Abstract
Huntington’s disease (HD) is a fatal neurodegenerative disorder caused by an expanded polyglutamine repeat in huntingtin (Htt) protein. Current management strategies temporarily relieve disease symptoms, but fail to affect the underlying disease progression. We previously demonstrated that calorie restriction ameliorated HD pathogenesis and slowed disease progression in HD mice1. We now report that overexpression of SIRT1, a mediator of beneficial metabolic effects of calorie restriction, protects neurons against mutant Htt toxicity, whereas reduction of SIRT1 exacerbates mutant Htt toxicity. Overexpression of SIRT1 significantly improves motor function, reduces brain atrophy, and attenuates mutant Htt-mediated metabolic abnormalities in both fragment and full-length HD mouse models. Further mechanistic studies suggest that SIRT1 prevents mutant Htt-induced decline in BDNF levels and its receptor Trk-B signaling, and restores medium spiny neuronal DARPP32 levels in the striatum. SIRT1 deacetylase activity is required for SIRT1-mediated neuroprotection in HD models. Notably, we demonstrate that mutant Htt interacts with SIRT1 and inhibits SIRT1 deacetylase activity. Inhibition of SIRT1 deacetylase activity results in hyperacetylation of SIRT1 substrates such as FOXO3a thereby inhibiting its prosurvival function. Overexpression of SIRT1 counteracts mutant Htt-induced deacetylase deficit, enhances deacetylation of FOXO3a, and facilitates cell survival. These findings demonstrate a neuroprotective role of SIRT1 in mammalian HD models, indicate key mediators of this protection, and open new avenues for the development of neuroprotective strategies in HD.
Previous studies have suggested that alteration of cellular metabolism plays an important role in the pathogenesis of Huntington’s disease (HD)2-4, raising a possibility of developing a class of therapeutic interventions in HD that activate the body’s own metabolic defenses. SIRT1 is one of seven identified sirtuins, an evolutionarily conserved proteins with NAD+-dependent deacetylase activity that participate in numerous cell activities including cellular metabolism5. Accumulating evidence indicates that SIRT1 has neuroprotective roles in neurodegenerative disorders 6-13. Whether SIRT1 has protective role in mammalian HD remains undefined, however, as results from lower organisms such as Caenorhabditis elegans 13 and Drosophila melanogaster 14 are contradictory; for example, overexpression of sir2, an ortholog of human SIRT1, suppressed mutant huntingtin (Htt)-induced neurotoxicity13, whereas overexpression of sir2 in drosophila showed no significant protection15. Therefore, we aimed to determine whether SIRT1 plays a neuroprotective role in mammalian models of HD. If so, increase in SIRT1 activity as a strategy to treat HD might not only rescue mutant Htt-mediated metabolic abnormalities, but also prevent or slow down the underlying neurodegenerative process.
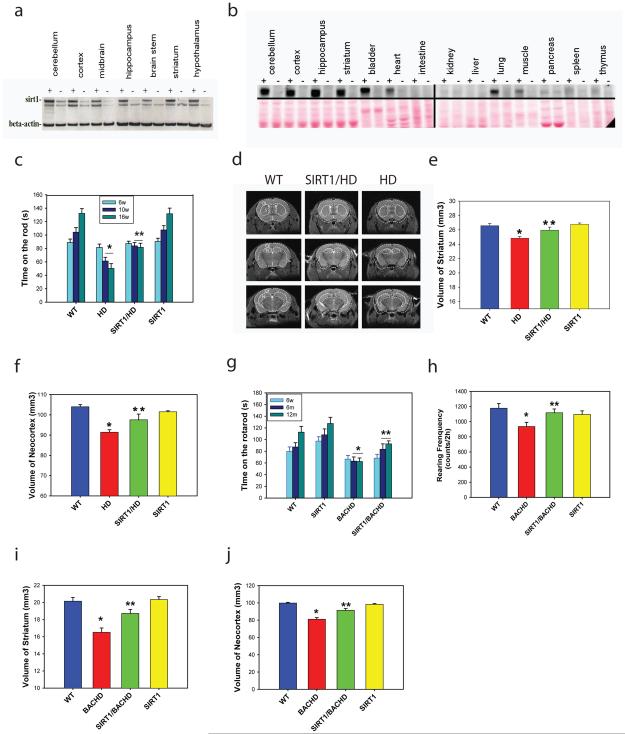
To determine the role of SIRT1 in mammalian HD models, we took advantage of a SIRT1 transgenic mouse model in which the SIRT1 (HA-tagged) is driven by a prion protein promoter (PrP)16. These mice, which are maintained on a C57BL/6 background, strongly overexpress SIRT1 in different brain regions including cortex, striatum, hippocampus as well as hypothalamus (Fig. 1a), although detectable levels of transgene are also observed in several peripheral tissues (Fig. 1b). As the first step, we crossed these SIRT1 transgenic mice with N171-82Q HD mice that express fragment mutant Htt driven by the PrP promoter17 and display progressive phenotypes resembling HD patients1. SIRT1 overexpression did not decrease mutant Htt levels (Supplementary Fig. 1a), but significantly delayed the onset and slowed the progression of motor deficits in HD mice (Fig. 1c). To determine whether improved motor function correlated with the progression of the neuropathological process, we performed in vivo structural magnetic resonance imaging (MRI) analysis, that has been shown to be an accurate measure of neuropathology in HD mice18 and is also translatable to human HD 19. Significant brain atrophy was detected in the striatum and neocortex of N171-82Q HD mice, resembling neuropathological changes in human HD patients. Importantly, overexpression of SIRT1 significantly attenuated the magnitude of brain atrophy in both striatum and neocortex (Fig. 1d-f). These results demonstrate that increased expression of SIRT1 attenuates neurodegeneration and improves motor function in HD-like mice.
Figure 1. SIRT1 improves the motor function and attenuates brain atrophy in N171-82Q HD mice and BACHD mice.
