processes was identified in this study. Pyrobaculum yellowstonensis strain WP30 is well-suited to hypoxic habitats (pH 6 to 9) containing elemental S, sulfide, and thiosulfate and is capable of utilizing the high levels of arsenate often observed in the geothermal systems of YNP.
FIG 8.
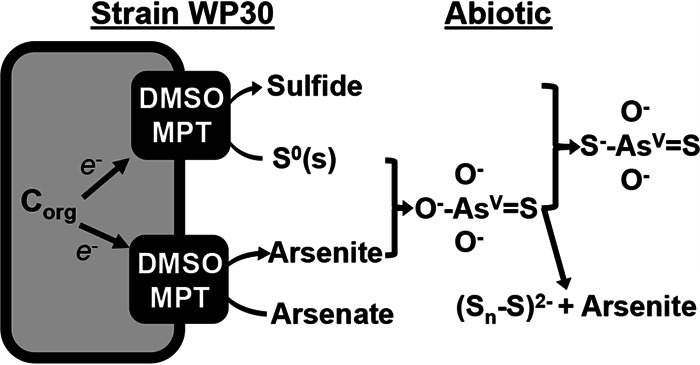
Proposed model of elemental sulfur and arsenate reduction by P. yellowstonensis strain WP30, which results in the formation of abiotic monothioarsenate, dithioarsenate, and polysulfide. Corg, organic carbon.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate support from the Pacific Northwest National Laboratory Foundational Science Focus Area (subcontract no. 112443), the U.S. Department of Energy (DOE)-Joint Genome Institute Community Sequencing Program (CSP 787081), and the NSF-IGERT (0654336). The work conducted by the Joint Genome Institute (DOE-AC02-05CH11231) and the Environmental Molecular Sciences Laboratory (EMSL) at the Pacific Northwest National Laboratory (Foundational Scientific Focus Area) is supported by the Genomic Science Program, Office of Biological and Environmental Research, DOE.
We thank C. Hendrix, S. Gunther, and D. Hallac (Center for Resources, YNP) for permitting this work in YNP (permits YELL-SCI-5068 and -5686) and Libor Kovarik for his expertise in the operation of the Titan S/TEM.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01095-15.
REFERENCES
- 1.Inskeep WP, Rusch DB, Jay ZJ, Herrgard MJ, Kozubal MA, Richardson TH, Macur RE, Hamamura N, Jennings RDM, Fouke BW, Reysenbach A-L, Roberto F, Young M, Schwartz A, Boyd ES, Badger JH, Mathur EJ, Ortmann AC, Bateson M, Geesey G, Frazier M. 2010. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One 5:e9773. doi: 10.1371/journal.pone.0009773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inskeep WP, Jay ZJ, Herrgard MJ, Kozubal MA, Rusch DB, Tringe SG, Boyd ES, Spear JR, Roberto FF. 2013. Phylogenetic and functional analysis of metagenome sequence from high-temperature archaeal habitats demonstrate linkages between metabolic potential and geochemistry. Front Microbiol 4:95–116. doi: 10.3389/fmicb.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer-Dombard DR, Swingley W, Raymond J, Havig J, Shock EL, Summons RE. 2011. Hydrothermal ecotones and streamer biofilm communities in the Lower Geyser Basin, Yellowstone National Park. Environ Microbiol 13:2216–2231. doi: 10.1111/j.1462-2920.2011.02476.x. [DOI] [PubMed] [Google Scholar]
- 4.Jay ZJ, Rusch DB, Tringe SG, Bailey C, Jennings RM, Inskeep WP. 2014. Predominant Acidilobus-like populations from geothermal environments in Yellowstone National Park exhibit similar metabolic potential in different hypoxic microbial communities. Appl Environ Microbiol 80:294–305. doi: 10.1128/AEM.02860-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozubal MA, Macur RE, Jay ZJ, Beam JP, Malfatti SA, Tringe SG, Kocar BD, Borch T, Inskeep WP. 2012. Microbial iron cycling in acidic geothermal springs of Yellowstone National Park: integrating molecular surveys, geochemical processes, and isolation of novel Fe-active microorganisms. Front Microbiol 3:109. doi: 10.3389/fmicb.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jennings RM, Whitmore LM, Moran JJ, Kreuzer HW, Inskeep WP. 2014. Carbon dioxide fixation by Metallosphaera yellowstonensis and acidothermophilic iron-oxidizing microbial communities from Yellowstone National Park. Appl Environ Microbiol 80:2665–2671. doi: 10.1128/AEM.03416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huber H, Huber R, Stetter KO. 2006. Thermoproteales, p 10–22. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes. Springer, New York, NY. [Google Scholar]
