Abstract
ATP analogs have been powerful tools in the study of kinase-catalyzed phosphorylation. However, the cell impermeability of ATP analogs has largely limited their use to in vitro lysate-based experiments. Here we report the first cell permeable ATP analog, ATP-polyamine-biotin (APB). APB promoted biotin labeling of kinase substrates in live cells. APB has future application in phosphoprotein purification and analysis. More generally, these studies provide a foundation for development of additional cell permeable ATP analogs for cell signaling research.
Keywords: Kinases, ATP, Cell-permeable, ATP analogs, Enzymes
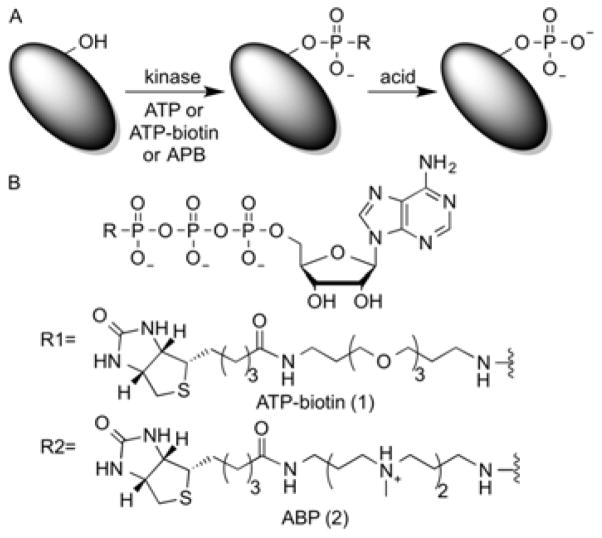
Living cells respond to extracellular conditions through signaling cascades, which are mediated by a variety of protein modification reactions. One ubiquitous protein modification that regulates many metabolic and cell signaling pathways is kinase-catalyzed protein phosphorylation (Figure 1A).[1] Alteration of pathways involving kinases and phosphorylation can lead to diseases, such as Parkinson’s,[2] cancer[3] and diabetes mellitus[4]. Therefore, studying protein kinases and their phosphorylated substrates is critical to understand cell signaling pathways in both diseased and healthy cells.
Figure 1.
(A) Kinase-catalyzed phosphorylation of a protein substrate (green) with (B) ATP (R = O−) or ATP analogs ATP-biotin (1), or APB (2).
With over 500 kinases and potentially thousands of phosphoproteins,[5] multiple complementary approaches are necessary to monitor the complex cellular phosphoproteome. One powerful approach exploits analogs of the universal co-substrate of kinases, adenosine-5′-triphosphate (ATP, Figure 1B). Multiple ATP analogs have been employed in kinase research, including base modified[6], sugar modified[7], and triphosphate modified analogs[8]. Our lab and others have utilized γ-phosphate modified ATP analogs to label kinase substrates for subsequent purification and analysis.[8e-j, 9] For example, ATP-biotin (1, Figure 1B) is promiscuously accepted as a cosubstrate by protein kinases to phosphorylbiotinylate substrates.[9b, 9d, 10] After kinase-catalyzed biotinylation with ATP-biotin, the biotin group facilitates analysis of phosphoproteins using various commercial streptavidin-conjugated reagents.[9b, 11] Unfortunately, due to the impermeability of ATP analogs,[12] ATP-biotin has been used in vitro only.[9d] The ability to utilize ATP-biotin in living cells would promote the study of protein kinases in more physiologically relevant conditions. Here, we describe the first cell permeable ATP-biotin analog for live cell kinase-catalyzed biotinylation.
Previous reports documented neutralizing the negative charge of phosphate groups to promote the cell permeability of compounds such as bisphosphonates[13] and phosphoinisitols[12, 14]. Building on these precedents, we replaced the PEG linker of ATP-biotin with a polyamine linker to create ATP-polyamine-biotin 2 (APB, Figure 1). Polyamines are known cell delivery vehicles for anionic nucleic acids.[15] In the case of APB, the polyamine linker will be positively charged under physiological conditions to partially neutralize the triphosphate charge and promote cell permeability. We chose spermine as the linker because its size mimics the original poly ethylene glycol (PEG) linker in ATP-biotin (Figure 1). In addition, we used methylated spermine to avoid possible side reactions of the nucleophilic secondary amines.
