Abstract
In vivo imaging at high spatiotemporal resolution holds the key to the fundamental understanding of complex biological systems. Integrating an optical phase-locked ultrasound lens into a conventional two-photon fluorescence microscope, we achieved microsecond scale axial scanning, which enabled high-speed volumetric imaging. We applied this system to multicolor volumetric imaging of fast processes, including calcium dynamics in the cerebral cortex of behaving mice, and transient morphology changes and trafficking of immune cells.
To understand the multi-scale signaling dynamics and cell-cell interactions that underlie the emergent behavior of complex biological systems1, 2, we need to observe large cell populations in vivo, while sampling near the natural spatiotemporal scales of the processes of interest. Therefore, high-speed volumetric imaging3–7 is desirable in biomedical fields such as neuroscience and immunology. Recent advances in imaging hardware have enabled fast wide-field image recording,6–8 which offers the highest degree of parallelism: millions of spatial points can be recorded simultaneously. It works well for small, highly transparent biological systems, such as C. elegans7 and zebrafishss6. However in large, turbid biological systems, such as mouse lymph node and brain, the optical aberration and scattering9 result in crosstalk between different spatial points in such parallel detection schemes, limiting the imaging depth8.
First reported in 1990, laser scanning two-photon microscopy (TPM) remains the gold standard for in vivo deep imaging in scattering samples10. Each spatial point is excited and recorded at a different time and therefore the signal is free from scattering-induced crosstalk. In TPM, imaging speed depends on the fluorescence signal intensity, which is affected by the excitation intensity, the fluorophores’ brightness, the fluorescence signal collection efficiency, etc. With sufficient signal, the imaging speed is limited by the 3D scanning methods. Conventional TPMs employ galvo scanners, resonant galvo scanners5 or polygon mirror scanners for transverse (2D) scanning. The translation of the objective lens with respect to the specimen, achieves axial scanning. Due to the large inertia of objective lenses, such axial scanning is typically slow. Novel strategies have emerged to achieve kHz rate axial scanning11. However, the fastest scanning along any direction is still on the order of ~10 kHz.
At least two types of biological activities of substantial interest require high-speed volumetric imaging. One is the functional imaging of neurons, in which the relative positions of the cells and subcellular structures do not change notably over time and the fluorescence variation is the meaningful signal. As the information of interest is not continuously distributed in space, there is a substantial speed advantage to only sparsely scan the relevant subvolume. Researchers have employed random access scanning (RAS)4, 11–13 to achieve fast 3D calcium imaging. Although the overall pixel rate is moderate (<100 kHz), RAS can rapidly acquire data from a priori defined points-of-interest in the volume by trading spatial sampling density for speed. In practice, however, RAS suffers from sample motion especially in behaving animal imaging. The other scenario requiring high-speed volumetric imaging is to observe the movement and interactions of various cell types and their morphology changes1, with a broad range of spatiotemporal scales, simultaneously within a volume. For example, monitoring how immune cells traffic to different locations in lymphoid tissues and how antigen presenting cells interact within lymph nodes will help to better understand the initiation of cell-mediated adaptive immune responses. In such studies, an observation of isolated points or planes becomes inadequate. One needs methods that perform extremely fast 3D scanning and recording over continuous space.
Here, we report a high-speed laser scanning TPM based on a precisely optical phase-locked ultrasound lens (OPLUL), which achieves microsecond scale axial scanning. Using a galvo scanner for transverse scanning, we achieved a cross-sectional frame rate of ~1 kHz, which enables rapid volumetric imaging. Taking advantage of a high-capacity field-programmable gate array (FPGA), our system can record, process and display the high-throughput data stream in real time from three color channels. We report the application of this system to both of the above-mentioned scenarios.
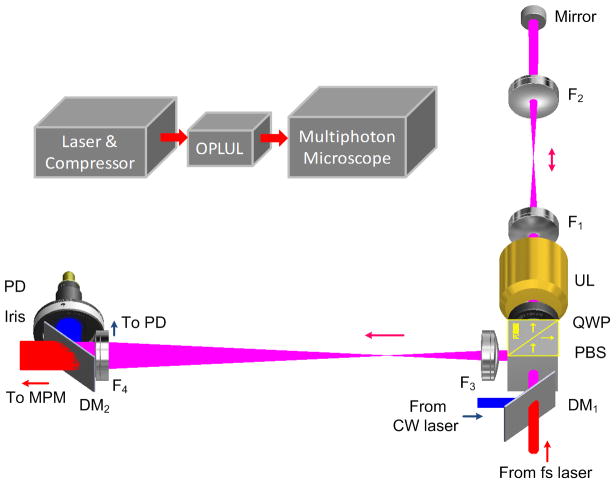
A conventional TPM consists of a femtosecond (fs) laser source, a pulse compressor and a scanning microscope. We inserted an OPLUL between the compressor and the scanning microscope (Fig. 1), which provided the rapid axial scanning. Although the ultrasound lens14 was invented several years ago, it has not been used for high-speed laser scanning TPM due to two challenges. One is the need for high-precision control of the lens oscillation to avoid image distortions. To this end, we introduced optical phase-locking to the system by adding a continuous wave (CW) diode laser to directly monitor the optical focusing of the ultrasound lens. With this optical feedback, we employed a phase-locked loop (PLL) to precisely drive the ultrasound oscillation at its resonance. With these two ingredients, we achieved high stability of both the oscillation amplitude and phase. The other challenge is the limited axial scan range of the ultrasound lens. We introduced a double pass configuration to double the z-scan range. Unlike the lossy and highly dispersive acousto-optic deflectors, the ultrasound lens can achieve nearly 100% light throughput with no spatial chirp and negligible temporal chirp. Moreover, the entire axial scanning system can become an add-on component to most existing TPMs.
