GARP (tethering factor)- and VAMP4 (v-SNARE)-dependent endosome-to-TGN retrograde transport is required for the efficient post-Golgi anterograde transport of cell-surface integral membrane proteins. Golgi-resident membrane proteins TMEM87A and TMEM87B are involved in endosome-to-TGN retrograde transport.
Abstract
The importance of endosome-to–trans-Golgi network (TGN) retrograde transport in the anterograde transport of proteins is unclear. In this study, genome-wide screening of the factors necessary for efficient anterograde protein transport in human haploid cells identified subunits of the Golgi-associated retrograde protein (GARP) complex, a tethering factor involved in endosome-to-TGN transport. Knockout (KO) of each of the four GARP subunits, VPS51–VPS54, in HEK293 cells caused severely defective anterograde transport of both glycosylphosphatidylinositol (GPI)-anchored and transmembrane proteins from the TGN. Overexpression of VAMP4, v-SNARE, in VPS54-KO cells partially restored not only endosome-to-TGN retrograde transport, but also anterograde transport of both GPI-anchored and transmembrane proteins. Further screening for genes whose overexpression normalized the VPS54-KO phenotype identified TMEM87A, encoding an uncharacterized Golgi-resident membrane protein. Overexpression of TMEM87A or its close homologue TMEM87B in VPS54-KO cells partially restored endosome-to-TGN retrograde transport and anterograde transport. Therefore GARP- and VAMP4-dependent endosome-to-TGN retrograde transport is required for recycling of molecules critical for efficient post-Golgi anterograde transport of cell-surface integral membrane proteins. In addition, TMEM87A and TMEM87B are involved in endosome-to-TGN retrograde transport.
INTRODUCTION
Protein trafficking in eukaryotic cells is mediated by bidirectional vesicular transport between cellular compartments (Brandizzi and Barlowe, 2013). Vesicular transport can be divided into two pathways: anterograde transport, which is the route from the endoplasmic reticulum (ER) to the plasma membrane, and retrograde transport, which is the opposite route. Each of these transport pathways consists of four steps: budding, movement, tethering, and uncoating and fusion (Bonifacino and Glick, 2004; Brandizzi and Barlowe, 2013). Anterograde transport from the ER to the ER–Golgi intermediate compartment is mediated by COPII-coated vesicles, and retrograde transport from the Golgi to the ER is mediated by COPI-coated vesicles. Inhibition of COPI-dependent retrograde transport disrupts COPII-mediated anterograde transport because recycling proteins, including vesicle–soluble N-ethylmaleimide–sensitive factor attachment protein receptors (v-SNAREs) and cargo receptors in the ER, are depleted (Pepperkok et al., 1993; Peter et al., 1993). Therefore protein recycling mediated by COPI-dependent retrograde transport is required for efficient anterograde transport. Anterograde and retrograde transport also take place between the TGN and the plasma membrane. Three retrograde transport pathways to the TGN have been identified: from early, recycling, and late endosomes (Chia et al., 2013). Retrograde transport carriers from early or recycling endosomes contain the v-SNARE vesicle-associated membrane protein 3 (VAMP3) or VAMP4. Fusion with the TGN membrane is mediated by SNARE complex assembly between VAMP4 and the TGN-localized target (t)-SNAREs syntaxin 6 (STX6), STX16, and Vti1 (Mallard et al., 2002; Hong and Lev, 2013). Retrograde transport to the TGN was first discovered through the transport routes of toxins (Mallard et al., 1998) and is now believed to act as a protein-recycling route from endosomes.
Previous studies showed that secretion of Wnt is dependent on endosome-to-TGN retrograde transport, based on the necessary recycling of its cargo receptor, wntless (Belenkaya et al., 2008; Franch-Marro et al., 2008; Harterink et al., 2011). These results highlighted the importance of protein recycling from endosomes. However, other studies showed that anterograde transport of thermosensitive vesicular stomatitis virus G protein (VSVGtsO45, hereafter VSVGts), which is more widely used as a model protein for testing anterograde transport, is not impaired in cells depleted of SNARE proteins involved in endosome-to-TGN retrograde transport (Choudhury et al., 2006; Nishimoto-Morita et al., 2009; Shitara et al., 2013). These results were quite different from the case of Wnt secretion and suggested that protein recycling to the TGN is less important for the anterograde transport of VSVGts. Therefore the relevance of endosome-to-TGN retrograde transport in the anterograde transport of various cargoes needs to be clarified.
Glycosylphosphatidylinositol (GPI) addition is a highly conserved posttranslational modification in eukaryotic cells and is used in the anchoring of proteins (Kinoshita et al., 2008). In mammalian cells, ∼150 proteins are anchored to the plasma membrane via GPI. The synthesis of GPI in the ER is followed by its transfer to proteins, generating immature GPI-anchored proteins (GPI-APs) in which GPI is then structurally remodeled by post-GPI attachment to proteins 1 (PGAP1) and PGAP5 (Tanaka et al., 2004; Fujita et al., 2009). Because structural remodeling is important for interaction with the p24 family complex, a cargo receptor of GPI-APs, the PGAP1 or PGAP5 mutant cells exhibit delayed anterograde transport of GPI-APs from the ER (Fujita et al., 2011). However, little is known about the molecular mechanism of post-Golgi anterograde transport of GPI-APs.
In this study, we carried out haploid genetic screening to identify genes necessary for the efficient anterograde transport of GPI-APs and thereby identified genes encoding subunits of the Golgi-associated retrograde protein (GARP) complex. The GARP complex is a multisubunit tethering factor localized in the TGN and involved in endosome-to-TGN retrograde transport (Bonifacino and Hierro, 2011). It is composed of four subunits—vacuolar protein sorting 51 (VPS51), VPS52, VPS53, and VPS54—each of which is essential for tethering (Conibear and Stevens, 2000; Siniossoglou and Pelham, 2002; Perez-Victoria et al., 2008, 2010a). Here we demonstrate that the GARP- and VAMP4-dependent endosome-to-TGN retrograde transport is required for recycling of factors necessary for the efficient post-Golgi anterograde transport of cell-surface integral membrane proteins such as GPI-anchored or transmembrane proteins.
RESULTS
Screening mutant haploid cells defective in GPI-AP transport
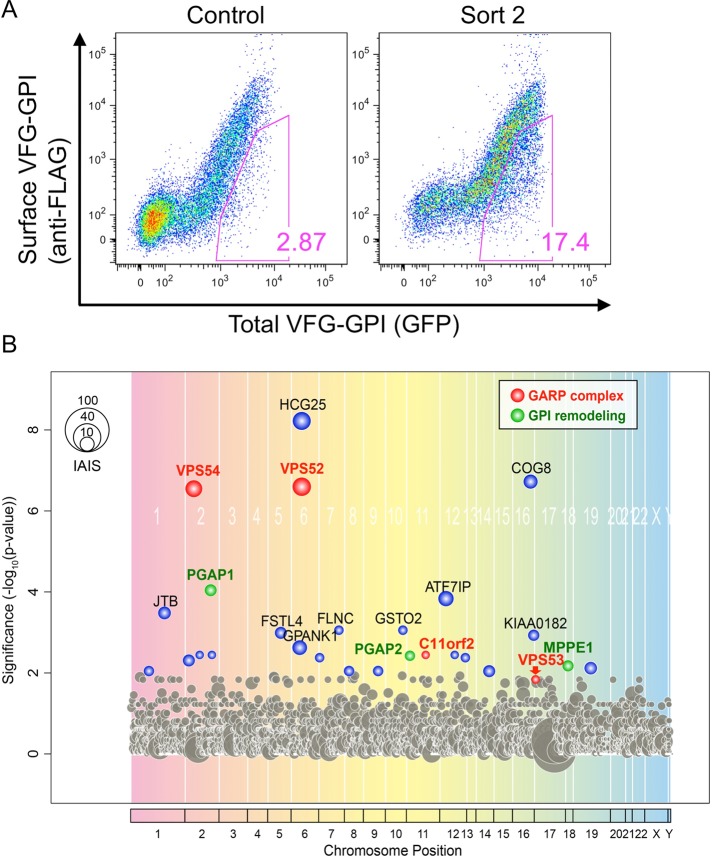
To identify genes necessary for the efficient anterograde transport of GPI-APs, we performed genetic screening in human haploid cells (HAP1; Carette et al., 2011b), using a GPI-anchored reporter protein to monitor the transport of GPI-APs previously developed in genetic screening with CHO cells (Maeda et al., 2008; Fujita et al., 2009). This reporter is a chimeric protein consisting of a luminal domain of VSVGts, a FLAG tag, an enhanced green fluorescent protein (EGFP), and a C-terminal GPI attachment signal (VFG-GPI). VSVGts causes accumulation of the reporter in the ER at the restrictive temperature of 40°C because of the protein's unfolding status and allows synchronized exit from the ER after rapid folding at the permissive temperature of 32°C. Therefore transport status at x minutes after the temperature shift can be measured by determining the ratio of surface (cell-surface FLAG tag determined by flow cytometry) to total (fluorescence of EGFP) VFG-GPI. Thus we first established a HAP1 cell line, HAP1FF9, with doxycycline (Dox)-inducible expression of VFG-GPI and mutagenized the cells using a retroviral gene-trapping vector. Control genomic DNA was extracted before the enrichment of transport mutants. Using a cell sorter, we collected mutant cells showing slow GPI-AP transport, as determined by decreased surface levels of VFG-GPI, 60 min after the temperature shift. The collected cells were cultured and then re-sorted to enrich mutants with delayed GPI-AP transport (sort 2, Figure 1A). Retroviral insertion sites in the genomic DNA from sort 2 cells and control, pre-enrichment cells were analyzed by deep sequencing. Enrichment rates were calculated for the respective genes by comparing the numbers of mapped independent-insertion sites of the respective gene between sort 2 and control cells. The genes PGAP1 and PGAP5 (MPPE1), whose mutations have been known to cause delayed transport of GPI-APs, were enriched in sort 2 (Figure 1B and Supplemental Table S1), indicating the usefulness of the method. In addition, the PGAP2 gene, which is required for GPI fatty acid remodeling in the Golgi (Tashima et al., 2006), was also enriched in sort 2. In the PGAP2 mutant cells, transport of GPI-APs is almost normal, but the surface expression of GPI-APs is greatly decreased because lyso form GPI-APs, the intermediate forms during the fatty acid remodeling, which harbor only a single acyl chain, are not reacylated due to the PGAP2 defect and are released easily from the membrane into medium soon after arrival at the cell surface.
FIGURE 1:
Haploid genetic screening of the factors required for the anterograde transport of GPI-APs. (A) Transport assay of VFG-GPI in HAP1FF9 wild-type cells (left) and the enriched cell population after sorting twice for delayed transport (right). (B) The significance of the enrichment of gene-trap insertions in an enriched population with delayed transport compared with the nonselected population was calculated and plotted as a bubble plot. The horizontal line shows the chromosomal position of the genes, and the vertical line shows the significance of enrichment of each gene (p value). The size of the bubble shows the number of inactivated insertion sites. Genes significantly enriched in sort 2 (p < 0.01) are colored. Red bubbles, genes encoding the GARP complex subunit; green bubbles, genes involved in GPI-AP remodeling. The bubble of VPS53 indicated by an arrow was close to the significance limit.
