Abstract
In multicellular organisms, temporal and spatial regulation of cell proliferation is central for generating organs with defined sizes and morphologies. For establishing and maintaining the post-mitotic quiescent state during cell differentiation, it is important to repress genes with mitotic functions. We found that three of the Arabidopsis MYB3R transcription factors synergistically maintain G2/M-specific genes repressed in post-mitotic cells and restrict the time window of mitotic gene expression in proliferating cells. The combined mutants of the three repressor-type MYB3R genes displayed long roots, enlarged leaves, embryos, and seeds. Genome-wide chromatin immunoprecipitation revealed that MYB3R3 binds to the promoters of G2/M-specific genes and to E2F target genes. MYB3R3 associates with the repressor-type E2F, E2FC, and the RETINOBLASTOMA RELATED proteins. In contrast, the activator MYB3R4 was in complex with E2FB in proliferating cells. With mass spectrometry and pairwise interaction assays, we identified some of the other conserved components of the multiprotein complexes, known as DREAM/dREAM in human and flies. In plants, these repressor complexes are important for periodic expression during cell cycle and to establish a post-mitotic quiescent state determining organ size.
Keywords: cell cycle regulation, cell differentiation, DREAM complex, G2/M phase, MYB transcription factors
See also: M Fischer & JA DeCaprio (August 2015)
Introduction
During organ development, cell proliferation and differentiation are regulated in a temporally and spatially coordinated manner. In general, there is a gradual decrease in cell division activity as organogenesis proceeds, and most, if not all, cells eventually stop dividing and differentiate. The scheduled cessation of cell division is critical for the formation of organs with genetically defined sizes and morphologies (Conlon & Raff, 1999; Potter & Xu, 2001; De Vos et al, 2012; Hepworth & Lenhard, 2014). Organ initiation and growth in plants are largely post-embryonic that relies almost entirely on the activity of meristems, which contain dividing undifferentiated cells and are maintained throughout the lifetime of plants (Scheres, 2007). As cells exit the meristematic zone, cell proliferation ceases which relies on the negative regulation of cell cycle progression, but the mechanisms are not fully understood (Gutierrez, 2005; de Jager et al, 2005; Inzé & De Veylder, 2006; Komaki & Sugimoto, 2012).
Leaf and sepal growth is determinate, and these organs represent the most studied models for the temporal and spatial regulation of cell proliferation. At initial stages of organ development, active cell division leads to a rapid increase in the number of cells within the primordia, which is followed by the gradual decrease in cell division activities (Beemster et al, 2005; Roeder et al, 2010; Andriankaja et al, 2012). In Arabidopsis, the cessation of cell division is associated with the onset of endoreduplication, in which DNA replication is repeated without intervening mitosis, leading to an increase in cellular DNA content (De Veylder et al, 2011; Fox & Duronio, 2013). As differentiation takes place, cells enter a quiescent state that is typically maintained for the rest of the plant’s life. Similar temporal changes in cell division activity occur during the indeterminate growth of the root, generating an apical–basal positional gradient of cell division activity (Vanstraelen et al, 2009; Ivanov & Dubrovsky, 2013).
In Arabidopsis, genome-wide expression profiling uncovered the dynamic transcriptional regulation during root and leaf development (Birnbaum et al, 2003; Beemster et al, 2005). The cluster of G2/M-phase-specific genes showed rapid and pronounced downregulation as cells differentiate which was correlated with the cessation of cell division. However, it is not clear whether such downregulation is an active process that is mediated by developmentally regulated transcriptional repression, or whether it is an indirect consequence of decreased cell proliferation activity.
It is widely accepted that transcriptional regulations are essential for the developmental control of cell division (Berckmans & De Veylder, 2009). One of the important mechanisms for such regulation is based on the retinoblastoma (RB)-E2F pathway, which regulates the expression of many genes required for cell proliferation. The conserved tumor suppressor RB, called RB-related (RBR) in Arabidopsis, is known to associate with three functionally distinct E2F transcription factors. RBR may repress cell proliferation through E2FB (Magyar et al, 2012), while E2FC acts as transcriptional repressor and is required for the timed cessation of cell division and occurrence of endoreduplication during leaf development (del Pozo et al, 2006; de Jager et al, 2009). Thus, both RBR1 and E2FC act as negative regulators for cell division and are required for scheduled exit from the mitotic cell cycle that might be important to set up the developmental gradient of cell division activities in growing organs.
In mammalian cells, DP1, the RB-related protein p130 and E2F4, together with the MuvB complex (containing LIN9, LIN37, LIN52, LIN54, and RBBP4), make a multiprotein complex known as DP, RB-like E2F, and MuvB (DREAM) complex (Litovchick et al, 2007; Sadasivam & DeCaprio, 2013). Current evidence suggests that this complex acts as a repressor of cell cycle-regulated genes during quiescence (Litovchick et al, 2007). When a quiescent mammalian cell is stimulated to enter the cell cycle, the complex releases p130, DP1, and E2F4 and instead recruits a member of the Myb transcription factors, B-MYB, to promoters of G2/M-specific genes. This recruitment is required for transcriptional activation of various genes essential for mitosis (Sadasivam et al, 2012). A similar multiprotein complex called Drosophila RBF, E2F2, and Myb (dREAM) is known in flies, which acts for repression of a variety of developmentally regulated genes and also for activation of the mitotic genes in proliferating cells (Korenjak et al, 2004; Georlette et al, 2007). The latter function of dREAM complex is attributed to dMYB, a single Myb gene in Drosophila (Beall et al, 2004; Lipsick, 2004). Accumulating evidence suggests that the conserved multiprotein complex has general roles as a global repressor and that Myb proteins counteract the repression (Sadasivam & DeCaprio, 2013). Although plants share the conserved members of E2F, DP, RB, Myb proteins (Vandepoele et al, 2002; Ito, 2005), and some MuvB components, the existence of DREAM/dREAM repressor complex and its possible functions are not known in plants.
There are more than a hundred Myb genes encoded in plant genomes. However, most of Myb proteins in plants contain only two Myb repeats in N-terminal DNA-binding domain, as opposed to the three Myb repeats in animals (Dubos et al, 2010). The plant-specific two-repeat, R2R3-Myb genes have diverse roles in plant development and environmental responses. On the other hand, a small number of three Myb repeat-containing plant proteins, called R1R2R3-Myb or MYB3R, are linked to the regulation of mitosis (Ito, 2005). We have previously shown that plant MYB3R proteins regulate many G2/M-specific genes such as CYCB1, CYCB2, and CDKB2, by binding to the common cis-acting elements that are known as MSA element (Ito et al, 1998, 2001; Kato et al, 2009; Haga et al, 2011). In Arabidopsis, there are five genes that encode MYB3R transcription factors (Dubos et al, 2010). Our previous studies showed that two structurally related MYB3R proteins, MYB3R1 and MYB3R4, act as transcriptional activators on many, if not on all, G2/M-specific genes (Haga et al, 2011). We also showed that they are required for cytokinesis via the transcriptional activation of a critical target gene, KNOLLE (KN), a gene essential for cell plate formation (Lukowitz et al, 1996; Haga et al, 2007).
Here, we report on the function of another pair of closely related MYB3R genes, MYB3R3 and MYB3R5, to be repressors of the transcription of G2/M-specific genes. The previously identified activator MYB3R1 can also have redundant repressor functions with MYB3R3 and MYB3R5 to inhibit the transcription of many G2/M-specific genes most pronouncedly in differentiated cells that have ceased to proliferate. The triple mutant of these three MYB3R genes shows hyperplasia, generating organs with increased sizes but also some developmental abnormalities and irregular cell divisions during embryogenesis. Genome-wide transcriptional profiling and chromatin immunoprecipitation experiments with MYB3R3 identified G2/M-specific target genes and show that MYB3R3 can also associate with promoters known to be E2F targets. However, the expression of these E2F target genes is not dependent on the repressor MYB3Rs. Accordingly, our biochemical data showed that MYB3R3 associates with E2FC and RBR1, while the activator MYB3R4 is found together with E2FB and RBR1. With mass spectrometry detection and pairwise interaction assays, we could also show other known DREAM/dREAM complex components together with MYB3R3, RBR1, and E2FB, but the exact composition of these complexes remains to be elucidated. We propose that the repressor MYB3R proteins may form complexes that are important for restricting the time window of mitotic gene expression in proliferating cells and for the maintenance of repressed states of G2/M-specific genes in post-mitotic cells.
Results
MYB3R1, MYB3R3, and MYB3R5 act redundantly as transcriptional repressors
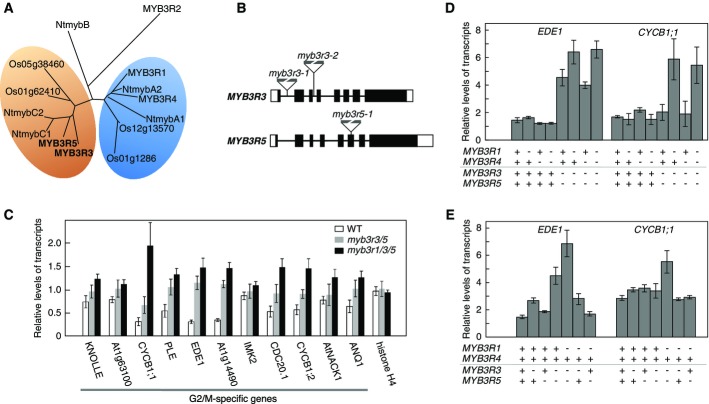
Phylogenetic analysis showed that there are two evolutionarily conserved groups in plant MYB3R family (Fig1A). One contains MYB3R1 and MYB3R4 (hereafter MYB3R1/4) from Arabidopsis, which were previously shown to act as transcriptional activators (Ito et al, 2001; Haga et al, 2007, 2011). The other group contains two Arabidopsis MYB3Rs, MYB3R3 and MYB3R5 (hereafter MYB3R3/5), whose function was addressed in this study. We analyzed T-DNA insertion alleles of these genes, myb3r3-1, myb3r3-2, and myb3r5-1, all of which resulted in complete loss of normal transcripts (Fig1B and Supplementary Fig S1). Both MYB3R3 alleles gave identical phenotypes when combined with the other myb3r mutants (see below), and thus are hereafter referred to as myb3r3. The myb3r1/4 double mutant was reported to have aberrant cytokinesis (Haga et al, 2007) suggesting that these two proteins positively regulate mitosis, a function which is not shared with MYB3R3/5, because combined triple and quadruple mutants do not influence these cytokinetic defects (Supplementary Fig S2). To analyze the roles for MYB3R3/5 in transcriptional regulation of G2/M-specific genes, we performed quantitative RT–PCR (qRT–PCR) analyses of seedlings with double myb3r3/5 mutation (Fig1C, gray bars) and found significant upregulation of many, but not all of the G2/M-specific genes with MSA element, which include those encoding mitotic regulators, CYCB1;1, CYCB1;2, CDC20.1, and also microtubule-associated proteins with cytokinetic functions, PLEIADE (PLE)/MAP65-3 (Müller et al, 2004) and ENDOSPERM DEFECTIVE1 (EDE1) (Pignocchi et al, 2009). This indicated a repressor function for MYB3R3/5.
