Abstract
The intestinal epithelium is remarkably robust despite perturbations and demand uncertainty. Here, we investigate the basis of such robustness using novel tracing methods that allow simultaneously capturing the dynamics of stem and committed progenitor cells (called enteroblasts) and intestinal cell turnover with spatiotemporal resolution. We found that intestinal stem cells (ISCs) divide “ahead” of demand during Drosophila midgut homeostasis. Their newborn enteroblasts, on the other hand, take on a highly polarized shape, acquire invasive properties and motility. They extend long membrane protrusions that make cell–cell contact with mature cells, while exercising a capacity to delay their final differentiation until a local demand materializes. This cellular plasticity is mechanistically linked to the epithelial–mesenchymal transition (EMT) programme mediated by escargot, a snail family gene. Activation of the conserved microRNA miR-8/miR-200 in “pausing” enteroblasts in response to a local cell loss promotes timely terminal differentiation via a reverse MET by antagonizing escargot. Our findings unveil that robust intestinal renewal relies on hitherto unrecognized plasticity in enteroblasts and reveal their active role in sensing and/or responding to local demand.
Keywords: EMT/MET, Escargot/Snail2–miR-8/miR-200, intestinal homeostasis, intestinal renewal, stemness
Introduction
The constancy of cell types and numbers and the epithelial integrity in adult tissues relies on the rapid and accurate handling of cell loss by their resident multipotent stem cells. Yet most adult stem cells divide relatively slowly and sparsely in normal physiology to reduce potential risks associated with high proliferation rate (Cairns, 1975; Fuchs, 2009; Biteau et al, 2011). As such, a long temporal separation exists between the decision to divide and commit to a specialized lineage and the generation of the fully differentiated cells that can replace an older or damaged differentiated cell. Therefore, taking into consideration this delay, to match supply to demand, two strategies can be considered. In one extreme, stem cells may divide only upon receiving a demand to replace cell loss, thereby precisely matching supply to the tissue needs. However, a failure to respond quickly may compromise the robust organization of the intestine, for example, diminishing an animal’s prospects to survive in a harsh environment. Therefore, faced with a constant demand for cell turnover, stem cells may divide continually to generate a continual supply of differentiating cells that can deal rapidly with the daily demand of the tissue. However, such behaviour implies that production of new cells is not based on the actual tissue requirement but rather in anticipation of future demand. Here, we investigate what cellular plasticity and factors may explain the apparent paradox of adult stem cell systems operating with both speed and precision even when confronted with unpredictable demand.
The Drosophila midgut represents a suitable model to investigate this important issue. Not only does the midgut undergo a high turnover of intestinal cells (Micchelli & Perrimon, 2006; Ohlstein & Spradling, 2006, 2007) but also intestinal stem cells (ISCs) are the only mitotic cells in this tissue and the committed progeny makes only two fate choices (Biteau et al, 2011). These simplifications, coupled with recent findings showing that conserved principles underline intestinal repair and regeneration in flies and mammals (Jiang & Edgar, 2012), provide an ideal opportunity to define the strategic decision-making processes in place to ensure that tissue homeostasis is not disrupted by constant cell turnover.
The ISCs in the midgut epithelium reside in a basal niche formed, at least in part, by the visceral muscle (Lin et al, 2008; Jiang et al, 2009; Buchon et al, 2010; Biteau & Jasper, 2011; O’Brien et al, 2011). The snail family gene, escargot, marks the ISCs and their committed progeny (Micchelli & Perrimon, 2006; Ohlstein & Spradling, 2006) and has recently been shown to sustain the undifferentiated state and self-renewing divisions of the ISCs (Korzelius et al, 2014; Loza-Coll et al, 2014). Under normal homeostasis, only 1–3 mitotic stem cells are detected in the entire midgut (Micchelli & Perrimon, 2006; Ohlstein & Spradling, 2006; Amcheslavsky et al, 2009; Jiang et al, 2009; Choi et al, 2011), reflecting that these cells divide sparesly and asyncronously. After ISC division, the two daughter cells adopt a stem cell or committed progenitor cell fate (enteroblasts, EB) under the tutelage of Delta–Notch signalling (Micchelli & Perrimon, 2006; Ohlstein & Spradling, 2006; Maeda et al, 2008; Perdigoto et al, 2011; de Navascues et al, 2012). Daughters of the ISCs retain the expression of the Snail gene escargot, but unlike the ISCs, have capacity for terminal differentiation, replacing a lost absorptive enterocyte or secretory enteroendocrine cell (Potten, 1998), through largely unknown mechanisms.
Here, we use novel lineage-tracing methods to decipher how individual ISCs cope with the fast-paced and changing needs for midgut cell replenishment. We found that physiological ISCs divide and generate lineage-committed descendants continually that are “stocked” in an undifferentiated state. More importantly, progenitor cells are highly dynamic sensors of local and physiological demands. Accordingly, while ISCs are small/apolar cells, the (enterocyte) lineage-committed progenitor cells have front–rear polarity and motility, and extend long-exploratory protrusions, while exercising a capacity to postpone their terminal differentiation for a long time interval in the absence of local demand. This cellular plasticity is linked to the epithelial-to-mesenchymal (EMT) programme mediated by the conserved escargot/Snail2 and zfh1/Zeb transcription factors. Finally, mechanical feedbacks, likely through adhesion between mature cells and their neighbouring mesenchymal/progenitor cells, help to coordinate cell loss with terminal differentiation via activation of the conserved microRNA miR-8/miR-200 in the “pausing” progenitor cell, in turn, directly silencing escargot and zfh1 and promoting the epithelial state. Due to the fact that a snail family gene is also involved in intestinal stem cell biology and tissue repair in mammals (Horvay et al, 2015), our findings revealing hitherto unrecognized divisional dynamic of stem cells and cellular plasticity of progenitor cells via an escargot__miR-8 axis should provide a basis for future therapies.
Results
ReDDM lineage method to analyse dynamics of precursor cells with simultaneous view of cell turnover
Tackling the temporal challenges associated with the fast turnover of intestinal cells by the slow cycling midgut, ISCs requires new lineage-tracing method configurations. To this aim, we devised a method “ReDDM” (Repressible Dual Differential stability cell Marker, Fig1A–D) that enables observations of single-cell to tissue-level dynamics of precursor cells with spatial and temporal resolution. This approach allows simultaneous quantification of precursor cell number and cell renewal, and is amenable for live-imaging and genetic analysis (Fig1D, and below).
Figure 1.
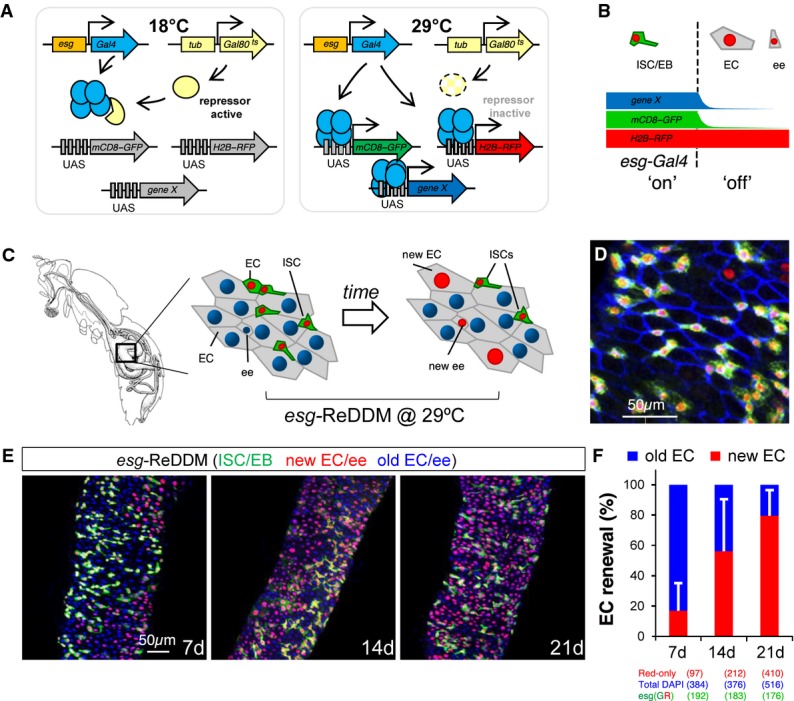
The ReDDM principle for marking and manipulating precursor cells and simultaneous view of the cell turnover at single-cell resolution
- Schematic illustration of the transgenes used in ReDDM (repressible dual differential stability markers) combined with the esg-Gal4 and a transgene (UAS-gene X) for mis/overexpression or downregulation via RNAi.
- The ReDDM relies on the differential protein stabilities of a pair of fluorescent proteins: the short-lived mCD8-GFP (green) serves as a morphological and an accurate temporal marker of the Gal4 activity (e.g. esg-Gal4), while the long-lived H2B-RFP (red) allows for tracing any newly differentiated progeny derived from the esg-Gal4 cells. The esg-Gal4 drives expression in the ISCs and enteroblasts (EBs) and is turned off in terminal differentiated EC (enterocyte) and ee (enteroendocrine) cells.
- Images illustrate the unlabelled (grey) adult Drosophila melanogaster gut and the esgReDDM-labelled midgut just after the temperature shift (left scheme) and 7 days later (right scheme).
- Representative confocal image from the midgut 7 days after temperature shift. Blue staining (anti-Discs-large-1, a-Dlg-1) outlines the intestinal epithelial cell membranes. Any newly generated differentiated progeny is highlighted by the nuclear H2B-RFP (red) label, while differentiated cells lasting in the midgut epithelium are unlabelled and visualized by counterstaining with DAPI or outlined by a-Dlg-1.
- Intestinal cell renewal (visualized as red-retaining labelling cells) in midguts 7, 14 and 21 days after the temperature shift.
- The graph shows the quantification of posterior midgut cell renewal (red/unlabelled EC and ee cells) ratio over time using ReDDM in the homeostatic midguts shown in (E). Red bars (new EC and ee cells) and blue bars (old EC and ee), detected by 4′,6-diamidino-2-phenylindole (DAPI, blue) counterstaining. Error bars represent standard deviation of the mean.
Source data are available online for this figure.
This method makes use of Gal4-responsive transgenes encoding fluorescent proteins with short (mCD8-GFP) and long (H2B-RFP) half-lives (Fig1B), and the temperature-sensitive Gal4 repressor, Gal80ts (tub-Gal80ts), to temporally restrict transgene expression to adult flies (Fig1A). ReDDM can then be combined with any stem and/or progenitor-selective Gal4 driver available and, as well, with transgenes for gene misexpression or downregulation via UAS-RNA interference (UAS-IR) constructs of particular genes.
