In meiosis I, the centromeres of sister chromatids are pulled, together, toward the same pole of the spindle. In mitotis, the opposite is true—sister centromeres are pulled apart. This study explores the manner in which mitotic outer kinetochores are shed to allow assembly of meiotic kinetochore structures.
Abstract
In mitosis, the centromeres of sister chromosomes are pulled toward opposite poles of the spindle. In meiosis I, the opposite is true: the sister centromeres move together to the same pole, and the homologous chromosomes are pulled apart. This change in segregation patterns demands that between the final mitosis preceding meiosis and the first meiotic division, the kinetochores must be restructured. In budding yeast, unlike mammals, kinetochores are largely stable throughout the mitotic cycle. In contrast, previous work with budding and fission yeast showed that some outer kinetochore proteins are lost in early meiosis. We use quantitative mass spectrometry methods and imaging approaches to explore the kinetochore restructuring process that occurs in meiosis I in budding yeast. The Ndc80 outer kinetochore complex, but not other subcomplexes, is shed upon meiotic entry. This shedding is regulated by the conserved protein kinase Ipl1/Aurora-B and promotes the subsequent assembly of a kinetochore that will confer meiosis-specific segregation patterns on the chromosome.
INTRODUCTION
Sexual reproduction relies on two key events: the formation of cells with haploid genomes (sperm and eggs) and the restoration of diploidy after their fusion. Meiosis is the specialized cell division program used to generate haploid gametes by halving the number of chromosomes. The proper execution of this process is essential because errors in meiotic segregation result in aneuploidy, which in humans is the leading cause of miscarriages and birth defects (Hassold and Hunt, 2001; Nagaoka et al., 2012). During meiosis, a single round of DNA replication is followed by two successive divisions: meiosis I, which segregates homologous chromosomes away from one another (reductional division), and meiosis II, which segregates the sister centromeres (equational division; Brar and Amon, 2008; Watanabe, 2012). The establishment of this meiotic segregation pattern requires major structural changes that affect the way in which the chromosomes interact with each other and with the spindle. First, homologous chromosomes need to become connected through a link, which is provided by reciprocal recombination. Second, sister chromatid cohesion is modified such that it can be removed in a stepwise way: along the chromosome arms in meiosis I and later at the centromeres in meiosis II. Third, the migration of sister chromatids to the same pole in meiosis I is accomplished through specific modifications of the kinetochores—the structure at the centromere that attaches to microtubules (Westermann et al., 2007; Cheeseman and Desai, 2008)—such that both sister centromeres become attached to microtubules emanating from the same pole (also called co- or mono-orientation). In the search for factors supporting meiosis I specific segregation, proteins have been identified that specifically load on chromosomes during meiosis I to modify cohesion and the ways in which the kinetochores interact with microtubules. Among them, investigators have discovered the conserved cohesin Rec8 (Watanabe and Nurse, 1999; Watanabe et al., 2001; Yokobayashi et al., 2003; Chelysheva et al., 2005; Tachibana-Konwalski et al., 2013), Moa1 from fission yeast (Yokobayashi and Watanabe, 2005), and the monopolin complex in budding yeast (Toth, Rabitsch, et al., 2000; Rabitsch, Petronczki, et al., 2003). In addition, important meiotic-specific roles in regulating these proteins have been identified for some non–meiosis- specific proteins (such as Dbf4-dependent protein kinase, Cdc5 kinase, casein kinase 1; Clyne et al., 2003; Lee and Amon, 2003; Petronczki et al., 2006; Lo et al., 2008; Matos, Lipp, et al., 2008). The simple expression of these factors in mitotic cells allows some portion of the chromosomes to exhibit meiosis-like segregation patterns but does not support the full conversion from mitotic to meiotic attachment (Watanabe and Nurse, 1999; Watanabe et al., 2001; Yokobayashi and Watanabe, 2005; Monje-Casas, Prabhu, et al., 2007; Miller, Ünal, et al., 2012). Therefore, beyond these changes, we can imagine that additional regulation is required to restructure the kinetochores.
The kinetochore can be thought of as an assembly of subcomplexes (for detailed review, see Lampert and Westermann, 2011). Briefly, the base of the kinetochore at the DNA interface includes a centromere-specific nucleosome and the Cbf3 complex. The Mif2, Ctf19, and Mtw1 complexes, each composed of multiple conserved components, provide the bridge to the outer kinetochore complexes (Spc105 and Ndc80) that contact microtubules. The Cnn1 complex may provide a separate or alternate DNA-to–outer kinetochore bridge (Bock, Pagliuca, et al., 2012; Schleiffer et al., 2012; Malvezzi et al., 2013).
In mammalian, chicken, and nematode mitosis, several of the outer kinetochore proteins are shed after chromosome segregation and then reassembled upon mitotic entry (Howe et al., 2001; Nabetani et al., 2001; Hori et al., 2003; Gascoigne and Cheeseman, 2011; Dorn and Maddox, 2012; Westhorpe and Straight, 2013). This refreshment is required each cell cycle in order to maintain genetic integrity. This shedding and reassembly does not occur in yeast mitosis but does in meiosis, as it does in mammalian meiosis (Asakawa et al., 2005; Hayashi et al., 2006; Parra et al., 2009; Sun et al., 2011), perhaps to facilitate the conversion from a mitotic to a meiotic structure.
Meiotic kinetochore shedding has been most extensively studied in fission yeast, in which meiosis typically occurs in the zygote formed from two mating haploid cells. These studies revealed that three outer kinetochore complexes (Ndc80/Mis12/Spc7) are shed upon meiotic entry and that this process is regulated (at least in part) by mating pheromone signaling (Asakawa et al., 2005; Hayashi et al., 2006). In budding yeast, outer kinetochore components, including the Nuf2/Ndc80 complex, are also shed (Asakawa et al., 2005; Miller et al., 2012; Kim et al., 2013), but the precise interface between the shed and maintained portions of the kinetochore is not known. Determining this interface will inform investigations into how the regulators of kinetochore shedding, which have not been identified for nonzygotic meioses, trigger this process.
RESULTS
Kinetochores undergo dynamic changes during meiotic prophase
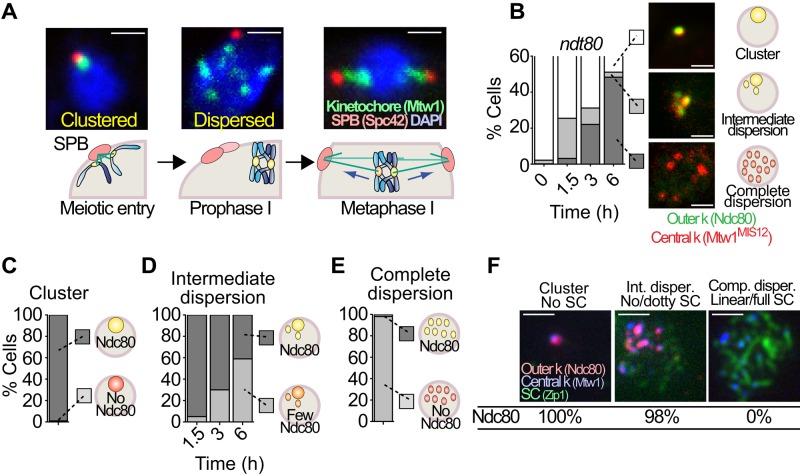
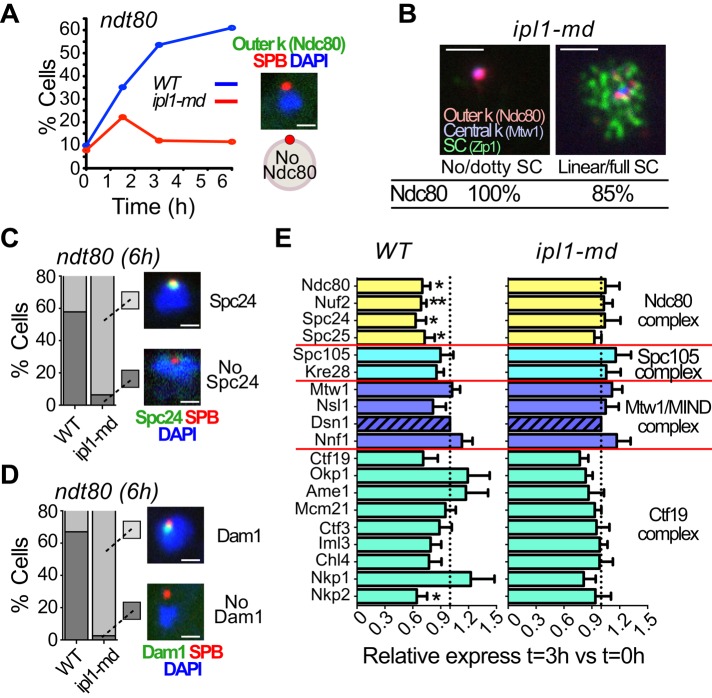
When yeast cells enter meiosis, centromeres are clustered near the single spindle pole body (SPB; Figure 1A). This cluster, termed the Rabl cluster, disperses as homologous chromosomes begin pairing (Hayashi et al., 1998; Jin et al., 1998). Approximately concomitant with the liberation of the centromeres from the SPBs, the chromosomes begin a series of rapid telomere-led movements that are believed to play critical roles in synapsis, detangling of chromosomes, and recombination (Conrad, Lee, et al., 2008; Koszul et al., 2008). After synapsis and the establishment of a recombination link between the homologues, chromosomes transition out of prophase, a spindle forms, and centromeres begin to reattach to microtubules (Figure 1A). It has been described that components of outer kinetochores are shed from centromeres between the entry into meiosis and prophase I, but the precise timing and duration of this shedding, its coordination with other meiotic events, and the boundary between the shed and retained portions of the kinetochore were unknown (Asakawa et al., 2005; Miller, Ünal, et al., 2012). The release of the outer kinetochore could be imagined to be the mechanism by which centromeres are released from their cluster at the SPBs. To explore this, we compared kinetochore disassembly dynamics to the timing of centromere dispersion. We tracked simultaneously a stable component of the kinetochore (Mtw1 tagged with red fluorescent protein [RFP]; Chuong and Dawson, 2010) and one protein that is known to be shed from the kinetochore (Ndc80 tagged with green fluorescent protein [GFP]; Kim et al., 2013). Using these two markers, we followed the cells from their entry (T = 0 h) into meiosis through prophase I (T = 6 h; Figure 1B). Cells were blocked from progressing beyond late prophase—pachytene—by deletion of NDT80, which promotes exit from pachytene. Cells were categorized as having clustered, intermediate, or dispersed kinetochores. When scoring the disposition of Ndc80 in these cells, we noticed several striking features. First, centromere dispersion and outer kinetochore disassembly started almost simultaneously as cells entered into meiosis (Figure 1B). Second, cells with clustered centromeres always exhibited strong Ndc80 staining (even at late time points; Figure 1C). Thus we saw no evidence that Ndc80 shedding significantly precedes dispersion of the centromeres. At early time points, cells showing partial dispersion of centromeres often still showed partial Ndc80 staining, but later in meiosis, cells with partial dispersion of centromeres had very little detectable Ndc80 (Figure 1, B and D). Quantification of the Ndc80-GFP fluorescence signals confirmed that in cells that do not release the kinetochore cluster upon transfer to meiosis-inducing (sporulation) medium, Ndc80 persists at the cluster and then gradually drops (Supplemental Figure S1, A and B). These cells with persisting clusters may represent those that remain in mitotic G1 and do not enter meiosis. In cells that exhibit dispersion of kinetochores, the Ndc80 fluorescence signal rapidly drops (Supplemental Figure S1C). The results are consistent with the notion that Ndc80 is lost from different kinetochores at different times concomitant with dispersion of centromeres from the Rabl cluster. When the dispersion of centromeres was complete, in cells arrested in prophase I, we failed to observe any signal for Ndc80 (Figure 1E and Supplemental Figure S1C). By pachytene, kinetochores have no detectable Ndc80 but retain Mtw1 (Figure 1F).
