Abstract
The centriole in eukaryotes functions as the cell's microtubule-organizing center (MTOC) to nucleate spindle assembly, and its biogenesis requires an evolutionarily conserved protein, SAS-6, which assembles the centriole cartwheel. Trypanosoma brucei, an early branching protozoan, possesses the basal body as its MTOC to nucleate flagellum biogenesis. However, little is known about the components of the basal body and their roles in basal body biogenesis and flagellum assembly. Here, we report that the T. brucei SAS-6 homolog, TbSAS-6, is localized to the mature basal body and the probasal body throughout the cell cycle. RNA interference (RNAi) of TbSAS-6 inhibited probasal body biogenesis, compromised flagellum assembly, and caused cytokinesis arrest. Surprisingly, overexpression of TbSAS-6 in T. brucei also impaired probasal body duplication and flagellum assembly, contrary to SAS-6 overexpression in humans, which produces supernumerary centrioles. Furthermore, we showed that depletion of T. brucei Polo-like kinase, TbPLK, or inhibition of TbPLK activity did not abolish TbSAS-6 localization to the basal body, in contrast to the essential role of Polo-like kinase in recruiting SAS-6 to centrioles in animals. Altogether, these results identified the essential role of TbSAS-6 in probasal body biogenesis and flagellum assembly and suggest the presence of a TbPLK-independent pathway governing basal body duplication in T. brucei.
INTRODUCTION
Trypanosoma brucei is an early branching microbial eukaryote and the causative agent of sleeping sickness in humans and nagana in cattle in sub-Saharan Africa. A trypanosome cell possesses a motile flagellum that is nucleated by the basal body, the cell's microtubule organizing center (MTOC), exits the cell body through the flagellar pocket, and is attached to the cell body along most of its length via a specialized cytoskeletal structure termed the flagellum attachment zone (FAZ). The flagellum is composed of a canonical 9 + 2 microtubule axoneme and an extra-axoneme structure termed the paraflagellar rod (PFR), and it is required for cell morphogenesis, cell motility, and cytokinesis (1, 2). Early in the cell cycle, a trypanosome cell possesses a basal body, which nucleates the flagellum axoneme, and an associated probasal body. The probasal body matures to a basal body when the cell proceeds to S phase of the cell cycle; subsequently, two probasal bodies are formed, each of which associates with each of the two mature basal bodies (3, 4). A new flagellum then is assembled from the newly matured basal body, which then undergoes a rotation toward the posterior of the old basal body. Following the elongation of the new flagellum, the new basal body/probasal body pair moves toward the posterior portion of the cell, which constitutes one of the first cytoskeletal events in the cell cycle of T. brucei (3, 4). Movement of the basal body is required for cell morphogenesis (4) and for segregation of the kinetoplast, the cell's mitochondrial genome that is attached to the basal body through a structure termed the tripartite attachment complex (5, 6).
The basal body adopts the same structure as the centriole, which is characterized by a 9-fold symmetrical array of microtubule triplets that emanates from the cartwheel located in the very proximal region of the centriole. The cartwheel has been well visualized by electron microscopy in some unicellular organisms, including Paramecium tetraurelia, Tetrahymena thermophila, and Chlamydomonas reinhardtii, and is composed of a central hub from which nine spokes emanate, radiate outwards, and connect with the A-microtubules of the microtubule triplets via a structure termed the pinhead (7–10). Previous work using Caenorhabditis elegans, Drosophila, and humans as the model systems has identified several key centriole components, including SPD-2/CEP192, ZYG-1/PLK4, SAS-5/Ana2/STIL, SAS-6, and SAS-4/CPAP, and delineated a conserved ZYG-1/PLK4-mediated regulatory pathway governing centriole assembly (11). In C. elegans, SPD-2 is required for centriole localization of ZYG-1, which in turn recruits the SAS-5–SAS-6 pair to centrioles, followed by the recruitment of SAS-4. In humans, PLK4 is required to recruit STIL and SAS-6 to centrioles, and the latter are needed for CPAP recruitment (11). Homologs of SAS-6 and SAS-4/CPAP are present in the majority of centriole-containing organisms, whereas homologs of SPD-2/CEP192, ZYG-1/PLK4, and SAS-5/Ana2/STIL appear to have been lost in many taxa (11–13). Although many additional components of centrioles have been identified in diverse organisms, a systematic bioinformatics analysis of 45 organisms discovered 14 ancestral centriole core components (13), and another study suggests that SAS-4/CPAP, SAS-6, and BLD10/CEP135 constitute the core ancestral module involved in centriole assembly (12). SAS-6 and BLD10/CEP135 form the cartwheel, with the former forming the cartwheel core (14, 15) and the latter forming the pinhead structure connecting the cartwheel spokes to the A-microtubules of the microtubule triplets (16, 17), whereas SAS-4/CPAP is involved in centriole microtubule elongation and stabilization (18–21).
Despite the essentiality of the basal body in T. brucei cell morphogenesis and cell division, our knowledge about its molecular composition and the regulatory pathway(s) governing its biogenesis still is limited. A few proteins have been localized to the basal body in T. brucei, including γ-tubulin (22, 23), TBBC (24), three centrins (23, 25, 26), T. brucei LRTP (TbLRTP) (27), and T. brucei NRKC (TbNRKC) (28). Previous bioinformatics analysis has identified the homologs of several conserved centriole components, including SAS-4/CPAP, SAS-6, and BLD10/CEP135, and also suggested the lack of homologs of SPD2/CEP192 and ZYG-1/PLK4 in T. brucei (12). Instead, T. brucei possesses a PLK1 homolog, TbPLK, which is localized to the basal body during early cell cycle stages (29, 30). However, whether TbPLK plays a role in basal body duplication is still controversial. One report suggested that TbPLK is required for basal body duplication (31), whereas other studies showed that TbPLK is required for segregation/rotation, but not duplication, of the basal body (30, 32).
We recently sought to identify the components of the basal body and to dissect the regulatory pathway(s) governing basal body duplication in T. brucei, and, as the first effort toward this goal, we characterized the T. brucei SAS-6 homolog, TbSAS-6, due to the fundamental role of its homologs from other organisms in establishing the cartwheel of centrioles. Our results identified an essential role of TbSAS-6 in probasal body biogenesis and flagellum formation and suggested the presence of a Polo-like kinase-independent pathway governing basal body biogenesis in T. brucei.
MATERIALS AND METHODS
Trypanosome cell culture and RNA interference (RNAi).
The procyclic 29-13 cell line was grown in SDM-79 medium supplemented with 10% fetal bovine serum (Atlanta Biologicals, Inc.), 15 μg/ml G418, and 50 μg/ml hygromycin. The procyclic 427 cell line was cultured at 27°C in SDM-79 medium supplemented with 10% fetal bovine serum. Cells were diluted once the cell density reaches 5 × 106 cells/ml.