- 8.Huber R, Kristjansson JK, Stetter KO. 1987. Pyrobaculum gen. nov., a new genus of neutrophilic, rod-shaped archaebacteria from continental solfataras growing optimally at 100°C. Arch Microbiol 149:95–101. doi: 10.1007/BF00425072. [DOI] [Google Scholar]
- 9.Feinberg LF, Srikanth R, Vachet RW, Holden JF. 2008. Constraints on anaerobic respiration in the hyperthermophilic archaea Pyrobaculum islandicum and Pyrobaculum aerophilum. Appl Environ Microbiol 74:396–402. doi: 10.1128/AEM.02033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cozen AE, Weirauch MT, Pollard KS, Bernick DL, Stuart JM, Lowe TM. 2009. Transcriptional map of respiratory versatility in the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. J Bacteriol 191:782–794. doi: 10.1128/JB.00965-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nunoura T, Sako Y, Wakagi T, Uchida A. 2003. Regulation of the aerobic respiratory chain in the facultatively aerobic and hyperthermophilic archaeon Pyrobaculum oguniense. Microbiology 149:673–688. doi: 10.1099/mic.0.26000-0. [DOI] [PubMed] [Google Scholar]
- 12.Nunoura T, Sako Y, Wakagi T, Uchida A. 2005. Cytochrome aa3 in facultatively aerobic and hyperthermophilic archaeon Pyrobaculum oguniense. Can J Microbiol 51:621–627. doi: 10.1139/w05-040. [DOI] [PubMed] [Google Scholar]
- 13.Fitz-Gibbon ST, Ladner H, Kim U-J, Stetter KO, Simon MI, Miller JH. 2002. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc Natl Acad Sci U S A 99:984–989. doi: 10.1073/pnas.241636498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos-Vera WH, Berg IA, Fuchs G. 2009. Autotrophic carbon dioxide assimilation in Thermoproteales revisited. J Bacteriol 191:4286–4297. doi: 10.1128/JB.00145-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramos-Vera WH, Weiss M, Strittmatter E, Kockelkorn D, Fuchs G. 2011. Identification of missing genes and enzymes for autotrophic carbon fixation in Crenarchaeota. J Bacteriol 193:1201–1211. doi: 10.1128/JB.01156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hügler M, Alber BE, Fuchs G. 2010. Autotrophic carbon fixation in archaea. Nat Rev Microbiol 8:447–460. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]
- 17.Berg IA, Ramos-Vera WH, Petri A, Huber H, Fuchs G. 2010. Study of the distribution of autotrophic CO2 fixation cycles in Crenarchaeota. Microbiology 156:256–269. doi: 10.1099/mic.0.034298-0. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y, Holden JF. 2006. Citric acid cycle in the hyperthermophilic archaeon Pyrobaculum islandicum grown autotrophically, heterotrophically, and mixotrophically with acetate. J Bacteriol 188:4350–4355. doi: 10.1128/JB.00138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inskeep WP, Ackerman GG, Taylor WP, Kozubal MA, Korf SL, Macur RE. 2005. On the energetics of chemolithotrophy in nonequilibrium systems: case studies of geothermal springs in Yellowstone National Park. Geobiology 3:297–317. doi: 10.1111/j.1472-4669.2006.00059.x. [DOI] [Google Scholar]
- 20.Macur RE, Jay ZJ, Taylor WP, Kozubal MA, Kocar BD, Inskeep WP. 2013. Microbial community structure and sulfur biogeochemistry in mildly-acidic sulfidic geothermal springs in Yellowstone National Park. Geobiology 11:86–99. doi: 10.1111/gbi.12015. [DOI] [PubMed] [Google Scholar]
- 21.Wolin EA, Wolin MJ, Wolfe RS. 1963. Formation of methane by bacterial extracts. J Biol Chem 238:2882–2886. [PubMed] [Google Scholar]
- 22.Pfennig N, Lippert KD. 1966. Über das Vitamin B12-Bedürfnis phototropher Schwefelbakterien. Arch Mikrobiol 55:245–256. doi: 10.1007/BF00410246. [DOI] [Google Scholar]
- 23.American Public Health Association. 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC. [Google Scholar]
- 24.Kamyshny A, Ekeltchik I, Gun J, Lev O. 2006. Method for the determination of inorganic polysulfide distribution in aquatic systems. Anal Chem 78:2631–2639. doi: 10.1021/ac051854a. [DOI] [PubMed] [Google Scholar]
