Abstract
Pharmacogenomics (PGx) offers the promise of utilizing genetic fingerprints to predict individual responses to drugs in terms of safety, efficacy and pharmacokinetics. Early-phase clinical trial PGx applications can identify human genome variations that are meaningful to study design, selection of participants, allocation of resources and clinical research ethics. Results can inform later-phase study design and pipeline developmental decisions. Nevertheless, our review of the clinicaltrials.gov database demonstrates that PGx is rarely used by drug developers. Of the total 323 trials that included PGx as an outcome, 80% have been conducted by academic institutions after initial regulatory approval. Barriers for the application of PGx are discussed. We propose a framework for the role of PGx in early-phase drug development and recommend PGx be universally considered in study design, result interpretation and hypothesis generation for later-phase studies, but PGx results from underpowered studies should not be used by themselves to terminate drug-development programs.
Keywords: clinical research, drug development, early-phase, PD, PGt, PGx, pharmacodynamics, pharmacogenetics, pharmacogenomics, pharmacokinetics, PK, POC, proof-of-concept
A genomic biomarker is defined as ‘a measurable DNA and/or RNA characteristic that is an indicator of normal biologic processes, pathogenic processes and/or response to therapeutic or other interventions’. Pharmacogenomics (PGx) is defined as ‘the study of variations of DNA and RNA characteristics as related to drug response’. Pharmacogenetics is a subset of PGx and is defined as ‘the study of variations in DNA sequence as related to drug response’ [1]. Another way of thinking about the difference between pharmacogenetics and PGx is that the former deals with single genes and the latter with the entire human genome [2,3].PGx focuses on the predictive outcome of drug interventions as opposed to genomic predictors of the natural course of illness, diagnostics and prognostics (‘genomics’). PGx is the study of the interaction between the drug and the individual in terms of drug efficacy, safety and pharmacokinetics (PK). It incorporates information from across the translational spectrum, from gene–disease relationships through confirmatory clinical studies, into the development and eventual application of new drugs [4]. PGx offers the promise of delivering personalized and targeted drug therapy to those most likely to benefit from it [5–8]. PGx may also contribute to the design and interpretation of clinical trials and improve effectiveness, safety and overall benefit:risk ratio of drugs in development [9,10]. It has the potential for earlier arrival at informed developmental decisions – an alternative to the ‘trial and error’ approach. Both the US FDA and NIH have identified PGx as a key tool in the drug-development armamentarium [11,12]. This review will start with a general background of PGx, proceed with a discussion of PGx applications in early-phase research and conclude with recommendations for future development of the field. PGx stakeholders to whom this review is addressed include drug developers, clinical and preclinical investigators, biostatisticians, translational science experts and drug-development consultants, clinical research operators (early phase), analytics operators and experts, business developers, healthcare payers, regulators and policy makers.
History
PGx considerations, even before the term was in use, can be traced to ancient times [13]. The principle of personalized treatment tailored to someone’s physical and physiological constitution is long-held [14,15].Pythagoras’ reluctance to pass through fava bean fields, possibly contributing to his death by captors, may have been due to his recognition of his own glucose-6-phosphate dehydrogenase (G6PD) deficiency and the hemolytic anemia associated with consumption of fava beans [16,17]. Treating someone based on their body type or other constitutional factors, as is the practice in ancient Ayurveda, Chinese, Tibetan and Iranian traditional medicine systems, incorporates genetic features into treatment considerations [13–15,18,19]. Evidence-based medicine may have yet to validate some claims of traditional medicine, but these examples demonstrate that the principles of PGx and related personalized medicine were long part of medical approaches and practice. More recently, at the dawn of a modern trend, the advent of PGx as a science and development tool was first conceptualized by Motulsky in 1957 [20–22]. A twin study of dicumarol pharmacokinetics provided the first evidence of genetic-based pharmacokinetics [23]. Another example of a PGx-guided individualized intervention that may not be thought of as such but has been practiced for decades is anesthesia, where the doses of the anesthetics are continuously adapted to the individual’s signs and symptoms [24].
Background
There are many interindividual differences in response to drug therapy, many may not matter, but some do and may pertain to the interaction of drug with genetically determined physiological and pathophysiological mechanisms. The differences that matter pertain to three main categories or domains of study in PGx: PK, beneficial pharmacodynamics (PD; beneficial, or efficacy), and adverse PD (adverse or toxicity). In every step of PK, or the processes that govern what the body does to the drug (principally absorption, distribution, metabolism and excretion), there could be genetic variability in the following: gastrointestinal tract environment and absorption, active transport across membranes, metabolism, protein binding. Similarly, genetic traits can impact a drug’s PD, or the mechanism(s) governing the drug’s actions at its targets (drug-related phenotypes [4]). The impact of interspecies genomic differences (between rhesus monkeys and humans) on a drug’s target binding and consequent dramatic clinical manifestations is exemplified by the first-in-man study of the anti-CD28 monoclonal antibody TGN1412 [25,26]. Genetic differences in response to drugs (e.g., due to CYP450 polymorphisms and G6PD deficiency) may be inter- or intra-ethnic [17,27–30]. PGx principles may inform drug developers’ marketing strategies and healthcare providers’ insurance policies [31–33].
Three main PGx approaches are applied in drug development:
-
▪
Candidate gene studies. Also called monogenetic (or simply genetic rather than the below genomic approaches) or targeted pathway analysis [10], these are studies of single genes known or hypothesized to impact specific pathways relevant to drug response (e.g., CYP2D6 metabolism) [34]. These studies are hypothesis-driven with predetermined and limited numbers of specific genetic associations between the pharmacological intervention and its outcomes. The disadvantage of the approach is that it may overlook associations with unknown and unanticipated genetic candidates. The advantage is the small number of analyses and the often large effect sizes [34,35].This has been the first approach to be used in PGx studies because it did not require access to the full human genome, available only since 2000 [23,34].
-
▪
Genome-wide association studies (GWAS) involve comparing two groups of individuals with differing characteristics (e.g., drug response) and identifying associations with many known genetic variants. Since the genetic variants are chosen without prior knowledge or suspicion of an association with the differing clinically relevant phenotypes (e.g., disease, responses to treatment), GWAS studies are considered an unbiased, ‘agnostic’ approach [36]. GWAS may assess associations of up to 1 million genetic variants with clinically relevant outcomes [37]. Associations found in GWAS do not necessarily imply causality of the identified genetic marker in the variation of the phenotype. Hence, follow-up studies on the impact of modifications in gene expression on the phenotype are required [4]. The multiple associations studied require adequate statistical corrections. With a few notable exceptions and notwithstanding the relatively small numbers tested so far using GWAS, it appears that PGx effect sizes using GWAS are usually small [34,35].
-
▪
Whole-exome and whole-genome sequencing – the most comprehensive approach that analyzes the entire human genetic material for variants relevant to drug response [35].It may help identify novel or very rare variants that would not be discovered via GWAS, but nevertheless may need to be combined with GWAS to evaluate associations with drug response. Although interpreting such data would be difficult. With reduced cost of whole-genome sequencing (at ~US$3000 in 2012 [4]) this approach is expected to become the dominant PGx approach.
Uses in clinical practice
Personalized medicine and the related term, individualized therapy, have been defined as the practice of adapting treatment to an individual’s medical condition, genetics, demographics, environment and lifestyle [38–40]. PGx addresses the genetic component of personalized medicine as it applies to drug therapy. Table 1 lists examples of successful applications of PGx in clinical care. The approach holds the promise of providing more effective, targeted treatments and quicker arrival at optimal care. This could translate into better public health benefits and eventually reduced hospitalizations and burden of illness, and increased productivity of affected individuals [33,41–43]. PGx has the potential to improve the health of vulnerable populations and neglected diseases [44]. Nevertheless, the application of PGx knowledge and principles in clinical practice has been slow [4,32,37,45–48]. PGx-driven clinical practice guidelines with decision-making algorithms informed by controlled-clinical trials may increase precision, accuracy and relevance of recommendations and subsequent applicability [28,49].
Table 1.
