Summary
Unrestrained receptor tyrosine kinase (RTK) signaling and epigenetic deregulation are root causes of tumorigenesis. We establish linkage between these processes by demonstrating that aberrant RTK signaling unleashed by oncogenic HRasG12V or loss of negative feedback through Sprouty gene deletion remodels histone modifications associated with active typical and super-enhancers. However, while both lesions disrupt the Ras-Erk axis, the expression programs, enhancer signatures, and transcription factor networks modulated upon HRasG12V-transformation or Sprouty deletion are largely distinct. Oncogenic HRasG12V elevates histone 3 lysine 27 acetylation (H3K27ac) levels at enhancers near the transcription factor Gata4 and the kinase Prkcb, as well as their expression levels. We show that Gata4 is necessary for the aberrant gene expression and H3K27ac marking at enhancers, and Prkcb is required for the oncogenic effects of HRasG12V-driven cells. Taken together, our findings demonstrate that dynamic reprogramming of the cellular enhancer landscape is a major effect of oncogenic RTK signaling.
Introduction
Enhancers are collections of DNA motifs that govern gene expression at long distances from transcriptional start sites, establishing cellular identity. Enhancers contain binding sites for sequence-specific transcription factors (TFs) and associated cofactors that assemble in a combinatorial manner to promote cell-type specific gene expression patterns (Spitz and Furlong, 2012). Enhancer dysfunction contributes to disease progression and occurs through mutation of enhancer regulatory factors, such as P300 and CBP in Rubinstein-Taybi syndrome, as well as through changes in the underlying enhancer DNA sequence, such as rearrangement of the IgH enhancer aberrantly activating c-MYC in Burkitt's Lymphoma (Smith and Shilatifard, 2014). This underscores the importance of understanding how enhancers function in normal development and disease.
Histone modification signatures can be used to classify enhancers. Primed and poised enhancers are identified by the presence of histone 3 lysine 4 mono-methylation (H3K4me1) and lack of histone 3 lysine 4 tri-methylation (H3K4me3), while active enhancers are marked with histone 3 lysine 27 acetylation (H3K27ac) and H3K4me1 (Creyghton et al., 2010; Heintzman et al., 2007; Rada-Iglesias et al., 2011). The deposition of H3K4me1 by MLL2 and MLL3, and H3K27ac by p300 and CBP, is dynamically regulated (Brown et al., 2014; Herz et al., 2012; Kaikkonen et al., 2013; Ostuni et al., 2013; Tie et al., 2009). For example, upon activation of NF-κB signaling, primed enhancers transition to an active state by gaining H3K27ac at regions with pre-existing H3K4me1. At a subset of unmodified regions, inducible deposition of H3K4me1 and H3K27ac occurs at latent or de novo enhancers (Kaikkonen et al., 2013; Ostuni et al., 2013). Super-enhancers (SE), which are similar to locus control regions or stretch enhancers, are an additional class of regulatory regions that contain clusters of typical enhancers (TE) and extend over several kilobases of the genome (Smith and Shilatifard, 2014). SEs are disproportionately marked with H3K27ac, are preferentially occupied by enhancer-associated factors including bromodomain and extra-terminal domain (BET) coactivator proteins such as BRD4, and control transcriptional regulators and fate-determining genes in normal and malignant cells (Loven et al., 2013; Whyte et al., 2013). During the inflammatory response, SEs are rapidly modified as NF-κB directs BRD4 redistribution at SEs (Brown et al., 2014). Although controlled pathway activation triggers dynamic chromatin remodeling at enhancers, how oncogenic signaling globally remodels enhancers has not been extensively studied.
While stimuli can elicit chromatin remodeling and TF assembly at enhancers and promoters, the rapid activation of signaling pathways precedes transcriptional responses. Receptor tyrosine kinase (RTK) signaling pathways are one example of a critical signaling network that is required for normal development and is misregulated in disease. Fibroblast growth factor (FGF)-mediated RTK activation triggers the Ras-ERK signaling cascade, dictating whether a cell will divide, survive, migrate, or differentiate (Turner and Grose, 2010). Mutations in signaling effectors including RAS, BRAF, and PIK3CA, which are among the most commonly mutated genes in cancer, unleash critical RTK pathways including the Ras-ERK and PI3K-AKT signaling cascades to promote tumorigenesis (Kandoth et al., 2013; Stephen et al., 2014). RTK signaling cascades are also regulated by feedback loops that promote or limit pathway activation. Sprouty genes (Spry1,2,3,4) encode RTK feedback inhibitors required for development of the kidney, inner ear, and other organs (Basson et al., 2005; Edwin et al., 2009; Shim et al., 2005). Spry proteins primarily silence the Ras-ERK pathway, while also antagonizing the PI3K-AKT and PLCγ-PKC pathways (Akbulut et al., 2010; Hacohen et al., 1998; Schutzman and Martin, 2012). Spry proteins have tumor suppressor activity and their expression levels are commonly downregulated in cancer, leading to aberrant amplification of RTK pathways, while their re-expression inhibits malignant growth (Masoumi-Moghaddam et al., 2014). As such, it is important to understand how unrestrained RTK signaling mediated by mutant oncogenes or Spry disruption coordinates changes in gene expression to promote malignant transformation.
The Ras-ERK signaling axis in part regulates gene expression through control of activating and repressive epigenetic mechanisms. Active ERK1/2 directly binds DNA, controls RNA polymerase II, and works in concert with TFs such as ELK1 to modulate gene expression (Goke et al., 2013; Hu et al., 2009; Tee et al., 2014). ERK1/2 indirectly impinges on chromatin by controlling the activity of MSK1/2, which is responsible for histone 3 serine 10 and serine 28 phosphorylation (H3S10ph and H3S28ph), and p300 (Chen et al., 2007; Soloaga et al., 2003). The Ras-Raf axis also activates the INK4A-ARF locus through upregulation of the histone demethylase JMJD3, leading to loss of histone 3 lysine 27 tri-methylation (H3K27me3) (Agger et al., 2009; Barradas et al., 2009). These examples of the interaction between RTK signaling and chromatin modifications, and their importance in tumorigenesis, led us to investigate whether unrestrained RTK signaling driven by loss of feedback regulation or mutant oncogene expression reprograms enhancer-associated chromatin modifications. In this study, we found that chronic Ras-Erk signaling mediated by Spry loss leads to inappropriate gene activation, which correlates with dynamic changes in H3K27ac at SEs and TEs. Constitutive HRasG12V, KRasG12V, or BRafV600E activation also leads to aberrant H3K27ac marking at SEs and TEs. However, the deregulated enhancers, target genes, and TF networks affected by oncogenic activation and loss of feedback regulation largely differ. Using the deregulated HRasG12V enhancer chromatin signature, we identified Gata4 as a key deregulated transcriptional regulator and Prkcb as a critical downstream kinase mediating the aberrant gene expression and oncogenic effects of HRasG12V-transformed cells, respectively. Our work suggests that unrestrained RTK activation modulates gene expression and contributes to malignant transformation through enhancer deregulation.
