Abstract
Chitin is an essential component of the fungal cell wall, providing rigidity and stability. Its degradation is mediated by chitinases and supposedly ensures the dynamic plasticity of the cell wall during growth and morphogenesis. Hence, chitinases should be particularly important for fungi with dramatic morphological changes, such as Ustilago maydis. This smut fungus switches from yeast to filamentous growth for plant infection, proliferates as a mycelium in planta, and forms teliospores for spreading. Here, we investigate the contribution of its four chitinolytic enzymes to the different morphological changes during the complete life cycle in a comprehensive study of deletion strains combined with biochemical and cell biological approaches. Interestingly, two chitinases act redundantly in cell separation during yeast growth. They mediate the degradation of remnant chitin in the fragmentation zone between mother and daughter cell. In contrast, even the complete lack of chitinolytic activity does not affect formation of the infectious filament, infection, biotrophic growth, or teliospore germination. Thus, unexpectedly we can exclude a major role for chitinolytic enzymes in morphogenesis or pathogenicity of U. maydis. Nevertheless, redundant activity of even two chitinases is essential for cell separation during saprophytic growth, possibly to improve nutrient access or spreading of yeast cells by wind or rain.
INTRODUCTION
Fungal cells are surrounded by a rigid cell wall that confers mechanical stability and protects the cell against environmental stress (1). The cell wall contains a layer of chitin, a polymer of β-1-4-linked N-acetylglucosamine (GlcNAc), which, through covalent linkage to glucans and mannoproteins, constitutes the structural scaffold of the cell (2). To maintain plasticity during growth, chitin is synthesized by chitin synthases (EC2.4.1.16) and cleaved by chitinases (EC3.2.1.14), which remodel or degrade the cell wall (1). All known fungal chitinases belong to glycosyl hydrolase (GH) family 18 (3) and cleave chitin polymers into shorter chito-oligosaccharides of a minimum chain length of n = 2. The resulting chitobioses can be further degraded into GlcNAc monomers by N-acetyl-glucosaminidases (NAG; EC 3.2.1.52) of the GH20 family.
The number of encoded GH18 chitinases is highly variable in fungi, ranging from one member in Schizosaccharomyces pombe to 20 members in Aspergillus nidulans (4). Although their importance as biocontrol agents and for biotechnological use is undisputed (5–7), functional characterization of chitinases so far has been difficult due to a high degree of redundancy. For example, deletion of 5 out of 18 chitinases in A. fumigatus only leads to a slight reduction in chitinolytic activity and no observable phenotype (8). Hence, knowledge of the biological role of GH18 chitinases mainly comes from organisms with a small repertoire of chitinase-encoding genes. ScCts1p in Saccharomyces cerevisiae and its functional homolog, CaCht3p from Candida albicans, are required for cytokinesis, and ScCts2p and CaCht4p are involved in sporulation (9–11). In addition, in C. albicans, CaCht1 and CaCht2 contribute to hyphal growth (10). Furthermore, fungal chitinases are important for utilization of chitin as a carbon source and during autolysis, for example, in A. nidulans, Penicillium chrysogenum, and Trichoderma spp. (reviewed in reference 12).
Ustilago maydis, the causal agent of corn smut, is a well-studied biotrophic phytopathogenic fungus (13). The switch from yeast to fast-growing hyphal cells is characteristic and essential for infection (13). Expansion at the apical growth cone and insertion of chitin-rich retraction septa at the basal pole of hyphae (14, 15) creates a need for new cell wall material to be incorporated and linked to the existing one. Thus, massive reorganization of chitin is expected during this morphological switch. Chitin synthases are essential and function at the tip as well as at retraction septa (16). In such rapidly elongating filaments, long-distance transport, including endosomal RNA transport, becomes important (13, 17). Previous studies on the RNA-binding protein Rrm4 identified the chitinase Cts1 as a potential target of active mRNA transport (18). Interestingly, Cts1 is unconventionally secreted (19). Even though Cts1 accumulation at the hyphal growth cone (18) makes Cts1 an ideal candidate for chitin remodeling in filaments, deletion of cts1 revealed no phenotypes during filamentous growth and pathogenicity. However, as described for other fungi, this lack of phenotypes could be due to functional redundancy, and we hypothesize that morphological changes and pathogenicity depend on redundant chitinase activities.