- 25.Rizkov D, Lev O, Gun J, Anisimov B, Kuselman I. 2004. Development of in-house reference materials for determination of inorganic polysulfides in water. Accred Qual Assur 9:399–403. [Google Scholar]
- 26.Schwedt G, Rieckhoff M. 1996. Separation of thio- and oxothioarsenates by capillary zone electrophoresis and ion chromatography. J Chromatogr A 736:341–350. doi: 10.1016/0021-9673(95)01319-9. [DOI] [Google Scholar]
- 27.Suess E, Scheinost AC, Bostick BC, Merkel BJ, Wallschläger D, Planer-Friedrich B. 2009. Discrimination of thioarsenites and thioarsenates by X-ray absorption spectroscopy. Anal Chem 81:8318–8326. doi: 10.1021/ac901094b. [DOI] [PubMed] [Google Scholar]
- 28.Planer-Friedrich B, London J, McCleskey RB, Nordstrom DK, Wallschläger D. 2007. Thioarsenates in geothermal waters of Yellowstone National Park: determination, preservation, and geochemical importance. Environ Sci Technol 41:5245–5251. doi: 10.1021/es070273v. [DOI] [PubMed] [Google Scholar]
- 29.Wallschläger D, Stadey CJ. 2007. Determination of (oxy)thioarsenates in sulfidic waters. Anal Chem 79:3873–3880. doi: 10.1021/ac070061g. [DOI] [PubMed] [Google Scholar]
- 30.Markowitz VM, Mavromatis K, Ivanova NN, Chen I-MA, Chu K, Kyrpides NC. 2009. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 31.Mavromatis K, Chu K, Ivanova N, Hooper SD, Markowitz VM, Kyrpides NC. 2009. Gene context analysis in the Integrated Microbial Genomes (IMG) data management system. PLoS One 4:e7979. doi: 10.1371/journal.pone.0007979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.955,10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matte-Tailliez O, Brochier C, Forterre P, Philippe H. 2002. Archaeal phylogeny based on ribosomal proteins. Mol Biol Evol 19:631–639. doi: 10.1093/oxfordjournals.molbev.a004122. [DOI] [PubMed] [Google Scholar]
- 38.Ostlund G, Schmitt T, Forslund K, Köstler T, Messina DN, Roopra S, Frings O, Sonnhammer ELL. 2010. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res 38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 40.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darling AE, Tritt A, Eisen JA, Facciotti MT. 2011. Mauve assembly metrics. Bioinformatics 27:2756–2757. doi: 10.1093/bioinformatics/btr451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Härtig C, Planer-Friedrich B. 2012. Thioarsenate transformation by filamentous microbial mats thriving in an alkaline, sulfidic hot spring. Environ Sci Technol 46:4348–4356. doi: 10.1021/es204277j. [DOI] [PubMed] [Google Scholar]
- 43.Härtig C, Lohmayer R, Kolb S, Horn MA, Inskeep WP, Planer-Friedrich B. 2014. Chemolithotrophic growth of the aerobic hyperthermophilic bacterium Thermocrinis ruber OC 14/7/2 on monothioarsenate and arsenite. FEMS Microbiol Ecol 90:747–760. doi: 10.1111/1574-6941.12431. [DOI] [PubMed] [Google Scholar]
- 44.Baumeister W, Wildhaber I, Phipps BM. 1989. Principles of organization in eubacterial and archaebacterial surface proteins. Can J Microbiol 35:215–227. doi: 10.1139/m89-034. [DOI] [PubMed] [Google Scholar]
- 45.Baumeister W, Lembcke G. 1992. Structural features of archaebacterial cell envelopes. J Bioenerg Biomembr 24:567–575. doi: 10.1007/BF00762349. [DOI] [PubMed] [Google Scholar]
- 46.Chan PP, Cozen AE, Lowe TM. 2011. Discovery of permuted and recently split transfer RNAs in Archaea. Genome Biol 12:R38. doi: 10.1186/gb-2011-12-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zillig W, Stetter KO, Schäfer W, Janekovic D, Wunderl S, Holz I, Palm P. 1981. Thermoproteales: a novel type of extremely thermoacidophilic anaerobic archaebacteria isolated from Icelandic solfataras. Zentralbl Bakteriol Mikrobiol Hyg Abt Orig C Allg Angew Ökol Mikrobiol 2:205–227. [Google Scholar]
