Complementation of a conditional KO of actin-depolymerizing factor (ADF) in Toxoplasma gondii demonstrates that ADF-dependent actin filament disassembly is essential for parasite development but not for cell motility. Furthermore, trans-genera complementation highlights genus-specific coevolution between ADF proteins and their native actins.
Abstract
Proteins of the actin-depolymerizing factor (ADF)/cofilin family have been shown to be crucial for the motility and survival of apicomplexan parasites. However, the mechanisms by which ADF proteins fulfill their function remain poorly understood. In this study, we investigate the comparative activities of ADF proteins from Toxoplasma gondii and Plasmodium falciparum, the human malaria parasite, using a conditional T. gondii ADF-knockout line complemented with ADF variants from either species. We show that P. falciparum ADF1 can fully restore native TgADF activity, demonstrating functional conservation between parasites. Strikingly, mutation of a key basic residue (Lys-72), previously implicated in disassembly in PfADF1, had no detectable phenotypic effect on parasite growth, motility, or development. In contrast, organelle segregation was severely impaired when complementing with a TgADF mutant lacking the corresponding residue (Lys-68). Biochemical analyses of each ADF protein confirmed the reduced ability of lysine mutants to mediate actin depolymerization via filament disassembly although not severing, in contrast to previous reports. These data suggest that actin filament disassembly is essential for apicomplexan parasite development but not for motility, as well as pointing to genus-specific coevolution between ADF proteins and their native actin.
INTRODUCTION
Apicomplexan parasites are obligate intracellular protozoa responsible for a diverse range of diseases in humans and other animals. The ancient phylum includes the widespread and opportunistic pathogen Toxoplasma gondii, which is responsible for toxoplasmosis, and the malaria parasite, Plasmodium falciparum, one of the leading causes of infant mortality in the developing world (White et al., 2014). Key to life cycle progression and pathobiology of these organisms is their ability to move and infect host cells, a process achieved by specialized motile stages termed -zoites (Baum et al., 2008).
Detailed studies over the past decade have revealed conserved molecular mechanisms underlying motility and invasion in T. gondii and both human and murine Plasmodium -zoites. Key conserved mechanisms include the engagement of an actomyosin motor complex that is unique to Apicomplexa (Dobrowolski et al., 1997a; Herm-Gotz et al., 2002; Meissner et al., 2002; Baum et al., 2006a) and is assembled in the alveolar space between the inner membrane complex and parasite plasma membrane (Bergman et al., 2003; Jones et al., 2006; Bullen et al., 2009; Frénal et al., 2010). Within this space, myosin is believed to engage short, highly dynamic actin filaments, thereby creating a rearward traction force that drives the -zoite forward (Kappe et al., 2004; Baum et al., 2006b).
A key feature of this model is the unconventional nature of apicomplexan actin, which is believed to exist predominantly in a monomeric state (G-actin) and is capable of forming only short and unstable microfilaments (F-actin; Dobrowolski et al., 1997b; Schmitz et al., 2005; Sahoo et al., 2006; Skillman et al., 2011). Treatment of parasites with either an actin filament–capping compound, such as cytochalasin D, or the F-actin–stabilizing drug jasplakinolide (JAS) inhibits motility and invasion, the latter inducing a prominent accumulation of actin in the periphery and apical region of Toxoplasma and Plasmodium -zoites (Shaw and Tilney, 1999; Mizuno et al., 2002; Angrisano et al., 2012). These observations not only highlight the presence of dynamic microfilaments within the alveolar space and apex, but they also emphasize the essential role that actin turnover (both polymerization and depolymerization) plays in facilitating efficient parasite motility and host cell invasion.
The spatiotemporal turnover of actin is highly regulated in the majority of eukaryotic cells (reviewed in Carlier and Pantaloni, 2007). Only a minimal set of actin-modulating proteins, however, has been identified in Apicomplexa, including members of the actin-depolymerizing factor (ADF)/cofilin family (Allen et al., 1997; Schüler et al., 2005; Schuler and Matuschewski, 2006; Baum et al., 2006a). ADF/cofilin proteins play a pivotal role in the morphogenesis and movement of eukaryotic cells and interact with both G- and F-actin, thus regulating the homeostasis by sequestering actin monomers and disassembling actin filaments (reviewed in Bamburg and Bernstein, 2010). Whereas most Apicomplexa encode only a single ADF protein, Plasmodium spp. express two isoforms, PfADF1 and PfADF2, the latter resembling conventional ADF (having an extended filament-binding or F-loop) but expressed minimally and possibly nonfunctional in -zoite stages (Schüler et al., 2005; Singh et al., 2011; Wong et al., 2011). Attempts to disrupt the adf1 locus in the rodent malaria parasite Plasmodium berghei have been unsuccessful, indicating a likely essential role for ADF1 in the asexual development of Plasmodium spp. (Schüler et al., 2005). Critically, down-regulation of the orthologue in T. gondii (TgADF) has proven to greatly impair cell motility and parasite propagation (Mehta and Sibley, 2011), strongly indicating a requirement for regulated actin disassembly in divergent apicomplexan cellular processes from motility to cell development.
The biochemical properties of PfADF1 and TgADF greatly differ from those of other eukaryotic ADF/cofilin proteins. Most strikingly, PfADF1 and TgADF have a low affinity for F-actin and preferentially bind to actin monomers, possibly reflecting the dominating G-actin environment within the apicomplexan cell (Schüler et al., 2005; Mehta and Sibley, 2010). Nevertheless, both are efficient in severing actin filaments in vitro (Mehta and Sibley, 2010; Wong et al., 2011), a central mechanism by which ADF/cofilin proteins mediate actin disassembly (Ono, 2007). They do this despite a significant reduction in the F-actin binding domain (F-loop) compared with other eukaryotic ADF/cofilin proteins (Mehta and Sibley, 2010; Singh et al., 2011; Wong et al., 2011). This domain is known to be critical for actin filament decoration and was postulated to be the mechanism by which severing occurs (Suarez et al., 2011). However, a study revealed a structurally conserved basic residue at the base of the F-loop to be important for PfADF1 severing activity in vitro, suggesting that severing may be independent of the F-loop (Wong et al., 2011).
Given the paucity of strategies for complementing an essential gene in Plasmodium parasites and based on the proposed high structural similarity between PfADF1 and TgADF (Yadav et al., 2011), we used a previously described TgADF conditional knockout (cKO) line (Mehta and Sibley, 2011) to examine whether trans-genera complementation of wild-type TgADF was possible with PfADF1. We further wanted to explore whether complementation could confirm the functional role of ADF proteins in the native context, including in vivo verification of the importance of the minimal F-actin binding motif at the base of the F-loop (Lys-72 in PfADF1 and Lys-68 in TgADF) in regulating actin filament turnover.
RESULTS
Native and trans-genera complementation of TgADF on a conditional knockout background
To explore the in vivo function of the Plasmodium ADF1 protein on a TgADF cKO background (Mehta and Sibley, 2011), we attempted complementation via random integration of the constructs tgadf-cmyc and pfadf1-cmyc, and tgadfK68A-cmyc and pfadf1K72A-cmyc, the last two carrying mutations in the homologous residues implicated in actin filament severing (Wong et al., 2011).
Parasites were initially selected in the presence of pyrimethamine and anhydrotetracycline (ATc) before cloning out the parental lines by limiting dilution. Several clones (24–56) per cell line were obtained 7–9 d thereafter, and from each cell line, four to six clones were initially selected to investigate transgene expression of the different cMyc-fusion proteins by immunoblot and immunofluorescence analysis using anti-cMyc and parasite-specific F-actin antibodies (Angrisano et al., 2012) as a loading control. Because no apparent differences in expression levels were observed, two clones of each cell line were chosen for further analysis to account for potential differences in parasite fitness.
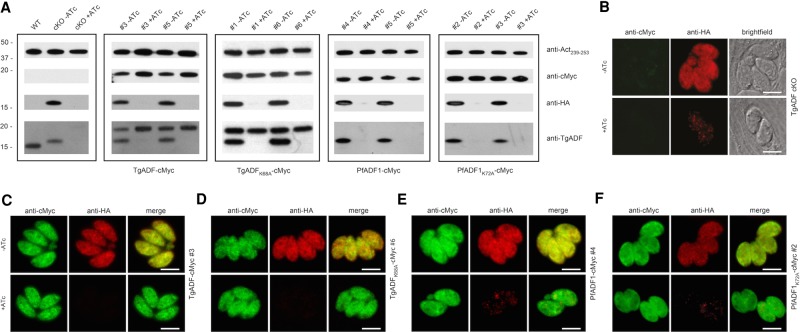
As previously shown (Mehta and Sibley, 2011), expression of the hemagglutinin (HA)-tagged TgADF replacement in the conditional knockout line was suppressed after growth in ATc for 48–50 h (Figure 1A, first panel, second and third lanes). Both clones of the respective TgADF-cMyc, PfADF1-cMyc, and mutant parasite lines expressed the corresponding fusion proteins at the predicted molecular weight of ∼18 kDa, where addition of ATc for 48–50 h did not affect the level of ectopic transgene expression (Figure 1A, second to fourth panels). TgADF antibodies, as expected, only recognized TgADF-cMyc– and TgADFK68A-cMyc–tagged proteins along with the inducible TgADF-HA copy (Figure 1A).