To computationally analyze the kinase compatibility of APB, docking studies with the PKA kinase crystal structure[16] were performed using the Autodock program.[17] Nearly identical PKA binding was observed with of APB (Figure 2A), ATP-biotin (Figure S1A), and ATP (Figure S1B). The biotin group protrudes from the active site, while the triphosphate is positioned in a close proximity to the co-crystallized peptide. The α-phosphate of APB is 3.8 Å from the catalytic amino acid K72, as compared to 3.7 and 3.2 Å with ATP or ATP-biotin, respectively (Figure S1C and D), suggesting that the three ATP molecules bind similarly in the active site. In contrast, the γ-phosphate of APB is 3.9 Å from K168 (Figure 2B), as compared to 2.4 Å with both ATP and ATP-biotin (Figure S1C and D). The docking studies suggest that APB is a potential kinase cosubstrate due to the similar active site binding. However, the long distance between the γ-phosphate of APB and K168 suggests that APB may be a less efficient cosubstrate compared to ATP or ATP-biotin.
Figure 2.
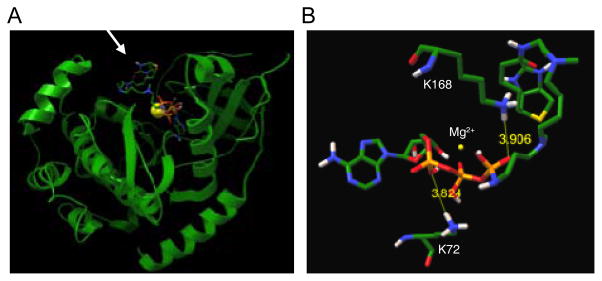
A) Docking of APB into the crystal structure of the catalytic active site of PKA kinase co-crystallized with peptide substrate (pdb: 4DH1)[16] using Autodock 4.2.[17] The arrow points to the solvent exposed biotin group. B) Enlarged view of the interaction of APB with the catalytic Mg2+ metal (yellow orb) and amino acids K72 and K168. The γ-phosphate of APB is positioned in a close proximity to K168 while the α-phosphate lies near K72. The APB atoms are color-coded (C = green; H = grey; N = blue; O = red; P = orange). For clarity, the atomic radius of Mg2+ was reduced to 0.5 Å.
To experimentally test APB as a kinase cosubstrate, it was first synthesized from commercially available spermine (Scheme 1). Spermine (3) was protected at the primary amines followed by reductive amination to give Boc-protected methylated spermine (4).[18] After deprotection,[18] the NHS-ester of biotin (Figure S2) was synthesized as reported[19] and coupled to give polyamine biotin (5).[20] Finally, polyamine-biotin (5) was coupled with ATP to obtain APB (2).[21]
Scheme 1.
