Abstract
Many tropical marine cyanobacteria are prolific producers of bioactive secondary metabolites with ecological relevance and promising pharmaceutical applications. One species of chemically rich, tropical marine cyanobacteria that was previously identified as Symploca hydnoides or Symploca sp. corresponds to the traditional taxonomic definition of Phormidium penicillatum. In this study, we clarified the taxonomy of this biomedically and ecologically important cyanobacterium by comparing recently collected specimens with the original type material and the taxonomic description of P. penicillatum. Molecular phylogenetic analyses of the 16S rRNA gene and the 16S-23S ITS regions showed that P. penicillatum formed an independent clade sister to the genus Symploca, and distantly related to Phormidium and Lyngbya. We propose the new genus Caldora for this clade, with Caldora penicillata comb. nov. as the type species and designate as the epitype the recently collected strain FK13-1. Furthermore, the production of bioactive secondary metabolites among various geographically dispersed collections of C. penicillata showed that this species consistently produced the metabolite dolastatin 10 and/or the related compound symplostatin 1, which appear to be robust autapomorphic characters and chemotaxonomic markers for this taxon.
Keywords: dolastatin 10, largazole, Lyngbya, natural products, Phormidium, phylogenetics, secondary metabolites, Symploca, symplostatin 1
INTRODUCTION
During the last three decades, natural products (NPs) discovery efforts have successfully exploited collections of tropical marine cyanobacteria for biologically active secondary metabolites with potential pharmaceutical properties (Nunnery et al. 2010, Tidgewell et al. 2010). Tropical marine cyanobacteria with morphological resemblance to the genus Symploca Kützing ex Gomont (1892) are prolific producers of NPs (Luesch et al. 2001, Thacker and Paul 2004, Taori et al. 2008, 2009, Engene et al. 2013). Collections of tropical marine cyanobacteria identified as S. hydnoides and Symploca sp. have yielded several important natural products with promising pharmaceutical potential, including the metabolites: symplostatin 1 (Harrigan et al. 1998), dolastatin 10 (Luesch et al. 2001), largazole (Taori et al. 2008), janthielamide A, kimbeamides A-C, and kimbelactone A (Nunnery et al. 2012).
The cyanobacterial genus Symploca was first described from terrestrial humid habitats, with Symploca meneghiniana Kützing ex Gomont (1892) later designated as the type species (Gardner 1932: 283), but also including two temperate marine species S. hydnoides Kützing ex Gomont (1892) and S. atlantica Gomont (1892), and later on S. funicularis Setchell & N.L. Gardner in Gardner (1918). Traditionally, Symploca spp. have been defined as fine filamentous cyanobacteria with isodiametric cells and filaments clustered into fasciculated bundles (Gomont 1892, Geitler 1932, Komárek and Anagnostidis 2005). However, this taxonomic definition is somewhat ambiguous and Symploca spp. usually overlap morphologically with other filamentous cyanobacteria genera within Oscillatoriales (Castenholz 2001, Komárek and Anagnostidis 2005), such as Microcoleus Desmazičres ex Gomont (1892) and Phormidium Kützing ex Gomont (1892). The widespread genus Phormidium (Ph. lucidum Kützing ex Gomont 1892 as generitype) was also described from terrestrial humid habitats and has been considered as possibly the most problematic genus within Oscillatoriaceae (Komárek et al. 2014). In particular, Symploca and Phormidium are primarily differentiated based on their growth morphologies; Symploca spp. form fasciculated, erect thalli, whereas Phormidium spp. form thin mats (Komárek and Anagnostidis 2005). A prime example of the confusing morphological overlap and complicated nomenclatural history between these two genera is Phormidium penicillatum Gomont (1893), which was first collected and described from coral reef habitats of the island of Réunion in the Indian Ocean. Despite being included in Phormidium, this species has a very characteristic upright growth form that corresponds better with the characterization of Symploca. Nearly a century later, Hoffmann and Compère (1990) transferred Phormidium penicillatum to Lyngbya C. Agardh ex Gomont (1892) as Lyngbya penicillata (Gomont) L.R. Hoffmann nom. illeg., for which a new name, Lyngbya penicilliformis P. C. Silva was proposed (Silva, Basson and Moe 1996), since Hoffman’s new combination was predated by Lyngbya penicillata (Kützing) Rabenhorst ex Forti (1907) based on a different type specimen.
The taxonomic confusion between the two genera Symploca and Phormidium highlights some common problems with cyanobacterial systematics. Firstly, the morphological characters used to define taxa are typically ambiguous and only a limited number of morphological characters are available for distinguishing different taxa (Komárek 2006, Sciuto et al. 2012, Dadheech et al. 2013). Many of these characters also show morphological plasticity and often vary depending on environmental conditions (Sumina 2006). As a result of these limitations in traditional mophology-based identification, modern cyanobacterial systematics is largely incorporating molecular data in order to construct monophyletic genera as a fundamental framework for biologically informative classification systems (Castenholz 2001, Hoffmann et al. 2005, Komárek and Anagnostidis 2005, Komárek 2006, Komárek et al. 2014). This polyphasic approach, combines genotypic (16S rRNA gene sequencing) and phenotypic data (morphological analysis, ultrastructural sections) (Sciuto et al. 2012, Dadheech et al. 2013, Strunecký et al. 2013, 2014), striving to find at least one autapomorphic character, in order to preliminarily characterize each genus (Komárek 2010).
However, a major obstacle with the use of a molecular phylogenetically-based system is to correlate the original type material used to describe a species with recently collected specimens used for genetic sequencing. Although cyanobacterial herbarium specimens have been used to obtain DNA for genetic sequencing (Palinska et al. 2006), many type specimens are lacking or no longer suitable for a complete polyphasic comparison (Sciuto et al. 2012,). As a consequence, reasonable presumptions are often necessary based on type locality and original descriptions to define reference strains. In regards to Lyngbya and Symploca, the cyanobacterial strains PCC 7419 (Castenholz et al. 2001a), and PCC 8002 (Castenholz et al. 2001b) have been used as reference strains for these genera, respectively, and CCALA 759 (Sciuto et al. 2012) has been designated as the epitype for Phormidium irriguum (Kützing) Anagnostidis & Komárek. Herein, we use molecular phylogenetic methods to resolve the phylogenetic relationships of the chemically rich tropical marine Symploca specimens that are responsible for several important NPs by comparing 16S rRNA gene sequences of recently collected specimens with sequences of the above-mentioned reference strains. We also show that these specimens are molecularly distinct (16S rRNA gene sequences with a similarity index of ±95% or less for generic definition; Komárek 2010) from Phormidium, Lyngbya, and Symploca specimens and instead form a separate lineage (previously described as Clade III in Engene et al. 2013). Furthermore, two of the analyzed sequences are highly similar (99-100% similarity on 16S rRNA gene sequences) to samples from the type locality of Phormidium penicillatum (Saint-Gilles, Réunion; Echenique-Subiabre et al. 2015). We conclude that those specimens traditionally identified in the NPs literature as Symploca spp., in fact, refer to Phormidium penicillatum Gomont (1893) and, consequently, we propose Clade III as the new genus Caldora, accommodating Caldora penicillata comb. nov. as the type species. Furthermore, we show that geographically dispersed collections of C. penicillata consistently produce the metabolite dolastatin 10 and/or the structurally related metabolite symplostatin 1. Thus, these compounds appear to act as autapomorphic characters and robust chemotaxonomic markers for the identification of this species.