An example of ReDDM with the ISC/EB-specific escargot (esg)-Gal4 driver (hereafter, esgReDDM) is presented in Fig1D–G. The esg-Gal4 is activated in ISCs and their committed progeny (called enteroblasts, EBs), and it is turned off in the newly differentiated enterocytes (EC) and enteroendocrine (ee) cells (Micchelli & Perrimon, 2006; Ohlstein & Spradling, 2006) (Fig1B). Although escargot is no longer active in differentiated cells, the stable H2B-RFP protein persists for at least 28 days (data not shown) allowing unequivocal labelling of any renewed cells derived from the labelled esg+ cells at single-cell resolution (Fig1D andG). Intestinal cells that have not yet been renewed remain colourless and can be detected by counterstaining with DAPI or outlined by the epithelial marker Discs-large-1 (Dlg-1, blue, Fig1D).
Spatiotemporal relationship of cell turnover and individual precursor cell dynamics
We used esgReDDM (Fig1G and H) to map homeostatic midgut cell turnover and found that complete midgut replenishment by new cells derived from labelled esg+ cells (red-retaining enterocytes and enteroendocrine cells, Fig1E) took 3 weeks (Fig1F) rather than 1 week as previously estimated by clonal analysis [see below and (Jiang et al, 2009)]. Furthermore, intestinal cell replenishment is not homogeneous as previously assumed. The distribution, size and shape of the renewed areas varied greatly from intestine to intestine in age-synchronized, co-cultured animals. Thus, it is clear that intestinal cell turnover events must be mapped to be able to make direct correlations of local demand with individual stem cell dynamics.
We also compared our method against several protocols that include a heat shock (37–38°C) to activate the flipase (FLP) transgene for FRT-mediated chromosomal recombination in clonal analysis (Golic & Lindquist, 1989; Xu & Rubin, 1993; Lee & Luo, 2001; Apidianakis & Rahme, 2011) and found that heat shock produces a significant increase in intestinal turnover rate and ISC mitosis (Supplementary Fig S1), which can explain the previous overestimation of cell turnover rate. Compared with the esgtsF/O method (Jiang et al, 2009) (esg-Gal4, tub-Gal80ts, UAS-Flp, act > stop > Gal4), the esgReDDM method has also the improvement that Gal4 is not continuing in the terminal differentiated cells, which could confound the interpretation of the phenotypes, and that ReDDM allows differential marking of esg+ and esg+-derived differentiated progeny.
For standardization, we describe the dynamics of adult esgReDDM-based guts (data pooled from guts obtained in at least three independent repeats) that have been renewed over a 1-week period after the temperature shift that activates the Gal4 and starting in adult mated females of 3–7 days of age. This avoids confounding variables of very young, virgin or old flies, where homeostasis is often breakdown (Jiang & Edgar, 2012). All guts that exhibit a breakdown of homeostasis (PH3 values are significantly above the homeostatic value) are ruled out from the analysis.
ReDDM revealed unsuspected cellular plasticity in commitment progenitor cells
By analysing thousands of cell turnover events (n > 100 intestines analysed) in esgReDDM-labelled midguts at day 7 (after the temperature shift), we observed that the majority (> 98%) of ISCs had undergone at least two divisions (as assessed by the number of cells in individual esg+ clusters) and many had divided 3–4 times even in the absence of any cell turnover in their surroundings (arrows in Fig2A). In these midguts, labelling by the antibody against phosphorylated histone H3 (PH3) to mark mitotic ISCs detected from 0 to 4 mitosis as reported previously (Micchelli & Perrimon, 2006; Ohlstein & Spradling, 2006). This reflects that PH3 provides a “snapshot” of the distribution of the slow cycling ISCs in mitosis but it is not alone a true kinetic measurement of “activated” ISCs. This indicates that, in normal physiology, fly ISCs divide slowly but regularly, likely to optimize their response by ensuring a stock of committed progenitors that can rapidly respond to a local demand.
Figure 2.
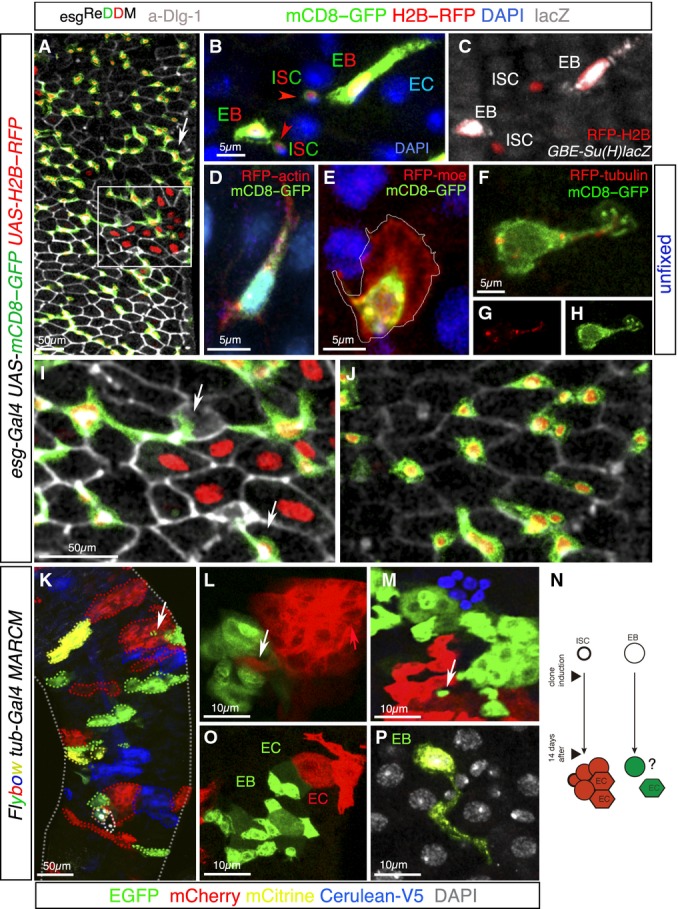
ReDDM-based analysis unveils hitherto unrecognized plasticity of progenitor cells
- A Representative confocal image of an esgReDDM adult midgut 7 days after the temperature shift. The large esg+ cell clusters (arrows) reflect substantial production by the individual ISCs in the absence of cell turnover in their local surroundings. Not-yet renewed cells (old mature cells) are detected by DAPI (not shown in the image) and outlined by the adherens junction protein marker Dlg-1 (grey).
- B, C A high magnification image (ISC, intestinal stem cell; EB, enteroblast, EC, enterocyte) in which the lower EB represents a middle stage, and the upper EB a typical late stage (enterocyte-committed, polyploid enteroblast). (C) Co-staining with GBE-Su(H)-lacZ (white) identified the enteroblasts from their mother ISCs. Note the stronger esg > GFP signal and larger size of Su(H)+ (EB) cells.
- D–H Confocal images of esg+ cells in unfixed wild-type adult midguts in homeostatic conditions. Enteroblasts are distinguished by their brighter GFP signal and larger size (see B). Cells are co-labelled by GFP (detecting esg > mCD8-GFP, green), RFP-actin (D), RFP-moesin (E) and RFP-tubulin (F–H). The image shows a single channel of RFP-tubulin (G) or mCD8-GFP (H).
- I Magnification from (A) showing the intestinal epithelial cells outlined by Dlg-1 labelling (grey) and the interstitial position of the enteroblasts (which lacks the epithelial marker Dlg-1). Enteroblasts extend long protrusions along the EC borders.
- J Magnification from (A) showing EBs in non-regenerated area.
- K–N Representative confocal images showing Flybow (FB2.0) clones 14 days ACI (after clone induction). (K) Clones are varied in size and elongated, round or irregular in shape revealing plastic patterns of tissue turnover. Neighbouring clones can have intercalated cells (K–M, arrows). (N) Scheme of the Flybow MARCM method (Lee & Luo, 2001; Hadjieconomou et al, 2011) of expected outcome (a multicellular clone or a single cell) depending on whether the labelled daughter cell after induction of FLP to activate FRT-mediated mitotic recombination is an ISC or a EB.
- O, P Images illustrate multicellular clones (O) and single-cell clone (P). The two neighbouring clones are composed of newly differentiated (hexagonal cells, EC) and undifferentiated (EB) cells, which tend to be strongly polarized. (P) A single-cell clone with long protrusion and large size typical of undifferentiated enteroblast surrounded by not-yet renewed cells. The old intestinal cells are colourless and visualized by DAPI nuclear counterstaining (white).
Since little is known about unique properties of stem cells and if their committed descendants rely on particular cellular architectures, we characterized and compared them using the membrane-bound mCD8-GFP in esgReDDM configuration. The descriptions relate to the prospective enteroblasts that give rise to the absorptive (polyploid) enterocyte cells, representing ∼90% of enteroblasts (O’Brien et al, 2011; de Navascues et al, 2012). ISCs [identified by stem cell marker Delta (Micchelli & Perrimon, 2006)] (Fig2B) are small cells with a high nuclear/cytoplasmic ratio and no obvious polarity, which may facilitate their continual division, and topologically restricted position in the base of the epithelium (Micchelli & Perrimon, 2006; Ohlstein & Spradling, 2006). In stark contrast, their newborn enteroblasts (identified by co-labelling with Su(H)-lacZ; Furriols & Bray, 2001; Micchelli & Perrimon, 2006; Fig2B and C) readily increased their size and adopted a spindle shape and extend long protrusions (up to 50 μm long, Fig2A and D) typical of mesenchymal cells (Heng & Koh, 2010; Lamouille et al, 2014).
Typically, during the epithelial-to-mesenchymal transition (EMT), cells undergo profound cell shape changes, cell elongation and front–rear polarity and form protrusive structures that are driven by remodelling of the actin and microtubule cytoskeletons (Waterman-Storer & Salmon, 1997; Wadsworth, 1999) and by de-assembly of epithelial junctions proteins (e.g. junctions protein Dlg-1; see Fig1) (Lamouille et al, 2014). The tendency to be damaged or lost after fixation requires that these protrusive actin-rich structures and the microtubule cytoskeleton are analysed in living, unfixed cells. Fig2D–H illustrates representative examples of enteroblasts in their native environment using an actin protein fused to RFP (RFP-actin; Fig2D), an actin-binding domain of moesin tagged at the N-terminal end with RFP (RFP-moe #B123; Ribeiro et al, 2004; Fig2E) and a β-tubulin fused to RFP to label the microtubule cytoskeleton (RFP-tubulin, Fig2F–H). The membranes of enteroblasts were marked by the membrane-tethered GFP under the control of esg-Gal4 (esg > mCD8-GFP). Ex vivo live imaging of whole midguts captured dynamic actin-containing membrane protrusions in enteroblasts. As previously reported for actin foci in migrating cells, both front and rear structures showed intense actin focus (RFP-actin, red in Fig2D). Lamellipodial-like actin-rich membrane structures typical of migrating cells, in which actin filaments inside the cytoplasm extend at the “front” leading edge, were seen in the enteroblasts (actin binding RFP-moe; Fig2E). The finger-like protrusions of live imaged enteroblasts also contained a dense network of microtubules, whereas the lamella-like structure was devoid of filamentous tubulin (β-tubulin-RFP in Fig2F–H), typical distribution of microtubules in migrating cells (Wadsworth, 1999).