Results
Spry loss persistently activates Erk signaling and deregulates gene expression
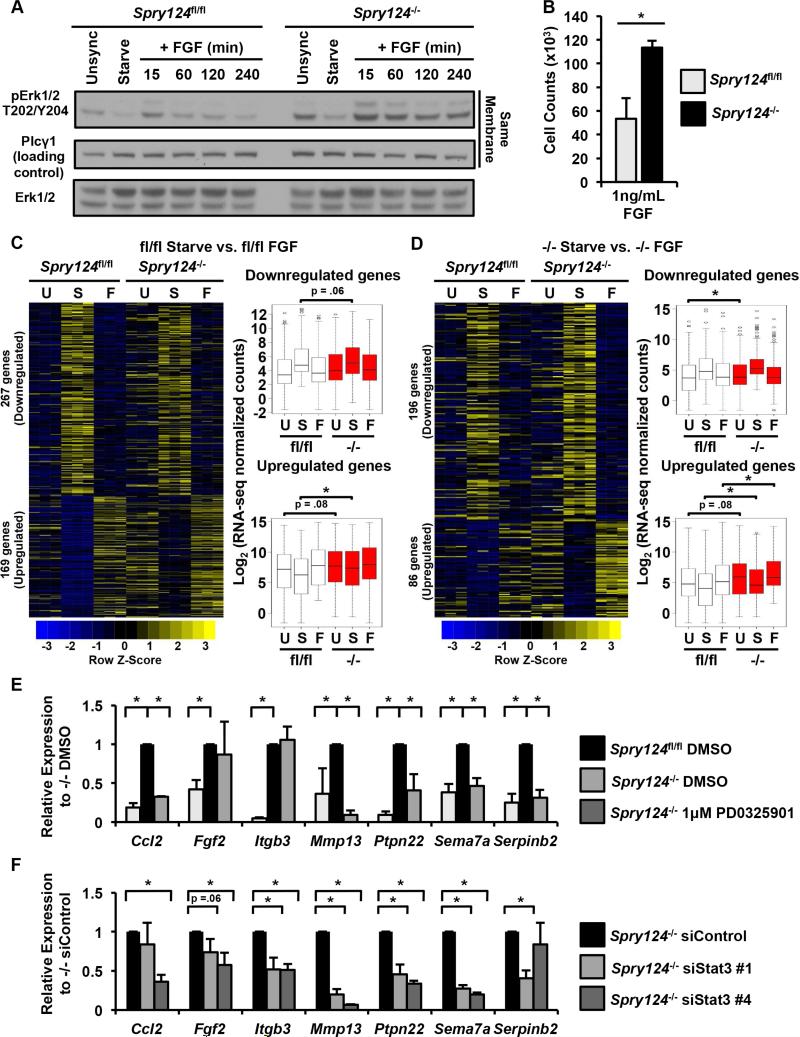
To assess the consequences of Spry1,2,4 loss on Ras-Erk signaling and gene expression, we compared immortalized mouse embryonic fibroblasts (MEFs) with wild-type Spry1,2,4 expression and genetically matched cells with Spry1,2,4 knocked out. This model was created by transducing Spry1,2,4flox/flox MEFs with either a control adenovirus empty vector (EV) Spry1,2,4 wild-type MEFs referred to as Spry124fl/fl) or an adenovirus expressing Cre to delete Spry1,2,4 (Spry1,2,4 deficient MEFs referred to as Spry124−/−) (Akbulut et al., 2010) (Figure S1A). We found that Spry124−/− MEFs exhibited elevated active, phosphorylated Erk at baseline in unsynchronized and serum starved states, relative to Spry124fl/fl MEFs (Figure 1A). Spry124−/− MEFs also displayed elevated Erk activation 15-60 minutes after FGF treatment, which persisted for 240 minutes, a time-point in which Erk activation returned to baseline levels in Spry124fl/fl MEFs. These molecular differences correlated with phenotypic characteristics of Spry124−/− MEFs, including more rapid proliferation in low FGF conditions and increased cell cycle entry in response to FGF after serum deprivation (Figures 1B and S1B). Consistent with previous data, our results indicate that Spry loss amplifies FGF-mediated Ras-Erk signaling (Hacohen et al., 1998; Shim et al., 2005).
Figure 1. Chronic Erk signaling is required for the aberrant gene expression upon Spry loss.
(A) Immunoblot analysis of Spry124fl/fl and Spry124−/− MEFs that were freely growing (Unsync), serum starved (Starve), or serum starved and treated with FGF over the time-course indicated.
(B) Cell counts of Spry124fl/fl and Spry124−/− MEFs cultured in the presence of FGF.
(C-D) RNA-seq analysis of Spry124fl/fl and Spry124−/− MEFs under unsynchronized (U), starved (S), and FGF (F) treated states. Heatmaps and boxplots depict the significantly differentially expressed genes comparing Spry124fl/fl (C) and Spry124−/− (D) MEFs starve and FGF treated conditions.
(E) Relative mRNA levels of Spry124−/− target genes upon treatment of Spry124−/− MEFs with DMSO or PD0325901. mRNA levels of Spry124fl/fl MEFs treated with DMSO are shown as a baseline reference.
(F) Relative mRNA levels of Spry124−/− target genes upon treatment of Spry124−/− MEFs with control or Stat3 siRNAs.
n = 3 for (A-F). In (A) a representative biological replicate is shown. In (B), (E), and (F) the values depict the mean + SD of biological replicates. In (C-D) each replicate per condition is shown in the heatmap, and the replicates are averaged in the boxplots. The p-values were calculated by a two-tailed t test in (B), (E), and (F) and wilcoxon rank sum test in (C-D); *p < 0.05.
To identify specific genes that may underlie the effects observed upon Spry loss, we performed RNA-sequencing (RNA-seq) comparing Spry124fl/fl and Spry124−/− MEFs. Given the increased baseline Erk activation in Spry124−/− MEFs, we first identified the differentially expressed genes in the unsynchronized states of Spry124fl/fl and Spry124−/− MEFs (Table S1). Ingenuity Pathway Analysis (IPA) revealed that these deregulated genes are involved in cancer and cardiovascular disease, and in cellular movement, morphology, and growth (Figure S1C). Gene set enrichment analysis (GSEA) also revealed that these deregulated genes are associated with breast cancer, signaling pathways, and chromatin regulators including PRC2 and MLL, suggesting that epigenetic deregulation is a consequence of persistent signaling upon Spry loss (Figure S1D).
To examine the signal-dependent transcriptional response in these cells, we identified the FGF-responsive genes in Spry124fl/fl MEFs and queried how they were regulated in Spry124−/− MEFs. In general, FGF-induced Spry124fl/fl genes were elevated in Spry124−/− MEFs in the unsynchronized and starved states, consistent with basal pathway activation prior to FGF treatment (Figure 1C and Table S1). We subsequently identified the FGF-responsive genes that were modulated in Spry124−/− MEFs (Figure 1D and Table S1). By contrast, FGF-induced Spry124−/− genes displayed significantly elevated expression levels in all cellular states in Spry124−/− MEFs. However, in both analyses, FGF-repressed Spry124fl/fl and Spry124−/− genes were largely overlapping in each comparison (Figures 1C-D). Therefore, we focused on Spry124−/− target genes that displayed both elevated baseline and FGF-induced activation. Many of these deregulated factors promote cellular migration (Itgb3, Fgf2, Mmp13, Serpinb2) and inflammation (Ccl2, Ptpn22, Sema7a). These seven Spry124−/− target genes displayed little to no activation in Spry124fl/fl MEFs, and were selected as a representative panel of Spry124−/− target genes for further assessment (Figure S1E). Treatment of Spry124−/− MEFs with a MEK inhibitor, PD0325901 (Barrett et al., 2008), led to a dose-dependent reduction in Erk activation, and reduced baseline expression of 5 out of 7 Spry124−/− target genes tested to Spry124fl/fl levels, indicating their dependence on Erk signaling (Figures 1E and S1F). We also noted that a number of Spry124−/− targets, including Ccl2 and Fgf2, are Stat3 target genes (Yu et al., 2009), suggesting that Stat3, an oncogenic TF activated by RTK signaling, may be aberrantly activated upon Spry loss. Accordingly, Spry124−/− MEFs displayed elevated constitutive phosphorylation of Stat3 tyrosine 705 and enhanced FGF-induced phosphorylation of Stat3 serine 727 (Figure S1G). Furthermore, Stat3 knockdown reduced the expression of all seven Spry124−/− target genes tested (Figures 1F and S1H). Collectively, these data indicate that a transcriptional program activated by Spry loss is driven by persistent Erk signaling and requires Stat3 activation.