Therefore, the aim of the current work was to investigate the function of all chitinolytic enzymes in U. maydis during its complete life cycle in a combination of a classical genetic analysis with enzyme activity measurements and protein localization studies.
MATERIALS AND METHODS
Plasmids, strains, and growth conditions.
For the generation of plasmids containing deletion constructs, standard cloning methods (20, 21) and the Golden Gate cloning technique (26) were used (see Table S1 in the supplemental material for plasmids and Table S2 for primers). The E. coli K-12 derivative Top10 (Invitrogen/Life Technologies) was used for cloning purposes. Bacterial cells were cultivated at 37°C with shaking at 200 rpm. U. maydis strains used in this study are shown in Table 1. U. maydis cultures were grown at 28°C with 200 rpm shaking. Cultures were grown in complete medium supplemented with 1% (wt/vol) glucose (CM-G) as described previously (22). For the induction of filamentous growth in the AB33 strain, cultures were grown to an optical density at 600 nm (OD600) of 0.5 in 50 ml CM-G and shifted to 50 ml nitrate minimal medium with 1% glucose (NM-G). After 6 h, cells were harvested by centrifugation at 10,000 × g for 15 min at 4°C (23).
TABLE 1.
Strains used in this studya
| Strain | Genotype | Reference or source |
|---|---|---|
| AB33 | a2 Pnar::bE1 Pnar::bW2 | 34 |
| SG200 | a1::mfa2 bE1 bW2 | 30 |
| FB1eGFP | a1b1 Potef::egfp cbxR | 73 |
| AB33 cts1Δ | a2 Pnar::bE1 Pnar::bW2 cts1Δ hygR | 18 |
| AB33cts1:eGFP | a2 Pnar::bE1 Pnar::bW2 cts1::egfp natR | 18 |
| SG200cts1Δ | a1::mfa2 bE1 bW2 cts1Δ hygR | 18 |
| AB33rrm4Δ cts1:eGFP | a2 Pnar::bE1 Pnar::bW2 cts1::egfp::natR rrm4Δ hygR | 18 |
| AB33cts2Δ | a2 Pnar::bE1 Pnar::bW2 cts2Δ hygR | This study |
| AB33cts3Δ | a2 Pnar::bE1 Pnar::bW2 cts3Δ hygR | This study |
| SG200cts2Δ | a1::mfa2 bE1 bW2 cts2Δ hygR | This study |
| SG200cts3Δ | a1::mfa2 bE1 bW2 cts3Δ hygR | This study |
| AB33cts1Δ cts2Δ | a2 Pnar::bE1 Pnar::bW2 cts1Δ cts2Δ natR hygR | This study |
| AB33cts1Δ cts3Δ | a2 Pnar::bE1 Pnar::bW2 cts1Δ cts3Δ hygR g418R | This study |
| SG200cts1Δ cts2Δ | a1::mfa2 bE1 bW2 cts1Δ cts2Δ hygR natR | This study |
| SG200cts1Δ cts3Δ | a1::mfa2 bE1 bW2 cts1Δ cts3Δ hygR g418R | This study |
| AB33cts2Δ cts3Δ | a2 Pnar::bE1 Pnar::bW2 cts2Δ cts3Δ natR hygR | This study |
| SG200cts2Δ cts3Δ | a1::mfa2 bE1 bW2 cts2Δ cts3Δ natR hygR | This study |
| AB33cts1Δ cts2Δ cts3Δ | a2 Pnar::bE1 Pnar::bW2 cts1Δ cts2Δ cts3Δ hygR natR g418R | This study |
| SG200cts1Δ cts2Δ cts3Δ | a1::mfa2 bE1 bW2 cts1Δ cts2Δ cts3Δ hygR natR g418R | This study |
| AB33cts4Δ | a2 Pnar::bE1 Pnar::bW2 cts4Δ hygR | This study |
| AB33cts2:eGFP | a2 Pnar::bE1 Pnar::bW2 cts2::egfp hygR | This study |
| SG200cts2:eGFP | a1::mfa2 bE1 bW2 cts2::egfp hygR | This study |
| AB33cts3:eGFP | a2 Pnar::bE1 Pnar::bW2 cts3::egfp hygR | This study |
| SG200cts3:eGFP | a1::mfa2 bE1 bW2 cts3::egfp hygR | This study |
| AB33cts1Δ cts2Δ cts3Δ cts4Δ | a2 Pnar::bE1 Pnar::bW2 cts1Δ cts2Δ cts3Δ cts4Δ hygR natR g418R cbxR | This study |
| SG200cts1Δ cts2Δ cts3Δ cts4Δ | a1::mfa2 bE1 bW2 cts1Δ cts2Δ cts3Δ cts4Δ hygR natR g418R cbxR | This study |
All strains additionally contain phleomycin resistance derived from the AB33 or SG200 background.