MATERIALS AND METHODS
Cyanobacterial collection
Cyanobacterial specimens identified as Lyngbya penicilliformis or Symploca spp. were collected by SCUBA or snorkeling from various marine habitats in Bonaire, Curaçao, Belize, Honduras, and the Florida Keys (see Table S1 in the Supporting Information for detailed collection information). Putative taxonomic identification and selection of the cyanobacterial specimens were performed in accordance with traditional classification systems (Geitler 1932, Castenholz 2001, Komárek and Anagnostidis 2005) and with reference to available field guides (Littler and Littler 2000, 2003). Specimens were cleaned immediately after collection under a dissecting microscope (Wild Heerbrugg, Heerbrugg, Germany) to remove other macroorganisms. All collections were preserved in RNAlater® (Ambion, Austin, TX, USA) for genetic analysis, in seawater with 5% formalin for morphological analysis, and frozen at -20 °C for chemical analysis. The Symploca reference strain PCC 8002 was obtained from the Pasteur Culture Collection (PCC) for phylogenetic comparison (a comprehensive morphological characterization of strain PCC 8002 is available in Porta et al. 2003). The type specimen of Phormidium penicillatum (PC0167627; see Figures S1 and S2 in the Supporting Information), as well as other P. penicillatum specimens deposited in the Cryptogamy collection (PC) were used for taxonomic revision and morphological comparison. In addition, syntypes of Lyngbya penicilliformis were obtained from UC, and from the Drouet collection at US. Herbarium acronyms follow Thiers (2015). All collected specimens were deposited in US, under the Algal Collection numbers: 217964, 217968-217983. Due to the troubled taxonomic history of this taxon and the inability to obtain representative DNA sequences from the type material of Phormidium penicillatum, in addition to the absence of voucher specimens of recently sequenced samples from the type locality (Saint-Gilles, Réunion; Echenique-Subiabre et al. 2015; see Discussion), we selected the recently collected strain FK13-1 (N. Engene, 10 June 2013, Looe Key, Florida Keys) as epitype for future reference, designated under the provisions of Art. 9.8 and in accordance with Art. 9.20 and 9.21 of the International Code of Nomenclature for algae, fungi, and plants (McNeill et al. 2012).
Microscopy
Light microscopy was performed using an Olympus IX51 Leica epifluorescent microscope (Olympus, Tokyo, Japan) equipped with a Nikon Coolpix camera (Olympus, Tokyo, Japan). Morphological measurements were determined as: mean ± standard deviation (SD). Filament and cell size averages (adjacent cells of ten filaments) were calculated based on ten measurements (Table S2 in the Supporting Information). Samples for SEM were placed on glass slides that had been coated with a drop of Tissue TAC slide adhesive (DADE®) to facilitate adhesion. Samples were then fixed in 2.5% glutaraldehyde in 1X phosphate-buffered saline for 30 min and a secondary fix of 2% (water) osmium tetroxide for 30 min. Dehydration was achieved with a graded (20%, 50%, 70%, 90%, 100%, 100%) ethanol series. Samples were then critical point dried and sputter coated (200 A) with gold palladium. A Hitachi S-4800 SEM (Chiyoda, Tokyo, Japan) was used to image the samples. Images of Caldora penicillata were taken from strain FK12-26, which was collected in the same exact coordinates (N. Engene, 13 August 2012, Looe Key, Florida Keys) as the designated epitype FK13-1.
DNA extraction, multiple displacement amplification, and gene sequencing
For genetic purposes, live, pigmented filaments were scrupulously selected. Genomic DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA), following manufacturer’s specifications. A modified DNA extraction protocol by Sauvage et al. (2013) was also used to attempt obtaining DNA from the herbarium specimens. DNA concentration and purity was measured on a ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA).
Whole-genome amplification was performed on bundles of filaments of the epitype strain FK13-1 that were isolated under a dissecting microscope followed by multiple displacement amplification (MDA). Filaments were washed twice in 2 μL filter-sterilized seawater and twice in 2 μL ddH2O before transfer into 0.2 mL PCR-tubes. DNA was amplified from the single-filaments by MDA using the REPLI-g® UltraFast Mini Kit (Qiagen, Valencia, CA, USA), following the manufacturer’s specifications. All MDA reactions were performed in 20 μL reaction volume for 1.5 h at 30°C.
The 16S rRNA gene and the 16S-23S ITS regions were PCR-amplified using the 106F primer (Nübel et al. 1997) and the 340 primer (Iteman et al. 2000), respectively. Various primer sets were used in attempts to PCR-amplify nitrogenase reductase (nifH) genes, including nifHf/nifHr(Gugger et al. 2005), CDHPnif53F/CDHPnif723R and nifH1/nifH2 (Steward et al. 2004), and MC-nifHDK primers (Bolhuis et al. 2010). PCR protocols and products purification follow Engene et al. (2013). Subcloning was performed using the pGEM®-T Easy Vector system (Promega), following the manufacturer’s specifications. Plasmid DNA was isolated using the QIAprep®Spin Miniprep Kit (Qiagen) and sequenced bidirectionally with M13 vector primers as well as the internal primers 359F, 785R, and 1509R (Nübe et al. 1997). Gene sequence anomalies, including chimeric sequences, were predicted using the Pintail software with the cut-off size set at >600 bp (Ashelford et al. 2005) and were manually confirmed by comparison of Neighbor Joining (NJ) phylogenetic trees for different regions (>300 bp) of the sequences. Genetic sequencing was performed at the Laboratories of Analytical Biology, NMNH. All gene sequences for the Caldora penicillata specimens are available in GenBank under the accession numbers (acc. nr.): KF746590-KF746608 (16S rDNA) and KF768777-KF768793 (16S-23S ITS regions).
Phylogenetic inference
The 16S rRNA gene (1,306 bp) and the 16S-23S ITS regions (480 bp) from a total of 16 specimens of Caldora penicillata were used for phylogenetic inference. Gene sequences from other cyanobacterial taxa were obtained from the National Center for Biotechnology Information (NCBI) web pages. DNA sequences of reference strains selected from Bergey’s Manual (Castenholz 2001) and of species identified as type species from CyanoDB (Komárek and Hauer 2011) and AlgaeBase (Guiry & Guiry 2015) were included in the analyses. The reference unicellular cyanobacterium Gloeobacter violaceus PCC 7421 (GenBank acc. nr. NC005125) was included as an evolutionarily distant outgroup. All gene sequences were aligned using MUSCLE (Edgar 2004). The 16S rRNA gene sequence alignment was visually compared and refined using the SSU secondary structures model of Escherichia coli J01695 (Cannone et al. 2002). Mutation types and domains of the 16S rRNA genes were predicted based on superimposing of the secondary structures on the E. coli strain J01695 SSU model. All multiple sequence alignments are available at TreeBASE (http://www.treebase.org) under the submission IDs: 14952 (16S rDNA) and 14953 (16S-23S ITS regions). The 16S-23S ITS regions were analyzed for transfer RNA and transfer-messenger RNA genes using ARAGORN v1.2.36 (Laslett et al. 2004). Pairwise sequence divergences were calculated in MEGA 5.1 without model selection (Tamura et al. 2011). Appropriate nucleotide substitution models were compared and selected using uncorrected/corrected Akaike Information Criterion (AIC), Bayesian Information Criterion (BIC), and the Decision-theoretic in jModeltest 0.1.1 (Posada 2008). Maximum likelihood (ML) inference was performed using PhyML (Guindon and Gascuel 2003) with the GTR+I+G model of evolution, assuming heterogeneous substitution rates and gamma substitution of variable sites (proportion of invariable sites (pINV) = 0.417, shape parameter (α) = 0.382, number of rate categories = 4). Bootstrap resampling was performed on 1,000 replicates. Bayesian inference (BI) was conducted using MrBayes 3.1 under the GTR+I+G model (Ronquist and Huelsenbeck 2003). Four Metropolis-coupled Markov chain Monte Carlo (one cold and three heated) were run for 1,000,000 generations and the first 100,000 generations (10%) were discarded as burn-in and the following data sets were sampled every 100 generations. Secondary metabolite screening. Algal biomass of each specimen was lyophilized and extracted twice with ethyl acetate: methanol (1:1). Extracts were analyzed for chemical compositions by low-resolution liquid chromatography electrospray ionization mass spectrometry (LC-ESI-MS) system with a linear trap quadrupole (LTQ) Advantage Max spectrometer (Thermo Finnigan, Waltham, MA, USA). Each sample (10 μL) was injected and separated on a reversed-phase HPLC column as outlined in Engene et al. (2013). MS and tandem MS (MS/MS) spectra and retention time of each peak were recorded using the positive and negative ion detection modes. Identification of secondary metabolites required support of predicted isotope patterns, corresponding MS/MS fragmentations, and conserved retention times that were compared with previously characterized secondary metabolites (Engene et al. 2013). Abundances of molecules were roughly estimated based on their ion abundance.