FIGURE 1:
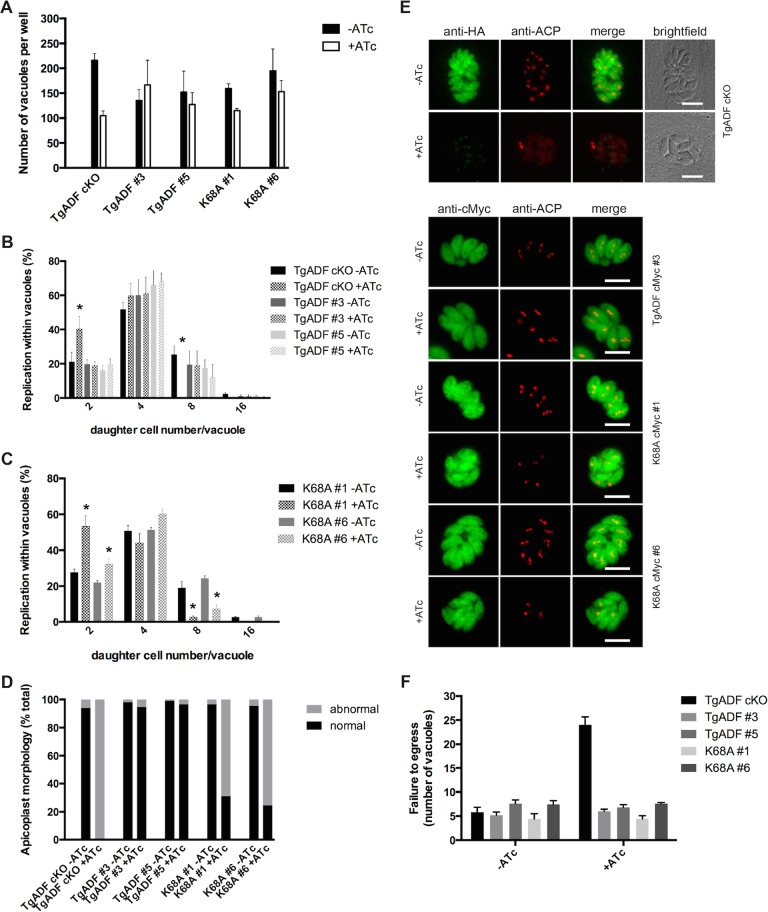
Ectopic expression of TgADF-cMyc, PfADF1-cMyc, and the corresponding Lys mutants in TgADF cKO parasites. (A) Western blot analysis of the parasite lines grown with or without ATc for 48–50 h. Parasite protein lysates were probed with anti-HA to control for down-regulation of the inducible TgADF-HA copy, as well as anti-cMyc and anti-TgADF to control for ectopic protein expression. Anti-Act239-253 was used as a loading control. (B–F) Immunofluorescence analysis of parasites grown with or without ATc demonstrates cytosolic localization of inducible TgADF-HA (A) and the complementing cMyc-tagged proteins (C–F). Scale bar, 5 μm (C–F).
Similarly, immunofluorescence analysis displayed down-regulation of TgADF-HA to barely detectable levels at 48–50 h in the presence of ATc as described before (Mehta and Sibley, 2011), whereas in the absence of ATc, the protein was evenly distributed throughout the parasite cytosol as detected by anti-HA antibodies (Figure 1B). ATc-dependent expression of the HA-tagged TgADF was retained for each of the respective TgADF-cMyc, PfADF1-cMyc, TgADFK68A-cMyc, and PfADF1K72A-cMyc lines, with the complementing protein being broadly cytosolic in both the absence and presence of ATc (Figure 1, C–F, and Supplemental Figure S2).
Wild-type and mutant PfADF1K72A can complement TgADF cKO loss of function but not TgADFK68A
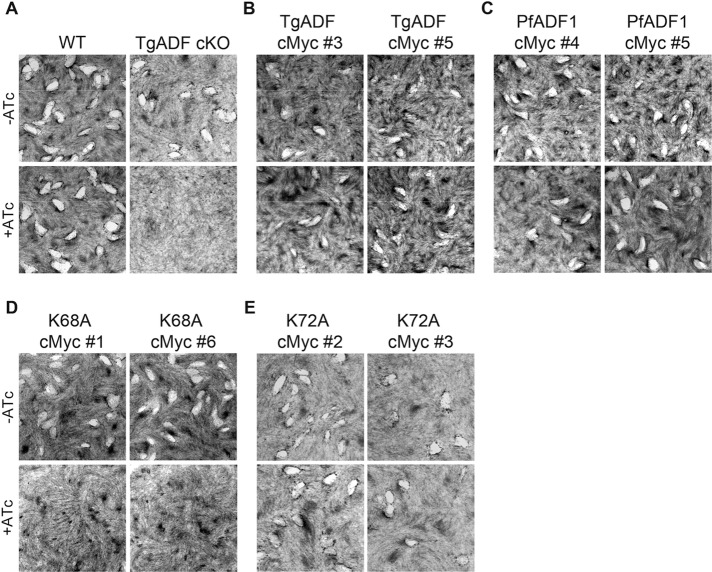
The inducible KO of TgADF has been shown to greatly impair host cell invasion and egress (Mehta and Sibley, 2011), which are important indicators of successful parasite propagation during the lytic cycle of T. gondii. Repetitive cycles of invasion, replication, and egress over several days lead to the formation of plaques due to lysis of the host cell monolayer. As such, plaque assays were performed to determine whether the KO phenotypes could be restored by the ectopic expression of wild-type TgADF-cMyc and PfADF1-cMyc.
No plaques were formed for TgADF cKO parasites grown in ATc (Figure 2A), as expected (Mehta and Sibley, 2011). Down-regulation of TgADF-HA in clones of the wild-type TgADF-cMyc and PfADF1-cMyc–expressing parasite lines, however, showed effectively wild-type levels of plaque formation, suggesting functional complementation (Figure 2, B and C). Surprisingly, no obvious differences in size and number of plaques were observed for the PfADF1K72A-cMyc mutant parasites grown in ATc (Figure 2E and Supplemental Table S1). In contrast, a mutant phenotype consistent with the full knockdown phenotype was seen with the TgADFK68A-cMyc–expressing clones (Figures 2D). Thus, by plaque assay alone, it appears that whereas the ectopically expressed wild-type proteins and the PfADF1K72A-cMyc mutant clones are functional with respect to successful parasite propagation, the loss of Lys-68 in TgADF severely affects the parasite at one or several points within the lytic cycle. The ability of the PfADF1K72A-cMyc mutant to complement native TgADF absence in contrast to the TgADFK68A mutant suggests major differences in either the activity of the ADFs or their interactions with native actin in the respective species.
FIGURE 2:
TgADFK68A-cMyc–expressing parasites cannot restore TgADF cKO loss of function. (A–E) Plaque assays of plated human fibroblasts grown with each parasite treated with or without ATc for 8 d. White plaques represent characteristic areas where host cells have been lysed due to parasite propagation. A growth defect is seen in A (bottom right) for the TgADF cKO line, as expected. A similar phenotype is seen in D (bottom for both clones), where no plaques have been formed for TgADFK68A-cMyc–expressing parasites grown in the presence of ATc. No obvious growth defect is detectable in other complementing lines.