Spry loss deregulates chromatin marking at enhancers
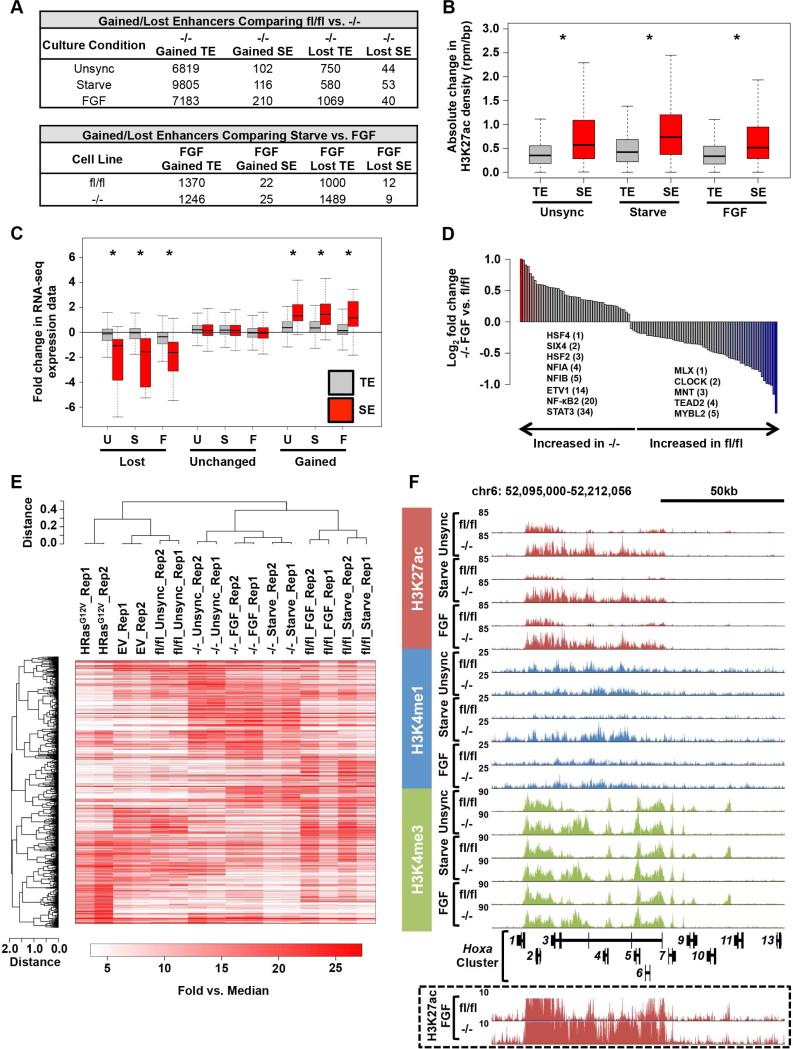
To explore the consequences of persistent Erk signaling on histone modifications associated with gene activation, we performed chromatin immunoprecipitation (ChIP) followed by next-generation sequencing (ChIP-seq) for the enhancer marks, H3K4me1 and H3K27ac, and the active promoter mark, H3K4me3. To identify TEs and SEs in each dataset, enhancer regions were rank ordered based on the extent of H3K27ac enrichment (Loven et al., 2013) (Figure S2A and Table S4). The median length and signal of H3K27ac at SEs is an order of magnitude greater than TEs. SEs accounted for 14-21% of the H3K27ac signal (Figure S2B). Comparing Spry124fl/fl and Spry124−/− MEFs in unsynchronized, starved or FGF-treated states, we observed significant gains and losses in H3K27ac at TEs and SEs (Figures 2A and S2C). The absolute change in H3K27ac signal between Spry124fl/fl and Spry124−/− MEFs was significantly greater at SEs than TEs (Figure 2B). Furthermore, gain or loss of a SE in Spry124−/− MEFs had a significantly greater effect on gene expression, than did changes at TEs (Figure 2C). This suggests that chromatin changes at SEs mediated the transcriptional effects of unrestrained signaling upon Spry loss. We therefore focused on the SE signatures, and examined the sequences at gained Spry124−/− SEs. This analysis revealed significant enrichment of binding sites for TFs such as NFIA/B (p < 10−6), ETS family members such as ETV1 (p < 0.001), NF-κB2 (p < 0.005), and STAT3 (p < 0.005) in Spry124−/− MEFs (Figure 2D and Table S6). This finding is consistent with the aberrant activation of NF-κB and Stat3 in Spry124−/− MEFs, requirement of Stat3 for expression Spry124−/− targets, and known role of NF-κB and STAT factors in enhancer remodeling (Brown et al., 2014; Ostuni et al., 2013) (Figures 1F, S1G, and S2D).
Figure 2. Spry loss globally reprograms enhancer-associated chromatin modifications.
(A) Table summarizing the number of H3K27ac defined enhancers significantly modulated between Spry124fl/fl and Spry124−/− MEFs in the indicated comparisons.
(B) Boxplot of the absolute change in H3K27ac density between Spry124fl/fl and Spry124−/− MEFs at TEs and SEs.
(C) Boxplot of RNA-seq expression for genes proximal to TEs and SEs that were gained, unchanged, or lost in Spry124−/− MEFs upon comparison between Spry124fl/fl and Spry124−/− MEFs under the indicated conditions.
(D) Bar plot of the ratio of TF motif density at gained Spry124−/− FGF SEs in the indicated comparison (p < 0.05). Colored bars represent TFs with p < 0.05 and fold change > 1.5. The fold change ranking of select TFs are indicated.
(E) Hierarchical clustering analysis of H3K27ac defined SE regions in the indicated MEFs. Each biological replicate (rep) is displayed.
(F) UCSC genome browser view of H3K27ac, H3K4me1, and H3K4me3 ChIP-seq binding density at the Hoxa cluster in Spry124fl/fl and Spry124−/− MEFs under the indicated culture conditions. H3K27ac binding density at an adjusted scale is depicted in the box.
n = 2 for (A-F). In (A-D) and (F) a representative biological replicate is shown. The p-values were calculated by a two-tailed t test; *p < 0.05.
We also examined whether enhancers were remodeled after FGF treatment (Figure 2A). Hierarchical clustering analyses of the SE signatures highlighted the high reproducibility among the biological replicates and demonstrated that the Spry124fl/fl and Spry124−/− SE signatures clustered separately with moderate changes across each cellular state (Figure 2E and Table S3). Baseline changes in SEs were largely maintained upon starvation and FGF treatment. For example, the oncogenic Hoxa cluster scored as a SE, displayed elevated H3K27ac enrichment in the presence or absence of FGF in Spry124−/− MEFs, which correlated with elevated expression of Hoxa3-5 and H3K4me3 promoter enrichment (Figure 2F and Table S1). Similarly, a broad region upstream of the inflammatory cytokine, Ccl2, displayed elevated H3K27ac enrichment in Spry124−/− MEFs and reached SE levels in most conditions, correlating with elevated expression of Ccl2 and the presence of H3K4me3 at its promoter (Figures S1E and S2E). While SEs were largely unresponsive to FGF treatment, there were significant changes at TEs associated with Spry124−/− target genes (Figures 2A and S2A). For example, in Spry124−/− MEFs, a TE downstream of Sema7a exhibited increased H3K27ac in the unsynchronized and starved states, which was further elevated upon FGF treatment. Again, these differences correlated with elevated Sema7a expression and increased H3K4me3 at its promoter (Figures S1E and S2E). These data indicate that Spry loss leads to significant changes in H3K27ac at critical enhancers in all cellular states.
