Abstract
Strict temporal control of cell cycle gene expression is essential for all eukaryotes including animals and plants. DREAM complexes have been identified in worm, fly, and mammals, linking several distinct transcription factors to coordinate gene expression throughout the cell cycle. In this issue of The EMBO Journal, Kobayashi et al (2015) identify distinct activator and repressor complexes for genes expressed during the G2 and M phases in Arabidopsis that can be temporarily separated during proliferating and post-mitotic stages of development. The complexes incorporate specific activator and repressor MYB and E2F transcription factors and indicate the possibility of the existence of multiple DREAM complexes in plants.
See also: K Kobayashi et al (August 2015)
During the past decade, an evolutionarily conserved multi-subunit protein complex comprised of several distinct transcription factors that serves as a master coordinator of cell cycle gene expression has been identified in a variety of animals. The ortholog complexes containing the retinoblastoma protein (RB), E2F and its dimerization partner DP, and MYB homologs were named dREAM (Drosophila RBF, E2F2, and MIP) or MMB (MYB–MuvB) in fly, DRM (DP, RB, and MuvB) in C. elegans, and DREAM or LINC (DP, RB-like, E2F, and MuvB) in mammals (Sadasivam & DeCaprio, 2013). The core complex of MIP (MYB-interacting proteins) or MuvB (multivulva class B proteins) contains LIN9, LIN37, LIN52, LIN53 (RBBP4), and LIN54. The E2F, MYB, and the Tesmin–CXC domain-containing LIN54 are each specific DNA-binding components of the DREAM complex. The animal DREAM complex represses most cell cycle-regulated genes during G0 when cells exit the cell cycle and enter quiescence. When cells reenter the cell cycle, activating E2Fs transactivate expression of genes required for DNA synthesis during the G1/S transition. As cells progress through S phase and enter G2, MYB bound to the MuvB complex contributes to transactivation of genes required for mitosis. While the DREAM complex contains at a minimum the MuvB complex and RB/E2F homologs, differences in the composition were found with the MYB component. In fly dREAM/MMB complex, MYB is present together in the same complex with RBF and the repressor E2F2. However, no MYB ortholog has been identified in worm. In contrast, the human B-MYB (MYBL2) is found exclusively in the MMB complex during S and G2 phases and is not present in the RB-related p107/p130- and E2F4/DP1-containing DREAM complex during quiescence. Conversely, p107, p130, and E2F4 are not components of the human MYB–MuvB (MMB) complex during S and G2 phases. MYB and MuvB serve important roles in the activation of late cell cycle genes during G2/M but not early cell cycle genes during G1/S. B-MYB may also have transcriptional repression functions in mammals (Sadasivam et al, 2012) and fly (Lewis et al, 2012).
Similar to other eukaryotes, plants share many conserved elements that control the cell cycle. The conserved entities in Arabidopsis thaliana include RBR1, an RB ortholog, three E2F transcription factors E2FA, E2FB, and E2FC, and two DP orthologs DPA and DPB. Previously, Magyar et al (2012) found that RBR1 and E2FA form a stable complex that contributes to proliferation and the post-mitotic endocycle. In Arabidopsis, three orthologs of LIN9 (ALY1, ALY2, and ALY3) and a family of Tesmin–CXC domain-containing proteins similar to LIN54 have been identified. Orthologs of the animal DREAM components LIN37 and LIN52 have not yet been identified in Arabidopsis. Interestingly, Arabidopsis has more than a hundred MYB proteins, with five members of the MYB3R (R1R2R3-MYB) family having cell cycle phenotypes. The Ito group previously reported that MYB3R1 and MYB3R4 were required for activation of late (G2/M) cell cycle genes that contain the MSA (mitosis-specific activator) element in their promoters (Haga et al, 2011).
In this issue of The EMBO Journal, the Ito group teamed up with Magyar and colleagues to focus on MYB3R3 and MYB3R5, a second subgroup of the MYB3R family (Kobayashi et al, 2015). The authors found that inactivating mutations of myb3r3/myb3r5 leads to increased levels of many G2/M genes suggesting that MYB3R3 and MYB3R5 act as transcriptional repressors. Unexpectedly, this effect of MYB3R3/MYB3R5 loss was enhanced by additional mutation of MYB3R1 but not MYB3R4. This led the authors to suggest a dual role for MYB3R1 in transcriptional activation and repression of the late cell cycle genes. By comparing leaves at different stages of development, the authors observed that loss of MYB3R1/MYB3R3/MYB3R5 resulted in several fold higher activation of G2/M genes in post-mitotic leaves at 9–15 days after germination (DAG) compared to proliferating leaves (five DAG). The authors confirmed this observation by employing G2/M promoter–reporter plasmids expressing YFP and found that myb3r1/3/5 mutant but not wild-type plants express YFP in differentiated cells.