FIGURE 1:
Centromere repositioning and outer kinetochore disassembly. (A) Representative pictures of diploid cells carrying markers for the kinetochore (MTW1-GFP) and the SPBs (SPC42-DsRed) progressing into meiosis. The cells enter meiosis with clustered centromeres, and centromeres disperse in prophase and then reattach, with sister chromatids attaching to microtubules from the same pole and homologues drawn to opposite poles. (B) A diploid strain with marked central and outer kinetochore proteins (MTW1-3xmCherry and NDC80-GFP) was released into meiosis, and cells with clustered centromeres and Ndc80 present, intermediate dispersion (<4 Mtw1 foci), or complete dispersion (>4 Mtw1 foci) and no Ndc80 were scored (n ≥ 100). Representative cells. Scale bar, 1 μm. (C) From B, the cells with clustered centromeres with (dark gray) or without Ndc80 (light gray) were scored. (D) From B, cells with intermediate dispersion and strong (dark gray) or weak (light gray) signals for Ndc80 were scored. (E) From B, cells with full dispersion and with (dark gray) or without Ndc80 (light gray) were scored. (F) In a diploid strain carrying the kinetochore markers MTW1-3xmCherry and NDC80-GFP, cells at different stages of synaptonemal complex (Zip1) assembly (none, dotty, or linear/full) were scored for the presence of Ndc80. Scale bar, 1 μm.
Tracking kinetochore components through meiotic prophase
The shedding of Ndc80 raises questions about what other proteins are shed from the kinetochore, how this process is controlled, and the role of kinetochore shedding in meiotic chromosome behavior. To track more specifically the changes in kinetochore structure during the earliest stage of meiosis, we took advantage of recently described methods that allow the purification of largely intact kinetochore particles from mitotic and meiotic yeast cells (Akiyoshi, Sarangapani, Powers, et al., 2010; Sarangapani, Duro, et al., 2014). We purified kinetochores from cells at meiotic entry and in late prophase. We used quantitative mass spectrometry to evaluate the relative levels of individual kinetochore proteins in the two samples, which would reveal the identities of proteins that were lost from kinetochores upon progression through prophase. Briefly, Dsn1, a component of the Mtw1 complex (Figure 2A), was FLAG tagged, and affinity purification was used to purify kinetochores from cells immediately before the switch to meiosis-inducing medium or 7 h later in a prophase arrest (ndt80). Mitotic kinetochore particles purified by this method contained almost all of the components of the complete kinetochore (except the Cbf3 complex; Akiyoshi, Sarangapani, Powers, et al., 2010)). We used mass spectrometry selective reaction monitoring (MS-SRM) for quantification of individual proteins in the purified kinetochore samples based on the liquid chromatography-tandem MS measurement of specific peptides formed by digestion of those proteins with the protease trypsin (see Materials and Methods; Kinter and Kinter, 2013). Quantities of each protein were normalized to the amount of Dsn1 seen in that experiment, and then the ratio of the amount of protein in late prophase versus meiotic entry was calculated. At the same time, we epitope tagged one or more representative components from each subcomplex and used immunofluorescence microscopy as a second way to monitor kinetochore disassembly in prophase. The GFP epitope tags did not result in significant growth defects and did not cause detectable meiotic chromosome segregation defects (Supplemental Figure S2A). As with Mtw1 and Ndc80 (Figure 1C), all epitope-tagged components of the mitotic kinetochore were present at the clustered kinetochores upon meiotic entry (Supplemental Figure S2, B and C), suggesting that there is no major loss of any of these components in the G1 phase that precedes meiotic induction.
FIGURE 2:
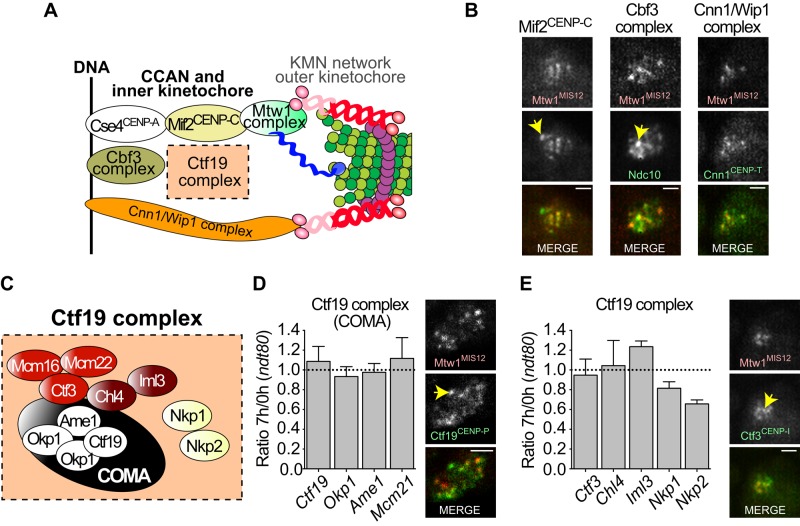
Behavior of the constitutive-centromere associated network during early stages of meiosis I. (A) Cartoon of the kinetochore microtubule interface. The major complexes of the CCAN are represented. (B) Representative pictures of prophase diploid cells carrying markers for the central kinetochore component Mtw1 (MTW1-3xmCherry) and Mif2, Ndc10 (for the Cbf3 complex), or Cnn1 tagged with GFP. The yellow arrow indicates SPBs marked by SPC42-CFP in the first two strains, Scale bar, 1 μm. (C) Cartoon of the Ctf19 complex. (D, E) Diploid cells expressing Dsn1-FLAG were switched to sporulation medium (T = 0 h). The strains were ndt80 mutants, so they fail to exit from pachytene. The relative amounts of Ctf19 complex components in purified kinetochores at pachytene arrest vs. the time of meiotic entry were determined by MS-SRM. Results are averages of three independent experiments. The levels of individual components on dispersed prophase centromeres (detected with Mtw1-3xmCherry) were examined by fluorescence microscopy. (D) COMA complex MS-SRM results and levels Ctf19-GFP. (E) Additional Ctf19 complex MS-SRM results and levels of Ctf3-GFP. The yellow arrow indicates SPBs (Spc42-CFP). Scale bars, 1 μm.
The inner/central kinetochore remains on chromosomes during meiotic prophase
To find the interface between the shed and nonshed portions of the kinetochore, we monitored the presence of proteins from inner and central subcomplexes on prophase kinetochores. Previous work identified several proteins (Ndc10, Mtw1, Ctf19, Chl4, and Iml3; Figure 2A) that remain at kinetochores through meiosis (Kamieniecki et al., 2000; Marston et al., 2004; Tsubouchi and Roeder, 2005; Gladstone et al., 2009). This suggests that the kinetochore–DNA interface and at least portions of the Ctf19 complex, which includes several subcomplexes, are not shed upon prophase entry. Indeed, immunofluorescence microscopy confirmed that when kinetochores are dispersed in prophase, Mif2, Ndc10, and Cnn1, representing three subcomplexes that are near the DNA–kinetochore interface, remain with the kinetochores (marked by Mtw1; Figure 2B). Using MS-SRM, we found that representatives of the COMA complex (Figure 2, C and D) and other Ctf19 subcomplexes (Figure 2, C and E) showed little variation in relative abundance in kinetochores purified from cells at meiotic entry and late prophase. Our immunofluorescence microscopy (Figure 2, D and E) and observations by others (Marston et al., 2004) supported this conclusion, although detection of components known to be at low levels on the kinetochore (Joglekar et al., 2006) were difficult to monitor by fluorescence microscopy (e.g., Ctf3), highlighting the complementarity of the MS-SRM approach. A potential exception to the stability of the inner/central kinetochore could be the Nkp1 and Nkp2 proteins (Figure 2E). These proteins were found at slightly lower levels in late prophase in the MS-SRM experiments but were not significantly underrepresented. Together these results suggest that the inner and most central kinetochore components are present on the kinetochore throughout meiotic prophase. Of note, this includes the Cnn1 complex. This complex was recently defined as an alternative bridge that can tether the Ndc80 complex to the inner kinetochore/centromeric DNA (Nishino et al., 2012; Schleiffer et al., 2012; Malvezzi et al., 2013), although in mitotic cells it is most abundant in anaphase. These results demonstrate that it is present at meiotic entry and remains on kinetochores throughout meiosis (Supplemental Figure S2B and Figure 2B).