Figure 1.
Identification of R1R2R3-Myb proteins with a repressor function
- A Phylogenetic analysis of R1R2R3-Myb proteins in plants. Protein names that begin with “Nt” are from tobacco, those that begin with “Os” are from rice, and MYB3R1–MYB3R5 are from Arabidopsis. Protein sequences within Myb domains were used to construct an unrooted phylogenetic tree.
- B Schematic structures of the MYB3R3 and MYB3R5 genes. The insertion sites of the T-DNA in each mutant allele are indicated. Exons are indicated by boxes, where untranslated regions and protein coding regions are shown in white and black colors, respectively.
- C Upregulation of G2/M-specific genes in the double myb3r3/5 and triple myb3r1/3/5 mutants. Transcript levels for a set of G2/M-specific genes were analyzed by qRT–PCR in wild-type (WT), myb3r3/5, and myb3r1/3/5 seedlings (10 DAG). Transcript level of histone H4 was also analyzed as a control. Expression levels of each transcript were normalized by the levels of ACT2 expression and are expressed as relative values with average levels of transcripts in all the plants analyzed being set to 1.0. Error bars represent standard deviation (SD) for n = 3.
- D, E MYB3R1, MYB3R3, and MYB3R5 act redundantly in the repression of G2/M-expressed genes. A qRT–PCR analysis of EDE1 and CYCB1;1 showed that MYB3R1, but not MYB3R4, acts as a repressor that is redundant with MYB3R3 and MYB3R5 (D), and that MYB3R1, MYB3R3, and MYB3R5 act redundantly with different contributions for repression of the G2/M-specific genes (E). The qRT–PCR was performed using 10-day-old seedlings with the indicated genotypes, where plus indicates the wild-type form and minus indicates homozygous mutation for each MYB3R gene. Expression levels are expressed as relative values that were normalized to the levels of ACT2 expression. Error bars represent SD for n = 3.
We then tested genetic interactions among MYB3Rs for regulating G2/M-specific genes, using qRT–PCR analysis. In the myb3r1/3/5 triple mutant, there is a further upregulation of G2/M-specific genes, EDE1 and CYCB1;1, compared to the double myb3r3/5 but not in the myb3r3/4/5 (Fig1D). This raised the unexpected possibility that MYB3R1, but not MYB3R4, has redundant functions both with activator- and repressor-type MYB3Rs. In the myb3r1/3/5 triple mutant, a large cohort of G2/M-specific genes are further upregulated in comparison to the double myb3r3/5 as shown by qRT–PCR (Fig1C, black bars). To gain insight whether this might also be the case on a genome-wide scale for all mitotic genes, we performed microarray expression profiling of seedlings (Supplementary Fig S3). Although this microarray analysis was done with a single biological replicate, the transcriptome data suggested that many genes annotated as “mitotic” or “G2/M-specific” were upregulated in myb3r1/3/5 seedlings as compared to wild type or the myb3r3/5 double mutant. It also suggested that this upregulation was specific for the gene sets annotated as “G2/M-specific” and “mitotic”, while genes related to other cell cycle phases were essentially unaffected (Supplementary Fig S4, list of each gene set is found in Supplementary Table S1).
To further investigate the genetic interactions between activator- and repressor-type MYB3Rs on mitotic gene regulation, we analyzed mitotic gene expression profiles in myb3r mutant combinations (Supplementary Fig S5, see quantitative expression data of individual mitotic genes in Supplementary Table S2). The single myb3r1 mutant had little impact on gene expression and myb3r4 showed minor downregulation, but in the double mutant, the downregulation was enhanced for a cohort of mitotic genes (Supplementary Fig S5). The single myb3r3 and myb3r5 mutants also displayed minor effects, but the double myb3r3/5 mutants showed upregulation for a distinct set of mitotic genes (Supplementary Fig S3). The upregulation of these genes was enhanced by introducing myb3r1 but not myb3r4. In the quadruple myb3r1/3/4/5 mutant, the upregulation and downregulation of these two cohorts of mitotic genes were combined (Supplementary Fig S5). The qRT–PCR analysis of EDE1 and CYCB1;1 in mutant combinations of myb3r1, myb3r3, and myb3r5 also confirmed that MYB3R1 can play redundant roles with repressor-type MYB3Rs and showed these repressor-type MYB3Rs act redundantly and contribute differently to the transcriptional repression (Fig1E). To validate the microarray results obtained from different mutant seedlings, we selected representative genes for up-, downregulated, and unchanged clusters and performed qRT–PCR analysis in biological triplicates (Supplementary Fig S6). This experiment fully confirmed the microarray data and our conclusion that MYB3R1 has redundant functions both with the activator MYB3R4 and the repressor MYB3R3/5. Hereafter, we refer to MYB3R1/3/5 as repressor MYBs (Rep-MYBs) and MYB3R1/4 as activator MYBs (Act-MYBs).
Repression of G2/M-specific genes in post-mitotic cells
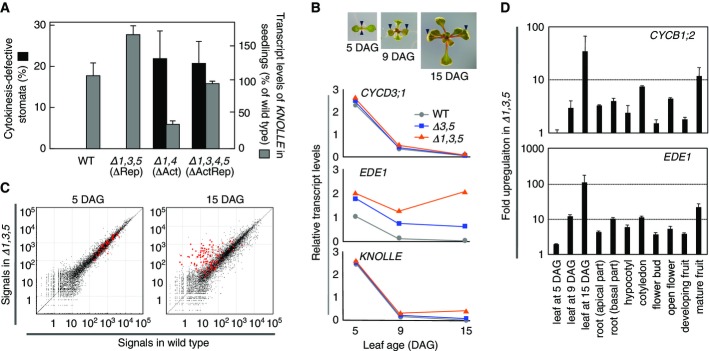
To address genetically whether or not the Act-MYB and Rep-MYB act antagonistically in the same cells, we aimed to correlate the expression of a critical mitotic target gene, KN and the cytokinetic phenotypes in the quadruple myb3r1/3/4/5 mutant. We found that the expression of KN, which is known to be downregulated in myb3r1/4 (Haga et al, 2007), but increased in myb3r1/3/5, came close to wild-type levels in the quadruple myb3r1/3/4/5 mutant, but the cytokinetic defect remained (Fig2A). This argues against that Act-MYB and Rep-MYB act in the same cell, but rather suggest that their functions are spatially and/or temporally separated. We reasoned that the loss of Rep-MYB might affect the expression of KN and other G2/M-specific genes in post-mitotic cells, which would not lead to the rescue of cytokinetic defects caused by the loss of Act-MYB in dividing cells.
Figure 2.
MYB3R1, MYB3R3, and MYB3R5 act as repressors in post-mitotic cells during organ development
- Genetic interactions between repressor and activator MYBs. The frequencies of cytokinesis-defective stomata (n = 6) and levels of KN transcripts (n = 3) were quantified using seedlings (9 DAG) with mutations in either repressor MYBs (ΔRep), activator MYBs (ΔAct), or both (ΔActRep). The qRT–PCR data were normalized by the levels of ACT2 expression. Error bars represent SD.
- Expression of G2/M-specific genes during leaf development. The first leaf pairs were harvested from wild-type, myb3r3/5, and myb3r1/3/5 plants at indicated times after germination and were used for qRT–PCR analysis to determine the transcript levels of EDE1 and KNOLLE. CYCD3;1 was also analyzed as a control with an expression that is dependent on cell division, but not directly regulated by MYB3Rs. Images of representative wild-type plants at indicated times are also shown, in which the first leaf pairs are indicated by arrowheads.
- Preferential upregulation of G2/M-specific genes in old leaves upon loss of repressor MYBs. Microarray analysis of the first leaf pair was conducted in wild-type and myb3r1/3/5 plants, and the expression signals are shown as scattered plots where red dots indicate G2/M-specific genes.
- Upregulation of G2/M-specific genes in various organs upon loss of repressor MYBs. Various organs at different developmental stages were harvested and used for qRT–PCR analysis to determine the expression levels of CYCB1;2 and EDE1. The qPCR data were normalized by the levels of ACT2 expression. Data are shown as fold upregulation in myb3r1/3/5 compared to wild-type plants. Error bars represent SD for n = 3.
Data Information: WT, wild type; Δ3,5, myb3r3/5 double mutant; Δ1,3,5, myb3r1/3/5 triple mutant.
To test this hypothesis, we determined at which developmental stages the Rep-MYBs act through time-course expression analysis of G2/M-specific genes during leaf development (Fig2B and Supplementary Fig S7A). In wild type, the transcript levels of G2/M-specific genes, EDE1 and KN, were initially high at 5 days after germination (DAG), showed rapid decrease by 9 DAG, and stayed at low levels until 15 DAG (Fig2B). It was clear that EDE1 and other G2/M-specific genes examined (except for IMK2) were upregulated in mature leaves (15 DAG) with stronger effects in the myb3r1/3/5 triple mutant than in the double myb3r3/5 mutant (Fig2B and Supplementary Fig S7A). KN expression was not affected by the myb3r3/5 double and myb3r1/3/5 triple mutations in proliferating stage of leaves at 5 DAG but was predominately upregulated at later leaf development stages (Fig2B). Consistent with the lack of phenotypic complementation in myb3r1/3/4/5 (Fig2A), expression of KN in young leaves was not recovered in the quadruple mutant in comparison with myb3r1/4 (Supplementary Fig S7B). To see whether cell proliferation is prolonged during development in these mutants, we determined CYCD3;1 expression, which is known to be correlated with cell division activities (Beemster et al, 2005), but CYCD3;1 showed age-dependent downregulation comparable with that in wild type (Fig2B).