Localization of F-actin in tachyzoites is unaffected by mutation of the exposed lysine residue of ADF
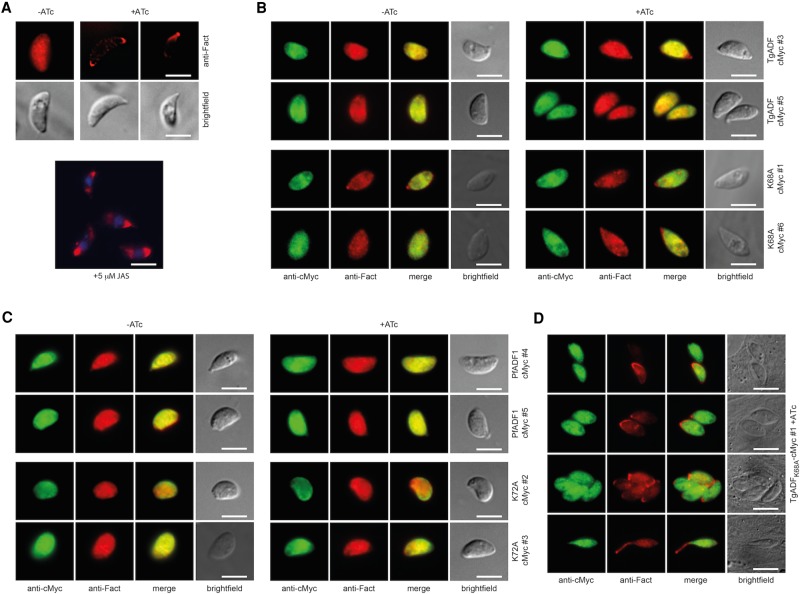
Suppression of TgADF has been shown to lead to pronounced filamentous structures or enhanced accumulation of actin at either pole of the extracellular parasite (Mehta and Sibley, 2011). To examine whether the defective phenotype of the TgADFK68A-cMyc–expressing parasites was due to perturbed interaction with actin, we performed immunofluorescence assays using a polyclonal antibody specific for apicomplexan F-actin (Angrisano et al., 2012) to analyze the distribution of actin in extracellular and intracellular parasites.
In contrast to the straight conditional knockout (Figure 3A), extracellular tachyzoites of neither the wild-type ADF-complementing lines nor the majority of parasites expressing the mutant proteins TgADFK68A and PfADF1K72A revealed obvious changes in the localization of actin in the presence of ATc (Figure 3, B and C). Similar results were found for the majority of intracellular stages, suggesting that substitution of the structurally conserved lysine to alanine does not greatly affect the overall subcellular distribution of apicomplexan actin in T. gondii parasites (Supplemental Figure S3). It is important to note that a filamentous actin distribution similar to the phenotype in TgADF-depleted or JAS-treated extracellular tachyzoites showing an apical concentration of actin (Figure 3A) was observed in ≤5% of intracellular TgADFK68A-cMyc clone #1 parasites (Figure 3D, top three rows). In addition, a prominent apical protrusion similar to apical extensions observed in JAS-treated wild-type (Shaw and Tilney, 1999) and yellow fluorescent protein-Act1–expressing tachyzoites (Wetzel et al., 2003) was exhibited by a minor population of parasites (Figure 3D, bottom row). The significance of these phenotypes in a minority population is unclear.
FIGURE 3:
Filamentous actin is distributed normally in the majority of complementing lines. (A) Parasites were grown with or without ATc for 68–72 h, and freshly released tachyzoites were fixed for immunofluorescence analysis. Suppression of TgADF-HA in the cKO line leads to an accumulation of actin at either (or both) of the two poles of the tachyzoite, which is similar to what is seen when tachyzoites are treated with JAS, a compound that stabilizes dynamic actin into filaments. (B, C) The majority of wild-type TgADF, mutant TgADFK68A, wild-type PfADF1, and PfADF1K72A-complementing clones display an even localization of F-actin throughout the cell in the absence and presence of ATc that is indistinguishable from what is observed in wild-type parasites. (D) A skewed distribution of actin is found in a small proportion (≤5%) of intracellular TgADFK68A-cMyc–expressing parasites, suggesting a polar accumulation of F-actin (as with JAS). Only very few parasites display an apical protrusion of actin (D, bottom). Scale bar, 5 μm.
Gliding motility in tachyzoites is unaffected by mutation of the exposed lysine residue of ADF
The ability to glide is a prerequisite for tachyzoite invasion and is based on the rapid turnover of apicomplexan actin. Although mutation of the minimal F-actin binding motif in TgADF and PfADF1 had no gross effect on the overall actin distribution in extracellular parasites, we next sought to investigate potential changes in parasite motility.
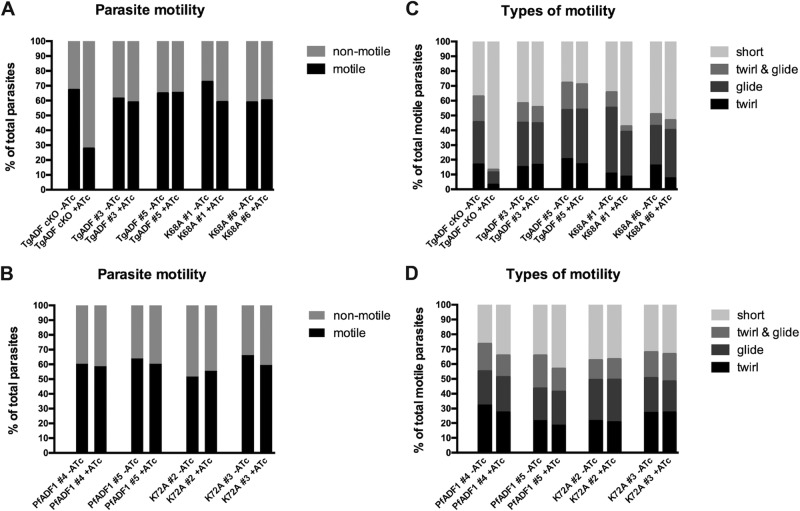
Videomicroscopy of gliding tachyzoites revealed that motility was greatly reduced upon down-regulation of TgADF in the TgADF cKO line, with ∼30% of the total parasite population displaying mainly short movements, including erratic rocking and flipping motions as described by Mehta and Sibley (2011; Figure 4A). In contrast, a broad repertoire of motility was observed for the wild-type TgADF and PfADF1-complementing lines grown in ATc, with levels of motility similar to parasites grown in the absence of ATc (Figure 4, A and B). Motility in these lines included twirling, helical gliding at similar rates of ∼0.8 μm/s (Table 1), and circular movements in different patterns (Supplemental Movie S1), as well as uncoordinated movement and short bursts of twirling or gliding motions (or both) over a brief period (mostly 10–40 s; Figure 4, C and D). The rocking and flipping motion (as predominantly described for the TgADF cKO phenotype in the presence of ATc) was also observed, but only to a much lower extent, for untreated parasites within all lines investigated, including the TgADF cKO parent line.
FIGURE 4:
Parasite gliding motility is independent of Lys68/72-mediated actin filament turnover. Parasite lines were cultured with or without ATc for 68–72 h, harvested, and allowed to glide on poly-l-lysine–coated slides. Motility was captured by time-lapse videomicroscopy. The average from two independent experiments is shown (number of parasites, on average 500 per line and clone). Only the TgADF cKO parasite demonstrates a significant reduction in motile parasites (A) or markedly aberrant motility (C) compared with the four other complementing lines (A–D).
TABLE 1:
Quantification of helical gliding motility from two independent experiments.
| Set | Helical gliding motility (μm/s)a | ||||
|---|---|---|---|---|---|
| Set 1 | TgADF cKO | TgADF #3 | TgADF #5 | TgADFK68A #1 | TgADFK68A #6 |
| −ATc | 0.82 ± 0.23 | 0.83 ± 0.20 | 0.81 ± 0.14 | 0.77 ± 0.23 | 0.75 ± 0.18 |
| +ATc | N/Ab | 0.80 ± 0.18 | 0.82 ± 0.23 | 0.74 ± 0.16 | 0.71 ± 0.15 |
| Set 2 | TgADF cKO | TgADF #3 | TgADF #5 | TgADFK68A #1 | TgADFK68A #6 |
| −ATc | 0.75 ± 0.15 | 0.79 ± 0.18 | 0.82 ± 0.12 | 0.74 ± 0.13 | 0.74 ± 0.18 |
| +ATc | N/Ab | 0.75 ± 0.18 | 0.79 ± 0.17 | 0.80 ± 0.20 | 0.74 ± 0.13 |
| Set 1 | PfADF1 #4 | PfADF1 #5 | PfADF1K72A #2 | PfADF1K72A #3 | |
| −ATc | 0.83 ± 0.38 | 0.84 ± 0.29 | 0.79 ± 0.17 | 0.83 ± 0.30 | |
| +ATc | 0.81 ± 0.19 | 0.81 ± 0.25 | 0.80 ± 0.15 | 0.84 ± 0.23 | |
| Set 2 | PfADF1 #4 | PfADF1 #5 | PfADF1K72A #2 | PfADF1K72A #3 | |
| −ATc | 0.79 ± 0.29 | 0.68 ± 0.16 | 0.77 ± 0.22 | 0.74 ± 0.16 | |
| +ATc | 0.78 ± 0.26 | 0.71 ± 0.20 | 0.76 ± 0.17 | 0.73 ± 0.18 | |
aMean speed ± SD measured over 60 s from n = 20 parasite trails per line, clone, and condition (± ATc).
bHelical gliding not observed for ATc-treated TgADF cKO parasites.