The authors performed ChIP-seq of GFP-tagged MYB3R3 and identified 398 genes significantly enriched for MYB3R3 binding. Motif enrichment analysis identified E2F and MSA elements within the MYB3R3 ChIP peaks. However, only the MSA genes with G2/M dependency were derepressed by myb3r1/3/5 mutation in the microarray expression profiling. The unexpected discovery of E2F motif enrichment in the MYB3R3 ChIP-seq experiment led Kobayashi and colleagues to search for a DREAM-like complex in Arabidopsis. Mass spectrometry analysis (LC-MS/MS) of immunoprecipitated GFP-tagged MYB3R3, RBR1, and E2FB from seven DAG seedlings revealed an association of repressor MYB3R3 with the LIN9 orthologs ALY2 and ALY3 and the LIN54 ortholog TCX5. The activator E2FB was found to associate with ALY3 and TCX5 in addition to RBR1, DPA, and DPB. The authors validated the mass spectrometry findings using immunoprecipitation–immunoblot assays in early (eight DAG) and late (14–15 DAG) stages of leaf development. The analyses confirmed that the repressor MYB3R3 did not bind to the activator E2FB but instead binds to RBR1 and the repressor E2FC at later stages of leaf development when cell proliferation rate is reduced. In contrast, the experiments showed that activator MYB3R4 binds to activator E2FB and RBR1 but not to repressor E2FC in early proliferating stages of leaf development. Thus, Arabidopsis appears to employ two distinct complexes for transcriptional regulation of G2/M genes that are temporarily separated during development (Fig1).
Figure 1.
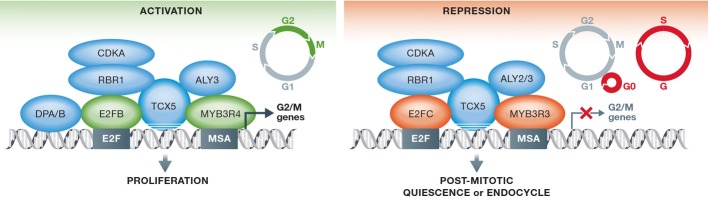
Distinct MYB activator and repressor complexes in Arabidopsis thaliana
Model based on data by Kobayashi et al (2015). Distinct MYB activator and repressor complexes regulate G2/M gene expression in Arabidopsis proliferating and post-mitotic endocycling cells. MYB3R3/4 may bind directly to MSA elements and E2FB/C to E2F elements. It is not known whether TCX5, the Arabidopsis ortholog of LIN54, binds directly to DNA.
Together, Kobayashi et al (2015) identified distinct roles for the MYB3R family members during the cell cycle of Arabidopsis and suggest the existence of at least two different DREAM-like complexes during proliferating and post-mitotic stages of organ development. The Arabidopsis complexes appear to incorporate several attributes from fly and mammalian DREAM including the co-occurrence of MYB and RB/E2F in one complex as seen in fly and the use of distinct temporarily separated repressor and activator complexes as in mammals. The Arabidopsis complexes include DREAM components such as RBR1, E2FB/C, DPA/B, ALY2/3, TCX5, and MYB3R3/4. Intriguingly, the authors found that the activator RBR1/E2FB/MYB3R4 complex and the repressor RBR1/E2FC/MYB3R3 complex regulate G2/M target genes via the MSA element. In contrast, the animal DREAM and MMB complexes were shown to regulate G2/M genes through CHR (cell cycle genes homology region) elements in mammals (Müller et al, 2014), worm (Tabuchi et al, 2011), and fly (Korenjak et al, 2012) with LIN54 likely providing direct DNA-binding specificity. Moreover, since LIN52 was shown to be crucial for the switch between repressor and activator DREAM and MMB complexes in human (Litovchick et al, 2011) and fly (Lewis et al, 2012), it will be interesting to identify the mechanism underlying the switch between the complexes reported here in Arabidopsis for which no LIN52 ortholog has been identified. It remains open for future investigations whether the complexes identified by Kobayashi and colleagues resemble plant versions of the animal DREAM and MMB complexes and whether the plethora of MYB homologs in plants can help to distinguish the repressor and activator roles of the DREAM complex.
References
- Haga N, Kobayashi K, Suzuki T, Maeo K, Kubo M, Ohtani M, Mitsuda N, Demura T, Nakamura K, Jürgens G, Ito M. Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 2011;157:706–717. doi: 10.1104/pp.111.180836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Suzuki T, Iwata E, Nakamichi N, Suzuki T, Chen P, Ohtani M, Ishida T, Hosoya H, Müller S, Leviczky T, Pettkó-Szandtner A, Darula Z, Iwamoto A, Nomoto M, Tada Y, Higashiyama T, Demura T, Doonan JH, Hauser MT, et al. Transcriptional repression by MYB3R proteins regulates plant organ growth. EMBO J. 2015;34:1992–2007. doi: 10.15252/embj.201490899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenjak M, Anderssen E, Ramaswamy S, Whetstine JR, Dyson NJ. RBF binding to both canonical E2F targets and noncanonical targets depends on functional dE2F/dDP complexes. Mol Cell Biol. 2012;32:4375–4387. doi: 10.1128/MCB.00536-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P, Sahoo D, Geng C, Bell M, Lipsick JS, Botchan MR. Drosophila lin-52 acts in opposition to repressive components of the Myb-MuvB/dREAM complex. Mol Cell Biol. 2012;32:3218–3227. doi: 10.1128/MCB.00432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Florens LA, Swanson SK, Washburn MP, DeCaprio JA. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 2011;25:801–813. doi: 10.1101/gad.2034211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, Horváth B, Khan S, Mohammed B, Henriques R, De Veylder L, Bakó L, Scheres B, Bögre L. Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 2012;31:1480–1493. doi: 10.1038/emboj.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller GA, Wintsche A, Stangner K, Prohaska SJ, Stadler PF, Engeland K. The CHR site: definition and genome-wide identification of a cell cycle transcriptional element. Nucleic Acids Res. 2014;42:10331–10350. doi: 10.1093/nar/gku696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013;13:585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi TM, Deplancke B, Osato N, Zhu LJ, Barrasa MI, Harrison MM, Horvitz HR, Walhout AJ, Hagstrom KA. Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet. 2011;7:e1002074. doi: 10.1371/journal.pgen.1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]