Examples of the successful application of pharmacogenomics.
| Drug | Disease | Phenotype | Type of impact (PK, toxicity, efficacy) |
Percentage of patients that could benefit |
Notes |
|---|---|---|---|---|---|
| Thiopurine Therapy |
Examples include acute lymphoblastic leukemia and ulcerative colitis |
TPMT intermediate activity | Toxicity (myelo- suppression) |
3–14% - those homozygous or heterozygous TPMT [28] |
Homozygous IP/WI-deficient patients (less than 0.5% of ndividuals) usually experience life- threatening myelosuppression |
| Codeine (but applies to many drugs) |
Pain | CYP2D6 conversion of codeine to morphine, the active metabolite |
PK (metabolism) and consequent efficacy, toxicity |
0–10% poor metabolizers; 1–2% ultrarapid metabolizers [83] |
Poor metabolizers have no analgesia; ultrarapid metabolizers risk morphine toxicity |
| Warfarin | Hypercoagulable states (e.g., thromboembolism) |
CYP2C9 metabolism of S-warfarin, the potent enantiomer [10] VKORC1 inhibition by warfarin (VKORC1 gene encodes the target enzyme of warfarin) |
PK (metabolism) and consequent efficacy (anticoagulation), as well as toxicity (lower chance of major hemorrhage) [10,33] |
Asian (86%) and Caucasian (55%) incidence of ‘low dose’ individuals from either VKORC1 or CYP2C9 variants (from at least one VKORC1 haplotype A or CYP2C9 variant allele) [91] |
Narrow therapeutic range Major bleeding in 10–16% [10] PGx findings led to US label updates in 2007 and 2010 |
| Clopidogrel | Hypercoagulable states (e.g., thromboembolism) |
LOF allele of CYP2C19 | PK(CYP2C19 metabolism to active drug) |
Up to 30% of study population [92]; 2–14% of the general population are poor CYP2C19 metabolizers [85] |
PGx findings led to US label updates in 2009 and 2010 |
| Gefitinib | NSCLC | EGFR | Efficacy | 12–18% [29] | PGx findings made during drug development; first EGFR-targeted category approved for NSCLC [29] |
| Trastuzumab | Breast cancer | HER2-positive tumors | Efficacy (overal survival and disease- free survival) |
20% of breast cancer patients | Cardiac toxicity an important consideration in benefit:risk analysis [93] |
| Abacavir | HIV | HLA-B*5701 | Safety (identifying serious adverse event: hypersensitivity reaction) |
5–8% of clinical trial patients [10,94] |
Postmarketing - leading to US label update in 2008 100% negative-predictive value for mmunologically confirmed hypersensitivity reaction |
| Ivacaftor (Kalydeco) |
Cystic fibrosis | G551D mutation in the CFTR gene | Efficacy [95] | The mutation exists in 4% of cystic fibrosis patients |
Development of the drug guided by PGx principles [37] |
| Irinotecan | Colorectal cancer | Lower capacity to metabolize SN-38 (active metabolite of irinotecan) in patients homozygous for the UGT1A1*28 allele |
Toxicity [96] |
UGT1A1*28 mutation in 26–31% of Caucasians, 42–56% of Africans and 9–16% of Asians, but may be higher in ndians [97] |
One of the first drugs to receive pharmagenomically guided labeling requirements by the US FDA in 2005 |
LOF: Loss of function; NSCLC: Non-small-cell lung cancer; PGx: Pharmacogenomics; PK: Pharmacokinetics; SN-38: 7-ethyl-10-hydroxy-camptothecin (the active metabolite of irinotecan).
PGx use in drug development
Understanding the science of PGx: the genotype–phenotype interactions that are relevant to drug response [50] are:
-
▪
Disease genotypes and phenotypes – understanding the genetic origins, mechanisms and manifestations of illnesses is crucial to cure, prevention or mitigation efforts, the processes that influence restoration of health, and the design of drugs to carry out these effects.
-
▪
Drug – the therapeutic agent whose actions are determined by the availability of genomic-driven targets and processes [51].
-
▪
Drug-relevant genotypes (genetic and epigenetic) – these are the genomic components (DNA and RNA) coding and determining drug-relevant phenotypes (see below). The genetic material may be germline (heritable, part of host constitution) or acquired (e.g., tumor mutations), and in either case may be modified by epigenetic and/or nongenetic elements (e.g., infections and toxins) temporarily or permanently [52].
-
▪
Drug-relevant phenotypes (biomarkers) – these are physiological and/or pathological phenotypes that are relevant to drug actions. These could be efficacy outcomes, adverse events, drug plasma concentrations or other PK/PD biomarkers. As the experience with gefitinib demonstrates, earlier identification of a predictive biomarker (e.g., the presence of EGFR mutation in the case of gefitinib; Table 1), understanding the connection between genetic markers and drug response can make drug-development programs more focused, effective and productive [29]. Membrane transporters and genetic variations in their expression will influence PK, efficacy and safety of drugs [53,54]. The drug may use normal physiological systems and processes to reach the target (e.g., chronotherapeutics – the impact of circadian rhythms on drug response [55]) and may impact normal physiological systems in a diseased individual.
Applications of PGx methods in clinical trials in early-phase clinical development
Application of PGx principles in drug development is a continuous process that starts with discovery and continues through the drug’s postmarketing period [3,31,56–68]. Most PGx discoveries to date have been made in post-marketing studies [10,37], but a consensus is being established over the utility and possibly the indispensable nature of PGx approaches as components of all stages of drug-development programs [4,35]. Some also recommend that all commonly used medications should undergo PGx investigation [35].Early-phase development is an important landmark in the overall R&D process since it presents the first introduction of the drug to humans. It is a phase characterized and limited by small sample sizes and trial durations due to ethical and economic reasons, as well as by uncertainties regarding doses, outcome measures, target populations, efficacy and toxicity parameters, their scope and magnitude.
Mechanistic pathways relevant to drug PD (efficacy and safety) affected by genetic polymorphisms with relevance to drug targets, and actions, can be determined before Phase I studies in humans [69].Similarly, genetic factors affecting drug metabolism, transporters and other PK parameters can be studied in vitro and in vivo prior to initiation of human studies [53,70].Such findings can then be incorporated in early-phase human study design. For example, genetic variations in HLA detected by in vitro tests may allow prediction of allergic and other idiosyncratic adverse events in the near future [69,71]. Drug targets may also be identified through their adverse event profile, and what may be undesirable or unexpected effects in one development program may prove desirable and beneficial in another [51]. PGx offers the potential of excluding patients at risk for adverse events from clinical trials and eventual clinical care. PGx identification and exclusion of those most likely to experience an adverse event, as in the case of abacavir hyper-sensitivity, have resulted in increased utilization and sales of this drug [72].
Utilization of PGx principles may be especially favorable in Phase II studies [52]. But even though O’Donnell et al. report only 19% positive PGx findings (defined as ‘worthy of additional follow-up and validation’) in Phase I compared with 70% in Phase II, such findings may still return the investment by contributing to more effective Phase II and Phase III designs and shorter time to critical developmental decisions (i.e., decisions influencing patent-life utilization of the drug under study or back-up candidates). A comprehensive review of PGx study designs is available elsewhere [69]. Randomized controlled trials in early-phase may be preferred for PGx studies as they provide more definitive information regarding predictive properties of biomarkers and subgroups than open-labeled or single-armed trials. Even though randomized controlled trials are more expensive, they may return the investment by providing more precise information about efficacy and safety of the drug in subpopulations of interest and facilitate faster and more focused later-phase development [29]. However, if clear association is demonstrated between genetic polymorphism and drug response in in vitro studies such randomization may be unethical (Box 1, point 3). Adaptive designs based on PGx data collected in the early part of the study may improve the efficiency and cost–effectiveness of early-phase PGx trials.
Box 1. Benefits of pharmacogenomics applications in drug development.