Figure 1.
Design of the high-speed volumetric imaging system. A fs laser beam (two-photon excitation beam) and a CW diode laser beam (reference beam), both horizontally polarized, are combined by a dichroic mirror (DM). The beams travel through a polarizing beam splitter (PBS), a quarter waveplate (QWP) and an ultrasound lens (UL). Two lenses (F1 and F2) form a 4f relay configuration to relay the UL onto an end mirror that directs the beam backward to the UL for a second pass. The beams exit the PBS and are relayed to the galvo scanning mirror by a second set of 4f relay lenses (F3 and F4). A second DM separates the CW beam from the fs beam. The CW beam is spatially filtered by an iris before entering a photodiode (PD). All of these elements form the OPLUL that is inserted between the light source and the laser scanning microscope to form the high-speed volumetric imaging system.
We drove the ultrasound lens near its ~455 kHz resonance and therefore a single axial scan took ~1.1 μs. By integrating the OPLUL into a TPM with galvo scanners, we achieved ~1 kHz cross-sectional frame rate. The volume rate can be flexibly varied by changing the number of 2D frames within the volume, according to desired volume size and resolution. We employed two configurations of different axial scan ranges, 40 μm and 130 μm (online methods and Supplementary Figs. 1–7).
Using this system, we performed calcium imaging of neuronal activity in mouse cerebral cortex. We injected GCaMP6s expressing adeno-associated virus (AAV2/1-Syn-GCaMP6s) to infect layer 2/3 (L2/3) neurons and employed our system to image visually-evoked calcium increases. Moving gratings were presented in eight directions to the contralateral eye of anesthetized mice, and the evoked fluorescence transients were recorded. With the capability of high-speed x-z plane scanning, we performed calcium imaging at 893 Hz frame rate (Supplementary Fig. 8 and Supplementary Video 1). We also imaged the transient calcium signals in the dendritic spines of V1 neurons to demonstrate volumetric imaging at high spatiotemporal resolutions. The spontaneous calcium events of the spines were recorded at 56 Hz volume rate (Fig. 2a, b and Supplementary Videos 2 and 3). We can also flexibly image a larger volume at a reduced rate. For example, we imaged V1 neuron ensemble during visual stimulation (Supplementary Fig. 9a and Supplementary Videos 4 and 5), and show the responses of three neurons (Supplementary Fig. 9b), which are one-orientation-selective, two-orientation-selective and two-direction-selective, respectively.
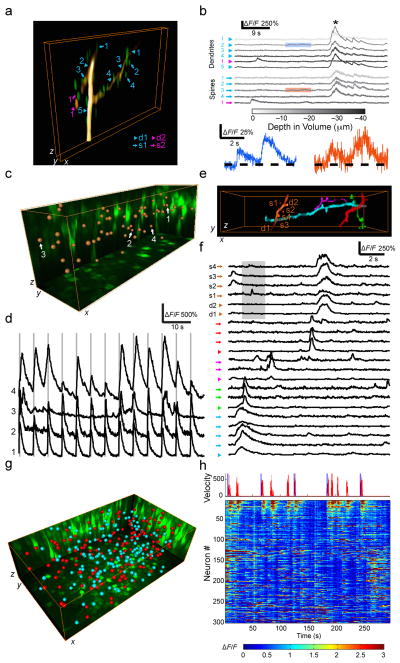
Figure 2.
High-speed 3D in vivo calcium imaging of neuronal networks. (a) Representative volume view of a GCaMP6s-expressing dendritic segment (of 3 datasets) in mouse V1 cortex (x × y × z: 60 × 3.75 × 40 μm3, at depth 107–147 μm under the dura). Dendritic spines are shown in green overlay. Several sections from two separate dendritic segments (cyan, magenta) are shown (thick arrows) as well as dendritic spines attached to these dendrites (thin arrows), with the legends: d1, dendrite 1; s1, spine 1; d2, dendrite 2; s2, spine 2. (b) ΔF/F traces (56 Hz) extracted from several ROIs. Traces are color coded according to the corresponding depth of the ROIs in the volume, with marks corresponding to those in a. Asterisk denotes the onset of a large global calcium signal. Colored boxes (orange, blue) are expanded below to highlight smaller calcium responses in a spine and dendrite, respectively. (c) Representative volume view of the S1 barrel cortex (375 × 112 × 130 μm3, at depth 115–245 μm under the dura) of awake mice (of 8 datasets). Maximum intensity projections (MIPs) are shown in green and the cell locations (63 in total) are indicated by brown spheres. (d) Examples of the sensory stimulation-evoked neuron activity (14 Hz) of the cells in c. The grey boxes show the time span of the air puff stimulation. (e) Representative volume view of the dendrites and spines in the S1 barrel cortex (78 × 20 × 40 μm3, at depth 40–80 μm under the dura) of awake mice (of 6 datasets), with the legends: d, dendrite; s, spine. (f) ΔF/F traces (14 Hz) extracted from each ROI for dendrites and spines in e. Traces are marked with their corresponding colors in e, thick arrows: dendritic segments, thin arrows: dendritic spines. The grey box highlights calcium responses that appear in a spine but not elsewhere along the dendrite. (g) Representative volume view of the M1 cortex (448 × 252 × 130 μm3, at depth 150–280 μm under the dura) of head-restrained behaving mice (of 10 datasets). MIPs are shown in green and the cell locations (304 in total) are indicated by spheres. The red spheres (146 in total) label the neuron cluster of significant (p<0.05, t-test) positive correlation with running, and the cyan spheres label the rest. (h) The mouse running velocity and the calcium dynamics (10 Hz) of neuron ensemble in g. In the upper panel, the blue bars show the time of air puff, and the red plot shows the running velocity (unit: mm/s).