The Ndc80 complex is shed upon meiotic entry
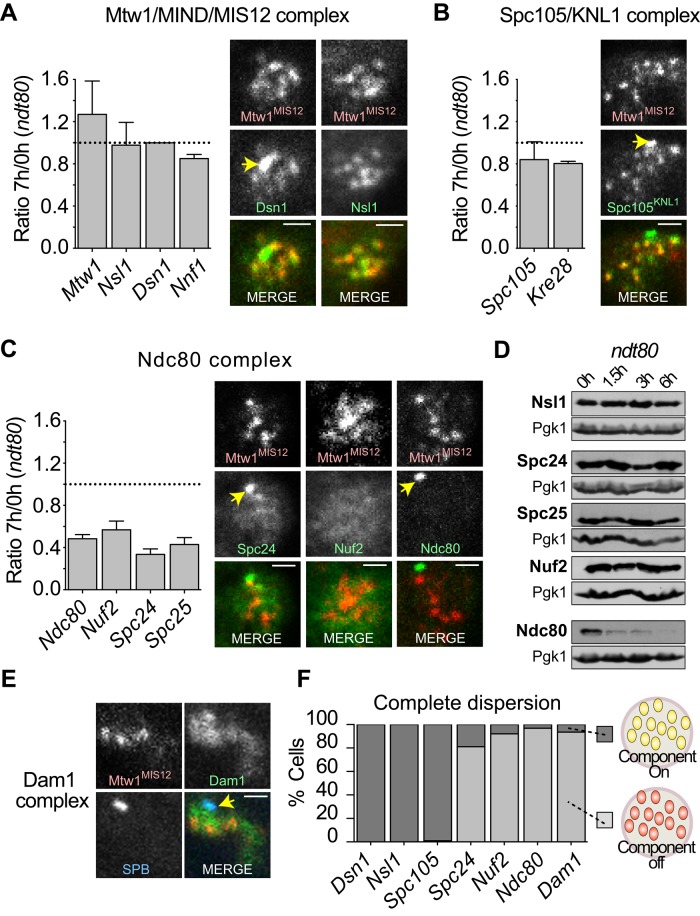
The Ndc80 and Spc105 complexes are located at the outer kinetochore (Supplemental Figure S1D). Ndc80 mediates kinetochore–microtubule interactions, and Spc105 has also been implicated in interacting with microtubules (reviewed in Lampert and Westermann, 2011). The Ndc80 and Spc105 complexes are anchored to the kinetochore through interactions with the Mtw1 complex. The Mtw1 complex persists on kinetochores through meiotic prophase (Figure 3A). Both components of the Spc105 complex were also found to remain on the kinetochores through meiotic prophase (Figure 3B). In contrast, MS-SRM revealed that all four components of the Ndc80 complex were lost from kinetochores by the end of prophase I (Figure 3C). The fact that ∼40% of the original level of Ndc80 components remains with the kinetochores reflects the fact that upon meiotic induction, typically ∼30–40% of the cells fail to enter meiosis and persist with clustered kinetochores (Figure 1B and Supplemental Figure S1). Immunofluorescence microscopy confirmed the loss of the Ndc80 complex, although the four proteins of the complex do not all behave in the same way. In cells with dispersed kinetochores, Ndc80 signal is virtually undetectable, whereas Spc24 and Nuf2 persist as a nuclear haze (Figure 3C). Western blots demonstrated that the levels of Ndc80 decline upon meiotic entry, as suggested by earlier work (Miller, Ünal, et al., 2012). In contrast, Nuf2, Spc24, and Spc25 levels are more stable (Figure 3D). Whether the shed Spc24, Spc25, and Nuf2 are later reincorporated into the meiotic kinetochores remains to be determined. Like the Ndc80 complex, Dam1 was not observed on dispersed prophase kinetochores using fluorescence microscopy (Figure 3E). We conclude that upon meiotic entry, there is a rather precise shedding of the Ndc80 complex from the outer kinetochore (Figure 3F). Other complexes are found at about the same relative abundance in late prophase as at meiotic entry, although we cannot eliminate the possibility that the small changes we observed for some proteins (increases in Mtw1, decreases in Nkp1, Nkp2, and the Spc105 complex) reflect real adjustments to the kinetochore as it converts from a mitotic to a prophase I structure.
FIGURE 3:
Behavior of the KMN network and Dam1 complex during early stages of meiosis I. (A–C) MS-SRM was used to determine the relative levels of proteins at pachytene relative to meiotic entry (three independent experiments), and fluorescence microscopy was used to visualize individual components tagged with GFP. Kinetochores were marked by Mtw1-3xmCherry, and SPBs by were marked by Spc42-CFP. (A) Mtw1 complex, Dsn1-GFP, and Nsl1-GFP. (B) Spc105 complex, Spc105-GFP. (C) Ndc80 complex, Spc24-GFP, Nuf2-GFP, and Ndc80-GFP. (D) Western blot analysis of the levels of GFP-tagged Ndc80 complex components. Samples were harvested from synchronous cultures, and extracts were prepared from samples harvested at the times indicated after meiotic induction. Blots were probed with antibodies against Pgk1 (loading control) and GFP. (E) Fluorescence microscopy examination of Dam1-GFP disposition in pachytene. (F) Quantification of fluorescence microscopy data (n ≥ 100). The yellow arrows indicate SPB marker when present. Scale bars, 1 μm.
Mitotic and meiotic centromeres are regulated differently
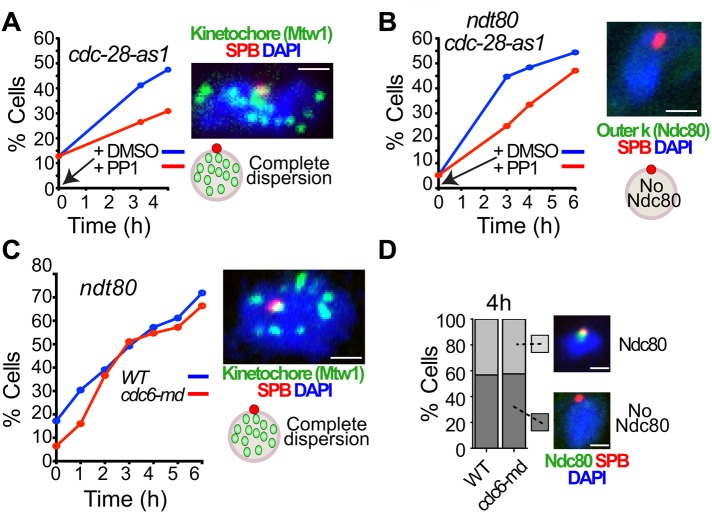
What is the regulatory mechanism that triggers the shedding of the mitotic outer kinetochore upon meiotic entry? Yeast cells enter the mitotic cycle with centromeres clustered at the SPBs. The centromeres disperse briefly in a manner that is dependent on DNA replication (Kitamura et al., 2007). We used two approaches to test whether meiotic centromere dispersion and kinetochore shedding depend upon DNA replication. First, cyclin-dependent kinase (CDK) is required for prophase events, including DNA replication and completion of meiotic recombination (Shuster and Byers, 1989; Xu et al., 1995; Benjamin et al., 2003). We tested whether Cdc28 was required for meiotic centromere dispersion and kinetochore shedding by using a conditional cdc28-as1 mutant that is sensitive to ATP-analogue chemical inhibitors (Bishop et al., 2000). The Cdc28-as1 inhibitor (1NM-PP1) was added to the cultures at the time of meiotic induction at a dose (5 μM) that blocks meiotic DNA replication and progression beyond prophase 1 (Benjamin et al., 2003). Inhibiting Cdc28-as1 did not prevent dispersion (Figure 4A) or kinetochore shedding (Figure 4B), although both processes were somewhat slowed when Cdc28-as1 was inhibited. As a second test, we monitored centromere dispersion and kinetochore shedding in cdc6-md (meiotic depletion) mutants, in which CDC6 is placed under the control of the SCC1 promoter, which is largely silent in meiosis. The cdc6-md mutants do not replicate their DNA but enter prophase nonetheless (Hochwagen et al., 2005a; Brar et al., 2009; Blitzblau et al., 2012). Although depletion of Cdc6 blocks both kinetochore dispersion and shedding in mitotic cells (Kitamura et al., 2007), we saw no reduction in either in meiosis (Figure 4, C and D). Thus long-term dispersion and kinetochore shedding in meiotic cells are regulated differently than the brief, replication-dependent dispersion and shedding in mitotic cells. Consistent with this, we find that only the Ndc80 complex is shed in meiotic cells, whereas the Cdc6-dependent short-term loss of mitotic kinetochores includes members of the Mtw1 and Ctf19 complexes as well (Kitamura et al., 2007).
FIGURE 4:
Blocking DNA replication does not block kinetochore disassembly. (A–D) Wild-type, cdc6-md, and cdc28-as1 diploid cells expressing a SPB marker (Spc42-DsRed) and either a central kinetochore marker, Mtw1-GFP (A, C), or an outer kinetochore marker, Ndc80-GFP (B, D), were sporulated. (B, C) The strains used were ndt80 mutants that arrest in pachytene. T = 0 h represents the time at which cells were switched to sporulation medium. (A, B) Cdc28-as1 was inhibited (red) or not (blue) by adding 5 μM 1NM-PP1 or DMSO, respectively, to the culture medium upon meiotic induction. The proportion of cells with dispersed kinetochores (A) or without outer kinetochores (B) was scored (n ≥ 100). Representative pictures of cell with dispersed kinetochores (A) or without the outer kinetochore (B). (C) The proportion of cells with dispersed kinetochores for wild-type (blue) or cdc6-md cells (red) was scored. A representative picture of a cell with dispersed kinetochores is shown (n ≥ 100/time point). (D) The proportion of cells at 4 h with (light gray) or without (dark gray) detectable outer kinetochores was scored in wild-type or cdc6-md cells (n ≥ 100). Representative pictures of cells with or without detectable Ndc80-GFP are shown. Inactivation of Cdc6 did not change the kinetics or level of dispersion and did not affect shedding of the outer kinetochore in detectable ways. Scale bars, 1 μm.
Ipl1/Aurora-B is necessary for the disassembly of outer kinetochore
Previous work showed that in ipl1 mutants, centromeres fail to disperse from their cluster around the SPBs when cells enter meiotic prophase (Meyer et al., 2013). The fact that Ipl1 is needed for centromere dispersion suggests a model in which Ipl1 triggers dispersion by promoting shedding of the Ndc80 complex. We tested this using the ipl1-md allele, in which the IPL1 promoter is replaced with the CLB2 promoter, which is expressed in mitotic but not meiotic cells (Grandin and Reed, 1993). Indeed, in ipl1-md cells, Ndc80 was retained on the clustered centromeres (Figure 5A) and present on pachytene kinetochores (Figure 5B). To confirm that this effect was not restricted to Ndc80, we also examined shedding of Spc24 and Dam1 (component of Dam1 complex). For both, we did not observe any significant disassembly (Figure 5, C and D). When we used MS-SRM to evaluate the levels of a wide range of kinetochore components after meiotic entry (T = 3 h) in wild-type and ipl1 mutant cells, it was clear the Ndc80 components and also Nkp2 of the Ctf19 complex were stabilized on the kinetochore when Ipl1 was absent (Figure 5E). There is less variation in the levels of many components upon meiotic entry in ipl1 mutants compared with what is observed in wild-type cells, leading us to wonder whether outer kinetochore shedding is accompanied by a global increase in the plasticity of the kinetochore structure that might be detected by more precise monitoring methods than those used here.