Membrane protrusions were also observed in fixed tissues of homeostatic midguts stained with anti-GFP to detect mCD8-GFP in the esgReDDM configuration (Fig2I and J). The dynamic nature of such structures was apparent by comparing enteroblast within an area being repaired (Fig2I) and a non-repairing area (Fig2J) in the same gut.
Enteroblast motility was shown by time-lapse video microscopy (Supplementary Movies S1 and S2), revealing occasional cellular movement and repositioning of enteroblasts (identified as esg+ cells with large size). Functional studies below indicate that an important role of the reorganization of actin structures in the mesenchymal/enteroblast cells is to acquire invasive capabilities and formation of long, thin actin-rich protrusions that might help to sense their surroundings. Indeed, leucocytes and other moving cells use filopodia to explore the surfaces of other cells and for sensing guidance cues (Wood & Martin, 2002) and mechanical input (Cai et al, 2014).
Flybow analysis reveals that committed progenitor cells defer their response in the absence of a local demand
The above findings open the question of how newborn progenitor cells after fate commitment can hold terminal differentiation for the time interval extending from their birth and the occurrence of cell loss in its vicinity in order to maintain the robust organization of the intestine. As clonal lineage information is fundamental in the reconstruction of individual cell dynamics, we used Flybow clonal analysis (Hadjieconomou et al, 2011) to label individual stem cells and/or their lineages in different colours (Fig2K–N).
When the midguts were analysed at 14 (Fig2K–P) and 21 days (data not shown) after marking ISCs or committed progenitors (n = 15 midguts and n > 100 clones scored), multicoloured clones displayed a variety of shapes and sizes, in agreement with the non-homogeneous nature of intestinal replenishment defined by ReDDM method (Figs1E and 2A). In the majority of clones, there was continuity between the same colour labelled cells (Fig2K), consistent with the individual descendants of a stem cell repairing their own area (Ohlstein & Spradling, 2007). However, Flybow also revealed the occasional intermingling of cells from two neighbouring lineages (arrows, Fig2K–M) and the fragmentation of some clones, highlighting that migration and the re-adjustment of enteroblast position are part of the renewal process.
Newly differentiated cells were unambiguously detected by their unique shape and residence in the epithelium, while enteroblasts were detected by their highly polarized shape (see Fig2O). A nearest-neighbour examination of Flybow clones determined that terminal differentiation is likely to be dictated by a local demand rather than to be scheduled by the birth time of the enteroblast (e.g. clones in Fig2O and P). To understand this phenomenon, it is necessary to consider that Flybow MARCM method generates over time a multicellular clone or a single-cell clone depending on whether the labelled cell was an ISC or an enteroblast (scheme in Fig2N). Accordingly, 14 days after clone induction, the presence of newly differentiated cells (as the neighbouring clones in Fig2O) illustrates the occurrence of division, commitment, and terminal differentiation of several cells within the clone growth time frame. Within the same midgut, the presence of single-cell clones with undifferentiated features, cell elongation, membrane protrusions and large (polyploid) nuclei (assessed by DAPI, grey in Fig2P) illustrates that enterocyte-committed enteroblasts born at the time of clone induction actively retained an undifferentiated state for up to 14 days. Thus, our Flybow data challenges the paradigm that cell division, terminal differentiation and replacement are a continuum. Flybow clones suggest that terminal differentiation and replacement are likely individual decision-making processes.
We observed a progressive underrepresentation of the single-cell clones in the guts from day 14 (∼20% of the clones, n > 100 clones scored) to day 21 (0% of single-cell clones), suggesting that single-cell clones terminally differentiate and were eventually turned over during normal homeostasis. This “postponement” of terminal differentiation can also be inferred from the ReDDM analysis (e.g. Fig2A,I and J), although only with clonal analysis one can make direct inference of birth time of the enteroblast. In sum, these findings unveil unrecognized cellular plasticity in committed progenitor cells and, against common belief, enteroblasts have active roles in sensing and responding to local demand.
Deferring differentiation is linked to the epithelial–mesenchymal programme mediated by escargot and antagonized by the microRNA miR-8
In order to understand how progenitor decision-making, to postpone or terminally differentiate, is achieved mechanistically, we set out to investigate whether this decision is linked to the epithelial-to-mesenchymal transition programme and its reverse, MET. In mammalian cell culture, the pro-epithelial microRNAs of the miR-200 family link EMT process with stemness (Shimono et al, 2009; Nieto, 2013) through direct repression of ZEB1,2 genes (Nieto, 2011). Given the expression of the snail gene escargot in the ISCs and enteroblasts, we focused our attention on escargot/Snail2, zfh1/Zeb and the microRNA miR-8/miR-200 to investigate the aforementioned cellular behaviours. For the sake of simplicity and given their genetic interactions, we present zfh1/Zeb data in Supplementary Fig S2. During the course of this work, two groups have reported the crucial role of escargot/Snail2 in sustaining stemness and undifferentiated state of Drosophila ISCs in the midgut (Korzelius et al, 2014; Loza-Coll et al, 2014). A similar requirement has also been found for murine Snail1 in the mouse intestinal epithelium (Horvay et al, 2015).
We used two independent transgenes for RNA interference (RNAi) knockdown (Dietzl et al, 2007; Ni et al, 2011) of escargot, and assayed overexpression using two transgenes (see Materials and Methods) escargot knock down phenotypes were validated using endogenous escargot mutations (esgL2 and esgG66B) (Fig3A–F). For the analysis of the microRNA miR-8, we examined midguts of adult flies null for mir-8 (mir-8Δ2/Δ3: Karres et al, 2007; Supplementary Fig S3) and for adult-restricted intestinal precursor cell-specific manipulations of the microRNA, we used the UAS-mir-8 transgene (Vallejo et al, 2011) and the miR-8 microRNA “sponge” (Loya et al, 2009) that faithfully mimics the mir-8−/− phenotypes (UAS-mir-8-sponge(SP)::GFP, Karres et al, 2007; Loya et al, 2009; Morante et al, 2013; Vallejo et al, 2011). The native and demand-induced pattern of endogenous mir-8 gene is presented in the next sections.
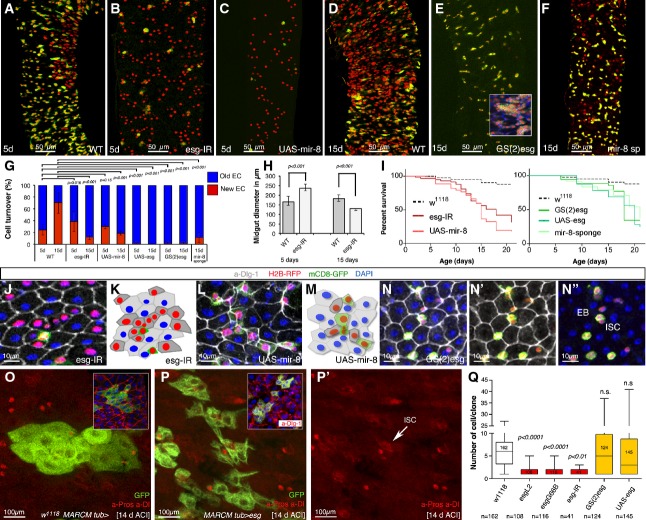
Figure 3.
The microRNA miR-8 and Escargot have opposing effects in controlling deferral versus terminal differentiation decision
- A–F Representative ReDDM in esg-Gal4 midguts of the indicated genotypes 5 or 15 days after the temperature shift that activates Gal4. (A) Intestinal renewal occurs in a patchy pattern indicating local demand. (B, C) Precocious differentiation of esg+ cells upon escargot depletion in esg+ cells (esgReDDM UAS-esg-IR, B) or upon misexpression of the mir-8 microRNA (esgReDDM UAS-mir-8, C). (D–F) ISCs and progenitor cells overexpressing escargot in the esg+ pattern (esgReDDM GS(2)esg, E) or with depleted mir-8 (esgReDDM UAS-mir-8-sponge, F) fail to exit the undifferentiated state, or their terminal differentiation was severely impaired, respectively. The control gut had renewed almost 75% of their enterocytes at the time point shown (2 weeks after temperature shift, D). Inset in (E) shows tumour-like accumulations of undifferentiated cells. The penetrance of all shown phenotypes is nearly 100%, and shown are representative images.
- G Quantification of intestinal cell replenishment as a ratio of new EC/old EC (red-only DAPI/”colourless” DAPI cells) in each genotype at the time point indicated. Error bars represent the standard deviations (SD) (n = 13 control guts scored for day 5 and n = 10 for day 15; n = 11 midguts (day 5) and n = 9 (day 15) for esg-IR; n = 6 (day 5) and n = 6 (day 15) for UAS-mir-8; n = 11 (day 5) and n = 11 (day 15) for GS(2)esg; n = 8 (day 5) and n = 7 (day 15) for UAS-mir-8-sponge). Unpaired t-test values are shown.
- H The graphs show the diameter of control (WT, n = 5 midguts (day 5) and n = 8 (day 15)) and escargot RNAi (esg > esg-IR: n = 8 (day 5) and n = 9 (day 15)) ReDDM-based midguts at the time point indicated. Error bars represent SD, and t-test values are shown.
- I Survival (as percentage of animals) over time of the indicated transgenes expressed under the esg-Gal4 control. Survival curves were constructed combining data from at least 10 vials, each with 10–15 flies, in a genotype group. Log-rank (Mantel–Cox) analysis indicated that escargot and mir-8 manipulations significantly reduced animal survival (P < 0.001).
- J–N (J) Precociously differentiated esg > esg-IR mutant ISC/EB cells are smaller than normal enterocytes, but integrate correctly into the epithelium. (L) Precociously differentiating mir-8 overexpressing ISC/EB intercalate incorrectly and amassed in the epithelium. Mature epithelial cells are marked by a-Dlg-1 (grey). (K, M) show schematic illustrations of the gut epithelium from the indicated genotypes. (N–N″) The escargot-overexpressing (esgReDDM GS(2)esg) progenitor cells have a more rounded shape than wild-type progenitor cells but can be distinguished from their mother ISCs by their larger size (N″).