Knockout of GARP complex subunits severely impairs anterograde transport of proteins
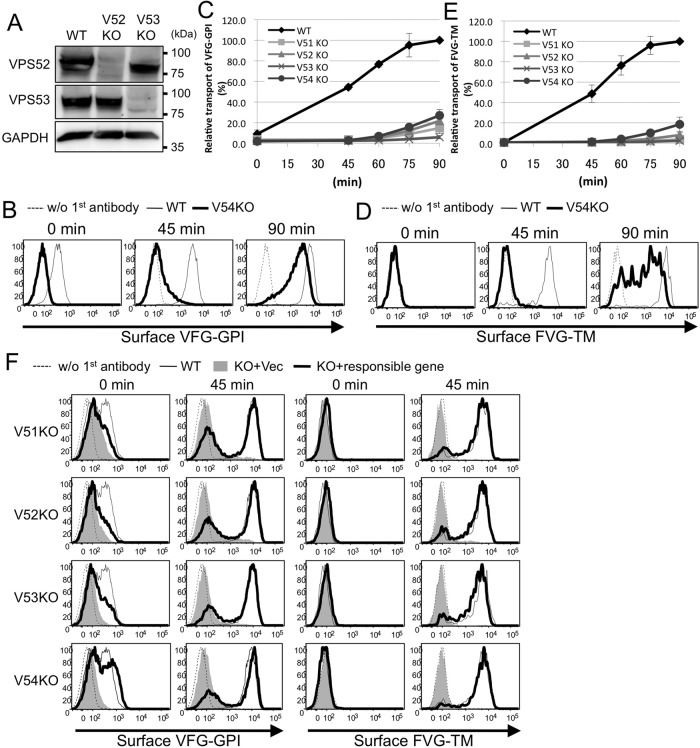
Three genes—C11orf2 (also known as VPS51 or Ang2), VPS52, and VPS54, encoding subunits of the GARP complex, a tethering factor—were significantly enriched in sort 2 cells (p < 0.01), and the fourth subunit—VPS53—was positioned close to the significance limit (Figure 1B and Supplemental Table S1). In addition, the COG8 gene, encoding a subunit of conserved oligomeric Golgi (COG) complex, another multisubunit tethering factor, was highly enriched. To investigate whether the GARP complex is involved in the anterograde transport of GPI-APs, we generated GARP-KO cells using the clustered, regularly interspaced, short palindromic repeat (CRISPR)–Cas9 system (Cong et al., 2013; Mali et al., 2013) in HEK293FF6 cells, bearing Dox-inducible VFG-GPI. All mutations caused frameshifts (Supplemental Table S2), indicating that all mutant cells had completely lost expression of the targeted genes. Indeed, VPS52 and VPS53 proteins nearly completely disappeared in V52KO and V53KO cells, respectively (Figure 2A). To compare the kinetics of anterograde transport of VFG-GPI, we conducted a transport assay using HEK293FF6 wild-type or GARP-KO cells. The relative expression of VFG-GPI in each KO cell line (V51KO, V52KO, V53KO, and V54KO) 90 min after the temperature shift was 14.7 ± 5.2, 21.7 ± 11.0, 5.9 ± 1.7, and 27.0 ± 3.5%, respectively (mean ± SD; Figure 2, B and C), suggesting that the anterograde transport of GPI-APs is severely impaired in GARP-KO cells. Note that the surface expression of VFG-GPI at 0 min was lower in GARP-KO cells than in wild-type cells, consistent with the delayed transport occurring from the very beginning in KO cells. To investigate whether delayed transport was specific to GPI-APs, we conducted a similar transport assay of a transmembrane-type reporter protein in GARP-KO cells. The reporter protein, FVG-TM, was a chimeric protein consisting of a FLAG tag, VSVGts (full length), and EGFP (Maeda et al., 2008; Fujita et al., 2009). Because the HEK293FF6 cell line does not have FVG-TM, its expression plasmid was transiently transfected into cells and its transport then assayed. After 90 min, the relative expression of FVG-TM in V51KO, V52KO, V53KO, and V54KO cells was only 3.7 ± 0.95, 8.0 ± 2.5, 2.4 ± 1.3, and 18.6 ± 7.0%, respectively (mean ± SD; Figure 2, D and E). Expression of the responsible gene in each KO cell restored the delayed transport of both VFG-GPI and FVG-TM (Figure 2F), thus ruling out off-target effects. These results indicated that the GARP complex is required for the efficient anterograde transport of both GPI-anchored and transmembrane proteins.
FIGURE 2:
Knockout (KO) of GARP complex subunits severely impairs the anterograde transport of proteins. (A) Western blotting of VPS52 and VPS53 in HEK293FF6 wild-type (WT), VPS52-KO (V52KO), and VPS53-KO (V53KO) cells, showing nearly complete reduction by specific knockout. GAPDH was used as a loading control. (B) Transport assay of VFG-GPI in wild-type (WT) and GARP-KO cells. Surface expression of VFG-GPI at the indicated time in WT and VPS54-KO (V54KO) cells is shown as an example. (C) Relative transport of VFG-GPI in GARP-KO cells. The geometric mean fluorescence of surface VFG-GPI, as shown in the histograms in B, was quantified for each time point, plotting the geometric mean of WT cells at 90 min as 100% relative transport. (D) Transport assay of FVG-TM in WT and GARP-KO cells. WT and GARP-KO cells were transiently transfected with FVG-TM. Surface expression of FVG-TM at the indicated time in WT and V54KO cells is shown as an example. (E) Relative transport of FVG-TM. The geometric mean fluorescence of surface FVG-TM, as shown in the histograms in D, was quantified similarly to C. (F) Rescue of transport delay in GARP-KO cells by the expression of the responsible genes. GARP-KO cells were transiently transfected with a VPS51, VPS52, VPS53, or VPS54 expression plasmid, after which the transport of VFG-GPI (left) was analyzed. For FVG-TM transport, cells were cotransfected with FVG-TM and indicated genes. The quantitative data are the means of three independent experiments. Error bars represent the SDs (n = 3).
Post-Golgi anterograde transport is defective in V54KO cells
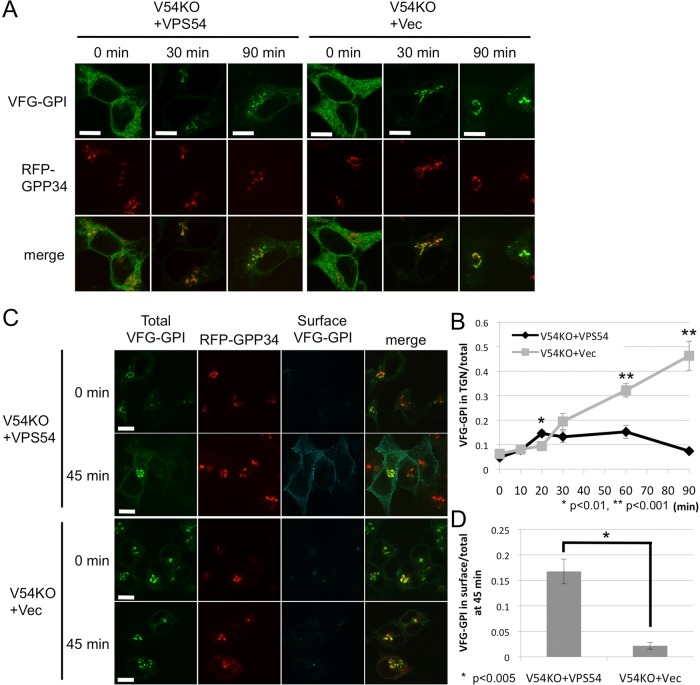
The step in anterograde transport that is affected by a defect in the GARP complex was identified using confocal microscopy in cells stably transfected with an empty vector or VPS54 (V54KO+Vec or V54KO+VPS54), together with red fluorescent protein (RFP)–GPP34 as a TGN marker. VFG-GPI transport was chased for the indicated time, after which the cells were fixed to terminate the transport. Ratios of VFG-GPI fluorescence intensity in the TGN to total cellular VFG-GPI fluorescence intensity were determined at each time point (Figure 3B). In V54KO+VPS54 cells, the amount of VFG-GPI that reached the TGN gradually increased for 20 min, remained constant for the next 60 min, and then finally decreased at 90 min (Figure 3, A and B), suggesting that after VFG-GPI reached the TGN, it was transported to the cell surface. In V54KO+Vec cells, the relative intensity in TGN was slightly increased at 20 min but further increased for the next 60 min (Figure 3, A and B), suggesting that after its arrival at the TGN, VFG-GPI accumulated in the TGN for at least 60 min. This result suggested that GARP complex is required for post-Golgi transport. Although ER-to-Golgi transport appeared to be slightly impaired in V54KO+Vec cells (0.15 ± 0.012 vs. 0.095 ± 0.0089 at 20 min; mean ± SEM), this might have been because of the indirect effect of impaired post-Golgi transport. To confirm that post-Golgi anterograde transport is impaired in V54KO cells, we allowed VFG-GPI to accumulate in the Golgi by culturing cells at restrictive temperature of 19°C and then synchronously releasing them by a temperature shift to 32°C. VFG-GPI that reached the cell surface was stained with anti-FLAG antibody under nonpermeabilized conditions. Ratios of VFG-GPI fluorescence intensity in the cell surface to total cellular VFG-GPI fluorescence intensity were quantified (Figure 3D). As shown in Figure 3C, at 0 min, most of the VFG-GPI was localized in the TGN in both V54KO+VPS54 and V54KO+Vec cells. By 45 min, although a large amount of the VFG-GPI reached the cell surface in V54KO+VPS54 cells (Figure 3, C and D), this was barely the case in V54KO+Vec cells, as VFG-GPI largely remained in the TGN (Figure 3, C and D). This result strongly suggested requirement of the GARP complex in post-Golgi anterograde transport. From these results, we concluded that the GARP complex is required for efficient post-Golgi anterograde transport.
FIGURE 3:
Post-Golgi anterograde transport is defective in V54KO cells. (A) VFG-GPI transport from the ER in V54KO or restored cells. Cells were cultured in medium containing 1 μg/ml Dox at 40°C for 24 h. VFG-GPI transport was chased at 32°C for the indicated time in medium containing 100 μg/ml CHX. Images at 0, 30, and 90 min. Green, VFG-GPI; red, RFP-GPP34 as a TGN marker. Scale bars, 10 μm. (B) The TGN localization of VFG-GPI was quantified based on A. The ratio of the VFG-GPI intensity in the TGN to the total fluorescence intensity was determined. Data are the means of 10 independent images. Error bars represent the SEM (n = 10). Similar results were obtained from two independent experiments. (C) Post-Golgi transport of VFG-GPI in V54KO or restored cells. Cells cultured in medium containing 1 μg/ml Dox for 24 h at 40°C were precultured in medium containing 100 μg/ml CHX for 3 h at 19°C, after which VFG-GPI transport was chased at 32°C for 0 or 45 min. To detect surface VFG-GPI, cells were stained with anti-FLAG M2 antibody under nonpermeabilized conditions. Green, total expression of VFG-GPI; red, RFP-GPP34; cyan, surface expression of VFG-GPI. Scale bars, 10 μm. Similar results were obtained from two independent experiments. (D) The ratio of surface arrived VFG-GPI was quantified based on C. Data are the means of five independent images. Error bars represent the SEM (n = 5). Similar results were obtained from two independent experiments.