Imaging neuronal activity in behaving animals is required to directly observe behavior or motor related neuronal activities and to avoid the negative effects of anesthesia. However, the motion of the brain that occurs in behaving animals introduces artifacts, especially for imaging fine structures such as the dendritic spines, which presents significant challenges for non-volumetric imaging strategies. As our system performs continuous scanning in 3D, motion artifact correction can easily be performed post hoc. To demonstrate such capabilities, we performed calcium imaging of the somatosensory cortex (S1) of awake mice under sensory stimulation (air puffs). We recorded the stimulation-evoked fluorescence transients (Fig. 2c and Supplementary Videos 6 and 7) of GCaMP6f-expressing S1 L2/3 neurons, and show the fluorescence traces from four neurons synchronized with the whisker stimulation (Fig. 2d). We also recorded the calcium transients of dendritic spines from the spontaneous action potentials in S1 cortex of awake mice (Fig. 2e, f and Supplementary Videos 8 and 9), in which the random motion was severe and would be very difficult for other methods to handle. In addition, we imaged neuron ensemble activity in the motor cortex (M1) while the head-restrained mice were running on a linear treadmill. The running induced fluorescence dynamics of GCaMP6s-expressing M1 L2/3 neurons (Fig. 2g, h) were recorded, based on which the neuron cluster with significant positive correlation (p<0.05, t-test) to running was identified (Supplementary Fig. 10 and Supplementary Videos 10 and 11).
The fundamental characteristic of the vertebrate immune system is the motility and migratory behavior of various cell types accompanied by rapid shape changes. Taking advantage of the ~1 kHz cross-sectional frame rate, we performed in vivo imaging of leukocytes in blood flow, a challenging task for in vivo flow cytometry15. During inflammatory responses, neutrophils16 are recruited to sites of tissue damage or infection via the circulatory system. We imaged neutrophils trafficking inside the vasculature of Lyz2gfp/+ B6.Albino mouse brain at a cross-sectional frame rate of 1 kHz (Fig. 3a and Supplementary Video 12). We imaged neutrophils trafficking through the pial veins of mouse brain at 39 Hz for a 112 × 38 × 40 μm3 volume. We show snapshots of a neutrophil (Fig. 3b). The peak velocity of these flowing neutrophils is 1172 ± 83 μm/s (mean ± standard deviation), determined from the 4D data (Supplementary Video 13). We recorded the neutrophils rolling along the vessel wall (Supplementary Video 14), and observed neutrophils passing through capillaries by quickly adapting their morphologies (Fig. 3c, Supplementary Fig. 11 and Supplementary Videos 15 and 16). We also recorded neutrophil trafficking in the ear vasculature and extravascular tissue (Supplementary Fig. 12 and Supplementary Videos 17–20). These experiments demonstrate our system’s capability of capturing the extremely fast leukocyte circulation within blood vessels and rolling along the interior endothelial lining of the vessel wall.
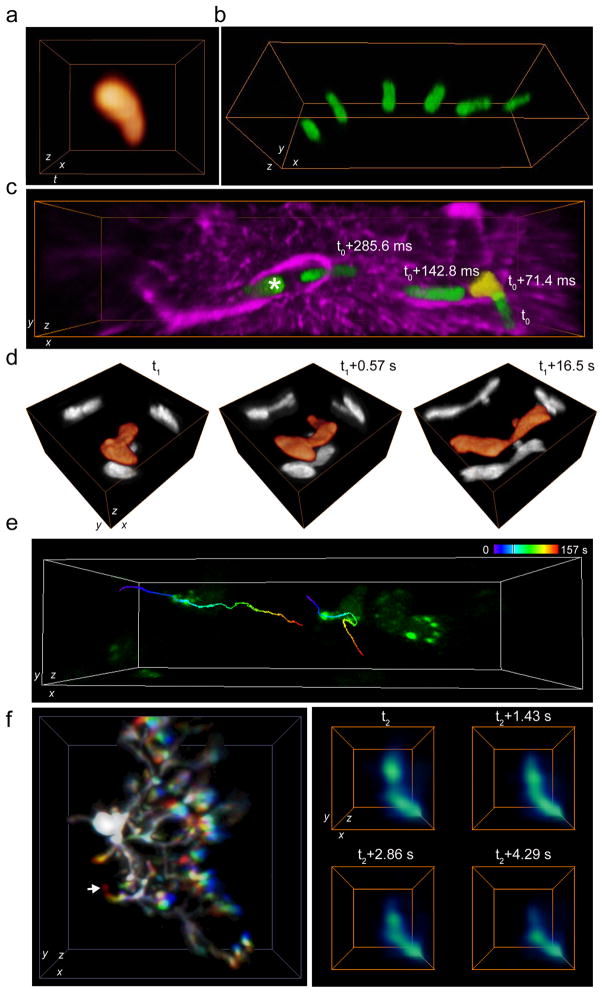
Figure 3.