(a) Transgene SIRT1 expression was detected in different brain regions of SIRT1 mice. “+” represents transgenic mice, “−” represents wild type mice. (b) SIRT1 expression in brain and peripheral tissues in SIRT1 transgenic mice; SIRT1 was detected by anti-HA antibody (SIRT1 transgene is HA-tagged). Note the highest expression of transgene SIRT1 is detected in brain. (c) SIRT1 overexpression slowed progression of motor deficits in N171-82Q HD mice. Mice were tested on an accelerating rotarod apparatus at 6, 10, and 16 weeks of age. Mean ± S.E.M., n=15. *p<0.05 compared with the value of WT mice; **p<0.05 compared with the value of HD group by two way ANOVA. (d) Representative MRI images at three different coronal levels in mice of indicated genotypes. (e-f) Quantification of brain regional volumes measured by structural MRI in indicated groups in 16-week-old N171-82Q mice. Values are mean ± S.E.M., n=5. *p<0.05 compared with the value of WT mice; **p<0.05 compared with the value of HD mice by Student’s t-tests. (g) Effects of SIRT1 overexpression on rotarod performance in BACHD mice at indicated ages. Values are mean ± S.E.M., n=15. *p<0.05 compared with the value of WT mice; **p<0.05 compared with the value of BACHD group by two way ANOVA. (h) Effects of SIRT1 on rearing frequency in open field assay in 15-month-old BACHD mice. Mean ± S.E.M., n=15. *p<0.05 compared with the value of WT mice; **p<0.05 compared with the value of BACHD mice by Student’s t-tests. (i-j) Effects of SIRT1 overexpression on brain atrophy measured by MRI in 15-month-old BACHD mice. Mean ± S.E.M., n=5. *p<0.05 compared with the value of WT mice; **p<0.05 compared with the value of BACHD mice by Student’s t-tests.
Having shown that overexpression of SIRT1 protects mutant Htt-induced neurodegeneration and improves motor function in a fragment HD mouse model, we then aimed to confirm these neuroprotective effects of SIRT1 in a full-length HD mouse model. We took advantage of BACHD mice that are well-characterized and display motor deficits and HD-like brain pathology relatively earlier than other available full-length HD mouse models 20,21. By crossing SIRT1 transgenic mice with BACHD mice, we obtained offsprings with four genotypes with the same strain background. Overexpression of SIRT1 did not decrease mutant Htt expression in BACHD mice (Supplementary Fig. 1b), but significantly attenuated motor deficits, as indicated in both accelerating rotarod tests and open field assays. (Fig. 1g-h). Notably, in vivo structural MRI scans in 15-month-old mice showed significant brain atrophy in the striatum and neocortex that was partially ameliorated by SIRT1 overexpression in BACHD mice (Fig. 1i-j), suggesting that SIRT1 overexpression also attenuates neurodegeneration in BACHD mice.
Abnormal energy metabolism has been suggested as a plausible pathogenic mechanism in HD22-24. We have previously demonstrated that N171-82Q HD mice exhibit metabolic abnormalities that were attenuated by calorie restriction (CR) 1. Therefore, we examined whether SIRT1 could modulate metabolic alterations in HD mice. Over expression of SIRT1 significantly attenuated hyperglycemia (Supplementary Fig. 2a), improved glucose tolerance (Supplementary Fig. 2 b-c) and attenuated weight loss in N171-82Q HD mice (Supplementary Fig. 2d). We then asked whether the attenuation of weight loss was due to the effect of SIRT1 on food intake and/or energy expenditure. Using Oxymax metabolic cages to measure energy expenditure continuously for a week and monitoring daily food intake, we found that SIRT1 overexpression mainly counteracted the negative effect of mutant Htt on food intake (Supplementary Fig. 2e) but did not affect energy expenditure (Supplementary Fig. 2f). Interestingly, insulin levels did not differ between N171-82Q HD mice and WT controls (315.4±51.5 pg/ml in WT mice vs 308.9 ± 34.4 pg/ml in HD mice, mean ± S.E.M., n=10), but HD mice exhibited hyperglycemia, suggesting insulin resistance in HD mice. Since insulin resistance constitutes a metabolic stressor that may contribute to neurological phenotype, we next examined the role of overexpression of SIRT1 on insulin levels. SIRT1 overexpression did not alter insulin levels in control mice (318.6 ±83.7 pg/ml in SIRT1 mice vs 315.4±51.5 pg/ml in WT mice, mean ± S.E.M., n=10), but significantly reduced insulin levels in HD mice (308.9 ± 34.4 pg/ml in HD mice vs 149.1 ± 6.8 pg/ml in SIRT1/HD mice, mean ± S.E.M., n=10). Since SIRT1 attenuated hyperglycemia and decreased insulin levels in HD mice, these results indicate that SIRT1 improves insulin sensitivity in HD mice. To determine whether overexpression of SIRT1 in brain and/or pancreas contributes to this effect, we examined transgene SIRT1 expression by immunohistochemistry. Whereas the transgene is highly expressed in various brain regions including the hypothalamus (Supplementary Fig. 3a), very low levels of expression were detected in the pancreatic islets of Langerhans (Supplementary Fig. 3b). Although these results suggest central action of SIRT1in mediating the metabolic rescue, the effects of SIRT1 on pancreas cannot be completely ruled out.
Intriguingly, SIRT1 overexpression did not extend the life span in N171-82Q HD mice (average life span is 152 ± 7 d in 19 N171-82Q HD mice versus 157 ± 6 d in 26 SIRT1/HD mice). Similar findings were noted in other studies of HD-like mice where reductions in neuropathology and improvements in motor function or normalization of glucose levels were observed in the absence of life span extension25-28. In addition, SIRT1 overexpression did not alter aggregation of mutant Htt (Supplementary Fig. 4), suggesting that aggregation does not play a major role in neuroprotective effects of SIRT1 in our models.
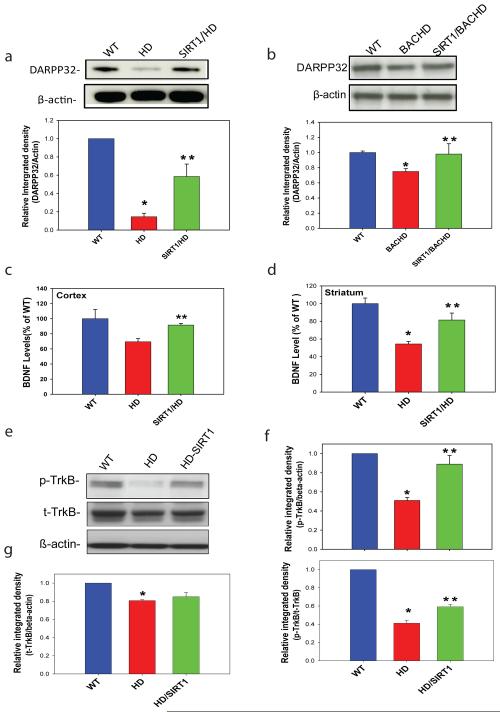
Next we determined the molecular basis underlying neuroprotection of HD by SIRT1. HD mice are severely impaired in their ability to regulate the physiological state of striatal neurons via dopamine29. DARPP-32 (dopamine- and cyclic AMP-regulated phosphoprotein of a molecular weight of 32 kDa) is a fundamental component of the dopamine-signaling cascade, 30,31 and HD pathology is marked by extensive loss of medium spiny striatal neurons that express high levels of DARPP32; therefore, DARPP32 can serve as a marker of neuronal loss as well as neuronal dysfunction in HD 32,33. We observed a significant reduction of DARPP-32 levels in both N171-82Q and BACHD mouse models (Fig. 2a-b), consistent with findings in other HD mice 29. Importantly, SIRT1 restored DARPP32 levels in both HD models (Fig. 2a-b), suggesting that SIRT1 protects striatal neurons against mutant Htt by preservation of DARPP32 levels in medium spiny neurons.