RESULTS
Variability in morphology
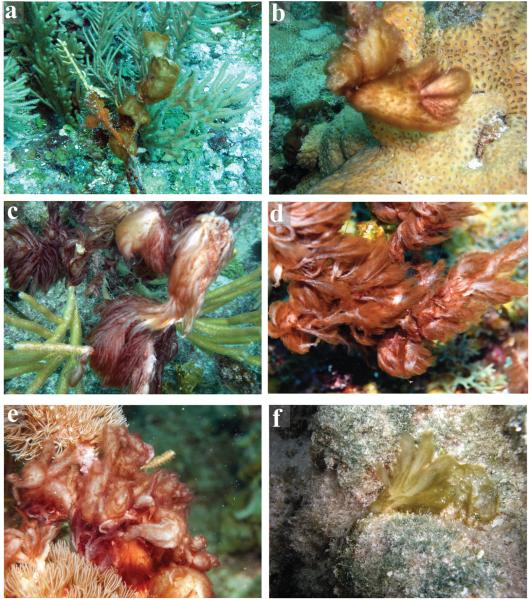
A total of 17 cyanobacterial specimens corresponding with the taxonomic definitions of Lyngbya penicilliformis and Symploca spp. were collected and analyzed from various geographically dispersed tropical and subtropical marine locations in Florida and the Caribbean Sea (Table S1). Collectively, the overall morphologies of Caldora penicillata specimens were soft and mucilaginous in appearance, erect and amorphic (Fig. 1). However, morphology varied from various puffball-shaped clumps (Fig. 1, a and b) to feathery shapes (Fig. 1, c-f). Size of Caldora penicillata specimens also varied from several centimeter sized clumps to extensive mats with up to 20 cm long finger-like projections. A shared feature among all specimens was a soft, mucilaginous internal portion, which was always colorless or whitish in color. The external portions varied widely in colors and ranged from various shades of orange (Fig. 1, a and b), red (Fig. 1, c-e), and green (Fig. 1f). These external colorations often reflected in the overall coloration of the specimens.
Fig. 1.
Environmental pictures illustrating morphological variability in shape and coloration between different Caldora penicillata specimens. Note external parts of specimens vary in color between reddish, greenish, to orange, while internal bases are always whitish or colorless. (a) Amorphic puffball-shaped specimen (BCBC11-25) growing on gorgonians of South Water Cay, Belize. (b) Small puffball-shaped tufts of C. penicillata attached to corals. (c) C. penicillata with feathery morphology and red coloration growing on shallow coral reef at Looe Key, FL (courtesy of B. Lapointe). (d) C. penicillata on coral reef at Carrie Bow Cay, Belize (courtesy of A. Wood). (e) C. penicillata on gorgonians of Miskito keys, Honduras (courtesy of Z. Foltz). (f) Greenish C. penicillata specimen (NAB11-29) on hard-bottom on a shallow-water reef of Lac Bay, Bonaire.
Specimens with dissimilarity in shape and coloration were collected from the same sites and compared to verify that the different morphotypes represented the same species. Specifically, strains NAB11-28 and NAB11-29 from Bonaire (Lac Bay) displayed puffball and feathery growth morphologies, respectively. Moreover, puffball-shaped specimens of the strains of NAC11-67 and NAC11-68 from Curaçao (Porto Marie) were greenish and red-orange, respectively. In these cases, the specimens were identical in gene sequences (see section below) and in the production of secondary metabolites (Table 1), highlighting the wide morphological variation in appearance in the field.
Table 1.
Production of bioactive secondary metabolites in Caldora penicillata specimens.
| Strain | Geographic location | Dolastatin 10 | Symplostatin 1 | Largazole |
|---|---|---|---|---|
| BCBC11-25 | Raph’s wall, Belize | + | ++ | − |
| BCBC11-38 | South Water Cay, Belize | ++ | +++ | + |
| BCBC12-2 | Carrie Bow Cay, Belize | +++ | + | ++ |
| NAB11-8 | Lac Bay, Bonaire | +++ | − | + |
| NAB11-21 | Playa Franz, Bonaire | ++ | +++ | − |
| NAB11-28 | Lac Bay, Bonaire | +++ | + | ++ |
| NAB11-29 | Lac Bay, Bonaire | +++ | + | ++ |
| NAB11-32 | Cali reef, Bonaire | + | − | − |
| NAC11-67 | Porto Marie, Curaçao | ++ | + | − |
| NAC11-68 | Porto Marie, Curaçao | ++ | + | − |
| FK12-2 | Wonderland, Florida Keys | ++ | + | + |
| FK12-18 | Big Pine Ledge, Florida Keys | +++ | + | ++ |
| FK12-20 | Wonderland, Florida Keys | ++ | ++ | + |
| FK12-26 | Looe Key, Florida Keys | ++ | ++ | + |
| FK13-1 | Looe Key, Florida Keys | ++ | ++ | + |
| HMC13-6 | Becerros, Honduras | ++ | − | − |
| HMC13-9 | Caratasca, Honduras | ++ | − | + |
(+++) main secondary metabolite, (++) major secondary metabolite (+) minor secondary metabolite or trace compound estimated based on liquid chromatographic electrospray ionization mass spectrometer (LC-ESI-MS) abundance, (−) not detected.
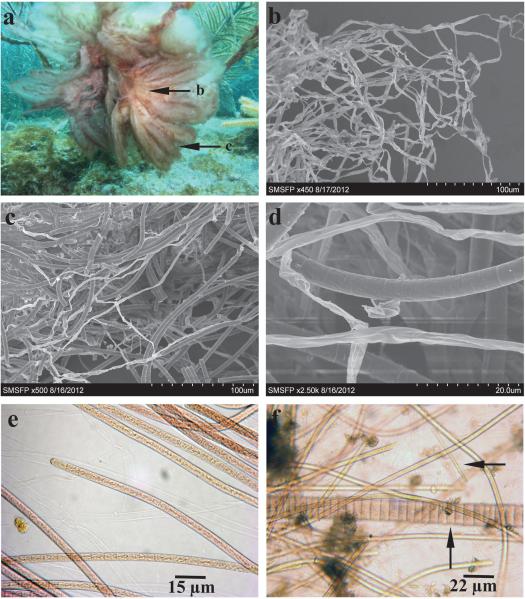
Sections of both the internal and external parts of strain FK12-26 were analyzed by both scanning electron microscopy (SEM) and light microscopy (Fig. 2, a-e). The internal base was shown to be composed primarily of residues of empty polysaccharide sheaths (Fig. 2b), and the external section contained mainly live and pigmented filaments (Fig. 2c). Filaments were thin (4-6 μm) and often entangled in bundles. Cells were barrel-shaped or occasionally cylindrical with no cross-wall constrictions and enclosed by thin, barely visible polysaccharide sheaths (Fig. 2, b-d). Notably, both the empty polysaccharide sheaths and the sheaths of live filaments were almost completely clean from associated biofilm-forming microorganisms (Fig. 2, b-d).