Abrogating Erk signaling diminishes H3K27ac at enhancers, while oncogenic Ras and Raf promotes H3K27ac at enhancers
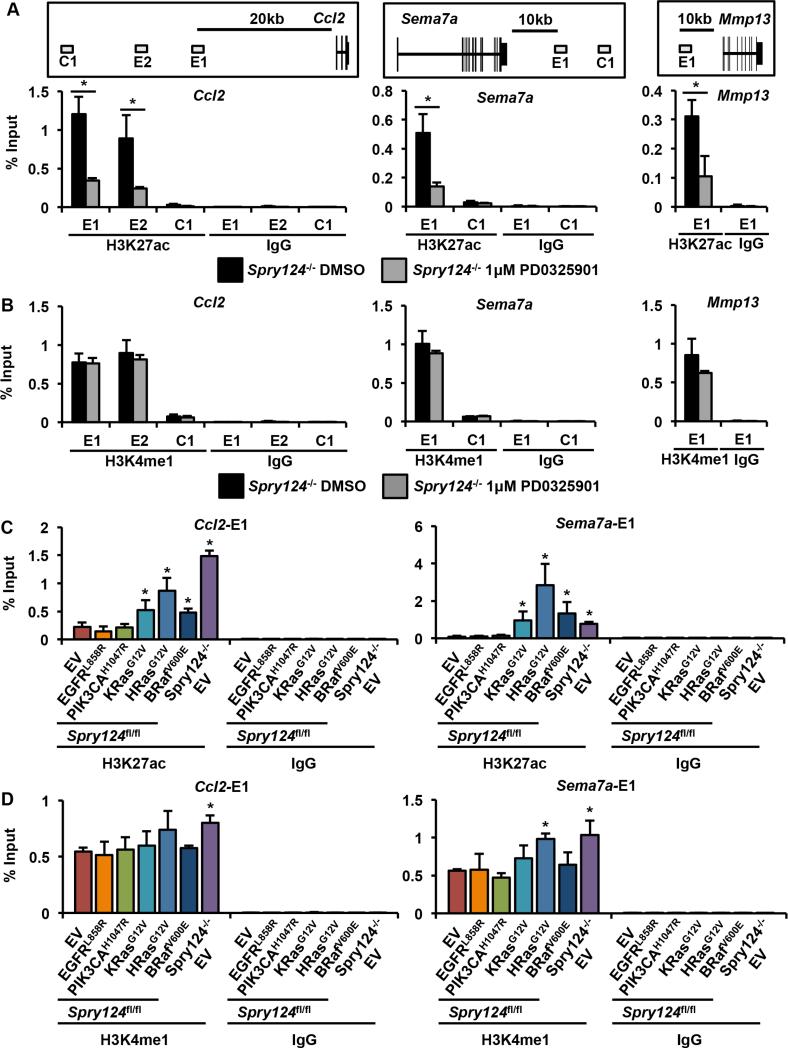
We next tested whether chromatin modifications at SEs and TEs were maintained after inhibiting aberrant Erk signaling. Treatment of Spry124−/− MEFs with PD0325901 significantly reduced H3K27ac levels at sites associated with Ccl2, Sema7a, and Mmp13, and decreased expression of these genes (Figures 1E and 3A). However, H3K4me1 was maintained at these sites, indicating that Erk signaling activates these primed enhancers by directing the deposition of H3K27ac (Figure 3B). Since Erk signaling was required to maintain H3K27ac, we predicted that BET bromodomain proteins and p300/CBP, which recognize and deposit H3K27ac, respectively, are necessary for the aberrant activation of Spry124−/− target genes. Accordingly, treatment of Spry124−/− MEFs with the BET bromodomain inhibitor, JQ1, which prevents association of BRD4 with TEs and SEs of oncogenes to inhibit cancer cell growth (Filippakopoulos et al., 2010; Loven et al., 2013), significantly reduced the elevated baseline and FGF-mediated expression of all seven Spry124−/− target genes tested to Spry124fl/fl levels (Figure S3A). In addition, treatment of Spry124−/− MEFs with the p300/CBP inhibitor, C646 (Bowers et al., 2010), significantly reduced the expression of 4 out of 7 Spry124−/− target genes tested upon serum deprivation, and significantly diminished FGF-mediated gene activation to Spry124fl/fl levels (Figure S3B). Collectively, our data indicates that aberrant deposition of H3K27ac at enhancers requires persistent Ras-Erk signaling and that BET bromodomain proteins and p300/CBP are necessary for aberrant gene activation upon Spry loss.
Figure 3. Inhibiting Erk signaling decreases H3K27ac at enhancers, while oncogenic activation of the Ras-Raf axis promotes H3K27ac at enhancers.
(A-B) ChIP-qPCR for H3K27ac (A) and H3K4me1 (B) at Ccl2, Sema7a, and Mmp13 enhancer (labeled E) and control (labeled C) regions upon treatment of Spry124−/− MEFs with DMSO or PD0325901. A schematic of primer regions is shown, which correspond to enriched Spry124−/− enhancer sites identified using ChIP-seq.
(C-D) ChIP-qPCR for H3K27ac (C) and H3K4me1 (D) at Ccl2 and Sema7a enhancer sites in the indicated MEFs. Schematic of the primer locations are shown in Figure 3A.
n = 3 for (A-D). In (A-D) the values depict the mean + SD of biological replicates. The p-values were calculated by a two-tailed t test; *p < 0.05.
To test the idea that Spry loss would resemble the effects of oncogenes that constitutively activate Ras-ERK and/or PI3K-AKT, we stably transduced Spry124fl/fl MEFs with an EV control or a panel of mutant oncogenes (EGFRL858R, PIK3CAH1047R, KRasG12V, HRasG12V, and BRafV600E) and surveyed the resulting expression and chromatin status of Spry124−/− target genes. Spry124−/− MEFs transduced with an EV served as a positive control. As expected, these oncogenes modulated Ras-Erk and PI3K-Akt signaling (Figure S3C). KRasG12V, HRasG12V, and BRafV600E significantly activated all seven Spry124−/− target genes tested (Figure S3D). Accordingly, KRasG12V, HRasG12V, and BRafV600E significantly increased H3K27ac levels at Spry124−/− activated enhancers (Figures 3C and S4A). The changes in enhancer marking were largely limited to