To determine genome-wide the cohort of genes that are regulated by Rep-MYB, we performed microarray analysis of leaves at two developmental stages, comparing wild type and the myb3r1/3/5 triple mutant. The results showed that the loss of Rep-MYB dramatically upregulated most of the G2/M-specific genes in mature leaves (15 DAG; Fig2C), while the upregulation was less pronounced in young leaves and was specific for a subset of genes, including EDE1 (5 DAG; Fig2C), which showed an increased expression both at early and late stages compared to wild type (Fig2B). In agreement with what we found in seedlings, the de-repression was specific to the genes annotated as “G2/M-specific” and “mitotic”, but not to genes related to other cell cycle phases such as “E2F target”, “replication”, and “histone” (Supplementary Fig S8). Statistical analysis of the microarray data obtained from biological triplicates of the 15-DAG leaf samples identified 72 and 5 genes that are significantly up- and downregulated, respectively, upon loss of Rep-MYB (Supplementary Table S3). The upregulated genes showed enrichment of gene ontology (GO) categories such as “cell cycle”, “cytokinesis,” and “M phase” (Supplementary Fig S9A). Venn diagram analysis of upregulated genes showed a striking overlap with G2/M-specific genes, but not with those related to other phases (Supplementary Fig S9B). These results suggest that Rep-MYB may have a role for selectively repressing G2/M-specific genes, especially in post-mitotic cells during organ development.
To validate the generality of this view, we analyzed various organs at different developmental stages. In all the organs examined, qRT–PCR analysis showed that upregulation of G2/M-specific genes due to the loss of Rep-MYB was more pronounced in organs at later stages of development than in young ones (Fig2D and Supplementary Fig S10).
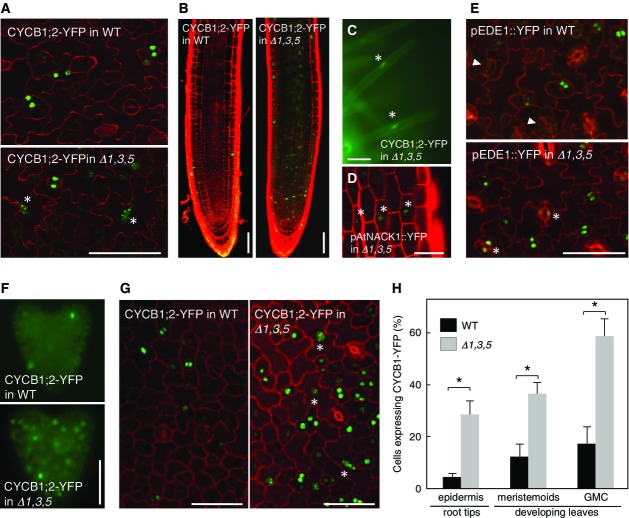
To determine the developmental position of cells that show this ectopic expression of mitotic genes in the myb3r1/3/5 triple mutant, we utilized fluorescent protein markers that are driven by promoters of the G2/M-specific genes, CYCB1;2, EDE1, and AtNACK1/HINKEL (Strompen et al, 2002). First, we used the CYCB1;2-YFP reporter, in which yellow fluorescent protein (YFP) fused to cyclin destruction box is driven by the CYCB1;2 promoter (Iwata et al, 2011). In wild-type plants, CYCB1:2-YFP expression is confined to dividing cells in leaf epidermis and root meristems. In triple myb3r1/3/5 mutants, however, there was a strong YFP expression in differentiated cells, such as large lobbed pavement cells in leaf epidermis (Fig3A), cells in the elongation zone of roots (Fig3B), and even in root hair cells (Fig3C), where CYCB1;2-YFP expression is not normally observed. Some YFP-expressing cells had enlarged nuclei, suggesting that the promoter remained active after the onset of endoreduplication (Fig3A and C, see also Supplementary Fig S11 for leaves at various developmental stages). For the proAtNACK1::YFP and proEDE1::YFP markers, a similar ectopic expression was observed in differentiated non-proliferating cells, including pavement cells and mature guard cells in leaves, and epidermal cells in hypocotyls (Fig3D and E, see also Supplementary Fig S12A). Taken together, our data strongly suggest that Rep-MYBs have a critical function in the general maintenance of the repressed state of G2/M-specific genes after the exit from cell proliferation.
Figure 3.
Ectopic expression of G2/M-specific genes in proliferating and post-mitotic quiescent cells upon loss of repressor MYBs
- Expression of CYCB1;2-YFP in the leaf epidermis of 9-day-old plants. Leaves from myb3r1/3/5 (Δ1,3,5) and wild-type (WT) plants were counterstained with propidium iodide for cell walls to visualize cell outlines and were analyzed by confocal microscopy. In myb3r1/3/5 leaves, CYCB1;2-YFP expression was often observed in cells with enlarged nuclei that had presumably undergone endoreduplication, as indicated by asterisks.
- Expression of CYCB1;2-YFP in roots of 3-day-old seedlings. CYCB1;2-YFP expression was expanded toward the basal zone of roots in myb3r1/3/5 seedlings.
- Ectopic expression of CYCB1;2-YFP in terminally differentiated root hair cells in myb3r1/3/5 seedlings (asterisks).
- Expression of proAtNACK1::YFP in epidermal non-dividing cells in myb3r1/3/5 hypocotyl (asterisks).
- Ectopic expression of proEDE1::YFP in maturing guard cells in myb3r1/3/5 leaves (asterisks). Such expression was absent in wild-type leaves (arrowheads).
- Expression of CYCB1;2-YFP in the developing embryo. In a myb3r1/3/5 embryo, a greater population of cells expressed CYCB1;2-YFP compared with a wild-type embryo.
- Expression of CYCB1;2-YFP in cotyledon from 3-day-old seedlings. Asterisks indicate YFP expression in endoreduplicated cells with enlarged nuclei.
- Quantitative comparison of CYCB1;2-YFP-expressing cells in myb3r1/3/5 and wild-type plants. Proportion of CYCB1;2-YFP-expressing cells was determined among epidermal cells in root tips of 5-day-old seedlings (n = 12), and meristemoids and guard mother cells (GMC) in first leaf pairs of 9-day-old seedlings (n = 8). Error bars represent SD. The asterisks in the graphs show differences that are statistically significant (t-test P-value < 0.05).
Data Information: WT, wild type; Δ1,3,5, myb3r1/3/5 triple mutant. Scale bars, 50 μm in (A–E, G), 200 μm in (F).
Roles of repressor MYBs in proliferating cells
Although mutations in Rep-MYB showed more pronounced upregulation in fully developed organs, we also observed increased transcript levels of some G2/M-specific genes in the developing young organs (Fig2B–D). To visualize how proliferating cells were affected by the loss of Rep-MYBs, we examined the expression of the CYCB1;2-YFP reporter in meristems and developing young organs. In wild-type root meristems, this reporter showed a characteristic patchy pattern of expression that was consistent with its previously reported G2/M specificity (Iwata et al, 2011). Compared to wild type, we observed a larger proportion of cells expressing the CYCB1;2-YFP marker in root meristems of the myb3r1/3/5 mutant (Fig3B). The early heart stage embryo is largely comprised of proli-ferating cells. In contrast to the typical patchy pattern of expression in wild type, the CYCB1;2-YFP expression was almost completely uniform in myb3r1/3/5 mutant (Fig3F). Similarly, a more uniform expression pattern was observed in proliferating cells of young cotyledons at 3 DAG (Fig3G) and leaves at 7 DAG in myb3r1/3/5 mutant (see Supplementary Fig S11). For quantitative comparison of CYCB1;2-YFP expression in myb3r1/3/5 and wild type, we determined the proportion of YFP-positive cells in epidermal layer of root meristems and in proliferating meristemoids and guard mother cells (GMC) of young developing leaves (Fig3H). The results show that a significantly larger proportion of proliferating cells are expressing CYCB1;2-YFP in the myb3r1/3/5 triple mutant, compared to wild type. Similar uniform expression was also observed for proAtNACK1::YFP and proEDE1::YFP in proliferating cells (Supplementary Fig S12). These data suggest that, in the myb3r1/3/5 triple mutant, either the CYCB1;2-YFP became ectopically expressed outside of its normal mitotic time window or there is a prolongation of or arrest in G2 phase. Cell cycle analysis by flow cytometry of young leaves and root meristems (see below) showed no evidence of increased G2 DNA content in the myb3r1/3/5 triple mutant. Therefore, our data are consistent with the idea that Rep-MYB is required to suppress transcription outside of the G2/M phase in proliferating cells.
To genetically demonstrate the functional relevance on the regulation of mitotic genes in proliferating cells, we studied the genetic interactions of myb3r1/3/5 mutant with weak mutant alleles of MAP65-3/PLE and EDE1, two genes that encode microtubule-associated proteins expressed specifically at the G2/M phase of the cell cycle and essential for mitosis and cytokinesis (Müller et al, 2004; Pignocchi et al, 2009). Both genes possess typical MSA motifs in their promoters and are upregulated after the loss of Rep-MYB (Fig1C and D, and Supplementary Fig S6), but in contrast to KN, these genes are upregulated both in proliferating cells and in cells that exited proliferation at late developmental stages as compared to wild type (Fig2B). Both ple-2 (Supplementary Fig S13B) and ede-1 (Supplementary Fig S15A) alleles have mutations at splice sites, producing mis-spliced transcripts that are likely to encode hypo-active proteins (Müller et al, 2002, 2004; Pignocchi et al, 2009). The introduction of ple-2 into myb3r1/3/5 backgrounds led to significant suppression of the cytokinetic defects seen in ple-2 roots (Fig4A). The reduced root growth phenotype of ple-2 was also markedly rescued in the myb3r1/3/5 mutant (Supplementary Fig S13A). This confirmed the idea that the reduced activity of PLE in the ple-2 hypoactive mutant was compensated by its increased expression due to the de-repression in the myb3r1/3/5 mutant. Consistent with this interpretation, we detected the upregulation of ple-2 incorrectly spliced transcripts upon loss of Rep-MYB (Supplementary Fig S13C). We further showed that increased ple-2 expression under the control of CDKA;1 promoter can partially rescue the ple-2 mutant root phenotype (Supplementary Fig S14A and B) and that the null allele of ple could not be rescued by the myb3r1/3/5 (Supplementary Fig S14C). Similar suppression by the loss of Rep-MYB was observed for the cytokinesis defect in ede1-1, which was also significantly recovered in the background of the myb3r3/5 and myb3r1/3/5 mutants (Fig4B, and Supplementary Fig S15C), and this phenotypic recovery was consistent with the upregulation of the ede1-1 transcript (Supplementary Fig S15B). The observed derepression of mitotic genes in meristematic tissues and the genetic data on the rescue of mitotic defects in proliferating cells when hypoactive alleles were upregulated in the myb3r1/3/5 mutant together suggest that Rep-MYBs do have roles in proliferating cells as well. Consistent with this notion, genes for Rep-MYBs, MYB3R1, MYB3R3, and MYB3R5, are expressed both in proliferating and maturing stages of leaves, whereas MYB3R4, without apparent repressive activity, is expressed specifically in proliferating stage (Supplementary Fig S16).