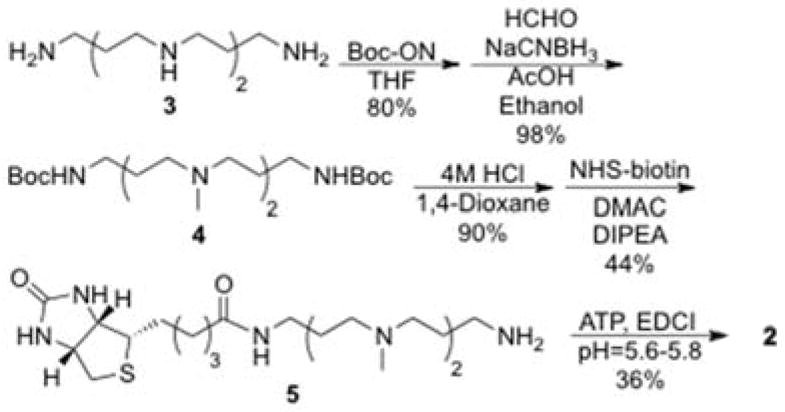
Synthesis of ATP-polyamine-biotin 2 (APB)
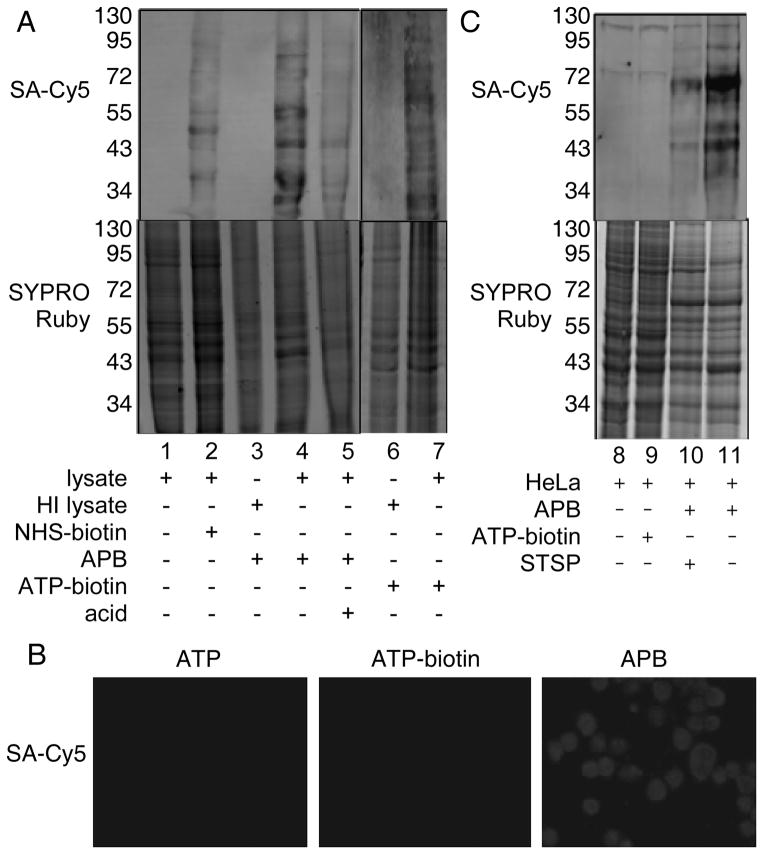
To test compatibility with kinase-catalyzed biotinylation, APB was incubated with PKA kinase and full-length protein substrate, myelin basic protein (MBP). Biotinylation was visualized after SDS-PAGE gel separation, transfer to PVDF membrane, and staining with a streptavidin-Cy5 conjugate (Figure 3A). Biotinylation was observed only in presence of kinase (Figure 3A, compare lanes 3 and 4). In addition, MBP biotinylation was lost in the absence of APB (Figure 3A, lane 2), in presence of the kinase inhibitor staursporine (Figure S11, compare lanes 2 and 3) or upon incubation with acid (Figure 3A, lane 5) due to cleavage of phosphoramidate bond (Figure 1A). To further confirm kinase-catalyzed biotinylation by APB, a mass spectrometric (MS) study was performed. In this case, the PKA peptide substrate kemptide was incubated with APB and PKA before MALDI-TOF MS analysis. Biotinylated kemptide product was observed only in the presence of APB cosubstrate (Figure S12, m/z 1332.490 (M+H)+). The combined gel and MS analyses confirm that APB is a kinase cosubstrate.
Figure 3.
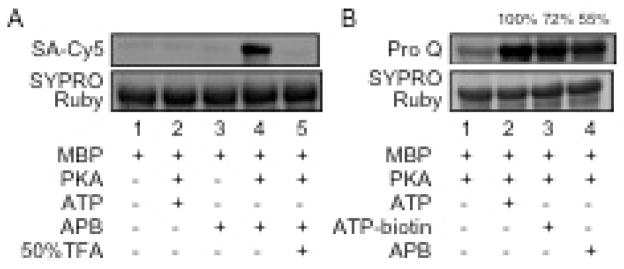
Kinase-catalyzed biotinylation with APB. A) Myelin basic protein (MBP) was incubated with or without PKA kinase in the presence of either ATP or APB. The labeling mixtures were separated by SDS-PAGE and visualized with SYPRO® Ruby total proteins stain (bottom), or streptavidin-Cy-5 (top). TFA (50% final concentration) was added after biotinylation labeling (lane 5). Full gel images are in Figure S10. B) Quantitative analysis of MBP phosphorylation was performed in the presence of PKA and ATP (lane 2), ATP-biotin (lane 3), or APB (lane 4). TFA was used to cleave the biotin group and produce the same phosphoprotein product with all analogs. The reaction mixtures were separated by SDS-PAGE and visualized with SYPRO® Ruby stain (bottom) or ProQ diamond phosphoprotein stain (top). The percentage phosphorylation was calculated by comparison with ATP (set as 100%). The gels are representative of at least three independent trials.