High-speed 3D in vivo imaging of cell dynamics. (a) A representative neutrophil image reconstructed from 2D cross sectional imaging of a pial vein in mouse brain at 1 kHz frame rate at depth 5–45 μm under the dura (of 6 datasets). The x-z frame size was 18 × 20 μm2 (25 frames in total). (b) Representative snapshots of a neutrophil trafficking in a pial vein of mouse brain (of 3 datasets). The volume was 112 × 38 × 40 μm3 (at depth 5–45 μm under the dura) and the imaging rate was 39 Hz. (c) Representative rapid morphological changes of a neutrophil trafficking through capillaries in mouse cerebral cortex (of 4 datasets). The volume was 151 × 38 × 23 μm3 (at depth 50–90 μm under the dura) and the imaging rate was 14 Hz. Green: neutrophil cell (the snapshot at t0+71.4 ms is in yellow), magenta: SR101 stained astrocytes. Another neutrophil crawling in the capillary is marked with asterisk. (d) Representative rapid morphological changes of a dendritic cell in the mouse popliteal lymph node (of 3 datasets). The volume was 35 × 35 × 40 μm3 at depth 55–95 μm under the surface. Orange: cell morphologies in 3D; grey: MIPs. (e) Representative spatiotemporal dynamics of the lymphocytes and intracellular fluorescent clusters in the mouse popliteal lymph node (of 5 datasets). The volume was 84 × 21 × 40 μm3 (at depth 100–140 μm under the surface). The time-color-coded traces show trajectories of two intracellular fluorescent clusters. (f) Left: representative transient subcellular structural changes of a microglial cell activated by the BBB disruption in the mouse cerebral cortex (of 2 datasets). The volume was 64 × 64 × 40 μm3 (at depth 100–140 μm under the dura). Three snapshots separated by 14.33 s are shown in red, green and blue. Right: transient structural changes of the microglia branch marked by the white arrow in the left image. The volume was 6 × 6 × 15 μm3.
Visualizing the cellular and intracellular dynamics in lymph nodes is critical to the study of immune system. We performed in vivo imaging of the popliteal lymph nodes in anesthetized CD11c-EYFP mice17 and Tbet:ZsGreen mice18. We recorded the 3D morphology changes of CD11c-EYFP bright cells, mainly antigen-presenting dendritic cells (Supplementary Video 21), and captured a cell that changed its morphology notably within 0.57 s (Fig. 3d and Supplementary Video 22). The fine cellular dendrites were constantly probing and sweeping (Supplementary Fig. 13 and Supplementary Video 23). With the Tbet:ZsGreen mice, we imaged the spatiotemporal dynamics (Fig. 3e and Supplementary Video 24) of both the lymphocytes (mostly memory T cells and natural killer cells19) and intracellular clusters (likely due to the overexpression of the ZsGreen proteins). These results demonstrate the substantial improvement of our method in 3D tracking of cellular and intracellular motions in tracking speed, precision and accurate identification16.
Microglia play a critical role in immune defense and tissue homeostasis in the central nervous system (CNS). Unlike neurons, microglia constantly survey the CNS in 3D with their fine branches. The study of these cells can thus benefit from high-speed continuous volumetric imaging. We imaged the dynamics of microglia in CX3CR1gfp/gfp mice20. In the resting state, microglial cells (Supplementary Figs. 14 and 15) continuously survey their microenvironment (Supplementary Video 25) while monocytes patrol around the cortex (Supplementary Fig. 16 and Supplementary Video 26). Once brain-blood-barrier (BBB) disruption or neuron damage occurs, activated microglia rapidly change their phenotypes (Fig. 3f and Supplementary Videos 27–29). These volumetric imaging results indicate that our system can be used for the in vivo study of fast 3D subcellular dynamics where dense spatiotemporal sampling is required.
Our system can also be used to observe both functional signals and rapid morphological changes in 3D simultaneously at high speed. We provide one example to show the potential of our system in studying neurovascular coupling (Supplementary Fig. 17 and Supplementary Videos 30 and 31).
In summary, we report a high-speed multicolor volumetric laser scanning microscope which employs an OPLUL to perform microsecond scale axial scanning. One useful feature of our system is that it can become an add-on to upgrade existing laser scanning microscopes for high-speed 3D imaging. A limitation in the current implementation is the moderate axial scanning range, limited by the spherical aberration induced by the ultrasound lens. While a more sophisticated implementation can potentially cancel the aberration (Supplementary Discussion 1), a straightforward solution would be replacing the lens liquid with a medium of higher acoustic velocity and greater acousto-optic figure of merit. Due to the aberration, the focus elongates toward the axial edges of the imaging volume (Supplementary Fig. 5). However, in many applications the resolution is still adequate. For example, the microglia’s fine processes were clearly resolved (Supplementary Fig. 14).
One powerful application of our method is 3D functional imaging in behaving animals at volume rates that match the time scales of the genetically encoded calcium indicators. The continuous high-speed 3D scanning enables post hoc motion artifact correction. Another important application is imaging moving cells with rapidly changing morphologies and intercellular interactions inside scattering tissues. The rapid cross-sectional scanning also enables in vivo imaging flow cytometry. Moreover, our system allows the simultaneous high-speed imaging of functional signals and transient morphology dynamics. With its high-speed imaging capacity and its compatibility with commonly used TPMs, we anticipate broad application of our system.
ONLINE METHOD
Setup
The key element is a precisely controlled ultrasound lens. To provide a reliable feedback of the lens oscillation, we combined a polarization-maintaining fiber coupled CW diode laser (LPS-PM785-FC, Thorlabs) and the fs laser (Chameleon, Coherent) with a dichroic mirror beam splitter (T810lpxr, Chroma). The CW laser beam went through the same path as the fs laser beam inside the OPLUL (Supplementary Fig. 1a). The ultrasound lens (TAG lens 2.0, TAG Optics) induced the laser beams to oscillate between converging and diverging states. We used a second dichroic mirror beam splitter to separate the CW beam from the fs beam and used an iris to spatially filter the CW beam. For converging beams, the optical power through the iris was higher. For diverging beams, the power was lower. By monitoring the beam intensity with the photodiode behind the iris, we precisely measured the ultrasound-induced lensing effect. To prevent the fs laser from entering the photodiode, we mounted a short-pass filter (FES0800, Thorlabs) on the photodiode. To maintain a consistent oscillation amplitude and phase, we employed a programmable digital PLL (HF2LI-PLL, Zurich Instruments) that used the photodiode signal as the feedback and drove the ultrasound transducer. Before locking the loop, we routinely measured the resonance peak (Supplementary Fig. 1b).