For RNAi of TbSAS-6, a 500-bp DNA fragment from the coding region of TbSAS-6 was cloned into the pZJM vector (33). The resulting plasmid was electroporated into the 29-13 cell line according to our published procedures (34). Successful transfectants were selected under 2.5 μg/ml phleomycin and cloned by limiting dilution in a 96-well plate in SDM79 medium containing 15% fetal bovine serum and 2.5 μg/ml phleomycin. At least three clonal cell lines were selected for further analysis. To induce RNAi, the clonal cell lines each were induced by incubation with 1.0 μg/ml tetracycline, and cell growth was monitored daily by counting the cells with a hemacytometer.
Purification of recombinant TbSAS-6 and antibody production.
A 1,002-bp fragment corresponding to the N-terminal coding region (amino acids [aa] 1 to 334) of TbSAS-6 was PCR amplified from the genomic DNA and cloned into the pET26 vector for expressing a hexahistidine-fused TbSAS-6 truncation protein in Escherichia coli. The construct was transformed into E. coli BL21 cells, and recombinant His-tagged TbSAS-6 truncation protein was induced with isopropyl-β-d-thiogalactopyranoside (IPTG), purified through a nickel column, and used for immunizing rabbit to produce anti-TbSAS-6 antibody at Cocalico Biologicals, Inc. (Reamstown, PA). Crude antiserum was used directly for immunofluorescence microscopy and Western blotting.
Overexpression of TbSAS-6.
The full-length coding sequence of TbSAS-6 was PCR amplified from genomic DNA and cloned into pLew100-3HA and pLew100 vectors for expressing triple hemagglutinin (3HA) epitope-tagged TbSAS-6 or nontagged TbSAS-6. The constructs then were linearized with NotI and transfected into the 29-13 cell line, and successful transfectants were selected under 2.5 μg/ml phleomycin. Cells then were cloned by limiting dilution in 96-well plates. To induce TbSAS-6 overexpression, cells were incubated with 0.1 μg/ml, 0.5 μg/ml, and 1.0 μg/ml tetracycline, and cell growth was monitored by daily counting the cell number with a hemacytometer.
In situ epitope tagging of TbSAS-6.
For endogenous epitope tagging of TbSAS-6, a 978-bp DNA fragment corresponding to the C-terminal coding region of TbSAS-6 was cloned into the pC-3HA-NEO vector. The resulting construct was linearized by digestion within the gene fragment with NcoI, electroporated into the 427 cell line, and selected with 40 μg/ml G418. A stable cell line was not obtained, because the transfectants all died 5 to 7 days after electroporation. Therefore, immunostaining was carried out with cells after transfection for 2 days, and localization of TbSAS-6::3HA to the mature basal body and the probasal body was observed.
TbPLK RNAi and inhibition by GW843682X.
RNAi of TbPLK was carried out as described previously (35), and knockdown of TbPLK was confirmed by Western blotting with anti-TbPLK antibody, which was raised by immunizing rabbits with purified recombinant His-tagged Polo-box domain of TbPLK at Cocalico Biologicals, Inc. (Reamstown, PA). The procyclic 427 cell line was treated with 5 μM GW843682X (Tocris Bioscience), which has been show to inhibit TbPLK activity in vitro and phenocopies TbPLK RNAi (36). TbPLK RNAi cells (24 h) and GW843682X-treated cells (24 h), as well as the noninduced and nontreated cells, then were coimmunostained with anti-TbSAS-6 polyclonal antibody and YL 1/2.
Immunofluorescence microscopy.
Cells were washed once with phosphate-buffered saline (PBS), settled onto glass coverslips for 20 min, fixed with cold methanol (−20°C) for 30 min, and rehydrated with PBS for 10 min. After blocking with 3% bovine serum albumin (BSA) in PBS for 1 h, coverslips were incubated with primary antibodies for 1 h. The following primary antibodies were used: fluorescein isothiocyanate (FITC)-conjugated anti-hemagglutinin (HA) monoclonal antibody (MAb) (1:400, Sigma-Aldrich), L8C4 (anti-PFR2 MAb; 1:50 dilution) (37), 20H5 (anti-centrin MAb; 1:400; Millipore) (38), anti-TbSAS-6 polyclonal antibody (1:400 dilution), and YL 1/2 (1:1,000 dilution). After washing three times, coverslips were incubated with FITC- or Alexa Fluor594-conjugated secondary antibody for 1 h. The coverslips then were washed three times and mounted with DAPI-containing VectaShield mounting medium (Vector Laboratories). Slides were examined under an inverted fluorescence microscope (Olympus IX71) equipped with a cooled charge-coupled-device (CCD) camera (model Orca-ER; Hamamatsu) and a PlanApo N 60×, 1.42-numeric-aperture differential inference contrast (DIC) objective. Images were acquired using Slidebook5 software (Intelligent Imaging Innovations).
RESULTS
The trypanosome SAS-6 homolog and its subcellular localization.
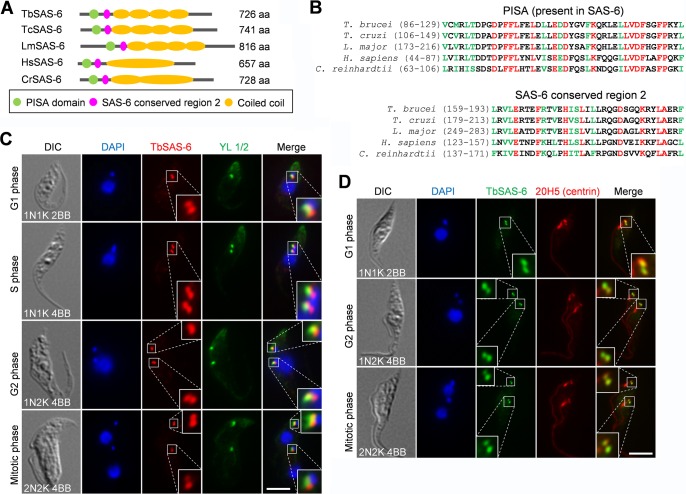
A comprehensive analysis of centriole components conserved across eukaryotic organisms (12, 13) showed that the T. brucei genome encodes a conserved SAS-6 homolog, which we named TbSAS-6 (Tb927.9.10550), in addition to a SAS-6-like protein (39). Despite a slightly larger size than its human homolog, TbSAS-6 contains two conserved motifs, a PISA (present in SAS-6) domain and a SAS-6 conserved domain 2, in its N terminus and four coiled-coil structures in the middle portion of the protein (Fig. 1A and B). The structure of the SAS-6 homolog in Leishmania major (LmSAS-6), another kinetoplastid parasite, has been determined, and it suggests that LmSAS-6 can form a 9-fold symmetric cartwheel hub-like structure (40). Given the high sequence similarity between TbSAS-6 and LmSAS-6, TbSAS-6 potentially also can form the cartwheel of the centriole/basal body, although this remains to be experimentally verified.
FIG 1.