Fig. 2.
Morphological characterization of external and internal sections of Caldora penicillata strain FK12-26. (a) Underwater picture of C. penicilla attached to gorgonians at depth of 7-8 m at Looe Key, FL (courtesy of B. Lapointe). Arrows highlight sections used for scanning electron microscopy (SEM) imaging in b-d. (b) Whitish, mucilaginous interior bases composed of empty polysaccharide sheath material. (c) Exterior section of cyanobacterial thallus, predominantly containing live, pigmented filaments. (d) Higher resolution SEM image of C. penicillata filaments. (e) Light microscopic image of C. penicillata filaments. (f) Consortium of C. penicillata filaments mixed with other filamentous cyanobacteria. Scale bars: (b) 100 μm, (c) 100 μm, (d) 20 μm, (e) 15 μm, and (f) 22 μm.
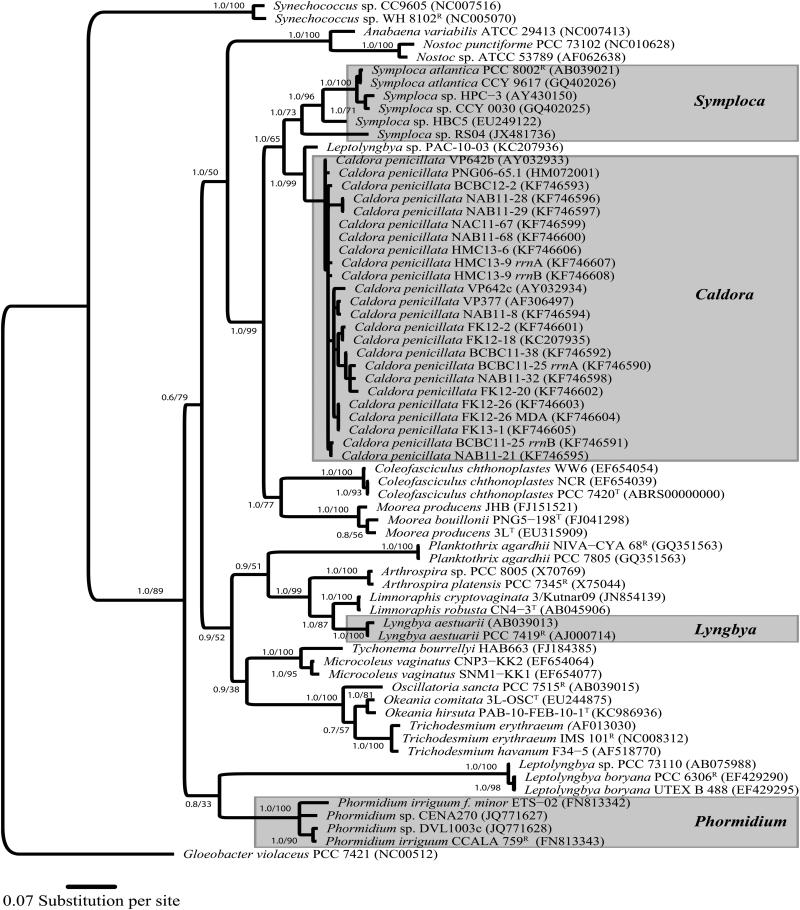
Evolutionary divergence
The SSU (16S) rRNA gene was sequenced from all recently collected specimens of Caldora penicillata and used for phylogenetic inferences (Fig. 3). Phylogenetic analysis revealed that C. penicillata sequences formed a monophyletic clade that was most closely related to those of the genera Symploca (p-distance = 4.5%), Coleofasciculus Siegesmund, J.R. Johansen & Friedl (2008) (p-distance = 6.5%), and Moorea Engene, Rottacker, Kastovsky, Byrum, Choi, Ellisman, Komárek & Gerwick (2012; p-distance = 6.6%). Moreover, C. penicillata sequences formed a tight clade with a mean intra-generic sequence divergence of only 0.9% (Fig. 3). The taxon that was most closely related to C. penicillata was a cyanobacterium originally identified as Leptolyngbya sp. (p-distance = 2.1%; strain PAC-10-03; GenBank acc. nr. KC207936) collected from the Coiba National Park of the Pacific coast of Panama (Medina et al. 2008, Engene et al. 2013).
Fig. 3.
Evolutionary tree based on SSU (16S) rRNA gene sequences with Caldora penicillata as well as Symploca, Lyngbya, and Phormidium clades highlighted with gray boxes. Phylogenetic inferences were performed using Bayesian Inference (BI) and Maximum Likelihood (ML) analyses. Support values are indicated at the nodes as posterior probability and bootstrap support. Reference strains are designated as (R) and type strains as (T). Unicellular Gloeobacter violaceus PCC 7421R strain (GenBank acc. nr. NC005125) was outgroup. Specimens indicated as species or strain; GenBank accession numbers in brackets. Scale bar depicts 0.04 expected nucleotide substitutions per site using GTR+I+G substitution model.
Interestingly, sequencing of multiple clones from libraries generated from the two strains BCBC11-25 and HMC13-9 showed that these two specimens contained two slightly genetically different copies of their 16S rRNA genes (Fig. 3). The highest degree of intra-genomic gene sequence divergence was observed in BCBC11-25, where the two gene copies (i.e. BCBC11-25 rrnA and BCBC11-25 rrnB in Figs. 3 and 4) varied in 16 nucleotide positions of 1319 nucleotides of the sequenced genes (1.2%). Secondary structure modeling revealed that the 16 nucleotide substitutions were all either: (i) located in loop-regions (7 bp), (ii) were C↔T substitutions located in stem-regions resulting in U-G bonds (3 bp), (iii) or resulted in compensatory substitutions in stem-regions (6 bp). Thus, these substitutions of the 16S rRNA gene were all likely true mutations, rather than PCR or sequencing artifacts, since the overall RNA structure was conserved in both gene copies.
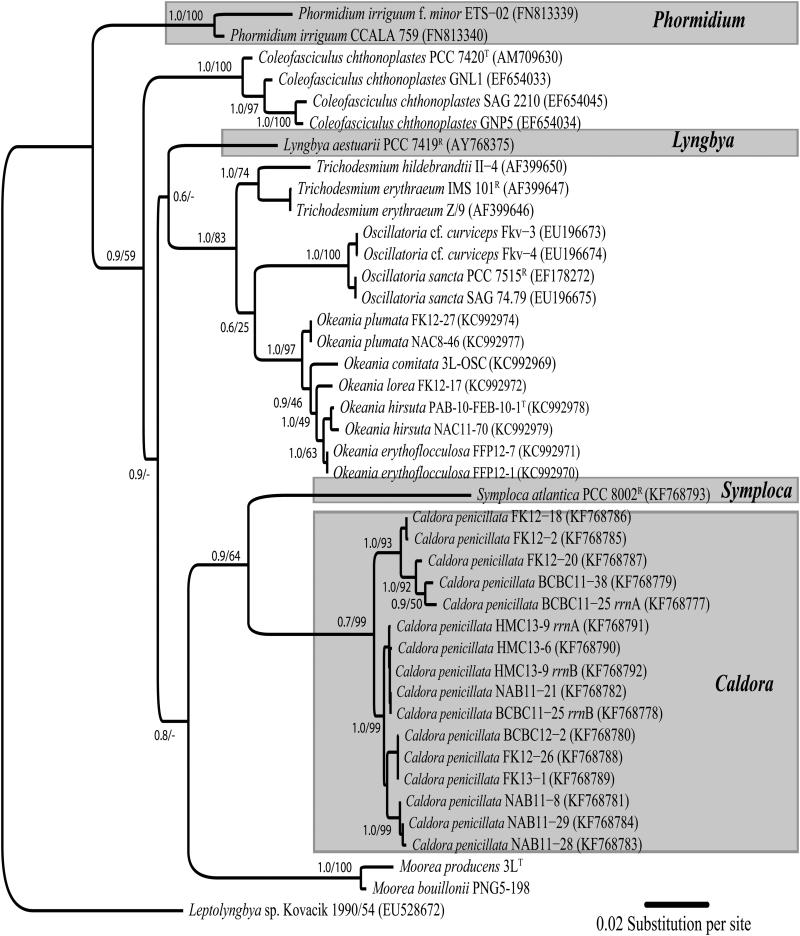
Fig. 4.