To investigate the efficiency of biotinylation using APB, both quantitative percentage conversion and kinetic studies were performed. For quantitative conversion studies, APB, ATP-biotin, and ATP were separately incubated with MBP and PKA, followed by cleavage of the phosphoramidate bond with acid to produce phosphoprotein products with all ATP analogs (Figure 1A), which allowed quantitative comparison. The reaction mixtures were then separated by SDS-PAGE with phosphoproteins visualized by ProQ diamond stain (Figure 3B), as previously reported.[9d] Quantification showed 55 ± 6 % conversion with APB compared to ATP (Figure 3B, lane 4), while ATP-biotin showed 72 ± 7 % conversion compared to ATP (Figure 3B, lane 3]. Biotinylation was less efficient with APB compared to ATP-biotin, as predicted by the docking studies. However, the observed quantitative analysis confirmed that APB is a kinase cosubstrate. Next, kinetic studies were performed by incubating APB or ATP (0.5–100 μM) with PKA and kemptide peptide substrate. APB showed a reduced kcat/KM (0.25 s−1μM−1) compared to ATP (0.52 s−1μM−1) (Figure S13). However, the kinetics are similar to those observed with other ATP analogs used for kinase studies, including the γ-phosphate modified ATP analog ATP-dansyl,[22] or the base-modified analogs N6-benzyl ATP or N6-(2-phenethyl) ATP.[6b, 23] In total, both quantitative conversion and kinetics studies confirm that APB is an efficient kinase cosubstrate with conversions and kinetics similar to other known ATP analogs.
To analyze the compatibility of APB with cellular kinases, HeLa cell lysates were incubated with APB, followed by SDS-PAGE analysis. Biotinylation of proteins was detected in the APB reaction (Figure 4A, lane 4), showing the promiscuity of cellular kinases for APB. Similar levels of labeling were observed comparing APB and ATP-biotin (Figure 4A, lane 4 versus 7). As a control, heat denatured lysates generated low levels of biotinylation with both APB and ATP-biotin (Figure 4A, lanes 3 and 6), which confirmed the kinase-dependence of biotinylation. Acid treatment also reduced biotinylation (Figure 4A, lane 5), which indicated labeling via the phosphoramidate bond in APB (Figure 1A). These studies in lysates further establish the compatibility of APB with a range of cellular kinases and substrates, similar to ATP-biotin.[9b, 9d]
Figure 4.
A) Kinase-catalyzed biotinylation of HeLa cell lysates (lysate) or heat-inactivated Hela cell lysates (HI lysate) with APB and ATP-biotin. Acid (50% trifluoroacetic acid final concentration) was added after biotin labeling to cleave the biotin tag (lane 5). NHS-biotin (Figure S2) was used to assess nonspecific biotinylation. B) Fluorescence microscopy images of Hela cells after treatment with ATP, ATP-biotin, or APB, fixation, and visualization with SA-Cy5 for detection of biotin (white). Enlarged images and DAPI nuclear staining are shown in Figure S14. C) In cellulo kinase-catalyzed biotinylation with ATP-biotin or APB in HeLa cells. As a control, kinase inhibitor staurosporine (STSP) was pre-incubated with cells to prevent kinase catalysis (lane 3). Reaction mixtures (A and C) were separated by SDS-PAGE and visualized with streptavidin-Cy5 (SA-Cy5, top gel) or SYPRO® Ruby total protein stain (bottom gels). The gels are representative of at least three independent trials.
Having confirmed the kinase compatibility of APB in vitro, we next sought to test kinase-catalyzed biotinylation of live cells. As a first step, fluorescence microscopy was used to confirm the cell permeability of APB. Hela cells were incubated with APB, followed by washing, fixation, and visualization with streptavidin-Cy5 to observe biotin. Cells treated with APB showed fluorescence corresponding to the presence of biotin (Figure 4B). As controls, untreated cells (Figure S14) or cells incubated with ATP or ATP-biotin showed no biotin signal (Figure 4B), which indicated that the polyamine linker in APB was required to promote cell permeability. These microscopy studies confirm that APB is cell permeable and validate the use of a polycationic groups to enhance the permeability of ATP analogs.