- O, P MARCM tub-Gal4 clones at day 14 ACI of control midguts (O) and overexpressing escargot cells (P). Insets show clones of the cells of the indicated genotype where mature cells are visualized by their labelling by Dlg-1 (red). (P′) Single-channel image illustrates that escargot-overexpressing clones contain enteroendocrine cells (nuclear red, a-Pros) and ISC (cytoplasmic red, a-Dl: arrowhead).
- Q Box plot of clonal size in MARCM tub-Gal4 of the indicated genotypes in midguts 7 days after clone induction. Median value is shown. Student’s t-test indicated that the number of cells (size) in escargot overexpression clones were not significantly (n.s.) different from that in control (w1118 MARCM tub-Gal4) clones, while clones with depleted escargot were significantly different from those in control as assessed using ANOVA. P-values and number of clones scored are indicated in the corresponding bars.
Source data are available online for this figure.
We combined the transgenes with esgReDDM in order to track esg+ cell loss through terminal differentiation and to be able to quantify cell turnover (ratio of red-only cells/total mature cells, Fig3G). Control and mutant ReDDM midguts were extensively analysed from days 5 to 21 postinduction of the transgenes (Fig3A–N, and data not shown), and at least 10 midguts from three independent crosses were quantified in each condition. We found that, after just 5 days of escargot depletion (Fig3B) or mir-8 overexpression (Fig3C), ∼80% of the esg+ pool was exhausted through accelerated differentiation through a mesenchymal–epithelial transition (see below). Importantly, while normal intestinal renewal follows a random, patchy pattern of tissue replenishment (e.g. Fig3A), escargot loss or the ectopic expression of the mir-8 microRNA resulted in a homogeneous, salt-and-pepper distribution of newly differentiated cells (Fig3B and C). Depleting zfh1 provoked phenotypes similar to that of escargot (Supplementary Fig S2A), however, pairwise combinations of escargot and zfh1 indicated that escargot is crucially important for the undifferentiated state acting downstream of zfh1 (Supplementary Fig S2B–D).
Reciprocal phenotypes were observed when escargot (Fig3D) (and zfh1, Supplementary Fig S2B) was overexpressed or the microRNA mir-8 was depleted through the mir-8-sponge (Fig3F). Thus, while in control ReDDM guts intestinal cell replenishment scales up progressively over time (see Fig3D and quantification in 3G), midguts in which escargot (Fig3E) or the mir-8-sponge (Fig3F) were overexpressed in the esg+ pattern showed no sign of differentiation or very little cell renewal at day 15, consistent with the recent finding that escargot maintains an undifferentiated state (Korzelius et al, 2014; Loza-Coll et al, 2014). See source data for Fig3 for details of statistics and number of midguts scored per genotype. Our data show that the escargot overexpression and loss of mir-8 retain an undifferentiated state accompanied by retention of a mesenchymal state (Fig3E and below).
Tumour-like accumulations of undifferentiated (small/round bicoloured) cells formed in 40% of esgReDDM > esg animals (n = 15 midguts, inset in Fig3E). The “undifferentiated” phenotype was evident in the majority of the midguts, although occasionally newly differentiated enteroendocrine cells were also observed (labelled by Pros). Thus, escargot and zfh1 promote deferral of terminal differentiation and the microRNA miR-8 opposes it.
We noted that 5 days after escargot depletion, the diameter of the gut increased due to the addition of unsolicited newly differentiated enterocytes (Fig3B, quantification in 3H and see 3J and K) but, after 2 weeks, the pool of esg+ cells had been completely exhausted through premature differentiation and the gut atrophied progressively due to the lack of cell replacement (Fig3G and H), diminishing animal survival (Fig3I) as recently reported. Precocious differentiation in mir-8 overexpression midguts did not increase gut diameter (see Fig3C), but the precociously differentiated cells were frequently intercalated incorrectly and amassed on the existing intestinal cells (Fig3L and M versus J). This difference may be explained by our finding further down that miR-8 directly represses both escargot and zfh1, suggesting that the microRNA resulted in a more abrupt transition from the mesenchymal to epithelial state resulting in a less effective integration in the epithelium. The impaired intestinal homeostasis in mir-8 mutants also diminished animal survival (Fig3I).
We complemented the studies of escargot using endogenous mutations and the tub-Gal4-driven MARCM clones (Fig3O–Q). Importantly, while escargot overexpression by esg-Gal4 causes most cells to retain rounded morphology (e.g. Fig3N), strong overexpression using the tub-Gal4 generated large, highly polarized enteroblast cells (Fig3P), significantly different from those of control (w1118, Fig3O) or the escargot overexpression by esg-Gal4 (Fig3N). tub-esg cells invariably failed to differentiate (compare Fig3O and P, and insets), again with the exception of rare enteroendocrine cells (Pros+, red nuclei, Fig3P). MARCM clonal analyses using endogenous esg mutations corroborated the results using the RNAi esg transgenes (Fig3Q). Quantification of the number of singletons and clone size (n for each genotype is indicated below Fig3Q) as percentage of total clonal events showed that in 7 days the majority (> 70%) of clones with esg loss are singletons consistent with escargot’s requirement for self-renewal. Together, these data support that enteroblast gains mesenchymal properties via escargot and suggests a model in which the mesenchymal/enteroblast state is determined when escargot surpasses a threshold level, below which escargot sustains a partial EMT, resulting in ISCs retaining a epithelioid/round shape that may facilitate cell division by limiting their motility. The fact that escargot antibodies are not available hampered the direct quantification of escargot protein and hence other explanations are also possible.
The biological relevance of the mir-8-sponge was confirmed by examining the midguts and renewal process of adult flies entirely null for mir-8 (mir-8Δ2/Δ3, Karres et al, 2007, Supplementary Fig S3). The intestinal integrity of adult mir-8−/− animals immediately after eclosion was fairly normal although enterocyte cell size was on average smaller than that in wild-type midgut, consistent with miR-8 promoting polyploidization during development (Loya et al, 2009; Morante et al, 2013). The integrity of the intestinal epithelium began to deteriorate few days (Supplementary Fig S3A), reflecting impaired intestinal homeostasis. Moreover, adult mir-8−/− flies survival was severely reduced following intestinal damage (Supplementary Fig S3E). Thus, endogenous mir-8 is an integral component of the intestinal repair and regeneration.
miR-8 triggers terminal differentiation and epithelial state by antagonizing escargot
The reciprocal phenotypes elicited by escargot and miR-8 suggest that miR-8 acts by antagonizing escargot either directly or indirectly. Indeed, when we tested pairwise combinations of loss and gain of the two genes, we found phenotypes that were consistent with miR-8 triggering terminal differentiation and MET by repressing escargot (Fig4A–D). For example, the failure to differentiation caused by depleting mir-8 using the mir-8-sponge (Fig3F) was fully suppressed by concomitant loss of escargot (mir-8-sp+esg-IR, Fig4B and C). Fig4C illustrates that RFP-retaining cells are precociously differentiated cells as detected by their acquisition of epithelial state (Dlg-1, grey staining) rather than simply undifferentiated precursors that have shut down the esg-Gal4 promoter. Conversely, the premature differentiation induced by ectopic overexpression of mir-8 (e.g. Fig3C) was fully blocked by concomitant overexpression of escargot (Fig4D), although a minority of cells still differentiated. Thus, miR-8 and escargot do not operate in isolation to one another, but are functioning in a common process, fine-tuning deferral versus terminal differentiation decision at individual cell level.
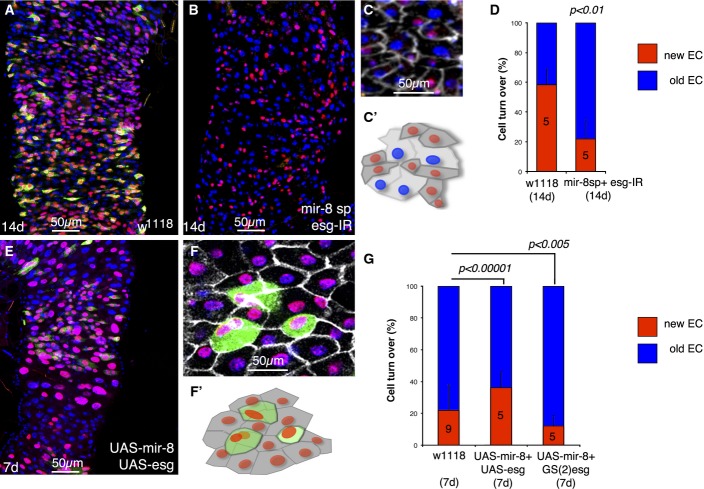
Figure 4.
Loss and gain of mir-8-induced intestinal precursor cell defects are rescued by loss and gain of escargot
- Control wild-type midgut 14 days after the temperature shift.
- Illustrative example of a midgut of 14 days with depleted mir-8 and escargot simultaneously (esgReDDM > mir-8-sp+esg-IR). Note that after 2 weeks, the pool of esg+ cells is completely depleted and no further renewal could be done.
- Dlg-1 staining (grey) illustrates that cells that turned off esg+ (GFP− RFP+ cells) are integrated into the epithelium (precociously differentiated enterocytes) and scheme below (C′).
- Graph shows quantification of replenishment (% of new EC, GFP− RFP+ cells) of control (w1118; esgReDDM >) and mutant esgReDDM > mir-8-sp+esg-IR midguts at the indicated time point. Error bars represent SD.
- Co-overexpression of mir-8 and escargot (esgReDDM > UAS-mir-8+UAS-esg) rescued in part the premature terminal differentiation, and after 7 days, esg+ undifferentiated cells are still seen.
- Dlg-1 staining (grey) in UAS-mir-8+UAS-esg midgut. Note the large size and rounded shaped of esg+ (GFP+) cells. (F′) is a scheme of (F).
- Graph shows quantification of the indicated genotypes at the indicated time point. The number of guts (n) scored is indicated in the graphs and statistical significance using ANOVA are shown. In the esgReDDM > UAS-mir-8+GS(2)esg genotype, most esg+ cells failed to terminally differentiate, further reflecting the phenotype involves a balance between miR-8 and escargot. Shown are representative images.
Source data are available online for this figure.
mir-8 expression is readily activated in late enteroblasts shortly before terminal differentiation
The above findings strongly suggest that expression of the conserved microRNA miR-8/miR-200 is the crucial step in triggering epithelial cell loss-driven terminal differentiation by turning off escargot. Therefore, we next examined mir-8 spatiotemporal expression pattern during intestinal homeostasis. We monitored activation of endogenous mir-8 promoter using the mir-8-Gal4 enhancer trap within mir-8 gene (Karres et al, 2007) that faithfully reflects the pattern of mir-8 in many other tissues (Vallejo et al, 2011; Morante et al, 2013). To provide temporal information, we used the short-lived mCD8-GFP protein under control of mir-8-Gal4 (mir-8 > mCD8-GFP, Fig5).