Figure 4.
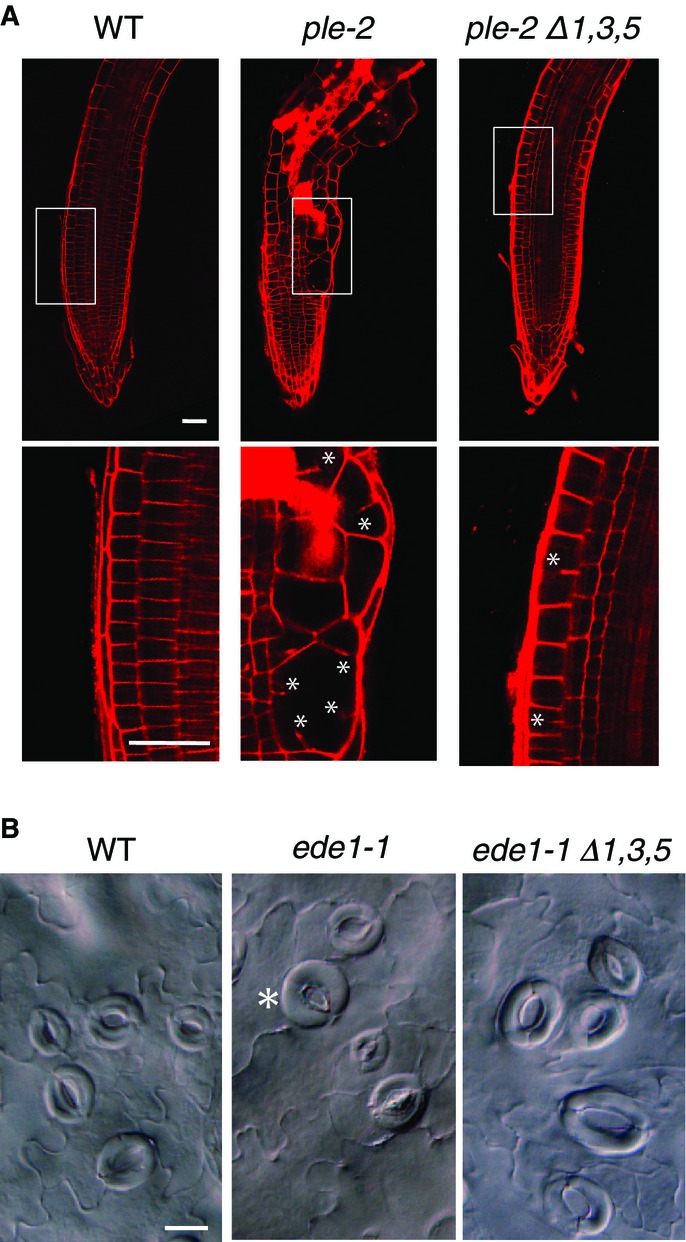
Genetic interactions between myb3r1/3/5 and ple-2 or ede1-1 reveal the functions of repressor MYBs in proliferating cells
- Cytokinesis defects in ple-2 roots were partially suppressed by combinational mutations in MYB3R1/3/5. The roots of plants with the indicated genotypes were stained at 7 DAG by propidium iodide to visualize cell outlines and were analyzed by confocal microscopy. Magnified views of the boxed area are provided below each image, to show cytokinesis defects, such as gapped cell walls and cell wall stubs (indicated by asterisks).
- Cytokinesis defects in ede1-1 cotyledons were partially suppressed by combinational mutations in MYB3R1/3/5. Cotyledons of plants (8 DAG) with the indicated genotypes were fixed, cleared, and observed by DIC microscopy. The ede1-1 mutation causes incomplete cytokinesis of guard mother cells, producing single-celled stomata (asterisk).
Data Information: WT, wild type; Δ3,5, myb3r3/5 double mutant; Δ1,3,5, myb3r1/3/5 triple mutant. Scale bars, 50 μm in (A), 20 μm in (B).
MYB3R3 associates with promoters of G2/M-specific genes and E2F target genes
Next, we asked whether Rep-MYBs associate physically with the MSA elements of target promoters. To test this in vivo, chromatin immunoprecipitation (ChIP) assays were performed using plants expressing a functional MYB3R3-GFP fusion that was driven by its own promoter (Supplementary Fig S17). Using anti-GFP antibodies, we tested binding to the promoters of 8 selected G2/M-specific genes and showed that MYB3R3-GFP was significantly enriched on all promoters that contained MSA motifs (Fig5A). As a negative control, we examined the CDKA;1 promoter, where no significant enrichment was quantified (Fig5A). To determine the genes bound by MYB3R3 on the genomic scale, we performed ChIP followed by deep sequencing (ChIP-seq). DNA libraries for deep sequencing were generated from the immunoprecipitated DNA fraction (ChIP DNA) and input DNA fraction and analyzed by Illumina Genome Analyzer IIx (Supplementary Fig S18A). This analysis identified 398 genes that were significantly enriched in ChIP DNA compared with input DNA fraction (Supplementary Tables S4 and S5). This set of genes significantly overlapped with the upregulated genes in myb3r1/3/5 (Supplementary Fig S18B) and showed extensive enrichment of GO categories such as “cell cycle”, “DNA replication” as well as “cytokinesis” (Supplementary Fig S18C, Supplementary Table S5). Venn diagram analysis showed significant overlaps not only with G2/M-specific genes as expected, but also with E2F target genes (Fig5B). Accordingly, we found that the ChIP DNA significantly overrepresented both E2F-like and MSA-like motifs (Fig5C, Supplementary Fig S18D). In most cases, binding of MYB3R3 was observed around transcriptional start sites as expected from the positions of the MSA elements (Fig5D).
Figure 5.
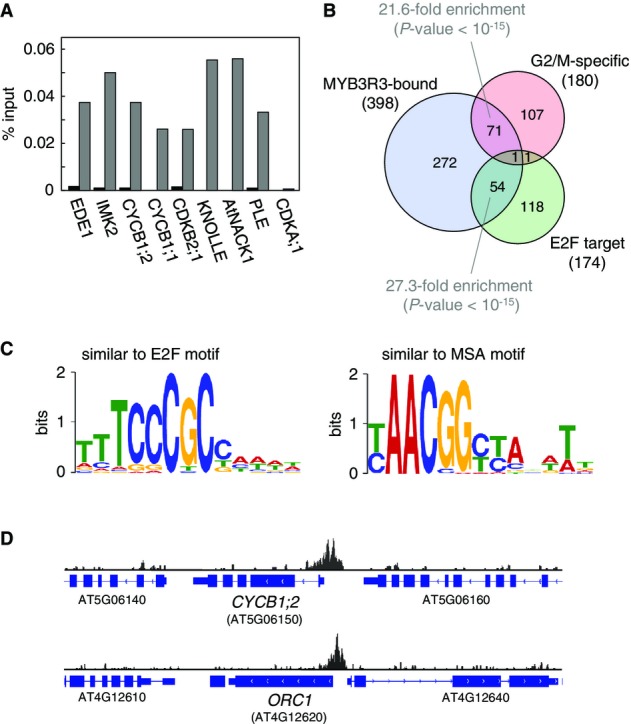
Genome-wide identification of MYB3R3-bound genes in vivo
- MYB3R3-GFP associates with promoters of G2/M-specific genes. ChIP–qPCR assays were performed using whole seedlings of MYB3R3-GFP, and the results are shown as the percentage of DNA fragments co-immunoprecipitated with anti-GFP antibody relative to input DNA. Grey and black bars indicate the results from MYB3R3-GFP and wild-type seedlings, respectively. Each measurement was performed twice and produced similar results.
- Both G2/M-specific genes and E2F target genes are enriched in MYB3R3-bound chromatin regions. Venn diagram analysis was conducted to compare MYB3R3-bound genes with the indicated gene categories. There are significant overlaps between MYB3R3-bound and G2/M-specific genes, and between MYB3R3-bound and E2F target genes with P-value < 10−15 in Fisher’s exact test. Numbers represent the gene number in each category.
- Enrichment of the MSA and E2F motifs in MYB3R3-bound chromatin regions. Enriched sequences in immunoprecipitated DNA faction were searched using motif-finding software MEME-ChIP web tool (http://meme.nbcr.net/meme/tools/meme-chip).
- Peak distributions around CYCB1;2 and ORC1 genes in the ChIP-seq analysis of MYB3R3-GFP plants. CYCB1;2 and ORC1 genes are shown as the representative examples of G2/M-specific and E2F target genes, respectively.
MYB3R3 and MYB3R4 are detected in complex with RBR1 and different E2F isoforms
Our ChIP-seq analysis showed association of MYB3R3 with E2F target genes, which indicated the presence of DREAM/dREAM-like complex in plants. To begin to investigate the existence of this complex, we performed pull-down experiments with available MYB3R3-GFP, RBR1-GFP, and E2FB-GFP plants using anti-GFP antibody and detected associated proteins with mass spectrometry (Supplementary Fig S19). We could find conserved components of the MuvB core of the plant DREAM/dREAM-like complex; ALY2, ALY3 and TCX5 with MYB3R3-GFP, and some of these were also present with RBR1-GFP and E2FB-GFP (see Supplementary Table S6 for the list of Arabidopsis homologs of DREAM/dREAM complex components). In these experiments, we could not find evidence for the association of MYB3R3 with RBR1, E2Fs, or DPs.