U. maydis mutants (Table 1) were obtained by transformation of protoplasts of progenitor strains with linearized plasmids (see Table S1 in the supplemental material). Endogenous egfp fusions and gene deletion mutants were generated by homologous recombination by following established protocols and described sources of antibiotics (21, 23, 24). Homologous recombination was confirmed by diagnostic PCR and Southern blot analysis.
To test sedimentation behavior and colony morphology of chitinase mutants, cells were grown to an OD600 of 1 (for budding cells) or to an OD600 of 0.5 (for filamentous growing cells) in 50 ml CM-G. Budding cells were either dropped on CM-G agar plates (5 μl) or transferred to reaction tubes (5 ml). To determine colony morphology, plates were incubated for 24 h at 28°C. Images were taken using a charge-coupled-device (CCD) camera combined with a stereoscope (Stemi 200 c; Zeiss). To analyze sedimentation behavior, 5 ml culture was transferred to a reaction tube and incubated for 5 min without shaking at room temperature. To analyze sedimentation behavior of filaments, cells were shifted to an of filaments OD600 of 0.5 in 5 ml NM-G in reaction tubes and incubated overnight at 28°C and 200-rpm shaking. Tubes then were transferred to room temperature without shaking and incubated for 5 min.
To analyze the stress tolerance of U. maydis chitinase mutants, strains were grown in CM-G to an OD600 of 1 and washed two times in H2O. Dilutions of the cell suspension were prepared, and 5 μl of each dilution was spotted on either CM-G (for budding cells) or NM-G (for filamentous growing cells) agar plates supplemented 150 μg/ml CW, 50 μg/ml CR, 1.5 mM H2O2, 100 μg/ml SDS, 1 M NaCl, or 1 M sorbitol. Growth was examined after 24 h.
Chitinase activity assays.
For dot-gel activity assays, 20 μg of total protein extracts was spotted onto a 12% acrylamide gel supplemented with 1% glycol chitin (25) and 200 mM sodium acetate at pH 5.3 and incubated overnight in a humid chamber at 28°C. The gel was stained with CW (0.01% CW, 0.5 M Tris-HCl, pH 7.5) for 10 min at room temperature, followed by three washing steps with water (1 h, room temperature, 50 rpm shaking). Chitinase activity was visualized under UV light at 254 nm using a Stratagene eagle-eye imaging system. To control for equal loading, the gel was stained with Coomassie brilliant blue.
Fluorogenic activity measurements were carried out with the substrates 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide, 4-methylumbelliferyl-β-d-N,N′-diacetyl-chitobiose, and 4-methylumbelliferyl-β-d-N,N′,N″-triacetyl-chitotriose (Sigma). Thirty microliters of U. maydis cell culture (OD600 of 1) or 10 μg of protein extracts (in 30 μl) was incubated with 70 μl of substrate solution (diluted 1:10 in KHM buffer, containing 110 mM potassium acetate, 20 mM HEPES, 2 mM MgCl2) at 28°C for 1 h. The reaction was stopped by adding 200 μl 1 M Na2CO3. Fluorescence intensity was determined with a monochromatic fluorescence spectrometer (Infinite 200; Tecan).