FIGURE 5:
Ipl1/Aurora-B is necessary for outer kinetochore disassembly. (A, C, D) Wild-type or ipl1-md diploid cells expressing Spc42-DsRed and outer kinetochore marker Ndc80-GFP (A), Spc24-GFP (C), or Dam1-GFP (D) were switched to sporulation medium (T = 0 h). (A–D) The strains used were ndt80 mutants that arrest in pachytene. (A) The proportion of cells with dispersed kinetochores was scored in wild-type (blue) or ipl1-md (red) diploid cells (n ≥ 100). A representative picture of a cell with dispersed kinetochores is shown. (B) The patterns of Ndc80-GFP staining were monitored in an ipl1-md diploid expressing Mtw1-3xmCherry and Ndc80-GFP. Cells were first categorized according to their stage in prophase by evaluating their Zip1 staining pattern (No/dotty or Linear/full). (C, D) The proportion of cells with (light gray) or without (dark gray) either Spc24 or Dam1 was scored (n ≥ 100). Representative pictures of cells with or without the designated kinetochore component are shown. (E) Wild-type or ipl1-md diploid cells carrying Dsn1-FLAG were sporulated. The quantification by mass spectrometry was done after purifying the kinetochore in five (ipl1-md) or seven (wild-type) independent experiments. The ratios represent the amount of each component at T = 3 h relative to the amount at T = 0 h. Scale bars, 1 μm.
The disassembly of the outer kinetochore facilitates the incorporation of monopolin
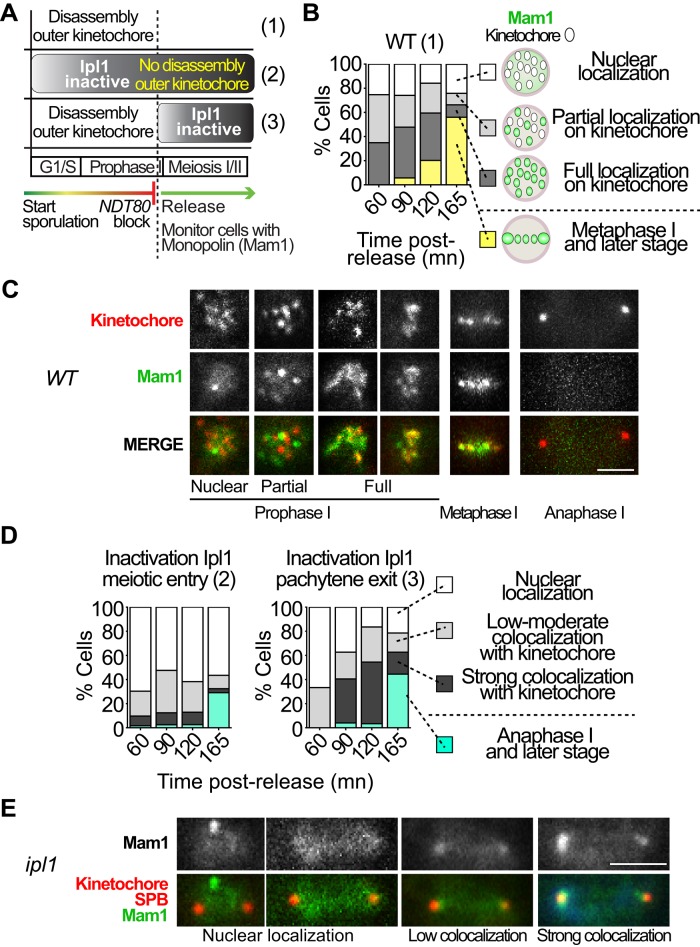
Why might yeast cells remove the outer kinetochore for an extended portion of meiosis but not mitosis? One explanation is that whereas mitotic sister chromatids are juxtaposed as soon as they are produced and thus are ready to be segregated, in meiosis, the pairing of the homologous partners to prepare them for segregation is a protracted enterprise. Eliminating outer kinetochores until homologues are paired prevents precocious segregation of unpaired partners (Kim et al., 2013). Kinetochore shedding might also contribute to one of the critical changes during meiosis I: the transition from a kinetochore that will support mitotic segregation to one constructed for separating homologues while keeping sister chromatids together. The mitotic-to-meiotic conversion is mainly supported in budding yeast by loading of the monopolin complex onto the centromere. This complex is composed of Hrr25 (casein kinase 1), Lrs4, Csm1, and Mam1 (Toth, Rabitsch, et al., 2000; Rabitsch, Petronczki, et al., 2003; Petronczki et al., 2006). The monopolin subunits Csm1 and Lrs4 form a V-shaped complex that might act by cross-linking sister kinetochores by forming a molecular clamp that enforces their co-orientation to the same pole (Corbett et al., 2010; Corbett and Harrison, 2012). Csm1 presents a conserved hydrophobic surface patch that binds the kinetochore protein Dsn1, a subunit of the Mtw1/MIND complex. The Ndc80 complex is dispensable for association of monopolin with the kinetochore (Sarkar, Shenoy, et al., 2013). Dsn1 is also the interface for binding of the Mtw1 complex with Spc24 and Spc25 of the Ndc80 complex, although binding studies suggest that the N-terminus of Dsn1 interacts with monopolin and the C-terminus interacts with Spc24/25 (Corbett et al., 2010; Corbett and Harrison, 2012; Malvezzi et al., 2013; Sarkar, Shenoy, et al., 2013). The close proximity of the monopolin and Ndc80 binding interfaces on the Mtw1 complex suggested to us that outer kinetochore shedding might expose the central kinetochore, making it more accessible for loading of the monopolin complex. Once monopolin had loaded, this could be followed by Ndc80 complex reloading on the now remodeled kinetochores. To test this idea, we monitored monopolin (Mam1-GFP) loading in ipl1-md mutants that never shed their kinetochores or ipl1-as5 mutants, in which kinetochores are shed before Ipl1-as5 is inactivated. In both scenarios, Ipl1 is inactive at the time of monopolin loading, but in one case, outer kinetochores have been shed, and in the other they, have not. In these experiments, the Ndt80 transcription factor was placed under the control of an estradiol-inducible promoter (Benjamin et al., 2003), allowing us to accumulate cells in late prophase and then synchronously release the cells out of prophase to monitor monopolin loading (Figure 6A).
FIGURE 6:
Loading of the monopolin complex. (A–E) Wild-type, ipl1-md, and ipl1-as5/ipl1-md diploid cells were sporulated and released from a pachytene arrest (PGAL1-NDT80 GAL4-ER) at 6 h by the addition of 5 μM β-estradiol. Ipl1-as5 was inhibited by the addition of 50 μM 1NA-PP1 at the time of the release. All strains expressed Mam1-GFP and Mtw1-3xmCherry and Spc42-DsRed (SPB) when indicated. The protocol is summarized in A. T = 0 h represents the time when cells were released from the arrest. (B) Wild-type cells, harvested at timed intervals after pachytene release, were categorized according to Mam1-GFP distribution. (C) Images of Mam1-GFP patterns observed in wild-type cells. (D) Distribution of Mam1-GFP in ipl1-md (2) and ipl1-as5/ipl1-md mutants (3) after release from pachytene arrest. n ≥ 100 for all time points. (E) Representative images of Mam1-GFP distribution in ipl1 mutants. Note the bright Mam1-GFP focus in the leftmost image. These foci were common in ipl1-md mutants (see Supplemental Figure S5). Scale bars, 2 μm.
In wild-type cells, after release from the prophase arrest, Mam1 begins to accumulate soon before spindle formation (Supplemental Figure S3A) and shows progressive colocalization with kinetochores—from a dispersed nuclear signal, to partial colocalization with dispersed kinetochores, to nearly complete overlap with kinetochores on the metaphase spindles (Figure 6, B and C). To monitor the effect of failed kinetochore shedding on monopolin loading, we inactivated Ipl1 at meiotic entry (ipl1-md) and then monitored Mam1 localization to kinetochores after release from the prophase arrest. The ipl1-md mutants show normal kinetics of progression from prophase through metaphase (Jordan et al., 2009), but inactivation of Ipl1 causes some cells to form spindles precociously, in prophase (Shirk et al., 2011; Kim et al., 2013; Newnham, Jordan, et al., 2013). Thus spindle formation alone is an unreliable marker for progression to metaphase in ipl1-md mutants. These precocious prophase spindles appear before Mam1 accumulates to detectable levels (Supplemental Figure S3, A and B). Thus, in our analysis of ipl1 mutants, we excluded those cells with bipolar spindles but without detectable Mam1. In the ipl1-md mutants, the kinetochores are nearly always associated with the SPBs (Meyer et al., 2013; Figure 6E). When we quantified the association of Mam1 with kinetochores in the ipl1-md strain, it was much reduced compared with the wild-type control (Figure 6D): instead of localizing with kinetochores, Mam1 showed a more general nuclear localization and in some cases an intense focus of staining that did not colocalize with the kinetochores (Figure 6E and Supplemental Figure S5). One explanation for the apparent defect in the ipl1-md mutants in loading monopolin is that the activity of Ipl1 might be necessary for efficient monopolin loading. To test this, we allowed outer kinetochores to be shed due to the presence of functional Ipl1-as5 and then inactivated Ipl1-as5 concomitant with the release from prophase (Figure 6A). In this regime, when Ipl1-as5 was inactivated after kinetochore shedding, monopolin loading occurred at near- wild-type levels (Figure 6D). To confirm the apparent loss of colocalization of Mam1 with kinetochores in the ipl1-md mutants, we quantified the amount of Mam1-GFP fluorescence signal that overlapped the kinetochore (Mtw1-RFP) signal in mutants with either early inactivation or late inactivation of Ipl1 (Supplemental Figure S5). Early loss of Ipl1 activity significantly reduced loading of Mam1 onto kinetochores. We have not tested whether the loading of other monopolin subunits is diminished in ipl1-md mutants. Together these approaches demonstrate that Ipl1 is dispensable for Mam1 loading after the release from prophase I and suggest that the deficiency of ipl1-md mutants in loading Mam1 (Figure 6D) must reflect a prophase function for Ipl1.