To address whether this is indeed the case, we used a more sensitive detection method of co-immunoprecipitation (Co-IP) assays using the MYB3R3-GFP plants. Immunopre cipitates were subjected to Western blot analysis with antibodies specific to RBR1, E2FB (Fig6A, see Supplementary Fig S20A for antibody specificity of E2FB), and E2FC (Fig6C). We found that MYB3R3-GFP did associate with RBR1 and E2FC, but not with E2FB (Fig6A and C, see also Supplementary Fig S20B and C). These associations were dependent on the developmental stage of leaves; MYB3R3-GFP exists in complex with RBR1 in both earlier and later stages (8 and 14 DAG) (Fig6A), whereas E2FC was only detected in the complex at later stages (15 DAG) of leaf development (Fig6C). The GFP-specific antibody repeatedly recognized two bands in Co-IP samples of MYB3R3-GFP, one of which (around 80 kDa) that corresponds with the calculated molecular weight of the fusion protein, and a faster migrating form that may represent a degradation product of MYB3R3-GFP (Fig6A).
Figure 6.
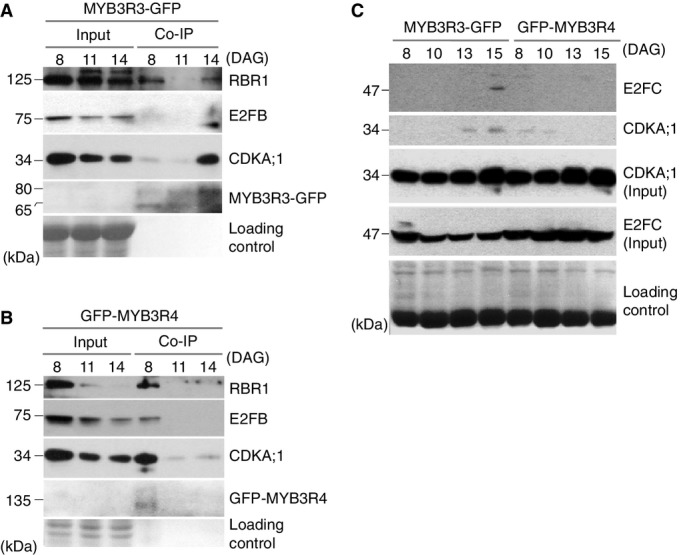
MYB3R3 and MYB3R4 both interact with RBR1 and differently associate with E2F isoforms
- A, B MYB3R3-GFP and GFP-MYB3R4 both interact with RBR1 and CDKA;1, but with a different E2F isoform in Arabidopsis leaves. IP was performed with anti-GFP antibodies from protein extracts prepared from first leaf pairs of MYB3R3-GFP or GFP-MYB3R4 transgenic plants at indicated days after germination (DAG). In these transgenic plants, expression of GFP fusion proteins was driven by the corresponding native promoters. Co-IP of RBR1 and E2FB was examined by Western blot analyses using corresponding antibodies. For detection of MYB3R3-GFP and CDKA;1, anti-GFP and anti-PSTAIRE (specific to CDKA;1) antibodies were used. As input, 1/10 of IP was loaded. Coomassie staining of the same membrane was used as a loading control.
- C MYB3R3-GFP interacts with E2FC, but GFP-MYB3R4 does not. IP was performed with anti-GFP antibodies from protein extracts prepared from first leaf pairs of MYB3R3-GFP or GFP-MYB3R4 transgenic plants at indicated days after germination (DAG). Co-IP of E2FC and CDKA;1 was examined by Western blot analyses using anti-E2FC and anti-PSTAIRE antibodies, respectively. As input, 1/16 of IP was loaded. Coomassie staining of the same membrane was used as a loading control.
Next, we conducted similar Co-IP assays for analyzing GFP-MYB3R4, which belongs to Act-MYB (Fig6B). The GFP-MYB3R4 was found to interact with RBR1, but unlike MYB3R3-GFP, it associates with E2FB and only at an early stage of leaf development (Fig6B, see also Supplementary Fig S20C–E), but not with E2FC at any stages (Fig6C). Our data suggest that the repressor MYB3R3 and activator MYB3R4 may exist in different complexes that contain different E2F isoforms with distinct functions and dynamic properties during development. In the Co-IP experiments, anti-GFP antibody could only detect MYB3R3-GFP and GFP-MYB3R4 when enriched in Co-IP samples, but not in crude extract (Fig6A and B), suggesting the low abundance of these proteins expressed under the control of their native promoters.
We previously showed that CDKA can regulate activator MYBs (Araki et al, 2004). Therefore, we tested whether CDKA can be recruited to the MYB3R3-GFP or GFP-MYB3R4 complexes. We found that CDKA;1 was present with GFP-MYB3R4 at earlier and with MYB3R3-GFP at later stages of leaf development (Fig6A and B).
To gain a third independent line of evidence for the existence of the DREAM-like complex in Arabidopsis, we synthesized the conserved Arabidopsis DREAM complex proteins by in vitro translation using wheat germ extracts and tested their pairwise interactions with in vitro-translated MYB3R3 and MYB3R4 proteins by luminescence proximity AlphaScreen assays (Takahashi et al, 2009). The repressor MYB3R3 showed clear interactions with the tested DREAM components, while the luminescence signal was much weaker with activator MYB3R4 (Supplementary Fig S21). Because the in vitro-synthesized test proteins were present together with endogenous wheat germ proteins in this assay, the interaction might not be direct, but use bridging proteins, or post-translational modifications provided from the wheat germ extract. Wheat germ cells are essentially post-mitotic, which might explain the difference in supporting complex formation around MYB3R3 but less with MYB3R4.
Taken together, the mass spectrometry data, the co-immunoprecipitation experiments, and the pairwise interaction assays in wheat germ extract show the existence of distinct DREAM-like complexes in proliferating and differentiated plant cells.
Loss of repressor MYBs led to increased organ growth
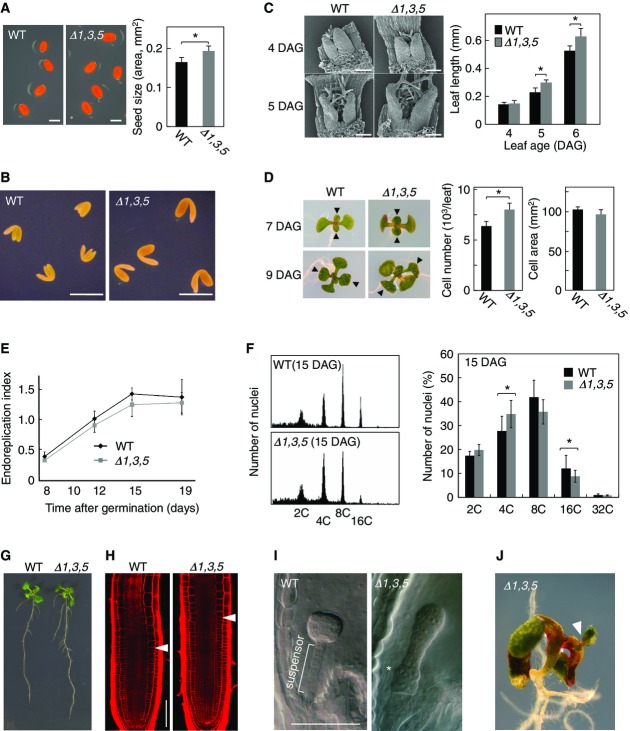
Derepression of mitotic regulators in the myb3r1/3/5 triple mutant may result in overproliferation or ectopic cell divisions. In line with this idea, the seeds (Fig7A) and embryos (Fig7B) of myb3r1/3/5 mutant are larger compared to the wild-type control. To study the reasons behind increased sizes, we examined in more details leaf development. Leaves were initiated at around the same time and had similar sizes at the earliest stage of 4 DAG but became visibly larger in the myb3r1/3/5 mutant than those of wild-type counterpart by 5 DAG (Fig7C). At 7 DAG, myb3r1/3/5 leaves contain 9.1% more cells compared with wild-type leaves, while cell sizes were not different, suggesting that the initial size increase in myb3r1/3/5 leaves is due to hypertrophy (Fig7D). To gain further insights into the processes leading to increased leaf growth, we quantified leaf growth parameters by kinematic analysis from the earliest stage (6 DAG onwards) (Beemster et al, 2005). We confirmed the initial increase in cell number at the earliest time point, but there was little difference in the cell division rates during the time window of 6–17 DAG between myb3r1/3/5 and wild-type plants (Supplementary Fig S22A). Difference in cell size was apparent after 9 DAG, with significantly larger mesophyll cells in myb3r1/3/5 leaves (Supplementary Fig S22A). As a result, myb3r1/3/5 seedlings attained significantly larger leaves that contained more cells with greater sizes as compared to wild type (Supplementary Fig S22A and B). There was only minor difference in the onset of and progression through endoreduplication between myb3r1/3/5 and wild type, somewhat decreased ploidy levels throughout leaf development, most prominently at 15 DAG (Fig7E and F, and Supplementary Fig S23).
Figure 7.
Loss of repressor MYBs causes enhanced organ growth and ectopic cell division
- Increased seed size of myb3r1/3/5 triple mutant. The average seed area after imbibition was determined (n = 25).
- Enlarged embryo of a myb3r1/3/5 triple mutant.
- Enhanced leaf growth at the initial stage of leaf development in myb3r1/3/5 triple mutant. SEM images of initiating first leaf pairs at 4 and 5 DAG are shown. A graph shows the average length of initiating leaves of wild-type and myb3r1/3/5 seedlings (n = 6).
- Increased cell number causes greater leaf size in myb3r1/3/5 triple mutants at the early seedling stage. Images show whole seedlings of wild-type and myb3r1/3/5 plants at 7 and 9 DAG. First leaf pairs are indicated by arrowheads. Graphs show that, at 7 DAG, the difference in the number of palisade cells per leaf, but not their size, is statistically significant (n = 5).
- Time-course changes of ploidy levels in first leaf pairs of wild-type and myb3r1/3/5 seedlings. The levels of ploidy are expressed by the value called “endoreduplication index,” which represents the average number of endoreduplication cycles that were experienced by the cells (n = 5).
- Ploidy distribution of first leaf pairs at 15 DAG. Representative results of flow cytometric analysis (left) and ploidy distributions (right) of wild-type and myb3r1/3/5 plants are shown (n = 5).
- Root phenotypes in wild-type and myb3r1/3/5 plants. Images show seedlings at 6 DAG.
- Increase in meristem size in myb3r1/3/5 roots. Images show propidium-iodide-stained roots at 5 DAG. Arrowheads indicate the basal end of the root meristem, which was determined by the position at which the cells start elongating.