Figure 2. SIRT1 preserves DARPP32 levels in medium spiny neurons, restores BDNF levels and facilitates TrkB activation in HD models.
(a-b) SIRT1 restored DARPP32 levels in N171-82Q HD mice (a) and BACHD mice (b). Top panels are representative Western blots and bottom panels are densitometry results. Mean ± S.E.M., n=4. (c-d) BDNF protein levels were measured by ELISA in cerebral cortex (c) or striatum (d) in 14-week-old mice. Mean ± S.E.M., n=3-6. (e) Representative Western blots of total Trk-B (t-TrkB) and phosphorylated TrkB (p-TrkB) levels in striatal cells expressing wild type (WT) huntingtin (SThdh Q7/Q7, WT) or mutant huntingtin (STHdh Q111/Q111, HD). Note that SIRT1 overexpression reduced mutant Htt-mediated dephosphorylation of Trk-B. (f) Quantification of p-TrkB levels from densitometry analysis of Western blots, Mean ± S.E.M., n=3. Top panel shows results as ratio of p-TrkB to β-actin; bottom panel shows results as ratio of p-TrkB to total TrkB (t-TrkB). (g) Quantification of total TrkB (t-TrkB) levels from densitometry analysis of Western blots, Mean ± S.E.M., n=3. *p<0.05, compared to the value of WT group; **p<0.05 compared to the value of HD group by Student’s t-tests.
Our previous study of CR in HD mice suggests that CR increases brain-derived neurotrophic factor (BDNF) levels, thereby protecting neurons from mutant Htt1. Moreover, reduced BDNF contributes to striatal dysfunction and degeneration in HD and increase of BDNF levels has been demonstrated to be neuroprotective in HD 34-42. BDNF signaling in the brain also plays an important role in regulating glucose metabolism 43 and administration of BDNF in brain reduces blood glucose levels in diabetic mice44. In addition, conditional deletion of BDNF from brain results in increased glucose levels45. BDNF is also required for normal ontogeny of DARPP-3246. BDNF levels were significantly decreased in our HD-like mice, whereas overexpression of SIRT1 markedly restored BDNF protein levels (Fig. 2 c-d). Moreover, we found that total and phosphorylated Trk B levels were significantly decreased in striatal cells expressing full-length mutant Htt (Fig. 2e-f). Overexpression of SIRT1 maintained the activated phospho-Trk-B (Fig. 2e-f) but not total TrkB levels (Fig. 2g). These experiments demonstrated that SIRT1 not only restores BDNF levels, but also enhances activation of BDNF receptor, suggesting that SIRT1-mediated restoration of BDNF levels represents an important mechanism for SIRT1 neuroprotection in HD.
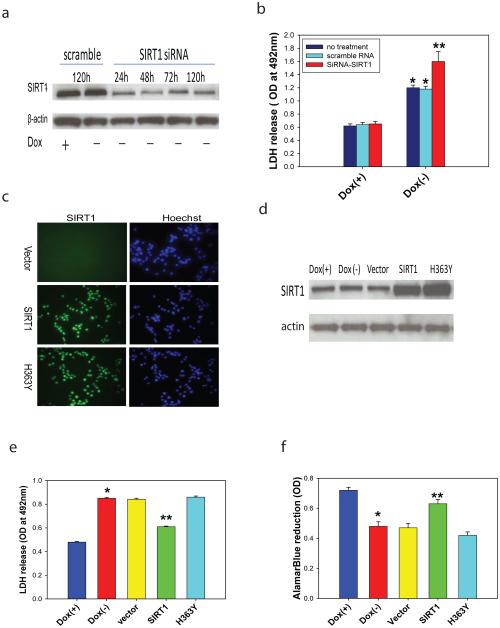
Since our data suggest that overexpression of SIRT1 has neuroprotective effects in HD models, we next sought to determine the role of ablation of endogenous SIRT1 in mutant Htt-induced toxicity. Using an siRNA approach, we showed that complete knockdown of SIRT1 led to cell death in the absence of mutant Htt (data not shown), further suggesting a role of SIRT1 in neuronal survival. Partial knockdown of SIRT1 (Fig. 3a) that was not toxic at baseline led to exacerbation of mutant Htt-induced toxicity (Fig. 3b). Together, these results indicate that endogenous SIRT1 plays a neuroprotective role and that deficiency of SIRT1 in neurons accelerates toxicity of mutant Htt.
Figure 3. Ablation of endogenous SIRT1 exacerbates mutant Htt toxicity and deacetylase activity of SIRT1 is required for its neuroprotection in HD.
(a-b) Reduction of SIRT1 exacerbates mutant Htt-induced toxicity. SIRT1 protein levels were reduced by siRNAs (25 nM) (a) and reduction of SIRT1 increased mutant Htt-induced toxicity (b). Values are mean ± S.D. from three independent experiments. *p<0.05 compared to the value of corresponding Dox(+)group, **p<0.05 compared to the value of scramble RNA-treated Dox(−) group by Student’s t-tests. (c-d) Overexpression of SIRT1 or deacetylase defect SIRT1 (H363Y) was introduced by retrovirus infection. Expression of SIRT1 was confirmed by immunostaining (c) and Western blot analysis (d). (e-f) Mutant Htt expression was induced by withdrawal of doxycycline (Dox) in NGF-differentiated PC12 cells. Overexpression of SIRT1 rescued mutant Htt-induced cell toxicity indicated by reduction of LDH release (e) and increase of AlamarBlue reduction (f). Values are mean ± S.E.M. from three independent experiments. *p<0.05 compared to the value of corresponding Dox(+)group, **p<0.05 compared to the value of vector-transfected Dox(−) group by Student’s t-tests.