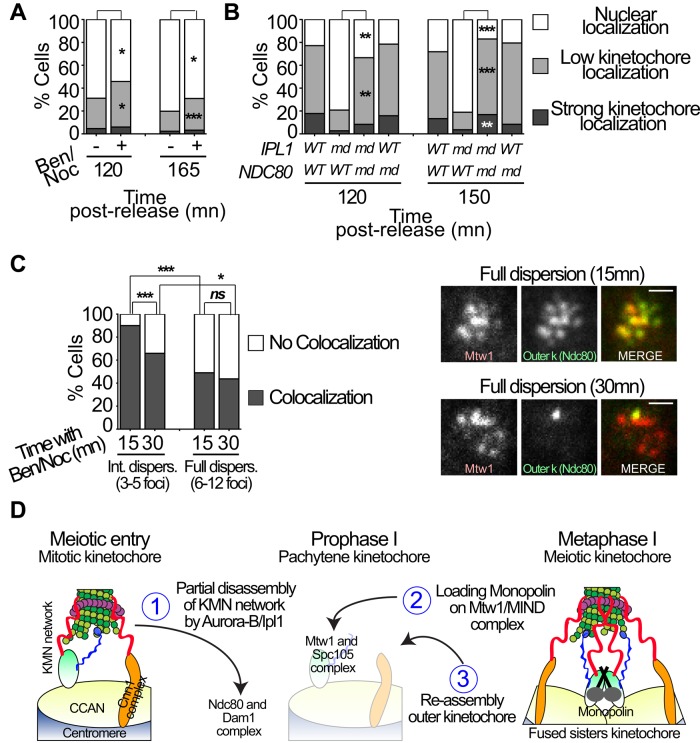
We imagine two different ways in which Ipl1 activity in prophase might promote monopolin loading. First, Ipl1's releasing of kinetochore-microtubule attachments might allow monopolin to load onto kinetochores. This possibility is suggested by the observation that when monopolin is expressed ectopically in mitotic cells, higher levels of meiosis I–like segregation are observed if microtubules are briefly depolymerized before mitosis (Miller, Ünal, et al., 2012). Second, removing the outer kinetochore might directly improve accessibility of the Mtw1 complex for association with monopolin, independently of kinetochore–microtubule association. To study the relative effect on monopolin loading of removing either kinetochore–microtubule interactions or the Ndc80 complex (and therefore any Ndc80 complex–mediated microtubule interactions as well), we used two complementary strategies. The first one disrupted kinetochore–microtubule associations (Figure 7A). The second approach was to remove the Ndc80 outer kinetochore complex (ndc80-md; Figure 7B). For the first approach, we used a combined treatment with benomyl and nocodazole that efficiently destabilized microtubules (Supplemental Figure 6, A–C). The ipl1-md cells, which do not shed outer kinetochores and exhibit persistent kinetochore–microtubule attachments, were released from a prophase arrest for 2½ h (corresponding to the time with maximum loading of Mam1; Figure 6, B and D) in the presence or absence of microtubule-depolymerizing drugs. Treatment with microtubule- destabilizing agents resulted in a small but significant increase in monopolin loading (Figure 7A). To test the effect of removing the outer kinetochore, we used ndc80-md strains in which NDC80 is placed under the control of the CLB2 promoter and is therefore not expressed in meiotic cells. We showed previously, using the ndc80-md mutation, that the lack of centromere dispersion in ipl1-md mutants could be rescued by removing the outer kinetochore complex (Meyer et al., 2013). The ndc80-md mutation resulted in a strong rescue of monopolin loading in the ipl1-md strain (Figure 7B; see representative cell in Supplemental Figure 6D). The much greater rescue of monopolin loading that is seen in the ipl1-md mutants when Ndc80 is removed compared to when microtubules are destabilized suggests that even in the absence of kinetochore–microtubule associations, the Ndc80 complex can interfere with monopolin loading, possibly by obstructing access to the Mtw1 complex. This predicts that Ndc80 remains on the detached kinetochores after treatment with microtubule-destabilizing agents. To test this, we treated cells with benomyl/nocodazole and identified those with dispersed kinetochores (Supplemental Figure 7). In these cells, we scored the localization of Ndc80-GFP with individual dispersed Mtw1-3xmCherry foci (Figure 7C). Cells were categorized as having either intermediate dispersion (3–5 Mtw1 foci) or full dispersion (6–12 foci). In cells with intermediate dispersion, most Mtw1 foci that had been released from the SPB showed colocalization with Ndc80. With increased exposure time to the microtubule-destabilizing agents and in cells with greater dispersion, the level of colocalization was reduced (Figure 7C). Together the results suggest that loss of kinetochore–microtubule attachments leads to the accumulation of kinetochores that lack Ndc80.
FIGURE 7:
Release of kinetochore–microtubule associations and shedding of outer kinetochores allow monopolin loading. (A) Monopolin loading on kinetochores with or without release of kinetochore–microtubule attachments. ipl1-md diploid cells were sporulated and released from a pachytene arrest (PGAL1-NDT80 GAL4-ER) at 6 h after the induction of meiosis by the addition of 5 μM β-estradiol (T = 0 h). Microtubules were destabilized by the addition of benomyl (30 μg/ml) and nocodazole (15 μg/ml), which were added at the time of release from the pachytene arrest (protocol is summarized in Figure 6A). At 120 and 165 min after release from pachytene arrest, cells were categorized according to their Mam1-GFP localization (nuclear, low kinetochore localization, strong kinetochore colocalization; see Supplemental Figure 5 for representative images). n ≥ 83 cells for each time point. (B) Monopolin loading with and without the Ndc80 complex on kinetochores. Wild-type, ipl1-md, ndc80-md, or ipl1-md ndc80-md diploid strains were induced to enter meiosis and then released from pachytene by the addition of 5 μM β-estradiol at 4.5 h after the induction of meiosis. Cells were scored as in A. n ≥ 59 cells for each time point. (C) ipl1-md diploid cells expressing Ndc80-GFP and Mtw1-3xmCherry were switched to sporulation medium. The strains used were ndt80 mutants that arrest in pachytene. Benomyl (30 μg/ml) and nocodazole (15 μg/ml) were added to the cells 6 h after induction of meiosis (T = 0 h). The colocalization of Ndc80-GFP with individual Mtw1-mCherry foci was scored in cells with 3–5 dispersed kinetochores (intermediate dispersion) or those with 6–12 kinetochores (full dispersion). Colocalization is indicated by dark gray, and Mtw1-mCherry foci with no colocalizing Ndc80-GFP signal are indicated in white. Representative cells are shown. Scale bars, 1 μm. n ≥ 41 Mtw1 foci for each time point. Fisher's exact tests were used to evaluate the significance of observed differences in A–C. *p < 0.05, **p < 0.01, ***p < 0.001. (D) A model representing the steps of kinetochore remodeling in meiosis I.
DISCUSSION
By monitoring the global changes in kinetochore structure that occur through meiosis I, we found that the Ndc80 complex disassembles from the kinetochores in budding yeast soon after meiotic entry and is only reloaded as cells are assembling the meiosis I spindle. This process has parallels to the loss of outer kinetochores in meiosis in fission yeast (Nabetani et al., 2001; Asakawa et al., 2005; Hayashi et al., 2006) and in mitosis in mammalian cells (Gascoigne and Cheeseman, 2013). Although there is kinetochore shedding in these situations, there are differences as well. Whereas only the Ndc80 complex is completely lost in budding yeast meiosis, in fission yeast, the Ndc80, Mis12/Mtw1, and Spc7/Spc105 complexes are lost (Asakawa et al., 2007). Similarly, in mammalian mitosis, the complete KMN network (Ndc80, Mis12, and KNL1 complexes) is removed as cells exit mitosis and reloaded upon mitotic entry (Gascoigne and Cheeseman, 2013). Our results are also consistent with centromere dispersion being the result of loss of the outer kinetochores, but the presence of some Ndc80 of dispersed kinetochores demonstrates that complete disassembly of the outer kinetochore is not required for the release of a centromere from the Rabl cluster.
We speculate that shedding the outer kinetochores may serve multiple purposes. First, shedding the outer kinetochores releases the centromeres from their tether to the SPBs, permitting the chromosomes to move freely during the rapid telomere-led chromosome movements in prophase that promote normal pairing and alignment (Conrad, Lee, et al., 2008; Koszul et al., 2008). Second, the restriction of reassembly of the kinetochores until after the pachytene checkpoint is satisfied ensures that segregation never precedes completion of homologue synapsis and recombination. Finally, shedding of the outer kinetochore appears to help prepare kinetochores for the conversion from the mitotic to meiotic form by enabling monopolin to load more efficiently. A similar function was also suggested by a series of experiments with fission yeast, in which ectopic meiosis was induced by inactivation of the Pat1 kinase (Yamamoto and Hiraoka, 2003; Chikashige et al., 2004; Asakawa et al., 2005; Hayashi et al., 2006). Under these conditions, the Ndc80 complex was found to remain on the kinetochores instead of being shed, and the centromeres remain associated with the SPB (instead of being released). Of interest, in the first meiotic division, the sister chromatids split (instead of staying together until the second division), consistent with the possibility that outer kinetochore shedding is important for the conversion from a mitotic to meiotic kinetochore region in fission yeast as well.
Experiments in budding yeast have also addressed the role played by kinetochore–microtubule attachments and outer kinetochore shedding in monopolin loading. The expression of the B-type cyclin Clb3 is normally restricted to meiosis II, but when it is ectopically expressed early in meiosis I, cells exhibit premature attachments of kinetochores to microtubules and reduced loading of monopolin on kinetochores. This reduced loading could be rescued by disrupting kinetochore–microtubule associations (Miller, Ünal, et al., 2012), suggesting a model in which one function of outer kinetochore shedding in early meiosis is to prevent persisting or premature kinetochore–microtubule attachments, which could block monopolin loading. We propose an additional function of shedding: facilitating the access of the monopolin more-interior parts of the kinetochore (e.g., the Mtw1 complex) by “opening” the kinetochore structure. Of course, these two processes are related: releasing kinetochore–microtubule connections appears to promote the loss of Ndc80 from the kinetochore (Figure 7C), and removing Ndc80 results in a loss of microtubule attachments.