CD59, an endogenous GPI-AP, was missorted to lysosomes in V54KO cells
The fact that post-Golgi anterograde transport was impaired in V54KO cells led us to investigate whether the cargo sorting at the TGN was impaired. To elucidate this point, we analyzed intracellular localization of endogenous GPI-AP, CD59. To eliminate cell-surface CD59, we treated cells with phosphatidylinositol-specific phospholipase C (PI-PLC) before fixation and analyzed them by confocal microscopy. In V54KO+VPS54 cells, CD59 was hardly detectable, whereas many CD59-positive vesicles were observed in V54KO+Vec cells (Figure 4A). These vesicles were largely colocalized with LAMP1 but not with EEA1 (Figure 4, A and B). This result suggested that CD59 was missorted to lysosomes in V54KO cells probably due to defective post-Golgi anterograde transport. If most of the GPI-APs are missorted to lysosomes and degraded, it is possible that surface expression of GPI-APs is reduced. However, flow cytometric analysis revealed that surface expression of CD59 was not reduced in V54KO+Vec cells compared with V54KO+VPS54 cells (Supplemental Figure S1), suggesting that the majority of GPI-APs are properly but slowly transported to the cell surface.
FIGURE 4:
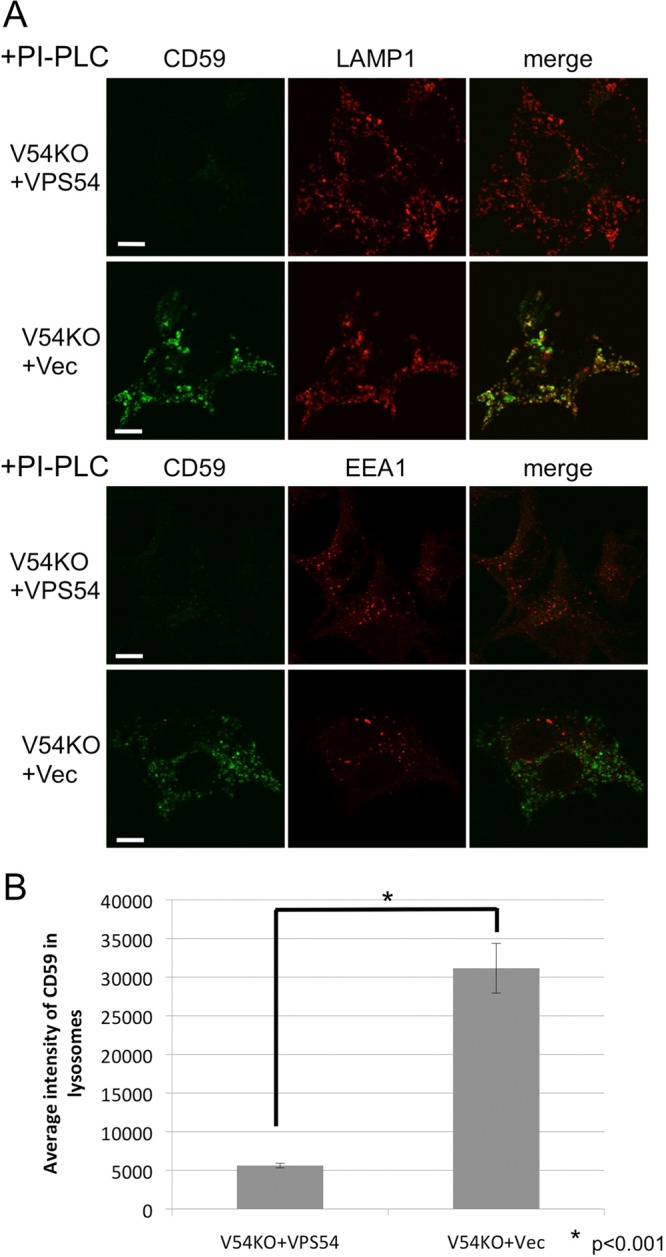
Missorting of CD59 to lysosomes in V54KO cells. (A) Intracellular localization of endogenous GPI-AP, CD59. Cells were treated with PI-PLC at 37°C for 1.5 h, after which they were fixed and double-stained with anti-CD59 and anti-LAMP1 or anti-EEA1. Green, CD59; red, LAMP1 (top) and EEA1 (bottom) as lysosome and early-endosome markers, respectively. Scale bars, 10 μm. (B) CD59 localized in lysosomes was quantified from the images of A. Data are the means of 10 independent images. Error bars represent the SEM (n = 10). Similar results were obtained in two independent experiments.
VAMP4-mediated retrograde transport is required for recycling of proteins critical for efficient anterograde transport
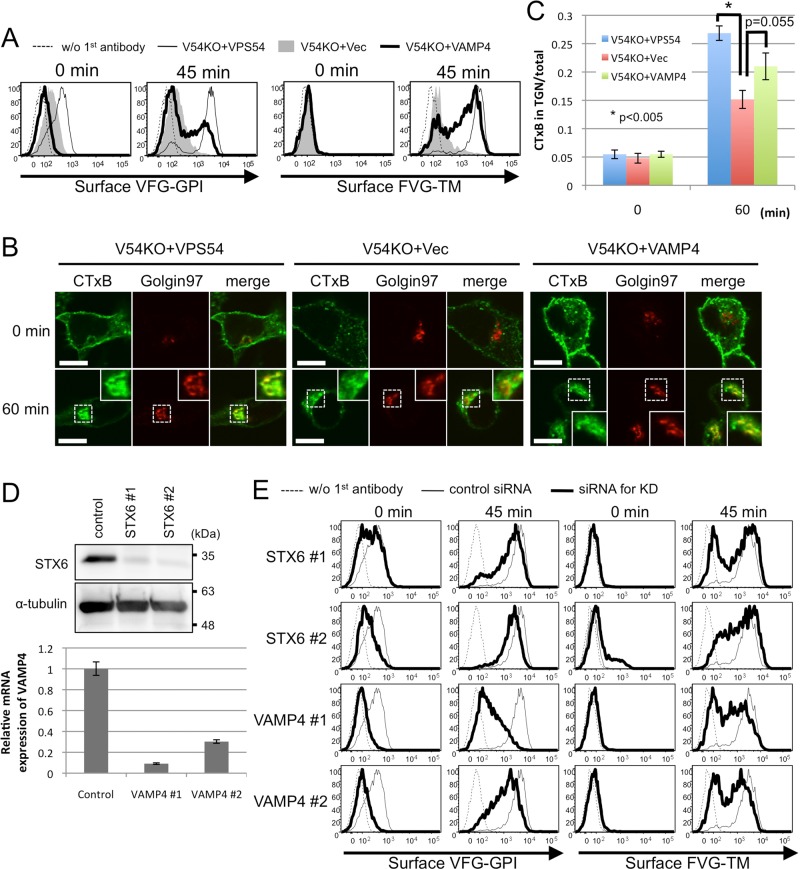
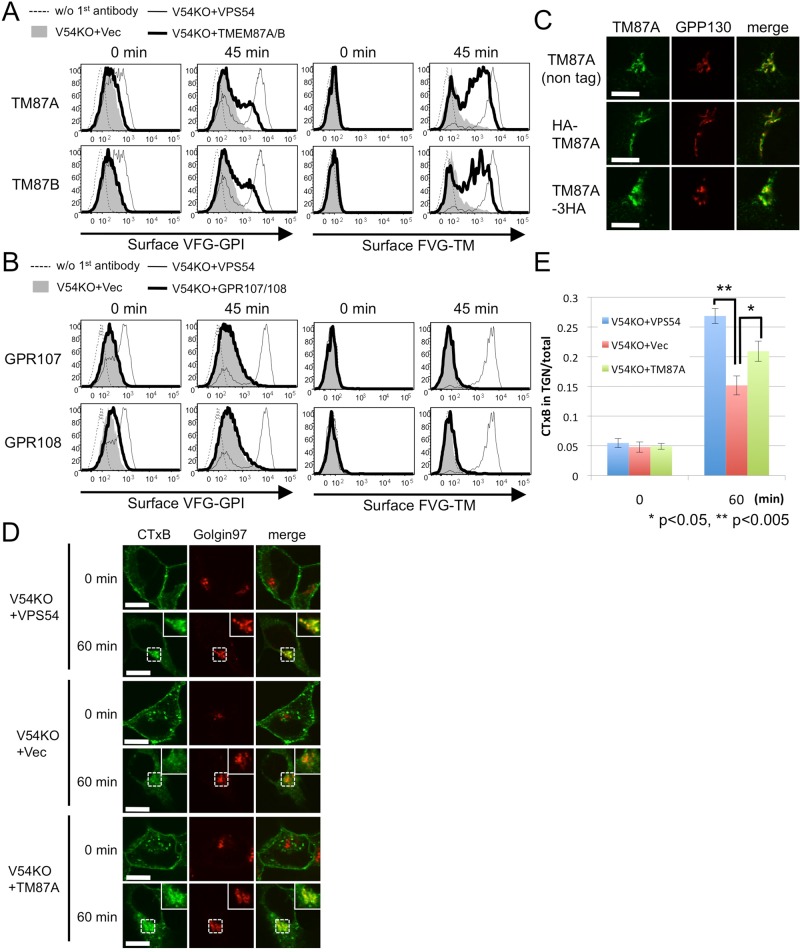
The GARP complex tethers endosome-derived transport carriers to the TGN, so endosome-to-TGN retrograde transport might contribute to post-Golgi anterograde transport by recycling the molecules involved. Thus, in V54KO cells, fewer of these molecules, such as SNARE proteins, would be found in their expected location. To examine this possibility, we overexpressed several SNARE proteins localized in the post-Golgi compartments in V54KO cells and then assessed restoration of anterograde transport . Overexpression of VAMP4 partially restored the delayed transport of both VFG-GPI and FVG-TM, whereas overexpression of VAMP2, VAMP3, VAMP7, STX1A, or STX6 did not (Figure 5A and Supplemental Figure S2A). To investigate the mechanism by which VAMP4 overexpression restored anterograde transport in V54KO cells, we analyzed plasma membrane–to-TGN retrograde transport using the cholera toxin B subunit (CTxB) as cargo. We prepared V54KO cells stably expressing VAMP4 (V54KO+VAMP4) and collected cells that restored anterograde transport of VFG-GPI (Supplemental Figure S2B). To assess CTxB retrograde transport, we bound Alexa 488–conjugated CTxB to the cell surface at 4°C, after which we incubated cells at 37°C for 60 min to allow the transport of CTxB. Because CTxB binding to the cell surface was decreased in V54KO cells (Supplemental Figure S2C), retrograde transport efficiency was determined by comparing the ratio of Golgi-associated CTxB to total CTxB. The ratio of CTxB localized in the TGN at 60 min in V54KO+Vec cells was lower than that in V54KO+VPS54 cells (Figure 5, B and C), indicating the impairment of endosome-to-TGN retrograde transport in V54KO cells, consistent with the findings of a previous study (Perez-Victoria et al., 2008). As shown in Figure 5, B and C, the amount of TGN-localized CTxB at 60 min was clearly higher in V54KO+VAMP4 cells than in V54KO+Vec cells (0.21 ± 0.024 vs. 0.15 ± 0.016; mean ± SEM). We interpreted these data as indicating that retrograde transport of CTxB was partially restored by VAMP4 overexpression and that the statistical insignificance (p = 0.055) may be accounted for by the presence in V54KO+VAMP4 cells of a population that did not restore the anterograde transport (Supplemental Figure S2B). These results and those on the restoration of anterograde transport strongly indicated that VAMP4-mediated retrograde transport is important for the recycling of molecules involved in the anterograde transport of both GPI-anchored and transmembrane proteins. This conclusion was confirmed by using small interfering RNA (siRNA) to abolish the expression of VAMP4 or STX6. For this experiment, we used a HEK293FF6 cell line for VFG-GPI transport and a HEK293TM10 cell line, in which expression of FVG-TM could be induced with Dox. As shown in Figure 5D, expression of both STX6 and VAMP4 was efficiently knocked down. Depletion of VAMP4 or STX6 impaired the anterograde transport of both VFG-GPI and FVG-TM (Figure 5E). These results complemented those from the VAMP4-overexpression experiments. We concluded that GARP- and VAMP4-mediated retrograde transport is required for recycling the factors necessary for the post-Golgi anterograde transport of both GPI-anchored and transmembrane proteins.