Next, we asked whether SIRT1 deacetylase activity is required for its neuroprotection in HD models. We took advantage of a cell model, in which inducible expression of mutant Htt caused significant cell toxicity 47. SIRT1 expression was introduced by retroviral transduction that achieves high transduction efficiency (>90%) in these cells (Fig. 3c-d). Overexpression of SIRT1 significantly reduced mutant Htt-induced toxicity as indicated by a decrease in LDH release (Fig. 3e) and an increase in AlamarBlue reduction (Fig. 3f). The deacetylase-deficient SIRT1 (H363Y) 10 completely eliminated these protective effects of SIRT1 (Fig 3e-f), demonstrating that SIRT1 deacetylase activity is required for neuroprotection.
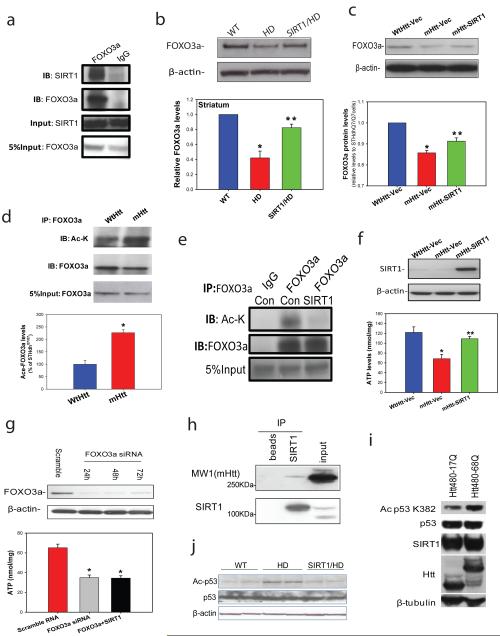
As a deacetylase, SIRT1 is known to deacetylate and modulate the activity of key transcription factors, such as PGC-1α48, p5349 and FOXO3a50. FOXO3a is a ubiquitously expressed mammalian forkhead transcription factor that is highly expressed in adult brain, and recent work indicates that FOXO3a has important roles in neuronal survival under both basal conditions and disease conditions 51-53. We detected an interaction between endogenous SIRT1 and FOXO3a in mouse brain (Fig. 4a). Interestingly, we found consistently decreased levels of FOXO3a protein in HD mouse brains and in cells expressing full-length mutant Htt (Fig. 4b-c). Importantly, levels of acetylated-FOXO3a that were significantly increased in cells expressing mutant Htt (Fig. 4 d) were returned to baseline levels by overexpression of SIRT1 (Fig. 4e). In addition, overexpression of SIRT1 restored FOXO3a levels in the striatum of HD mice (Fig 4 b) and striatal cells expressing mutant Htt (Fig.4c). To further examine whether FOXO3a contributes to neuroprotection of SIRT1 in HD, we employed immortalized striatal cells expressing full-length mutant Htt (STHdh Q111/Q111) or normal Htt (STHdh Q7/Q7). STHdh Q111/Q111 cells were more vulnerable to serum withdrawal compared to STHdh Q7/Q7, as indicated by reduced ATP levels as well as decreased ATP/ADP ratio (Fig. 4f, Supplementary Fig. 6a). SIRT1 overexpression protected STHdh Q111/Q111 cells from mutant Htt, indicated by recovery of ATP levels (Fig. 4f). The direct neuroprotective effect of SIRT1 was further confirmed in primary cortical neurons (Supplementary Fig. 5). Notably, reduction of FOXO3a by siRNA compromised the protective effect of SIRT1 in HD cells (Fig. 4g), suggesting that expression of FOXO3a is at least in part required for SIRT1-mediated neuroprotection. Having shown that FOXO3a partially mediates the neuroprotection by SIRT1 in HD models, we asked whether restoring FOXO3a levels could protect cells from mutant Htt. Although restoring FOXO3a levels preserved ATP levels and ATP/ADP ratio in HD cells (Supplementary Fig. 6b), co-expression of FOXO3a and SIRT1 did not have an additive effect (Supplementary Fig. 6c). Next we examined a possible link between FOXO3a levels and mutant Htt-induced deficits of DARPP32 and BDNF. In HD cells, overexpression of FOXO3a increased levels of BDNF and DARPP32, whereas knockdown of FOXO3a further decreased DARPP-32 levels (Supplementary Fig. 7a-c), suggesting that FOXO3a deficiency may contribute, either directly or indirectly, to mutant Htt-induced abnormalities in BDNF and DARPP-32.
Figure 4. SIRT1 counteracts mutant Htt-induced hyperacetylation of FOXO3a and p53 and protects cells against mutant Htt-mediated energy deficits.
(a) Immunoprecipitation in mouse striatum indicates interactions between endogenous SIRT1 and FOXO3a. (b) FOXO3a levels were determined by Western blotting analysis in striatum of 14-week-old mice. Mean ± S.E.M., n=4. *p<0.05 vs wild type (WT) group; **p<0.05 vs HD group by Student’s t-tests. (c) FOXO3a levels were measured in cell lysates from striatal cells expressing wild type Htt (STHdhQ7/Q7, WtHtt) or mutant Htt (STHdhQ111/Q111, mHtt) transfected with vector (Vec), and mutant Htt expressing cells transfected with SIRT1(mHtt-SIRT1). Mean ± S.E.M. from three independent experiments. *p<0.05 vs WtHtt-Vec group; **p<0.05 vs mHtt-Vec group by Student’s t-tests. (d) Acetylated-FOXO3a levels are increased in cells expressing mutant Htt. Cell lysates were immunoprecipitated with FOXO3a antibody, and blotted with anti-acetylated lysine antibody. Upper panel shows representative blots and bottom panel shows the quantification data from three individual samples. *p<0.01 vs STHdh Q7/Q7 cells by Student’s t-tests. (e) SIRT1 deacetylates FOXO3a in striatal cells. STHdhQ111/Q111 cells were transfected with Vector (Con) or SIRT1 cDNAs for 24 h. Cell lysates were immunoprecipitated with FOXO3a antibody, and blotted with anti-acetylated lysine antibody. (f) SIRT1 protects striatal cells against mutant Htt-induced energy deficits. Striatal cells were cultured in DMEM for 24 h, SIRT1 was transfected into cells. Serum was removed 48 h after transfection, and ATP levels were measured at 24 h after serum withdrawal. The data are mean ± S.E.M. from three independent experiments. *p<0.05 vs the values of STHdhQ7/Q7 cells; **p<0.05 vs the values of vector-transfected STHdh Q111/Q111 cells by Student’s t-tests. (g) Knockdown of FOXO3a (top panel) enhanced mutant Htt-induced energy deficits and compromised the protective effects of SIRT1 (bottom panel). STHdhQ111/Q111 cells were transfected with FOXO3a siRNA (50 nM) or co-transfected with SIRT1, ATP levels were measured at 72 h after transfection. The data are mean ± S.E.M. from three independent experiments. *p<0.05 vs scrambled RNA transfected cells.*p<0.05 vs the values of scrambled RNA-treated cells by Student’s t-tests. (h) SIRT1 interacts with mutant huntingtin. Co-immunoprecipitation was performed with anti-SIRT1 antibody and probed for MW1 antibody which recognizes mutant huntingtin in mouse brain tissue. Beads were used to assess nonspecific binding. (i) Mutant Htt inhibits SIRT1 deacetylase activity which was assessed by p53 deacetylation at Lys 382. HEK T/17 cells transfected with SIRT1 and Htt480-17Q or Htt480-68Q were treated with 500 nM TSA and 100 µM etoposide to block class I and II HDAC activities and induce p53 expression. SIRT1 deacetylase activity was evaluated by monitoring endogenous p53 deacetylation at Lys 382. Equal expression of total p53, SIRT1, wild-type and mutant Htt was ensured by Western blotting with respective antibodies. β-Tubulin was used as loading control. Three independent experiments were performed and a representative blot is shown. (j) Acetyl-p53 and total p53 level were detected by Western blotting in cerebral cortex of 18-week-old mice of indicated genotypes.