Identifying drug targets – those relevant to the drug’s efficacy, safety, pharmacokinetics
-
Study design [98]:
-
Outcomes that could be impacted by genotype variability:
Pharmacokinetics – area under the curve, Cmax, Tmax, clearance, volume of distribution, half-life, trough drug concentrations [10]
Pharmacodynamics – biomarkers related to efficacy and/or toxicity [99]
Drug–drug interactions
Clinical outcomes – response, remission and other global measures not directly associated with specific biomarkers
Dose selection – influenced by existing population and subpopulation information on dose–response and concentration–response relationships of genes relevant to the drug or disease under study [10,100]
Therapeutic window for the drug: identifying drug plasma concentrations that are between toxic levels (upper limit) and non-effective levels (lower limit)
Selection of participants (i.e., contributing to more meaningful inclusion/exclusion criteria) – for example: inclusion of ethnic or other subpopulations with known genetic variants relevant to drug response (e.g., apoE [101])
Prospective selection of covariates to understand gene-covariate impact on variability [10]
Statistics – exploratory and hypothesis-generating approach is the rule in early-phase studies
-
Ethics: adhering to pharmacogenomic principles would enable more ethical study designs by limiting the testing of new medications to those most likely to benefit and least likely to experience adverse outcomes
Acquisition of information and expertise required to design future studies (i.e., Phase III and Phase IV)
Potential for genotype-specific regulatory approvals and product labeling [29,69,102]
Drug ‘rescue’ and ‘repurposing’
Vulnerable population and rare disease drug development [44]
Genetic profiling of subgroups at risk for adverse events may allow reintroduction of effective drugs that were withdrawn by regulators owing to rare but serious adverse events in the postmarketing period. Such ‘rescuing’, ‘resuscitation’ and ‘repurposing’ of drugs has been proposed by the NIH as a key initiative in the fight against stagnation in drug development [11,69]. Finally, PGx technologies may facilitate development of drugs for vulnerable populations and rare diseases, reduce ethnic disparities in applications of research findings and contribute to improved global public health [44].
Challenges & recommendations for PGx applications in early-phase trials
Box 2 outlines the challenges to PGx applications in early-phase development and Box 3 outlines recommendations aimed at increasing the likelihood of success of such applications. To take full advantage of the potential contribution of PGx approaches, study designs and data to early-phase drug development, PGx strategies should be considered well before entry into human trials, preferably at least 2 years before anticipated first-in-man studies. This will allow sufficient time to gather available drug-specific and disease-specific genomic data (e.g., from target validation, in vitro and in vivo preclinical studies), consultations with subject-matter experts and regulators, and validation of assays, methods and models involved [73,74]. It is further recommended that all drugs in development be considered for application of PGx principles in their early-phase clinical development, and especially those meeting the criteria outlined in Box 4. Toxicity and PK PGx biomarkers may have advantage over efficacy biomarkers in Phase I, even in oncology studies where efficacy Phase I studies are routine [52].
Box 2. Challenges to methodology and applications of pharmacogenomics in early-phase clinical trials.
Limited knowledge of genotypes and phenotypes relevant to drug response (efficacy/safety/pharmacokinetics). This may be particularly applicable to the development of drugs with new targets and/or mechanisms of action (applies to items next two points below as well)
Limited knowledge regarding disease phenotypes and genotypes
Limited knowledge regarding target populations and ethnic subpopulations relevant to drug response
Limited availability of technology and expertise to design and interpret PGx studies
Methodological hurdles – PGx signals may be too weak for the limited power of early-phase studies. Frueh argued ‘you cannot be personalized if you randomize’ [103] implying the goals of personalized medicine and the current gold standard of clinical research might be conceptually and methodologically in conflict
Regulatory hurdles to the standardization of PGx study methodologies and use of PGx data in drug labels and clinical practice [86,88,104]
Ethical hurdles – PGx studies require a definition of meaningful efficacy and toxicity thresholds. However, such thresholds are challenging to define [105]. Equity of access to expensive diagnostics and treatments is another ethical issue relevant to PGx applications [106]. Comprehension and confidentiality hurdles can limit healthy volunteer and patient participation in PGx studies [107]
-
Business and financial considerations:
Limited knowledge, at earlier phases of development, about a drug’s eventual market value, extent and scope of developmental program
Limited adoption of PGx principles by drug developers. Only 27% of studies that include PGx outcomes are carried out by industry according to our ana lysis of the clinicaltrials.gov database
Healthcare payers – although enthusiastic about PGx, are still struggling to translate study findings into effective policies [32,33]
Cost – the cost of whole-genome sequencing was approximately US$3000 in 2012 [4]. PGx cost is trending down while speed and efficiency are increasing. Whole-genome sequencing has the advantage that it only needs to be done once per lifetime
PGx: Pharmacogenomics.
Box 3. Recommendations for the application of pharmacogenomics in early-phase clinical trials.
-
General (including PGx applications during preclinical development)
-
Logistics:
Early planning – 2 years before human trials
PGx considered in all POC clinical trials
Drug/disease genomic data gathered
Experts identified and consulted
Regulators engaged
Validation of genotyping and phenotyping
-
Methodology:
Exploratory and hypothesis-generating
Study types: pharmacokinetics/pharmacodynamics in healthy volunteers and/or patients; dose-response
Outcomes: phenotypes, genotypes of drug response, subpopulations, high-risk, outliers
PGx data collected from all participants
Active metabolites considered as targets
Validation of genotyping and phenotyping assays
Statistics: PGx-specific statistical methods
-
Operational:
Routine collection of biological samples
Long-term storage of DNA, RNA and tissues
Identify technological platforms (e.g., diagnostics)
Incorporation of PGx language into informed consents
-
Business development & pipeline:
Drug labeling
Drug ‘rescue’ and ‘repurposing’
-
-
Phase 0 (microdosing): PGx can be used to study:
Pharmacokinetics (ADME – absorption, distribution, metabolism and excretion)
Transporter polymorphisms
Imaging biomarkers
-
Phase I: In addition to the above, PGx can be used to study:
Pharmacokinetics in MAD/SAD studies
Active metabolites
Toxicity
Drug–drug interactions
Efficacy (e.g., oncology) or proxy-efficacy biomarkers
Healthy volunteers (unless too toxic)
Validity and utility of preclinical data
Inform Phase II and later-phase study design
-
Phase II: In addition to the above, PGx can be used to study:
Focus on efficacy biomarkers and associated genomic parameters
Follow-up on any signals generated in earlier studies
Dose range and ‘therapeutic window’
-
Validation & follow-up
PGx signals validated in adequately powered studies
Low-frequency genotypes may preclude follow-up studies
PGx relevance to regulatory submissions/labeling
New indications, guidelines, pharmacovigilance
MAD: Multiple ascending dose; PGx: Pharmacogenomics; POC: Proof-of-concept; SAD: Single ascending dose.
Box 4. Criteria favoring inclusion of pharmacogenomics design in early-phase clinical trials.
Outcomes suggesting clear and meaningful differences among subgroups: exceptional efficacy, severe toxicity and outlier pharmacokinetics
Known drug-response phenotypes
Drug-response phenotypes are associated with known genomic markers
Existing assays for the phenotypic biomarkers and genomic markers
Methodological PGx considerations should take into account the small, short and exploratory nature of early-phase trials. PGx findings in such studies are likely to be hypothesis-generating rather than hypothesis testing, but nevertheless capable of providing mechanistic support of PGx effects relevant to drug response. If identified, these effects will need to be confirmed in definitive, larger studies in later stages of development [10]. PGx study designs including participant selection criteria and outcomes should be tailored to the drug and disease under consideration [10,69]. Genetic material should be collected from all participants if possible to increase power in these usually small studies, reduce selection bias and optimize the opportunity to identify and study outliers. Samples may be analyzed at a later stage of drug development when they can be combined with other sources of information to increase the likelihood of detecting meaningful signals. Potential relevance of active metabolites to efficacy and toxicity (as in the case of codeine, tamoxifen and clopidogrel) should be considered when designing PGx studies. PGx-specific statistical methods should be used in study design and analyses of the data [75].
Operational standardization is crucial for the applicability, success and generalizability of PGx studies. Prospective and routine collection of biological samples (germline: e.g., blood, buccal cells; acquired mutations: e.g., tumor tissue and metastases) from healthy volunteers or patients in all clinical trials should be practiced [10,76]. This should be accompanied by proper, long-term storage of DNA, RNA and tissues relevant to drug response. Routine PGx language should be incorporated into informed consents and obtained, preferably, from all trial participants at screening to avoid introduction of selection bias by those who discontinue the study. Technological platforms (e.g., analytic and diagnostic) suited to the drug, disease and design under consideration should be identified and validated prior to study initiation.
Business development and pipeline decision-making and considerations may drive PGx approaches. PGx data may drive and contribute to drug labeling and related business development and marketing strategies. PGx findings may play an important part in decisions to rescue or repurpose drugs based on promising PGx signals in subgroup analyses.