- 48.Chun J, Rainey FA. 2014. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int J Syst Evol Microbiol 64:316–324. doi: 10.1099/ijs.0.054171-0. [DOI] [PubMed] [Google Scholar]
- 49.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mende DR, Sunagawa S, Zeller G, Bork P. 2013. Accurate and universal delineation of prokaryotic species. Nat Methods 10:881–884. doi: 10.1038/nmeth.2575. [DOI] [PubMed] [Google Scholar]
- 52.Burggraf S, Larsen N, Woese CR, Stetter KO. 1993. An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum. Proc Natl Acad Sci U S A 90:2547–2550. doi: 10.1073/pnas.90.6.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Itoh T, Suzuki K, Sanchez PC, Nakase T. 1999. Caldivirga maquilingensis gen. nov., sp. nov., a new genus of rod-shaped crenarchaeote isolated from a hot spring in the Philippines. Int J Syst Bacteriol 49:1157–1163. doi: 10.1099/00207713-49-3-1157. [DOI] [PubMed] [Google Scholar]
- 54.Itoh T, Nomura N, Sako Y. 2003. Distribution of 16S rRNA introns among the family Thermoproteaceae and their evolutionary implications. Extremophiles 7:229–233. [DOI] [PubMed] [Google Scholar]
- 55.Nakayama H, Morinaga Y, Nomura N, Nunoura T, Sako Y, Uchida A. 2003. An archaeal homing endonuclease I-PogI cleaves at the insertion site of the neighboring intron, which has no nested open reading frame. FEBS Lett 544:165–170. doi: 10.1016/S0014-5793(03)00497-6. [DOI] [PubMed] [Google Scholar]
- 56.Sugahara J, Kikuta K, Fujishima K, Yachie N, Tomita M, Kanai A. 2008. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order Thermoproteales. Mol Biol Evol 25:2709–2716. doi: 10.1093/molbev/msn216. [DOI] [PubMed] [Google Scholar]
- 57.Morinaga Y, Nomura N, Sako Y. 2002. Population dynamics of archaeal mobile introns in natural environments: a shrewd invasion strategy of the latent parasitic DNA. Microbes Environ 17:153–163. doi: 10.1264/jsme2.17.153. [DOI] [Google Scholar]
- 58.Stoddard BL. 2005. Homing endonuclease structure and function. Q Rev Biophys 38:49–95. [DOI] [PubMed] [Google Scholar]
- 59.Jay ZJ, Inskeep WP. 2015. The distribution, diversity, and importance of 16S rRNA gene introns in the order Thermoproteales. Biol Direct 10:35. doi: 10.1186/s13062-015-0065-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lykke-Andersen J, Garrett RA. 1994. Structural characteristics of the stable RNA introns of archaeal hyperthermophiles and their splicing junctions. J Mol Biol 243:846–855. doi: 10.1006/jmbi.1994.1687. [DOI] [PubMed] [Google Scholar]
- 61.Fabbri S, Fruscoloni P, Bufardeci E, Negri EDN, Baldi MI, Attardi DG, Mattoccia E, Tocchini-Valentini GP. 1998. Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science 280:284–286. doi: 10.1126/science.280.5361.284. [DOI] [PubMed] [Google Scholar]
- 62.Xue S, Calvin K, Li H. 2006. RNA recognition and cleavage by a splicing endonuclease. Science 312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- 63.Hirata A, Fujishima K, Yamagami R, Kawamura T, Banfield JF, Kanai A, Hori H. 2012. X-ray structure of the fourth type of archaeal tRNA splicing endonuclease: insights into the evolution of a novel three-unit composition and a unique loop involved in broad substrate specificity. Nucleic Acids Res 40:10554–10566. doi: 10.1093/nar/gks826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elferink MGL, Albers S-V, Konings WN, Driessen AJM. 2001. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol Microbiol 39:1494–1503. doi: 10.1046/j.1365-2958.2001.02336.x. [DOI] [PubMed] [Google Scholar]