On the basis of these observations, we hypothesized that mutant Htt may influence SIRT1 deacetylase activity and thereby increase acetylation of its substrates such as FOXO3a. To examine this possibility, we first tested whether SIRT1 was able to interact with mutant Htt. As shown in Fig 4h, co-immunoprecipitation experiments revealed that SIRT1 specifically interacted with mutant Htt. We then measured SIRT1 deacetylase activity in the presence of mutant Htt. Using a specific antibody that detects acetylated p53 at lysine 382, we found increased levels of acetylated-p53 in cells expressing mutant Htt compared to those expressing wild-type Htt, suggesting that the mutant protein interfered with SIRT1 deacetylase activity (Fig. 4i). In support of this notion, acetylated p53 was also increased in HD mouse brains, whereas overexpression of SIRT1 led to decreased acetylation of p53 (Fig. 4j). Together, these experiments demonstrate that mutant Htt inhibits deacetylase activity of SIRT1, and overexpression of SIRT1 partially corrects hyperacetylation of SIRT1 substrates.
In conclusion, we demonstrate a neuroprotective role of SIRT1 in cell and mouse models of HD. Using both fragment and full-length Htt transgenic mouse models, we found that SIRT1 partially prevents neurodegeneration and ameliorates metabolic abnormalities in HD. Our data indicate that SIRT1 restores BDNF levels and facilitates BDNF prosurvival signals by activating BDNF receptor Trk-B. Importantly, mutant Htt interacts with SIRT1 and inhibits SIRT1 deacetylase activity that in turn leads to hyperacetylation of SIRT1 substrates, such as FOXO3a and p53. Overexpression of SIRT1 partially reverses these inhibitory effects of mutant Htt and reduces the acetylation of FOXO3a and p53. Although a subset of SIRT1 substrates was examined in our models, it is possible that inhibition of SIRT1 deacetylation by mutant Htt affects other unknown or known substrates such as PGC-1α, which has been previously implicated in HD pathogenesis 54-56. Nonetheless, our data indicate that at least one of these substrates, FOXO3a, plays an important role in mediating SIRT1 protection. FOXO3a protein has been also shown to control insulin sensitivity and influence energy metabolism and longevity 57-59. Thus, modulation of FOXO3a by SIRT1 may also affect alterations in energy metabolism observed in HD models. Our findings suggest that pharmacological targeting SIRT1 to enhance its deacetylase activity may provide a novel therapeutic opportunity for HD. Small compounds with SIRT1 activation have been shown to be beneficial in experimental animals prone to metabolic diseases 60. Some of these compounds are already in phase II clinical trials and could be tested in HD. Our study suggests that development of SIRT1 activators with high specificity and bioavailability may represent a promising approach for the treatment of HD.
ONLINE METHODS
Animals
N171-82Q HD mice expressing N-terminal fragment Htt with 82 polyglutamine repeat were maintained by breeding HD males with C3B6F1 females (Taconic). As we reported previously, there are significant gender-dependent differences; therefore, we used all male N171-82Q mice for the study. BACHD mice expressing full-length mutant Huntingtin were bred in FVB background and we used both males and females for the study. DNA was obtained from tails of the offspring for determination of the genotype and CAG repeat size by PCR assay, which was performed by Laragen Inc. genotyping service (Los Angeles, CA, USA). SIRT1 transgenic mouse line (line 1)16 was generated by overexpression of HA-tagged Sirt1 transgene driven by the mouse prion protein promoter (PrP). Sirt1 transgene was constructed by inserting a 2.3-kb fragment of the mouse Sirt1 cDNA into the vector carrying the mouse PrP promoter, after eliminating three NotI sites without changing the SIRT1 amino acid sequence and adding the HA tag to the 3' end of the Sirt1 coding sequence61. The PrP-Sirt1-HA transgene was linearized, purified, and microinjected into C57BL/6J × CBA hybrid blastocysts in the Washington University Mouse Genetic Core Facility. Transgenic mice were identified by PCR genotyping with tail DNA. Transgenic mice were backcrossed to C57BL/6 mice (Jackson Laboratories) for 6–7 generations before being imported to Johns Hopkins University. Nontransgenic littermates were used as controls. Mice were generated by crossing SIRT1 heterozygotes with HD heterozygotes, therefore producing four genotypes: wild type (WT) mice; SIRT1 transgenic mice; HD mice; and double transgenic mice (SIRT1/HD) for the current study. The mice were housed in groups of 3-5 with access to food and water ad libitum and a 12-h light/dark cycle. All animal procedures were performed according to procedures approved by Institutional Animal Care and Use Committee at Johns Hopkins University.
In vivo MRI acquisition
In vivo MRI studies were performed on a horizontal 11.7 T MR scanner (Bruker Biospin, Billerica, MA, USA) with a triple-axis gradient and an animal imaging probe. The scanner was also equipped with a physiological monitoring system (EKG, respiration, and body temperature). Mice were anesthetized with isoflurane (1%) together with oxygen and air at 1:3 ratio via a vaporizer and a facial mask. We used a 40-mm diameter birdcage coil for the radiofrequency transmitter and receiver. Temperature was maintained by a heating block built into the gradient system. Respiration was monitored throughout the entire scan. Images were acquired with a three-dimensional (3D) T2-weighted fast spin echo sequence, with the following parameters: echo time (TE)/repetition time (TR) = 40/700 ms, resolution = 0.1 mm × 0.1 mm × 0.1 mm, echo train length = 4, number of average = 2, flip angle = 40°. Mice recovered quickly once the anesthesia was turned off, and all mice survived the 50-min imaging sessions. Imaging resolution and contrast were sufficient for automatic volumetric characterization of mouse brain and substructures. Detailed image analysis was described in our previous study18.