PGx in Phase 0 (microdosing) studies can be used to study PK parameters (absorption, distribution, metabolism and excretion), transporter polymorphisms and imaging biomarkers [76–79]. An example is the utilization of PET to image receptor binding of the drug labeled with positron-emitting nuclides. Doses needed to image receptor binding are typically in the microdose levels, thus benefiting from regulatory leniency at the pre-investigational new drug phase of development [80,81]. Such imaging of receptor binding could help identify subpopulations exhibiting variability in binding to receptors of interest.
During Phase I, PGx can be used to study, in addition to the above, toxicity parameters in single and multiple ascending dose studies, drug–drug interactions, conversion to metabolically active compounds and, in some cases (e.g., oncology), drug efficacy or proxy-efficacy biomarkers (i.e., biomarkers suggesting efficacy even in healthy volunteers [e.g., reduction in blood pressure with antihypertensive drugs]). Phase I PGx studies can be used to explore magnitude and relevance of preclinical PGx signals pertaining to drug targets and mechanism of action and PK parameters [69]. Phase I PGx findings can inform Phase II and later-phase study design, including stratification of subpopulations, choice of doses, biomarkers and diagnostics [69].
In addition to the above (Phase 0 and Phase I) Phase II studies should focus on efficacy bio-markers, follow-up on any signals generated in earlier studies (including preclinical) and help identify the range and therapeutic window of dosing regimens (especially useful for drugs with a narrow therapeutic index) [82]. These in turn should be followed by confirmation in large, adequately-powered and long-term studies where possible, and results incorporated, after regulatory approval, into product labeling, practice guidelines, repurposing of approved drugs (new indications), healthcare policies and pharmacovigilance programs [11,28,83,84].
Regulatory role of the application of PGx in clinical trials & drug-development prgrams
Regulatory guidelines
Regulators are promoting the utilization and standardization of PGx principles in drug development. At the time of writing, the regulatory guideline in effect for PGx is the International Conference on Harmonization (ICH) E15 guideline: ‘Definitions for Genomic Biomarkers, Pharmacogenomics, Pharmacogenetics, Genomic Data and Sample Coding Categories’ [1]. The guidelines have been adopted by Europe’s EMA, the US FDA and Japan’s Ministry of Health, Labour and Welfare (MHLW) in 2007, 2008 and 2008, respectively. Application of PGx principles and regulations has led to inclusion of PGx information and recommendations in drug labels and treatment guidelines [28,83–87].
Recently, the FDA approved two new drugs, vemurafenib and crizotinib, whose development was strongly enhanced by application of PGx principles. Both anticancer drugs were tested in subpopulations identified by positive genomic markers and specially developed diagnostic tests, the BRAFV600E mutation and anaplastic lymphoma kinase tests, respectively [4].
Regulatory agencies may classify a drug as first-line treatment based on PGx-derived efficacy and safety data (e.g., gefitinib and the EGFR mutation test [29]).
Early engagement with regulatory agencies is recommended in order to become current with the latest developments in this rapidly evolving field and take full advantage of the flexibility available in the regulations, scientific discoveries, technologies and statistical methodologies relevant to PGx in early-phase clinical development [81,88].
Economics of PGx applications in early-phase drug development
Will PGx applications improve the economics of drug development? Utilization of PGx principles in drug development may lower drug costs by reducing size and length of clinical trials, permit earlier arrival at developmental decisions and increase the postapproval patent-protection period [89]. Hence, the impact of expensive PGx methodology and smaller eventual market may be mitigated by smaller and shorter studies, more efficient and cost-effective drug-development programs. Importantly, drug-development programs may ground to a halt and effective drugs may not reach regulatory approval if subpopulations experience prohibitive levels of adverse events or efficacy signals are diluted by inclusion of non-responsive subpopulations. In addition, an ethical challenge presents itself regarding the administration of non-effective or potentially toxic treatments in face of PGx technologies that permit identification and preventing such untoward healthcare management. Finally, cost–effectiveness of PGx applications may depend on the quality of patient care provided by the healthcare system and needs to be studied and demonstrated before widespread implementation [32,47,84].
Conclusion
The process of developing a new treatment involves progressive reduction of uncertainties regarding efficacy, safety, and, in the case of drugs, PK. PGx is potentially a powerful tool in reducing such uncertainties. Currently, PGx application in early-phase clinical research is minimal (at less than 1% of studies), stationary, and facing a host of challenges. We believe the promise of safe, effective, specific and personal treatments will serve as a powerful driver of PGx utilization in clinical development in general and early-phase research in particular and help overcome the challenges facing the PGx field. Familiarity with PGx methodologies and technologies, wider application and eventual reduced costs will lead to more efficient, cost-effective and productive drug-development programs and translation of genomic findings into meaningful improvements in patient care. We proposed a framework for the role of PGx in early-phase drug development and recommend PGx be universally considered in study design, result interpretation and hypothesis generation for later-phase studies, but PGx results from underpowered studies should not be used by themselves to terminate drug-development programs.
Future perspective
Notwithstanding the low and stationary rate of PGx utilization in early-phase clinical trials in the past decade, we predict a growing and steady increase in utilization for the upcoming 5–10 year period (2013–2023) and beyond. The main reason for this prediction is the belief that an approach that has the potential to provide valuable information for the development of effective, safe, specific and personal treatments will drive the approach past its challenges. Each of the challenges we have identified (limited knowledge about targets, genes and outcomes, limited familiarity with PGx, regulatory and ethical hurdles, methodological challenges and high costs) is manageable and improving [38,47]. In addition, current regulatory guidelines and efforts [10], pressures on industry to make drug development a more efficient and informed process [86,89], pressures from insurers to develop specific and cost-effective treatments [32] and public appeal for personalized therapeutics [90] will all serve as incentives to grow and establish PGx as an indispensable component of the clinical research and development process.
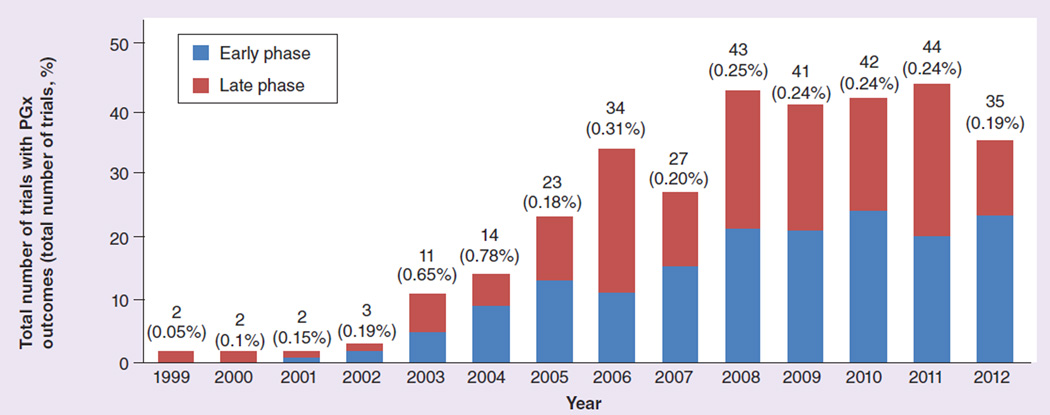
Figure 1. Total number of trials with pharmacogenomics outcomes (and percentage of total trials) per year in the clinicaltrials.gov database.
Studies are divided into early phase (Phases 0, I and II) and late phase (Phases III and IV) 1999–2012. PGx: Pharmacogenomics.