- 65.Siebers B, Klenk H-P, Hensel R. 1998. PPi-dependent phosphofructokinase from Thermoproteus tenax, an archaeal descendant of an ancient line in phosphofructokinase evolution. J Bacteriol 180:2137–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hansen T, Schönheit P. 2001. Sequence, expression, and characterization of the first archaeal ATP-dependent 6-phosphofructokinase, a non-allosteric enzyme related to the phosphofructokinase-B sugar kinase family, from the hyperthermophilic crenarchaeote Aeropyrum pernix. Arch Microbiol 177:62–69. doi: 10.1007/s00203-001-0359-1. [DOI] [PubMed] [Google Scholar]
- 67.Berg IA, Kockelkorn D, Buckel W, Fuchs G. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 68.Siebers B, Zaparty M, Raddatz G, Tjaden B, Albers S-V, Bell SD, Blombach F, Kletzin A, Kyrpides N, Lanz C, Plagens A, Rampp M, Rosinus A, von Jan M, Makarova KS, Klenk H-P, Schuster SC, Hensel R. 2011. The complete genome sequence of Thermoproteus tenax: a physiologically versatile member of the Crenarchaeota. PLoS One 6:e24222. doi: 10.1371/journal.pone.0024222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.García-Horsman JA, Barquera B, Rumbley J, Ma J, Gennis RB. 1994. The superfamily of heme-copper respiratory oxidases. J Bacteriol 176:5587–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sousa FL, Alves RJ, Pereira-Leal JB, Teixeira M, Pereira MM. 2011. A bioinformatics classifier and database for heme-copper oxygen reductases. PLoS One 6:e19117. doi: 10.1371/journal.pone.0019117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jünemann S. 1997. Cytochrome bd terminal oxidase. Biochim Biophys Acta 1321:107–127. doi: 10.1016/S0005-2728(97)00046-7. [DOI] [PubMed] [Google Scholar]
- 72.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. 2011. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta 1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hensel M, Hinsley AP, Nikolaus T, Sawers G, Berks BC. 1999. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol Microbiol 32:275–287. doi: 10.1046/j.1365-2958.1999.01345.x. [DOI] [PubMed] [Google Scholar]
- 74.Duval S, Ducluzeau A-L, Nitschke W, Schoepp-Cothenet B. 2008. Enzyme phylogenies as markers for the oxidation state of the environment: the case of respiratory arsenate reductase and related enzymes. BMC Evol Biol 8:206. doi: 10.1186/1471-2148-8-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chan PP, Cozen AE, Lowe TM. 2012. Reclassification of Thermoproteus neutrophilus Stetter and Zillig 1989 as Pyrobaculum neutrophilum comb. nov. based on phylogenetic analysis. Int J Syst Evol Microbiol 63:751–754. doi: 10.1099/ijs.0.043091-0. [DOI] [PubMed] [Google Scholar]
- 76.Blumentals II, Itoh M, Olson GJ, Kelly RM. 1990. Role of polysulfides in reduction of elemental sulfur by the hyperthermophilic archaebacterium Pyrococcus furiosus. Appl Environ Microbiol 56:1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kamyshny A Jr, Gun J, Rizkov D, Voitsekovski T, Lev O. 2007. Equilibrium distribution of polysulfide ions in aqueous solutions at different temperatures by rapid single phase derivatization. Environ Sci Technol 41:2395–2400. doi: 10.1021/es062637+. [DOI] [PubMed] [Google Scholar]
- 78.Planer-Friedrich B, Fisher JC, Hollibaugh JT, Suess E, Wallschläger D. 2009. Oxidative transformation of trithioarsenate along alkaline geothermal drainages—abiotic versus microbially mediated processes. Geomicrobiol J 26:339–350. doi: 10.1080/01490450902755364. [DOI] [Google Scholar]
- 79.Helz GR, Tossell JA. 2008. Thermodynamic model for arsenic speciation in sulfidic waters: a novel use of ab initio computations. Geochim Cosmochim Acta 72:4457–4468. doi: 10.1016/j.gca.2008.06.018. [DOI] [Google Scholar]
- 80.Jormakka M, Yokoyama K, Yano T, Tamakoshi M, Akimoto S, Shimamura T, Curmi P, Iwata S. 2008. Molecular mechanism of energy conservation in polysulfide respiration. Nat Struct Mol Biol 15:730–737. doi: 10.1038/nsmb.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.