To analyze chitinase reaction products of U. maydis cell extract, defined chito-oligosaccharides were incubated with 5 μg of total protein extracts overnight at 28°C. Samples were purified by filtration using a 3-kDa polyethersulfone column (13,000 × g, 15 min, room temperature; VWR) and freeze-dried for storage. The freeze-dried samples were dissolved in 10 μl H2O, and 1 μl of each sample was used for the hydrophilic interaction liquid chromatography-evaporative light-scattering detector-electrospray ionization-mass spectroscopy (HILIC-ELSD-ESI-MS) analysis using a method described previously (26). The chito-oligomers were quantified by standard calibration curves of each chitin oligomer (see Fig. S1 in the supplemental material) using the peak areas of ELSD. As a standard, the chitin oligomer (GlcNAc)1 was purchased from Sigma-Aldrich (Munich, Germany), and the chitin oligomers (GlcNAc)2–6 were from Megazyme (Bray, Ireland). GlcNAc1–6 oligomers containing equal weights (50 to 500 ng) were used to prepare the standard curves, which have a power function of f(x) = axb, where the peak area of the ELSD signal is f(x), x is the amount of the chitin oligomers (in nanograms), and a and b are substrate- and device-specific variables (27, 28, 29).
Microscopy, image processing, and staining procedures.
Microscopic analysis was performed using the following wide-field microscope setup from Visitron Systems (Munich, Germany). A Zeiss (Oberkochen, Germany) Axio Imager M1 was equipped with a Spot Pursuit CCD camera (Diagnostic Instruments, Sterling Heights, MI) and Plan Neofluar 40× (numeric aperture [NA], 1.3) and 63× (NA, 1.25) objective lenses. The excitation of fluorescently labeled proteins was carried out using an HXP metal halide lamp (LEj, Jena, Germany) in combination with a filter set for green fluorescent protein (GFP) (ET470/40BP, ET495LP, and ET525/50BP), red fluorescent protein (RFP), and mCherry (ET560/40BP, ET585LP, and ET630/75BP; Chroma, Bellow Falls, VT), and 4′,6-diamidino-2-phenylindole (DAPI) (HC387/11BP, BS409LP, and HC 447/60BP; AHF Analysentechnik, Tübingen, Germany). All parts of the microscope system were controlled by the software package MetaMorph (version 7; Molecular Devices, Sunnyvale, CA), which was also used for image processing, including the adjustment of brightness and contrast. Maximal projections are shown for localization studies. To visualize fungal cell walls and septa, 1 ml of cell culture was stained with CW (1 μg/ml) directly prior to microscopy. The U. maydis plasma membrane was visualized with 0.8 μM FM4-64 (Invitrogen).
Protein preparation and Western blot analysis.
U. maydis cells were grown to an OD600 of 1 and harvested by centrifugation (10,000 × g, 5 min, 4°C). For Western blot analysis, cells were resuspended in 2 ml of lysis buffer (100 mM sodium phosphate buffer, 10 mM Tris-HCl, pH 8.0, 8 M urea, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 2.5 mM benzamidine, and 2× complete protease inhibitor cocktail; Roche). For native protein extraction, yeast cells or filaments were resuspended in 2 ml native lysis buffer (7.9 mM Na2HPO4, 14.5 mM KH2PO4, 0.5 mM MgCl2, 2.7 mM KCl, 137 mM NaCl, 1 mM reduced DTT, 1 mM PMSF, 2.5 mM benzamidine, and 2× complete protease inhibitor cocktail; Roche). The cell suspension was frozen in liquid nitrogen and crushed in a pebble mill (5 min, 30 Hz; Retsch). After centrifugation (8,000 × g, 20 min, 4°C), the protein concentration of the supernatant was determined by Bradford assay (Bio-Rad).
Ten micrograms of total protein was used for SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. Enhanced green fluorescent protein (EGFP) fusion proteins were detected using an α-GFP antibody (mixture of the two mouse monoclonal antibodies 7.1 and 13.1; Roche) as well as a goat α-mouse IgG horseradish peroxidase (HRP) conjugate (Promega). α-Tubulin was detected as a loading control using an α-Tub1 antibody (DM1A; Sigma), as well as a goat α-mouse IgG HRP conjugate (Promega).
Pathogenicity assay.