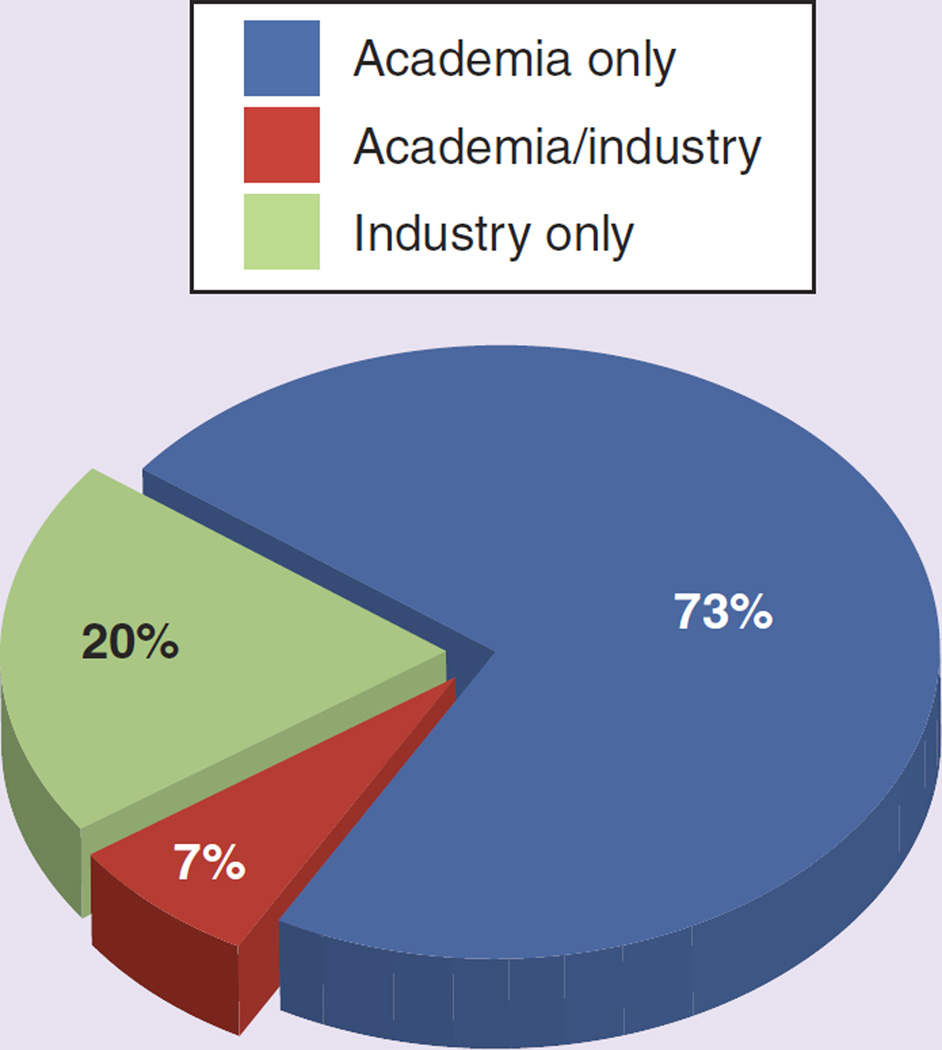
Figure 2. The percentage of pharmacogenomics studies sponsored by industry and academia.
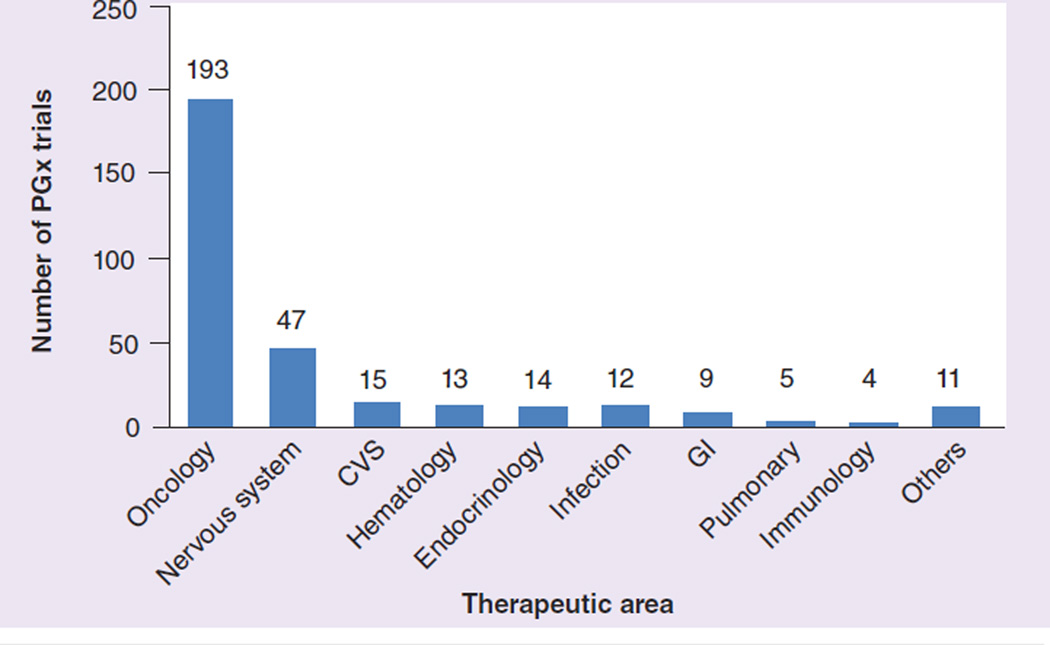
Figure 3. Number of pharmacogenomics studies per therapeutic area.
CVS: Cardiovascular system; GI: Gastrointestinal; PGx: Pharmacogenomics.
Analysis of clinicaltrials.gov database for pharmacogenomics in clinical trials.
Purpose
To assess the scope of application of PGx principles and outcomes in clinical trials as reflected in trials registered on clinicaltrials.gov.
Methodology
Clinicaltrials.gov database [201] was accessed on 1 January 2013. The ‘search for studies’ feature was used to conduct the search with ‘pharmacogenomics’ as the keyword (also returning entries for ‘pharmacogenomic’). Each study entry was reviewed by the two authors and information on phase, sponsor, therapeutic area, objectives, study start date, sample size and end points captured.
Results
A total of 323 studies included PGx as outcome (primary or secondary) in the entire 14 years (1999 to 2012) available in the clinicaltrials.gov database. This represents 0.23% of the total 138,416 studies registered during that period (mean: 341.4 participants per study; standard deviation: 878.2). 164 were early-phase (Phase 0, I and II) studies comprising 0.32% of the total 50,994 early-phase studies. PGx study numbers increased gradually from 2003 but have plateaued since 2008. Out of the total, 258 (80%) have been sponsored by academic institutions, 27% by industry and 7% were industry–academic collaborations. Oncology, at 193 studies (59.7%), was the therapeutic area with most PGx, followed by CNS (45 studies; 13.9%). See Figures 1–3.
Conclusion
Over the 14-year period of the clinicaltrial.gov database, PGx applications in clinical trials (both general and early phase) remained limited with minimal growth.
PGx: Pharmacogenomics.
Executive summary.
Background & uses in clinical practice
-
▪
Pharmacogenomics (PGx) is the study of variations of DNA and RNA characteristics as related to drug response. PGx offers the promise of matching the person and the illness with the optimal treatment. There is a growing number of clinical applications of PGx in routine healthcare and a large number of potential future applications.
Clinicaltrials.gov analysis
-
▪
Less than 1% of clinical trials registered at clinicaltrials.gov included PGx outcomes. The rate has been stationary for the past 6 years and was similar for early- and late-phase trials. Oncology accounted for approximately 60% and academia for 80% of PGx trials.
Application in early-phase clinical development
-
▪
PGx can substantially contribute to early-phase study design, selection of outcomes, stratification of participants, allocation of resources and improvement of clinical research ethics. Results can inform later-phase study design and pipeline developmental decisions.
Challenges & recommendations for PGx application in early-phase trials
-
▪
Challenges include: limited knowledge about targets, genes and outcomes, limited familiarity with PGx, regulatory and ethical hurdles, methodological challenges and high costs. We recommend PGx be universally considered in study design, result interpretation and hypothesis generation for later-phase studies, but PGx results from underpowered studies should not be used by themselves to terminate drug-development programs.