Finally, motility behavior similar to that seen for the wild-type lines also was observed for TgADFK68A-cMyc– and PfADF1K72A-cMyc–expressing parasites grown in ATc, with no major differences in the form or level of movement (Figure 4, A–D, and Table 1). Parasites of the TgADFK68A-cMyc clone #1 line, however, displayed a ∼20% increase of shorter twirling and gliding motions in the presence of ATc, which might reflect the protruding actin phenotype observed by immunofluorescence analysis in a small population of parasites (Figure 3D). Again, the significance of this minor population in these instances is unclear.
In summary, our findings demonstrate that wild-type PfADF1 and the mutant TgADFK68A and PfADF1K72A proteins can functionally restore the basic role of TgADF in tachyzoite motility. Because disassembly is expected to be reduced in mutants for the single exposed lysine residue, this would suggest that actin filament disassembly may not be as critical for regulating actin-dependent tachyzoite gliding motility as previously believed.
Parasite replication and apicoplast segregation are compromised in TgADFK68A mutant parasites
To further elucidate the growth defect found with the TgADFK68A-cMyc–expressing parasites by plaque assay (Figure 2), we next independently examined the capacity of wild-type and mutant TgADF parasites to invade, replicate, and egress from host cells.
We performed invasion assays by culturing parasites in the absence and presence of ATc for 48 h before allowing freshly released tachyzoites to invade host cells for 1 h, removing extracellular forms, and growing intracellular parasites for another 24 h. As previously described (Mehta and Sibley, 2011), suppression of the TgADF-HA protein led to ∼50% reduction in the number of intracellular parasites, whereas the efficiency of invasion was fully restored in wild-type TgADF-complementing lines grown in ATc (Figure 5A). For both clones expressing the mutant TgADFK68A-cMyc protein, the total number of parasite vacuoles was slightly reduced, by ∼25% for K68A #1 and ∼20% for K68A #6 (Figure 5A). Extending the incubation of host cells with ATc-pretreated extracellular tachyzoites to 4–5 h, however, restored mutant TgADF parasite invasion rates to levels of invasion in the absence of ATc, but the number of parasite vacuoles was still reduced by ∼50% in the straight TgADF cKO line (unpublished data).
FIGURE 5:
Daughter cell formation and apicoplast segregation are impaired in TgADFK68A-cMyc–expressing parasites. (A) ATc-treated TgADF mutant parasite lines are slightly impaired in host cell invasion. Vacuoles were counted ∼24 h postinvasion and were visualized by using GAP45 antibodies by immunofluorescence analysis (IFA). Average of three independent experiments (± range). (B, C) TgADFK68A mutant parasites grown in ATc display a replication defect like that of the parent knockdown line (in which an increase in low vs. high nuclei number can be observed in the presence of ATc; asterisks). Daughter cells per vacuole were counted for each line and clone from the invasion experiments shown and normalized to the total vacuole number per condition (n = on average 300 vacuoles, ± range shown). (D, E) In parallel to the IFA carried out for invasion analysis (24 h postinvasion), IFA using anti-ACP antibodies was performed to investigate apicoplast segregation. Segregation of the apicoplasts as determined by distribution and number of plastid foci of fluorescence compared with no ATc (TgADF cKO) was greatly impaired in most TgADFK68A-cMyc–expressing parasites (clones #1 and #6 shown). Scale bar, 5 μm (E). (F) Egress (as determined by number of vacuoles in replicate assays) was not affected by mutation of the critical lysine residue. Average of five independent image panels from a representative egress assay.
To analyze parasite replication, we determined the number of daughter cells per parasite vacuole, finding an impaired development of daughter cells in ATc-treated TgADF cKO parasites. This was demonstrated by ∼50% increase of vacuoles containing two cells, with a concomitant decrease in eight cells per vacuole compared with TgADF cKO parasites grown in the absence of ATc and parasites expressing the complementing TgADF-cMyc wild-type protein (Figure 5B, asterisk). An increase of two cell stages (and four cell stages for clone K68A #6) matched by a concomitant decrease of eight cells per vacuole could also be observed for the TgADFK68A-cMyc–expressing parasites, albeit with slight differences in the percentages between the two clones investigated (Figure 5C, asterisks). In addition to a general defect in replication, daughter cells within the vacuoles of the ATc-treated TgADF cKO line appeared to be predominantly disorganized, which also was observed for the TgADFK68A-cMyc lines but to a lower extent.
A defect in daughter cell formation was previously described for TgMyoF-depleted parasites in which apicoplast segregation was severely affected (Jacot et al., 2013). Furthermore, apicoplast inheritance had also been found to be impaired in the inducible knockouts of T. gondii actin (Andenmatten et al., 2013), as well as for T. gondii profilin and the TgADF cKO line as a result of perturbed actin dynamics (Jacot et al., 2013). Apicoplast segregation coincides with daughter cell formation during endodyogeny, and manipulation of this organelle ultimately leads to a delayed parasite death (Dahl and Rosenthal, 2008). In light of the previous findings, we therefore performed immunofluorescence analysis of intracellular parasites using the apicoplast marker acyl carrier protein (ACP). Apicoplast morphology and segregation were perturbed in ∼70% of TgADFK68A-cMyc–expressing parasites in the presence of ATc (Figure 5D). In accordance with the observations made by Jacot et al. (2013), apicoplast numbers were greatly reduced in the four- to eight-cell stage of TgADF cKO and TgADFK68A-cMyc–expressing parasites, whereas the wild-type TgADF-complementing lines displayed normal organelle numbers and distribution (Figure 5E). These findings not only corroborate previous observations indicating involvement of TgADF and therefore actin in apicoplast segregation (Jacot et al., 2013), they also suggest that this dependence on actin and ADF is linked to the disassembly of actin filaments.
Finally, no egress defect could be observed for any of the complementing parasite lines (neither for wild-type nor for mutant TgADF) grown in the presence of ATc compared with parasites cultured in the absence of ATc (Figure 5F). In contrast, on average, fivefold more vacuoles were counted for parasites lacking TgADF expression, which is indicative of unsuccessful (and/or inefficient) tachyzoite exit as described by Mehta and Sibley (2011).
The combined cellular data therefore demonstrate a negligible role of ADF-dependent actin disassembly in tachyzoite motility (including invasion, gliding, and egress) but, remarkably, reveal a critical role for ADF-mediated disassembly, and the lysine residue in particular, in daughter cell division, specifically organelle segregation.
TgADF is a more potent actin-depolymerizing factor than PfADF1
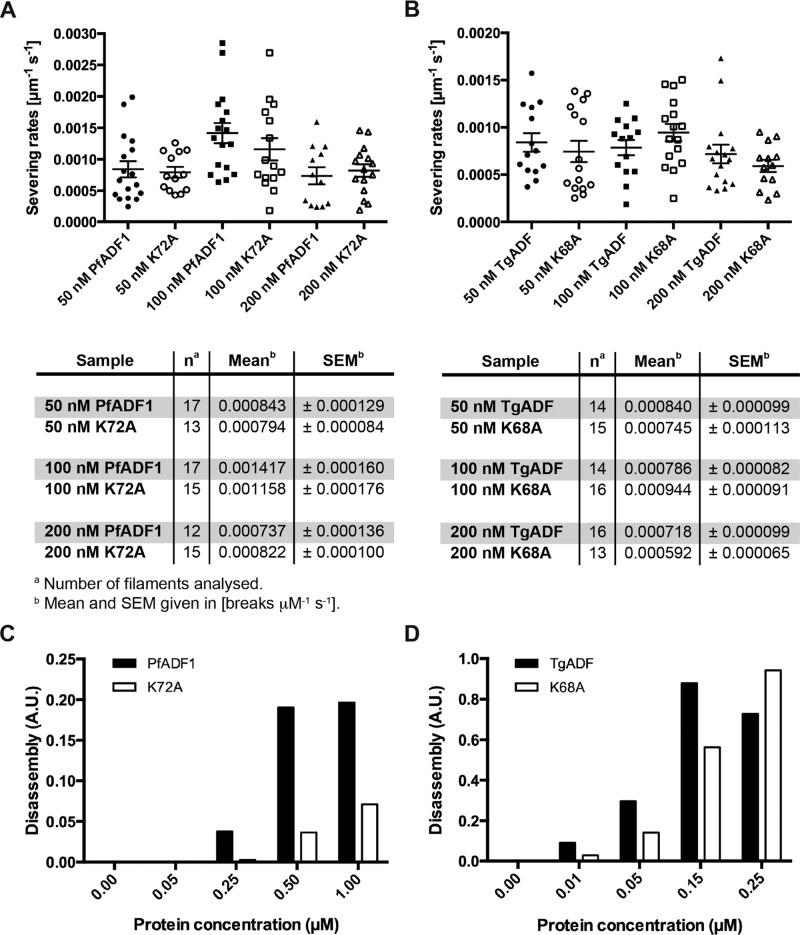
We previously showed that PfADF1 (at 100 nM) severs preformed filaments at an average rate of 0.0007 break μm-1 s-1, whereas a mutant form carrying the K72A substitution at the base of the much-reduced F-loop severs at half this rate (Wong et al., 2011). We therefore sought to reassess these findings for TgADF versus its respective lysine mutant (K68A) and used total internal reflection fluorescence microscopy (TIRFM) to measure its ability to sever actin filaments. We found similar severing rates (∼0.0008 break μm-1 s-1) for PfADF1 at protein concentrations of 50 and 200 nM and a slightly elevated rate of ∼0.001 break μm-1 s-1 when using 100 nM of the wild-type protein. Despite a perceived reduction in severing at 100 nM between the wild-type and mutant PfADF1 proteins (and in line with previous observations), however, the corresponding severing rates were not significantly different (Figure 6A). Furthermore, at 50 and 200 nM, the values for wild-type and mutant K72A were almost identical (Figure 6A). Thus, although we are unable to explain the discrepancy between past and present measures, these results suggest that mutant PfADF1 most likely retains its full capacity for severing actin filaments under the present in vitro conditions.