Figure 5.
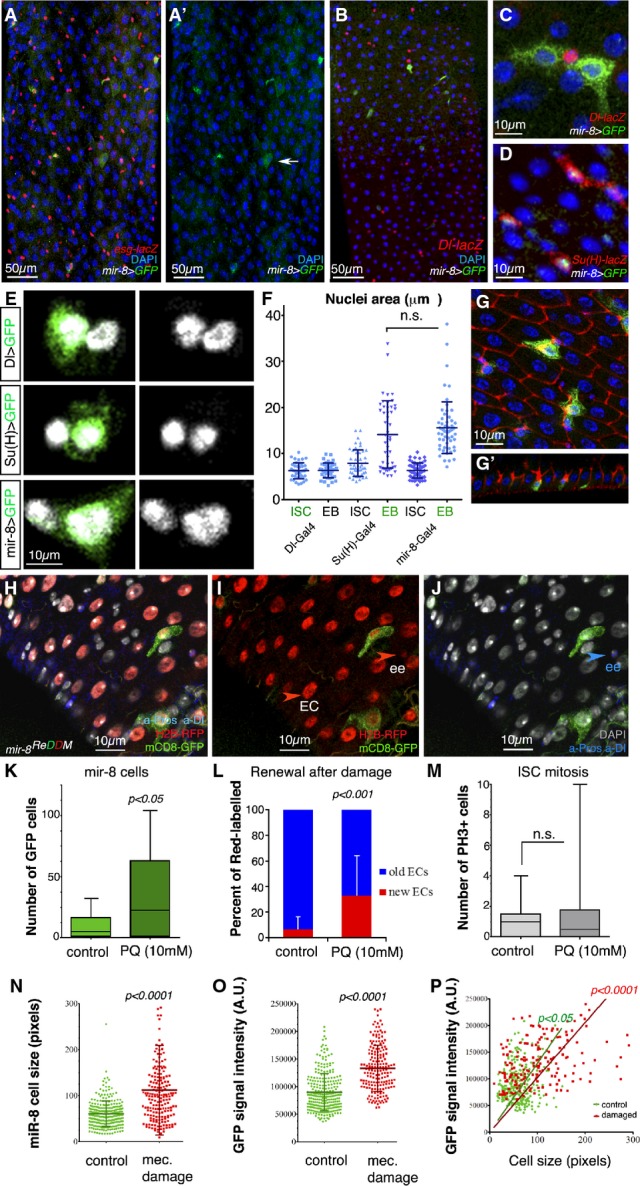
Endogenous microRNA mir-8 is readily activated in late-stage enteroblasts before terminal differentiation
- A–D Representative confocal images and single-channel image (A′) of an adult mir-8-Gal4 enhancer trap midgut stained with antibodies against GFP and esg-lacZ (A), the ISC marker Dl-lacZ (B, C) and the enteroblast marker Su(H)-lacZ (D). Note that in (D) GFP is nuclear (mir-8 > GFPnls GBE-Su(h)-lacZ).
- E mir-8+ cells are late-stage enteroblasts (as deduced by their large polyploid nuclei).
- F Histogram of the quantification of the nuclear size (μm2) of DAPI-stained GFP+ cells driven by the indicated Gal4 lines and that of neighbouring non-GFP cells. The data are represented as the mean ± SD. There are no statistically significant differences in GFP and non-GFP labelled cells in Dl-Gal4 and GBE-Su(H)-Gal4 midguts (t-test, n = 45 and 47 for each genotype).
- G Tangential confocal sections show that mir-8+ enteroblasts (green) are integrating into the epithelium (outlined by Dlg-1, in red, and note Dlg-1 accumulation).
- H–J Lineage tracing of mir-8-Gal4 enteroblasts using ReDDM shows that both enterocytes and enteroendocrine cells (ee: red-retaining, Pros+) are derived from mir-8+ progenitor cells. Old ee cells are detected as Pros-positive cells (nuclear staining). DAPI staining in grey, while cytoplasmic Dl staining marks ISCs (white arrowhead).
- K, L (K) Histogram of number of mir-8+ cells in midguts after 4 h of Paraquat (PQ)-ingestion and sibling control and (L) quantification of ratio of cell turnover (red-only cells/old cells (DAPI) in the same midguts as (K) (n = 13 and 12 WT and damaged midguts scored, respectively). Error bars represent SD.
- M Quantification of mitotic ISCs assessed by labelling with PH3 (n = 13 and 12 of each condition). Error bars represent SD.
- N, O Scatter dot plots showing average cell size (n) and average pixel (GFP) intensity signal (O: mean value per cell, arbitrary units, A.U.) with population mean and standard deviation. Number of cells counted, n = 280 in wild-type guts, and n = 188 in damaged guts.
- P Linear regression analysis of GFP signal and cell size from the same data as in (N, O). Best fitting line with (0;0) origin is shown for both populations.
Source data are available online for this figure.
GFP signal was found in few esg+ cells in basal homeostatic condition (esg-lacZ, mir-8 > mCD8-GFP, Fig5A and A′), and signal was rapidly turned off in terminally differentiated cells. The mir-8+ cells were enteroblasts, not ISCs, as shown by co-labelling with ISC marker Delta-lacZ and enteroblast marker Su(H)-lacZ (Fig5B–D). mir-8+ enteroblasts appear to represent late-stage enteroblasts shortly before their terminal differentiation as judged by: (i) their large nuclei size (see average size in Fig5E and F) and (ii) the observation that most mir-8+ cells are undertaking integration into the epithelium (see vertical confocal section in Fig5G and G′). Occasionally diploid cells mir-8+ were seen which might represent enteroendocrine-committed precursors undergoing terminal differentiation. In agreement with the notion that mir-8+ cells reflect a temporal phase of enteroblasts, lineage tracing using mir-8-Gal4 and ReDDM showed that both newly differentiated enterocytes and enteroendocrine cells in the renewing midgut were derived from mir-8+ cells (mir-8ReDDM, Fig5H–J). Altogether, these data define miR-8 as an instructive factor timing terminal differentiation. The weak and transient expression of mir-8 as detected by the weak GFP signal (mir-8-Gal4 > mCD8-GFP) did not allow mir-8 dynamics to be defined by in situ hybridization with single-cell resolution.
As a means to correlate activation of the endogenous mir-8 promoter (mir-8-Gal4) with demand to replace cell loss, we next assayed mir-8-Gal4 > mCD8-GFP expression and intestinal cell turnover rate during damage/regeneration induced by two different approaches. Previously, it has been established that ingestion of toxins or non-lethal pathogenic bacterium induces a dramatic remodelling of the midgut, which occurs initially through the immediate differentiation of a pool of undifferentiated progenitors. There is a latency period of ∼20–24 h from the time of injury or bacterial infection until the high proliferation of ISCs is observable in the damaged midgut (Buchon et al, 2010). Therefore, in this first assay, synchronized adults were fed with Paraquat (1,1′-dimethyl-4,4′-bipyridylium dichloride), and 4 h after ingestion, midguts were dissected to quantify any increase in mir-8+ cells and/or cell renewal independent of the late response involving increased ISC proliferation. The experiments were repeated twice. We observed a significant increase in the number of mir-8+ cells (Fig5K), along with an increased intestinal cell turnover (Fig5L). Consistent with previous work (Buchon et al, 2010), 4 h after Paraquat poisoning, the average ISC mitosis was still within the homeostatic range (Fig5M). This confirmed that the increased number of mir-8+ cells reflects activation of the mir-8 promoter in pre-existing enteroblasts to match the increased cell loss/demand.
Similarly, we found that a short pulse of mechanical force provoked an increased number of mir-8+ cells in the midgut accompanied by increased intestinal cell turnover (Fig5N–P) without increased cell division (data not shown). By plotting the size of mir-8+ cells (mir-8-Gal4 > mCD8-GFP, Fig5N) against GFP fluorescence intensity measured by confocal microscopy image analysis (Fig5O), we obtained a high correlation both in steady-state midguts and in mechanically stressed midguts (Fig5P, P < 0.05 in homeostatic midgut (control) and P < 0.0001 in damaged/regenerating midguts, see source data for Fig5). This linear correlation supports the notion that the microRNA mir-8 expression levels increases in correlation with maturation and repair. Moreover, as mir-8+ enteroblast cell numbers increase in response to cell loss/damage, these data further define miR-8 as an instructive “timer” factor of terminal differentiation. More speculative, the mechanical stress-induced mir-8 expression opens the possibility that physiological mechanical forces during the clearance of apoptotic cells (Teng & Toyama, 2011) serve as a stimuli to drive expression of mir-8 in enteroblasts that are in direct contact with the dying cell. Mechanical feedback through direct cell–cell adhesion between the leading edge-like of mesenchymal/progenitor cells and individual mature cells is also consistent with our time-lapse data which show individual enteroblasts re-adjusting their positioning, while the neighbours are relatively static (see Supplementary Movies S1 and S2).
Mutual opposing regulation of escargot and mir-8
In line with functional and expression data, we found that miR-8 directly represses escargot and also zfh1/Zeb via mir-8 seed sequences within their 3′ untranslated regions in cell culture (Fig6A–D). In agreement with these data, endogenous expression levels of escargot and zfh1 were significantly increased in mir-8 null animals (Fig6E and F).
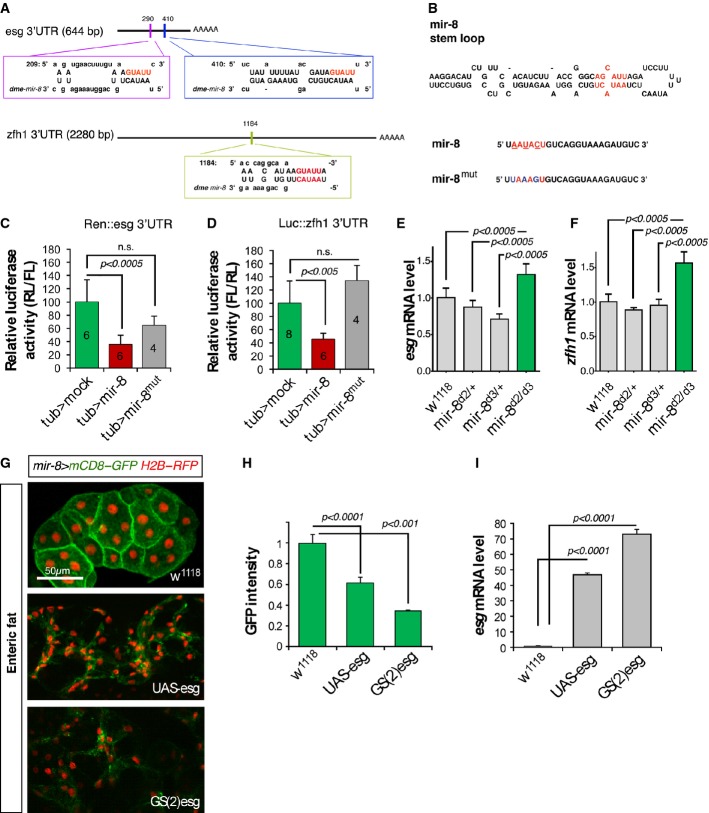
Figure 6.