Acknowledgements
The authors would like to thank J Sundy and D Voora for their review and valuable comments made to the manuscript.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
- 1.ICH. E15 Definitions for Genomic Biomarkers, Pharmacogenomics, Pharmacogenetics, Genomic Data and Sample Coding Categories. US Department of Health and Human Services, US FDA, Center for Drug Evaluation and Research (CDER), Center for Biologics Evaluation and Research (CBER) 2008 [Google Scholar]
- 2.Kalow W. Human pharmacogenomics: the development of a science. Hum. Genomics. 2004;1(5):375–380. doi: 10.1186/1479-7364-1-5-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinshilboum RM, Wang L. Pharmacogenetics and pharmacogenomics: development, science, and translation. Annu. Rev. Genomics Hum. Genet. 2006;7:223–245. doi: 10.1146/annurev.genom.6.080604.162315. [DOI] [PubMed] [Google Scholar]
- 4.Crews KR, Hicks JK, Pui CH, Relling MV, Evans WE. Pharmacogenomics and individualized medicine: translating science into practice. Clin. Pharmacol. Ther. 2012;92(4):467–475. doi: 10.1038/clpt.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286(5439):487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 6.Ohashi W, Tanaka H. Benefits of pharmacogenomics in drug development-earlier launch of drugs and less adverse events. J. Med. Syst. 2010;34(4):701–707. doi: 10.1007/s10916-009-9284-7. [DOI] [PubMed] [Google Scholar]
- 7.Powanda MC, Moyer ED. Some applications of pharmacogenomics and epigenetics in drug development and use in pursuit of personalized medicine. Inflammopharmacology. 2012;20(5):245–250. doi: 10.1007/s10787-012-0145-5. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein DB, Tate SK, Sisodiya SM. Pharmacogenetics goes genomic. Nat. Rev. Genet. 2003;4(12):937–947. doi: 10.1038/nrg1229. [DOI] [PubMed] [Google Scholar]
- 9.Milos PM, Seymour AB. Emerging strategies and applications of pharmacogenomics. Hum. Genomics. 2004;1(6):444–455. doi: 10.1186/1479-7364-1-6-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.US FDA. Guidance for Industry Clinical Pharmacogenomics: Premarketing Evaluation in Early Phase Clinical Studies. US Department of Health and Human Services, US FDA, Center for Drug Evaluation and Research (CDER), Center for Biologics Evaluation and Research (CBER), Center for Devices and Radiological Health (CDRH) 2011 [Google Scholar]
- 11.Collins FS. Reengineering translational science: the time is right. Sci. Transl. Med. 2011;3(90) doi: 10.1126/scitranslmed.3002747. 90cm17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.US FDA. Innovation or Stagnation: Challenge and Opportunity on the Critical Path to New Medical Products. US Department of Health and Human Services, US FDA. 2004 [Google Scholar]
- 13.Lesko LJ, Schmidt S. Individualization of drug therapy: history, present state, and opportunities for the future. Clin. Pharmacol. Ther. 2012;92(4):458–466. doi: 10.1038/clpt.2012.113. [DOI] [PubMed] [Google Scholar]
- 14.Roberti di Sarsina P, Ottaviani L, Mella J. Tibetan medicine: a unique heritage of person-centered medicine. EPMA J. 2011;2(4):385–389. doi: 10.1007/s13167-011-0130-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sumantran VN, Tillu G. Insights on personalized medicine from Ayurveda. J. Altern. Complement. Med. 2013;19(4):370–375. doi: 10.1089/acm.2011.0698. [DOI] [PubMed] [Google Scholar]
- 16.Meletis J, Konstantopoulos K. Favism - from the ‘avoid fava beans’ of pythagoras to the present. Haema. 2004;7(1):17–21. [Google Scholar]
- 17.Sukumar S, Colah R, Mohanty D. G6PD gene mutations in India producing drug-induced haemolytic anaemia. Br. J. Haematol. 2002;116(3):671–672. doi: 10.1046/j.0007-1048.2001.03328.x. [DOI] [PubMed] [Google Scholar]
- 18.Nasser M, Tibi A, Savage-Smith E. Ibn Sina’s canon of medicine: 11th century rules for assessing the effects of drugs. J. R. Soc. Med. 2009;102(2):78–80. doi: 10.1258/jrsm.2008.08k040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghodke Y, Joshi K, Patwardhan B. Traditional medicine to modern pharmacogenomics: ayurveda prakriti type and C Y P2 C19 gene polymorphism associated with the metabolic variability. Evid. Based Complement. Alternat. Med. 2011;2011:249528. doi: 10.1093/ecam/nep206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motulsky AG. Drug reactions enzymes, and biochemical genetics. J. Am. Med. Assoc. 1957;165(7):835–837. doi: 10.1001/jama.1957.72980250010016. [DOI] [PubMed] [Google Scholar]
- 21.Motulsky A. From pharmacogenetics and ecogenetics to pharmacogenomics. Med. Secoli. 2002;14(3):683–705. [PubMed] [Google Scholar]
- 22.Gurwitz D, Motulsky AG. ‘Drug reactions, enzymes, and biochemical genetics’: 50 years later. Pharmacogenomics. 2007;8(11):1479–1484. doi: 10.2217/14622416.8.11.1479. [DOI] [PubMed] [Google Scholar]
- 23.Vesell ES, Page JG. Genetic control of dicumarol levels in man. J. Clin. Invest. 1968;47(12):2657–2663. doi: 10.1172/JCI105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemmens HJ, Stanski DR. Individualized dosing with anesthetic agents. Clin. Pharmacol. Ther. 2012;92(4):417–419. doi: 10.1038/clpt.2012.131. [DOI] [PubMed] [Google Scholar]
- 25.Suntharalingam G, Perry MR, Ward S, et al. Cytokine storm in a Phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N. Engl. J. Med. 2006;355(10):1018–1028. doi: 10.1056/NEJMoa063842. [DOI] [PubMed] [Google Scholar]
- 26.Kenter MJ, Cohen AF. Establishing risk of human experimentation with drugs: lessons from TGN1412. Lancet. 2006;368(9544):1387–1391. doi: 10.1016/S0140-6736(06)69562-7. [DOI] [PubMed] [Google Scholar]
- 27.Myrand SP, Sekiguchi K, Man MZ, et al. Pharmacokinetics/genotype associations for major cytochrome P450 enzymes in native and first- and third-generation Japanese populations: comparison with Korean, Chinese, and Caucasian populations. Clin. Pharmacol. Ther. 2008;84(3):347–361. doi: 10.1038/sj.clpt.6100482. [DOI] [PubMed] [Google Scholar]
- 28.Relling MV, Gardner EE, Sandborn WJ, et al. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin. Pharmacol. Ther. 2011;89(3):387–391. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armour AA, Watkins CL. The challenge of targeting EGFR: experience with gefitinib in nonsmall cell lung cancer. Eur. Respir. Rev. 2010;19(117):186–196. doi: 10.1183/09059180.00005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kahn J. Exploiting race in drug development: BiDil’s interim model of pharmacogenomics. Soc. Stud. Sci. 2008;38(5):737–758. doi: 10.1177/0306312708091928. [DOI] [PubMed] [Google Scholar]
- 31.Ginsburg GS, Konstance RP, Allsbrook JS, Schulman KA. Implications of pharmacogenomics for drug development and clinical practice. Arch. Intern. Med. 2005;165(20):2331–2336. doi: 10.1001/archinte.165.20.2331. [DOI] [PubMed] [Google Scholar]
- 32.Wong WB, Carlson JJ, Thariani R, Veenstra DL. Cost effectiveness of pharmacogenomics: a critical and systematic review. Pharmacoeconomics. 2010;28(11):1001–1013. doi: 10.2165/11537410-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 33.Epstein RS, Moyer TP, Aubert RE, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study) J. Am. Coll. Cardiol. 2010;55(25):2804–2812. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Weinshilboum RM. Pharmacogenomics: candidate gene identification, functional validation and mechanisms. Hum. Mol. Genet. 2008;17(R2):R174–R179. doi: 10.1093/hmg/ddn270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harper AR, Topol EJ. Pharmacogenomics in clinical practice and drug development. Nat. Biotechnol. 2012;30(11):1117–1124. doi: 10.1038/nbt.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bush WS, Moore JH. Chapter 11: genome-wide association studies. PLoS Comput. Biol. 2012;8(12):e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giacomini KM, Yee SW, Ratain MJ, Weinshilboum RM, Kamatani N, Nakamura Y. Pharmacogenomics and patient care: one size does not fit all. Sci. Transl. Med. 2012;4(153) doi: 10.1126/scitranslmed.3003471. 153ps118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reynolds KS. Achieving the promise of personalized medicine. Clin. Pharmacol. Ther. 2012;92(4):401–405. doi: 10.1038/clpt.2012.147. [DOI] [PubMed] [Google Scholar]