FIGURE 6:
TgADF is more efficient in disassembling F-actin than PfADF1. (A, B) TIRF microscopy analysis of cofilin-mediated F-actin severing by wild-type and mutant ADF proteins. Filaments were assembled from 1 μM G-actin (10% Oregon green–labeled, 0.1% biotinylated) in flow chambers and attached to biotin-PEG (0.1%)–coated glass slides by streptavidin. Plots of the normalized severing rates (break μm-1 s-1) in the presence of different protein concentrations show the individual data points, as well as the mean ± SEM. (C, D) F-actin disassembly assays using pyrene-labeled actin, induced by vitamin D–binding protein. A range of wild-type and mutant ADF protein concentrations was tested on preassembled 1.5 μM F-actin (10% pyrene-actin; Chaudhry et al., 2013). Disassembly rates were determined by measuring the slope in the linear region of the original data set shown in Supplemental Figure S4.
Severing rates for the orthologous TgADF proteins (at identical protein concentrations) were comparable to those of PfADF1 and K72A, again with no major difference in activity between the wild-type TgADF and mutant K68A protein across all concentrations tested (Figure 6B). These findings confirm the ability of apicomplexan ADF proteins to sever actin filaments but suggest caution about the importance of the minimal F-actin binding motif in actin-filament severing, certainly under in vitro conditions.
Severing efficiency by TIRFM is, by convention, conducted at F-actin concentrations well above the critical concentration for filament disassembly (i.e., at saturating levels), which will bias the reaction toward severing events but not disassembly. We therefore set out to investigate the activity of the wild-type and mutant ADF orthologues at lower concentrations of actin, at which disassembly (as opposed to severing) is favored, with the goal of explaining the TgADFK68A in vivo phenotype. Measurements of pyrene-actin disassembly in the presence of the different ADF variants (Figure 6, C and D), initiated by the addition of 3 μM vitamin D–binding protein (Chaudhry et al., 2013), revealed not only that TgADF is more efficient in disassembling F-actin in a concentration-dependent manner, but also that it does so at significantly lower protein concentrations (reaching saturating levels at a protein concentration of ∼150 nM, with activity decreasing at 250 nM). Indeed, an ∼15-fold higher disassembly rate was observed for TgADF at 250 nM compared with PfADF1 (Figure 6, C and D), whereas PfADF1 is able to disassemble preformed F-actin only at much higher protein concentrations (reaching saturating levels at ∼1 μM) and with lower efficiency (Figure 6, C and D, and Supplemental Figure S4). Mutation of the conserved lysine residue for both ADF proteins, however, strikingly leads to a marked reduction in disassembly activity, with ≥50% decrease of depolymerizing activity in PfADF1K72A and ∼30–50% reduction in disassembly for TgADFK68A (Figures 6, C and D). This supports our previous observations of a role for the conserved basic residue in F-actin disassembly, although not necessarily via severing. Furthermore, this adds support to the interpretation that the phenotype of daughter cell segregation defects is due to a diminished ability of ADF to disassemble actin filaments.
The absence of a concordant phenotype between the TgADFK68A-cMyc and orthologous PfADF1K72A-cMyc lines points to what may be a far greater degree of coevolutionary alignment between native ADF and actin in apicomplexan cells than previously recognized.
DISCUSSION
In this study, we used T. gondii as a cellular model to investigate and compare the activities of PfADF1 and TgADF variants in vivo and address the role of ADF-mediated disassembly, specifically by a conserved lysine residue (Lys-72 in P. falciparum and Lys-68 in T. gondii), as previously implicated in actin filament severing in vitro (Wong et al., 2011).
TgADF shares ∼75% homology to PfADF1, and so the biochemical and structural properties are expected to be very similar (Singh et al., 2011; Supplemental Figure S1). Indeed, we demonstrate that complementation with wild-type PfADF1 can fully rescue the conditional knockdown phenotype of TgADF to similar levels as for the complementing T. gondii orthologue, supporting their conservation. In contrast, substitution of a key exposed residue, Lys-68, to alanine in TgADF had a significant detrimental effect on parasite development, most dramatically demonstrated with impaired daughter cell formation and apicoplast segregation. Host cell entry, however, was only mildly affected, and parasite motility and egress were comparable to those of the wild-type ADF-expressing parasite lines. On reassessment of ADF activity in vitro, this shows that actin filament disassembly appears to be critical for regulation of T. gondii parasite development but, remarkably, not cell motility.
Biochemical analysis of the native ADF and those carrying a mutated conserved lysine residue, implicated in actin-filament severing (Wong et al., 2011), demonstrate that its function may instead be limited to a broader effect on filament disassembly. Although we cannot be certain about the mechanism of action, measurements of the total disassembly rates from TIRF movies for the Toxoplasma ADF proteins (Supplemental Figure S4C) showed an enhanced rate of disassembly for the wild-type protein compared with the K68A lysine mutant. Although this is consistent with the notion that impairments seen in depolymerization may be related to pointed-end disassembly (Carlier et al., 1997), given the spatial resolution limits of TIRF microscopy measurements (1 pixel = 100 nm) and other technical caveats with regard to the assay, we cannot rule out alternative possibilities, such as that a portion of measured depolymerization may arise from severing of fragments below the resolution limit. Combined with awareness of the emerging complexity of ADF/cofilin interactions with the actin filament (Wong et al., 2014), the precise mechanism of action here, and thus the effect of the conserved lysine residue, therefore remains elusive.
ADF activity has also been implicated in regulating actin-based cytoskeletal rearrangements during the morphological remodeling of the rodent malaria parasite P. berghei. Parasites lacking the PbADF2 isoform, which more resembles a conventional ADF having an extended F-loop, display significant defects in ookinete-to-oocyst transformation and the differentiation of sporozoites to extraerythrocytic forms. Parasite motility, however, was unaffected and suggests a possible activation of PbADF1 to govern motility in the absence of the PbADF2 (Doi et al., 2010). Combined with observations that apicoplast inheritance is impaired in the inducible knockouts of T. gondii actin (Andenmatten et al., 2013), T. gondii myosin F, profilin, and ADF (Jacot et al., 2013), these data suggest a growing sense that actin filament dynamics are critical for intracellular development and organelle division. However, at the same time, they bring into question our understanding of how ADF proteins, and actin filament disassembly specifically, govern apicomplexan gliding motility.
Perhaps most surprisingly, no defect in parasite propagation was observed for the PfADF1K72A mutant parasite clones. This points toward an incomplete understanding of potential genus-specific differences in the coevolution of apicomplexan ADF proteins, their activities, and/or specific interactions with each respective actin and possibly other actin-binding proteins. Hence this highlights the limitations of trans-genera complementation strategies to address fundamental questions while at the same time pointing to the potential for real cellular differences between the often-assumed similarities between Plasmodium and Toxoplasma parasites.