Another reason that cells might shed the Ndc80 complex is to prevent the precocious association of kinetochores with microtubules before the homologous chromosomes have fully aligned with their homologous partners. An apparent contradiction to this idea is that Spc105/KNL1 complex might have some ability to associate with microtubules (Cheeseman et al., 2006), and it remains on the kinetochore during prophase I. The Spc105/KNL1 complex is insufficient for mediating persistent kinetochore–microtubule attachments, however, as these are not seen in the absence of the Ndc80 complex (Kim et al., 2013). Why then shed the Ndc80 complex and retain the Spc105 complex? Previous work has shown that Spc105/Spc7 is important for the association of Bub1 kinase with the kinetochore (London et al., 2012; Shepperd et al., 2012). We speculate that retaining Spc105 might be important to promote the early loading of essential critical actors of the spindle checkpoint to promote and monitor the early stages of kinetochore–microtubule attachment.
The Mtw1/Mis12/MIND complex also remained on kinetochores during prophase I. This complex has been defined as a central hub of the kinetochore, docking to centromeric chromatin and serving as a multivalent receptor for protein complexes that interact with microtubules (Ndc80, Spc105/KNL1). More recently, it has been also described as a receptor for the monopolin complex (Corbett et al., 2010; Sarkar, Shenoy, et al., 2013). Taken together, our results and those of others (Yamamoto and Hiraoka, 2003; Chikashige et al., 2004; Asakawa et al., 2005, 2007; Hayashi et al., 2006; Monje-Casas, Prabhu, et al., 2007; Corbett et al., 2010; Corbett and Harrison, 2012; Miller, Ünal, et al., 2012; Sarkar, Shenoy, et al., 2013) suggest the following model (Figure 7D). Ipl1/Aurora-B promotes the shedding of Dam1/Ndc80 complexes upon meiotic entry. This partial disassembly of the KMN network serves two purposes. First, it releases centromeres from the SPBs, liberating chromosomes to reposition themselves in the homologous pairing process. Second, removing the Ndc80 complex exposes the Mtw1 complex. This then facilitates the loading of the monopolin complex and allows the cross-bridging of Mtw1 complexes originating from the two sister kinetochores. To avoid premature bipolar attachment of the sister chromatids, this cross-bridging needs to occur before the chromosomes reattach. Thus we speculate that Ndc80 complexes only reload on kinetochores when the foundation of the kinetochore is already modified.
Although there are clear similarities in kinetochore restructuring among different organisms, the regulation shows some clear differences. Ipl1 is required for the shedding of outer kinetochores upon meiotic entry but appears dispensable for reassembly in meiotic prometaphase. Conversely, CDK seems to play minor roles in kinetochore shedding (its role in reassembly is not known). In contrast, in mammals, mitotic shedding seems to require CDK activity (Gascoigne and Cheeseman, 2013), and reassembly is promoted by Aurora kinase (Emanuele et al., 2008). In budding yeast, an obvious unanswered question involves the different behaviors of outer kinetochores in meiosis and mitosis. Ipl1 is present in mitotic cells, yet there is no long-term loss of the outer kinetochores. How is this behavior restricted to meiotic prophase?
Our results also demonstrate that throughout meiosis, the inner-central kinetochore components stay on the kinetochore. As suggested by previous work (Brar and Amon, 2008), by remaining on the chromosome, these complexes probably serve as a platform for multiple centromeric functions. These include centromere coupling and pairing (Kemp et al., 2004; Mehta et al., 2014), centromeric crossover repression (Lambie and Roeder, 1986), and centromeric DNA replication and pericentromeric cohesion establishment (Ghosh et al., 2004; Marston et al., 2004; Fernius and Marston, 2009; Fernius et al., 2013; Eckert et al., 2007; Ng et al., 2009; Natsume et al., 2013). These results suggest that the Ndc80 complex is dispensable for these functions. Indeed, Ctf19/COMA and Iml3/Chl4 subcomplexes have been shown to be critical for the particular role of establishing pericentromeric cohesion, and we find that these persist on kinetochores through meiosis I. It will be interesting to determine whether outer kinetochore shedding is necessary for the proper execution of some of those meiotic centromeric functions.
MATERIALS AND METHODS
Yeast strains and culture conditions
The strains used in these experiments are isogenic derivatives of two strains termed X and Y, which are rapidly sporulating strains from primarily S288C and W303 ancestry, derived in the R. E. Esposito, University of Chicago, laboratory (Dresser et al., 1994). Strain genotypes are listed in Supplemental Tables S1 and S2 We used standard yeast culture methods (Burke et al., 2000). To induce meiosis, cells were grown at 30°C until saturation and switched in yeast extract/peptone (YP)-acetate at ∼2 × 106 cells/ml, grown to (4–4.5) × 107 cells/ml, and then shifted to 1% potassium acetate at 108 cells/ml. The inhibitor of Cdc28-as1 (1NM-PP1; Calbiochem, San Diego, CA; 5 mM stock in dimethyl sulfoxide [DMSO]) was added when indicated at a concentration of 5 μM. For disrupting kinetochore–microtubule associations, benomyl (#45339; Sigma-Aldrich, St Louis, MO) was used alone at 120 μg/ml (Hochwagen et al., 2005b) or in combination with nocodazole (#M1404; Sigma-Aldrich; 30 μg/ml for benomyl and 15 μg/ml for nocodazole), as previously described (Fernius and Marston, 2009).
Strain construction
PCR-based methods were used to create complete deletions of open reading frames (ndt80::LEU2), epitope tags (DAM1-yeGFP-TRP1, MAM1-yeGFP-TRP1, SPC24-yeGFP-TRP1, NUF2-yeGFP-TRP1, NDC80-eGFP-TRP1, NSL1-yeGFP-TRP1, DSN1-yeGFP-TRP1, MTW1-GFP-URA3, CNN1-yeGFP-TRP1, CTF19-yeGFP-TRP1, CTF3-yeGFP-TRP1, CHL4-yeGFP-TRP1, NDC10-yeGFP-TRP1, MIF2-yeGFP-TRP1), and promoter insertions (natNT2::PGAL1-NDT80, KanMX::PGAL1-NDT80; Longtine et al., 1998; Janke et al., 2004). The MTW1-3xmCherry-hphNT1, PGPD1-GAL4(848)-ER-URA3::hphNT1, SPC42-DsRed-URA3, PCLB2-3HA-NDC80 (ndc80-md), PCLB2-3HA-IPL1 (ipl1-md), and ipl1ΔKAN:ipl1-as5-MYC:HIS3:LEU2 (ipl1-as5) strains were described previously (Meyer et al., 2013). The cdc28-as1 strain was constructed by two-step gene replacement (Burke et al., 2000) using OPL148. OPL148 was built by ligating cdc28-as1 (excised from pDrive-cdc28-as1, a gift from the Haase lab, Duke University) into pRS406. cdc28-as1 was cloned into the pDrive vector by Laura Simmons Kovacs, Duke University, using a strain from David Morgan's laboratory, University of California, San Francisco. Dsn1-3xFLAG was constructed using a PCR-based integration system with primers OPR693, OPR694, and template OPL167 (p2L-3XFLAG-kanMX, a gift from Toshio Tsukiyama, Fred Hutchinson Cancer Research Center). For the meiotic depletion of Cdc6 (cdc6-md), the PSCC1-CDC6 strain was constructed by a one-step PCR-based gene replacement method using as a template KanMX6-PSCC1 plasmid (p502; a gift from Angelika Amon, Massachusetts Institute of Technology), which carries the mitosis-specific SCC1 promoter (−1000 to +6 of the SCC1 gene).
Generation and analysis of postpachytene cultures
To examine postpachytene cells, we eliminated the asynchrony caused by the variation in timing of entry into the meiotic program by reversibly arresting cells in pachytene using an inducible allele of NDT80 (Benjamin et al., 2003; Carlile and Amon, 2008). With the use of the PGAL1-NDT80 GAL4-ER system, cells were allowed to exit pachytene by addition of 5 μM β-estradiol (Sigma-Aldrich; 5 mM, stock in ethanol) to the medium at 6 h. Inactivation of Ipl1-as5 (for the late inactivation) was done by adding 50 μM of the inhibitor 1NA-PP1 (Tocris, Minneapolis, MN; 10 mM stock in DMSO) to the medium at the same time.
Fluorescence microscopy
Images were collected using a Roper CoolSNAP HQ2 camera on a Zeiss Axio Imager 7.1 with a 100×/1.4 numerical aperture (NA) objective or a Zeiss Axioplan 2ie fitted with a 63×/1.4 NA objective microscope. Images were processed and analyzed using AxioVision software. Gamma setting adjustments of the entire image were made to enhance the visibility of weak signals (e.g., GFP- and 3xmCherry-tagged kinetochore components). The localization of kinetochore components tagged with GFP/RFP (eGFP, yeGFP, or 3xmCherry) were performed after fixing cells for 5 min in 2% formaldehyde and washing one time in phosphate-buffered saline (PBS). These cells were stained with 4′,6-diamidino-2-phenylindole, washed once with PBS, and mounted for viewing. Meiotic nuclear spreads were prepared according to Dresser and Giroux (1988) with the following modifications. Cells were spheroplasted using 20 mg/ml Zymolyase 100T for ∼30 min. Spheroplasts were briefly suspended in MEM (100 mM 2-(N-morpholino)ethanesulfonic acid, 10 mM EDTA, 500 uM MgCl2) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), fixed with 4% paraformaldehyde plus 0.1% Tween-20, and spread onto poly-l-lysine–coated slides (Fisherbrand Superfrost Plus). Slides were blocked with 4% nonfat dry milk in PBS for at least 30 min and incubated overnight at 4°C with primary antibodies. Indirect immunofluorescence was performed to detect the following epitopes: Mtw1-13MYC with primary antibody mouse anti-MYC (9E10; a gift from Susannah Rankin, Oklahoma Medical Research Foundation; 1:1000 dilution), Zip1 with preadsorbed primary antibody (rabbit anti-Zip1; SC 33733, Santa Cruz Biotechnology, Dallas, TX; 1:1000 dilution), and Ndc80-eGFP with preadsorbed primary antibody (chicken anti-GFP; AB16901, Chemicon/Millipore; 1:500 dilution). Secondary antibodies were Alexa Fluor 488–conjugated goat anti-chicken, Alexa Fluor 568–conjugated goat anti-mouse, and Alexa Fluor 647–conjugated goat anti-rabbit (all from Molecular Probes, Grand Island, NY; dilution 1:1000).