With the cell permeability of APB confirmed, live cell biotinylation was performed. HeLa cells were incubated with APB, washed to remove excess analog, lysed, and then analyzed by SDS-PAGE. Cells incubated with APB showed protein biotinylation (Figure 4C, lane 11), which is consistent with cell permeability. Biotinylation was absent with ATP-biotin under the same conditions (Figure 4C, lane 9), further confirming that the polyamine linker is necessary to enhance cell permeability. Also, pre-treating cells with the kinase inhibitor staursporine reduced biotinylation (Figure 4C, lane 10), indicating that the labeling is kinase-dependent. As a final control, ATP-biotin was incubated with HeLa cells in the presence of lysates containing kinase activity and no biotin signal was observed (Figure S15A), which assures that biotinylation is independent of cell surface protein labeling. Treatment with APB was accompanied by a modest loss of total protein (Figure 4C, lane 10 and 11, bottom gel) compared with controls (Figure 4C, lane 8 and 9, bottom gel), which is similar to the levels of protein loss observed in previous cell permeability studies,[24] including experiments with the widely used cationic-based permeabilization reagents.[25] To assess APB cytotoxicity, a dose-dependent cell viability assay was performed. APB showed a cytotoxicity EC50 value of 19 ± 1 mM (Figure S16A). Importantly, 96 ± 1 % cell viability was observed with the 5 mM concentration of APB used in the cell labeling assay (Figure S16B). The cell-based studies show that APB is cell permeable and nontoxic at low mM concentrations, with cell penetration and labeling dependent on the polyamine linker.
To assess the quality of biotinylation in the various labeling reactions, lysates were incubated with the non-specific biotinylation reagent, NHS-biotin (Figure S2). Different biotinylated protein bands were observed with NHS-biotin treatment compared to kinase-catalyzed reactions with APB or ATP-biotin (Figure 4A and C, compare lane 2 to lanes 4, 7 and 11), suggesting kinase-selective biotinylation of both lysates and cells. Importantly, a comparison of APB labeling reactions in lysates and cells revealed different biotinylated protein products (Figure 4A and C, lanes 4 versus 11), which indicated that in cellulo labeling is distinct from labeling in lysates. We speculate that the difference in live cell versus lysates labeling may be due to compartmentalization inside the cell, which suggests that in cellulo labeling studies will better interrogate the phosphoproteome for cell signaling studies.
In conclusion, we report the first cell permeable ATP analog compatible with kinase-catalyzed biotinylation. APB acted as a cosubstrate with protein kinases in vitro and in cellulo. While the percentage conversion and kinetic efficiency was reduced compared to ATP or ATP-biotin in vitro, APB was able to label phosphoproteins in live cells. Importantly, different biotinylated proteins were observed with in cellulo compared to lysate studies, which argue that labeling in cellulo will better reflect the cellular phosphoproteome. These results lay the foundation for future work using APB and kinase-catalyzed biotinylation as tools to identify and isolate phosphoproteins from cells, which will enhance cell signaling research. More generally, these studies establish that cationic groups attached to ATP analogs promote cell permeability, which provides a general strategy for creation of other ATP analogs for live cell labeling studies.
Experimental Section
APB (2) was synthesized and characterized by 1H, 13C, and 31P NMR spectroscopy, UV spectroscopy, ESI mass spectroscopy, and MALDI spectrometry, as discussed in the supporting information (Figures S3–S8). For in cell kinase-catalyzed biotinylation, Hela cells were grown in 12 well plates for 2 days in growth media (F-12 containing 10% FBS, 9 units penicillin, and 9 units streptomycin). The growth media was removed, replaced with fresh growth media containing APB (5 mM) or ATP-biotin (5 mM), and incubated at 37°C for 1 hour in a CO2 incubator to allow kinase-catalyed labeling. As a control, staurosporine (1 μM) in fresh growth media was added to the cells for one hour before adding APB. After washing the cells, the reaction mixtures were separated by SDS-PAGE and visualized with SYPRO® Ruby total protein stain. Where indicated, the gel was stained with ProQ Diamond Phosphoprotein stain (Invitrogen), or the proteins were transferred onto a PVDF membrane (Immobilon-P, Milipore) and visualized with streptavidin-Cy5 reagent (Life Technologies) to detect biotinylated proteins. Detailed experimental procedures and data are supplied as supporting information.