Mutual antagonism between miR-8 and Escargot
- A Schematic drawing of the 3′UTR regions of escargot and zfh1/zeb genes highlighting the mir-8 seed sites and that of the microRNA miR-8.
- B Schematic mir-8 pri-miRNA structure and the mutated sites (below).
- C, D Luciferase assay in Drosophila Schneider (S2) cells co-transfected with the empty vector (green bars), tub > mir-8 (red bars) or the mutated version of the microRNA tub > mir-8mut (grey bars) together with a sensors containing the escargot 3′UTR (Hartl et al, 2011) (C) or zfh1 3′UTR (D). Firefly luciferase activity was measured 48 h after transfection and normalized against Renilla luciferase. The values represent the mean ± SD, and the biological repeats of empty vector, mir-8, and mir-8mut, respectively, are indicated in the bars.
- E, F Differences in escargot and zfh1 and mRNA expression assessed by real-time qPCR in animals null for mir-8. Values represent the mean ± SD of three independent repeats and n = 40 animals in each condition and P < 0.0005 (Student’s t-test).
- G mir-8 is expressed in adult fat polyploid cells surrounding the midgut (enteric fat cells: green staining).
- H, I Overexpression of escargot using the UAS-esg and GS(2)esg in enteric fat cells converted epithelial fat cells into spindle-shaped cells. (H) Quantification of GFP (mir-8 > mCD8-GFP) intensity measured as mean pixel value and expressed as fold change to control. (I) Real-time quantitative PCR of escargot mRNA levels driven by two different transgenes (UAS-esg and GS(2)esg) and measured after 1-h heat shock in hsp70-Gal4 animals with or without the indicated transgenes. Error bars represent SD.
Source data are available online for this figure.
Finally, it did not escape our notice that miR-8 and escargot have also opposite effects in the control of polyploidy/diploidy (Fuse et al, 1994; Hyun et al, 2009; Loya et al, 2009; Morante et al, 2013). The vertebrate homolog of escargot, Snail (also known as Slug), is also implicated in maintaining diploid state, while repressing polyploidy during early development (Nieto, 2011). In agreement with these interactions being more generally required, we found that ectopic expression of mir-8 dramatically increases enteroblast nuclei size (Supplementary Fig S3F–H), whereas animals null for mir-8 have reduced enterocyte nuclei size compared to wild-type midguts (Supplementary Fig S3B and C). Conversely, ectopic expression of escargot in mir-8 expression pattern (mir-8-Gal4 > esg) readily converted adult epithelial (endopolyploid) cells into spindle-shaped (diploid-like) cells (Fig6G) and the transformation was associated with decreased mir-8 (mir-8-Gak4 > GFP, Fig6H and I). Together, these data mechanistically link transition from diploid to polyploid via EMT and stemness/undifferentiated state to an Escargot/Snail2-miR-8/miR-200 circuit.
Discussion
We have identified that the robustness of intestinal cell renewal relies on cellular plasticity in committed progenitor cells and a rather loose regulation of ISCs proliferation. One key finding is that stem cells divide continually and generate a “stock” of committed progenitor cells that do not terminally differentiate right away but postpone their final differentiation for long time intervals in the absence of a local epithelial cell loss. Accordingly, one noticeable change in newborn progenitor cells after their (enterocyte) fate commitment is their transformation from rounded cells to spindle-shaped cells that appear to actively monitor their surroundings by extending long membrane actin-rich protrusions that make cell–cell contact with mature epithelial cells and their mother ISCs. Timely terminal differentiation with epithelial cell loss is orchestrated by activation of a conserved pro-epithelial microRNA, in turn, directly repressing the repressors of differentiation. A microRNA-induced repression of the repressors of differentiation provides a faster mechanism than one involving a transcriptional regulator since synthesizing a miRNA likely requires less time than synthesizing a protein. Importantly, mutual antagonism between the microRNA (MiR-8/miR-200) and its targets (Escargot/Snail2 and Zfh1/ZEB) (Karres et al, 2007; Krejci & Bray, 2007; Burk et al, 2008; Wellner et al, 2009; this study) may serve to slow down the mesenchymal-to-epithelial process inside individual mesenchymal/progenitor cells until they are successfully integrated in the epithelium. Consistently, abrupt transition as in mir-8 overexpressing midguts results in erroneous tissue repair.
Supply–demand: make to order or make to stock strategy?
Supply and demand in business production involves frequently two alternative solutions called “make-to-stock” and “make-to-order”. In “make-to-stock” or MTS, production is continuous so that response to customers can be supplied immediately. However, as production is not based on actual demand, the MTS solution is not robust against fluctuations in demand and errors in forecasting can result in shortages (if there is insufficient residual stock) or overproduction. In “make-to-order”, or MTO, production only starts upon receiving a customer’s order, thereby precisely matching production to demand. However, the MTO generates a delay in the response and can be less efficient and competitive than the MTS paradigm. The dynamics of stem cells and committed progenitor cells in the midgut suggests a hybrid solution between MTS and MTO—reminiscent to the business solution known as delayed differentiation (Gupta & Benjaafar, 2004). Thus, in basal homeostasis, production of new cells to replace cell loss occurs in two stages: (i) a “make-to-stock” stage where committed progenitor cells are continually generated and “stocked” in an “undifferentiated” state; and (ii) a “make-to-order” stage where terminal differentiation takes place only in response to a local demand (model in Fig7). In mice and humans, the rapid turnover that occurs in the small intestinal epithelium is thought to be the result of continual shedding of superficial cells (Ren et al, 2010) balanced by the continual stem cell production. The mechanism described here may be more general than expected and could account for how murine cells after fate commitment like the secretory-committed cells defer for long periods their terminal differentiation (Buczacki et al, 2013).
Figure 7.
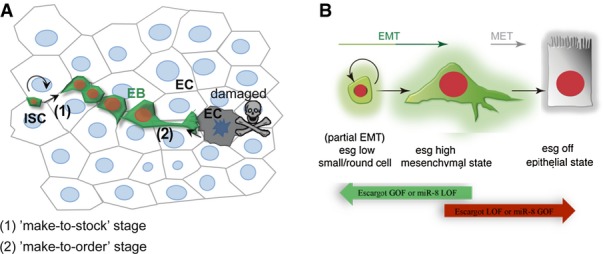
Model
- ISCs divide regularly (“make-to-stock”) and progenitor cells are flexible in delaying their terminal differentiation until detecting a local demand to replace an older or dying epithelial cell. Mechanical input or a chemical signal from the dying cell communicates its state to the neighbouring mesenchymal/progenitor cells that activates mir-8 expression (“make-to-order” stage).
- The balance between escargot (and zfh1) and miR-8 triggers the mesenchymal-to-epithelial transition (MET) and terminal differentiation to replace a lost enterocyte or enteroendocrine cell.
Uncoupling EMT and stemness?
Escargot/Snail2 sustains the undifferentiated state and self-renewing divisions of midgut intestinal stem cells (Korzelius et al, 2014; Loza-Coll et al, 2014) (this study). However, the committed progenitor cells also express escargot and apparently at higher levels than the stem cells (Fig2B and C). We hypothesize that below a certain threshold level, Escargot maintains stemness and a partial EMT that may facilitate regular cell division and a topologically confined position at the base of the intestinal epithelium (Micchelli & Perrimon, 2006; Ohlstein & Spradling, 2006). Conversely, when Escargot surpasses a certain threshold level, it promotes a full EMT that confers invasive properties and motility for the successful response and integration of the newly differentiated cells in the pre-existing epithelium (Fig7). Intriguingly, the enteroendocrine cells appear to escape from this block in terminal differentiation and differentiate at the normal rate in the absence of escargot. We have not yet an explanation for the behaviour of these progenitor cells.
Mechanistically, the different levels of escargot could be achieved via Notch signalling pathway, which is prominently activated in enterocyte-committed progenitors. Notch signalling activates directly zfh1 gene (Krejci & Bray, 2007) and Zfh1, a homolog of the mammalian stemness and EMT-determinant Zeb1,2 (Postigo et al, 1999; Wellner et al, 2009), and binds to the escargot promoter region (Negre et al, 2011), and we show that Zfh1 acts genetically upstream of escargot (Supplementary Fig S2). Thus, progenitor cells receiving Notch signalling might enhance escargot transcriptional levels via Notch-induced zfh1 transcription. Such regulatory mechanism would explain, for example, that loss of Notch results in stem-like/round cells (Micchelli & Perrimon, 2006; Ohlstein & Spradling, 2007).
In mammalian cell culture, the EMT process has been linked to the acquisition of stem-like nature (e.g. Mani et al, 2008; Shimono et al, 2009; Wellner et al, 2009) via an interplay between the ZEB1,2 and Snail transcription factors and the microRNAs of the miR-200 family. Moreover, EMT determinants often regulate each other to promote EMT (Lamouille et al, 2014). Thus, the interactions between Escargot/Snail2, zfh1/Zeb and miR-8/miR-200 that were identified here exemplify the conservation of the regulatory mechanisms involved in EMT/MET and stemness in an in vivo context and a normal physiology of an adult organism. However, we show that escargot__zfh1 promotes stemness and full EMT/invasive properties in distinct cell populations and likely at different concentration levels, highlighting the utility of Drosophila midgut as a model to dissect out mechanisms linking physiological EMT to cellular plasticity and stemness (Nieto, 2002) as well as provide novel insights linking polyploidy and EMT towards stemness.