H3K27ac, as a significant increase in H3K4me1 was only observed at the Mmp13 enhancer (Figures 3D and S4B). These changes also appeared to be specifically mediated by Ras-Erk signaling. EGFRL858R and PIK3CAH1047R, which preferentially activated Akt, did not stimulate changes in the expression or H3K27ac levels at Spry124−/− target genes. This data suggests that while oncogenic Ras and Raf modulate H3K27ac at shared subsets of Spry124−/− enhancers, oncogenes that primarily activate AKT may mediate their effects on cell fate and gene expression through other sets of enhancers.
HRasG12V-transformation modulates the enhancer landscape
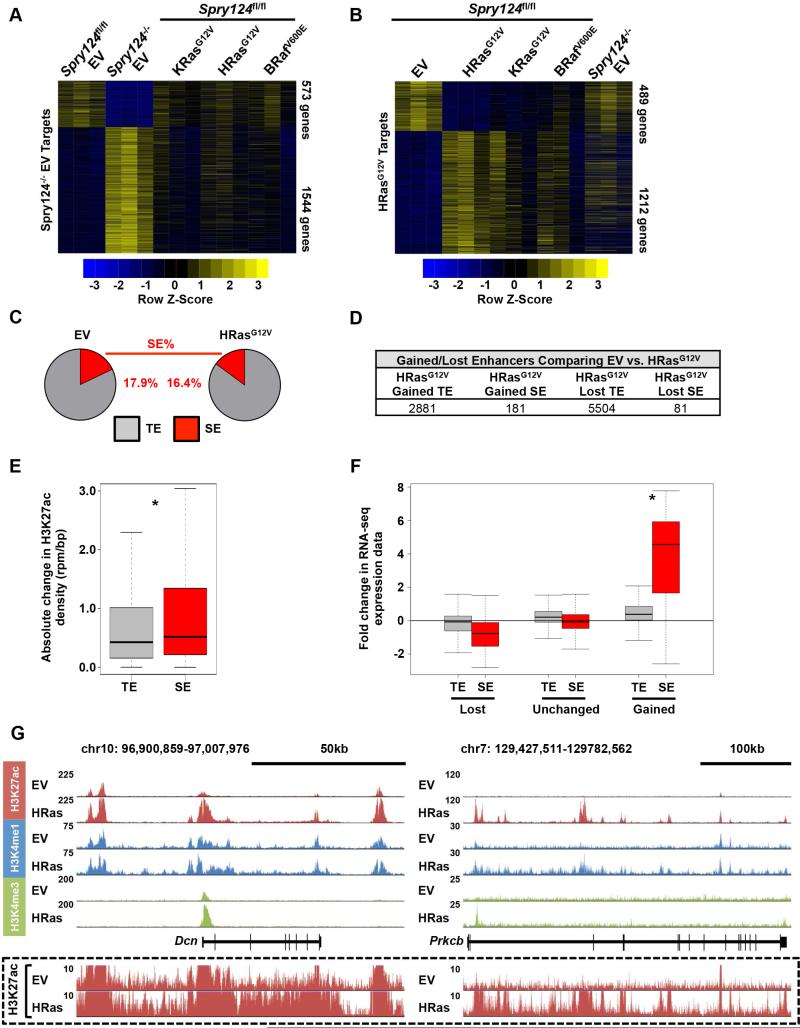
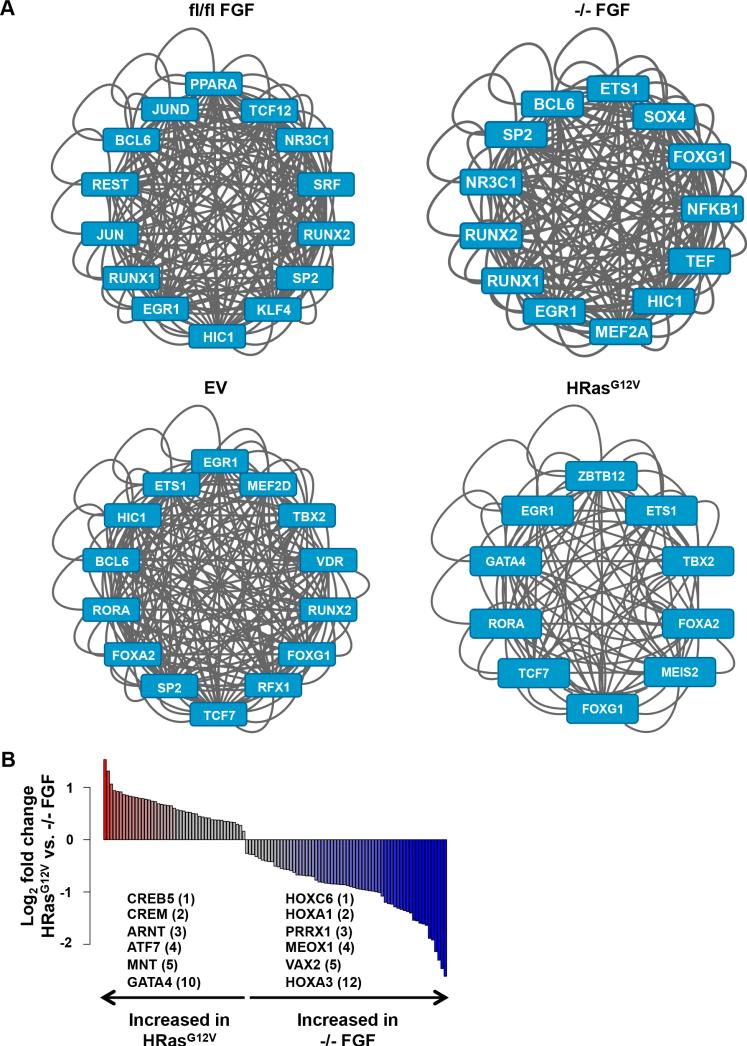
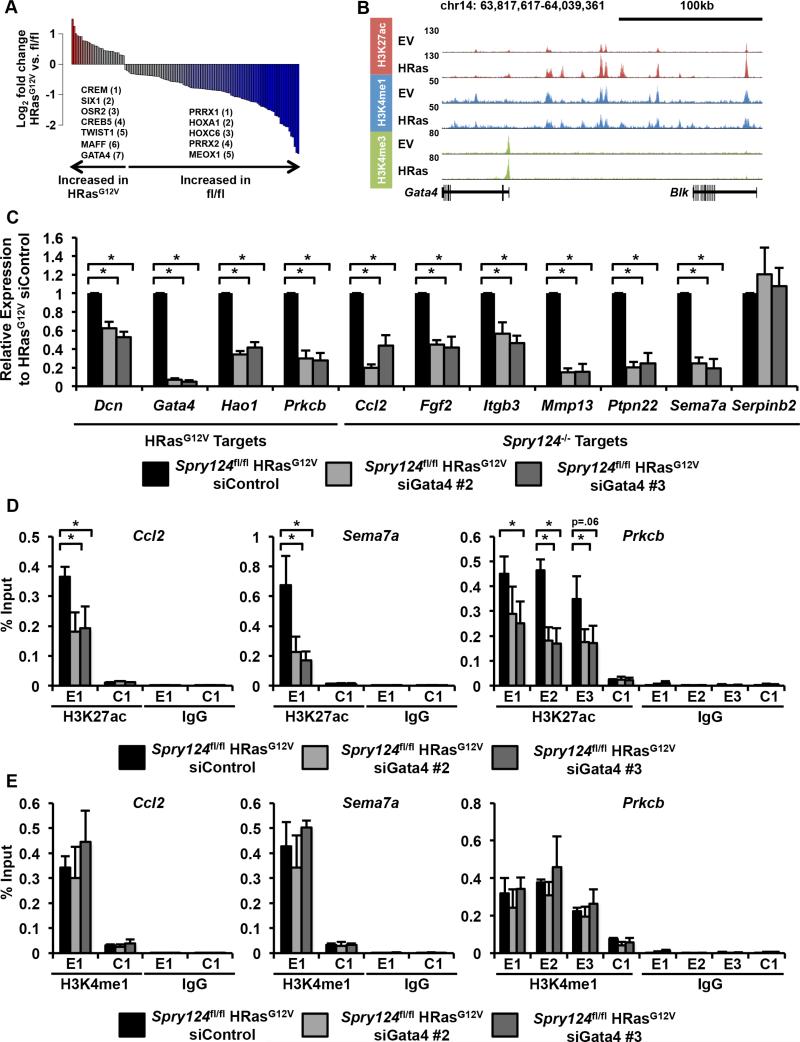
To determine the overlap of genes deregulated upon Spry loss and those modulated by oncogenes, we performed RNA-seq to identify genes differentially expressed in KRasG12V, HRasG12V, BRafV600E, and Spry124−/− EV MEFs, compared to Spry124fl/fl EV MEFs (Table S2). Surprisingly, the expression pattern of genes modulated in Spry124−/− EV MEFs was largely different from those altered by KRasG12V, HRasG12V or BRafV600E (Figure 4A). There was a high degree of overlap of genes deregulated in response to KRasG12V, HRasG12V, and BRafV600E (for example, KRasG12V targets overlapped with 94% of HRasG12V and 59% of BRafV600E targets), with each of these gene sets having relatively small overlap with Spry124−/− EV target genes (Spry124−/− EV targets overlapped with 23% of KRasG12V, 34% of HRasG12V, and 19% of BRafV600E targets) (Figure 4B and Table S2). We noted a core set of 290 deregulated target genes that overlapped in all datasets, which include the validated Spry124−/− target genes (Figure S4C). However, IPA revealed that many of the same top classes of genes such as cancer-associated genes and pathways such as cellular movement and proliferation, were shared among the oncogenic Ras, Raf, and Spry124−/− gene sets (Figure S4D). This suggests that despite the differences in target genes affected, all four lesions affected common core processes.
Figure 4. HRasG12V promotes global changes in chromatin marking at enhancers.
(A-B) Heatmaps from RNA-seq analyses representing the significantly differentially expressed genes comparing Spry124fl/fl EV and Spry124−/− EV MEFs (A) or Spry124fl/fl EV and HRasG12V MEFs (B).
(C) Pie charts displaying H3K27ac signal at TEs and SEs upon comparison between Spry124fl/fl EV and HRasG12V MEFs.
(D) Table summarizing the number of H3K27ac defined enhancers significantly modulated between Spry124fl/fl EV and HRasG12V MEFs.
(E) Boxplot of the absolute change in H3K27ac density between Spry124fl/fl EV and HRasG12V MEFs at TEs and SEs.
(F) Boxplot of RNA-seq expression for genes proximal to TEs and SEs that were gained, unchanged, or lost in HRasG12V MEFs upon comparison between Spry124fl/fl EV and HRasG12V MEFs.
(G) UCSC genome browser view of H3K27ac, H3K4me1, and H3K4me3 ChIP-seq binding density at the Dcn (left) and Prkcb (right) loci in Spry124fl/fl EV and HRasG12V MEFs. H3K27ac binding density at an adjusted scale is depicted in the box.
n = 3 for (A-B); n = 2 for (C-G). In (A-B) each biological replicate is shown. In (C-G) a representative biological replicate is shown. The p-values were calculated by a two-tailed t test; *p < 0.05.