FIGURE 5:
VAMP4-dependent retrograde transport is required for recycling of proteins critical for the post-Golgi anterograde transport. (A) Transport assay of VFG-GPI (left) and FVG-TM (right) in V54KO cells transiently transfected with the indicated genes. (B) Retrograde transport assay using CTxB. Cells incubated with Alexa Fluor 488–conjugated CTxB for 30 min on ice, followed by a chase at 37°C for 0 or 60 min, were analyzed by confocal microscopy after staining with anti-golgin97 antibody. Green, CTxB; red, golgin97 as a TGN marker. Scale bars, 10 μm. Similar results were obtained in two independent experiments. (C) CTxB in the TGN was quantified. Data are the means of 10 independent images. Error bars represent the SEM (n = 10). Similar results were obtained in two independent experiments. (D) Estimation of knockdown efficiency. HEK293FF6 was transfected with indicated siRNAs, and the expression of STX6 or VAMP4 was analyzed by Western blotting or qRT-PCR, respectively. For Western blotting, α-tubulin was used as a loading control. For qRT-PCR, HPRT1 was used as a control. Here #1 and #2 are siRNAs targeting different sites. (E) Effect of STX6 or VAMP4 knockdown on anterograde transport. Transport assay of VFG-GPI (left) or FVG-TM (right) in HEK293FF6 or HEK293TM10 cells, respectively, transfected with indicated siRNAs.
Potential role for TMEM87A and TMEM87B in endosome-to-TGN retrograde transport
We reasoned that overexpression of a protein involved in VAMP4-mediated endosome-to-TGN retrograde transport would rescue the V54KO transport defect. Therefore we next tried to isolate suppressor genes that, when overexpressed, rescued the transport delay in V54KO cells. A cDNA library derived from human brain was introduced into V54KO cells, and the cDNAs in the transport-restored cell populations were sequenced. From this screening, we obtained TMEM87A, a multipass transmembrane protein belonging to the LU7TM family (from Uniprot; Supplemental Table S3). Overexpression of TMEM87A partially restored the defect in anterograde transport of both VFG-GPI and FVG-TM in V54KO cells (Figure 6A; geometric means of cell surface levels of VFG-GPI and FVG-TM were 1.92 ± 0.22 and 4.45 ± 1.1 times, respectively, those in V54KO+Vec; n = 3). Overexpression of TMEM87B, a close homologue of TMEM87A and a member of the same family, also restored the defect in V54KO cells (Figure 6A; geometric means of cell surface levels of VFG-GPI and FVG-TM were 1.85 ± 0.22 and 3.42 ± 0.99 times, respectively, those in V54KO+Vec; n = 3), suggesting its functional redundancy with TMEM87A. Recently a role for GPR107—another LU7TM family member—was shown in retrograde transport of toxins such as exotoxin A of Pseudomonas aeruginosa and ricin (Carette et al., 2011a; Elling et al., 2011; Tafesse et al., 2014; Zhou et al., 2014). Unlike TMEM87A and TMEM87B, overexpression of GPR107 or GPR108, a member of LU7TM family closely related to GPR107, did not restore the delayed transport of either VFG-GPI or FVG-TM in V54KO cells (Figure 6B; geometric means of cell surface levels of VFG-GPI and FVG-TM in V54KO+GPR107 were 1.21 ± 0.38 and 0.98 ± 0.11 times, respectively, those in V54KO+Vec [n = 2], and those in V54KO+GPR108 were 1.23 ± 0.14 and 1.15 ± 0.027 times, respectively, those in V54KO+Vec [n = 2]). These results suggested the specific function of TMEM87A and TMEM87B. Because these two proteins have not been characterized, we first investigated their localization in mammalian cells. Because endogenous TMEM87A protein could not be detected by immunoblotting and immunofluorescence analysis in HEK293 cells, expression plasmids of N-terminally hemagglutinin (HA)-tagged or C-terminally 3×HA-tagged TMEM87A and TMEM87B were constructed and their functions confirmed based on their ability to restore the V54KO phenotype. Overexpression of N- or C-terminally tagged TMEM87A restored the impairment of anterograde transport of VFG-GPI to an extent similar to that of the nontagged version (Supplemental Figure S3A), indicating that both HA-tagged TMEM87As were functional; by contrast, neither of the tagged TMEM87Bs was functional. Nontagged or N- or C-terminally HA-tagged TMEM87A was transiently expressed in wild-type cells, and the localization of the proteins was analyzed by confocal microscopy. As shown in Figure 6C, nontagged or N- or C-terminally HA-tagged TMEM87A predominantly colocalized with GPP130, indicating its Golgi localization.
FIGURE 6:
Potential role for TMEM87A and TMEM87B in endosome-to-TGN retrograde transport. (A) Overexpression of TMEM87A (TM87A) and TMEM87B (TM87B) partly rescues the delayed transport of proteins in V54KO cells. Transport assay of VFG-GPI (left) or FVG-TM (right) in V54KO cells transiently transfected with the indicated genes. (B) Overexpression of GPR107 and GPR108 does not rescue delayed transport in V54KO cells. Transport assay of VFG-GPI (left) or FVG-TM (right) in V54KO cells transfected with the indicated genes. (C) Subcellular localization of TMEM87A. Wild-type cells were transfected with nontagged or HA-tagged TMEM87A and double-stained with anti-TMEM87A (nontagged version) or anti-HA7 and anti-GPP130, a Golgi marker. Green, TMEM87A; red, GPP130 as a Golgi marker. Scale bars, 10 μm. (D) CTxB retrograde transport at 0 and 60 min. Green, CTxB; red, golgin97. Scale bars, 10 μm. Similar results were obtained in two independent experiments. (E) Quantitative data were obtained as described in the legend to Figure 5C. Data are the means of 10 independent images. Error bars represent SEM (n = 10). Similar results were obtained in two independent experiments.
To investigate the mechanism by which TMEM87A overexpression restored anterograde transport in V54KO cells, retrograde transport of CTxB was examined in TMEM87A-overexpressing cells. Thus N-terminally HA-tagged TMEM87A was stably expressed in V54KO cells (V54KO+TM87A), and cells in which anterograde transport of VFG-GPI was partially restored were collected (Supplemental Figure S3B). At 60 min, TGN-localized CTxB levels were significantly higher in V54KO+TM87A cells than in V54KO+Vec cells (Figure 6, D and E), which indicated that the V54KO phenotype could be restored by the restoration of retrograde transport in V54KO cells. Finally, we investigated whether a loss of TMEM87A and B affects retrograde transport of CTxB. For this, we generated TMEM87A and TMEM87B double-KO (TM87A, B-DKO) cell lines using HEK293 cells. HEK293 cells expressed both genes at levels comparable to VPS51 and VPS54 as assessed by quantitative reverse-transcription PCR (qRT-PCR; Supplemental Figure S4A), although TMEM87A protein was not detected by monoclonal anti-TMEM87A antibody and TMEM87B protein could not be tested due to a lack of anti-TMEM87B antibody. Retrograde transport of CTxB was not affected in TM87A, B-DKO cell lines (data with clone #2 shown in Supplemental Figure S4B). It seemed that TMEM87A and B were not directly involved in retrograde transport of CTxB or that yet another protein was redundantly involved. It is therefore likely that overexpression of TMEM87A or B in V54KO cells enhanced retrograde transport efficiency in general. Taken together, our data provide strong evidence that TMEM87A and TMEM87B are Golgi-localized proteins involved in endosome-to-TGN retrograde transport.
DISCUSSION
The aim of this study was to identify genes necessary for the efficient anterograde transport of GPI-APs. Haploid-genetic screening identified genes encoding subunits of the GARP complex. Depletion of each component of this complex revealed its requirement in the anterograde transport of both GPI-anchored and transmembrane proteins. In V54KO cells, post-Golgi anterograde transport was severely delayed, consistent with localization of the GARP complex within the TGN (Perez-Victoria et al., 2008). The delay in anterograde transport in V54KO cells was partially restored by the overexpression of VAMP4 but not VAMP3, which also functions in endosome-to-TGN retrograde transport (Mallard et al., 2002). VAMP4 overexpression also repaired the endosome-to-TGN retrograde transport defect. According to these results, the defective anterograde transport in GARP-KO cells was caused by impaired endosome-to-TGN retrograde transport. In addition, knockdown of VAMP4 or STX6 resulted in the defective anterograde transport of both GPI-anchored and transmembrane proteins. These results support a model in which endosome-to-TGN retrograde transport mediated by VAMP4 and GARP is required for recycling of molecular elements involved in post-Golgi anterograde transport (Supplemental Figure S5). Because anterograde transport of GPI-AP was more efficiently restored than retrograde transport of CTxB in V54KO+VAMP4 cells (Supplemental Figure S2B and Figure 5C), VAMP4 might be one of the factors that are GARP-dependently recycled and essential for the post-Golgi anterograde transport. Indeed, it was reported that the localization of VAMP4 was impaired in GARP-depleted cells (Perez-Victoria and Bonifacino, 2009). Although the possibility that the GARP complex is directly involved in anterograde transport cannot be ruled out, this is unlikely because the overexpression of VAMP2, which forms a complex with plasma membrane–localized t-SNAREs and is required for the post-Golgi anterograde transport of proteins (Calakos et al., 1994; Olson et al., 1997; Chen and Scheller, 2001), could not restore delayed anterograde transport in V54KO cells (Supplemental Figure S2A).