- Ectopic cell division during myb3r1/3/5 embryo development. A cell clump was formed in suspensor of myb3r1/3/5 embryo (asterisk).
- An ectopic shoot apical meristem generated in a myb3r1/3/5 seedling.
Data Information: WT, wild type; Δ1,3,5, myb3r1/3/5 triple mutant. Scale bars: 500 μm in (A) and (B), and 100 μm in (C), (H), and (I). In all panels, error bars represent SD. The asterisks in the graphs show differences that are statistically significant (t-test P-value < 0.05).
The growth of primary roots was also significantly enhanced in the myb3r1/3/5 mutant (Fig7G and Supplementary Fig S22C). We also quantified root growth by kinematic analysis and found an elevated cell production rate as cells progress toward the elongation zone where cells exit proliferation in the root meristem (Supplementary Fig S22D). In agreement, myb3r1/3/5 roots had increased size of meristems (Fig7H and Supplementary Fig S22F), suggesting a delayed exit from cell proliferation. Flow cytometry measurements in root meristematic zones showed significant reduction in the proportion of cells with 4C DNA content in myb3r1/3/5 mutant, which is consistent with decreased duration of G2 phase (Supplementary Fig S22E). The formation of lateral roots was also significantly enhanced in myb3r1/3/5 seedlings (Fig7G, see also Supplementary Fig S22G). In conclusion, Rep-MYBs function to negatively regulate organ growth, mainly by inhibiting cell proliferation.
In addition to enhanced growth, the myb3r1/3/5 triple mutants exhibited ectopic cell divisions during embryo development. The suspensors, which connect the embryo proper to maternal tissues, normally comprises a single file of 6–8 cells and is derived from the basal daughter cell of the zygote that undergoes a few rounds of horizontal divisions. In most of the myb3r1/3/5 embryos, however, we observed irregularly oriented division planes, which created multiple files of cells or clumps of cells, within the suspensor (Fig7I and Supplementary Fig S22H). Albeit at a very low frequency, we also observed the generation of ectopic meristems, which may have been due to unscheduled division of differentiated cells (Fig7J). Therefore, Rep-MYBs may also contribute to the maintenance of the quiescent state of post-mitotic cells in different developmental contexts.
Discussion
In plants, populations of rapidly dividing cells are restricted to areas of the apical and lateral meristems and organ primordia. Strikingly, we showed that plants have mechanisms to restrict the expression of mitotic genes to meristematic tissues by the active and continuous repression of these genes outside the meristems. We identified the R1R2R3-Myb proteins, MYB3R1/3/5, to be required for the stable repression of G2/M-specific genes during plant development. Loss of Rep-MYB resulted in the upregulation of G2/M-specific genes that was most pronounced in cells that exited proliferation in organs at later stages of development. The global and long-term transcriptional repression of cell proliferation genes in differentiated cells might be important to maintain quiescence during organ development.
In meristematic cells, the loss of Rep-MYB led to nearly uniform expression of G2/M-specific genes in all cells, rather than the normal patchy pattern due to G2/M-phase-restricted expression. This was not due to an arrest in G2 phase, and therefore indicated that in proliferating cells, Rep-MYBs narrow the expression window of their target genes by inhibiting promoter activities outside the mitotic time window. We suggest that the G2/M-phase-specific transcription in plant cells is achieved by the interplay of G2/M-phase-specific promoter activation mediated by the Act-MYBs, and repression by Rep-MYB along the cell cycle except G2/M phase (Supplementary Fig S24).
One of the Rep-MYBs, MYB3R1, did not conform to the neat separation of activator and repressor functions. In the myb3r1 single mutant, the expression of mitotic genes were largely unaffected, but combining myb3r1 mutant with myb3r4 led to downregulation of G2/M-specific genes confirming our previous reports (Haga et al, 2007, 2011), while with myb3r3/5 to their upregulation (Supplementary Figs S5 and S6). These genetic data indicated that MYB3R1 can act redundantly both with activator and with repressor MYB3Rs, but this might happen in developmentally distinct cells or time window of the cell cycle. This is consistent with the idea that Rep-MYBs including the MYB3R1 have broad expression domains and play roles in differentiated cells and proliferating cells outside the G2/M phase, whereas the Act-MYB, MYB3R4, is exclusively expressed in mitotic cells and solely function during the G2/M phases. Thus, activators and repressors may work in coordination rather than in competition with each other. There are other reports of transcription factors that act both as activators and repressors. For example, the auxin response factors, ARF5, activates transcription of target genes in the presence of auxin, while in the absence it is involved in transcriptional repression by binding to AUX/IAA family proteins that are able to associate with TOPLESS corepressors (Tiwari et al, 2004; Szemenyei et al, 2008). Dual functionality of MYB3R1 may also be provided by the dynamic changes in the composition of protein complexes during the cell cycle or during plant development.
We have previously shown that mutations in Act-MYBs result in the downregulation of many, but not all G2/M-specific genes (Haga et al, 2011). Similarly, here, we found that most, but not all G2/M-specific genes were upregulated when Rep-MYBs were mutated. We noticed that the genes upregulated by the loss of Rep-MYB were largely unaffected by the loss of Act-MYB and conversely, genes that were downregulated by the loss of Act-MYB were unaffected by the loss of Rep-MYB (Supplementary Fig S5). There is no apparent difference in the core MSA sequences between downregulated and unaffected genes in myb3r1/4 (Haga et al, 2011), and between upregulated and unaffected genes in myb3r1/3/5 (Supplementary Fig S25). Therefore, these differential sensitivities to the presence or absence of Act-MYBs or Rep-MYBs might depend on the context of promoters of individual G2/M-specific genes. Depending on the promoter contexts, some genes may be mainly regulated by repression outside the G2/M phase, whereas others may be regulated through activation at the G2/M phase.
We found that the loss of Rep-MYB was associated with hypertrophy both during embryonic and post-embryonic development. The enlargement of leaves at the earliest proliferative growth stage was traced to increased cell numbers soon after leaf emergence. Overproliferation was previously reported in plants with reduced expression of the cell cycle repressors, E2FC and RBR1 (del Pozo et al, 2006; Borghi et al, 2010). Other similarities between myb3r1/3/5 mutant and the E2FC knockdown seedlings include the increased lateral root formation and upregulation of the CYCB1;1 gene (del Pozo et al, 2006). However, unlike myb3r1/3/5 mutants, overall organ growth was decreased in the E2FC and RBR1 knockdown plants, mainly due to reduced cell sizes. However, the recently identified samba mutant shows abnormalities that closely resemble the myb3r1/3/5 phenotype of increased organ size, enlarged mature embryos, enhanced root growth, and increased number of lateral roots (Eloy et al, 2012). It should also be noted that both myb3r1/3/5 and samba mutants have elevated numbers of cells as well as increased cell size in leaves. SAMBA encodes an activator protein of the anaphase-promoting complex/cyclosome (APC/C), which is an evolutionarily conserved ubiquitin–ligase complex that has a critical function in the degradation of mitotic regulators, including cyclins of CYCB and CYCA classes. Thus, the commonalities between the myb3r1/3/5 and samba mutants in respect to enlarged organ size might be due to the elevated levels of mitotic regulators either through their increased transcription or their decreased degradation, respectively. Correspondingly, it was reported that overexpression of CYCB1;1 can increase lateral root growth (Doerner et al, 1996). Our results further underline that cell cycle regulation at the G2/M phase can have a strong impact on plant growth and morphogenesis.
The conserved DREAM/dREAM repressor complexes have important roles in the coordination of cell proliferation with the developmentally imposed quiescence and the periodic expression of cell cycle genes in animal cells (Sadasivam & DeCaprio, 2013). We could identify by mass spectrometry the presence of orthologous proteins; LIN54 and LIN9 named as TCX5 and ALY2, ALY3 in Arabidopsis in association with both MYB3R3 and RBR1 or E2FB, suggesting that the MuvB core of the DREAM/dREAM-like complex might also link the RBR-E2F and MYB3R functions in Arabidopsis. We also confirmed the interaction of MYB3R3 with RBR1 and specifically with E2FC while MYB3R4 with RBR1 and E2FB. Pairwise interaction assays in wheat germ extracts of in vitro translated MYB3R3 with conserved plant components of the DREAM/dREAM further support the existence of similar complexes in Arabidopsis. Analogous to the roles of Rep-MYBs on mitotic genes in Arabidopsis, the mammalian DREAM complexes are required for repression of many cell cycle-regulated genes during quiescence (Litovchick et al, 2007). However, in mammalian cells, B-MYB is only recruited to the DREAM complex at the cell cycle entry, which coincides with the release of the repressive E2F4, DP1, and the Rb-related p130, leading to the activation of G2/M-specific genes (Sadasivam et al, 2012). In contrast, we showed that, in differentiated plant cells, the Rep-MYB can coexist in the same protein complex with RBR1 and/or E2FC. In this respect, this is more similar to the Drosophila dREAM complex formed around RBF, E2F2, and Myb (Korenjak et al, 2004; Georlette et al, 2007). The only Myb protein in Drosophila, dMYB, is required both for the transcriptional activation and for the repression (Georlette et al, 2007). In Arabidopsis, there are two distinct classes of Myb proteins: Act-MYBs responsible for activation in mitotic cells, while Rep-MYBs acting as global repressors in post-mitotic cells and during the cell cycle progression outside mitosis. Unlike animal Myb proteins that generally act as activators by antagonizing the DREAM/dREAM complexes (Georlette et al, 2007; Sadasivam et al, 2012; DeBruhl et al, 2013), Rep-MYBs may act exclusively as repressors on mitotic genes in Arabidopsis. Moreover, our data suggested that Arabidopsis has separate complexes containing either the repressor MYB3R3 and E2FC or the activator MYB3R4 and E2FB. Thus, the Arabidopsis MYB3R complexes may differ from the conserved animal DREAM/dREAM complexes in terms of their composition and heterogeneity. Further biochemical characterization of the dynamic changes in protein composition of the plant DREAM/dREAM-like complexes during development and cell cycle progression, and genetic studies of the constituents may provide new insights into the molecular basis and the plant-specific nature how cell proliferation is actively restricted to meristems and how the quiescent state is maintained during post-mitotic organ development.