The differences in gene expression patterns upon Spry loss and oncogene gain, led us to investigate the genome-wide consequences of oncogenic mutations of the Ras-Raf axis on enhancer marking. We performed ChIP-seq for H3K27ac, H3K4me1, and H3K4me3, comparing Spry124fl/fl EV and HRasG12V MEFs due to the potent effect of this oncogene on gene expression and H3K27ac at Spry124−/− activated enhancers (Figures 3C and S3D). Comparison of Spry124fl/fl EV and HRasG12V MEFs revealed that HRasG12V-transformation significantly remodeled SEs and TEs (Figures 4C-D, S4E-F, and Table S5). Consistent with the ChIP-qPCR, HRasG12V-transformation modified TEs of shared Spry124−/− target genes including Sema7a (Figures 3C and S5A). However, the absolute change in H3K27ac signal between Spry124fl/fl EV and HRasG12V MEFs was significantly greater at SEs than TEs (Figure 4E). Furthermore, genes that gained SEs upon HRasG12V-transformation had significantly increased expression levels, compared to genes that gained only TEs (Figure 4F). This result suggests that chromatin changes at SEs generated changes in gene expression upon HRasG12V-transformation. For example, HRasG12V-transformation resulted in significantly elevated H3K27ac levels at regions associated with the proteoglycan Dcn, the glycolate oxidase Hao1, and the kinase Prkcb, correlating with their elevated expression levels (Figures 4G, S5B, and S7A). We selected these factors as a representative set of HRasG12V target genes with deregulated SEs for further assessment, and focused on the SE signatures in the subsequent analyses. Collectively, this data demonstrates that HRasG12V globally remodels enhancer marking.
We next compared SEs remodeled upon Spry loss and HRasG12V-transformation, as well as the TF networks deployed by these lesions. Hierarchical clustering analyses revealed that the HRasG12V SE signature was distinct from the Spry124fl/fl EV, Spry124fl/fl, and Spry124−/− SE signatures, with only small subsets of SEs found in common (Figure 2E). By examining the SE sequences in these cell lines, we identified the top core regulatory TF network that is likely to bind and activate gene expression in each dataset (Figure 5A). ETS1 was identified as a core TF in both the Spry124−/− FGF and HRasG12V networks, consistent with the role of ETS factors as mediators of Ras-ERK signaling (Charlot et al., 2010). However, there were significant differences in the TF networks upon Spry loss or HRasG12V-transformation. For example, NF-κB1 and MEF2A were identified as core Spry124−/− FGF TFs, while GATA4 and MEIS2 were identified as core HRasG12V TFs. We also observed significant differences in TF binding sites enriched at activated SEs upon comparison of Spry124−/− FGF and HRasG12V MEFs (Figure 5B and Table S6). In particular, we observed enrichment of binding sites for ARNT (p < 0.01) and GATA4 (p < 0.001) upon HRasG12V-transformation, while binding sites for HOXA1-3 (p < 0.0001) were overrepresented upon Spry loss. To potentially explain the differences in gene expression upon Spry loss and HRasG12V-transformation, we examined the levels of phosphotyrosine species in these cells. Spry124−/− EV MEFs displayed increased baseline intensity of a ~60 kDa band, and a unique ~150-185 kDa FGF-induced band (Figure S5C). The larger band may represent phosphorylated FGFR, indicating that Spry loss may impact receptor activation at levels above Ras. There were also differences in the intensity of activated Erk and Akt in Spry124−/− EV MEFs (Figure S3C). Collectively, our data suggests that differences in the expression profiles and SE signatures upon Spry loss and HRasG12V-transformation may be due to quantitative and qualitative changes in the activated signaling molecules leading to modulation of different TF networks.
Figure 5. Spry loss and oncogenic HRasG12V modify distinct TF networks.
(A) The top ranking regulatory TF network identified at SEs in Spry124fl/fl FGF, Spry124−/− FGF, Spry124fl/fl EV, and Spry124fl/fl HRasG12V MEFs. Each TF depicted represents a node in the network.
(B) Bar plot of the ratio of TF motif density at gained HRasG12V SEs in the indicated comparison (p < 0.05). Colored bars represent TFs with p < 0.05 and fold change > 1.5. The fold change ranking of select TFs are indicated.
n = 2 for (A-B). In (A-B) a representative biological replicate is shown.
Gata4 is required for H3K27ac marking at enhancers
To identify TFs responsible for the expression and SE signatures upon HRasG12V-transformation, we identified TF binding sites enriched at activated HRasG12V SEs (Figure 6A and Table S6). Significant enrichment of binding sites for TFs such as TWIST1 (p < 0.01) was detected upon HRasG12V-transformation, consistent with the known role of oncogenic Ras in regulating and cooperating with Twist1 in oncogenesis (De Craene and Berx, 2013). We also noted significant enrichment of binding sites for GATA4 (p < 0.001) and identified GATA4 as a core HRasG12V TF (Figures 5A, 6A, and Table S6). Furthermore, in HRasG12V MEFs, we observed elevated Gata4 expression and a marked increase in H3K27ac at a broad TE upstream of Gata4, indicating that Gata4 may mediate the oncogenic Ras program (Figures 6B and S7A). Gata4, which was not expressed to any significant level in Spry124fl/fl MEFs, encodes a TF required for heart development, and Gata4 mutations cause cardiac dysfunction (Garg et al., 2003). Gata4 knockdown significantly reduced the expression of all four HRasG12V target genes and 6 out of 7 Spry124−/− target genes tested (Figures 6C and S6A). Gata4 knockdown led to a significant reduction in H3K27ac but not H3K4me1 at enhancers (Figures 6D-E and S6B). Consistent with the known role of Gata4 as a pioneer factor at enhancers (Cirillo et al., 2002), our data indicates that Gata4 is necessary for maintaining the aberrant H3K27ac marking and gene expression upon HRasG12V-transformation. Although Stat3 binding sites were not enriched at activated HRasG12V SEs, a Stat3 expression signature was detected in the HRasG12V RNA-seq datasets (Figure S6C). Stat3 knockdown significantly reduced the expression of all four HRasG12V target genes and 6 out of 7 Spry124−/− target genes tested, indicating that Stat3 is also required for aberrant gene activation (Figures S6D-E). Together, our data indicates that HRasG12V-driven modulation of H3K27ac at enhancers is dependent on Gata4 and Stat3.
Figure 6. Gata4 is required for HRasG12V expression programs and H3K27ac marking at enhancers.
(A) Bar plot of the ratio of TF motif density at gained HRasG12V SEs in the indicated comparison (p < 0.05). Colored bars represent TFs with p < 0.05 and fold change > 1.5. The fold change ranking of select TFs are indicated.
(B) UCSC genome browser view of H3K27ac, H3K4me1, and H3K4me3 ChIP-seq binding density at the Gata4 locus in Spry124fl/fl EV and HRasG12V MEFs.
(C) Relative mRNA levels of HRasG12V and Spry124−/− target genes upon treatment of HRasG12V MEFs with control or Gata4 siRNAs.
(D-E) ChIP-qPCR for H3K27ac (D) and H3K4me1 (E) at Ccl2, Sema7a, and Prkcb enhancer (labeled E) and control (labeled C) regions upon treatment of HRasG12V MEFs with control or Gata4 siRNAs. Schematic of the primer locations are shown in Figures 3A and S6B.
n = 2 for (A-B); n = 3 for (C-E). In (A-B) a representative biological replicate is shown. In (C-E) the values depict the mean + SD of biological replicates. The p-values were calculated by a two-tailed t test; *p < 0.05.
HRasG12V enhancer signature identifies Prkcb as a key downstream target gene
The enhancer dysfunction due to HRasG12V-transformation led us to investigate whether chemical inhibition of chromatin regulators or target genes with aberrantly marked enhancers could relieve the oncogenic effects of Ras. Consistent with our observations in Spry124−/− MEFs, treatment of HRasG12V MEFs with PD0325901 or JQ1 significantly reduced the expression of all four HRasG12V and all seven Spry124−/− target genes tested to Spry124fl/fl EV levels, indicating their dependence on Erk signaling and BET bromodomain activity (Figure S7A). Accordingly, JQ1 treatment significantly diminished the clonogenicity of HRasG12V MEFs (Figure S7B). JQ1 treatment reduced the viability of HRasG12V MEFs, although Spry124fl/fl EV MEFs were also sensitive to inhibition by JQ1 alone. However, a moderate synergistic effect was observed in HRasG12V MEFs upon co-treatment of PD0325901 and JQ1 at doses of 0.1-0.5 μM (Figure S7C). C646 treatment alone also significantly reduced the viability of HRasG12V MEFs, while combinations of C646 and PD0325901 did not reveal a synergistic effect on cell viability (Figure S7D). Our data indicates that BET bromodomain proteins are necessary for the aberrant gene expression, clonogenicity, and viability HRasG12V-transformed cells.