Recently it was reported that members of GARP complex form another protein complex, termed endosome-associated recycling protein (EARP), which is the multisubunit tethering factor required for endocytic recycling (Schindler et al., 2015). In addition to VPS51, VPS52, and VPS53, EARP complex contains syndetin instead of VPS54. We produced VPS51, 52, 53, and 54 KO cells. In the VPS51, 52, or 53 KO cells, both GARP and EARP functions would be impaired. In this study, however, VPS54 KO cells showed defects in anterograde protein transport from the Golgi, suggesting that GARP function itself for tethering of endosome-derived vesicles to the Golgi is important for the transport.
The COG8 gene, encoding a subunit of the COG complex, was identified in this screening (Figure 1B). Similar to GARP complex, COG complex is a multisubunit tethering factor composed of eight subunits (Ungar et al., 2006). COG complex acts as a tethering complex required for intra-Golgi or endosome-to-TGN retrograde transport (Oka et al., 2004; Laufman et al., 2011). Either or both of these functions of COG might contribute to post-Golgi anterograde transport of integral membrane proteins.
Our results (Figure 5, A–C) are consistent with previous reports showing that both tethering factors and SNAREs, especially v-SNAREs, contribute to vesicle docking and that overexpression of v-SNAREs compensates for tethering defects (VanRheenen et al., 1998; Wiederkehr et al., 2004; Laufman et al., 2011). As discussed by Laufman et al. (2011), overexpression of VAMP4 could bypass the requirement for tethering complexes (GARP in our case) because endosome-derived transport vesicles bearing highly concentrated v-SNARE proteins may directly interact with the t-SNARE complex, leading to their fusion (Laufman et al., 2011). Unlike v-SNAREs, however, t-SNAREs have no such activity (Supplemental Figure S2A; Laufman et al., 2011). There are at least three different t-SNARE complexes in the TGN or the Golgi: STX6-STX16-Vti1, STX10-STX16-Vti1, and STX5-Ykt6-GS28. Although distinct functions for these complexes have been proposed (Mallard et al., 2002; Tai et al., 2004; Ganley et al., 2008), there may be partial overlap such that the total amount of t-SNAREs is what allows vesicle tethering and fusion. Consistent with this speculation, the transport delay in cells with a knocked down STX6 was milder than in cells with a knocked down VAMP4 (Figure 5E).
In previous studies, knockdown of VAMP4 or STX6 did not cause delayed anterograde transport of VSVGts (Choudhury et al., 2006; Shitara et al., 2013). However, we found that the anterograde transport of GPI-anchored and transmembrane reporter proteins was clearly affected by the knockdown of VAMP4 or STX6 (Figure 5E). The discrepancy might be because of the different analytical methods; in the cited work, microscopy-based analyses were used to quantify transport efficiency, whereas we used a flow cytometry–based transport assay. This allowed the simultaneous analysis of >10,000 cells, in addition to being a more sensitive and quantitative method than microscopy. Therefore flow cytometric analysis might be advantageous in the detection of small changes in transport efficiency, such as those seen with STX6 knockdown.
Mutations in the GARP complex have been described in the literature. For example, the wobbler mouse, used as a model for amyotrophic lateral sclerosis, has a point mutation in VPS54, leading to neurodegeneration (Schmitt-John et al., 2005; Moser et al., 2013). The wobbler mutation causes VPS54 destabilization and leads to impaired endosome-to-TGN retrograde transport (Perez-Victoria et al., 2010a; Karlsson et al., 2013). However, the pathogenic mechanism has yet to be elucidated. More recently, mutations in VPS53 were identified in patients with progressive cerebello-cerebral atrophy type 2 (Feinstein et al., 2014). These patients display cerebellar and cerebral neuronal atrophy before the age of 1 yr. The molecular mechanism leading to progressive cerebello-cerebral atrophy type 2 in patients carrying a mutation in VPS53 is also not understood. Of interest, despite the different phenotypes of VPS53 and VPS54 mutations, motor neurons are affected in both cases (Brunet and Sacher, 2014). In humans, it was reported that mutations in genes involved in anterograde transport of proteins cause motor neuronal diseases (Maruyama et al., 2010; Novarino et al., 2014). Our study revealed the importance of the GARP complex in the post-Golgi anterograde transport of GPI-anchored and transmembrane proteins. In addition to the retrograde transport defect itself, defective anterograde transport resulting from inefficient protein recycling from the endosome to the TGN might contribute to the pathogenic mechanism of disorders caused by VPS53 and VPS54 mutations.
We further screened genes whose overexpression restored delayed anterograde transport in V54KO cells and identified an as-yet-uncharacterized gene, TMEM87A, a member of the LU7TM family. Both TMEM87A and its close homologue TMEM87B restored delayed anterograde transport in V54KO cells, suggesting their functional redundancy. Microscopy analysis demonstrated the Golgi localization of TMEM87A, indicating its function at this site. This result is consistent with a previous report in which Ptm1p, the yeast homologue of TMEM87A, was identified in Tlg2p (yeast homologue of STX16)-containing compartments (Inadome et al., 2005). The retrograde transport of CTxB provided proof that transport was partially restored in V54KO+TM87A cells. Thus restoration of the V54KO phenotype by the overexpression of TMEM87A resulted from the restoration of retrograde transport in V54KO cells and suggested the involvement of TMEM87A in endosome-to-TGN retrograde transport (Supplemental Figure S5). Retrograde transport might be a common function of LU7TM family proteins, since it is shared by GPR107, another member of this family. However, our overexpression experiments (Figure 6, A and B) evidenced two functional subfamilies, such that TMEM87A/B and GPR107/108 might be involved in different retrograde transport pathways.
Several questions regarding the precise functions of these LU7TM family proteins and how these proteins are involved in retrograde transport remain unresolved. One possibility is that G protein–coupled receptors (GPCRs) are needed to maintain Golgi homeostasis. In pharmacological studies, Golgi-localized Gβγ was shown to be required for post-Golgi anterograde transport (Stow et al., 1991; Irannejad and Wedegaertner, 2010). However, the GPCRs associated with Golgi-localized Gβγ are not known. Tafesse et al. (2014) proposed GPR107 as one such GPCR. In our study, TMEM87A/B modulated both endosome-to-TGN retrograde transport and post-Golgi anterograde transport. Thus TMEM87A and TMEM87B are also good candidate GPCRs. Further studies are needed to elucidate the molecular function of these proteins.
In conclusion, this study showed that GARP- and VAMP4-dependent endosome-to-TGN retrograde transport is required for recycling of molecules critical for the efficient post-Golgi anterograde transport of GPI-anchored and transmembrane proteins. GPI-APs lack a cytoplasmic domain, so their efficient transport probably relies on putative transmembrane cargo receptors. Identification of cargo receptors that recognize GPI-APs would confirm this molecular mechanism.
MATERIALS AND METHODS
Reagents and antibodies
Lipofectamine 2000 and Lipofectamine RNAi MAX were purchased from Life Technologies (Carlsbad, CA). As primary antibodies, we used mouse monoclonal anti-CD59 (clone 5H8; Hirata et al., 2013), anti-FLAG (clone M2), anti-HA (clone HA7; Sigma-Aldrich, St. Louis, MO), anti-STX6 (Stressgen, San Diego, CA), anti–α-tubulin, anti-Golgin97 (clone CDF4; Life Technologies), anti-TMEM87A (clone #772807; R&D systems, Minneapolis, MN), rabbit monoclonal anti-LAMP1 (clone D2D11; Cell Signaling Technology, Danvers, MA), rabbit polyclonal anti-VPS53, VPS52 (a kind gift from Chris Schindler and Juan Bonifacino, National Institutes of Health, Bethesda, MD; Perez-Victoria et al., 2008), anti-GPP130 (Covance, Princeton, NJ), and anti-EEA1. The secondary antibodies were phycoerythrin (PE)-conjugated goat anti-mouse immunoglobulin G (IgG; BioLegend, San Diego, CA), Alexa Fluor 594–conjugated goat anti-mouse IgG, and Alexa Fluor 647–conjugated goat anti-mouse IgG (Life Technologies). Alexa Fluor 488–conjugated CTxB was purchased from Life Technologies.
Plasmid construction
A DNA fragment of VSVGex-FF-mEGFP-GPI (VFG-GPI), which encodes a GPI reporter protein, was amplified from pME-Neo2dH-VSVGex-FF-mEGFP-GPI (Maeda et al., 2008). The amplified fragment was cloned into the NotI and MluI sites of pRetroX-Tight–puromycin N-acetyl-transferase (Pur; Clontech, Palo Alto, CA) using the In-fusion cloning reagents (Takara, Shiga, Japan) to generate pRetroX-VFG-GPI-Pur. A DNA fragment of Flag-VSVGfull-EGFP (FVG-TM), encoding a transmembrane reporter protein, was amplified from pME-FLAG-VSVGfull-EGFP (Maeda et al., 2008). The amplified fragment was cloned into pRetroX-Tight-Pur using the same method as used to generate pRetroX-FVG-TM-Pur. A retroviral gene-trap vector containing an adenoviral splice acceptor (SA) site, a PGK polyA signal, a PGK promoter (PGKpro), the blasticidin-S deaminase (BSD) gene, and human growth hormone (hGH) polyA was constructed as follows. The adenoviral SA site and hGH polyA sequence were amplified from pCMT-SAhygpA-NP21 (GenBank AB609713; Horie et al., 2011), which was kindly provided by Kyoji Horie (Nara Medical University, Nara, Japan). The primers (SA site) GTTCCTATTCTCTAGAAAGTATAGGAACTTCAGTG and ttGCGGCCGCTCAggTCAgTCAGAATTCCGGCGGCTAGCGATACCGTC, and (hGH polyA) ccTGAGCGGCCGCaaaaATCGATtCTGTGCCTTCTAGTTGCCAGC and TGGACCATCCTCTAGACTGCC, were used in the amplification. The amplified fragments were cloned into pCMT-SAhygpA-NP21 at the two XbaI sites using the In-fusion cloning reagents, generating pCMT-SA. The PGK polyA and PGKpro sequences were amplified from pCMT-SAhygpA-NP21 using primers CTGACCTGAGCGGCCGCAGAAATTGATGATCTATTAAACAATAAAGA and ggtggACGCGTAGGTCGAAAGGCCCGGAGAT. The BSD gene was amplified from pLIB2-pgkBSD (Maeda et al., 2008) using primers acctACGCGTccaccATGCCTTTGTCTCAAGAAGAATC and GAAGGCACAGAATCGATTTAGCCCTCCCACACATAACC. The resulting PGK polyA-PGKpro and BSD fragments were cloned into the NotI and ClaI sites of pCMT-SA using In-fusion cloning reagents to generate pCMT-SApA-pgkBSD. The CRISPR/Cas9 expression plasmid, pX330 (Cong et al., 2013), was obtained from Addgene and used to generate pX330-hVPS51, -hVPS52, -hVPS53, -hVPS54#1, and -hVPS54#2; targeting sequences (described later) were cloned into pX330 plasmids digested with BbsI. To construct pX330-mEGFP plasmid, a PCR fragment amplified from a template plasmid, pME-(G4S)2-mEGFP, using primers (gagaGGCCGGCCAaGCtAAgAAaAAgAAaATGACGCGTGGCGGAG and gagaGGCCGGCCTTcTTgGTtGCgGCaGGtCTcTTtccCTTGTACAGCTCGTCCA) was digested with Fsel and cloned into the same site of pX330 vector. To generate pX330-mEGFP-hTMEM87A, -hTMEM87B#1, and -hTMEM87B#2, targeting sequences (described later) were cloned into pX330-mEGFP digested with BbsI. To express the GARP complex subunits, pEF6-BSD-VPS51, VPS52, VPS53, and mVPS54-V5-6His were kind gifts by from Juan Bonifacino and Chris Schindler (Perez-Victoria et al., 2008, 2010b). To construct the corresponding expression plasmids, VAMP2, VAMP3, VAMP4, VAMP7, STX1A, and STX6 were PCR-amplified from a human brain cDNA library and GPR107 and GPR108 from HeLa and Hep3B cell cDNA libraries, respectively. The products were cloned into pME-Zeo plasmid digested with EcoRI and NotI. TMEM87A and TMEM87B cDNAs were amplified from HeLa and Hep3B cell cDNA libraries, respectively, and inserted into pME-Zeo. To construct the N-terminally HA-tagged expression plasmids, TMEM87A and TMEM87B, except for their signal sequences (ss), were PCR-amplified and the XhoI- and NotI-digested products cloned into pME-ssHA plasmid, which harbors an ss derived from human CD59 upstream of the HA tag. To construct C-terminally 3×HA-tagged expression plasmids, TMEM87A and TMEM87B, except for their stop codons, were PCR-amplified, and the products were cloned into XhoI- and MluI-digested pME-3xHA plasmid. To construct pLIB-BSD-VAMP4, pME-Zeo-VAMP4 was digested with EcoRI and NotI and cloned into pLIB-BSD digested with the same enzymes. Plasmid pLIB-BSD-ssHA-TMEM87A was constructed by PCR-amplifying a cDNA of ssHA-tagged TMEM87A, followed by digestion with SalI and NotI and cloning into pLIB2-BSD. To construct pLIB-Hyg-RFP(S158T)-GPP34, pME-RFP(S158T)-GPP34 was digested with EcoRI and NotI and cloned into pLIB-Hyg.