T. brucei SAS-6 homolog, TbSAS-6, and its subcellular localization. (A) Schematic drawing of conserved domains in TbSAS-6 and its homologs from Trypanosoma cruzi, Leishmania major, H. sapiens, and Chlamydomonas reinhardtii. (B) Alignment of the PISA (present in SAS-6) motif and the SAS-6 conserved region 2 among T. brucei, T. cruzi (TcSAS-6), L. major (LmSAS-6), H. sapiens (HsSAS-6), and C. reinhardtii (CrSAS-6). Identical residues are highlighted in red, and homologous residues are in green. (C) Subcellular localization of TbSAS-6 in procyclic trypanosomes. Cells were coimmunostained with anti-TbSAS-6 polyclonal antibody and YL 1/2, which labels the transition fibers in the mature basal body. Cells were counterstained with DAPI for nucleus and kinetoplast DNA. Scale bar, 5 μm. (D) Coimmunostaining of procyclic trypanosomes with anti-TbSAS-6 and 20H5, which detects centrins in both the mature basal body and the probasal body in T. brucei. Scale bar, 5 μm.
To determine the subcellular localization of TbSAS-6, we generated a rabbit polyclonal antibody against recombinant TbSAS-6. Immunofluorescence microscopy showed that anti-TbSAS-6 detected two fluorescence spots in G1 cells, one of which partly overlapped the fluorescence spot of YL 1/2 antibody (Fig. 1C), which stains TbRP2 in the transition fibers radiating from the mature basal body (41) and has served as a marker for the mature basal body since its first use in trypanosomes (42). During S, G2, and mitotic phases, anti-TbSAS-6 labeled two pairs of fluorescence spots, with one spot in each pair partly overlapping YL 1/2 (Fig. 1C). In all of the cells examined, TbSAS-6 was found to be proximal to the YL 1/2 signal (Fig. 1C). These results suggest that TbSAS-6 localizes to both the mature basal body and the probasal body. To further confirm this observation, we coimmunostained the cells with anti-TbSAS-6 and 20H5, which was raised against Chlamydomonas centrin (38, 43) and labels both the mature basal body and the probasal body as well as the bilobe structure and the flagellum in trypanosomes (23). The results showed that TbSAS-6 colocalizes with 20H5 in both the mature and probasal bodies in all (n > 1,000) of the cells examined (Fig. 1D). Moreover, we also tagged TbSAS-6 with a triple HA epitope at one of its endogenous loci, and immunostaining with anti-HA antibody showed a localization pattern identical to that of anti-TbSAS-6 (data not shown). Therefore, TbSAS-6 is localized to both the mature basal body and the probasal body throughout the cell cycle.
TbSAS-6 is essential for cell viability.
To investigate the function of TbSAS-6 in trypanosomes, RNAi was carried out in the procyclic form of T. brucei. Western blotting with anti-TbSAS-6 antibody showed that upon RNAi induction with tetracycline, the TbSAS-6 level was significantly decreased, although it was not completely depleted from the cell (Fig. 2A). Knockdown of TbSAS-6 resulted in significant growth inhibition and eventual cell death after 4 days of RNAi induction (Fig. 2B), suggesting that TbSAS-6 is essential for cell viability. At least three clonal cell lines were examined, and all of the three cell lines showed similar growth defects (data not shown). Flow cytometry analysis showed that after TbSAS-6 RNAi for 2 days, there was an accumulation of cells with 8C DNA content (Fig. 2C). To further characterize the defects, we stained the noninduced control cells and RNAi-induced cells with DAPI and counted the cells with different numbers of nuclei and kinetoplasts. There was an initial accumulation of cells with two nuclei and one kinetoplast (2N1K) after 1 day of RNAi and a subsequent accumulation of cells with multiple (>2) nuclei and one kinetoplast (XN1K) after 2 to 3 days of RNAi (Fig. 2D). The XN1K cells were likely to be derived from the 2N1K cells. These results suggest that depletion of TbSAS-6 inhibited kinetoplast segregation and cell division.
FIG 2.
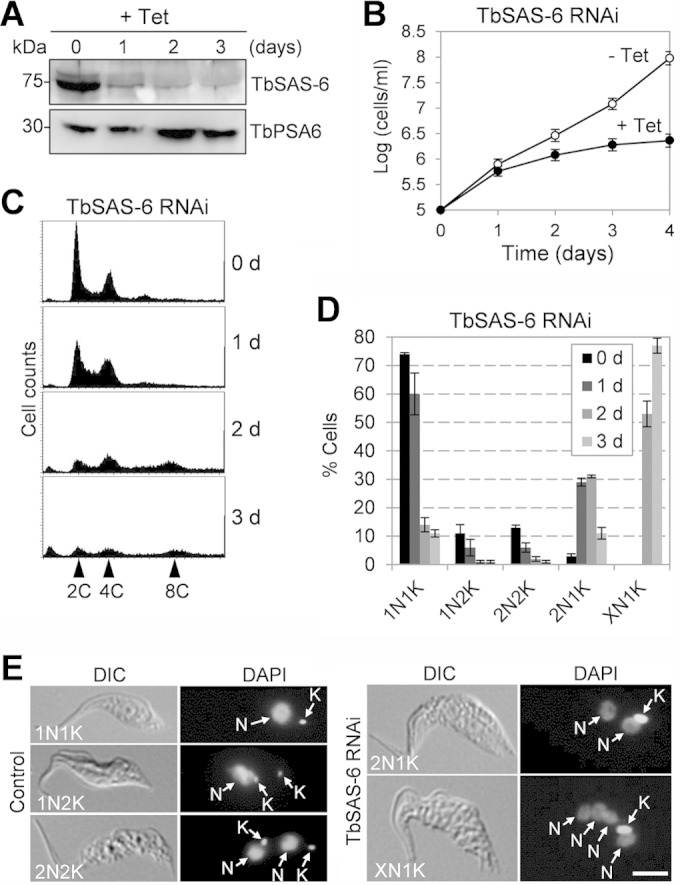
RNAi of TbSAS-6 inhibits kinetoplast segregation and cell division. (A) TbSAS-6 level in control and RNAi cell line detected by Western blotting with anti-TbSAS-6 antibody. The level of the proteasome α6 subunit, detected with anti-TbPSA6 polyclonal antibody (48), was included as the loading control. Tet, tetracycline. (B) RNAi of TbSAS-6 inhibited cell proliferation. (C) Flow cytometry analysis of TbSAS-6 RNAi cells. (D) RNAi of TbSAS-6 inhibited kinetoplast duplication/segregation. About 300 cells were counted for the numbers of nuclei and kinetoplasts for each time point, and error bars represented standard deviations (SD) calculated from three independent experiments. (E) Morphology of TbSAS-6 RNAi cells induced for 1 day. Shown are the 2N1K and XN1K cells that accumulated after RNAi induction. Nucleus (N) and kinetoplast (K) are indicated. Scale bar, 5 μm.
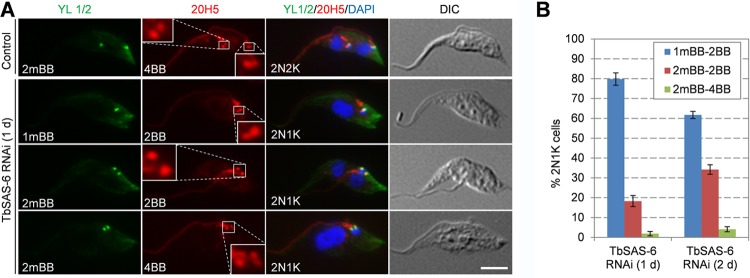
TbSAS-6 is required for probasal body biogenesis.