We drove the lens near its ~455 kHz acoustic resonance peak and therefore a single axial scan took 1.1 microseconds (two axial scans per oscillation period). In this work, we only used the central area of the lens aperture (1.7 mm in diameter) whose refractive index profile was similar to that of a simple lens. As our galvo scanner had a 5 mm aperture, we used a 4f relay lens pair (AC254-100-B and AC253-300-B, Thorlabs) to magnify the laser beam by a factor of three. The objective lens used in all the imaging experiments was the Nikon 16x NA 0.8 water-dipping lens. The z scan range was ~40 microns. For applications requiring even longer axial range, we had a second configuration to achieve a ~130 microns range, in which we used a different relay lens pair (AC254-150-B and AC254-250-B, Thorlabs). In this configuration, the beam magnification was less and the beam did not completely fill the objective pupil. Therefore, we achieved a longer scan range at the cost of resolution.
We inserted the OPLUL between the light source and the scanning microscope whose line rate (x scan) was ~1 kHz. Therefore, we achieved a ~1 kHz frame rate in the x-z plane. Using a resonant galvo for the x scan, we can potentially achieve a frame rate of 10–30 kHz.
The FPGA based DAQ system can simultaneously record and process data from three color channels: blue (filter ET460/36, Chroma); green (filter FF01-520/70, Semrock); red (filter FF01-625/90, Semrock).
Data acquisition and image reconstruction
As shown in the flow chart (Supplementary Fig. 2), we used a high-speed data acquisition card to record both the PMT signals (from multiple channels of different colors) and the photodiode signal (optical feedback). As the required sampling rate was high, we used the fs laser’s pulse train as the external sample clock for the card. A high-speed comparator converted the analog pulse train signal into digital TTL pulses that were transmitted by a USB cable to the card. The start of each data recording was triggered by the laser scanning microscope’s line trigger.
The photodiode signal contained the information of the axial scan position. We used a sinusoidal fitting of the photodiode signal to extract the phase and frequency of the axial scanning, from which we determined the exact axial position for every data point. We then applied a 2D linear interpolation to map the raw non-uniformly sampled x-z image onto a regularly spaced grid. The processed x-z frame contains 1024 × 40 voxels. We repeated the same procedure for different y positions and obtained the 3D image stack. For each channel, the sampling rate was 80 MHz and the effective voxel rate was 40 MHz. On average, two laser pulses contribute to the signal at one voxel. As an example, we show the raw data recorded for a 1 μm fluorescence bead (Supplementary Fig. 3), from which we reconstructed the 3D volume images. All data shown is not averaged along the time axis unless otherwise specified.
In the development stage, we used ScanImage to control the laser scanning microscope, and recorded data with a high-speed data acquisition card (ATS9440, AlazarTech). The recorded data was quickly streamed to data storage. We tested a RAM disk (DataRAM), internal SSD (840pro, SAMSUNG) and an external RAID system (CX3250, Colfax International); all provided sufficient throughput for the data streaming. The data processing (curve fitting and 2D interpolation) was done on a 2500-core cluster. Currently, we control the microscope by a custom Labview program and use a high-speed FPGA card (PXIe-7962R, NI) for data acquisition and processing in real time.
Code availability
The custom codes for hardware control and data analysis are available upon request from the authors.
System calibration
The OPLUL induced a slight degree of optical aberration. Using a recently demonstrated wavefront correction method9, we measured and compensated this system aberration (Supplementary Fig. 4a, b). After the aberration correction, we used 1 μm fluorescence beads embedded in 1.5% agarose to measure the axial scan range (Supplementary Fig. 4c, d). We used 0.2 μm fluorescence beads embedded in 1.5% agarose to measure the spatial resolution (Supplementary Fig. 5). We further quantified the system performance over depth (Supplementary Fig. 6) by in vivo deep imaging of fine structures, such as the dendrites and dendritic spines in S1 cortex of thy1-YFP (H line) transgenic mice. The spines can be resolved at up to ~400 μm depth (103 mW at 935 nm). Due to the ultrasound lens induced additional aberration, the axial resolution decreased when the focus was shifted away from the center. However, this did not substantially affect the resolution over the specified axial scanning ranges (Supplementary Fig. 14). We compared the volume images recorded by the objective lens translation and by the OPLUL (Supplementary Fig. 18), which shows the advantage of high-speed volume imaging in reducing the artifact caused by fast events.
The signal-to-noise ratio (SNR) of each voxel depends on the excitation power, the brightness of the fluorophores, the imaging depth, the objective’s NA, the signal collection efficiency, the quantum efficiency of PMT, etc. We tested SNR with thy1-YFP (H line) transgenic mice in vivo and the typical single voxel SNR was ~5 in the fast volume imaging of fine structures such as the dendrites in the cortex (laser power ~ 100 mW at 935 nm, Nikon 16x NA 0.8 objective, PMT of ~50% quantum efficiency, imaging depth ~50–100 μm beneath the dura). With similar parameters, we also imaged the brains of CX3CR1gfp/gfp transgenic mice; here the single voxel SNR was typically ~9.5 for fast volume imaging of microglial cell bodies (imaging depth ~100 μm).