Establishment of HAP1FF9, HEK293FF6, and HEK293TM10 cells
Retroviruses were produced in PLAT-GP packaging cells by cotransfection of the VSVG plasmid (pLC-VSVG) and pRetroX-Tet-On Advanced (Clontech) with pRetroX-VFG-GPI-Pur or pRetroX-FVG-TM-Pur. HAP1FF9 cells were established from HAP1 cells, kindly provided by Thijn R. Brummelkamp (Netherlands Cancer Institute, Amsterdam, Netherlands; Carette et al., 2011b). HAP1 cells were infected with retroviruses bearing pRetroX-VFG-GPI-Pur and pRetroX-Tet-On Advanced, followed by selection with puromycin and G418. A HAP1FF9 cell clone was obtained after limiting dilution and karyotype analysis.
HEK293FF6 and HEK293TM10 cells were established from HEK293 cells by infection with retroviruses bearing pRetroX-Tet-On Advanced and pRetroX-VFG-GPI-Pur or pRetroX-FVG-TM-Pur, respectively, followed by selection with puromycin and G418 and limiting dilution.
Cell culture
HAP1-FF9 cells were cultured in Iscove's modified Dulbecco's medium supplemented with 10% fetal calf serum (FCS), 600 μg/ml G418, and 6 μg/ml puromycin. HEK293FF6, HEK293TM10, GARP-KO cell lines and TMEM87A and B double-KO (DKO) cell line established from HEK293FF6 were cultured in DMEM supplemented with 10% FCS, 600 μg/ml G418, and 1 μg/ml puromycin. V54KO cells stably expressing empty vector, mVPS54-V5-His, hVAMP4, or ssHA-hTMEM87A (V54KO+Vec, V54KO+VPS54, V54KO+VAMP4, and V54KO+TM87A, respectively) were cultured in DMEM supplemented with 10% FCS, 600 μg/ml G418, 1 μg/ml puromycin, and 10 μg/ml blasticidin. V54KO+Vec and V54KO+VPS54 stably expressing RFP(S158T)-GPP34 were cultured in DMEM supplemented with 10% FCS, 600 μg/ml G418, 1 μg/ml puromycin, 10 μg/ml blasticidin, and 500 μg/ml hygromycin B. RFP(S158T)-GPP34, a fusion protein of enhanced RFP and TGN protein GPP34, was used as a TGN marker (Shaner et al., 2008).
Enrichment of transport-delayed HAP1FF9 mutant cells
A gene-trap virus was produced by transfection of PLAT-GP cells in eight 15-cm dishes using Lipofectamine 2000 and a mixture of pCMT-SApA-BSD and pLC-VSVG. The virus-containing supernatant was concentrated five times using PEG-it solution and then mixed with 8 μg/ml Polybrene. HAP1FF9 cells (6 × 107 cells in total) prepared in six-well plates containing 2.5 × 106 cells/well were infected by centrifugation at 2500 rpm for 2 h at 32°C. Two days after infection, the cells were selected with 6 μg/ml blasticidin for 1 wk. HAP1FF9 cells mutagenized by gene-trap vectors were cultured in medium containing 1 μg/ml Dox at 40°C for 24 h to induce and accumulate the VFG-GPI reporter protein in the ER (Maeda et al., 2008; Fujita et al., 2009). The cells were then quickly harvested with trypsin-EDTA and incubated in medium at 32°C for 60 min, followed by staining with an M2 anti-Flag antibody and PE-conjugated goat anti-mouse IgG. Cells with delayed surface expression of the reporter protein were enriched twice by cell sorting using a FACSAria (BD Bioscience, Franklin Lakes, NJ).
Sequence analysis of the gene-trap insertion sites
Genomic DNA was isolated from 3 × 107 cells using the Wizard Genomic DNA purification kit (Promega, Fitchburg, WI) according to the manufacturer's protocol. Genomic DNA (15 μg) was digested with HaeIII, followed by ligation with the splinkerette adaptor (Horie et al., 2011), which consists of two oligonucleotides: Spl-top-HaeIII (CGAATCGTAACCGTTCGTACGAGAATTCGTACGAGAATCGCTGTCCTCTCCAACGAGCCAAGG) and SplB-BLT-HaeIII (CCTTGGCTCGTTTTTTTTTGCAAAAA). After ligation, the DNA fragments were digested with PvuII, which cleaves the vector sequence between the 3′-long terminal repeat (LTR) and the upstream HaeIII site, to prevent unwanted vector amplification. After column purification, the fragments were used as templates for splinkerette PCR using the SPl-P1 primer (CGAATCGTAACCGTTCGTACGAGAA) and the first LTR primer (AGTGTATGTAAACTTCTGACCCACTGG). The resulting DNA fragments were further amplified by nested PCRs using the SPl-P2 primer (TCGTACGAGAATCGCTGTCCTCTCC), the second LTR primer (CTTGTGTCATGCACAAAGTAGATGTCC), Rd1Tru-LTR (ACACTCTTTCCCTACACGACGCTCTTCCGATCTGCTAGCTTGCCAAACCTACAGGTGGG), and Rd2Tru-Splink (GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGCTGTCCTCTCCAACGAGCCAAGG). The underlined sequences indicate the Illumina sequencing primers. The resulting DNA products contained the end of the 5′LTR retroviral sequence, followed by the genomic DNA sequence flanking the insertion site ending at the HaeIII restriction site and part of the splinkerette adaptor sequence. Illumina P5 (AATGATACGGCGACCACCG) and P7 (CAAGCAGAAGACGGCATACGA) adapters and barcode sequences were attached to the products by six cycles of PCR with 10 ng of each of initial PCR product as the template. Paired-end sequencing (151 base pairs × 2) was performed on the MiSeq (Illumina, San Diego, CA) system. The number of reads obtained from control and Sort 2 cells was ∼1.4 million and 1.2 million, respectively.
Analysis of gene-trap insertions
FASTQ data files were analyzed using CLC Genomic Workbench software version 7.0.4 (CLC Bio) according to a previously described method (Carette et al., 2011a, b). Briefly, after quality trimming and removal of the common LTR sequence, the 50–base pair reads were mapped onto the human genome (hg19). To exclude ambiguous alignments, mismatch reads were not allowed, and all nonspecific matched reads were ignored. To eliminate PCR amplification bias and determine the unique insertion sites, duplicate reads were removed and counted as one read (a unique insertion site). The independent insertion sites were further classified as being in the sense or antisense orientation compared with the gene. The total number of inactivating insertions, which consisted of all the sense or antisense orientations in the exons of the genes and the number of inactivating insertions per individual gene, were counted. The amount of enrichment of a particular gene in the screen was calculated by comparing the selected with the unselected population. For each gene, a p value and a p value corrected for the false-discovery rate (FDR) were calculated by the one-sided Fisher exact test using R software. A bubble plot was created using R software.
Establishment of knockout cell lines
GARP-KO cell lines and TM87A, B-DKO cell lines were generated using the CRISPR-Cas9 system. For the generation of GARP-KO cell lines, HEK293FF6 cells were transiently transfected with pX330 plasmids bearing the targeting sequence for VPS51 (GGGAGGCTCCGGAGCGTCGG), VPS52 (GGCCCGGGAACTGGTGTTGC), VPS53 (GCTCACGCCCGAGGTGCAGC), and VPS54 (#1; GAGGCACTGGTGAAGAACTG and #2; GGACACACATCTGGCAGTGA). After ∼10 d of culture, cells with delayed VFG-GPI transport were sorted using a FACSAria cell sorter. The collected population was subjected to limiting dilution. To generate TM87A, B-DKO cell lines, HEK293FF6 cells were transiently transfected with pX330-mEGFP plasmids bearing the targeting sequence for hTMEM87A (GGAACAAACCTTACCTTTAT). After 3 d, cells with EGFP were sorted using a FACSAria cell sorter. The collected cells were subjected to limiting dilution. A clone that had no wild-type allele was picked up and used for next-round knockout process. A TMEM87A KO cell line was transiently transfected with pX330-mEGFP plasmids bearing the targeting sequence for hTMEM87B#1 or #2 (#1; GCCGCCGCTGCTTTCCCGCC, #2; GACCCCGGCGGCTGTGCGCG). After cell sorting, collected cells were subjected to limiting dilution. DNA sequences were analyzed by the Sanger method. The DNA sequence of each KO cell line is given in Supplemental Table S2.