The fact that TbSAS-6 RNAi inhibited kinetoplast segregation and that TbSAS-6 is localized to basal bodies suggests that basal body duplication/segregation was affected. To test whether RNAi of TbSAS-6 affected basal body duplication or segregation, we coimmunostained the cells with the YL 1/2 antibody, which serves as the marker of the mature basal body, and the 20H5 antibody, which serves as the marker for the mature basal body and the probasal body. In noninduced control 2N2K cells, two YL 1/2 spots and four 20H5 spots were clearly detected (Fig. 3A), indicating that the 2N2K cells contain two mature basal bodies, each of which is accompanied by a probasal body. For TbSAS-6 RNAi cells, we focused on the 2N1K cells at the early time point (day 1) of RNAi induction, since these cells emerged first upon RNAi induction, which most likely represented the direct effect of TbSAS-6 depletion. This 2N1K population consists of cells containing one YL 1/2 spot and two 20H5 spots (Fig. 3A and B), indicating that they possess a mature basal body and an associated probasal body (1mBB2BB; ∼80% of the population), cells containing two YL 1/2 spots and two 20H5 spots, indicating two mature basal bodies lacking new probasal bodies (2mBB2BB; ∼18% of the population), and cells containing two YL 1/2 spots and four 20H5 spots, indicating two mature basal bodies, each with an associated probasal body (2mBB4BB; ∼2% of the population). These results suggest that biogenesis of the new probasal bodies was inhibited, but it is unclear whether maturation of the old probasal body also was affected. Therefore, we examined the basal bodies in the 2N1K cells collected from day 2 of TbSAS-6 RNAi. Compared to the 2N1K cells from day 1 of RNAi, the 2N1K cells containing one mature basal body and one probasal body (1mBB2BB) decreased to about 62%, whereas the 2N1K cells containing two mature basal bodies and two basal bodies (2mBB2BB) increased to ∼34% of the total 2N1K population (Fig. 3B). It appeared that the old probasal body continued to mature in these cells, but biogenesis of new probasal bodies still was inhibited. Taken together, these results suggest that TbSAS-6 is required for the biogenesis of new probasal bodies in procyclic trypanosomes.
FIG 3.
RNAi of TbSAS-6 inhibits probasal body biogenesis. (A) Immunostaining of control and TbSAS-6 RNAi cells (day 1) with YL 1/2 for mature basal body and 20H5 for both mature and probasal bodies. Scale bar, 5 μm. (B) Quantitation of cells with different numbers of basal bodies in 2N2K cells from the noninduced control and 2N1K cells from TbSAS-6 RNAi induced for 1 and 2 days. About 200 cells were counted for each time point, and error bars represented SD calculated from three independent experiments.
TbSAS-6 is required for new flagellum biogenesis.
The basal body in trypanosomes is known to nucleate flagellum microtubule assembly, and biogenesis of the new flagellum is initiated from the newly matured basal body (3, 4). Since TbSAS-6 is not required for maturation of the existing probasal body (Fig. 3), we asked whether RNAi of TbSAS-6 affected new flagellum biogenesis. We coimmunostained the cells with L8C4, which recognizes the paraflagellar rod protein PFR2 in the flagellum (37), and YL 1/2 to label the mature basal body. We then tabulated the cells with different numbers of flagella and mature basal bodies in TbSAS-6-depleted 2N1K cells and the noninduced control 2N2K cells. While the noninduced 2N2K cells all contained two mature basal bodies and two full-length flagella (2mBB2F) (Fig. 4A), the 2N1K cells from TbSAS-6 RNAi contained various numbers of mature basal bodies and flagella (Fig. 4A and B). After RNAi induction for 1 day, ∼98% of the 2N1K cells contained only one flagellum and either one mature basal body (1mBB1F; ∼80%) or two mature basal bodies (2mBB1F; ∼18%). The rest (∼2%) of the 2N1K cells contained two flagella and two mature basal bodies (2mBB2F), but the new flagellum appeared to be shorter than that in the noninduced control 2N2K cells (Fig. 4A, arrow). After RNAi for 2 days, about 14% of the 2N1K cells contained one mature basal body but no flagellum (1mBB0F) (Fig. 4A and B). It is unclear how these cells were produced. Presumably they could be derived from the 1N1K cells lacking flagella (1mBB0F), which were detected 1 day after TbSAS-6 RNAi and accumulated to about 20% of the total 1N1K population after 2 days (Fig. 4C to E). In addition, there was an accumulation of 1N1K cells with a short flagellum (1mBB1sF; ∼27%) (Fig. 4C to E). Both types of these 1N1K cells were smaller in size (Fig. 4C and E), likely due to premature cytokinesis.
FIG 4.
TbSAS-6 RNAi impairs new flagellum assembly. (A) Immunostaining of 2N2K cells from noninduced control and 2N1K cells from TbSAS-6 RNAi cells (days 1 and 2) with L8C4 and YL 1/2 for flagellum and mature basal body, respectively. The white arrow showed a short new flagellum in a 2N1K cell with two mature basal bodies. Scale bar, 5 μm. (B). Quantitation of cells with different numbers of flagella and mature basal bodies in 2N1K cells from TbSAS-6 RNAi induced for 1 to 2 days and XN1K cells from TbSAS-6 RNAi induced for 2 days. About 200 cells were counted for each cell type, and the error bars represented SD calculated from three independent experiments. (C) Immunostaining of 1N1K cells from control and TbSAS-6 RNAi cells (2 days) with L8C4 and YL 1/2. Scale bar, 5 μm. (D) Quantitation of 1N1K cells with different numbers of mature basal bodies and flagella. About 200 1N1K cells were counted for each time point, and error bars represent SD calculated from three independent experiments. (E) Measurement of flagellum length and cell length of the 1N1K cells with one flagellum from control and TbSAS-6 RNAi (2 days) cells. About 120 1N1K cells from each sample were measured and plotted. Those circled within the dotted oval indicate the cells with a short flagellum.
In the XN1K cell, ∼81% of them possessed only one flagellum and either one mature basal body (1mBB1F; ∼61%), two mature basal bodies (2mBB1F; ∼14%), or multiple (>2) mature basal bodies (XmBB1F; ∼6%) (Fig. 4B). Altogether, given that the majority of the 2N1K and XN1K cells contained only one flagellum, these results suggest that TbSAS-6 RNAi impaired the formation of the new flagellum.
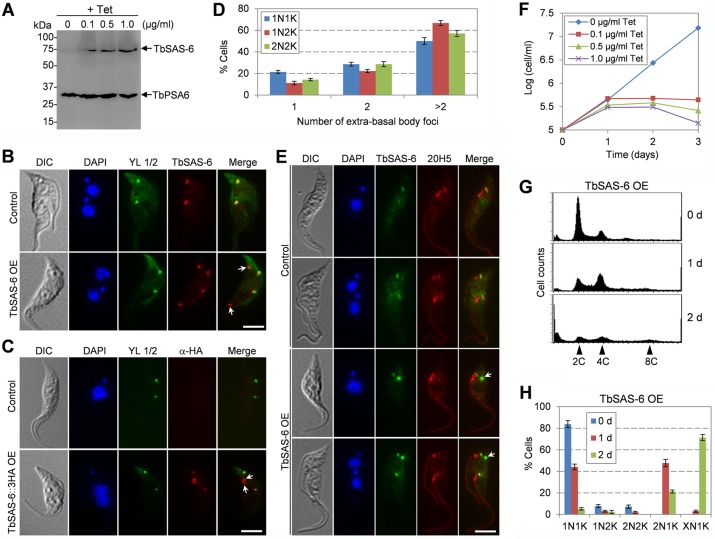
Overexpression of TbSAS-6 impairs biogenesis of probasal body and flagellum.