One reason for a lack of phenotype with PfADF1K72A could lie in subtle differences between the apicomplexan actin orthologues and their intrinsic properties (Skillman et al., 2011, 2013; Vahokoski et al., 2014) by which the interaction of PfADF1 with T. gondii actin may not be affected by mutating the conserved lysine residue (which is not identically placed in structural alignments; Supplemental Figure S1). Another possible explanation emerges from our poor knowledge about how apicomplexan ADF proteins are regulated. Among other mechanisms, ADF/cofilin activity is inhibited upon phosphorylation of a conserved N-terminal serine residue at position 2 or 3 (reviewed in Mizuno, 2013). Along these lines, in vitro analysis of the corresponding phosphomimetic in TgADF (TgADFS3E) resulted in an inactive protein (Mehta and Sibley, 2010). In this regard, it is noteworthy that despite the high degree of sequence conservation between PfADF1 and TgADF, the N-terminal region of TgADF more closely resembles that of Acanthamoeba castellani actophorin as well as plant ADFs (Allen et al., 1997). The solution structure of TgADF and its corresponding electrostatic surface features several positively charged residues (Yadav et al., 2011), which are also found in the N-terminus of Arabidopsis thaliana ADF1 (AtADF1; Bowman et al., 2000). In contrast, only half of these residues are present in PfADF2, and just one residue is conserved in the N-terminal region of PfADF1 (Supplemental Figure S1). Investigations of AtADF1 reveal an important role of the N-terminus and its intramolecular interactions in regulating AtADF1 activity (Dong et al., 2013). Similar to AtADF1, the N-terminus of TgADF is a poorly defined but highly dynamic structure, possibly allowing for conformational changes in response to phosphorylation (Yadav et al., 2011). It will be interesting to address the N-terminal genus-specific differences in future studies.
In summary, our investigations reveal the need for a reassessment of how ADF proteins regulate actin dynamics that drive parasite cell motility, pointing to a previously unappreciated complexity in the role of actin filament turnover for different developmental processes within apicomplexan cells.
MATERIALS AND METHODS
Generation of complementation constructs
Complementation was attempted via random integration of the respective constructs tgadf-cmyc, pfadf1-cmyc, tgadfK68A-cmyc, and pfadf1K72A-cmyc into the TgADF cKO line (Mehta and Sibley, 2011). Therefore gBlock fragments (Integrated DNA Technologies) of full-length wild-type and mutant tgadf were synthesized in-frame with a 3′ triple-cmyc tag and additional restriction sites (5′ BglII/AatII and 3′ AvrII-3xcMyc-EcoRV) and cloned into pDTGDRE (C.J.T., unpublished data) via BglII/EcoRV. Wild-type pfadf1 and the K72A mutant gene were amplified from existing plasmids using oligos PfADF1-F1 (5′GgacgtcATGATCTCTGGCATCCGCGTCAAC) and PfADF1-R1 (5′GgatatccctaggCTTGAGGTCCGCAACGTCCTGC), thereby introducing AatII/AvrII restriction sites to replace the tgadf gene.
T. gondii culture and transfection
TgADF cKO parasites were grown in human foreskin fibroblasts (HFFs) and maintained in DMEM supplemented with 1% fetal calf serum (LifeTechnologies, Carlsbad, CA) and 1% (vol/vol) glutaMAX (LifeTechnologies). HFFs were maintained in DMEM supplemented with 10% Cosmic Calf Serum (Thermo Scientific , Waltham, MA). For the generation of transformants, ∼1 × 107 freshly released TgADF cKO parasites were transfected by electroporation with 50 mg of plasmid DNA and selected in the presence of 4 mM pyrimethamine and ATc at 1 mg/ml. Clonal lines were established via limiting dilution of one parasite per well in a 96-well format.
Immunofluorescence analysis
For immunofluorescence analysis, HFF cells were grown on coverslips and inoculated with T. gondii parasites in the absence and presence of ATc (at 1 mg/ml) for 48–50 h. To investigate the distribution of actin in extracellular parasites, growth in ATc was extended to 68–72 h, and freshly released tachyzoites were harvested. Cells were fixed with 4% (wt/vol) paraformaldehyde and 0.0075% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA) in phosphate-buffered saline (PBS) for 10 min before permeabilization with 1% (vol/vol) Triton X-100 in PBS for 10 min, followed by blocking in 3% (wt/vol) bovine serum albumin (Sigma-Aldrich, St. Louis, MO) in PBS for 1 h. Primary antibodies were diluted in blocking solution and used in the following concentrations: rabbit anti-HA clone C29F4 (Cell Signaling Technology, Danvers, MA), 1:4000; mouse anti-cMyc clone 9E10 (Sigma-Aldrich), 1:2000; and rabbit anti-Act239-253 (Angrisano et al., 2012), 1:4000. Secondary antibodies used were Alexa Fluor 488 goat anti-mouse and Alexa Fluor 594 goat anti-rabbit (Life Technologies) in 1:4000 dilutions.
Fluorescence microscopy was performed using a DeltaVision Microscopy System (Applied Precision, Seattle, WA) and an Olympus UPlanSApo 60×/numerical aperture (NA) 1.4 or 100×/1.4 NA oil objective.
Western blot analysis
For immunoblot analysis, parasites were grown in the absence and presence of ATc (1 mg/ml) for 48–50 h. Approximately 3 × 106 freshly released parasites were harvested and resuspended in reducing sample buffer. Equal amounts of protein lysate were separated using NuPAGE 4–12% Bis-Tris gels in MES buffer (Life Technologies) and transferred onto nitrocellulose membranes using the iBlot system (Life Technologies). Membranes were blocked in PBS with 5% skim milk (wt/vol) for 1 h before incubation with one of the primary antibodies (in blocking solution) rabbit anti-HA clone C29F4 (Cell Signaling Technology), 1:4000; mouse anti-cMyc clone 9E10 (Sigma-Aldrich), 1:1000; rabbit anti-Act239-253 (Angrisano et al., 2012), 1:4000; and rabbit anti-TgADF (kindly provided by D. Sibley, Washington University, St. Louis), 1:4000. Secondary antibodies used were goat anti-rabbit horseradish peroxidase (HRP) and donkey anti-mouse HRP (Life Technologies) in 1:4000 and 1:2000 dilutions, respectively. Gels were run in duplicate, and membranes were stripped, blocked, and reprobed for multiple analyses.
Plaque assays
Confluent monolayers of HFF cells were grown in six-well plates and inoculated with 200 parasites/well in the presence and absence of ATc (at 1 mg/ml). Infected monolayers were fixed with 70% EtOH at 8 d postinfection and stained with 0.1% crystal violet. Assays were repeated four times in duplicate per cell line and clone.
Gliding motility assays
Parasites were grown in the presence and absence of ATc for 68–72 h, needle-passed, and filtered before resuspension in warm Ringer's solution. Tachyzoites were then applied to poly-l-lysine (Electron Microscopy Sciences)–coated eight-well slides and allowed to settle for 2 min. Motion was captured every 2 s over a total time of 2 min using a Zeiss LSM 5 Live microscope and a 20× objective. Duplicate movies of each cell line and clone were processed and analyzed from two independent experiments using FIJI software (Schindelin et al., 2012). To measure the rate of helical gliding, parasite trails were visualized and overlaid with the respective time-lapse video using FIJI software (Supplemental Movie S1). Speed was calculated by measuring the parasite's displacement (multiples of parasite body length ∼7 μm) over a total time of 60 s.
On average ∼500 parasites per cell line, clone, condition (±ATc), and experiment were counted and forms of motility categorized using standards established in the literature (Wetzel et al., 2003; Mehta and Sibley, 2011): “twirling” parasites remained attached to the substrate; “gliding” parasites displayed different (and sometimes repetitive) helical and circular patterns, whereas some combined twirling and gliding; and “short” movements were characterized by twirling, gliding, or uncoordinated motion over a shorter period (usually 10–40 s) and included the “rocking” and “flipping” motions as described for the TgADF cKO phenotype.
Invasion assays and parasite replication analysis
To investigate the ability to invade, parasites were initially grown in the absence and presence of ATc for 48 h. Infected host cells were then scraped off, needle-passed, and filtered to release tachyzoites and inoculate 5 × 105 parasites per condition (±ATc), line, and clone onto HFF monolayers (in duplicate for multiple immunofluorescence analyses; see later description). Parasites were allowed to invade for 1 or 4–5 h (±ATc), respectively, extracellular parasites were removed (followed by several PBS washes), and intracellular parasites were incubated in the absence and presence of ATc for another 24 or 48 h, respectively.