ELISA analysis of BDNF protein levels
BDNF protein levels were quantified with a commercially available kit (Chemicon) as described previously62. Briefly, samples were processed by acidification and subsequent neutralization. Wells of 96-well plates were coated with anti-BDNF antibody, incubated in the presence of “ block and sample” buffer, and washed in TBST (Tris-buffered saline with Tween-20 ). Samples (300 µg of protein) were added to triplicate wells, and serial dilutions of BDNF standard (0 - 500 pg/ml) were added to wells in order to generate a standard curve. Wells were washed five times with TBST, and a hydrogen peroxide solution was added together with a peroxidase substrate, and plates were incubated for 10 min. Reactions were stopped by adding 100 μl of 1 M phosphoric acid, and absorbance was measured at 450 nm by using a plate reader. The concentrations of BDNF in each sample were determined in triplicate, and the average of the three values was used as the value for that mouse.
Inducible PC12 cells expression of mutant Htt and cell toxicity assay
PC12 cells inducibly expressing mutant Htt were generated as we described47. Cells were maintained in the presence of doxycycline (Dox, 200 ng/ ml) and medium was changed every 48 h. SIRT1 siRNA or SIRT1 virus was introduced after cells attached to the plate, and cell toxicity and viability were measured at 72 h after mutant Htt induction. Cytotoxicity Detection Kit (Roche) was used for measurement of lactate dehydrogenase (LDH). AlamarBlue reduction was used for the cell viability assay.
SIRT1 deacetylase activity
For p53 deacetylation assay, HEK T/17 cells co-transfected with SIRT1 and Htt480-17Q or Htt480-68Q were treated with 500 nM TSA and 100 μM etoposide for 6 h. At 24 h post-transfection, cell were harvested and analyzed for p53 acetylation by Western blotting as previously described.
Immunoprecipitation
Acetylated-FOXO3a levels were analyzed by immunoprecipitation of FOXO3a followed by Western blotting with anti-acetyl-lysine antibody. Cell extracts were obtained by incubating with 10 mM nicotinamide, 1μM TSA, 500 µM H2O2 for 1 h and lysed in RIPA buffer (Sigma) containing deacetylase inhibitors (10 mM nicotinamide, 1 µM TSA), protease and phosphatase inhibitors (Sigma). FOXO3a was immunoprecipitated with anti-FOXO3a (1:2000, Sigma) antibody. Levels of total FOXO3a and acetylated FOXO3a were detected using specific antibodies for FOXO3a or monoclonal anti-acetyl-lysine (1:1000 dilution; gift from Dr. D. Krainc at Harvard University). For FOXO3a and SIRT1 interaction assay, mouse striatal samples were lysed in RIPA buffer (Sigma) containing protease inhibitors (Sigma). Tissue lysates were immunoprecipitated using anti-FOXO3a antibody for 12 h, then washed extensively with RIPA buffer, and membranes were probed with anti-SIRT1 antibody (1:10,000, Upstate) or anti-FOXO3a.
Western blot analysis
Solubilized proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was incubated with primary antibodies (Bcl-XL,1:200, Santa Cruz; SIRT1, 1:10,000, Upstate; DARPP32, 1:2000, Chemicon; p-TrkB, 1:100, Santa Cruz; FOXO3a, 1:2000, Sigma and anti-β-actin, 1:5000, Sigma). The membrane was then exposed for 1 h to HRP-conjugated secondary antibody (1:3000; Jackson ImmunoResearch Labs Inc.) and proteins were visualized by using a chemiluminescence-based detection kit (ECL kit; Amersham Corp.).
Nucleotide extraction and HPLC assay
Conditionally immortalized striatal cell lines, STHdhQ7/Q7 cells and STHdhQ111/Q111 cells, which were a gift from Dr. M. MacDonald, were used in this study. Cells were cultured in DMEM (Gibco) supplemented with 4% fetal bovine serum (HyClone), 2 mM l-glutamine (Mediatech Inc.), and 100 units/ml penicillin and 100 units/ml streptomycin (Mediatech Inc.). The cells were grown at 33 °C in a humidified atmosphere containing 5% CO2. ATP levels were determined by HPLC analysis with UV detection. Briefly, the cell pellets (about one million cells) were washed twice in cold PBS (pH 7.4). Nucleotides were extracted with 50 μl of ice-cold acetonitrile followed by 150 μl of cold water 63,64, and then centrifuged at 14000 × g for 10 min at −2 °C. The supernatant fraction was transferred into a new 1.5 ml tube, kept on ice, and the solvent partially evaporated with N2 for 15~20 min to remove the acetonitrile. The cell pellets were solubilized and the protein content was analyzed using the BCA protein assay kit (Pierce). HPLC analysis were performed on a Shimadzu system using a SCL-10A VP system controller and ZORBAX 3.5 μm SB-C18 column (4.6 × 150 mm) with flow rate of 0.8 ml/min. The samples were analyzed using an isocratic mobile phase consisting of 0.05 mol/L NH4H2PO4 (pH 5.7) and an injection volume of 20 μl. The detection wavelength was set at 257 nm. HPLC-grade nucleotide standards were used to calibrate the signals, the recovery rate exceeded 90% for ATP, ADP, and AMP. All data were quantified by LCsolution software (Shimadzu). The ATP levels were normalized by protein content.
Statistical analyses
We used two-way (genotype and age) repeated analysis of variance to calculate differences in longitudinal behavioral results and body weight data, and one-way or two way ANOVA with Scheffė post hoc test to calculate differences in all other data among groups. All the cell culture experiments were repeated independently at least three times. Significance between groups was determined by standard Student’s t-test between two groups. Data were analyzed with SIGMASTAT version 3.1 software.