Transport assay of reporter proteins using fluorescence-activated cell sorting
The transport assay was conducted as previously described (Maeda et al., 2008; Fujita et al., 2009). Briefly, cells were cultured in medium supplemented with 1 μg/ml Dox at 40°C for 24 h, harvested using trypsin-EDTA (Sigma-Aldrich), and then incubated in complete medium at 32°C for the required time. The cells were stained with an M2 anti-FLAG antibody and PE-conjugated goat anti-mouse IgG and then analyzed by FACSCantoII (BD). For the KD experiments, 0.5 × 106 cells were transfected with siRNA oligonucleotides purchased from Life Technologies using the Lipofectamine RNAi MAX reagent. Two days later, the cells were retransfected with the same siRNA oligos. On the next day, they were cultured in complete medium supplemented with 1 μg/ml Dox at 40°C for 24 h, after which transport was analyzed as described earlier. The siRNA sequences used were as follows: STX6#1, GCAACUGAAUUGAGUAUAATT (siRNA ID s19959); STX6#2, GCAGUUAUGUUGGAAGAUUTT (siRNA ID s19960); VAMP4#1, CAAACAACUUCGAAGGCAATT (siRNA ID s16525); and VAMP4#2, GAUUUGGACCUAGAAAUGATT (siRNA ID s16526). For the transient expression of FVG-TM, 0.5 × 106 cells were transfected with pME-FLAG-VSVGfull-EGFP using the Lipofectamine 2000 reagent. On the next day, the incubation temperature was shifted to 40°C for 48 h. If required, expression plasmids alone or together with pME-FLAG-VSVGfull-EGFP were transfected into the cells described.
Western blotting
Cell lysate was prepared by incubation in buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 5 mM EDTA, and 1× protease inhibitor cocktail (Roche, Basel, Switzerland), followed by centrifugation at 15,000 rpm for 15 min. Supernatant was recovered, mixed with SDS sample buffer, and boiled at 95°C for 5 min. Samples were run on 10–20% SDS–PAGE gels and transferred to polyvinylidene fluoride membranes. VPS52, VPS53, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), STX6, and α-tubulin were detected by anti-VPS52, anti-VPS53, anti-GAPDH, anti-STX6, and anti–α-tubulin antibodies.
Quantitative reverse-transcription PCR
qRT-PCR was performed and normalized by HPRT1 expression as described previously (Theiler et al., 2014).
Immunofluorescence microscopy
The cells were fixed with phosphate-buffered saline (PBS) containing 4% paraformaldehyde for 20 min at room temperature, followed by washing with 40 mM NH4Cl for 10 min. The cells were double-stained with anti-TMEM87A or anti-HA7 and anti-GPP130 antibody followed by Alexa 488–conjugated anti-mouse goat secondary antibody and Alexa 594–conjugated anti-rabbit goat secondary antibody dissolved in staining buffer A (PBS containing 1% bovine serum albumin [BSA], 0.1% NaN3, and 0.1% Triton X-100). For intracellular localization of CD59, cells were pretreated with 1 unit/ml PI-PLC (Life Technologies) for 1.5 h at 37°C. The cells were double-stained with anti-CD59 and anti-LAMP1 or anti-EEA1 as primary antibodies followed by secondary antibodies as described in staining buffer B (PBS containing 1% BSA, 0.1% NaN3, and 0.1% saponin). Confocal images were acquired on a FluoView FV1000 (Olympus, Tokyo, Japan).
Flow cytometric analysis of surface expression of CD59
Surface expression of CD59 was determined as previously described (Hirata et al., 2013).
Transport assay of reporter proteins based on immunofluorescence microscopy analysis
Cells stably expressing RFP(S158T)-GPP34 were cultured in medium supplemented with 1 μg/ml Dox at 40°C for 24 h. On the next day, they were washed with cold PBS and chased with warmed complete medium at 32°C containing 100 μg/ml cycloheximide (CHX) for the indicated time, followed by fixation. For post-Golgi anterograde transport analysis, the cells were precultured at 19°C for 3 h with complete medium containing 100 μg/ml CHX and then chased at 32°C. After fixation, the cells were stained with M2 anti-FLAG antibody as the primary antibody and Alexa 647–conjugated anti-mouse goat as the secondary antibody; the antibodies were dissolved in staining buffer without detergent. Images were obtained as described.
Retrograde transport assay of CTxB
The cells were washed twice with PBS and then cultured in cold medium supplemented with 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid and 1 μg/ml CTxB-Alexa 488 for 30 min on ice. After incubation, the cells were washed with ice-cold PBS once and incubated in warmed complete medium at 37°C for 60 min, followed by fixation and staining with combinations of anti-golgin97 antibody as primary antibody and Alexa 594–conjugated anti-mouse goat secondary antibody dissolved in staining buffer A. Images were obtained as described.
Quantification of immunofluorescence images
Fluorescence intensities were quantified using MetaMorph 3.0 (Molecular Devices, Sunnyvale, CA). The VFG-GPI, CD59, and CTxB regions and the TGN (RFP-GPP34– or golgin97-positive regions), cell surface (FLAG-positive regions), and lysosomes (LAMP1 positive regions) in each fluorescence image were assigned a threshold by MetaMorph 3.0. The fluorescence intensities of total VFG-GPI, CD59, and CTxB (I(t)) and of VFG-GPI, CD59, and CTxB that colocalized with markers (I(m)) were measured, and their ratios were expressed as the percentage of I(m) in I(t). Means and standard errors were calculated and plotted from 10 independent fluorescence images.
Expression cloning
A retrovirus-based cDNA library from human brain was stably transfected into V54KO cells. Those cells with increased transport were sorted using a cell sorter. The culture and sorting steps were repeated once more. Genomic DNA was isolated from pre-enrichment control cells and the sorted cells as described earlier. Genomic DNA (15 μg) was digested with Covaris S2 (M&S, Osaka, Japan) using the 800–base pair protocol, followed by repair of the ends of the products and ligation with the aforementioned splinkerette adaptor. After ligation, the DNA fragments were used as templates for splinkerette PCR using the primers SPl-P2 and pLIB-F0 (CTCCCTTTATCCAGCCCTCACTCC). The resulting DNA fragments were further amplified by nested PCRs using Rd1Tru-pLIB (ACACTCTTTCCCTACACGACGCTCTTCCGATCTCGCCGGAATTCGTTAGGCCATTA) and Rd2Tru-Splink (GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGCTGTCCTCTCCAACGAGCCAAGG). The underlined sequences indicate the Illumina sequencing primers. Illumina P5 and P7 adapters and barcode sequences were attached as described, and single-end sequencing (151 base pairs) was performed on the HiSeq2500 (Illumina) system. The number of reads obtained from control and sorted cells was ∼26 million and 7.2 million, respectively.
Sequence analysis of recovered cDNAs
FASTQ data files were analyzed using CLC Genomic Workbench software, version 7.0.4. After quality trimming and removal of the common pLIB sequence, the reads were mapped to the human genome (hg19). For each gene, test statistics, a p value, and the p value corrected for the FDR of Kal's Z test were calculated by comparing the number of mapped reads using CLC Genomic Workbench software, version 7.0.4 (Kal et al., 1999). Supplemental Table S3 lists the highest to lowest test statistics of Kal's Z test.
Supplementary Material
Acknowledgments
We thank T. R. Brummelkamp and the Whitehead Institute for Biomedical Research for HAP1 cells; K. Horie for pCMT-SAhygpA-NP21; C. Schindler and J. S. Bonifacino for rabbit polyclonal anti-VPS52 and anti-VPS53 and expression plasmids of VPS51, VPS52, VPS53, and VPS54; Noriyuki Kanzawa, Romina Theiler, Gun-Hee Lee, and Jihyoun Seong for helpful discussions; Keiko Kinoshita, Yukari Onoe, and Kana Miyanagi for technical help; and Kohjiro Nakamura and Yuko Kabumoto for assistance with cell sorting. This work was supported by grant-in-aids from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to T.K. and M.F.), the National Natural Science Foundation for Young Scientists of China (31400693), and the Natural Science Foundation of Jiangsu Province (BK20140141) (to M.F.). T.H. was supported by the Japan Society for the Promotion of Science Research Fellowship.
Abbreviations used:
- BSD
blasticidin-S deaminase
- CHX
cycloheximide
- COG
conserved oligomeric Golgi
- CRISPR
clustered, regularly interspaced, short palindromic repeat
- CTxB
cholera toxin B subunit
- Dox
doxycycline
- GARP
Golgi-associated retrograde protein
- GPCR
G protein–coupled receptor
- GPI-AP
glycosylphosphatidylinositol-anchored protein
- PI-PLC
phosphatidylinositol-specific phospholipase C
- Pur
puromycin N-acetyl-transferase
- RFP
red fluorescent protein
- VPS
vacuolar protein sorting
- VSVG
vesicular stomatitis virus G protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1568) on July 8, 2015.
*These authors contributed equally.