During the course of work of overexpressing TbSAS-6::3HA fusion protein in the procyclic form, we found that this overexpression appeared to result in defects similar to that caused by TbSAS-6 RNAi, i.e., formation of 2N1K and XN1K cells and growth inhibition (see below). To rule out the possibility that epitope tagging affects TbSAS-6 function, we overexpressed TbSAS-6 without an epitope tag and observed similar defects. Therefore, we further characterized the defects caused by TbSAS-6 overexpression. Overexpression of TbSAS-6 was induced with different concentrations (0.1 μg/ml, 0.5 μg/ml, and 1.0 μg/ml) of tetracycline and was confirmed by Western blotting with anti-TbSAS-6 antibody (Fig. 5A). Interestingly, immunofluorescence with anti-TbSAS-6 antibody detected multiple TbSAS-6 foci, some of which overlapped with the basal bodies stained by YL 1/2 (Fig. 5B). However, it was unclear whether the basal body-localizing TbSAS-6 was native TbSAS-6 or overexpressed TbSAS-6. To clarify this, we examined the cells overexpressing TbSAS-6::3HA and performed immunofluorescence microscopy with anti-HA antibody and YL 1/2. The results showed that multiple TbSAS-6::3HA foci were detected, some of which overlapped with the YL 1/2-labeled mature basal bodies (Fig. 5C). These results suggest that overexpressed TbSAS-6::3HA was able to be recruited to the basal body and additionally was able to form extrabasal body structures. The extrabasal body structures appeared to vary in number in the TbSAS-6 overexpressed cells, with the majority of the cells containing more than 2 extrabasal body foci regardless of the cell cycle stages (Fig. 5D). Moreover, coimmunostaining with TbSAS-6 and 20H5 showed that the extrabasal body structure was not able to recruit centrins (Fig. 5E), suggesting that these TbSAS-6-containing structures are not functional basal bodies.
FIG 5.
Overexpression of TbSAS-6 inhibits kinetoplast segregation and cell division. (A) Western blotting to monitor the overexpression of TbSAS-6 induced with different concentrations of tetracycline. The level of TbPSA6 was detected in the same Western blot as the loading control. (B) Subcellular localization of overexpressed TbSAS-6. Noninduced control and tetracycline-induced cells (4 h) were immunostained with anti-TbSAS-6 antibody and YL 1/2 and counterstained with DAPI for DNA. Arrows showed TbSAS-6-containing extrabasal body structures. OE, overexpression. Scale bar, 5 μm. (C) Subcellular localization of overexpressed TbSAS-6::3HA. Noninduced control and tetracycline-induced cells (4 h) were coimmunostained with FITC-conjugated anti-HA antibody and YL 1/2 and counterstained with DAPI for DNA. Arrows show TbSAS-6::3HA-containing extrabasal body structures. Scale bar, 5 μm. (D) Quantification of TbSAS-6-containing extrabasal body foci in different cell types after TbSAS-6 overexpression for 16 h. Cells were immunostained with anti-TbSAS-6 and 20H5, and about 200 cells for each cell type (1N1K, 1N2K, and 2N2K) were counted. Error bars represent SD calculated from three independent experiments. (E) Coimmunostaining of control and TbSAS-6 overexpression cells with TbSAS-6 and 20H5. The arrow indicates the TbSAS-6-containing extrabasal body structure. Scale bar, 5 μm. (F) Overexpression of TbSAS-6 caused growth inhibition. (G) Flow cytometry analysis of TbSAS-6 overexpression cells. Cells were induced with 0.5 μg/ml tetracycline for up to 2 days and analyzed by flow cytometry. (H) Quantitation of cells with different numbers of nucleus and kinetoplast in noninduced control and TbSAS-6 overexpression cells. About 300 cells from each time point were counted, and error bars represent SD calculated from three independent experiments.
Induction of TbSAS-6 overexpression with all three concentrations of tetracycline resulted in growth inhibition and eventual cell death (Fig. 5F). Flow cytometry showed that after induction for 1 day, there was an accumulation of cells with 4C DNA content, and after induction for 2 days, cells with 8C DNA content started to emerge (Fig. 5G). This notion was further confirmed by tabulating the cells with different numbers of nuclei and kinetoplasts, which showed that 2N1K cells emerged to about 43% of the total population after induction for 1 day and XN1K cells accumulated to about 65% after induction for 2 days (Fig. 5H). Together, these results suggest that overexpression of TbSAS-6 inhibited kinetoplast segregation and cell division.
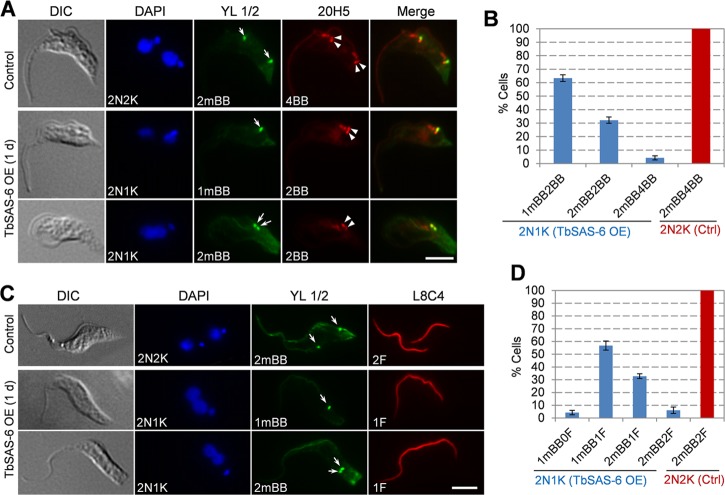
We next immunostained the cells with YL 1/2 and 20H5 to label the mature basal body and both the mature basal body and probasal body, respectively. We found that ∼95% of the 2N1K cells contained either one mature basal body with an accompanying probasal body (1mBB2BB; ∼63%) or two mature basal bodies without accompanying probasal bodies (2mBB2BB; ∼32%) (Fig. 6A and B), suggesting that probasal body biogenesis was compromised in TbSAS-6 overexpression cells. Moreover, in these 2N1K cells, ∼94% of them contained either one mature basal body and one flagellum (1mBB1F; ∼61%) or two mature basal bodies and one flagellum (2mBB1F; ∼33%) (Fig. 6C and D). These results suggest that new flagellum biogenesis also was inhibited when TbSAS-6 was overexpressed.
FIG 6.