Materials and Methods
Plant materials and growth condition
Arabidopsis thaliana Columbia (Col) was used as the wild-type plant. All mutants and transgenic lines used in this study were in a Col background. The mutant alleles myb3r3-1 (SALK_041111), myb3r3-2 (GABI 546A07), and myb3r5-1 (SALK_031972) were identified from the SALK and GABI-KAT T-DNA collections (Alonso et al, 2003; Rosso et al, 2003) and were used to generate multiple mutant combinations. Other mutant and transgenic lines, myb3r1-1, myb3r4-1, ede1-1, ple-2, CYCB1;1-GUS, CYCB1;2-YFP, RBR1-GFP, and E2FB-GFP, were described previously (Colón-Carmona et al, 1999; Müller et al, 2002; Haga et al, 2007; Pignocchi et al, 2009; Iwata et al, 2011; Magyar et al, 2012). The sterilized seeds were germinated on one-half-strength Murashige and Skoog (1/2 MS) medium containing 2% (w/v) sucrose and 0.6% (w/v) agar. Plants were grown on 1/2 MS agar medium or soil under continuous light at 22°C. For the analysis of root phenotypes, plants were grown on a vertical surface of 1/2 MS medium containing 1.0% (w/v) agar.
Plasmid construction and transformation
For the construction of proAtNACK1::YFP and proEDE1::YFP, upstream regions with a length of 2.1 and 1.1 kb, respectively, were amplified by PCR from wild-type (Col) genomic DNA and cloned into the pENTR/D-TOPO vector (Invitrogen). The upstream fragments were then transferred into the pBGYN binary vector (Kubo et al, 2005) using LR Clonase II (Invitrogen), to create the fusion constructs between promoters and nuclear-localized YFP. To create the MYB3R3-GFP expression construct, a genomic fragment of MYB3R3 containing 1.3 kb of upstream region and the complete coding sequence (CDS) was amplified by PCR from Col genomic DNA and cloned into the pENTR/D-TOPO vector to create pENTR-MYB3R3. The GFP fragment was prepared by PCR and inserted into the pENTR-MYB3R3 plasmid using the In-fusion cloning system (Clontech), to generate an MYB3R3-GFP fusion construct in which GFP is fused in frame to the C-terminus of MYB3R3 in the native genomic context. The fusion construct was then transferred using LR Clonase II to the pGWB501 destination vector (Nakagawa et al, 2007), to create proMYB3R3::MYB3R3-GFP. proMYB3R4::GFP-MYB3R4 was similarly prepared using MYB3R4 genomic fragment containing 1.5 kb upstream region. The primers used for the construction of these plasmids are listed in Supplementary Table S7.
RNA extraction and qRT–PCR
Extraction of total RNA and synthesis of first-strand cDNA were performed as described previously (Haga et al, 2007). Real-time qRT–PCR was performed using the SYBR Premix Ex Taq (Perfect Real Time) Kit (TaKaRa Biomedicals) on a LightCycler480 machine (Roche Diagnostics). See Supplementary Table S7 online for a list of the primers used for qRT–PCR.
Microarray analysis
Microarray analyses were performed using an ATH1 GeneChip® (Affymetrix). A 10-μg aliquot of total RNA was reverse-transcribed, labeled with the One-Cycle Target Labeling and Control Reagents Kit (Affymetrix), and used for hybridization to the chip according to the supplier’s protocol. Data analysis was performed using Microarray Suite ver. 5 (Affymetrix) and GeneSpring 7.1 (Agilent Technologies). Per-chip normalization was performed using the 50th percentile of all measurements, to adjust total signal intensity in each chip. For transcriptome profiling in various myb3r mutant combinations, whole seedlings at 9 DAG were analyzed with single biological replicate. To compare transcript levels in leaves from 15-day-old plants, three biological replicates for wild type and myb3r1/3/5 were analyzed. Genes with a false discovery rate (FDR) < 0.05 were defined as upregulated genes in myb3r1/3/5 compared with wild-type plants. Heat maps of G2/M-specific genes were created using log2-tranformed values of fold change levels that were calculated from the normalized data.
Microscopic observation
Microscopic observations with differential interference contrast (DIC) and fluorescent optics were as described previously (Haga et al, 2007). Confocal microscopy of live tissues was carried out using an FV1000 Olympus Confocal Microscope. To counterstain cell outlines, tissues were placed in a solution of 10 μM FM4-64 (Molecular Probes) or 0.1 mg ml−1 propidium iodide for 1–2 min. GFP was excited at 473 nm and fluorescence was detected at 485–545 nm, and FM4-64 and propidium iodide were excited at 559 nm and fluorescence was detected at 570–670 nm.
Clearing of plant materials and histochemical GUS assay were performed as described previously (Haga et al, 2007, 2011).
For scanning electron microscopy (SEM), seedlings were fixed overnight in FAA (3.7% formaldehyde, 5% acetic acid, and 50% ethanol) at 4°C. The samples were treated with 50, 70, 90, and 100% ethanol and were dried with a critical point drier (HCP-2, Hitachi, Tokyo, Japan). They were coated with Pt-Pd in an ion sputter (E-1030, Hitachi) on an aluminum SEM sample holder and then observed by SEM (S-3000N, Hitachi).
Other phenotypic analyses
Ploidy analysis was performed as described previously (Haga et al, 2011). Briefly, nuclei were extracted from whole leaves using the High Resolution Kit for Plant DNA (Partec). After filtration, nuclei were stained by adding 4′,6-diamidino-2-phenylindole (DAPI) solution and analyzed using a PAS flow cytometer (Partec) according to the supplier’s instructions. The proportion of nuclei of each ploidy level was estimated as described previously (Imai et al, 2006). The endoreduplication index, which represents the average number of endoreduplication cycles per nucleus, was calculated as described previously (Lammens et al, 2008).
To measure seed size, photographs were taken under a stereomicroscope (MZ16, Leica), and seed area was measured using the ImageJ software. The sizes of root meristems were determined as described previously (Takahashi et al, 2013).
ChIP analysis
The ChIP-qPCR assay was performed as described previously (Nakamichi et al, 2010). Briefly, the wild-type and transgenic plants carrying proMYB3R3::MYB3R3-GFP were grown for 9 days in liquid 1/2 MS medium supplemented with 2% (w/v) sucrose, with gentle agitation. Approximately 0.8 g (FW) of whole seedlings were fixed and used in the ChIP assay. An anti-GFP antibody (ab290; Abcam) was used for immunoprecipitation of chromatin complexes bound to MYB3R3-GFP. The amount of each precipitated DNA and input DNA was determined by real-time PCR using specific primers (Supplementary Table S7).
For ChIP-seq analysis, 3 g of MYB3R3-GFP-expressing plants at 9 DAG were fixed as in ChIP-qPCR analysis. Isolation of nuclei, ChIP with anti-GFP antibody, and library construction were performed as described previously (Nakamichi et al, 2012). The resulting ChIP and input DNA libraries were sequenced with an Illumina Genome Analyzer IIx (GAIIx).
ChIP-seq data analysis
Basecalls of sequence reads were done by the Illumina GA II pipeline. To map sequence reads on the Arabidopsis genome, ChIP and input DNA sequence data in the FASTQ format were analyzed by Bowtie (Langmead et al, 2009) against the reference genome TAIR10. Peaks significantly appearing in ChIP DNA compared to the input (FDR q < 10−50) were annotated as binding loci of MYB3R3 (a total of 398 loci) by Model-based Analysis of ChIP-Seq (MACS2) (Zhang et al, 2008). MACS2 was also used for validating forward- and reverse-peak distribution. The data for MACS2 were drawn by R (http://www.r-project.org/). Peaks from forward and reverse strands were within 200 bp, indicating that DNA fragment sizes in the ChIP library were acceptable and that MYB3R3 associates within a very close region of these peaks (Supplementary Fig S18A). Mapping reads (bam format from CLC-bio data analysis) were visualized with Integrative Genomics Viewer (http://www.broadinstitute.org/igv/).
Co-IP experiments
For Co-IP experiments, total protein was extracted from the first leaf pairs of MYB3R3-GFP or GFP-MYB3R4 lines according to Henriques et al (2010). Equal amount of proteins (between 400 and 800 μg) was precipitated by using 10 μl of GFP-Trap coupled to magnetic beads (ChromoTek). Immunoprecipitated proteins were eluted from the magnetic beads by SDS-sample buffer and separated on 10% SDS–PAGE together with equal loading of 20–25 μg of total protein extract as input material and blotted to PVDF membrane. Specific antibodies against RBR1 (Horvath et al, 2006), E2FB (Magyar et al, 2005), E2FC (López-Juez et al, 2008), CDKA;1 (anti-PSTAIRE, Sigma), and GFP (Roche) were used in immunoblotting experiments.
Data deposition
The microarray and ChIP-seq data have been submitted to Gene Expression Omnibus and assigned to the identifier accession: GSE52298 and GSE60554, respectively. The data can be viewed from the following Web site: http://www.ncbi.nlm.nih.gov/geo.
Acknowledgments
We thank C. Kotani, M. Kurata, C. Ohno, K. Takahashi, K. Kato, M. Miyake, and N. Ono for technical assistance, and Y. Machida, Y. Yoshioka, and Y. Mizukami for helpful discussion. This work was supported in part by JSPS KAKENHI (grant numbers, 25119710 and 26113509), MEXT KAKENHI (grant number 26291058), and JST, CREST given to M.I., János Bolyai Research Scholarship of the Hungarian Academy of Sciences given to A.P.-S., Hungarian Scientific Research Found (OTKA 105816) given to Z.M., and by the EU FP7 program AGRON-OMICS network (037704) given to J.D.