Phylogenetic inference of Caldora penicillata specimens based on 16S-23S internal transcribed spacer (ITS) regions highlighted in gray box, as well as Symploca, Lyngbya, and Phormidium clades. Support values indicated as posterior probability (BI) and bootstrap support (ML). Scale bar indicates 0.02 expected nucleotide substitutions per site using GTR+G substitution model.
Despite the general absence of associated heterotrophic bacteria as shown by SEM (Fig. 2d), Caldora penicillata specimens were shown microscopically and genetically to often form assemblages with other filamentous cyanobacteria (Fig. 2f). These associated filamentous cyanobacteria varied greatly in morphologies and likely belonged to a variety of taxonomic groups. Two of the predominant cyanobacterial taxa associated with C. penicillata were cf. Spirulina sp. (e.g., GenBank acc. nr. KJ439051) and Hormoscilla spongeliae (Gomont) Anagnostidis & Komárek (e.g., GenBank acc. nr. KJ439052). Moreover, these cyanobacterial consortia varied greatly in complexity and overall proportions among the different cyanobacterial components. Although cyanobacterial complexity was only determined from representative microscopic sections, all specimens analyzed appeared to have C. penicillata as the dominant component, and all the clone libraries generated from the C. penicillata specimens contained gene sequences belonging to the C. penicillata clade. Whole-genome amplification was performed by MDA to pinpoint the gene sequences that corresponded to the correct species filaments in the epitype strain FK13-1. A bundle of morphologically uniform C. penicillata filaments were micromanipulated from the strain FK13-1 and their genomic-DNA amplified by MDA. The 16S rRNA gene PCR-amplified from the MDA-amplified DNA (GenBank acc. nr. KF746604) was identical to the gene sequence obtained from the genomic DNA (GenBank acc. nr. KF746603), which verified the identity of the specimens of C. penicillata (i.e., FK13-1 MDA in Fig. 3).
The 16S-23S internal transcribed spacer (ITS) regions were also sequenced and independently used to infer the phylogenetic relationships among C. penicillata specimens (Fig. 4). The 16S-23S ITS regions varied in sequence length between 429 and 480 nucleotides among the specimens. However, the gene regions all included a conserved 79-nucleotide gene stretch encoding tRNA-Il. In addition to the 16S-23S ITS regions available for strains of Moorea and Coleofasciculus, the Symploca reference strain PCC 8002 was obtained from PCC and its ITS region was sequenced. Phylogenetic analyses of the 16S-23S ITS regions were congruent with the 16S rRNA gene in placing C. penicillata as an independent clade, moderately supported, and sister to Symploca (p-distance = 30.6%) and Moorea (p-distance = 32.0%), with Coleofasciculus somewhat distantly related (p-distance = 32.6%).
Several unsuccessful attempts were also made to PCR-amplify nitrogenase reductase (nifH) genes from C. penicillata specimens as well as the SSU (16S) rRNA gene from the Lyngbya penicilliformis herbarium specimens using various nonspecific primers.
Characteristic secondary metabolites
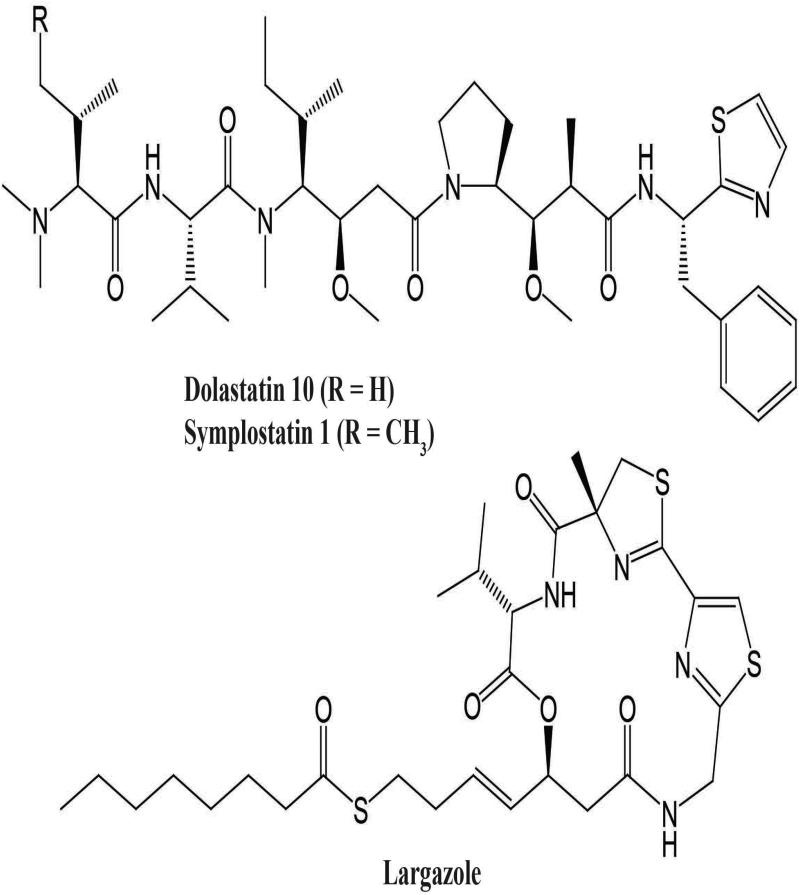
All C. penicillata collections were extracted and analyzed by ESI-LC-MS for their production of bioactive secondary metabolites, and abundance of each molecule was estimated based on its ion abundance (Table 1). Notably, all 16 specimens produced dolastatin 10, and this secondary metabolite often formed the major component of the crude extracts. The presence of dolastatin 10 was often complemented by the structurally related analog symplostatin 1, which typically was present in lower abundance. In addition, 10 of the specimens also produced the metabolite largazole (Table 1; Fig. 5).
Fig. 5.
Molecular structures of bioactive secondary metabolites dolastatin 10, symplostatin 1, and largazole; distribution of compounds among Caldora penicillata specimens shown in Table 1.
DISCUSSION
In this study, we applied a combined molecular, morphological and chemical approach to evaluate the taxonomy of the marine cyanobacteria specimens traditionally identified as Lyngbya penicilliformis and Symploca spp. (Luesch et al. 2001, Thacker and Paul 2004, Taori et al. 2008, 2009, Engene et al. 2013), which are herein assigned to the new genus Caldora, designating C. penicillata as generitype (accommodating Phormidium penicillatum; see below under Taxonomic conclusions).
In many coral reef habitats surveyed, Caldora penicillata was one of the most common and abundant cyanobacteria present. Caldora penicillata often formed large populations attached to different solid substrata, such as hard bottoms, dead corals, or gorgonians. Our field observations suggest that the most suitable habitats for C. penicillata are exposed reefs with high water circulation. The depth range varied greatly from 1-30 m, but C. penicillata appeared to be most prevalent at depths of 3-12 m. Although C. penicillata was found all year round in tropical marine locations, a greater abundance was typically observed during seasons with warmer water temperatures (> 25°C).