Overexpression of TbSAS-6 impairs the biogenesis of probasal body and flagellum. (A) Effect of TbSAS-6 overexpression on basal body duplication. Noninduced control and TbSAS-6 overexpression (day 1) cells were immunostained with 20H5 for both the mature basal body and probasal body (arrowheads) and with YL 1/2 for the mature basal body (arrows). Scale bar, 5 μm. (B) Quantitation of cells with different numbers of basal bodies. About 300 2N1K cells from TbSAS-6 overexpression cells and 200 2N2K cells from the noninduced control (Ctrl) cells were counted. Error bars represent SD calculated from three independent experiments. (C) Effect of TbSAS-6 overexpression on flagellum biogenesis. Noninduced control and TbSAS-6 overexpression (day 1) cells were immunostained with L8C4 for the flagellum and with YL 1/2 for the mature basal body (arrows). Scale bar, 5 μm. (D) Quantitation of cells with different numbers of flagella and mature basal bodies. About 300 cells from each cell type (2N1K cells from TbSAS-6 overexpression and 2N2K cells from the noninduced control) were counted. Error bars represent SD calculated from three independent experiments.
TbPLK is not required for TbSAS-6 localization to the basal body.
In C. elegans, a Polo-like kinase-related kinase, ZYG-1, interacts with SAS-6 and recruits the latter to centrioles, but the kinase activity of ZYG-1 appears to be nonessential for SAS-6 recruitment (44). In another report, however, ZYG-1 is found to phosphorylate SAS-6, and this phosphorylation is required to maintain SAS-6 at centrioles (45). T. brucei expresses an essential Polo-like kinase, TbPLK, that is localized to the basal body during early cell cycle stages (29, 30). Intriguingly, like TbSAS-6 RNAi and overexpression, RNAi of TbPLK and overexpression of TbPLK also caused similar defects in inhibiting basal body duplication/segregation (31), raising the question of whether TbPLK regulates TbSAS-6. To investigate whether TbPLK is required for TbSAS-6 localization to the basal body, we first treated the cells with GW843682X, which inhibits TbPLK activity in vitro and phenocopies TbPLK RNAi (36). Similar to the previous report (36), we also found that GW843682X treatment of procyclic trypanosomes resulted in an initial accumulation of 2N1K cells with a detached flagellum after 1 day and subsequent accumulation of XN1K cells after 2 days (data not shown). We focused on the 2N1K cells with a detached new flagellum, because these cells are typical of TbPLK-deficient cells (30, 31). In all of the 2N1K cells examined, TbSAS-6 was still detected at the four basal bodies that were not well segregated (Fig. 7A), suggesting that TbPLK activity is not required for TbSAS-6 localization to the basal body.
FIG 7.
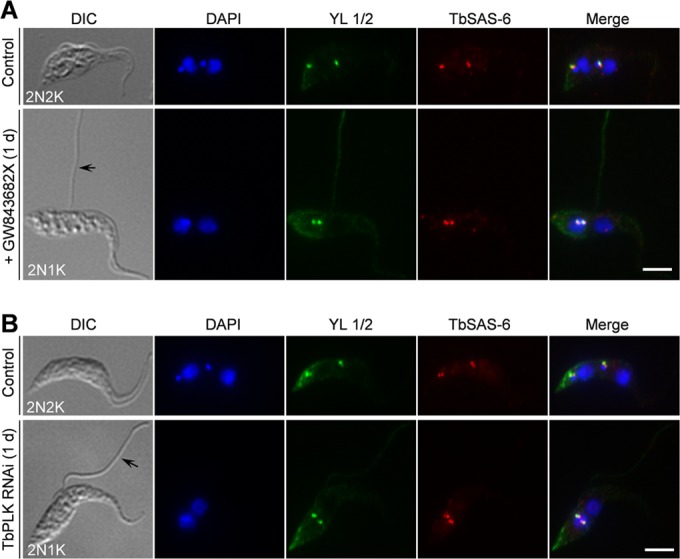
Polo-like kinase is not required for recruitment of TbSAS-6 to the basal body. (A) Inhibition of TbPLK activity by a small-molecule inhibitor, GW843682X, did not affect TbSAS-6 localization to the basal body. Control and GW843682X-treated cells were immunostained with anti-TbSAS-6 antibody and YL 1/2. The black arrow showed the detached flagellum in a GW843682X-treated cell (2N1K cell). Scale bar, 5 μm. (B) Effect of TbPLK depletion on TbSAS-6 localization to the basal body. TbPLK RNAi was induced for 1 day. Noninduced control and RNAi cells were immunostained with anti-TbSAS-6 antibody and YL 1/2. The black arrow pointed to the detached flagellum in a TbPLK RNAi cell (2N1K cell). Scale bar, 5 μm.
We further tested whether depletion of TbPLK interferes with TbSAS-6 localization. Depletion of TbPLK by RNAi in the procyclic form, confirmed by Western blotting with anti-TbPLK antibody, also resulted in an initial accumulation of 2N1K cells with a detached flagellum and subsequent accumulation of XN1K cells after longer RNAi (data not shown), similar to previous reports (30, 31). When TbPLK was depleted, TbSAS-6 was still localized to the basal bodies in all of the 2N1K cells examined (Fig. 7B), similar to GW843682X treatment (Fig. 7A). These results suggest that, unlike in animals, the Polo-like kinase in T. brucei is not required for TbSAS-6 recruitment to the basal body. Thus, a distinct regulatory pathway controlling SAS-6 recruitment to the basal body might be operating in T. brucei, which remains to be explored.
DISCUSSION
Basal body duplication represents the first cytoskeletal event in the cell division cycle of T. brucei and is crucial for segregation of other single-copy organelles and for morphogenesis of the daughter cell (4), but our knowledge about the molecules and the signaling pathway controlling its biogenesis remains very limited. In this paper, we reported the functional characterization of the SAS-6 homolog in the procyclic form of T. brucei and demonstrated that TbSAS-6 is required for probasal body biogenesis (Fig. 3). Our results also suggest that TbSAS-6 is not needed for maturation of the existing probasal body, although there appeared to be a delay in the maturation process, resulting in the accumulation of a large proportion of 2N1K cells with one mature basal body and one probasal body, which then became mature, as evidenced by the gradual increase of the 2N1K cells with two mature basal bodies after RNAi for longer times (Fig. 3). The requirement of TbSAS-6 for probasal body biogenesis in T. brucei is analogous to the function of SAS-6 in regulating procentriole formation in animals (11), suggesting a conserved role of SAS-6 in this early branching protozoan.
Our data also suggest that TbSAS-6 RNAi caused defective assembly of the new flagellum, as shown by the accumulation of ∼80% of the 2N1K cells with one mature basal body and one flagellum (1mBB1F) after RNAi for 1 day (Fig. 4A and B). This conclusion is not unexpected, because it is known that the new flagellum is assembled from the newly matured basal body (3, 4), and these 2N1K cells did not have the new mature basal body. However, it is surprising that almost 20% of the 2N1K cells contained two mature basal bodies but only one flagellum (2mBB1F) (Fig. 4A and B), which suggests that in these cells, either the newly matured basal body was unable to nucleate flagellum assembly or biogenesis of the new flagellum was significantly delayed. The fact that a small proportion of the 2N1K cells with two mature basal bodies did assemble a short new flagellum (Fig. 4A and B) suggested a delay in new flagellum biogenesis. However, it should be noted that RNAi was induced using an asynchronous cell population which contained some 1N1K-2mBB cells that have already assembled a short new flagellum (Fig. 4D). Therefore, these 2N1K-2mBB2F cells might be derived from the 1N1K-2mBB2F cells in the absence of probasal body duplication and kinetoplast segregation caused by TbSAS-6 RNAi.