Considering that the power applied in our experiments is generally higher than that of 2D imaging, we performed photodamage quantification by 10 min continuous recording of calcium dynamics of deep somata and dendrites (Supplementary Fig. 7). It is known that the photodamage is associated with an increase in basal fluorescence21. Despite the long time continuous imaging, the raw data (Supplementary Fig. 7b, d) shows no variation in the basal fluorescence of somata or small and dim structures such as dendrites.
Calcium imaging of mouse V1 neurons
All procedures involving mice were approved by the Animal Care and Use Committees of HHMI Janelia Research Campus and National Institute of Allergy and Infectious Diseases, National Institutes of Health. Mice approximately 6–12 weeks old of both sexes were used without randomization.
The procedures related to the calcium imaging of C57BL/6 mouse V1 neurons with visual stimulation, including the V1 neuron labeling, the window surgery for acute experiments, the visual stimuli and the imaging of mouse V1 neurons, were conducted as in a previous report22 with the following exceptions. (1) The visual stimuli were generated by a MATLAB program and displayed in blue color on a 7 inch LCD monitor. (2) Each stimulus trial included a 7 s blank period followed by a 4 s drifting sinusoidal grating. (3) The images were recorded at a higher speed. For the 2D cross-sectional calcium imaging of mouse V1 cortex (Supplementary Fig. 8), the frame rate was 893 Hz with ~85 mW laser power (λ=935 nm). For the 3D calcium imaging of mouse visual cortex (Supplementary Fig. 9), the volume was 375 × 375 × 130 μm3, and the volume rate was 5.6 Hz with ~100 mW laser power (λ=935 nm). Both the soma ensembles in the above results were at 100–250 μm beneath the dura.
To monitor the spontaneous activity of V1 neurons, the mice were kept in the dark. During imaging, the mice were sedated with chlorprothixene (10–20 μL at 0.33 mg ml−1, intramuscular), kept anesthetized (isoflurane, 0.5–2%), and were placed on a regulated heating pad (37 °C). For the volumetric imaging of dendritic segment in mouse V1 cortex (Fig. 2a), the volume size was 60 × 3.75 × 40 μm3 and the volume rate was 56 Hz (85 mW at 935 nm). The imaging depth was 100–250 μm beneath the dura.
Calcium imaging in mouse S1 cortex
Surgeries were conducted on adult C57BL/6 mice under anesthesia (1.5–2% isoflurane). To reduce potential inflammation, 5 mg kg−1 ketofen was injected subcutaneously. 0.1 mg kg−1 buprenorphine was injected intraperitoneally to provide general analgesia. To label neurons of barrel cortex at layer 2/3, a small area craniotomy was performed over the left S1. Virus expressing GCaMP6f (AAV2/1-Syn-GCaMP6f) was injected slowly (30 nL in 5 mins) through the sharp tips of glass pipettes. After 1–2 weeks of expression, a craniotomy was performed above the former injection position, and a round imaging window made of two layers of microscope coverglass was mounted for chronic imaging experiments. Imaging was carried out 2–4 weeks after the craniotomy. Awake mice were placed on a regulated heating pad (37 °C) during the imaging. Puffs of compressed air (8 psi), with 500 ms duration and 5 s interval, were applied to the contralateral whiskers through a 1-mm-diameter tube placed ~15 mm away. The mice were tail-vein injected with Qtracker® 655 (blood plasma staining, Life Technologies) to label the blood vessels as references for motion registration. The laser power was ~60 mW at 935 nm. The imaging depth was 100–250 μm beneath the dura for the soma ensemble, and 0–100 μm beneath the dura for the dendrite network.
Calcium imaging in mouse M1 cortex
Surgeries were conducted on adult C57BL/6 mice under anesthesia (1.5–2% isoflurane). 5 mg kg−1 ketofen was injected subcutaneously to reduce potential inflammation, and 0.1 mg kg−1 buprenorphine was injected intraperitoneally to provide general analgesia. To label neurons of motor cortex at layer 2/3, a small area craniotomy was performed at 1.75 mm lateral and 0.13 mm rostal of bregma. Virus expressing GCaMP6s (AAV2/1-Syn-GCaMP6s) or the mixture of diluted AAV2/1-Syn-Cre and AAV2/1-Flex-Syn-GCaMP6f were injected slowly (30 nL in 5 mins) through the sharp tips of glass pipettes. After 1–2 weeks, the imaging window was installed above the former injection position after the craniotomy. The mice were trained to run on a linear treadmill after 1–2 weeks’ recovery. Imaging was carried out 2–4 weeks after the craniotomy. For the volumetric imaging of neuron ensemble in mouse M1 cortex (Fig. 2g, h), puffs of compressed air (6–8 psi), with 200 ms duration and 60 s interval, were applied to the contralateral whiskers (through a 1-mm-diameter tube placed ~15 mm away) of the head-restrained mice on a custom linear treadmill. The motion was monitored by an optical encoder mechanically coupled with the treadmill. The laser power was ~66 mW at 935 nm, and the imaging depth was 150–280 μm beneath the dura. For the photodamage quantification through calcium imaging of soma ensemble (Supplementary Fig. 7a), the head-restrained mice were running freely with no air puff applied. The laser power was ~125 mW at 935 nm, and the imaging depth was 370–500 μm beneath the dura. For the photodamage quantification through calcium imaging of neuron ensemble (Supplementary Fig. 7c), the head-restrained mice were kept standing in a plastic tube. The laser power was ~103 mW at 935 nm, and the imaging depth was 350–390 μm beneath the dura.