Cells were fixed as described before and immunolabeled with mouse anti-HA clone 12CA5 (1:1000) and rabbit anti-TgGAP45 (1:1000, kindly provided by C.J.T.) to control for the inducible TgADF-HA copy and vacuole formation. In parallel, colabeling with rabbit anti-TgACP (1:1000; C.J.T.) in combination with either mouse anti-cMyc or mouse anti-HA was performed to investigate apicoplast segregation. Invasion rates were determined by counting five consecutive fields using a 20×/0.75 NA objective and calculated as a percentage of untreated parasites normalized to 100%.
Egress assays
Parasites were grown in the absence and presence of ATc for ∼58–60 h, and egress was stimulated by adding 2 μM calcium ionophore A23187 for 10 min (Mehta and Sibley, 2011). Parasites were fixed as described and immunolabeled with rabbit anti-TgGAP45 (1:1000) in addition to either mouse anti-HA clone 12CA5 (1:1000) or mouse anti-cMyc clone 9E10 (1:1000). To determine the number of parasite vacuoles, indicative of unsuccessful egress events, five independent panels of 10 × 10 fields (∼1.2 mm2) were imaged and stitched using the softWoRx software per parasite line, clone, and condition.
Recombinant protein expression and purification
Wild-type and mutant adf genes from T. gondii were cloned into pGEX4T (GE Healthcare, Uppsala, Sweden) and expressed as glutathione S-transferase fusion proteins in Escherichia coli strain BL21 (DE3; Life Technologies). Recombinant proteins were purified as previously described (Wong et al., 2011) and dialyzed against PBS before cleavage with thrombin protease (GE Healthcare). Untagged ADF proteins were purified by size exclusion chromatography using a Superdex 200, GL10/300 column (GE Healthcare) and eluted in 20 mM MES/10 mM NaCl, pH 7.0, for downstream biochemical analysis.
Pyrene F-actin disassembly assays
F-actin disassembly rates were monitored using the method of Chaudhry et al. (2013). In brief, 100 μl of preformed F-actin (1.5 μM final, 10% pyrene-labeled) and 30 μl of control buffer or ADF variant in the same buffer was prepared using a range of concentrations for wild-type and mutant ADF proteins. Immediately before the experiment, 20 μl of 3 μM vitamin D–binding protein/Gc-globulin from human plasma (VDBP; Sigma-Aldrich) was mixed with the ADF protein mix. At time zero, disassembly was induced by addition of the VDBP/ADF mix to F-actin. Fluorescence was monitored at 365-nm excitation and 407-nm emission in a Safire2 fluorescence spectrophotometer (Tecan M200 Pro; Tecan Trading AG, Männedorf, Switzerland).
TIRFM
For all experiments, 24 × 40 mm coverslips (Fisher Scientific, Leicestershire, UK) were Piranha acid–cleaned as described previously (Winkelman et al., 2014) and coated for 18 h with a polyethylene glycol (PEG)–silane (5000 molecular weight at 1 mg/ml in 95% EtOH; Creative PEGWorks, Chapel Hill, NC) solution (pH 2.0) supplemented with 0.1% (vol/vol) Biotin-PEG-silane (Laysan Bio, Arab, AL). Parallel strips of double-sided tape were placed on the coverslip to create multiple flow chambers. Flow chambers were stored in a wet chamber at 4°C. TIRFM images were collected at 5-s intervals with an iXon electron-multiplying charge-coupled device camera (Andor Technology, Belfast, UK) using an Olympus IX-71 microscope equipped with a plan-apochromatic through-the-objective TIRFM illumination lens (100×/1.45 NA oil). Flow chambers were incubated for 3 min with 50 μg/ml streptavidin; this was followed by a 5-min incubation with Ca-buffer G (2 mM Tris-Cl, pH 8.0, 0.2 mM ATP, 0.5 mM dithiothreitol, 0.1 mM CaCl2) containing 0.5% bovine serum albumin. Flow chambers were washed with one chamber volume Ca-buffer G. Actin filaments were assembled from a mixture of 1 μM final concentration Mg-ATP actin (10% Oregon green–labeled) supplemented with 0.1% biotin-actin until they reached ∼10 μm in length. Cofilin-mediated severing rates were determined as previously reported (Wong et al., 2011). Final protein concentrations are indicated in the figure legends. Five filaments per TgADF protein (at 100 nM) were measured to assess the total disassembly rates (three control filaments) over time from two independent movies (i.e., different days). Because filaments sever, one filament becomes multiple ends over time, with total ends measured given as the n number (Supplemental Figure S4C). Median values were calculated from the individual disassembly slopes per filament.
Supplementary Material
Acknowledgments
We acknowledge L. David Sibley and his laboratory for generous provision of the conditional TgADF line and antibodies, as well as Connie Li Wai Suen for statistical advice. Experimental data presented here were made possible through Victorian State Government Operational Infrastructure Support and Australian Government National Health and Medical Research Council of Australia Independent Research Institute Infrastructure Support. The research was directly supported by a National Health and Medical Research Council of Australia Project Grant (APP1024678 to J.B.) and Human Frontier Science Program Young Investigator Program Grant (RGY0071/2011; J.B. and D.R.K.). M.A.O. is supported through a National Health and Medical Research Council of Australia Dora Lush Scholarship (APP1018002), with work here supported through an Overseas Research Experience Scholarship and travel award provided by the University of Melbourne and the Australian Society of Parasitology, respectively. W.W. is supported through an Early Career Fellowship (APP1053801) from the National Health and Medical Research Council of Australia. J.B. was supported through a Future Fellowship (FT100100112) from the Australian Research Council and is currently supported by a New Investigator Award from the Wellcome Trust (100993/Z/13/Z).
Abbreviations used:
- ADF
actin-depolymerizing factor
- TIFRM
total internal reflection fluorescence microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-10-1427) on July 8, 2015.
*These authors contributed equally to this work.
The authors declare no conflict of interest.
REFERENCES
- Allen ML, Dobrowolski JM, Muller H, Sibley LD, Mansour TE. Cloning and characterization of actin depolymerizing factor from Toxoplasma gondii. Mol Biochem Parasitol. 1997;88:43–52. doi: 10.1016/s0166-6851(97)00069-8. [DOI] [PubMed] [Google Scholar]
- Andenmatten N, Egarter S, Jackson AJ, Jullien N, Herman JP, Meissner M. Conditional genome engineering in Toxoplasma gondii uncovers alternative invasion mechanisms. Nat Methods. 2013;10:125–127. doi: 10.1038/nmeth.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrisano F, Riglar DT, Sturm A, Volz JC, Delves MJ, Zuccala ES, Turnbull L, Dekiwadia C, Olshina MA, Marapana DS, et al. Spatial localisation of actin filaments across developmental stages of the malaria parasite. PLoS One. 2012;7:e32188. doi: 10.1371/journal.pone.0032188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg JR, Bernstein BW. Roles of ADF/cofilin in actin polymerization and beyond. F1000 Biol Rep. 2010;2:62. doi: 10.3410/B2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J, Gilberger T-W, Frischknecht F, Meissner M. Host-cell invasion by malaria parasites: insights from Plasmodium and Toxoplasma. Trends Parasitol. 2008;24:557–563. doi: 10.1016/j.pt.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Baum J, Papenfuss AT, Baum B, Speed TP, Cowman AF. Regulation of apicomplexan actin-based motility. Nat Rev Microbiol. 2006a;4:621–628. doi: 10.1038/nrmicro1465. [DOI] [PubMed] [Google Scholar]
- Baum J, Richard D, Healer J, Rug M, Krnajski Z, Gilberger T-W, Green JL, Holder AA, Cowman AF. A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites. J Biol Chem. 2006b;281:5197–5208. doi: 10.1074/jbc.M509807200. [DOI] [PubMed] [Google Scholar]
- Bergman LW, Kaiser K, Fujioka H, Coppens I, Daly TM, Fox S, Matuschewski K, Nussenzweig V, Kappe SHI. Myosin A tail domain interacting protein (MTIP) localizes to the inner membrane complex of Plasmodium sporozoites. J Cell Sci. 2003;116:39–49. doi: 10.1242/jcs.00194. [DOI] [PubMed] [Google Scholar]
- Bowman GD, Nodelman IM, Hong Y, Chua NH, Lindberg U, Schutt CE. A comparative structural analysis of the ADF/cofilin family. Proteins. 2000;41:374–384. doi: 10.1002/1097-0134(20001115)41:3<374::aid-prot90>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Bullen HE, Tonkin CJ, O'Donnell RA, Tham WH, Papenfuss AT, Gould S, Cowman AF, Crabb BS, Gilson PR. A novel family of Apicomplexan glideosome-associated proteins with an inner membrane-anchoring role. J Biol Chem. 2009;284:25353–25363. doi: 10.1074/jbc.M109.036772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Pantaloni D. Control of actin assembly dynamics in cell motility. J Biol Chem. 2007;282:23005–23009. doi: 10.1074/jbc.R700020200. [DOI] [PubMed] [Google Scholar]