The virulence of U. maydis chitinase mutants was tested in established pathogenicity assays as described before (30). For infection of the host plant Zea mays (Early Golden Bantam), strains were grown to an OD600 of 0.8 in CM-G, washed three times with H2O, and resuspended to an OD600 of 3 in H2O. The cell suspension was injected into 7-day-old maize seedlings. Three days postinfection (dpi), fungal hyphae were stained with wheat germ agglutinin-fluorescein isothiocyanate (WGA-FITC; 1 μg/ml) and observed microscopically. Plants at 7 dpi were scored for symptom formation. Categories for disease rating were the following: 1, no symptoms; 2, chlorosis; 3, anthocyanin accumulation; 4, small tumors (<1 mm); 5, medium tumors (>1 mm) and heavy tumors associated with bending of the stem.
RESULTS
Chitinolytic potential of U. maydis.
The genome of U. maydis (http://pedant.helmholtz-muenchen.de) contains four genes annotated as chitinolytic enzymes. Three of them are predicted chitinases (cts1 [um10419], cts2 [um02758], and cts3 [um06190]) containing a GH18 (PF00704) domain, whereas cts4 (um00695) is a predicted N-acetyl-beta-d-glucosaminidase with a GH20 (PF00728) domain (Fig. 1A). The catalytically active residues DxxDxDxE and DE (http://www.cazy.org/) (31) are conserved, suggesting that all four genes encode active enzymes (Fig. 1B). Predicted secretion signals for conventional secretion (32) are present in Cts2, Cts3, and Cts4 but are absent from Cts1. Consistently, previous studies experimentally showed unconventional secretion of Cts1, which bypasses glycosylation in the endoplasmic reticulum (ER) (18, 19). Further domains known from other chitinases, such as chitin-binding domains, glycosylphosphatidylinositol (GPI) anchors, or transmembrane domains, are absent. BLAST searches (33) with GH domains and their active sites showed no additional chitinolytic enzymes and revealed that the genomes of two close relatives, U. hordei and Sporisorium reilianum, contain the same set of chitinases, suggesting evolutionary conservation in the chitinase gene family within smut fungi. Previous phylogenetic analysis placed Cts1 in the bacterial-type chitinases, while Cts2 is a plant-like and Cts3 a basidiomycete-specific chitinase (18). In summary, we predict that the chitinolytic repertoire of U. maydis consists of three chitinases, Cts1, Cts2, and Cts3, and the N-acetyl-glucosaminidase Cts4.
FIG 1.

Chitinolytic repertoire of U. maydis. (A) SMART (http://smart.embl-heidelberg.de/) domain architecture and SignalP prediction of U. maydis chitin-degrading enzymes. The U. maydis genome encodes three chitinases (GH-18 domain), Cts1, Cts2, Cts3, and one N-acetyl-glucosaminidase (GH-20 domain), Cts4. Cts1 lacks a conventional, N-terminal signal peptide (red). (B) The consensus sequence (gray) with the catalytic residues (black) characteristic for GH-18 and GH-20 glycosyl hydrolase are conserved (www.cazy.org). The experimentally verified active sites of S. cerevisiae Cts1 (ScCts1p; P29029) and Trichoderma harzianum NAG1 (ThNag1; P87258) are provided for comparison.
Cts1 and Cts2 are active chitinases with different modes of action.
Based on our bioinformatics analysis, the catalytic center is conserved in all U. maydis chitinases, implying that they should possess enzymatic activity. However, in previous studies, deletion of cts1 led to the loss of chitinase activity toward a commercial, short-chain fluorescent substrate (18), indicating that the other two do not contribute to activity. To test the chitinolytic activity in detail, we employed three different chitinase activity assays on chitinase deletion mutants in the background strain, AB33. This strain (34) enables the induction of filamentous growth without prior mating, thereby allowing the analysis of the yeast as well as the filamentous growth stage. Deletion mutants lacking one, two, three, or all four chitinase genes (Table 1) were generated by gene replacement via homologous recombination (21, 23, 24). Chitinase deletions, even in the quadruple mutant, were readily obtained, and recombination rates of 30 to 50% are indicative of nonessential functions of the enzymes.