In this study, we sampled cyanobacterial specimens from the Florida Keys and various Caribbean Sea locations such as Belize, Netherlands Antilles (Bonaire and Curaçao), and Honduras. In addition, specimens corresponding morphologically to Caldora penicillata and with <1% 16S rRNA gene sequence divergence to the designated epitype strain FK13-1have also been reported from several Pacific locations, including Papua New Guinea (strain PNG06-65.1; GenBank acc. nr. HM072001; Choi et al. 2010), Guam (strain VP377; GenBank acc. nr. AF306497; Harrigan et al. 1998), and Palau (strain VP642b; GenBank acc. nr. AY032933; Luesch et al. 2001) (Fig. 3). Furthermore, recent studies have reported 99% and 100% similarity of our C. penicillata 16S rRNA gene sequences from Palau (VP642b) and Papua New Guinea (PNG06-65.1), respectively, with OTU03 from Réunion Island, corresponding to samples collected in the type locality of Phormidium penicillatum (Saint-Gilles) and nearby (ca. 25 km from Saint-Gilles; Etang Salé) (Echenique-Subiabre et al. 2014). Molecular results of our collections from various Caribbean and Pacific locations (at depths ranging between 2-20 m), together with the above-mentioned molecular records from Réunion indicate that this taxon is widely distributed in tropical and subtropical ocean basins globally. The tropical marine, coral reef habitats of the collected specimens also agree well with the environment of the type material of Phormidium penicillatum. Moreover, our collections correspond phenotypically with the original type material and taxonomic description of P. penicillatum by Gomont (1893), which shows the distinctive barrel-shaped filament cells (see Fig. S2) as in the specimens of Caldora penicillata used in this study. Thus, based on molecular, ecological and phenotypic characteristics we find it most likely that our collected specimens correspond with the originally description of Phormidium penicillatum.
Phormidium penicillatum has undergone several nomenclatural changes, including a taxonomic revision and transfer from the genus Phormidium to the genus Lyngbya (Hoffmann and Compère 1990). This transfer was followed by a name replacement to Lyngbya penicilliformis (Silva et al. 1996). However, our DNA-based phylogenetic analyses reveal that specimens traditionally identified in the natural products literature as L. penicilliformis and Symploca spp. are evolutionarily unrelated to the reference strains of both Lyngbya and Phormidium. Instead, these specimens form a separate lineage sister to Symploca and closely related to Moorea and somewhat to Coleofasciculus. The uncorrected 16S rRNA gene sequence divergences between these groups were approximately 5%. This value corresponds with the threshold recommended for distinguishing genera in cyanobacteria (Komárek 2006, Tindall et al. 2010). Thus, based on evolutionary and ecological divergence, we propose the new genus Caldora (see below under Taxonomic conclusions). It is also likely that additional species will be included in this genus. For example, the closely related specimen, a cyanobacterium originally identified as a Leptolyngbya sp., shared many morphological features with C. penicillata (Medina et al. 2008). This specimen also produced the bioactive secondary metabolite coibamide A that shares the N,N-dimethyl valine motif with dolastatin 10 (Medina et al. 2008). Further research is needed to elucidate whether the coibamide A-producing cyanobacterium should also be included in the genus Caldora.
All our collected specimens of Caldora penicillata, despite coming from geographically dispersed regions, produced the secondary metabolite dolastatin 10, a potent microtubule inhibitor effective against various types of tumors (Luesch et al. 2001). The majority of C. penicillata specimens also produced the analog symplostatin 1, and structural similarities between these molecules suggest that the same genetic pathway is involved in the biosynthesis of these two metabolites (Harrigan et al. 1998). The stable production of dolastatin 10 and/or symplostatin 1 in all analyzed specimens highlights the potential of these metabolites as reliable chemotaxonomic markers for rapid and robust identification of C. penicillata. Several specimens also produced the cytotoxic metabolite largazole (Taori et al. 2008). Interestingly, this compound was irregularly produced in specimens from the same geographic regions, suggesting that environmental factors might affect its production. At this point, it is unclear if largazole is produced by associated microbes or if the biosynthetic pathway encoding this secondary metabolite shows differential gene expression. It should also be noted that several secondary metabolites, including janthielamide A, kimbeamides A-C, and kimbelactone A (Nunnery et al. 2012), have also been reported from geographically localized collections of C. penicillata as verified by gene sequence data. However, none of these metabolites were detected in any of our C. penicillata specimens. Future genomic sequencing efforts will likely give better insights into the metabolic origins of these metabolites.
Taxonomic conclusions
The diagnosis of the new genus Caldora and a detailed description of Caldora penicillata follows:
Caldora Engene, Tronholm et V.J. Paul, gen. nov.
Diagnosis
Thalli habit from amorphic feathery or wispy to puffball clumps, sometimes mat-like. Coloration varies from red to orange and green. Each filament composed of entangled, thin, unbranched filaments; trichomes cylindrical, lacking heterocysts and other specialized cells, surrounded by barely visible polysaccharide sheaths. Cells barrel-shaped or sometimes cylindrical.
Etymology
From the Latin word caldor, meaning “heat, warmth” referring to the pantropical distribution of the genus.
Type species
Caldora penicillata Engene, Tronholm et V.J. Paul, comb. nov.
Basionym
Phormidium penicillatum Gomont, Bull. Soc. Bot. France, ser. 2, 15: LXXXVIII (1893).
Synonym
Lyngbya penicilliformis P. C. Silva in Silva, Basson & Moe. Univ. Calif. Publ. Bot. 79: 39 (1996).
Epitype
strain FK13-1, US 217964. Looe Key, Florida (24°35’400”N, 81°31’100”W), 10 June 2013, epizoic on corals and epilithic, 10 m depth, leg. N. Engene.
Caldora penicillata Engene, Tronholm et V.J. Paul, comb. nov.; Figures 1 and 2
Description
Thalli vary in shape from amorphic feathery, undulating or wispy to puffball-shaped clumps, often forming thick, undulating, digital projections; specimens up to 20 cm long; basal parts composed of empty polysaccharide sheaths, colorless or whitish, and mucilaginous; exterior composed of living, pigmented, filaments that vary in coloration such as red, orange, and green; filaments unbranched with individual sheaths, typically entangled into fasciculated bundles; filaments (4) 5-6 (7) μm in diameter; trichomes cylindrical, surrounded by thin (>0.5 μm), barely visible, polysaccharide sheaths; cells barrel-shaped or cylindrical (4) 5-6 (7) μm broad and (5) 6-8 (10) μm long, with no constrictions at cross-walls; terminal cells of filaments rounded; trichomes lack heterocysts or other specialized cells; producing secondary metabolites dolastatin 10 and/or symplostatin 1, and sometimes largazole; acc. nr. KF746590-KF746608 (16S rDNA), KF768777-KF768793 (16S-23S ITS regions).