Neutrophil imaging in mouse cerebral cortex and mouse ear
To image neutrophils trafficking in mouse cerebral cortex, the Lyz2gfp/+ B6.Albino mice16 were first anesthetized (isoflurane, 1.5–2%), and a small area craniotomy was performed over the left S1. Astrocytes were stained by applying SR101 solution23 for 2–3 mins before window installation. The mice were kept warm and anesthetized during the imaging. The laser power was ~90 mW (at 935 nm). The imaging of neutrophils in pial veins was performed at 0–60 μm beneath the dura, and the imaging of neutrophils in the capillaries of mouse cortex was performed at 50–150 μm beneath the dura.
For neutrophil imaging in mouse ear, the Lyz2gfp/+ B6.Albino mice were first anesthetized (isoflurane, 1.5–2%), and tail-vein injected with Qtracker® 655 to label the blood vessels. Another mouse strain, DsRed+/− Lyz2gfp/+ B6.Albino mice16 were used to visualize the blood vessel structure, without injecting blood tracer. Mild tissue damage was induced in the ear pinnae of the anesthetized mice before they were immobilized on a homemade stage. The mice were kept warm and anesthetized during the imaging. For the imaging of neutrophil trafficking in the ear vasculature of a Lyz2gfp/+ B6.Albino transgenic mouse (Supplementary Fig. 12 and Supplementary Video 17), the volume rate was 37 Hz (90 mW at 935 nm). For the 3D imaging of neutrophils rolling along the mouse ear vasculature of a Lyz2gfp/+ B6.Albino transgenic mouse (Supplementary Video 18), the volume rate was 19 Hz (100 mW at 935 nm). For the 3D imaging of neutrophils rolling along the mouse ear vasculature and extravascular tissue (Supplementary Videos 19 and 20), the volume rate was 3.5 Hz (95 mW at 935 nm). The vascular systems in the above data sets were 50–250 μm beneath the mouse ear surface.
Dendritic cell and lymphocyte cell imaging in mouse lymph nodes
CD11c-EYFP17 or Tbet:ZsGreen18 transgenic mice (for dendritic cell and lymphocyte cell imaging, respectively), both from Taconic-NIH mouse exchange program, were anesthetized by continuous inhalation of isoflurane and immobilized on a homemade stage, after which the popliteal lymph node was carefully exposed surgically. During the imaging, the mice were kept warm by a temperature-regulated heating pad and under continuous anesthesia. The laser power was ~100 mW (at 935 nm). The motility and fluorescence of the labeled cells did not exhibit apparent alteration during the imaging that typically lasted a few minutes. The imaging depth was 10–250 μm beneath the surface of the popliteal lymph nodes.
Microglia imaging
Craniotomy was performed on the skull (2 mm posterior and 2 mm lateral to the Bregma point) of adult CX3CR1gfp/gfp mice20 under isoflurane anesthesia (1.5–2%). To image the microglia and astrocyte simultaneously, astrocyte was stained with SR101 solution for 2–3 mins before window installation. Then the mice were placed on a regulated heating pad (37 °C) and kept anesthetized while we monitored the dynamics of microglia. To visualize the blood flow, the mice were tail vein injected with Qtracker® 655 (Life Technologies) for blood plasma staining. To study the microglia’s response to neuron damage, R-GECO (Addgene) was injected 2~3 weeks before the craniotomy (same as the protocol of GCaMP6 virus injection, except for the injection position). The neuron was damaged via laser ablation24. The laser power for imaging microglia was ~100 mW at 935 nm. The microglia ensembles activated by BBB disruption (Fig. 3f) were 100–140 μm beneath the dura. The volumetric imaging of microglia and neuron ensembles (Supplementary Videos 25–29) was 60–200 μm beneath the dura.
Image analysis
To correct motion artifacts, motion registrations were carried out with a cross correlation algorithm25. We manually chose the stacks with no apparent motion, and used the average intensity projections of these stacks along the time axis as the reference. Generally, we registered the MIPs of the volume (in x-y and x-z planes) at each time point with respect to MIPs of the reference, then aligned the volume accordingly.
To define the region-of-interest in the imaging of dendrites and dendritic spines, we computed the standard deviation in time for every pixel in the volume. Reconstruction was performed in a semiautomated fashion using the Simple Neurite Tracer plugin in ImageJ. Reconstructions were manually inspected for accuracy. The three-dimensional masks for each spine and dendritic segment were generated after the reconstructions. We then computed the average ΔF/F in each mask-defined volume.
To measure the neuronal activity of the soma ensemble, we performed MIP along the time axis of the 4D data after the image alignment. We detected and segmented the cells, as in the detection of mGRASP26. Each cell was segmented based on the seeded region growing method27. Using the cell’s mask, we were able to calculate the cell’s time domain response.
We further processed the calcium dynamics of the neuron ensemble in M1 cortex using the following procedures. We performed meta-k-means clustering and Pearson’s correlation coefficient calculations (between any two calcium traces of interest), to identify the neurons of strong correlation with each other and with running (correlation coefficient > 0.7). Then we calculated the correlations of the mean activity trace of these neurons with that of each neuron, to distinguish the neurons with significant (p < 0.05, t-test) positive correlation to running. We sorted the neurons first into their respective clusters and then within each cluster by their correlation to the cluster’s mean activity28 (Supplementary Fig. 10c).
For tracking the dendritic cells in lymph nodes, we selected a seed point for each cell of interest. Then we segmented the cell body around the seed point by means of the seeded region growing method27.
We used Amira (FEI visualization sciences group) and Imaris (Bitplane) to prepare the figures and supplementary videos.