- 39.Kirchheiner J, Seeringer A. Clinical implications of pharmacogenetics of cytochrome P450 drug metabolizing enzymes. Biochim. Biophys. Acta. 2007;1770(3):489–494. doi: 10.1016/j.bbagen.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 40.Kirchheiner J, Nickchen K, Bauer M, et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol. Psychiatry. 2004;9(5):442–473. doi: 10.1038/sj.mp.4001494. [DOI] [PubMed] [Google Scholar]
- 41.Seeringer A, Kirchheiner J. Pharmacogenetics-guided dose modifications of antidepressants. Clin. Lab. Med. 2008;28(4):619–626. doi: 10.1016/j.cll.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Slamon D, Eiermann W, Robert N, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N. Engl. J. Med. 2011;365(14):1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stehle S, Kirchheiner J, Lazar A, Fuhr U. Pharmacogenetics of oral anticoagulants: a basis for dose individualization. Clin. Pharmacokinet. 2008;47(9):565–594. doi: 10.2165/00003088-200847090-00002. [DOI] [PubMed] [Google Scholar]
- 44.Olivier C, Williams-Jones B. Pharmacogenomic technologies: a necessary ‘luxury’ for better global public health? Global Health. 2011;7(1):30. doi: 10.1186/1744-8603-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirchheiner J, Fuhr U, Brockmoller J. Pharmacogenetics-based therapeutic recommendations - ready for clinical practice? Nat. Rev. Drug Discov. 2005;4(8):639–647. doi: 10.1038/nrd1801. [DOI] [PubMed] [Google Scholar]
- 46.Kirchheiner J, Seeringer A, Viviani R. Pharmacogenetics in psychiatry - a useful clinical tool or wishful thinking for the future? Curr. Pharm. Des. 2010;16(2):136–144. doi: 10.2174/138161210790112728. [DOI] [PubMed] [Google Scholar]
- 47.Johnson JA, Burkley BM, Langaee TY, Clare-Salzler MJ, Klein TE, Altman RB. Implementing personalized medicine: development of a cost-effective customized pharmacogenetics genotyping array. Clin. Pharmacol. Ther. 2012;92(4):437–439. doi: 10.1038/clpt.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frueh FW, Amur S, Mummaneni P, et al. Pharmacogenomic biomarker information in drug labels approved by the United States Food and Drug Administration: prevalence of related drug use. Pharmacotherapy. 2008;28(8):992–998. doi: 10.1592/phco.28.8.992. [DOI] [PubMed] [Google Scholar]
- 49.Relling MV, Klein TE. CPIC: Clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin. Pharmacol. Ther. 2011;89(3):464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012;92(4):414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takarabe M, Kotera M, Nishimura Y, Goto S, Yamanishi Y. Drug target prediction using adverse event report systems: a pharmacogenomic approach. Bioinformatics. 2012;28(18) doi: 10.1093/bioinformatics/bts413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Donnell PH, Stadler WM. Pharmacogenomics in early-phase oncology clinical trials: is there a sweet spot in Phase II? Clin. Cancer Res. 2012;18(10):2809–2816. doi: 10.1158/1078-0432.CCR-11-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giacomini KM, Huang SM, Tweedie DJ, et al. Membrane transporters in drug development. Nat. Rev. Drug Discov. 2010;9(3):215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan Q. Membrane transporters and drug development: relevance to pharmacogenomics, nutrigenomics, epigenetics, and systems biology. Methods Mol. Biol. 2010;637:1–21. doi: 10.1007/978-1-60761-700-6_1. [DOI] [PubMed] [Google Scholar]
- 55.Delezie J, Challet E. Interactions between metabolism and circadian clocks: reciprocal disturbances. Ann. NY Acad. Sci. 2011;1243:30–46. doi: 10.1111/j.1749-6632.2011.06246.x. [DOI] [PubMed] [Google Scholar]
- 56.Bartfai T. Pharmacogenomics in drug development: societal and technical aspects. Pharmacogenomics J. 2004;4(4):226–232. doi: 10.1038/sj.tpj.6500249. [DOI] [PubMed] [Google Scholar]
- 57.Eisenberg RS. Will pharmacogenomics alter the role of patents in drug development? Pharmacogenomics. 2002;3(5):571–574. doi: 10.1517/14622416.3.5.571. [DOI] [PubMed] [Google Scholar]
- 58.Ferentz AE. Integrating pharmacogenomics into drug development. Pharmacogenomics. 2002;3(4):453–467. doi: 10.1517/14622416.3.4.453. [DOI] [PubMed] [Google Scholar]
- 59.Guo Y, Shafer S, Weller P, Usuka J, Peltz G. Pharmacogenomics and drug development. Pharmacogenomics. 2005;6(8):857–864. doi: 10.2217/14622416.6.8.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iqbal O. Pharmacogenomics in anticoagulant drug development. Pharmacogenomics. 2002;3(6):823–828. doi: 10.1517/14622416.3.6.823. [DOI] [PubMed] [Google Scholar]
- 61.Johnson K, Thompson J, Power A. Pharmacogenomics: integration into drug discovery and development. Curr. Top. Med. Chem. 2005;5(11):1039–1046. doi: 10.2174/156802605774297047. [DOI] [PubMed] [Google Scholar]
- 62.Kirk RJ, Hung JL, Horner SR, Perez JT. Implications of pharmacogenomics for drug development. Exp. Biol. Med. 2008;233(12):1484–1497. doi: 10.3181/0805-S-150. [DOI] [PubMed] [Google Scholar]
- 63.Ledley FD. Can pharmacogenomics make a difference in drug development? Nat. Biotechnol. 1999;17(8):731. doi: 10.1038/11614. [DOI] [PubMed] [Google Scholar]
- 64.Lee W, Lockhart AC, Kim RB, Rothenberg ML. Cancer pharmacogenomics: powerful tools in cancer chemotherapy and drug development. Oncologist. 2005;10(2):104–111. doi: 10.1634/theoncologist.10-2-104. [DOI] [PubMed] [Google Scholar]
- 65.Leeder JS. Translating pharmacogenetics and pharmacogenomics into drug development for clinical pediatrics and beyond. Drug Discov. Today. 2004;9(13):567–573. doi: 10.1016/S1359-6446(04)03129-0. [DOI] [PubMed] [Google Scholar]
- 66.Lindpaintner K. Pharmacogenetics and pharmacogenomics in drug discovery and development: an overview. Clin. Chem. Lab. Med. 2003;41(4):398–410. doi: 10.1515/CCLM.2003.063. [DOI] [PubMed] [Google Scholar]
- 67.Liou SY, Stringer F, Hirayama M. The impact of pharmacogenomics research on drug development. Drug Metab. Pharmacokinet. 2012;27(1):2–8. doi: 10.2133/dmpk.dmpk-11-rv-093. [DOI] [PubMed] [Google Scholar]
- 68.Ross JS, Symmans WF, Pusztai L, Hortobagyi GN. Pharmacogenomics and clinical biomarkers in drug discovery and development. Am. J. Clin. Pathol. 2005;124(Suppl.):S29–S41. doi: 10.1309/XYQAFANAPYNC6X59. [DOI] [PubMed] [Google Scholar]
- 69.Stingl Kirchheiner JC, Brockmoller J. Why, when, and how should pharmacogenetics be applied in clinical studies?: current and future approaches to study designs. Clin. Pharmacol. Ther. 2011;89(2):198–209. doi: 10.1038/clpt.2010.274. [DOI] [PubMed] [Google Scholar]
- 70.Stormer E, von Moltke LL, Greenblatt DJ. Scaling drug biotransformation data from cDNA-expressed cytochrome P-450 to human liver: a comparison of relative activity factors and human liver abundance in studies of mirtazapine metabolism. J. Pharmacol. Exp. Ther. 2000;295(2):793–801. [PubMed] [Google Scholar]
- 71.Daly AK, Donaldson PT, Bhatnagar P, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat. Genet. 2009;41(7):816–819. doi: 10.1038/ng.379. [DOI] [PubMed] [Google Scholar]
- 72.Ingelman-Sundberg M. Pharmacogenomic biomarkers for prediction of severe adverse drug reactions. N. Engl. J. Med. 2008;358(6):637–639. doi: 10.1056/NEJMe0708842. [DOI] [PubMed] [Google Scholar]