Supplementary Material
Acknowledgments
We thank Wayne State University and NIH (GM079529) for funding, R. Donovan, Dr. M. Ammesou, members of M. Greenberg lab, and Dr. X.-D. Zhang for technical support and usage of the fluorescence microscope, and T. Anthony, D. M. Embogama, P. Dedigama-Arachichige for comments on the manuscript.
References
- 1.Johnson SA, Hunter T. Nat Meth. 2005;2:17–25. doi: 10.1038/nmeth731. [DOI] [PubMed] [Google Scholar]
- 2.a) Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, Tomiyama H, Nakashima K, Hasegawa K, Obata F, Yoshikawa T, Kawakami H, Sakoda S, Yamamoto M, Hattori N, Murata M, Nakamura Y, Toda T. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]; b) Dachsel JC, Farrerl MJ. Arch Neurol. 2010;67:542–547. doi: 10.1001/archneurol.2010.79. [DOI] [PubMed] [Google Scholar]
- 3.Cohen P. Nat Rev Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 4.Koya D, King GL. Diabetes. 1998;47:859–866. doi: 10.2337/diabetes.47.6.859. [DOI] [PubMed] [Google Scholar]
- 5.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 6.a) Shah K, Liu Y, Deirmengian C, Shokat KM. Proc Natl Acad Sci USA. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Liu Y, Shah K, Yang F, Witucki L, Shokata KM. Bioorganic & Medicinal Chemistry. 1998;6:1219–1226. doi: 10.1016/s0968-0896(98)00099-6. [DOI] [PubMed] [Google Scholar]; c) Couzens AL, Gill RM, Scheid MP. BMC Biotechnology. 2014;14:2. doi: 10.1186/1472-6750-14-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni Q, Shaffer J, Adams JA. Protein science : a publication of the Protein Society. 2000;9:1818–1827. doi: 10.1110/ps.9.9.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Hastie CJ, McLauchlan HJ, Cohen P. Nat Protocols. 2006;1:968–971. doi: 10.1038/nprot.2006.149. [DOI] [PubMed] [Google Scholar]; b) Eckstein F. Annu Rev Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]; c) Wang Z, Lee J, Cossins AR, Brust M. Anal Chem. 2005;77:5770–5774. doi: 10.1021/ac050679v. [DOI] [PubMed] [Google Scholar]; d) Wang Z, Levy R, Fernig DG, Brust M. J Am Chem Soc. 2006;128:2214–2215. doi: 10.1021/ja058135y. [DOI] [PubMed] [Google Scholar]; e) Song H, Kerman K, Kraatz HB. Chem Commun (Camb) 2008:502–504. doi: 10.1039/b714383d. [DOI] [PubMed] [Google Scholar]; f) Martić S, Gabriel M, Turowec JP, Litchfield DW, Kraatz H-B. J Am Chem Soc. 2012 doi: 10.1021/ja302586q. [DOI] [PubMed] [Google Scholar]; g) Wilke KE, Francis S, Carlson EE. J Am Chem Soc. 2012;134:9150–9153. doi: 10.1021/ja3041702. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Lee SE, Elphick LM, Anderson AA, Bonnac L, Child ES, Mann DJ, Gouverneur V. Bioorg Med Chem Lett. 2009;19:3804–3807. doi: 10.1016/j.bmcl.2009.04.028. [DOI] [PubMed] [Google Scholar]; i) Allen JJ, Lazerwith SE, Shokat KM. J Am Chem Soc. 2005;127:5288–5289. doi: 10.1021/ja050727t. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Allen JJ, Li M, Brinkworth CS, Paulson JL, Wang D, Hubner A, Chou WH, Davis RJ, Burlingame AL, Messing RO, Katayama CD, Hedrick SM, Shokat KM. Nat Methods. 2007;4:511–516. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Senevirathne C, Green KD, Pflum MKH. Current Protocols in Chemical Biology. Vol. 4. John Wiley & Sons, Inc; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Green KD, Pflum MH. J Am Chem Soc. 2007;129:10–11. doi: 10.1021/ja066828o. [DOI] [PubMed] [Google Scholar]; c) Suwal S, Senevirathne C, Garre S, Pflum MK. Bioconjug Chem. 2012;23:2386–2391. doi: 10.1021/bc300404s. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Senevirathne C, Pflum MK. ChemBioChem. 2013;14:381–387. doi: 10.1002/cbic.201200626. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Suwal S, Pflum MH. Angew Chem Int Ed Engl. 2010;49:1627–1630. doi: 10.1002/anie.200905244. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Garre S, Senevirathne C, Pflum MK. Bioorg Med Chem. 2014;22:1620–1625. doi: 10.1016/j.bmc.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao X, Schutz-Geschwender A, Hardwidge PR. Biotechnol Lett. 2009;31:113–117. doi: 10.1007/s10529-008-9824-0. [DOI] [PubMed] [Google Scholar]
- 11.Dunn JD, Reid GE, Bruening ML. Mass Spectrometry Reviews. 2010;29:29–54. doi: 10.1002/mas.20219. [DOI] [PubMed] [Google Scholar]
- 12.Laketa V, Zarbakhsh S, Morbier E, Subramanian D, Dinkel C, Brumbaugh J, Zimmermann P, Pepperkok R, Schultz C. Chemistry & Biology. 2009;16:1190–1196. doi: 10.1016/j.chembiol.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Webster MR, Zhao M, Rudek MA, Hann CL, Freel Meyers CL. Journal of medicinal chemistry. 2011;54:6647–6656. doi: 10.1021/jm200521a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mentel M, Laketa V, Subramanian D, Gillandt H, Schultz C. Angew Chem Int Ed Engl. 2011;50:3811–3814. doi: 10.1002/anie.201007796. [DOI] [PubMed] [Google Scholar]
- 15.a) Duan SY, Ge XM, Lu N, Wu F, Yuan W, Jin T. International journal of nanomedicine. 2012;7:3813–3822. doi: 10.2147/IJN.S33101. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shah S, Solanki A, Sasmal PK, Lee KB. J Am Chem Soc. 2013;135:15682–15685. doi: 10.1021/ja4071738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovalevsky AY, Johnson H, Hanson BL, Waltman MJ, Fisher SZ, Taylor S, Langan P. Acta crystallographica Section D, Biological crystallography. 2012;68:854–860. doi: 10.1107/S0907444912014886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García J, Pereira R, de Lera AR. Tet Lett. 2009;50:5028–5030. [Google Scholar]
- 19.Chaturvedi DN, Knittel JJ, Hruby VJ, Castrucci AMdL, Hadley ME. J Med Chem. 1984;27:1406–1410. doi: 10.1021/jm00377a005. [DOI] [PubMed] [Google Scholar]
- 20.Albarella JP, Minegar RL, Patterson WL, Dattaguptal N, Carlson E. Nucl Acids Res. 1989;17:4293–4308. doi: 10.1093/nar/17.11.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parang K, Kohn JA, Saldanha SA, Cole PA. FEBS Lett. 2002;520:156. doi: 10.1016/s0014-5793(02)02778-3. [DOI] [PubMed] [Google Scholar]
- 22.Green KD, Pflum MH. ChemBioChem. 2009;10:234–237. doi: 10.1002/cbic.200800393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulrich SM, Kenski DM, Shokat KM. Biochemistry. 2003;42:7915–7921. doi: 10.1021/bi030042a. [DOI] [PubMed] [Google Scholar]
- 24.a) Kelner KL, Morita K, Rossen JS, Pollard HB. Proc Natl Acad Sci U S A. 1986;83:2998–3002. doi: 10.1073/pnas.83.9.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sarafian T, Aunis D, Bader MF. The Journal of biological chemistry. 1987;262:16671–16676. [PubMed] [Google Scholar]
- 25.Hoyer J, Schatzschneider U, Schulz-Siegmund M, Neundorf I. Beilstein J Org Chem. 2012;8:1788–1797. doi: 10.3762/bjoc.8.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.