The production of 1N1K-1mBB cells without a flagellum and 1N1K-1mBB cells with a short flagellum (Fig. 4C to E) after TbSAS-6 RNAi suggests that a premature cytokinesis occurred in some cells. These two types of cells were very likely to be the daughters of 2N1K-2mBB cells without a new flagellum and 2N1K-2mBB cells with a short new flagellum, respectively. If this was the case, then the two mature basal bodies might still be able to separate to some extent, resulting in the segregation of the duplicated kinetoplasts. However, it should be noted that aberrant cytokinesis occurred in only a small proportion of the 2N1K cells, because most of the 2N1K cells likely had become XN1K cells (Fig. 2D).
An unexpected but intriguing observation made in this report is the similar defects caused by overexpression of TbSAS-6 and RNAi of TbSAS-6 (Fig. 2 to 6). Unlike in humans, overexpression of HsSAS-6 and knockdown of HsSAS-6 exert opposite effects on centriole duplication: overexpression results in centriole overduplication, whereas knockdown inhibits centriole duplication (46, 47). The HsSAS-6 level is tightly regulated during the cell cycle, which ensures that centriole duplication occurs only once per cell cycle. This is achieved through degradation of HsSAS-6 after centriole duplication by APCcdh1-mediated ubiquitination of HsSAS-6 and subsequent degradation of HsSAS-6 by the 26S proteasome (47). The fact that overexpression of TbSAS-6 and RNAi of TbSAS-6 both inhibit basal body duplication suggests that the TbSAS-6 protein level also is tightly regulated in order to maintain a certain level of TbSAS-6 in the cell, although the control mechanism remains unclear. Therefore, we envisage that after the basal body has duplicated or when the TbSAS-6 level has exceeded a threshold, a feedback pathway is invoked, which may directly downregulate the TbSAS-6 level or activate a negative regulator of TbSAS-6. Overexpression of TbSAS-6 may activate this feedback pathway, thereby inhibiting probasal body biogenesis. Another explanation for the defects caused by TbSAS-6 overexpression is that overexpressed TbSAS-6 titrates some of its interacting factors away from the basal body, which can be recruited to the extrabasal body structures, thereby causing defects in basal body biogenesis.
Our results also suggest that the Polo-like kinase homolog in T. brucei, TbPLK, is not required for recruiting TbSAS-6 to the basal body because depletion of TbPLK by RNAi or inhibition of TbPLK activity did not abolish TbSAS-6 localization to the basal bodies (Fig. 7). This is in striking contrast to the ZYG-1/SAK/PLK4-mediated pathway that governs centriole duplication by regulating SAS-6 assembly/maintenance in the centriole, which appears to be conserved in animals (11). Although TbPLK is localized to the basal body at early cell cycle stages (30), our results confirmed that it is required for basal body segregation but not basal body duplication (Fig. 7), which provides another line of evidence to support that TbPLK is not required for targeting TbSAS-6 to the basal body. Unlike ZYG-1/SAK/PLK4, which possesses one Polo-box domain (PBD) and a cryptic PBD, TbPLK possesses two PBDs and is grouped into the PLK1 clade but not the SAK/PLK4 clade (12, 13). It has been proposed that an ancestral PLK triggers centriole/basal body biogenesis in the last eukaryotic common ancestor (LECA) (12), but phylogenetic analysis of the PLKs from 45 organisms showed that although PLK was present in the LECA, it has been lost several times during evolution (13), suggesting that in those organisms that have lost PLK, centriole biogenesis either does not need PLK or is regulated by other kinase(s). Our results suggest that although T. brucei possesses a canonical PLK that localizes to the basal body, basal body duplication in T. brucei likely employs a PLK-independent pathway.
ACKNOWLEDGMENTS
We are grateful to George Cross from Rockefeller University for providing the 29-13 cell line and the pLew100 vector, Paul Englund from Johns Hopkins University for providing the pZJM vector, Arthur Gunzl from University of Connecticut Health Center for providing the pC-HA-BSD vector, and Keith Gull from University of Oxford for providing the L8C4 antibody.
This work was supported by NIH grants AI101437 and AI108657.
We have no competing financial interests to declare.
REFERENCES
- 1.Gull K. 1999. The cytoskeleton of trypanosomatid parasites. Annu Rev Microbiol 53:629–655. doi: 10.1146/annurev.micro.53.1.629. [DOI] [PubMed] [Google Scholar]
- 2.Ralston KS, Kabututu ZP, Melehani JH, Oberholzer M, Hill KL. 2009. The Trypanosoma brucei flagellum: moving parasites in new directions. Annu Rev Microbiol 63:335–362. doi: 10.1146/annurev.micro.091208.073353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherwin T, Gull K. 1989. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos Trans R Soc Lond B Biol Sci 323:573–588. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- 4.Lacomble S, Vaughan S, Gadelha C, Morphew MK, Shaw MK, McIntosh JR, Gull K. 2010. Basal body movements orchestrate membrane organelle division and cell morphogenesis in Trypanosoma brucei. J Cell Sci 123:2884–2891. doi: 10.1242/jcs.074161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson DR, Gull K. 1991. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352:731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- 6.Ogbadoyi EO, Robinson DR, Gull K. 2003. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol Biol Cell 14:1769–1779. doi: 10.1091/mbc.E02-08-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dippell RV. 1968. The development of basal bodies in paramecium. Proc Natl Acad Sci U S A 61:461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen RD. 1969. The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J Cell Biol 40:716–733. doi: 10.1083/jcb.40.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavalier-Smith T. 1974. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J Cell Sci 16:529–556. [DOI] [PubMed] [Google Scholar]
- 10.Geimer S, Melkonian M. 2004. The ultrastructure of the Chlamydomonas reinhardtii basal apparatus: identification of an early marker of radial asymmetry inherent in the basal body. J Cell Sci 117:2663–2674. doi: 10.1242/jcs.01120. [DOI] [PubMed] [Google Scholar]
- 11.Gonczy P. 2012. Towards a molecular architecture of centriole assembly. Nat Rev Mol Cell Biol 13:425–435. doi: 10.1038/nrm3373. [DOI] [PubMed] [Google Scholar]
- 12.Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. 2010. Stepwise evolution of the centriole-assembly pathway. J Cell Sci 123:1414–1426. doi: 10.1242/jcs.064931. [DOI] [PubMed] [Google Scholar]