- 73.Dickinson GL, Rezaee S, Proctor NJ, Lennard MS, Tucker GT, Rostami-Hodjegan A. Incorporating in vitro information on drug metabolism into clinical trial simulations to assess the effect of CYP2D6 polymorphism on pharmacokinetics and pharmacodynamics: dextromethorphan as a model application. J. Clin. Pharmacol. 2007;47(2):175–186. doi: 10.1177/0091270006294279. [DOI] [PubMed] [Google Scholar]
- 74.Yengi LG, Xiang Q, Shen L, Chandrasekaran A, Kao J, Scatina J. Application of pharmacogenomics in drug discovery and development: correlations between transcriptional modulation and preclinical safety observations. Drug Metab. Lett. 2007;1(1):41–48. doi: 10.2174/187231207779814274. [DOI] [PubMed] [Google Scholar]
- 75.Grady BJ, Ritchie MD. Statistical optimization of pharmacogenomics association studies: key considerations from study design to ana lysis. Curr. Pharmacogenomics Person. Med. 2011;9(1):41–66. doi: 10.2174/187569211794728805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N. Engl. J. Med. 2011;364(12):1144–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ieiri I, Doi Y, Maeda K, et al. Microdosing clinical study: pharmacokinetic, pharmacogenomic (SLCO2B1), and interaction (grapefruit juice) profiles of celiprolol following the oral microdose and therapeutic dose. J. Clin. Pharmacol. 2012;52(7):1078–1089. doi: 10.1177/0091270011408612. [DOI] [PubMed] [Google Scholar]
- 78.Ieiri I, Nishimura C, Maeda K, et al. Pharmacokinetic and pharmacogenomic profiles of telmisartan after the oral microdose and therapeutic dose. Pharmacogenet. Genomics. 2011;21(8):495–505. doi: 10.1097/FPC.0b013e3283489ce2. [DOI] [PubMed] [Google Scholar]
- 79.Maeda K, Sugiyama Y. Novel strategies for microdose studies using non-radiolabeled compounds. Adv. Drug Deliv. Rev. 2011;63(7):532–538. doi: 10.1016/j.addr.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 80.Wagner CC, Langer O. Approaches using molecular imaging technology - use of PET in clinical microdose studies. Adv. Drug Deliv. Rev. 2011;63(7):539–546. doi: 10.1016/j.addr.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.US FDA. Guidance for Industry, Investigators, and Reviewers. Exploratory IND Studies. US Department of Health and Human Services, US FDA, Center for Drug Evaluation and Research (CDER) 2006 [Google Scholar]
- 82.Paugh SW, Stocco G, McCorkle JR, Diouf B, Crews KR, Evans WE. Cancer pharmacogenomics. Clin. Pharmacol. Ther. 2011;90(3):461–466. doi: 10.1038/clpt.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin. Pharmacol. Ther. 2012;91(2):321–326. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Verhoef TI, Redekop WK, van Schie RM, et al. Cost-effectiveness of pharmacogenetics in anticoagulation: international differences in healthcare systems and costs. Pharmacogenomics. 2012;13(12):1405–1417. doi: 10.2217/pgs.12.124. [DOI] [PubMed] [Google Scholar]
- 85.US FDA. Drug Safety Communication: Reduced effectiveness of Plavix (clopidogrel) in patients who are poor metabolizers of the drug. 2010 [Google Scholar]
- 86.Piana C, Surh L, Furst-Recktenwald S, et al. Integration of pharmacogenetics and pharmacogenomics in drug development: implications for regulatory and medical decision making in pediatric diseases. J. Clin. Pharmacol. 2012;52(5):704–716. doi: 10.1177/0091270011401619. [DOI] [PubMed] [Google Scholar]
- 87.Surh LC, Pacanowski MA, Haga SB, et al. Learning from product labels and label changes: how to build pharmacogenomics into drug-development programs. Pharmacogenomics. 2010;11(12):1637–1647. doi: 10.2217/pgs.10.138. [DOI] [PubMed] [Google Scholar]
- 88.Salerno RA, Lesko LJ. Pharmacogenomics in drug development and regulatory decision-making: the Genomic Data Submission (GDS) Proposal. Pharmacogenomics. 2004;5(1):25–30. doi: 10.2217/14622416.5.1.25. [DOI] [PubMed] [Google Scholar]
- 89.Cook J, Hunter G, Vernon JA. The future costs, risks and rewards of drug development: the economics of pharmacogenomics. Pharmacoeconomics. 2009;27(5):355–363. doi: 10.2165/00019053-200927050-00001. [DOI] [PubMed] [Google Scholar]
- 90.Hamburg MA, Collins FS. The path to personalized medicine. N. Engl. J. Med. 2010;363(4):301–304. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 91.Marsh S, King CR, Porche-Sorbet RM, Scott-Horton TJ, Eby CS. Population variation in VKORC1 haplotype structure. J. Thromb. Haemost. 2006;4(2):473–474. doi: 10.1111/j.1538-7836.2006.01759.x. [DOI] [PubMed] [Google Scholar]
- 92.Mega JL, Close SL, Wiviott SD, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N. Engl. J. Med. 2009;360(4):354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 93.Moja L, Tagliabue L, Balduzzi S, et al. Trastuzumab containing regimens for early breast cancer. Cochrane Database Syst. Rev. 2012;4 doi: 10.1002/14651858.CD006243.pub2. CD006243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mallal S, Phillips E, Carosi G, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N. Engl. J. Med. 2008;358(6):568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 95.Ramsey BW, Davies J, McElvaney NG, et al. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N. Engl. J. Med. 2011;365(18):1663–1672. doi: 10.1056/NEJMoa1105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O’Dwyer PJ, Catalano RB. Uridine diphosphate glucuronosyltransferase (UGT) 1A1 and irinotecan: practical pharmacogenomics arrives in cancer therapy. J. Clin. Oncol. 2006;24(28):4534–4538. doi: 10.1200/JCO.2006.07.3031. [DOI] [PubMed] [Google Scholar]
- 97.Balram C, Sabapathy K, Fei G, Khoo KS, Lee EJ. Genetic polymorphisms of UDP-glucuronosyltransferase in Asians: UGT1A1*28 is a common allele in Indians. Pharmacogenetics. 2002;12(1):81–83. doi: 10.1097/00008571-200201000-00012. [DOI] [PubMed] [Google Scholar]
- 98.Meisel C, Gerloff T, Kirchheiner J, et al. Implications of pharmacogenetics for individualizing drug treatment and for study design. J. Mol. Med. 2003;81(3):154–167. doi: 10.1007/s00109-002-0417-4. [DOI] [PubMed] [Google Scholar]
- 99.Lai Y, Varma M, Feng B, et al. Impact of drug transporter pharmacogenomics on pharmacokinetic and pharmacodynamic variability - considerations for drug development. Expert Opin. Drug Metab. Toxicol. 2012;8(6):723–743. doi: 10.1517/17425255.2012.678048. [DOI] [PubMed] [Google Scholar]
- 100.van Schie RM, Wadelius MI, Kamali F, et al. Genotype-guided dosing of coumarin derivatives: the European pharmacogenetics of anticoagulant therapy (EU-PACT) trial design. Pharmacogenomics. 2009;10(10):1687–1695. doi: 10.2217/pgs.09.125. [DOI] [PubMed] [Google Scholar]
- 101.Laskowitz DT, Vitek MP. Apolipoprotein E and neurological disease: therapeutic potential and pharmacogenomic interactions. Pharmacogenomics. 2007;8(8):959–969. doi: 10.2217/14622416.8.8.959. [DOI] [PubMed] [Google Scholar]
- 102.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin. Pharmacol. Ther. 2008;83(3):460–470. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 103.Frueh FW. Back to the future: why randomized controlled trials cannot be the answer to pharmacogenomics and personalized medicine. Pharmacogenomics. 2009;10(7):1077–1081. doi: 10.2217/pgs.09.62. [DOI] [PubMed] [Google Scholar]
- 104.Prasad K. Role of regulatory agencies in translating pharmacogenetics to the clinics. Clin. Cases Miner. Bone Metab. 2009;6(1):29–34. [PMC free article] [PubMed] [Google Scholar]
- 105.Fleck LM. Pharmacogenomics and personalized medicine: wicked problems, ragged edges and ethical precipices. N. Biotechnol. 2012;29(6):757–768. doi: 10.1016/j.nbt.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 106.Wertz DC. Ethical, social and legal issues in pharmacogenomics. Pharmacogenomics J. 2003;3(4):194–196. doi: 10.1038/sj.tpj.6500188. [DOI] [PubMed] [Google Scholar]
- 107.Peterson-Iyer K. Pharmacogenomics, ethics, and public policy. Kennedy Inst. Ethics J. 2008;18(1):35–56. doi: 10.1353/ken.0.0004. [DOI] [PubMed] [Google Scholar]
Website
- 201.Clinicaltrials.gov database. www.clinicaltrials.gov.