Supplementary Material
ACKNOWLEDGMENTS
This research project was funded by the Smithsonian Marine Science Network (NE) and in part by the NIH (R01CA172310). Field collections were supported by the Carmabi Research Station (Curaçao), Carrie Bow Cay Field Station (Belize), Mote Marine Laboratory (Summerland Key, FL), Smithsonian Marine Station (Ft. Pierce, FL), and the Council on International Educational Exchange Research Station (Bonaire). We acknowledge the governments of Curaçao, Belize, Honduras, and Bonaire for permits to collect cyanobacteria. Research activities in the Florida Keys National Marine Sanctuary occurred under permit FKNMS-2013-023 (VP) and FKNMS-2011-007 (B. Lapointe). Field collections were partly supported by a NMNH Small Grant (VP) for Bonaire and Curaçao, Mote Protect our Reefs Grants for the Florida Keys (VP), and by the Summit Foundation through the Centre for Marine Studies for Honduras (S. Box). We thank R. Ritson-Williams, S. Reed, and B. Lapointe for help with sampling, as well as A. Wood, B. Lapointe and Z. Foltz for underwater photography. We thank K. A. Miller and the late P. C. Silva at the University and Jepson Herbaria of the University of California at Berkeley for lending isotypes, W. Gerwick and T. Byrum for sharing the Symploca reference strain, and K. McPhail for providing the Leptolyngbya sp. strain PAC-10-03. We are very grateful to L. Le Gall and L. Kervran at PC for the loan of specimens of Phormidium penicillatum. We also thank B. Brooks for help with deposition of specimens at US. We are also grateful to A. Economou-Amilli for help with botanical nomenclature. We thank M. Zubia, O. De Clerck and T. Sauvage for providing us with valuable information about samples from Réunion. We gratefully acknowledge H. Reichardt and the staff at the SMSFP for general lab assistance. All SEM imaging was performed at the USDA Horticultural research laboratory with the assistance of J. Piraino. All gene sequencing was performed at the Laboratories of Analytical Biology of the NMNH thanks to J. Hunt and L. Weight. We thank A. Wright and P. Winder at Harbor Branch Oceanographic Institute at Florida Atlantic University for usage of LCMS and NMR, and S. Gunasekera for analytical assistance. SMSFP contribution no. 994 and CCRE contribution no. 974.
Abbreviations
- ITS
internal transcribed spacer
- PKS
polyketide synthases
- NRPS
nonribosomal peptide synthetases
Contributor Information
Niclas Engene, Department of Biological Sciences, Florida International University, Miami, FL 33199, USA.
Ana Tronholm, Smithsonian Marine Station at Fort Pierce, Fort Pierce, FL 34949, USA.
Lilibeth A. Salvador-Reyes, Marine Science Institute, University of the Philippines, Diliman, Quezon City 1101, Philippines
Hendrik Luesch, Department of Medicinal Chemistry and Center for Natural Products, Drug Discovery and Development, University of Florida, Gainesville, FL 32610, USA.
Valerie J. Paul, Smithsonian Marine Station at Fort Pierce, Fort Pierce, FL 34949, USA
REFERENCES
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman A. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Env. Microbiol. 2005;71:7724–36. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis H, Severin I, Confurius-Guns V, Wollenzien UIA, Stal LJ. Horizontal transfer of the nitrogen fixation gene cluster in the cyanobacterium Microcoleus chthonoplastes. ISMEJ. 2010;4:121–30. doi: 10.1038/ismej.2009.99. [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanin S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Schang Z, Yu N, Gutell RR. The Comparative RNA Web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:1471–2105. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castenholz RW. Phylum BX. Cyanobacteria. Oxygenic photosynthetic bacteria. In: Boone DR, Castenholz RW, Garrity GM, editors. Bergey’s Manual of Systematic Bacteriology. Springer; New York, USA: 2001. pp. 473–87. [Google Scholar]
- Castenholz RW, Rippka R, Herdman M. Phylum BX. Cyanobacteria. Subsection III. Form-genus VII. Lyngbya Agardh 1824 (sensu Anagnostidis and Komárek 1988) In: Boone DR, Castenholz RW, Garrity GM, editors. Bergey’s Manual of Systematic Bacteriology. Springer; New York, USA: 2001a. pp. 547–8. [Google Scholar]
- Castenholz RW, Rippka R, Herdman M. Phylum BX. Cyanobacteria. Subsection III. Form-genus XV. Symploca Kützing 1843. In: Boone DR, Castenholz RW, Garrity GM, editors. Bergey’s Manual of Systematic Bacteriology. Springer; New York, USA: 2001b. pp. 559–60. [Google Scholar]
- Choi H, Pereira AR, Cao Z, Shuman CF, Engene N, Byrum T, Matainaho T, Murray TF, Mangoni A, Gerwick WH. The hoiamides, structurally-intriguing neurotoxic lipopeptides from Papua New Guinea marine cyanobacteria. J. Nat. Prod. 2010;73:1411–21. doi: 10.1021/np100468n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadheech PK, Casamatta DA, Casper P, Krienitz L. Phormidium etoshii sp. nov. (Oscillatoriales, Cyanobacteria) described from the Etosha Pan, Namibia, based on morphological, molecular and ecological features. Fottea. 2013;13:235–44. [Google Scholar]
- Echenique-Subiabre I, Villeneuve A, Golubic S, Turquet J, Humbert JF, Gugger M. Influence of local and global environmental parameters on the composition of cyanobacterial mats in a tropical lagoon. Microb. Ecol. 2015;69:234–44. doi: 10.1007/s00248-014-0496-0. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engene N, Gunasekera SP, Gerwick WH, Paul VJ. Phylogenetic inferences reveal large extent of novel biodiversity in chemically rich tropical marine cyanobacteria. Appl. Environ. Microbiol. 2013;79:1882–8. doi: 10.1128/AEM.03793-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engene N, Rottacker EC, Kaštovský J, Byrum T, Choi H, Ellisman MH, Komárek J, Gerwick WH. Moorea producensgen. nov., sp. nov. and Moorea bouillonii comb. nov., tropical marine cyanobacteria rich in bioactive secondary metabolites. Int. J. Syst. Evol. Microbiol. 2012;62:1171–8. doi: 10.1099/ijs.0.033761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner NL. New Pacific coast marine algae III. Univ. Calif. Publ. Bot. 1918;6:455–486. [Google Scholar]
- Gardner NL. The Myxophyceae of Porto Rico and the Virgin Islands. In: Anon, editor. Scientific Survey of Porto Rico and the Virgin Islands. Vol. 8. New York Academy of Sciences; New York: 1932. pp. 249–311. [Google Scholar]
- Geitler L. Cyanophyceae. In: Rabenhorst L, editor. Kryptogamen-Flora von Deutschland, Österreich und der Schweiz, vol. XIV. Akademische Verlag; Koeltz Scientific Books; Leipzig: Königstein: 1932. pp. 1027–68. 1985 reprint. [Google Scholar]
- Gomont M. Monographie des Oscillariées (Nostocacées homocystées) Ann. Sci. Nat. Bot. Ser. 1892;16:91–264. [Google Scholar]
- Gomont M. Sur quelques Phormidium à thalle rameux. B. Soc. Bot. Fr. 1893;40:86–90. [Google Scholar]
- Gugger M, Molica R, Le Berre B, Dufour P, Bernard C, Humbert J-F. Genetic diversity of Cylindrospermopsis strains (Cyanobacteria) isolated from four continents. Appl. Environ. Microbiol. 2005;71:1097–100. doi: 10.1128/AEM.71.2.1097-1100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. System. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guiry MD, Guiry GM. AlgaeBase. 2015 World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org; searched on 25 March 2015.