Author contributions
KK, ToS, and MI generated plants with multiple mutations in MYB3R genes and performed their genetic and molecular characterizations. TD and MO performed microarray analysis. NN, TaS, and TH conducted ChIP experiments and data processing. EI, PC, and MU contributed to the phenotypic characterization of myb3r1/3/5. TI and KS generated and analyzed transgenic plants expressing MYB3R3-GFP and GFP-MYB3R4. JHD provided the ede1-1 mutant and contributed to its genetic characterization. SM and MTH provided the ple-2 mutant and contributed to its genetic characterization. AI and HH performed kinematic analysis of roots. ZM, TL, and LB performed Co-IP experiments. APS and ZD conducted mass spectrometry analysis. MN and YT contributed to the Alphascreen interaction assays. KK, ToS, LB, and MI wrote the manuscript with contributions from all coauthors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figures, Supplementary Tables S6 and S7, Supplementary Methods
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Review Process File
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GT, Inzé D. Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell. 2012;22:64–78. doi: 10.1016/j.devcel.2011.11.011. [DOI] [PubMed] [Google Scholar]
- Araki S, Ito M, Soyano T, Nishihama R, Machida Y. Mitotic cyclins stimulate the activity of c-Myb-like factors for transactivation of G2/M phase-specific genes in tobacco. J Biol Chem. 2004;279:32979–32988. doi: 10.1074/jbc.M403171200. [DOI] [PubMed] [Google Scholar]
- Beall EL, Bell M, Georlette D, Botchan MR. Dm-myb mutant lethality in Drosophila is dependent upon mip130: positive and negative regulation of DNA replication. Genes Dev. 2004;18:1667–1680. doi: 10.1101/gad.1206604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GT, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M. Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 2005;138:734–743. doi: 10.1104/pp.104.053884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans B, De Veylder L. Transcriptional control of the cell cycle. Curr Opin Plant Biol. 2009;12:599–605. doi: 10.1016/j.pbi.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Borghi L, Gutzat R, Fütterer J, Laizet Y, Hennig L, Gruissem W. Arabidopsis retinoblastoma-related is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell. 2010;22:1792–1811. doi: 10.1105/tpc.110.074591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 1999;20:503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A. Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 2011;16:624–634. doi: 10.1016/j.tplants.2011.07.001. [DOI] [PubMed] [Google Scholar]
- De Vos D, Dzhurakhalov A, Draelants D, Bogaerts I, Kalve S, Prinsen E, Vissenberg K, Vanroose W, Broeckhove J, Beemster GT. Towards mechanistic models of plant organ growth. J Exp Bot. 2012;63:3325–3337. doi: 10.1093/jxb/ers037. [DOI] [PubMed] [Google Scholar]
- DeBruhl H, Wen H, Lipsick JS. The complex containing Drosophila Myb and RB/E2F2 regulates cytokinesis in a histone H2Av-dependent manner. Mol Cell Biol. 2013;33:1809–1818. doi: 10.1128/MCB.01401-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner P, Jørgensen JE, You R, Steppuhn J, Lamb C. Control of root growth and development by cyclin expression. Nature. 1996;380:520–523. doi: 10.1038/380520a0. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Eloy NB, Gonzalez N, Van Leene J, Maleux K, Vanhaeren H, De Milde L, Dhondt S, Vercruysse L, Witters E, Mercier R, Cromer L, Beemster GT, Remaut H, Van Montagu MC, De Jaeger G, Ferreira PC, Inzé D. SAMBA, a plant-specific anaphase-promoting complex/cyclosome regulator is involved in early development and A-type cyclin stabilization. Proc Natl Acad Sci USA. 2012;109:13853–13858. doi: 10.1073/pnas.1211418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ. Endoreplication and polyploidy: insights into development and disease. Development. 2013;140:3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georlette D, Ahn S, MacAlpine DM, Cheung E, Lewis PW, Beall EL, Bell SP, Speed T, Manak JR, Botchan MR. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev. 2007;21:2880–2896. doi: 10.1101/gad.1600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. Coupling cell proliferation and development in plants. Nat Cell Biol. 2005;7:535–541. doi: 10.1038/ncb0605-535. [DOI] [PubMed] [Google Scholar]
- Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, Suzuki K, Müller I, Voss U, Jürgens G, Ito M. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development. 2007;134:1101–1110. doi: 10.1242/dev.02801. [DOI] [PubMed] [Google Scholar]
- Haga N, Kobayashi K, Suzuki T, Maeo K, Kubo M, Ohtani M, Mitsuda N, Demura T, Nakamura K, Jürgens G, Ito M. Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 2011;157:706–717. doi: 10.1104/pp.111.180836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, Szabados L, de la Torre C, Koncz C, Bögre L. Arabidopsis S6 kinase mutants display chromosome instability and altered RBR-E2F pathway activity. EMBO J. 2010;29:2979–2993. doi: 10.1038/emboj.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth J, Lenhard M. Regulation of plant lateral-organ growth by modulating cell number and size. Curr Opin Plant Biol. 2014;17:36–42. doi: 10.1016/j.pbi.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Horvath BM, Magyar Z, Zhang Y, Hamburger AW, Bako L, Visser RG, Bachem CW, Bögre L. EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J. 2006;25:4909–4920. doi: 10.1038/sj.emboj.7601362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai KK, Ohashi Y, Tsuge T, Yoshizumi T, Matsui M, Oka A, Aoyama T. The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell. 2006;18:382–396. doi: 10.1105/tpc.105.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D, De Veylder L. Cell cycle regulation in plant development. Annu Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- Ito M, Iwase M, Kodama H, Lavisse P, Komamine A, Nishihama R, Machida Y, Watanabe A. A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell. 1998;10:331–341. doi: 10.1105/tpc.10.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A. G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell. 2001;13:1891–1905. doi: 10.1105/TPC.010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M. Conservation and diversification of three-repeat Myb transcription factors in plants. J Plant Res. 2005;118:61–69. doi: 10.1007/s10265-005-0192-8. [DOI] [PubMed] [Google Scholar]
- Ivanov VB, Dubrovsky JG. Longitudinal zonation pattern in plant roots: conflicts and solutions. Trends Plant Sci. 2013;18:237–243. doi: 10.1016/j.tplants.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Iwata E, Ikeda S, Matsunaga S, Kurata M, Yoshioka Y, Criqui MC, Genschik P, Ito M. GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell. 2011;23:4382–4393. doi: 10.1105/tpc.111.092049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager SM, Maughan S, Dewitte W, Scofield S, Murray JA. The developmental context of cell-cycle control in plants. Semin Cell Dev Biol. 2005;16:385–396. doi: 10.1016/j.semcdb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- de Jager SM, Scofield S, Huntley RP, Robinson AS, den Boer BGW, Murray JAH. Dissecting regulatory pathways of G1/S control in Arabidopsis: common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol Biol. 2009;71:345–365. doi: 10.1007/s11103-009-9527-5. [DOI] [PubMed] [Google Scholar]
- Kato K, Gális I, Suzuki S, Araki S, Demura T, Criqui MC, Potuschak T, Genschik P, Fukuda H, Matsuoka K, Ito M. Preferential up-regulation of G2/M phase-specific genes by overexpression of the hyperactive form of NtmybA2 lacking its negative regulation domain in tobacco BY-2 cells. Plant Physiol. 2009;149:1945–1957. doi: 10.1104/pp.109.135582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki S, Sugimoto K. Control of the plant cell cycle by developmental and environmental cues. Plant Cell Physiol. 2012;53:953–964. doi: 10.1093/pcp/pcs070. [DOI] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binné UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens T, Boudolf V, Kheibarshekan L, Zalmas LP, Gaamouche T, Maes S, Vanstraelen M, Kondorosi E, La Thangue NB, Govaerts W, Inzé D, De Veylder L. Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc Natl Acad Sci USA. 2008;105:14721–14726. doi: 10.1073/pnas.0806510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsick JS. synMuv verite–Myb comes into focus. Genes Dev. 2004;18:2837–2844. doi: 10.1101/gad.1274804. [DOI] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- López-Juez E, Dillon E, Magyar Z, Khan S, Hazeldine S, de Jager SM, Murray JA, Beemster GT, Bögre L, Shanahan H. Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell. 2008;20:947–968. doi: 10.1105/tpc.107.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Magyar Z, De Veylder L, Atanassova A, Bako L, Inzé D, Bögre L. The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell. 2005;17:2527–2541. doi: 10.1105/tpc.105.033761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, Horváth B, Khan S, Mohammed B, Henriques R, De Veylder L, Bakó L, Scheres B, Bögre L. Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 2012;31:1480–1493. doi: 10.1038/emboj.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Fuchs E, Ovecka M, Wysocka-Diller J, Benfey PN, Hauser MT. Two new loci, PLEIADE and HYADE, implicate organ-specific regulation of cytokinesis in Arabidopsis. Plant Physiol. 2002;130:312–324. doi: 10.1104/pp.004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Smertenko A, Wagner V, Heinrich M, Hussey PJ, Hauser MT. The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr Biol. 2004;14:412–417. doi: 10.1016/j.cub.2004.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, Watanabe Y, Nakamura K, Kimura T, Ishiguro S. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem. 2007;71:2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA. 2012;109:17123–17128. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignocchi C, Minns GE, Nesi N, Koumproglou R, Kitsios G, Benning C, Lloyd CW, Doonan JH, Hills MJ. ENDOSPERM DEFECTIVE1 is a novel microtubule-associated protein essential for seed development in Arabidopsis. Plant Cell. 2009;21:90–105. doi: 10.1105/tpc.108.061812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Xu T. Mechanisms of size control. Curr Opin Gene Dev. 2001;11:279–286. doi: 10.1016/s0959-437x(00)00191-x. [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C. The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell. 2006;18:2224–2235. doi: 10.1105/tpc.105.039651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AH, Chickarmane V, Cunha A, Obara B, Manjunath BS, Meyerowitz EM. Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol. 2010;8:e1000367. doi: 10.1371/journal.pbio.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013;13:585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B. Stem-cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol. 2007;8:345–354. doi: 10.1038/nrm2164. [DOI] [PubMed] [Google Scholar]
- Strompen G, El Kasmi F, Richter S, Lukowitz W, Assaad FF, Jürgens G, Mayer U. The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr Biol. 2002;12:153–158. doi: 10.1016/s0960-9822(01)00655-8. [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Nozawa A, Seki M, Shinozaki K, Endo Y, Sawasaki T. A simple and high-sensitivity method for analysis of ubiquitination and polyubiquitination based on wheat cell-free protein synthesis. BMC Plant Biol. 2009;9:39. doi: 10.1186/1471-2229-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Kajihara T, Okamura C, Kim Y, Katagiri Y, Okushima Y, Matsunaga S, Hwang I, Umeda M. Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr Biol. 2013;23:1812–1817. doi: 10.1016/j.cub.2013.07.051. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell. 2004;16:533–543. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell. 2002;14:903–916. doi: 10.1105/tpc.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Baloban M, Da Ines O, Cultrone A, Lammens T, Boudolf V, Brown SC, De Veylder L, Mergaert P, Kondorosi E. APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc Natl Acad Sci USA. 2009;106:11806–11811. doi: 10.1073/pnas.0901193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures, Supplementary Tables S6 and S7, Supplementary Methods
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Review Process File