- 13.Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. 2010. Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci 123:1407–1413. doi: 10.1242/jcs.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakazawa Y, Hiraki M, Kamiya R, Hirono M. 2007. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol 17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, Glover DM. 2007. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr Biol 17:1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 16.Hiraki M, Nakazawa Y, Kamiya R, Hirono M. 2007. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol 17:1778–1783. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 17.Matsuura K, Lefebvre PA, Kamiya R, Hirono M. 2004. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J Cell Biol 165:663–671. doi: 10.1083/jcb.200402022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelletier L, O'Toole E, Schwager A, Hyman AA, Muller-Reichert T. 2006. Centriole assembly in Caenorhabditis elegans. Nature 444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- 19.Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, Dynlacht BD, Khodjakov A, Gonczy P. 2009. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol 19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. 2009. Control of centriole length by CPAP and CP110. Curr Biol 19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. 2009. CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol 11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 22.Scott V, Sherwin T, Gull K. 1997. Gamma-tubulin in trypanosomes: molecular characterisation and localisation to multiple and diverse microtubule organising centres. J Cell Sci 110(Part 2):157–168. [DOI] [PubMed] [Google Scholar]
- 23.He CY, Pypaert M, Warren G. 2005. Golgi duplication in Trypanosoma brucei requires Centrin2. Science 310:1196–1198. doi: 10.1126/science.1119969. [DOI] [PubMed] [Google Scholar]
- 24.Dilbeck V, Berberof M, Van Cauwenberge A, Alexandre H, Pays E. 1999. Characterization of a coiled coil protein present in the basal body of Trypanosoma brucei. J Cell Sci 112(Part 24):4687–4694. [DOI] [PubMed] [Google Scholar]
- 25.Selvapandiyan A, Kumar P, Morris JC, Salisbury JL, Wang CC, Nakhasi HL. 2007. Centrin1 is required for organelle segregation and cytokinesis in Trypanosoma brucei. Mol Biol Cell 18:3290–3301. doi: 10.1091/mbc.E07-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi J, Franklin JB, Yelinek JT, Ebersberger I, Warren G, He CY. 2008. Centrin4 coordinates cell and nuclear division in T. brucei. J Cell Sci 121:3062–3070. doi: 10.1242/jcs.030643. [DOI] [PubMed] [Google Scholar]
- 27.Morgan GW, Denny PW, Vaughan S, Goulding D, Jeffries TR, Smith DF, Gull K, Field MC. 2005. An evolutionarily conserved coiled-coil protein implicated in polycystic kidney disease is involved in basal body duplication and flagellar biogenesis in Trypanosoma brucei. Mol Cell Biol 25:3774–3783. doi: 10.1128/MCB.25.9.3774-3783.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pradel LC, Bonhivers M, Landrein N, Robinson DR. 2006. NIMA-related kinase TbNRKC is involved in basal body separation in Trypanosoma brucei. J Cell Sci 119:1852–1863. doi: 10.1242/jcs.02900. [DOI] [PubMed] [Google Scholar]
- 29.de Graffenried CL, Ho HH, Warren G. 2008. Polo-like kinase is required for Golgi and bilobe biogenesis in Trypanosoma brucei. J Cell Biol 181:431–438. doi: 10.1083/jcb.200708082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda KN, de Graffenried CL. 2012. Polo-like kinase is necessary for flagellum inheritance in Trypanosoma brucei. J Cell Sci 125:3173–3184. doi: 10.1242/jcs.101162. [DOI] [PubMed] [Google Scholar]
- 31.Hammarton TC, Kramer S, Tetley L, Boshart M, Mottram JC. 2007. Trypanosoma brucei Polo-like kinase is essential for basal body duplication, kDNA segregation and cytokinesis. Mol Microbiol 65:1229–1248. doi: 10.1111/j.1365-2958.2007.05866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozano-Nunez A, Ikeda KN, Sauer T, de Graffenried CL. 2013. An analogue-sensitive approach identifies basal body rotation and flagellum attachment zone elongation as key functions of PLK in Trypanosoma brucei. Mol Biol Cell 24:1321–1333. doi: 10.1091/mbc.E12-12-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Morris JC, Drew ME, Englund PT. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem 275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- 34.Wei Y, Hu H, Lun ZR, Li Z. 2014. Centrin3 in trypanosomes maintains the stability of a flagellar inner-arm dynein for cell motility. Nat Commun 5:4060. doi: 10.1038/ncomms5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar P, Wang CC. 2006. Dissociation of cytokinesis initiation from mitotic control in a eukaryote. Eukaryot Cell 5:92–102. doi: 10.1128/EC.5.1.92-102.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Umeyama T, Li Z, Wang CC. 2010. Polo-like kinase guides cytokinesis in Trypanosoma brucei through an indirect means. Eukaryot Cell 9:705–716. doi: 10.1128/EC.00330-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohl L, Sherwin T, Gull K. 1999. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J Eukaryot Microbiol 46:105–109. doi: 10.1111/j.1550-7408.1999.tb04592.x. [DOI] [PubMed] [Google Scholar]
- 38.Lingle WL, Salisbury JL. 1997. Centrin and the cytoskeleton of the protist Holomastigotoides. Cell Motil Cytoskeleton 36:377–390. doi:. [DOI] [PubMed] [Google Scholar]
- 39.de Leon JC, Scheumann N, Beatty W, Beck JR, Tran JQ, Yau C, Bradley PJ, Gull K, Wickstead B, Morrissette NS. 2013. A SAS-6-like protein suggests that the Toxoplasma conoid complex evolved from flagellar components. Eukaryot Cell 12:1009–1019. doi: 10.1128/EC.00096-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Breugel M, Wilcken R, McLaughlin SH, Rutherford TJ, Johnson CM. 2014. Structure of the SAS-6 cartwheel hub from Leishmania major. eLife 3:e01812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephan A, Vaughan S, Shaw MK, Gull K, McKean PG. 2007. An essential quality control mechanism at the eukaryotic basal body prior to intraflagellar transport. Traffic 8:1323–1330. doi: 10.1111/j.1600-0854.2007.00611.x. [DOI] [PubMed] [Google Scholar]
- 42.Sherwin T, Schneider A, Sasse R, Seebeck T, Gull K. 1987. Distinct localization and cell cycle dependence of COOH terminally tyrosinolated alpha-tubulin in the microtubules of Trypanosoma brucei brucei. J Cell Biol 104:439–446. doi: 10.1083/jcb.104.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salisbury JL, Baron AT, Sanders MA. 1988. The centrin-based cytoskeleton of Chlamydomonas reinhardtii: distribution in interphase and mitotic cells. J Cell Biol 107:635–641. doi: 10.1083/jcb.107.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lettman MM, Wong YL, Viscardi V, Niessen S, Chen SH, Shiau AK, Zhou H, Desai A, Oegema K. 2013. Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev Cell 25:284–298. doi: 10.1016/j.devcel.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kitagawa D, Busso C, Fluckiger I, Gonczy P. 2009. Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryos. Dev Cell 17:900–907. doi: 10.1016/j.devcel.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol 7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- 47.Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell 13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z, Zou CB, Yao Y, Hoyt MA, McDonough S, Mackey ZB, Coffino P, Wang CC. 2002. An easily dissociated 26 S proteasome catalyzes an essential ubiquitin-mediated protein degradation pathway in Trypanosoma brucei. J Biol Chem 277:15486–15498. doi: 10.1074/jbc.M109029200. [DOI] [PubMed] [Google Scholar]