Additional methods
Detailed methods, including mutant Htt inclusion assay, blood glucose measurement, energy expenditure, behavioral tests and survival study are described in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Tsai for providing SIRT1 and H363Y retrovirus constructs; S. Imai for providing sir2 cDNAs; M. Macdonald for providing STHdhQ7/Q7 and STHdhQ111/Q111 cells; S. Li and X. Li for providing EM48 antibody; E. Waldron, C. Berlinicke, Y. Cheng, and J. Jin for their technical assistance. This work was supported by Hereditary Disease Foundation (W.D.), CHDI A-2120 (W.D.), NIH NS 16375 (C.A.R.), NIH NS35902 (D.M.H.); NIA Intramural Research Program (B.M., M.P.M.); NIH R01NS051303 (D.K), NIH EB003543 and NIH ES012665 (S.M.) and NIH NS 065306 (J.Z.)
Footnotes
AUTHOR CONTRIBUTIONS
M. J. designed and performed experiments in N171-82Q mice and striatal cells, and co-wrote the manuscript. J. W. designed and performed experiments in BACHD mice. J. F. designed and performed experiments in inducible PC12 cell model. L.D. and R.C performed HPLC assay and data analysis. H. J. and D. K contributed SIRT1 deacetylase activity assay in cells. T. W. and D. M. H. generated SIRT1 transgenic mice and performed Western blots on tissue SIRT1 expression. H.C. performed immunohistochemistry in the pancreas. L. X. performed behavioral tests and glucose measurements in N171-82Q mice. Q. P. performed cross-breeding and genotyping work, and was also involved in behavioral tests and data analysis. Z. H., J. Z. and S. M. performed structural MRI scans and data analysis. T. S. and R. L-C. performed BDNF assays. N. A. and C. A. R. performed primary culture experiments and contributed to discussion. S. Z. supervised and co-performed co-immunoprecipitation experiments. K. L. K. T. and T. H. M. performed CT scans for body mass. S. A. performed Oxymax experiments and data analysis. B.M., S. M. and M. P. M. performed insulin measurements and contribute to discussion. K. S. counted cell numbers with Htt inclusions. J. Q. performed immunohistochemistry in brain. W. Y. provided BACHD mice. D.K. contributed to discussion and helped with the revision and writing of the paper. W. D. designed, directed and coordinated the project, and wrote the paper.
References
- 1.Duan W, et al. Dietary restriction normalizes glucose metabolism and BDNF levels, slows disease progression, and increases survival in huntingtin mutant mice. Proc Natl Acad Sci U S A. 2003;100:2911–2916. doi: 10.1073/pnas.0536856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross CA, Thompson LM. Transcription meets metabolism in neurodegeneration. Nat Med. 2006;12:1239–1241. doi: 10.1038/nm1106-1239. [DOI] [PubMed] [Google Scholar]
- 3.McGill JK, Beal MF. PGC-1alpha, a new therapeutic target in Huntington's disease? Cell. 2006;127:465–468. doi: 10.1016/j.cell.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Browne SE, Beal MF. Oxidative damage in Huntington's disease pathogenesis. Antioxid Redox Signal. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 5.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Oliveira RM, Pais TF, Outeiro TF. Sirtuins: common targets in aging and in neurodegeneration. Curr Drug Targets. 2010;11:1270–1280. doi: 10.2174/1389450111007011270. [DOI] [PubMed] [Google Scholar]
- 7.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Gan BQ, Tang BL. Sirt1's beneficial roles in neurodegenerative diseases - a chaperonin containing TCP-1 (CCT) connection? Aging Cell. 2010;9:924–929. doi: 10.1111/j.1474-9726.2010.00597.x. [DOI] [PubMed] [Google Scholar]
- 9.Albani D, Polito L, Forloni G. Sirtuins as Novel Targets for Alzheimer's Disease and Other Neurodegenerative Disorders: Experimental and Genetic Evidence. J Alzheimers Dis. 2009 doi: 10.3233/JAD-2010-1215. [DOI] [PubMed] [Google Scholar]
- 10.Kim D, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, et al. SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-kappaB signaling. J Biol Chem. 2005;280:40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi Y, et al. SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int J Mol Med. 2005;16:237–243. [PubMed] [Google Scholar]
- 13.Parker JA, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 14.Pallos J, et al. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington's disease. Hum Mol Genet. 2008;17:3767–3775. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pallas M, et al. Modulation of SIRT1 expression in different neurodegenerative models and human pathologies. Neuroscience. 2008;154:1388–1397. doi: 10.1016/j.neuroscience.2008.04.065. [DOI] [PubMed] [Google Scholar]
- 16.Satoh A, et al. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J Neurosci. 2010;30:10220–10232. doi: 10.1523/JNEUROSCI.1385-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schilling G, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, et al. Longitudinal characterization of brain atrophy of a Huntington's disease mouse model by automated morphological analyses of magnetic resonance images. Neuroimage. 2010;49:2340–2351. doi: 10.1016/j.neuroimage.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohanna I, Georgiou-Karistianis N, Hannan AJ, Egan GF. Magnetic resonance imaging as an approach towards identifying neuropathological biomarkers for Huntington's disease. Brain Res Rev. 2008;58:209–225. doi: 10.1016/j.brainresrev.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Gray M, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28:6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menalled L, et al. Systematic behavioral evaluation of Huntington's disease transgenic and knock-in mouse models. Neurobiol Dis. 2009;35:319–336. doi: 10.1016/j.nbd.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner C, Schapira AH. Mitochondrial matters of the brain: the role in Huntington's disease. J Bioenerg Biomembr. 2010;42:193–198. doi: 10.1007/s10863-010-9290-y. [DOI] [PubMed] [Google Scholar]
- 23.Oliveira JM. Nature and cause of mitochondrial dysfunction in Huntington's disease: focusing on huntingtin and the striatum. J Neurochem. 2010;114:1–12. doi: 10.1111/j.1471-4159.2010.06741.x. [DOI] [PubMed] [Google Scholar]
- 24.Petersen A, Hult S, Kirik D. Huntington's disease - new perspectives based on neuroendocrine changes in rodent models. Neurodegener Dis. 2009;6:154–164. doi: 10.1159/000225377. [DOI] [PubMed] [Google Scholar]
- 25.Li M, Huang Y, Ma AA, Lin E, Diamond MI. Y-27632 improves rotarod performance and reduces huntingtin levels in R6/2 mice. Neurobiol Dis. 2009;36:413–420. doi: 10.1016/j.nbd.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 26.Chopra V, et al. A small-molecule therapeutic lead for Huntington's disease: preclinical pharmacology and efficacy of C2-8 in the R6/2 transgenic mouse. Proc Natl Acad Sci U S A. 2007;104:16685–16689. doi: 10.1073/pnas.0707842104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou SY, et al. CGS21680 attenuates symptoms of Huntington's disease in a transgenic mouse model. J Neurochem. 2005;93:310–320. doi: 10.1111/j.1471-4159.2005.03029.x. [DOI] [PubMed] [Google Scholar]