REFERENCES
- Belenkaya TY, Wu Y, Tang X, Zhou B, Cheng L, Sharma YV, Yan D, Selva EM, Lin X. The retromer complex influences Wnt secretion by recycling wntless from endosomes to the trans-Golgi network. Dev Cell. 2008;14:120–131. doi: 10.1016/j.devcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Hierro A. Transport according to GARP: receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol. 2011;21:159–167. doi: 10.1016/j.tcb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat Rev Mol Cell Biol. 2013;14:382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S, Sacher M. In sickness and in health: the role of TRAPP and associated proteins in disease. Traffic. 2014;15:803–818. doi: 10.1111/tra.12183. [DOI] [PubMed] [Google Scholar]
- Calakos N, Bennett MK, Peterson KE, Scheller RH. Protein-protein interactions contributing to the specificity of intracellular vesicular trafficking. Science. 1994;263:1146–1149. doi: 10.1126/science.8108733. [DOI] [PubMed] [Google Scholar]
- Carette JE, Guimaraes CP, Wuethrich I, Blomen VA, Varadarajan M, Sun C, Bell G, Yuan B, Muellner MK, Nijman SM, et al. Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat Biotechnol. 2011a;29:542–546. doi: 10.1038/nbt.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011b;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Scheller RH. SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- Chia PZ, Gunn P, Gleeson PA. Cargo trafficking between endosomes and the trans-Golgi network. Histochem Cell Biol. 2013;140:307–315. doi: 10.1007/s00418-013-1125-6. [DOI] [PubMed] [Google Scholar]
- Choudhury A, Marks DL, Proctor KM, Gould GW, Pagano RE. Regulation of caveolar endocytosis by syntaxin 6-dependent delivery of membrane components to the cell surface. Nat Cell Biol. 2006;8:317–328. doi: 10.1038/ncb1380. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elling U, Taubenschmid J, Wirnsberger G, O'Malley R, Demers SP, Vanhaelen Q, Shukalyuk AI, Schmauss G, Schramek D, Schnuetgen F, et al. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein M, Flusser H, Lerman-Sagie T, Ben-Zeev B, Lev D, Agamy O, Cohen I, Kadir R, Sivan S, Leshinsky-Silver E, et al. VPS53 mutations cause progressive cerebello-cerebral atrophy type 2 (PCCA2) J Med Genet. 2014;51:303–308. doi: 10.1136/jmedgenet-2013-101823. [DOI] [PubMed] [Google Scholar]
- Franch-Marro X, Wendler F, Guidato S, Griffith J, Baena-Lopez A, Itasaki N, Maurice MM, Vincent JP. Wingless secretion requires endosome-to-Golgi retrieval of Wntless/Evi/Sprinter by the retromer complex. Nat Cell Biol. 2008;10:170–177. doi: 10.1038/ncb1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Maeda Y, Ra M, Yamaguchi Y, Taguchi R, Kinoshita T. GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell. 2009;139:352–365. doi: 10.1016/j.cell.2009.08.040. [DOI] [PubMed] [Google Scholar]
- Fujita M, Watanabe R, Jaensch N, Romanova-Michaelides M, Satoh T, Kato M, Riezman H, Yamaguchi Y, Maeda Y, Kinoshita T. Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. J Cell Biol. 2011;194:61–75. doi: 10.1083/jcb.201012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Espinosa E, Pfeffer SR. A syntaxin 10-SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J Cell Biol. 2008;180:159–172. doi: 10.1083/jcb.200707136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harterink M, Port F, Lorenowicz MJ, McGough IJ, Silhankova M, Betist MC, van Weering JR, van Heesbeen RG, Middelkoop TC, Basler K, et al. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat Cell Biol. 2011;13:914–923. doi: 10.1038/ncb2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T, Fujita M, Kanzawa N, Murakami Y, Maeda Y, Kinoshita T. Glycosylphosphatidylinositol mannosyltransferase II is the rate-limiting enzyme in glycosylphosphatidylinositol biosynthesis under limited dolichol-phosphate mannose availability. J Biochem. 2013;154:257–264. doi: 10.1093/jb/mvt045. [DOI] [PubMed] [Google Scholar]
- Hong W, Lev S. Tethering the assembly of SNARE complexes. Trends Cell Biol. 2013;24:35–43. doi: 10.1016/j.tcb.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Horie K, Kokubu C, Yoshida J, Akagi K, Isotani A, Oshitani A, Yusa K, Ikeda R, Huang Y, Bradley A, Takeda J. A homozygous mutant embryonic stem cell bank applicable for phenotype-driven genetic screening. Nat Methods. 2011;8:1071–1077. doi: 10.1038/nmeth.1739. [DOI] [PubMed] [Google Scholar]
- Inadome H, Noda Y, Adachi H, Yoda K. Immunoisolaton of the yeast Golgi subcompartments and characterization of a novel membrane protein, Svp26, discovered in the Sed5-containing compartments. Mol Cell Biol. 2005;25:7696–7710. doi: 10.1128/MCB.25.17.7696-7710.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irannejad R, Wedegaertner PB. Regulation of constitutive cargo transport from the trans-Golgi network to plasma membrane by Golgi-localized G protein betagamma subunits. J Biol Chem. 2010;285:32393–32404. doi: 10.1074/jbc.M110.154963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kal AJ, van Zonneveld AJ, Benes V, van den Berg M, Koerkamp MG, Albermann K, Strack N, Ruijter JM, Richter A, Dujon B, et al. Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol Biol Cell. 1999;10:1859–1872. doi: 10.1091/mbc.10.6.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson P, Droce A, Moser JM, Cuhlmann S, Padilla CO, Heimann P, Bartsch JW, Fuchtbauer A, Fuchtbauer EM, Schmitt-John T. Loss of vps54 function leads to vesicle traffic impairment, protein mis-sorting and embryonic lethality. Int J Mol Sci. 2013;14:10908–10925. doi: 10.3390/ijms140610908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Fujita M, Maeda Y. Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J Biochem. 2008;144:287–294. doi: 10.1093/jb/mvn090. [DOI] [PubMed] [Google Scholar]
- Laufman O, Hong W, Lev S. The COG complex interacts directly with Syntaxin 6 and positively regulates endosome-to-TGN retrograde transport. J Cell Biol. 2011;194:459–472. doi: 10.1083/jcb.201102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Ide T, Koike M, Uchiyama Y, Kinoshita T. GPHR is a novel anion channel critical for acidification and functions of the Golgi apparatus. Nat Cell Biol. 2008;10:1135–1145. doi: 10.1038/ncb1773. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard F, Antony C, Tenza D, Salamero J, Goud B, Johannes L. Direct pathway from early/recycling endosomes to the Golgi apparatus revealed through the study of shiga toxin B-fragment transport. J Cell Biol. 1998;143:973–990. doi: 10.1083/jcb.143.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard F, Tang BL, Galli T, Tenza D, Saint-Pol A, Yue X, Antony C, Hong W, Goud B, Johannes L. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J Cell Biol. 2002;156:653–664. doi: 10.1083/jcb.200110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- Moser JM, Bigini P, Schmitt-John T. The wobbler mouse, an ALS animal model. Mol Genet Genomics. 2013;288:207–229. doi: 10.1007/s00438-013-0741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto-Morita K, Shin HW, Mitsuhashi H, Kitamura M, Zhang Q, Johannes L, Nakayama K. Differential effects of depletion of ARL1 and ARFRP1 on membrane trafficking between the trans-Golgi network and endosomes. J Biol Chem. 2009;284:10583–10592. doi: 10.1074/jbc.M900847200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, Abdellateef M, Rosti B, Scott E, Mansour L, et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343:506–511. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Ungar D, Hughson FM, Krieger M. The COG and COPI complexes interact to control the abundance of GEARs, a subset of Golgi integral membrane proteins. Mol Biol Cell. 2004;15:2423–2435. doi: 10.1091/mbc.E03-09-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson AL, Knight JB, Pessin JE. Syntaxin 4, VAMP2, and/or VAMP3/cellubrevin are functional target membrane and vesicle SNAP receptors for insulin-stimulated GLUT4 translocation in adipocytes. Mol Cell Biol. 1997;17:2425–2435. doi: 10.1128/mcb.17.5.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperkok R, Scheel J, Horstmann H, Hauri HP, Griffiths G, Kreis TE. Beta-COP is essential for biosynthetic membrane transport from the endoplasmic reticulum to the Golgi complex in vivo. Cell. 1993;74:71–82. doi: 10.1016/0092-8674(93)90295-2. [DOI] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Abascal-Palacios G, Tascon I, Kajava A, Magadan JG, Pioro EP, Bonifacino JS, Hierro A. Structural basis for the wobbler mouse neurodegenerative disorder caused by mutation in the Vps54 subunit of the GARP complex. Proc Natl Acad Sci USA. 2010a;107:12860–12865. doi: 10.1073/pnas.1004756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Bonifacino JS. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-golgi network. Mol Cell Biol. 2009;29:5251–5263. doi: 10.1128/MCB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Mardones GA, Bonifacino JS. Requirement of the human GARP complex for mannose 6-phosphate-receptor-dependent sorting of cathepsin D to lysosomes. Mol Biol Cell. 2008;19:2350–2362. doi: 10.1091/mbc.E07-11-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Schindler C, Magadan JG, Mardones GA, Delevoye C, Romao M, Raposo G, Bonifacino JS. Ang2/fat-free is a conserved subunit of the Golgi-associated retrograde protein complex. Mol Biol Cell. 2010b;21:3386–3395. doi: 10.1091/mbc.E10-05-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter F, Plutner H, Zhu H, Kreis TE, Balch WE. Beta-COP is essential for transport of protein from the endoplasmic reticulum to the Golgi in vitro. J Cell Biol. 1993;122:1155–1167. doi: 10.1083/jcb.122.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Chen Y, Pu J, Guo X, Bonifacino JS. EARP is a multisubunit tethering complex involved in endocytic recycling. Nat Cell Biol. 2015;17:639–650. doi: 10.1038/ncb3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt-John T, Drepper C, Mussmann A, Hahn P, Kuhlmann M, Thiel C, Hafner M, Lengeling A, Heimann P, Jones JM, et al. Mutation of Vps54 causes motor neuron disease and defective spermiogenesis in the wobbler mouse. Nat Genet. 2005;37:1213–1215. doi: 10.1038/ng1661. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitara A, Shibui T, Okayama M, Arakawa T, Mizoguchi I, Sakakura Y, Takuma T. VAMP4 is required to maintain the ribbon structure of the Golgi apparatus. Mol Cell Biochem. 2013;380:11–21. doi: 10.1007/s11010-013-1652-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR. Vps51p links the VFT complex to the SNARE Tlg1p. J Biol Chem. 2002;277:48318–48324. doi: 10.1074/jbc.M209428200. [DOI] [PubMed] [Google Scholar]
- Stow JL, de Almeida JB, Narula N, Holtzman EJ, Ercolani L, Ausiello DA. A heterotrimeric G protein, G alpha i-3, on Golgi membranes regulates the secretion of a heparan sulfate proteoglycan in LLC-PK1 epithelial cells. J Cell Biol. 1991;114:1113–1124. doi: 10.1083/jcb.114.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafesse FG, Guimaraes CP, Maruyama T, Carette JE, Lory S, Brummelkamp TR, Ploegh HL. GPR107, a G-protein-coupled receptor essential for intoxication by Pseudomonas aeruginosa exotoxin A, localizes to the Golgi and is cleaved by furin. J Biol Chem. 2014;289:24005–24018. doi: 10.1074/jbc.M114.589275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai G, Lu L, Wang TL, Tang BL, Goud B, Johannes L, Hong W. Participation of the syntaxin 5/Ykt6/GS28/GS15 SNARE complex in transport from the early/recycling endosome to the trans-Golgi network. Mol Biol Cell. 2004;15:4011–4022. doi: 10.1091/mbc.E03-12-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Maeda Y, Tashima Y, Kinoshita T. Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J Biol Chem. 2004;279:14256–14263. doi: 10.1074/jbc.M313755200. [DOI] [PubMed] [Google Scholar]
- Tashima Y, Taguchi R, Murata C, Ashida H, Kinoshita T, Maeda Y. PGAP2 is essential for correct processing and stable expression of GPI-anchored proteins. Mol Biol Cell. 2006;17:1410–1420. doi: 10.1091/mbc.E05-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theiler R, Fujita M, Nagae M, Yamaguchi Y, Maeda Y, Kinoshita T. The alpha-helical region in p24gamma2 subunit of p24 protein cargo receptor is pivotal for the recognition and transport of glycosylphosphatidylinositol-anchored proteins. J Biol Chem. 2014;289:16835–16843. doi: 10.1074/jbc.M114.568311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D, Oka T, Krieger M, Hughson FM. Retrograde transport on the COG railway. Trends Cell Biol. 2006;16:113–120. doi: 10.1016/j.tcb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- VanRheenen SM, Cao X, Lupashin VV, Barlowe C, Waters MG. Sec35p, a novel peripheral membrane protein, is required for ER to Golgi vesicle docking. J Cell Biol. 1998;141:1107–1119. doi: 10.1083/jcb.141.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A, De Craene JO, Ferro-Novick S, Novick P. Functional specialization within a vesicle tethering complex: bypass of a subset of exocyst deletion mutants by Sec1p or Sec4p. J Cell Biol. 2004;167:875–887. doi: 10.1083/jcb.200408001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GL, Na SY, Niedra R, Seed B. Deficits in receptor-mediated endocytosis and recycling in cells from mice with Gpr107 locus disruption. J Cell Sci. 2014;127:3916–3927. doi: 10.1242/jcs.135269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.