Supplementary Material
Acknowledgments
L.K. and M.C. thank W. Gan and H. Dana for many helpful discussions, instructions and suggestions on mouse cerebral cortex imaging, T.-W. Chen for helps on data analysis in visual cortex experiments, S. Peron and N.-L. Xu (Janelia) for providing GCaMP6 labeled animals during system calibration, K. Ritola (Janelia) for preparing GCaMP6 virus, K. Morris and J. Rouchard for animal surgeries and GCaMP6 virus injections, J. Liu for help on cluster computing, P. Keller for suggestions on data storage, B.-C. Chen and W. Legant for suggestions on data visualization, V. Iyer for support on ScanImage, D. Milkie for developing the custom Labview program, S. Sawtelle and L. Ramasamy for designing and making the low noise high speed comparator, V. Goncharov for testing high speed photo-detectors, J. Freeman and F. Amat for help on data analysis, Howard Hughes Medical Institute for funding the project. This work was supported in part by the Intramural Program of National Institute of Allergy and Infectious Diseases, US National Institutes of Health (to R.N.G.), and US National Institutes of Health P41 E8015903-02S1 (to C.P.L.).
Footnotes
Author Contributions
M.C. devised the high speed volumetric imaging method based on OPLUL and designed both the hardware and software. L.K. provided critical feedback to the system design. L.K. and M.C. built and calibrated the system. L.K. and J.P.L. designed the calcium imaging of dendrites of V1 cortex. L.K. designed and performed calcium imaging of mouse cerebral cortex. J.T., L.K., T.L. and R.N.G. designed neutrophil experiments. C.P.L. designed the blood flow imaging. J.T. and R.N.G. designed the lymph node imaging. J.T. and L.K. performed lymph node and neutrophil imaging. L.K. designed and performed microglia imaging. L.K., Y.Y., J.T. and J.P.L. performed 3D data rendering and analysis. All authors contribute to the writing of the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Germain RN, Miller MJ, Dustin ML, Nussenzweig MC. Nat Rev Immunol. 2006;6:497–507. doi: 10.1038/nri1884. [DOI] [PubMed] [Google Scholar]
- 2.Grewe BF, Helmchen F. Curr Opin Neurobiol. 2009;19:520–529. doi: 10.1016/j.conb.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Göbel W, Kampa BM, Helmchen F. Nat Methods. 2006;4:73–79. doi: 10.1038/nmeth989. [DOI] [PubMed] [Google Scholar]
- 4.Grewe BF, Langer D, Kasper H, Kampa BM, Helmchen F. Nat Methods. 2010;7:399–405. doi: 10.1038/nmeth.1453. [DOI] [PubMed] [Google Scholar]
- 5.Cheng A, Gonçalves JT, Golshani P, Arisaka K, Portera-Cailliau C. Nat Methods. 2011;8:139–142. doi: 10.1038/nmeth.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Nat Methods. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- 7.Schrodel T, Prevedel R, Aumayr K, Zimmer M, Vaziri A. Nat Methods. 2013;10:1013–1020. doi: 10.1038/nmeth.2637. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard MB, et al. Nat Photonics. 2015;9:113–119. doi: 10.1038/nphoton.2014.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang J, Germain RN, Cui M. Proc Natl Acad Sci USA. 2012;109:8434–8439. doi: 10.1073/pnas.1119590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denk W, Strickler JH, Webb WW. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 11.Botcherby EJ, et al. Proc Natl Acad Sci USA. 2012;109:2919–2924. doi: 10.1073/pnas.1111662109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katona G, et al. Nat Methods. 2012;9:201–208. doi: 10.1038/nmeth.1851. [DOI] [PubMed] [Google Scholar]
- 13.Reddy GD, Kelleher K, Fink R, Saggau P. Nat Neurosci. 2008;11:713–720. doi: 10.1038/nn.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLeod E, Hopkins AB, Arnold CB. Opt Lett. 2006;31:3155–3157. doi: 10.1364/ol.31.003155. [DOI] [PubMed] [Google Scholar]
- 15.Fan Z, et al. Nat Med. 2010;16:718–722. doi: 10.1038/nm.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lämmermann T, et al. Nature. 2013;498:371–375. doi: 10.1038/nature12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindquist RL, et al. Nat Immunol. 2004;5:1243–1250. doi: 10.1038/ni1139. [DOI] [PubMed] [Google Scholar]
- 18.Zhu J, et al. Immunity. 2012;37:660–673. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kastenmüller W, Torabi-Parizi P, Subramanian N, Lämmermann T, Germain RN. Cell. 2012;150:1235–1248. doi: 10.1016/j.cell.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung S, et al. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otsu Y, et al. J Neurosci Methods. 2008;173:259–270. doi: 10.1016/j.jneumeth.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 22.Chen TW, et al. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nimmerjahn A, Kirchhoff F, Kerr JN, Helmchen F. Nat Methods. 2004;1:31–37. doi: 10.1038/nmeth706. [DOI] [PubMed] [Google Scholar]
- 24.Nimmerjahn A, Kirchhoff F, Helmchen F. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 25.Guizar-Sicairos M, Thurman ST, Fienup JR. Opt Lett. 2008;33:156–158. doi: 10.1364/ol.33.000156. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, et al. Nat Methods. 2012;9:96–102. [Google Scholar]
- 27.Adams R, Bischof L. IEEE Trans Pattern Anal Mach Intell. 1994;16:641–647. [Google Scholar]
- 28.Dombeck DA, Graziano MS, Tank DW. J Neurosci. 2009;29:13751–13760. doi: 10.1523/JNEUROSCI.2985-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.