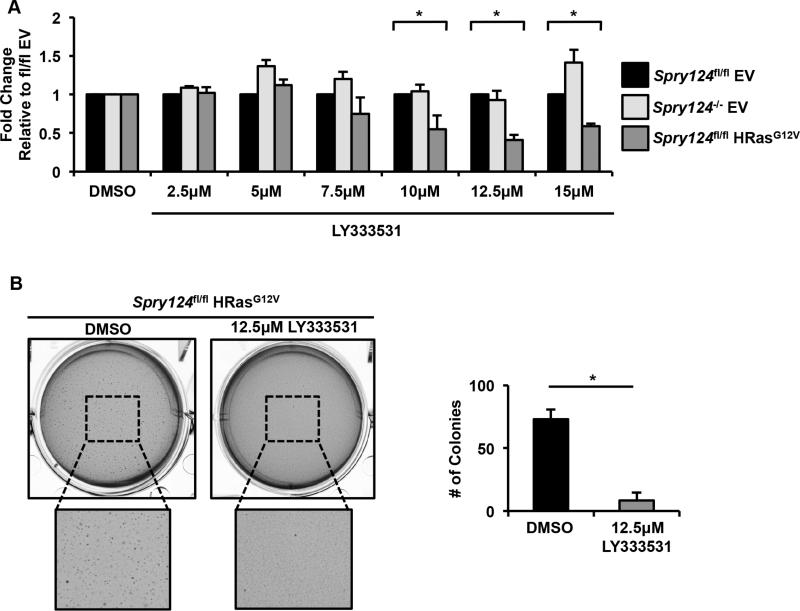
Finally, we explored whether enhancer signatures might reveal dependencies of HRasG12V-transformed cells. In particular, the Prkcb locus had dramatically elevated H3K27ac levels upon HRasG12V-transformation, correlating with significant overexpression (Figures 4G and S7A). Prkcb is frequently amplified in many malignancies and encodes a kinase that is considered a potential therapeutic target in cancer, diabetes, and heart disease (Mochly-Rosen et al., 2012). We hypothesized that HRasG12V MEFs would be more sensitive to LY333531, a clinically relevant Prkcb inhibitor also known as Ruboxistaurin (Jirousek et al., 1996), than cell lines lacking H3K27ac at the Prkcb locus. Indeed, HRasG12V MEFs were significantly more sensitive to doses of LY333531 ranging from 10-15 μM, compared to Spry124fl/fl EV or Spry124−/− EV MEFs (Figures 7A and S7E). Furthermore, LY333531 treatment significantly reduced the clonogenicity of HRasG12V MEFs (Figure 7B). Collectively, these data highlight that enhancer signatures can aid in the identification of key deregulated chromatin regulators and target genes that are contributing to the oncogenic phenotypes of HRasG12V-transformed cells.
Figure 7. HRasG12V-transformed cells are sensitive to Prkcb inhibition.
(A) Viability of Spry124fl/fl EV, Spry124fl/fl HRasG12V, and Spry124−/− EV MEFs treated with DMSO or LY333531.
(B) Colony formation analysis of HRasG12V MEFs treated with DMSO or LY333531.
n = 3 for (A-B). In (A) the values depict the mean + SD of biological replicates. In (B) a representative well is shown and quantification represents counts per field from mean + SD of biological replicates. The p-values were calculated by a two-tailed t test; *p < 0.05 and fold change > 1.5 (A); *p < 0.05 (B).
Discussion
Epigenetic deregulation and aberrant activation of RTK signaling pathways drives tumorigenesis. However, the relationship between unrestrained RTK signaling and chromatin modifications at cis-regulatory elements remains to be fully elucidated. In this study, we contrasted the effects of loss of feedback regulation and oncogenic RTK signaling on changes in gene expression and enhancer-associated chromatin modifications. We found that Spry loss led to Erk-dependent changes in gene expression and H3K27ac deposition at enhancers of genes with key roles in oncogenesis such as the Hoxa cluster and Ccl2. We and others previously showed that Spry loss alters developmental processes such as branching morphogenesis of organs in response to RTK signaling (Basson et al., 2005; Edwin et al., 2009). Furthermore, decreased Spry expression, particularly Spry1 and Spry2, has been documented in many malignancies suggesting that it may play a role in pathogenesis by removing restraints on RTK signaling (Masoumi-Moghaddam et al., 2014). Our data suggests that Spry loss and persistent Erk signaling reprograms enhancer-associated chromatin modifications to deregulate gene expression.
Using Spry deficient cells, we aimed to identify general mechanisms relevant to oncogenic Rasand Raf driven cancers. Indeed, expression of KRasG12V, HRasG12V, and BRafV600E led to aberrant activation and H3K27ac marking at a subset of Spry124−/− targets. Early work showed that HRasG12V induces a more relaxed chromatin conformation (Laitinen et al., 1990), which is consistent with the increased H3K27ac at enhancers that we observed in response to oncogenic Ras and Raf. However, our study revealed that the majority of deregulated target genes, reprogrammed SEs, and deployed TF networks altered upon HRasG12V-transformation are distinct from those modulated upon Spry loss. This disparity may result from differences in the strength and duration of Ras-Erk activation in response to these perturbations. Alternatively, RTK and non-RTK pathways may be differentially activated in the contexts of Spry loss and HRasG12V-transformation. In accordance with the latter idea, we detected a unique FGF-induced ~150-185kda phosphotyrosine species in Spry deficient cells, which may reflect hyperactivation of the FGFR upon Spry loss. Spry proteins limit signaling at many points downstream of the RTK pathway, including the activation of Ras and Raf (Edwin et al., 2009), which may influence the resulting output of chromatin deregulation and aberrant gene expression. Oncogenic EGFRL858R and PIK3CAH1047R potently activated Akt but did not induce changes in the expression or chromatin marking at enhancers of Spry124−/− target genes, suggesting that EGFRL858R and PIK3CAH1047R regulate distinct subsets of genes and enhancers. It remains to be determined if aberrant PI3K-AKT signaling results in global chromatin changes, or whether our observations are specific to the Ras-ERK axis.
Due to the high frequency of RAS mutations in almost all forms of cancer and the lack of therapies targeting cancers driven by oncogenic Ras, the study of this oncogene has become a centerpiece of new basic and translational research efforts (Stephen et al., 2014). Tumors commonly harbor mutations in both RTK pathway components and chromatin regulators, and recent studies indicate that oncogenic Ras signaling mediated by loss of Nf1 cooperates with disruption chromatin regulators including Mll3 and Suz12 to accelerate oncogenesis (Chen et al., 2014; De Raedt et al., 2014; Kandoth et al., 2013). The BET bromodomain inhibitor JQ1 is highly efficacious in treating pre-clinical cancer models driven by aberrant Ras activation including non-small cell lung cancer, acute myeloid leukemia, and malignant peripheral nerve sheath tumors (Chen et al., 2014; De Raedt et al., 2014; Shimamura et al., 2013). In accordance with these studies, we found that JQ1 treatment repressed the aberrant transcriptional responses and clonogenicity of HRasG12V-transformed cells. In viability assays, control cells were more sensitive to JQ1 treatment than HRasG12V-transformed cells, which may be due to the high level of amplified RTK signaling and redistribution of H3K27ac at key target genes upon HRasG12V-transformation. Combining low doses of PD0325901 with JQ1 was moderately more potent than using each compound alone in HRasG12V-transformed cells. The lack of a more potent effect when combining these compounds may be due to their convergence on a shared set of target genes, or their sharp dose response curves such that suboptimal doses of the drugs fail to effectively inhibit gene expression. Investigation into the functional role of BET bromodomain proteins in oncogenic Ras-driven cancers will provide further insight into the mechanisms driving oncogenesis.