- Harrigan GG, Luesch H, Yoshida WY, Moore RE, Nagle DG, Paul VJ, Mooberry SM, Corbett TH, Valeriote FA. Symplostatin 1: a dolastatin 10 analogue from the marine cyanobacterium Symploca hydnoides. J. Nat. Prod. 1998;61:1075–7. doi: 10.1021/np980321c. [DOI] [PubMed] [Google Scholar]
- Hoffmann L, Komárek J, Kaštovský J. System of cyanoprokaryotes (cyanobacteria) state in 2004. Algol. Stud. 2005;117:95–115. [Google Scholar]
- Hoffmann L, Compère P. Nomenclatural notes in the Cyanophyceae. Taxon. 1990;39:308–10. [Google Scholar]
- Iteman I, Rippka R, Tandeau de Marsac N, Herdman M. Comparison of conserved structural and regulatory domains within divergent 16S rRNA–23S rRNA spacer sequences of cyanobacteria. Microbiology. 2000;146:1275–86. doi: 10.1099/00221287-146-6-1275. [DOI] [PubMed] [Google Scholar]
- Komárek J. Cyanobacterial taxonomy: current problems and prospects for the integration of traditional and molecular approaches. Algae. 2006;21:349–75. [Google Scholar]
- Komárek J. Recent changes (2008) in cyanobacteria taxonomy based on a combination of molecular background with phenotype and ecological consequences (genus and species concept) Hydrobiologia. 2010;639:245–59. [Google Scholar]
- Komárek J, Anagnostidis K. Süsswasserflora von Mitteleuropa. 19/2 Elsevier/Spektrum; Heidelberg, Germany: 2005. p. 759. [Google Scholar]
- Komárek J, Hauer T. CyanoDB.cz - On-line database of cyanobacterial genera. 2011 - Word-wide electronic publication, Univ. of South Bohemia & Inst. of Botany AS CR, http://www.cyanodb.cz.
- Komárek J, Kaštovský J, Mareš J, Johansen JR. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia. 2014;86:295–335. [Google Scholar]
- Laslett D, Canback B. ARAGORN, a program for the detection of transfer RNA and transfer-messenger RNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–6. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littler DS, Littler MM. Offshore Graphics; Washington, DC: 2000. Caribbean Reef Plants: An Identification Guide to the Reef Plants of the Caribbean, Bahamas, Florida and Gulf of Mexico; p. 542. [Google Scholar]
- Littler DS, Littler MM. Offshore Graphics; Washington, DC: 2003. South Pacific Reef Plants: A Divers’ Guide to the Plant Life of South Pacific Coral Reefs; p. 331. [Google Scholar]
- Luesch H, Moore RE, Paul VJ, Mooberry SL, Corbett TH. Isolation of Dolastatin 10 from the marine cyanobacterium Symploca species VP642 and total stereochemistry and biological evaluation of its analogue symplostatin 1. J. Nat. Prod. 2001;64:907–10. doi: 10.1021/np010049y. [DOI] [PubMed] [Google Scholar]
- McNeill J, Barrie FR, Buck WR, Demoulin V, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Marhold K, Prado J, Prud'homme van Reine WF, Smith GF, Wiersema JH, Turland N. Koeltz Scientific Books; Königstein: 2012. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code), adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011; p. 140. [Google Scholar]
- Medina RA, Goeger DE, Hills P, Mooberry SL, Huang N, Romero LI, Ortega-Barria E, Gerwick WH, McPhail KL. Coibamide A, a potent antiproliferative cyclic depsipeptide from the Panamanian marine cyanobacterium Leptolyngbya sp. J. Am. Chem. Soc. 2008;130:6324–5. doi: 10.1021/ja801383f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 1997;63:3327–32. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnery JK, Mevers E, Gerwick WH. Biologically active secondary metabolites from marine cyanobacteria. Curr. Opin. Biotech. 2010;21:1–7. doi: 10.1016/j.copbio.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnery JK, Engene N, Byrum T, Cao Z, Sairam J, Pereira A, Teatulohi M, Murray TF, Gerwick WH. Biosynthetically-intriguing chlorinated lipophilic metabolites from geographically distant tropical marine cyanobacteria. J. Nat. Prod. 2012;77:4198–208. doi: 10.1021/jo300160e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palinska KA, Thomasius CF, Marquardt J, Golubic S. Phylogenetic evaluation of cyanobacteria preserved as historic herbarium exsiccate. Int. J. Syst. Evol. Microbiol. 2006;56:2253–63. doi: 10.1099/ijs.0.64417-0. [DOI] [PubMed] [Google Scholar]
- Porta D, Hernandez-Marine M, Herdman M, Rippka R. Structural and ultrastructural characterization of Symploca atlantica Gomont, strain PCC 8002 (Oscillatoriales, Cyanophyta, Cyanobacteria) Algol. Stud. 2003;109:509–24. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–6. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxf) 2003;12:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sauvage T, Payri C, Draisma SGA, Prud'homme van Reine WF, Verbruggen H, Belton GS, Gurgel CFD, Gabriel D, Sherwood AR, Fredericq S. Molecular diversity of the Caulerpa racemosa–Caulerpa peltata complex (Caulerpaceae, Bryopsidales) in New Caledonia, with new Australasian records for C. racemosa var. cylindracea. Phycologia. 2013;52:6–13. [Google Scholar]
- Sciuto K, Andreoli C, Rascio N, La Rocca N, Moro I. Polyphasic approach and typification of selected Phormidium strains (Cyanobacteria) Cladistics. 2012;28:357–74. doi: 10.1111/j.1096-0031.2011.00386.x. [DOI] [PubMed] [Google Scholar]
- Siegesmund M, Johansen JR, Karsten U, Friedl T. Coleofasciculus gen. nov. (Cyanobacteria): morphological and molecular criteria for revision of the genus Microcoleus Gomont. J. Phycol. 2008;44:572–85. doi: 10.1111/j.1529-8817.2008.00604.x. [DOI] [PubMed] [Google Scholar]
- Silva PC, Basson PW, Moe RL. Catalogue of the benthic marine algae of the Indian Ocean. Univ. Calif. Publ. Bot. 1996;79:1–39. [Google Scholar]
- Steward GF, Jenkins BD, Ward BB, Zehr JP. Development and testing of a DNA macroarray to assess nitrogenase (nifH) gene diversity. Appl. Environ. Microbiol. 2004;70:1455–65. doi: 10.1128/AEM.70.3.1455-1465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunecky O, Komárek J, Johansen J, Lukešova A, Elster J. Molecular and morphological criteria for revision of the genus Microcoleus and its relation to Phormidium autumnale, Cyanobacteria. J. Phycol. 2013;49:1167–80. doi: 10.1111/jpy.12128. [DOI] [PubMed] [Google Scholar]
- Strunecky O, Komarek J, Šmarda J. Kamptonema (Microcoleaceae, Cyanobacteria), a new genus derived from the polyphyletic Phormidium on the basis of combined molecular and cytomorphological markers. Preslia. 2014;86:193–207. [Google Scholar]
- Sumina EL. Behavior of filamentous cyanobacteria in laboratory culture. Microbiology. 2006;75:459–64. [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taori K, Paul VJ, Luesch H. Structure and activity of largazole, a potent antiproliferative agent from the Floridian marine cyanobacterium Symploca sp. J. Am. Chem. Soc. 2008;130:1806–7. doi: 10.1021/ja7110064. [DOI] [PubMed] [Google Scholar]
- Taori K, Liu Y, Paul VJ, Luesch H. Combinatorial strategies by marine cyanobacteria: symplostatin 4, an antimitotic natural dolastatin 10/15 hybrid that synergizes with the coproduced HDAC inhibitor largazole. Chembiochem. 2009;6:1634–9. doi: 10.1002/cbic.200900192. [DOI] [PubMed] [Google Scholar]
- Thacker RW, Paul VJ. Morphological, chemical, and genetic diversity of tropical marine cyanobacteria Lyngbya spp. and Symploca spp. (Oscillatoriales) Appl. Env. Microbiol. 2004;70:3305–12. doi: 10.1128/AEM.70.6.3305-3312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiers B. Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden's Virtual Herbarium. [continuously updated] http://sweetgum.nybg.org/ih/
- Tidgewell K, Clark BR, Gerwick WH. The natural products chemistry of cyanobacteria. In: Mander L, Lui H-W, editors. Comprehensive Natural Products II Chemistry and Biology. Elsevier; Oxford, UK: 2010. pp. 141–88. [Google Scholar]
- Tindall BJ, Rosello-Mora R, Busse H-J, Ludwig W, Kampfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010;60:249–66. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.