- 28.Simmons DA, Mehta RA, Lauterborn JC, Gall CM, Lynch G. Brief ampakine treatments slow the progression of Huntington's disease phenotypes in R6/2 mice. Neurobiol Dis. 2011;41:436–444. doi: 10.1016/j.nbd.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bibb JA, et al. Severe deficiencies in dopamine signaling in presymptomatic Huntington's disease mice. Proc Natl Acad Sci U S A. 2000;97:6809–6814. doi: 10.1073/pnas.120166397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greengard P, Allen PB, Nairn AC. Beyond the dopamine receptor: the DARPP-32/protein phosphatase-1 cascade. Neuron. 1999;23:435–447. doi: 10.1016/s0896-6273(00)80798-9. [DOI] [PubMed] [Google Scholar]
- 31.Fienberg AA, et al. DARPP-32: regulator of the efficacy of dopaminergic neurotransmission. Science. 1998;281:838–842. doi: 10.1126/science.281.5378.838. [DOI] [PubMed] [Google Scholar]
- 32.Vonsattel JP, et al. Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 33.de la Monte SM, Vonsattel JP, Richardson EP., Jr Morphometric demonstration of atrophic changes in the cerebral cortex, white matter, and neostriatum in Huntington's disease. J Neuropathol Exp Neurol. 1988;47:516–525. doi: 10.1097/00005072-198809000-00003. [DOI] [PubMed] [Google Scholar]
- 34.Strand AD, et al. Expression profiling of Huntington's disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J Neurosci. 2007;27:11758–11768. doi: 10.1523/JNEUROSCI.2461-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sari Y. Huntington's Disease: From Mutant Huntingtin Protein to Neurotrophic Factor Therapy. Int J Biomed Sci. 2011;7:89–100. [PMC free article] [PubMed] [Google Scholar]
- 36.Sari Y. Potential drugs and methods for preventing or delaying the progression of Huntington's disease. Recent Pat CNS Drug Discov. 2011;6:80–90. doi: 10.2174/157488911795933884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bithell A, Johnson R, Buckley NJ. Transcriptional dysregulation of coding and non-coding genes in cellular models of Huntington's disease. Biochem Soc Trans. 2009;37:1270–1275. doi: 10.1042/BST0371270. [DOI] [PubMed] [Google Scholar]
- 38.Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol. 2009;5:311–322. doi: 10.1038/nrneurol.2009.54. [DOI] [PubMed] [Google Scholar]
- 39.Mostert JP, Koch MW, Heerings M, Heersema DJ, De Keyser J. Therapeutic potential of fluoxetine in neurological disorders. CNS Neurosci Ther. 2008;14:153–164. doi: 10.1111/j.1527-3458.2008.00040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan W, et al. Paroxetine retards disease onset and progression in Huntingtin mutant mice. Ann Neurol. 2004;55:590–594. doi: 10.1002/ana.20075. [DOI] [PubMed] [Google Scholar]
- 41.Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in Huntington's disease. Prog Neurobiol. 2007;81:294–330. doi: 10.1016/j.pneurobio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington's disease. Nat Rev Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa T, et al. Brain-derived neurotrophic factor (BDNF) regulates glucose and energy metabolism in diabetic mice. Diabetes Metab Res Rev. 2002;18:185–191. doi: 10.1002/dmrr.290. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa T, et al. Brain-derived neurotrophic factor regulates glucose metabolism by modulating energy balance in diabetic mice. Diabetes. 2000;49:436–444. doi: 10.2337/diabetes.49.3.436. [DOI] [PubMed] [Google Scholar]
- 45.Rios M, et al. Conditional deletion of brain-derived neurotrophic factor in the postnatal brain leads to obesity and hyperactivity. Mol Endocrinol. 2001;15:1748–1757. doi: 10.1210/mend.15.10.0706. [DOI] [PubMed] [Google Scholar]
- 46.Ivkovic S, Polonskaia O, Farinas I, Ehrlich ME. Brain-derived neurotrophic factor regulates maturation of the DARPP-32 phenotype in striatal medium spiny neurons: studies in vivo and in vitro. Neuroscience. 1997;79:509–516. doi: 10.1016/s0306-4522(96)00684-7. [DOI] [PubMed] [Google Scholar]
- 47.Wang W, et al. Compounds blocking mutant huntingtin toxicity identified using a Huntington's disease neuronal cell model. Neurobiol Dis. 2005;20:500–508. doi: 10.1016/j.nbd.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 48.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 49.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 50.Morris BJ. A forkhead in the road to longevity: the molecular basis of lifespan becomes clearer. J Hypertens. 2005;23:1285–1309. doi: 10.1097/01.hjh.0000173509.45363.dd. [DOI] [PubMed] [Google Scholar]
- 51.Kops GJ, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 52.Peng K, et al. Knockdown of FoxO3a induces increased neuronal apoptosis during embryonic development in zebrafish. Neurosci Lett. 2010;484:98–103. doi: 10.1016/j.neulet.2010.07.068. [DOI] [PubMed] [Google Scholar]
- 53.Mojsilovic-Petrovic J, et al. FOXO3a is broadly neuroprotective in vitro and in vivo against insults implicated in motor neuron diseases. J Neurosci. 2009;29:8236–8247. doi: 10.1523/JNEUROSCI.1805-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 55.Chaturvedi RK, et al. Impairment of PGC-1alpha expression, neuropathology and hepatic steatosis in a transgenic mouse model of Huntington's disease following chronic energy deprivation. Hum Mol Genet. 2010;19:3190–3205. doi: 10.1093/hmg/ddq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weydt P, et al. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 57.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 58.Tran H, et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 59.Puigserver P, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 60.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramadori G, et al. Brain SIRT1: anatomical distribution and regulation by energy availability. J Neurosci. 2008;28:9989–9996. doi: 10.1523/JNEUROSCI.3257-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duan W, et al. Sertraline slows disease progression and increases neurogenesis in N171-82Q mouse model of Huntington's disease. Neurobiol Dis. 2008;30:312–322. doi: 10.1016/j.nbd.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gines S, et al. Specific progressive cAMP reduction implicates energy deficit in presymptomatic Huntington's disease knock-in mice. Hum Mol Genet. 2003;12:497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- 64.Au JL, Su MH, Wientjes MG. Extraction of intracellular nucleosides and nucleotides with acetonitrile. Clin Chem. 1989;35:48–51. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.