Kinetochore protein purification
Kinetochore particle isolation was carried out by affinity purifying Dsn1-3XFLAG using a purification protocol adapted from Sue Biggin's lab (Akiyoshi, Sarangapani, Powers, et al., 2010). Diploid cells were grown in YP-adenine/dextrose overnight at 30°C until saturation, switched to YP-acetate for presporulation, and finally shifted into 1% potassium acetate at 108 cells/ml. Samples were harvested at the desired time points. Cells were then pelleted by centrifugation. Pellets were washed once with distilled, deionized H2O supplemented with 0.2 mM PMSF and repelleted. To prepare cell lysates, equal volumes of zirconium oxide beads and buffer H/PI/PI (Akiyoshi, Sarangapani, Powers, et al., 2010) were added to the cell pellet. Cells were homogenized in a Blue Bullet Blender 50 (Averill Park, NY) for 15 min at speed 10 in the cold (4°C) and then put on ice for 5 min and homogenized for an additional 6 min. The supernatant was then transferred to a Beckman centrifuge tube (Ultra-Clear, 344057; Brea, CA). Ultracentrifugation was done at 24,000 rpm for 90 min at 4°C in an SW55ti Beckman rotor. Clear extract was isolated with a syringe needle inserted through the tube wall at the bottom of the clear layer.
A 60-μl amount of homogeneously suspended Dynabeads coupled to protein G (#100.04D; LifeTech, Grand Island, NY) was washed twice with citrate phosphate buffer, pH 5.0. A 30-μl amount of anti-FLAG M2 (F1804; Sigma-Aldrich) diluted in 70 μl of PBS/Tween (PBST), pH 7.4, was then added to the washed beads and incubated overnight at 4°C with gentle rotation. Beads were washed twice with PBST and once with buffer H/0.15 (25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 8.0, 2 mM MgCl2, 0.1 mM EDTA, 0.5 mM ethylene glycol tetraacetic acid, 0.1% NP-40, 150 mM KCl, 15% glycerol; Akiyoshi, Sarangapani, Powers, et al., 2010). Finally, cell extract (∼4 ml) was added to the beads and incubated at 4°C for 4 h. Beads were then washed four times with buffer H/0.15 supplemented with protease inhibitors (10 μl/ml leupeptin, 10 μl/ml pepstatin, 10 μl/ml chymostatin, 0.2 mM PMSF), phosphatase inhibitors (1 mM sodium pyrophosphate, 2 mM Na-β-glycerophosphate, 0.1 mM NA3VO4, 5 mM NaF, 100 mM microcystin-LR), and 2 mM dithiothreitol (DTT) and twice with buffer H/0.15 containing protease inhibitors. Finally, associated proteins were eluted off the beads with ∼55 μl of elution buffer (0.5 mg/ml 3XFLAG peptide in buffer H/0.15 with protease inhibitors added).
Mass spectrometry
The eluate containing the purified kinetochores was mixed with 8 pmol of bovine serum albumin (BSA) as an internal standard in 50 μl of 1% SDS. The sample was heated at 80°C for 15 min to equilibrate and cooled to room temperature, and 1 ml of ice-cold acetone was added to precipitate the protein overnight. The precipitated protein was pelleted by microcentrifuge at 10,000 × g for 10 min, the supernatant gently poured away, and residual acetone removed in a SpeedVac. The protein pellet was dissolved in 20 μl of Laemmli buffer containing DTT and heated at 80°C for 15 min. The entire sample was loaded into a 12.5% Criterion Gel (Bio-Rad, Hercules, CA) and the gel run for 15 min at 150 V. This short-run gel electrophoresis moved the proteins ∼1.5 cm into the gel with some separation. The gel was fixed with 50% ethanol/10% acetic acid in water for 30 min, equilibrated into water, and stained briefly (5 min) to visualize the proteins. The gel was then soaked in water overnight with several changes to wash away all salts and the 3XFLAG peptide.
Each lane was cut from the gel as a single sample. These large gel pieces, ∼1.5 cm × 0.7 cm, were cut into 10–12 smaller pieces and completely destained in 50% ethanol/10% acetic acid in water at 50°C with multiple changes as needed. The proteins were reduced with DTT (10 mM) and alkylated with iodoacetamide (50 mM) before digestion with 1 μg trypsin (Promega, Madison, WI) in 200 μl of 10 mM ammonium bicarbonate overnight at room temperature. The peptides produced by the digestion were extracted from the sample with 2 × 200 μl of 50% methanol/10% formic acid in water. The extracts were evaporated to dryness in a SpeedVac and reconstituted in 50 μl of 1% acetic acid for the LC-tandem MS analysis.
The samples were analyzed on an LTQ-Vantage triple quadrupole mass spectrometer system (ThermoScientific, Tewksbury, MA) coupled to an Eksigent NanoLC. A 10 cm × 75 μm reversed-phase column was used (self-packed with Phenomonex Jupiter C18). The column was eluted by a linear gradient of acetonitrile in 0.1% formic acid (3–43% in 40 min) at 150 nl/min. The mass spectrometer was operated in the selected reaction (SRM) mode, recording the elution of 62 peptides for 21 proteins, including the BSA internal standard and two trypsin autolysis peptides. Time scheduling was used to help maximize the dwell time. The specific peptides that were monitored were determined by a series of method development experiments designed to use the three peptides that gave the best response for each protein (Kinter and Kinter, 2013). The three peptides that were monitored to estimate the abundance of each kinetochore protein monitored are listed in Supplemental Table S3. Overall a single multiplexed assay monitored 419 reactions for the 62 peptides used to determine the amounts of these proteins. The program Pinpoint was used to integrate the chromatographic peak area for all peptides. Representative chromatographic peaks for each peptide are shown in Supplemental Figure S4. Data were analyzed by using the total area of all peptides for a given protein to calculate the total protein response (Kinter and Kinter, 2013). These responses were normalized to the response for Dsn1, with normalization to the BSA used as confirmation.
Western blots
Cells harvested from meiotic time courses (1 ml/time point) were disrupted in cold 16.6% trichloroacetic acid in a Bullet Blue Blender following the manufacturer's instructions. Protein precipitates were washed in 95% ethanol, pelleted in a microcentrifuge, and resuspended in SDS-PAGE buffer. Equal volumes of each sample were loaded on the gel. Blotted proteins were detected using primary antibodies against PGK1 (22C5; Molecular Probes; 1:10,000) and GFP (12600500; Roche, Basel, Switzerland; 1:2000).
Supplementary Material
Acknowledgments
We thank Saili Moghe, Jingrong Chen, and Susannah Rankin for help with Western blots, members of the Dawson laboratory, past and present, for the use of shared strains and reagents, and members of the Dawson laboratory and the Program in Cell Cycle and Cancer Biology for their many contributions to the development of this project. We thank Elçin Ünal for helpful comments on the data. We thank Sue Biggins for essential reagents and advice on kinetochore purification and Toshio Tsukiyama, David Morgan, Mark Chee, Steve Haase, and Angelika Amon for plasmids. This project was supported by National Institutes of Health Grant R01GM087377 and National Science Foundation Grant MCB 0950005.
Abbreviations used:
- MS-SRM
mass spectrometry selective reaction monitoring
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-01-0032) on July 8, 2015.
REFERENCES
Boldface names denote co–first authors.
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa H, Haraguchi T, Hiraoka Y. Reconstruction of the kinetochore: a prelude to meiosis. Cell Div. 2007;2:17. doi: 10.1186/1747-1028-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa H, Hayashi A, Haraguchi T, Hiraoka Y. Dissociation of the Nuf2-Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast. Mol Biol Cell. 2005;16:2325–2338. doi: 10.1091/mbc.E04-11-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin KR, Zhang C, Shokat KM, Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Blitzblau HG, Chan CS, Hochwagen A, Bell SP. Separation of DNA replication from the assembly of break-competent meiotic chromosomes. PLoS Genet. 2012;8:e1002643. doi: 10.1371/journal.pgen.1002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock LJ, Pagliuca C, Kobayashi N, Grove RA, Oku Y, Shrestha K, Alfieri C, Golfieri C, Oldani A, Dal Maschio M, et al. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat Cell Biol. 2012;14:614–624. doi: 10.1038/ncb2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Amon A. Emerging roles for centromeres in meiosis I chromosome segregation. Nat Rev Genetics. 2008;9:899–910. doi: 10.1038/nrg2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Hochwagen A, Ee LS, Amon A. The multiple roles of cohesin in meiotic chromosome morphogenesis and pairing. Mol Biol Cell. 2009;20:1030–1047. doi: 10.1091/mbc.E08-06-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D Dawson D, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Carlile TM, Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Marquez-Lema A, Bhatt AM, Horlow C, et al. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci. 2005;118:4621–4632. doi: 10.1242/jcs.02583. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Kurokawa R, Haraguchi T, Hiraoka Y. Meiosis induced by inactivation of Pat1 kinase proceeds with aberrant nuclear positioning of centromeres in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2004;9:671–684. doi: 10.1111/j.1356-9597.2004.00760.x. [DOI] [PubMed] [Google Scholar]
- Chuong H, Dawson DS. Meiotic cohesin promotes pairing of nonhomologous centromeres in early meiotic prophase. Mol Biol Cell. 2010;21:1799–1809. doi: 10.1091/mbc.E09-05-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133:1175–1187. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- Corbett KD, Harrison SC. Molecular architecture of the yeast monopolin complex. Cell Rep. 2012;1:583–589. doi: 10.1016/j.celrep.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Yip CK, Ee LS, Walz T, Amon A, Harrison SC. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010;142:556–567. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn JF, Maddox PS. Kinetochore dynamics: how protein dynamics affect chromosome segregation. Curr Opin Cell Biol. 2012;24:57–63. doi: 10.1016/j.ceb.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Dresser ME, Ewing DJ, Harwell SN, Coody D, Conrad MN. Nonhomologous synapsis and reduced crossing over in a heterozygous paracentric inversion in Saccharomyces cerevisiae. Genetics. 1994;138:633–647. doi: 10.1093/genetics/138.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresser ME, Giroux CN. Meiotic chromosome behavior in spread preparations of yeast. J Cell Biol. 1988;106:567–573. doi: 10.1083/jcb.106.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert CA, Gravdahl DJ, Megee PC. The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 2007;21:278–291. doi: 10.1101/gad.1498707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Lan W, Jwa M, Miller SA, Chan CS, Stukenberg PT. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol. 2008;181:241–254. doi: 10.1083/jcb.200710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernius J, Marston AL. Establishment of cohesion at the pericentromere by the Ctf19 kinetochore subcomplex and the replication fork-associated factor, Csm3. PLoS Genet. 2009;5:e1000629. doi: 10.1371/journal.pgen.1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernius J, Nerusheva OO, Galander S, Alves Fde L, Rappsilber J, Marston AL. Cohesin-dependent association of scc2/4 with the centromere initiates pericentromeric cohesion establishment. Curr Biol. 2013;23:599–606. doi: 10.1016/j.cub.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Cheeseman IM. Kinetochore assembly: if you build it, they will come. Curr Opin Cell Biol. 2011;23:102–108. doi: 10.1016/j.ceb.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Cheeseman IM. CDK-dependent phosphorylation and nuclear exclusion coordinately control kinetochore assembly state. J Cell Biol. 2013;201:23–32. doi: 10.1083/jcb.201301006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SK, Sau S, Lahiri S, Lohia A, Sinha P. The Iml3 protein of the budding yeast is required for the prevention of precocious sister chromatid separation in meiosis I and for sister chromatid disjunction in meiosis II. Curr Genet. 2004;46:82–91. doi: 10.1007/s00294-004-0516-6. [DOI] [PubMed] [Google Scholar]