H3K27ac enhancer signatures are useful in the identification of aberrant oncogenic transcriptional programs and classification of malignant tissue (Chapuy et al., 2013). HRasG12V SE signatures and TF motif analyses highlighted Gata4 as a candidate TF promoting the oncogenic HRasG12V program. We demonstrate that Gata4 is required for the aberrant H3K27ac enhancer marking and expression changes upon HRasG12V-transformation. Previous studies showed that Gata4 is regulated by post-translational modifications including phosphorylation by kinases downstream of ERK (Liang et al., 2001). Our data suggests that Gata4 may be a misregulated pioneer factor downstream of oncogenic HRasG12V, activated through a feed-forward loop due to aberrant enhancer marking. We also identified Prkcb as a HRasG12V target susceptible to chemical inhibition, which promotes the viability and clonogenicity of HRasG12V-transformed cells. One limitation of our study is that the experiments were performed exclusively in MEFs. Therefore, further work will be required to evaluate the functional role of Gata4 and clinical significance of BET bromodomain and Prkcb inhibition in oncogenic Ras-driven epithelial cancers using appropriate cell and mouse models. In sum, our study shows that examination of histone modification signatures and identification of oncogene-regulated enhancers can yield insight into the key processes and targets that drive malignancy. Our data support a model in which unrestrained RTK signaling modulates gene expression through coordinated regulation of enhancers.
Experimental Procedures
Cell lines, culture conditions, and FGF treatment
Spry124fl/fl and Spry124−/− MEFs were cultured in DMEM containing 10% FBS as previously described (Akbulut et al., 2010). Unsynchronized MEFs were maintained in DMEM containing 10% FBS for 24 hours. Serum starved MEFs were maintained in DMEM containing 0.1% FBS for 24 hours. FGF-treated MEFs were maintained in DMEM containing 0.1% FBS for 20 hours after which 10 ng/mL FGF (Life Technologies) was directly added to the media for an additional 4 hours, unless otherwise noted. Additional details related to retroviral transduction, inhibitor treatments, siRNA transfection, protein and RNA isolation, and biological assays can be found in the Supplemental Experimental Procedures.
ChIP-qPCR and ChIP-seq
ChIP experiments for histone modifications were performed as described previously (Martinez-Garcia et al., 2011). ChIP-seq analyses were performed as described previously (Loven et al., 2013). Antibodies, primers, and detailed ChIP-seq analyses are described in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgements
This work was initiated under grants from the NIH (CA59998) and the Lynn Sage and Northwestern Memorial Foundations, and supported by a NCI Physical Sciences-Oncology Center grant (U54CA143869) (J.D.L.). This work was also supported by a NIH T32 training grant (GM08061) (B.N.), a Northwestern University Malkin Scholars Award (B.N.), a NIGMS T32 training grant (GM007288) (K.S.), and a Department of Defense CDMRP (CA120184) (C.Y.L.). We thank members of the Licht laboratory and Suzanne Nabet for helpful discussions and critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
B.N. and J.D.L designed the research and interpreted the data.
B.N. performed the experiments and received assistance from C.M.W., R.P., and T.E. P.O., K.S., and A.A.G. performed the RNA-seq analyses and contributed to the ChIP-seq analyses.
J.M.R., C.Y.L., and J.E.B. performed the ChIP-seq analyses.
B.N. and J.D.L. wrote the manuscript.
Accession Number
The GEO accession number for all the ChIP-seq and RNA-seq data is GSE64195.
References
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbulut S, Reddi AL, Aggarwal P, Ambardekar C, Canciani B, Kim MK, Hix L, Vilimas T, Mason J, Basson MA, et al. Sprouty proteins inhibit receptor-mediated activation of phosphatidylinositol-specific phospholipase C. Mol. Biol. Cell. 2010;21:3487–3496. doi: 10.1091/mbc.E10-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, Maertens G, Banck M, Zhou MM, Walsh MJ, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SD, Bridges AJ, Dudley DT, Saltiel AR, Fergus JH, Flamme CM, Delaney AM, Kaufman M, LePage S, Leopold WR, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg. Med. Chem. Lett. 2008;18:6501–6504. doi: 10.1016/j.bmcl.2008.10.054. [DOI] [PubMed] [Google Scholar]
- Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, Shakya R, Gross I, Martin GR, Lufkin T, McMahon AP, et al. Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev. Cell. 2005;8:229–239. doi: 10.1016/j.devcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem. Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, Bair S, Newton G, Lichtman AH, Kung AL, et al. NF-kappaB Directs Dynamic Super Enhancer Formation in Inflammation and Atherogenesis. Mol. Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlot C, Dubois-Pot H, Serchov T, Tourrette Y, Wasylyk B. A review of post-translational modifications and subcellular localization of Ets transcription factors: possible connection with cancer and involvement in the hypoxic response. Methods Mol. Biol. 2010;647:3–30. doi: 10.1007/978-1-60761-738-9_1. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Rappaport AR, Kitzing T, Schultz N, Zhao Z, Shroff AS, Dickins RA, Vakoc CR, Bradner JE, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25:652–665. doi: 10.1016/j.ccr.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Wang YN, Chang WC. ERK2-mediated C-terminal serine phosphorylation of p300 is vital to the regulation of epidermal growth factor-induced keratin 16 gene expression. J. Biol. Chem. 2007;282:27215–27228. doi: 10.1074/jbc.M700264200. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, Helin K, Hornick JL, Mautner V, Kehrer-Sawatzki H, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514:247–251. doi: 10.1038/nature13561. [DOI] [PubMed] [Google Scholar]
- Edwin F, Anderson K, Ying C, Patel TB. Intermolecular interactions of Sprouty proteins and their implications in development and disease. Mol. Pharmacol. 2009;76:679–691. doi: 10.1124/mol.109.055848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Goke J, Chan YS, Yan J, Vingron M, Ng HH. Genome-wide kinase-chromatin interactions reveal the regulatory network of ERK signaling in human embryonic stem cells. Mol. Cell. 2013;50:844–855. doi: 10.1016/j.molcel.2013.04.030. [DOI] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirousek MR, Gillig JR, Gonzalez CM, Heath WF, McDonald JH, 3rd, Neel DA, Rito CJ, Singh U, Stramm LE, Melikian-Badalian A, et al. (S)-13-[(dimethylamino)methyl]-10,11,14,15-tetrahydro-4,9:16, 21-dimetheno-1H, 13H-dibenzo[e,k]pyrrolo[3,4-h][1,4,13]oxadiazacyclohexadecene-1,3(2H)-d ione (LY333531) and related analogues: isozyme selective inhibitors of protein kinase C beta. J. Med. Chem. 1996;39:2664–2671. doi: 10.1021/jm950588y. [DOI] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen J, Sistonen L, Alitalo K, Holtta E. c-Ha-rasVal 12 oncogene-transformed NIH-3T3 fibroblasts display more decondensed nucleosomal organization than normal fibroblasts. J. Cell Biol. 1990;111:9–17. doi: 10.1083/jcb.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Wiese RJ, Bueno OF, Dai YS, Markham BE, Molkentin JD. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol. Cell. Biol. 2001;21:7460–7469. doi: 10.1128/MCB.21.21.7460-7469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L, Heffner A, Will C, Lamy L, Staudt LM, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117:211–220. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoumi-Moghaddam S, Amini A, Morris DL. The developing story of Sprouty and cancer. Cancer Metastasis Rev. 2014;33:695–720. doi: 10.1007/s10555-014-9497-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochly-Rosen D, Das K, Grimes KV. Protein kinase C, an elusive therapeutic target? Nat Rev Drug Discov. 2012;11:937–957. doi: 10.1038/nrd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutzman JL, Martin GR. Sprouty genes function in suppression of prostate tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 2012;109:20023–20028. doi: 10.1073/pnas.1217204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim K, Minowada G, Coling DE, Martin GR. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev. Cell. 2005;8:553–564. doi: 10.1016/j.devcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Shimamura T, Chen Z, Soucheray M, Carretero J, Kikuchi E, Tchaicha JH, Gao Y, Cheng KA, Cohoon TJ, Qi J, et al. Efficacy of BET bromodomain inhibition in Kras-mutant non-small cell lung cancer. Clin. Cancer Res. 2013;19:6183–6192. doi: 10.1158/1078-0432.CCR-12-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. Enhancer biology and enhanceropathies. Nat. Struct. Mol. Biol. 2014;21:210–219. doi: 10.1038/nsmb.2784. [DOI] [PubMed] [Google Scholar]
- Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nature reviews. Genetics. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Tee WW, Shen SS, Oksuz O, Narendra V, Reinberg D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 2014;156:678–690. doi: 10.1016/j.cell.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.