- Chaudhry F, Breitsprecher D, Little K, Sharov G, Sokolova O, Goode BL. Srv2/cyclase-associated protein forms hexameric shurikens that directly catalyze actin filament severing by cofilin. Mol Biol Cell. 2013;24:31–41. doi: 10.1091/mbc.E12-08-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl EL, Rosenthal PJ. Apicoplast translation, transcription and genome replication: targets for antimalarial antibiotics. Trends Parasitol. 2008;24:279–284. doi: 10.1016/j.pt.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Dobrowolski JM, Carruthers VB, Sibley LD. Participation of myosin in gliding motility and host cell invasion by Toxoplasma gondii. Mol Microbiol. 1997a;26:163–173. doi: 10.1046/j.1365-2958.1997.5671913.x. [DOI] [PubMed] [Google Scholar]
- Dobrowolski JM, Niesman IR, Sibley LD. Actin in the parasite Toxoplasma gondii is encoded by a single copy gene, ACT1 and exists primarily in a globular form. Cell Motil Cytoskeleton. 1997b;37:253–262. doi: 10.1002/(SICI)1097-0169(1997)37:3<253::AID-CM7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Doi Y, Shinzawa N, Fukumoto S, Okano H, Kanuka H. ADF2 is required for transformation of the ookinete and sporozoite in malaria parasite development. Biochem Biophys Res Commun. 2010;397:668–672. doi: 10.1016/j.bbrc.2010.05.155. [DOI] [PubMed] [Google Scholar]
- Dong CH, Tang WP, Liu JY. Arabidopsis AtADF1 is functionally affected by mutations on actin binding sites. J Integr Plant Biol. 2013;55:250–261. doi: 10.1111/jipb.12015. [DOI] [PubMed] [Google Scholar]
- Frénal K, Polonais V, Marq J-B, Stratmann R, Limenitakis J, Soldati-Favre D. Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe. 2010;8:343–357. doi: 10.1016/j.chom.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Herm-Gotz A, Weiss S, Stratmann R, Fujita-Becker S, Ruff C, Meyhofer E, Soldati T, Manstein DJ, Geeves MA, Soldati D. Toxoplasma gondii myosin A and its light chain: a fast, single-headed, plus-end-directed motor. EMBO J. 2002;21:2149–2158. doi: 10.1093/emboj/21.9.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot D, Daher W, Soldati-Favre D. Toxoplasma gondii myosin F, an essential motor for centrosomes positioning and apicoplast inheritance. EMBO J. 2013;32:1702–1716. doi: 10.1038/emboj.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ML, Kitson EL, Rayner JC. Plasmodium falciparum erythrocyte invasion: a conserved myosin associated complex. Mol Biochem Parasitol. 2006;147:74–84. doi: 10.1016/j.molbiopara.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Kappe SH, Buscaglia CA, Bergman LW, Coppens I, Nussenzweig V. Apicomplexan gliding motility and host cell invasion: overhauling the motor model. Trends Parasitol. 2004;20:13–16. doi: 10.1016/j.pt.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Mehta S, Sibley LD. Toxoplasma gondii actin depolymerizing factor acts primarily to sequester G-actin. J Biol Chem. 2010;285:6835–6847. doi: 10.1074/jbc.M109.068155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Sibley LD. Actin depolymerizing factor controls actin turnover and gliding motility in Toxoplasma gondii. Mol Biol Cell. 2011;22:1290–1299. doi: 10.1091/mbc.E10-12-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner M, Schluter D, Soldati D. Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science. 2002;298:837–840. doi: 10.1126/science.1074553. [DOI] [PubMed] [Google Scholar]
- Mizuno K. Signaling mechanisms and functional roles of cofilin phosphorylation and dephosphorylation. Cell Signal. 2013;25:457–469. doi: 10.1016/j.cellsig.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Mizuno Y, Makioka A, Kawazu S, Kano S, Kawai S, Akaki M, Aikawa M, Ohtomo H. Effect of jasplakinolide on the growth, invasion, and actin cytoskeleton of Plasmodium falciparum. Parasitol Res. 2002;88:844–848. doi: 10.1007/s00436-002-0666-8. [DOI] [PubMed] [Google Scholar]
- Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int Rev Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- Sahoo N, Beatty W, Heuser J, Sept D, Sibley LD. Unusual kinetic and structural properties control rapid assembly and turnover of actin in the parasite Toxoplasma gondii. Mol Biol Cell. 2006;17:895–906. doi: 10.1091/mbc.E05-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz S, Grainger M, Howell S, Calder LJ, Gaeb M, Pinder JC, Holder AA, Veigel C. Malaria parasite actin filaments are very short. J Mol Biol. 2005;349:113–125. doi: 10.1016/j.jmb.2005.03.056. [DOI] [PubMed] [Google Scholar]
- Schuler H, Matuschewski K. Regulation of apicomplexan microfilament dynamics by a minimal set of actin-binding proteins. Traffic. 2006;7:1433–1439. doi: 10.1111/j.1600-0854.2006.00484.x. [DOI] [PubMed] [Google Scholar]
- Schüler H, Mueller A-K, Matuschewski K. A Plasmodium actin-depolymerizing factor that binds exclusively to actin monomers. Mol Biol Cell. 2005;16:4013–4023. doi: 10.1091/mbc.E05-02-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw MK, Tilney LG. Induction of an acrosomal process in Toxoplasma gondii: visualization of actin filaments in a protozoan parasite. Proc Natl Acad Sci USA. 1999;96:9095–9099. doi: 10.1073/pnas.96.16.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK, Sattler JM, Chatterjee M, Huttu J, Schüler H, Kursula I. Crystal structures explain functional differences in the two actin depolymerization factors of the malaria parasite. J Biol Chem. 2011;286:28256–28264. doi: 10.1074/jbc.M111.211730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skillman KM, Diraviyam K, Khan A, Tang K, Sept D, Sibley LD. Evolutionarily divergent, unstable filamentous actin is essential for gliding motility in apicomplexan parasites. PLoS Pathog. 2011;7:e1002280. doi: 10.1371/journal.ppat.1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skillman KM, Ma CI, Fremont DH, Diraviyam K, Cooper JA, Sept D, Sibley LD. The unusual dynamics of parasite actin result from isodesmic polymerization. Nat Commun. 2013;4:2285. doi: 10.1038/ncomms3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough BR, Reymann A-C, Guérin C, Martiel J-L, De La Cruz EM, Blanchoin L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahokoski J, Bhargav SP, Desfosses A, Andreadaki M, Kumpula EP, Martinez SM, Ignatev A, Lepper S, Frischknecht F, Siden-Kiamos I, et al. Structural differences explain diverse functions of Plasmodium actins. PLoS Pathog. 2014;10:e1004091. doi: 10.1371/journal.ppat.1004091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel DM, Håkansson S, Hu K, Roos D, Sibley LD. Actin filament polymerization regulates gliding motility by apicomplexan parasites. Mol Biol Cell. 2003;14:396–406. doi: 10.1091/mbc.E02-08-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NJ, Pukrittayakamee S, Hien TT, Faiz MA, Mokuolu OA, Dondorp AM. Malaria. Lancet. 2014;383:723–735. doi: 10.1016/S0140-6736(13)60024-0. [DOI] [PubMed] [Google Scholar]
- Winkelman JD, Bilancia CG, Peifer M, Kovar DR. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc Natl Acad Sci USA. 2014;111:4121–4126. doi: 10.1073/pnas.1322093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, Skau CT, Marapana DS, Hanssen E, Taylor NL, Riglar DT, Zuccala ES, Angrisano F, Lewis H, Catimel B, et al. Minimal requirements for actin filament disassembly revealed by structural analysis of malaria parasite actin-depolymerizing factor 1. Proc Natl Acad Sci USA. 2011;108:9869–9874. doi: 10.1073/pnas.1018927108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, Webb AI, Olshina MA, Infusini G, Tan YH, Hanssen E, Catimel B, Suarez C, Condron M, Angrisano F, et al. A mechanism for actin filament severing by malaria parasite actin depolymerizing factor 1 via a low affinity binding interface. J Biol Chem. 2014;289:4043–4054. doi: 10.1074/jbc.M113.523365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R, Pathak PP, Shukla VK, Jain A, Srivastava S, Tripathi S, Krishna Pulavarti SV, Mehta S, David Sibley L, Arora A. Solution structure and dynamics of ADF from Toxoplasma gondii. J Struct Biol. 2011;176:97–111. doi: 10.1016/j.jsb.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.