Enteroblasts extend long membrane protrusions and response to the repair needs around them
Although midgut mesenchymal/progenitor cells have motility (Supplementary Movies S1 and S2), most of them maintain their own local area as clearly defined by Flybow clonal analysis (Fig2). This situation is similar to the leading edge mesenchymal cells during collective cell migration (Nieto, 2011). Midgut enteroblasts retain contact via E-cadherin with their mother ISC (Maeda et al, 2008), a process that might be regulated by escargot as in tracheal cells (Tanaka-Matakatsu et al, 1996; Ribeiro et al, 2004). Cell–cell contact is crucial to sustain Notch signalling in committed progenitor cells and likely to help to stabilize polarity of enteroblasts and their membrane protrusions that contact mature cells. Through these protrusions, mesenchymal/enteroblasts might actively monitor their surroundings. When a protrusion detects changes in tension and mechanical forces generated during the elimination of a dying cells (Teng & Toyama, 2011; Allena, 2013), a positive input might be created that triggers the activation of expression of the microRNA mir-8 in the particular progenitor cell which, in turn, promotes the epithelial state and integration of the newly differentiated cell in the epithelium (Fig7, model). Adhesion via E-cadherin (Renkawitz & Sixt, 2010; Cai et al, 2014) could facilitate communication between an epithelial cells and a mesenchymal/progenitor cell in its vicinity so that a single, newly differentiated cell fills the gap left by the cleared cell.
Dynamic pseudopodia in migrating cells have been proposed as a mechanism for temporal and spatial sensing during cell migration (Allena, 2013). Direction sensing (Teng & Toyama, 2011; Allena, 2013; Cai et al, 2014) is also consistent with our time-lapse data showing individual progenitor cells re-adjusting position in the homeostatic midguts (Supplementary Movies S1 and S2). Transduction of mechanical cues via YAP and TAZ (called Yorkie in flies) (Dupont et al, 2011) is functionally involved in differentiation of mesenchymal stem cells. Hence, Drosophila Hippo/Yorkie-YAP in mature enterocytes (Li et al, 2014) is a primary candidate pathway for a potential transduction of mechanical cues activating mir-8 in response to cell death.
In summary, the miR-8–escargot–zfh1 axis and the EMT/MET programme provides a conceptual shift of the current stem cell-centred view of tissue renewal and offers a starting point for investigating how mature cells speak with neighbouring committed progenitor cells to ensure that epithelial cell loss and cell addition are kept in balance.
Materials and Methods
Genetics and fly husbandry
The GS(2)101M2 line upstream of escargot locus (GS(2)esg) was isolated in a genetic screen for enhancer/suppressors of a large eye phenotype caused by Delta overexpression (Ferres-Marco et al, 2006) (see below). The mir-8-Gal4, mir-8Δ2 and mir-8Δ3 mutants were from S.M. Cohen and described in Karres et al (2007); UAs-mir-8 was described previously in Vallejo et al (2011); UAS-mir-8 sponge in Loya et al (2009); and UAS-zfh1 Postigo et al (1999). The other fly stocks used were from the Bloomington Stock Centre: w1118, esgL2, esgG66B, esg-Gal4, UAS-mCD8-GFP and UAS-H2B-RFP, Dl-Gal4 UAS-esg. The RNAi transgenic flies for escargot (P[GD1437]) and zfh1 (P[KK109931]) were obtained from the Vienna Drosophila RNAi Centre (Dietzl et al, 2007). The other escargot RNAi transgenic construct (P[TRiP.JF03134]) was from the Bloomington Stock Centre (Ni et al, 2011).
The ReDDM-based crosses were kept at 18°C to ensure Gal80ts repression until adult eclosion, and 7-day-old adult mated female flies were shifted to 29°C to inactive the Gal80, and they were supplied with new food every 7 days. Standard “Iberian” fly food was made by mixing 15 l of water, 0.75 kg of wheat flour, 1 kg of brown sugar, 0.5 kg of yeast, 0.17 kg of agar, 130 ml of a 5% nipagin solution in ethanol and 130 ml of propionic acid.
Flybow MARCM clones
For the Flybow MARCM clones, we used yw hsp70-FLP1.22; FB2.0 260b/hsp-mFLP5; tub-GAL4 FRT82B tub-GAL80/FRT82B flies (a kind gift of Iris Salecker) (Hadjieconomou et al, 2011). The guts were analysed at the time points indicated.
Gain- and loss-of-function tub-Gal4 MARCM clones were generated in adult midgut stem and progenitor cells by crossing 2- to 5-day-old adult female virgins of the hsp-70-Flp; tub-Gal4 UAS-mCD8-GFP/CyO; tub-Gal80 FRT82B/+ genotype to FRT82B UAS-esg (or UAS-zfh1) males. MARCM clones of esg-RNAi were induced by crossing female virgins tub-Gal80 FRT40A; repo-Flp1C, tub-Gal4 UAS-GFPnls/TM6 (a gift from C. Klambt) to males FRT40A UAS-esg-RNAi. Clones were induced by a single heat stock of 45 min at 37°C, adult midguts were analysed at 7, 14, or 21 days after clone induction (ACI), and at least 10 clones/midgut were examined in n = 10 adult midguts.
Time-lapse imaging of adult midguts
The esg+ cells were labelled by membrane-bound mCD8-GFP and nuclear H2B-RFP driven by esg-Gal4. Committed progenitor cells were clearly detected by video microscopy through their strong GFP signal, and adult midgut mature epithelial cells were defined by DE-cadherin-GFP. Time-lapse microscopy was performed using a Confocal microscope (Leica Microscopy), and images were acquired every 5 min over 90 min using A-Plan 10×/0.45 Ph1 or Plan-Apochromat 40×/0.95 Korr M27 objectives in fluorescence field (GFP filters: EX BP 470/40; at 350 ms) and with a CoolSNAPHQ2 monochrome camera (Photometrics). Images were stitched and processed with ImageJ (http://imagej.nih.gov/ij/) software and with the specific tracking plugin MTrackJ. The fluorescence signal was measured as the difference of the maximum and minimum intensity within a defined region of interest (ROI) around each cell.
Image processing and analysis
Confocal images were obtained with a Leica TCS SP5 inverted confocal microscope, using a 1,024 × 1,024 pixel size. Stacks were typically collected every 1 μm, and the images were reconstructed using max projection. Images were adjusted, evaluated and scaled using ImageJ. A self-written script optimized for the ReDDM method that recognizes quality (size, colour) and quantity (events) was used to count undifferentiated cells (stem and progenitor cells), replaced enterocytes and enteroendocrine (ee) cells, the total number of enterocytes and ee and the total area. Enterocyte cell (EC) density was calculated as total EC/total area and tissue replenishment as new cells (red-only cells)/total mature cells (DAPI).
GS-element mapping
Genomic DNA flanking the P-element insertions in the GS(2)101M2 (GS(2)esg) line was recovered by inverse PCR (http://www.fruitfly.org/about/methods) and sequenced. A BLAST search with the sequence produced perfect matches to the genomic region at 35D1 (2L chromosome), with an insertion point at 2L:15331910…15336067 chromosome upstream of escargot gene.
miRNA-mRNA 3′UTR alignment
Binding sites for miR-8 in the escargot 3′UTR were identified using TargetScanFly database and those in zfh1 3′UTR were identified using the BiBiServ server (Rehmsmeier et al, 2004). Seed sequences with G:U wobbles or mismatches within the 7–6-mer seed sequence were not considered, and only miRNA:target-mRNA duplexes with a free energy above −15 kcal/mol were considered.
Constructs for sensor and luciferase reporter assays
The escargot 3′UTR sensor has been described before (Hartl et al, 2011). The zfh1-3′UTR was inserted into the 3′ end of tub-luciferase to construct the tub-luc:Zfh1-3′UTR. For Drosophila Schneider (S2) cells (Invitrogen) luciferase assays, cells were co-transfected in 24-well plates with the tub-mir-8 plasmid (250 ng), the luciferase-zfh1-UTR construct (25 ng) and the Renilla luciferase plasmid (25 ng) for normalization. The relative luciferase activity was measured 60 h post-transfection, and luciferase activity was measured using Dual-Glo Luciferase Assay (Promega).
Real-time qPCR
RNAi knockdown efficiency was determined by heat-shock-dependent expression over 30–60 min at 37°C in wandering L3 larvae (hs-Gal4) and over 30 min at RT for target gene knockdown. All tissue samples were stored at −80°C in RNAlater Tissue Protect Tubes (Qiagen) until total RNA from at least 10 whole larvae was extracted using RNeasy Mini Kit (Qiagen), from which cDNAs were prepared with SuperScript First-Strand Synthesis System (Invitrogen) using oligo-dT primers. Quantitative PCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems) in a 7500 Real-Time PCR System (Applied Biosystems) using rp49 as a housekeeping control gene. All qPCRs were performed in triplicate, and the relative expression was calculated using the comparative Ct method.
Sequence of the primers:
| Forward 5′–3′ | Reverse 5′–3′ | |
|---|---|---|
| zfh1 | CCCTATGTGTGCGATCAGTG | GTTGACCGGAATGCTCGTAT |
| esg | ATATGTCGCCCGAAACTATGCCGA | CGGGCAATGGAACTGCTGATGTTT |
| rp49 | TGTCCTTCCAGCTTCAAGATGACCATC | CTTGGGCTTGCGCCATTTGTG |
Paraquat and mechanical damage/regeneration assays
For Paraquat poisoning, adult female flies of 3–7 days of age carrying the mir-8-Gal4 and UAS-mCD8-GFP transgenes were starved for 4 h without water and then transferred to vials containing filter paper soaked in 5% sucrose with or without Paraquat (10 mM, methyl viologen, Sigma-Aldrich). Four hours later, midguts were dissected from the poisoned and control females.
Adult female flies of 3–7 days of age carrying the mir-8-Gal4 and UAS-mCD8-GFP transgenes were pinched with tweezers in the abdomen and let recover for 4–6 h before dissection. As controls, non-pinched flies from the same cohort (sibling) were used.
mir-8 mutagenesis
To validate in mass in silico predicted targets of the microRNA mir-8, we employed a strategy of mutating the microRNA mir-8 instead of individual 3′ UTR of the various candidate target genes. To mutagenize mir-8 sequence in the tub > mir-8 plasmid (JB-25_miR8), we used the QuikChange® II XL Site-Directed Mutagenesis Kit (Stratagene) according to manufacturer protocol. Gradient optimization of PCR with different temperatures for elongation were used (55.1–57.2–59.8–62.5–64.6°C). For transformation, we used the XL10 Gold Ultracompetent cells. Mutagenesis was verified by sequencing. Used primers are listed below.