- Gladstone MN, Obeso D, Chuong H, Dawson DS. The synaptonemal complex protein Zip1 promotes bi-orientation of centromeres at meiosis I. PLoS Genet. 2009;5:e1000771. doi: 10.1371/journal.pgen.1000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Reed SI. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol Cell Biol. 1993;13:2113–2125. doi: 10.1128/mcb.13.4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genetics. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Hayashi A, Asakawa H, Haraguchi T, Hiraoka Y. Reconstruction of the kinetochore during meiosis in fission yeast Schizosaccharomyces pombe. Mol Biol Cell. 2006;17:5173–5184. doi: 10.1091/mbc.E06-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi A, Ogawa H, Kohno K, Gasser SM, Hiraoka Y. Meiotic behaviours of chromosomes and microtubules in budding yeast: relocalization of centromeres and telomeres during meiotic prophase. Genes Cells. 1998;3:587–601. doi: 10.1046/j.1365-2443.1998.00215.x. [DOI] [PubMed] [Google Scholar]
- Hochwagen A, Tham WH, Brar GA, Amon A. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell. 2005a;122:861–873. doi: 10.1016/j.cell.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Hochwagen A, Wrobel G, Cartron M, Demougin P, Niederhauser-Wiederkehr C, Boselli MG, Primig M, Amon A. Novel response to microtubule perturbation in meiosis. Mol Cell Biol. 2005b;25:4767–4781. doi: 10.1128/MCB.25.11.4767-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Haraguchi T, Hiraoka Y, Kimura H, Fukagawa T. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J Cell Sci. 2003;116:3347–3362. doi: 10.1242/jcs.00645. [DOI] [PubMed] [Google Scholar]
- Howe M, McDonald KL, Albertson DG, Meyer BJ. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J Cell Biol. 2001;153:1227–1238. doi: 10.1083/jcb.153.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jin Q-W, Trelles-Sticken E, Scherthan H, Loidl J. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J Cell Biol. 1998;141:21–29. doi: 10.1083/jcb.141.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Copsey A, Newnham L, Kolar E, Lichten M, Hoffmann E. Ipl1/Aurora B kinase coordinates synaptonemal complex disassembly with cell cycle progression and crossover formation in budding yeast meiosis. Genes Dev. 2009;23:2237–2251. doi: 10.1101/gad.536109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamieniecki RJ, Shanks RM, Dawson DS. Slk19p is necessary to prevent separation of sister chromatids in meiosis I. Curr Biol. 2000;10:1182–1190. doi: 10.1016/s0960-9822(00)00723-5. [DOI] [PubMed] [Google Scholar]
- Kemp B, Boumil RM, Stewart MN, Dawson DS. A role for centromere pairing in meiotic chromosome segregation. Genes Dev. 2004;18:1946–1951. doi: 10.1101/gad.1227304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Meyer R, Chuong H, Dawson DS. Dual mechanisms prevent premature chromosome segregation during meiosis. Genes Dev. 2013;27:2139–2146. doi: 10.1101/gad.227454.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinter M, Kinter CS. Application of Selected Reaction Monitoring to Highly Multiplexed Targeted Quantitative Proteomics: A Replacement for Western Blot Analysis. New York: Springer; 2013. [Google Scholar]
- Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188–1201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie EJ, Roeder GS. Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics. 1986;114:769–789. doi: 10.1093/genetics/114.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F, Westermann S. A blueprint for kinetochores— new insights into the molecular mechanics of cell division. Nat Rev Mol Cell Biol. 2011;12:407–412. doi: 10.1038/nrm3133. [DOI] [PubMed] [Google Scholar]
- Lee BH, Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- Lo HC, Wan L, Rosebrock A, Futcher B, Hollingsworth NM. Cdc7-Dbf4 regulates NDT80 transcription as well as reductional segregation during budding yeast meiosis. Mol Biol Cell. 2008;19:4956–4967. doi: 10.1091/mbc.E08-07-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N, Ceto S, Ranish JA, Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr Biol. 2012;22:900–906. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Malvezzi F, Litos G, Schleiffer A, Heuck A, Mechtler K, Clausen T, Westermann S. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 2013;32:409–423. doi: 10.1038/emboj.2012.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Tham WH, Shah H, Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, Junqueira M, Shevchenko A, Zachariae W. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell. 2008;135:662–678. doi: 10.1016/j.cell.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Mehta GD, Agarwal M, Ghosh SK. Functional characterization of kinetochore protein, Ctf19 in meiosis I: an implication of differential impact of Ctf19 on the assembly of mitotic and meiotic kinetochores in Saccharomyces cerevisiae. Mol Microbiol. 2014;91:1179–1199. doi: 10.1111/mmi.12527. [DOI] [PubMed] [Google Scholar]
- Meyer RE, Kim S, Obeso D, Straight PD, Winey M, Dawson DS. Mps1 and Ipl1/Aurora B act sequentially to correctly orient chromosomes on the meiotic spindle of budding yeast. Science. 2013;339:1071–1074. doi: 10.1126/science.1232518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MP, Ünal E, Brar GA, Amon A. Meiosis I chromosome segregation is established through regulation of microtubule-kinetochore interactions. Elife. 2012;1:e00117. doi: 10.7554/eLife.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 2001;110:322–334. doi: 10.1007/s004120100153. [DOI] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nature reviews. Genetics. 2012;13:493–504. doi: 10.1038/nrg3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T, Muller CA, Katou Y, Retkute R, Gierlinski M, Araki H, Blow JJ, Shirahige K, Nieduszynski CA, Tanaka TU. Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol Cell. 2013;50:661–674. doi: 10.1016/j.molcel.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnham L, Jordan PW, Carballo JA, Newcombe S, Hoffmann E. Ipl1/Aurora kinase suppresses S-CDK-driven spindle formation during prophase I to ensure chromosome integrity during meiosis. PLoS One. 2013;8:e83982. doi: 10.1371/journal.pone.0083982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TM, Waples WG, Lavoie BD, Biggins S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol Biol Cell. 2009;20:3818–3827. doi: 10.1091/mbc.E09-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra MT, Gomez R, Viera A, Llano E, Pendas AM, Rufas JS, Suja JA. Sequential assembly of centromeric proteins in male mouse meiosis. PLoS Genet. 2009;5:e1000417. doi: 10.1371/journal.pgen.1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- Sarangapani KK, Duro E, Deng Y, Alves Fde L, Ye Q, Opoku KN, Ceto S, Rappsilber J, Corbett KD, Biggins S, Marston AL, Asbury CL. Sister kinetochores are mechanically fused during meiosis I in yeast. Science. 2014;346:248–251. doi: 10.1126/science.1256729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Shenoy RT, Dalgaard JZ, Newnham L, Hoffmann E, Millar JB, Arumugam P. Monopolin subunit Csm1 associates with MIND complex to establish monopolar attachment of sister kinetochores at meiosis I. PLoS Genet. 2013;9:e1003610. doi: 10.1371/journal.pgen.1003610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol. 2012;14:604–613. doi: 10.1038/ncb2493. [DOI] [PubMed] [Google Scholar]
- Shepperd LA, Meadows JC, Sochaj AM, Lancaster TC, Zou J, Buttrick GJ, Rappsilber J, Hardwick KG, Millar JB. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr Biol. 2012;22:891–899. doi: 10.1016/j.cub.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirk K, Jin H, Giddings TH, Jr, Winey M, Yu HG. The Aurora kinase Ipl1 is necessary for spindle pole body cohesion during budding yeast meiosis. J Cell Sci. 2011;124:2891–2896. doi: 10.1242/jcs.086652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster EO, Byers B. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics. 1989;123:29–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC, Zhang DX, Lee SE, Xu YN, Kim NH. Ndc80 regulates meiotic spindle organization, chromosome alignment, and cell cycle progression in mouse oocytes. Microsc Microanal. 2011;17:431–439. doi: 10.1017/S1431927611000274. [DOI] [PubMed] [Google Scholar]
- Tachibana-Konwalski K, Godwin J, Borsos M, Rattani A, Adams DJ, Nasmyth K. Spindle assembly checkpoint of oocytes depends on a kinetochore structure determined by cohesin in meiosis I. Curr Biol. 2013;23:2534–2539. doi: 10.1016/j.cub.2013.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- Tsubouchi T, Roeder GS. A synaptonemal complex protein promotes homology-independent centromere coupling. Science. 2005;308:870–873. doi: 10.1126/science.1108283. [DOI] [PubMed] [Google Scholar]
- Watanabe Y. Geometry and force behind kinetochore orientation: lessons from meiosis. Nat Rev Mol Cell Biol. 2012;13:370–382. doi: 10.1038/nrm3349. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yokobayashi S, Yamamoto M, Nurse P. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature. 2001;409:359–363. doi: 10.1038/35053103. [DOI] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Westhorpe FG, Straight AF. Functions of the centromere and kinetochore in chromosome segregation. Curr Opin Cell Biol. 2013;25:334–340. doi: 10.1016/j.ceb.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 2003;22:2284–2296. doi: 10.1093/emboj/cdg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Watanabe Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 2005;123:803–817. doi: 10.1016/j.cell.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Yokobayashi S, Yamamoto M, Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol. 2003;23:3965–3973. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.