Pairs of primer used for mir-8 mutagenesis and sequencing.
| Use | Oligo Name | Sequence (5′–3′) |
|---|---|---|
| Mutagenesis | miR-8-mut_up | GATCCTTTTTATAACTCTTAAAGTGTCAGGTAAAGATGTCGTCCG |
| miR-8-mut_low | CGGACGACATCTTTACCTGACACTTTAAGAGTTATAAAAAGGATC | |
| Sequencing | M8short-up | AAGGGGGCCAATGTTCTAAG |
| M8short-low | CCGCTTGTCTTCGCATTATC |
Immunohistochemistry
Adult Drosophila midguts were dissected and fixed for 40 min in 4% paraformaldehyde and stained using the following antibodies: rabbit anti-PH3 (1:2,000, Upstate), mouse anti-Dlg-1 (1:100, Hybridoma Bank), mouse anti-Pros (1:100, Hybridoma Bank), mouse anti-Dl (1:100, Hybridoma Bank), sheep anti-GFP (1:1,000, Biogenesis), chicken anti-β-Gal (1:1,000 Abcam), rabbit anti-HA::tag (1:500, Abcam), mouse anti-V5 (1:500, Abcam). Secondary antibodies were used at 1:400 (Alexa), the nuclei were counterstained with DAPI (Sigma), and the tissue was mounted in Fluoromount-G (Southern Biotech).
Survival curves
Crosses were reared at 18°C. Females of the appropriate genotype and of 3–7 days of age were then shifted to 29°C. No more than 15 flies were kept per tubes to avoid overcrowding stress, and flies were transferred to new tubes every 3 days to exclude influence of bacterial contamination in the food on gut homeostasis. Flies were daily checked for survival until natural death or experimental checkpoint (21 days). At least 12 tubes per genotype were scored (n > 100 flies scored per genotype). Survival curves were plotted using Prism GraphPad software and analysed by the log-rank test.
Acknowledgments
We thank Dr. S. Cohen, Dr. S. Hou, Dr. I. Salecker, Dr. A. Postigo, and the Bloomington Stock Center and the Vienna Drosophila Research Centre for fly stocks; Dr. IC. Grunwald Kadow for the escargot sensor-luciferase plasmid. We also thank A. Nieto for comments and the suggestion that ISCs are epithelioid cells and the members of the laboratory for helpful comments, and I. Oliveira and L. Mira for technical assistance. T.R. was funded by a postdoctoral Deutsche Forschungsgesellschaft (DFG) fellowship and by the Foundation Botin, and Z.A.A. is a fellow from MEC-CONSOLIDER. M.D. is funded by Spanish National Grants (BFU2009-09074, SAF2012-35181 and MEC-CONSOLIDER CSD2007-00023), Generalitat Valenciana Grant (PROMETEO II/2013/001) and Fundación Botin.
Author contributions
ZAA, TR and MD conceived the study and designed experiments. ZAA, TR and EBI performed experiments, and ZAA, TR and MD analysed the data. MD wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Movie S1
Supplementary Movie S2
Supplementary Legends
Source Data for Supplementary Figure S1
Source Data for Supplementary Figure S3
Review Process File
Source Data for Figure 1
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
References
- Allena R. Cell migration with multiple pseudopodia: temporal and spatial sensing models. Bull Math Biol. 2013;75:288–316. doi: 10.1007/s11538-012-9806-1. [DOI] [PubMed] [Google Scholar]
- Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell. 2009;4:49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apidianakis Y, Rahme LG. Drosophila melanogaster as a model for human intestinal infection and pathology. Dis Model Mech. 2011;4:21–30. doi: 10.1242/dmm.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell. 2011;9:402–411. doi: 10.1016/j.stem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Jasper H. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development. 2011;138:1045–1055. doi: 10.1242/dev.056671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Kuraishi T, Lemaitre B. Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 2010;8:1741–7007. doi: 10.1186/1741-7007-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Chen SC, Prasad M, He L, Wang X, Choesmel-Cadamuro V, Sawyer JK, Danuser G, Montell DJ. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell. 2014;157:1146–1159. doi: 10.1016/j.cell.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- Choi NH, Lucchetta E, Ohlstein B. Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proc Natl Acad Sci USA. 2011;108:18702–18707. doi: 10.1073/pnas.1109348108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:4179–4183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Ferres-Marco D, Gutierrez-Garcia I, Vallejo DM, Bolivar J, Gutierrez-Avino FJ, Dominguez M. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439:430–436. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furriols M, Bray S. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr Biol. 2001;11:60–64. doi: 10.1016/s0960-9822(00)00044-0. [DOI] [PubMed] [Google Scholar]
- Fuse N, Hirose S, Hayashi S. Diploidy of Drosophila imaginal cells is maintained by a transcriptional repressor encoded by escargot. Genes Dev. 1994;8:2270–2281. doi: 10.1101/gad.8.19.2270. [DOI] [PubMed] [Google Scholar]
- Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Gupta D, Benjaafar S. Make-to-order, make-to-stock, or delay product differentiation? A common framework for modeling and analysis. IIE Trans. 2004;36:529–546. [Google Scholar]
- Hadjieconomou D, Rotkopf S, Alexandre C, Bell DM, Dickson BJ, Salecker I. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat Methods. 2011;8:260–266. doi: 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- Hartl M, Loschek LF, Stephan D, Siju KP, Knappmeyer C, Kadow IC. A new Prospero and microRNA-279 pathway restricts CO2 receptor neuron formation. J Neurosci. 2011;31:15660–15673. doi: 10.1523/JNEUROSCI.2592-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng YW, Koh CG. Actin cytoskeleton dynamics and the cell division cycle. Int J Biochem Cell Biol. 2010;42:1622–1633. doi: 10.1016/j.biocel.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Horvay K, Jarde T, Casagrande F, Perreau VM, Haigh K, Nefzger CM, Akhtar R, Gridler T, Berx G, Haigh JJ, Barker N, Polo JM, Hime GR, Abud HE. Snai1 regulates cell lineage allocation and stem cell maintenance in the mouse intestinal epithelim. EMBO J. 2015;34:1319–1335. doi: 10.15252/embj.201490881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139:1096–1108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Edgar BA. Intestinal stem cell function in Drosophila and mice. Curr Opin Genet Dev. 2012;22:354–360. doi: 10.1016/j.gde.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Korzelius J, Naumann SK, Loza-Coll MA, Chan JS, Dutta D, Oberheim J, Glasser C, Southall TD, Brand AH, Jones DL, Edgar BA. Escargot maintains stemness and suppresses differentiation in Drosophila intestinal stem cells. EMBO J. 2014;33:2967–2982. doi: 10.15252/embj.201489072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci A, Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Li Q, Li S, Mana-Capelli S, Roth Flach RJ, Danai LV, Amcheslavsky A, Nie Y, Kaneko S, Yao X, Chen X, Cotton JL, Mao J, McCollum D, Jiang J, Czech MP, Xu L, Ip YT. The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Dev Cell. 2014;31:291–304. doi: 10.1016/j.devcel.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Xu N, Xi R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455:1119–1123. doi: 10.1038/nature07329. [DOI] [PubMed] [Google Scholar]
- Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza-Coll MA, Southall TD, Sandall SL, Brand AH, Jones DL. Regulation of Drosophila intestinal stem cell maintenance and differentiation by the transcription factor Escargot. EMBO J. 2014;33:2983–2996. doi: 10.15252/embj.201489050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Takemura M, Umemori M, Adachi-Yamada T. E-cadherin prolongs the moment for interaction between intestinal stem cell and its progenitor cell to ensure Notch signaling in adult Drosophila midgut. Genes Cells. 2008;13:1219–1227. doi: 10.1111/j.1365-2443.2008.01239.x. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Morante J, Vallejo DM, Desplan C, Dominguez M. Conserved miR-8/miR-200 defines a glial niche that controls neuroepithelial expansion and neuroblast transition. Dev Cell. 2013;27:174–187. doi: 10.1016/j.devcel.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Navascues J, Perdigoto CN, Bian Y, Schneider MH, Bardin AJ, Martinez-Arias A, Simons BD. Drosophila midgut homeostasis involves neutral competition between symmetrically dividing intestinal stem cells. EMBO J. 2012;31:2473–2485. doi: 10.1038/emboj.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, Li Z, Ishii H, Spokony RF, Chen J, Hwang L, Cheng C, Auburn RP, Davis MB, Domanus M, Shah PK, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- O’Brien LE, Soliman SS, Li X, Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147:603–614. doi: 10.1016/j.cell.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Perdigoto CN, Schweisguth F, Bardin AJ. Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development. 2011;138:4585–4595. doi: 10.1242/dev.065292. [DOI] [PubMed] [Google Scholar]
- Postigo AA, Ward E, Skeath JB, Dean DC. zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol Cell Biol. 1999;19:7255–7263. doi: 10.1128/mcb.19.10.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS. Stem cells in gastrointestinal epithelium: numbers, characteristics and death. Philos Trans R Soc Lond B Biol Sci. 1998;353:821–830. doi: 10.1098/rstb.1998.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Wang B, Yue T, Yun EY, Ip YT, Jiang J. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc Natl Acad Sci USA. 2010;107:21064–21069. doi: 10.1073/pnas.1012759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz J, Sixt M. Mechanisms of force generation and force transmission during interstitial leukocyte migration. EMBO Rep. 2010;11:744–750. doi: 10.1038/embor.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C, Neumann M, Affolter M. Genetic control of cell intercalation during tracheal morphogenesis in Drosophila. Curr Biol. 2004;14:2197–2207. doi: 10.1016/j.cub.2004.11.056. [DOI] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM, Somlo G, Pera RA, Lao K, Clarke MF. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Uemura T, Oda H, Takeichi M, Hayashi S. Cadherin-mediated cell adhesion and cell motility in Drosophila trachea regulated by the transcription factor Escargot. Development. 1996;122:3697–3705. doi: 10.1242/dev.122.12.3697. [DOI] [PubMed] [Google Scholar]
- Teng X, Toyama Y. Apoptotic force: active mechanical function of cell death during morphogenesis. Dev Growth Differ. 2011;53:269–276. doi: 10.1111/j.1440-169X.2011.01251.x. [DOI] [PubMed] [Google Scholar]
- Vallejo DM, Caparros E, Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. EMBO J. 2011;30:756–769. doi: 10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth P. Regional regulation of microtubule dynamics in polarized, motile cells. Cell Motil Cytoskeleton. 1999;42:48–59. doi: 10.1002/(SICI)1097-0169(1999)42:1<48::AID-CM5>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon ED. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J Cell Biol. 1997;139:417–434. doi: 10.1083/jcb.139.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- Wood W, Martin P. Structures in focus—filopodia. Int J Biochem Cell Biol. 2002;34:726–730. doi: 10.1016/s1357-2725(01)00172-8. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Movie S1
Supplementary Movie S2
Supplementary Legends
Source Data for Supplementary Figure S1
Source Data for Supplementary Figure S3
Review Process File
Source Data for Figure 1
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6