To test activity on long-chain chitin, the chitinase activity of AB33 and the chitinase mutants was analyzed with a gel-based dot activity assay (35). Protein extracts of yeast and filamentous AB33 cultures degraded polymeric chitin (Fig. 2A). Yeast cells lacking cts1Δ or cts2Δ had reduced chitinolytic activity, and this activity was completely abolished in extracts from the cts1/2Δ double mutant (Fig. 2A, upper). This shows that Cts1 and Cts2 are the active chitinases in yeast cells, in which Cts3 activity could not be detected. Interestingly, cell extracts of filamentous cultures of the cts1Δ mutant had no detectable chitinase activity, while in this growth stage deletion of cts2Δ did not impair chitin degradation (Fig. 2A, lower). Hence, Cts1 is the main chitinase acting in concert with Cts2 during yeast growth, but it is on its own during filamentous growth. This fits nicely with expression data showing equal expression of cts1 and cts2 in yeast cells and downregulation of cts2 in filaments, while cts3 is not expressed in either growth stage (see Fig. S2 in the supplemental material).
FIG 2.
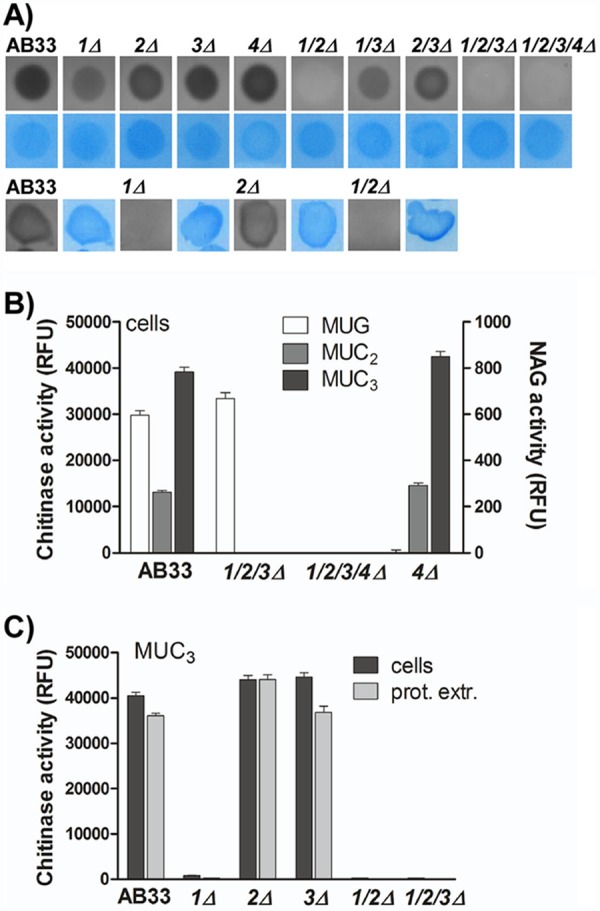
Cts1 and Cts2 contribute to chitin degradation during yeast growth, but only Cts1 is active in filaments. (A) Dot-gel activity assay for chitinase activity with protein extracts of AB33 and chitinase-deficient strains. Chitinase activity is observed as a dark halo. Equal loading was controlled by Coomassie staining (blue). Cts1 and Cts2 contribute to chitin degradation in yeast-like cells (upper), while only Cts1 is active in filaments (lower). (B) Enzymatic activity of yeast cells toward the commercial, fluorogenic substrates for chitinases (MUC2 and MUC3) and N-acetyl-glucosaminidases (NAG) (MUG). Cts4 is the only NAG that does not act on the chitinase substrates, while complete deletions of chitinases do not affect NAG activity. RFU, relative fluorescence units. (C) MUC3 degradation depends on the presence of cts1 but is not affected by deletion of the other chitinases. No difference is detected when intact cells or protein extracts (prot. extr.) are used. Error bars indicate standard deviations from three technical replicates.
The gel-based activity assay is not suitable to detect the predicted N-acetyl-glucosaminidase activity of Cts4. Therefore, we tested its enzymatic activity using short-chain fluorescent substrates. 4-Methylumbelliferyl-N-acetyl-β-d-glucosaminide (MUG) is a substrate for N-acetyl-glucosaminidases such as Cts4, and 4-methylumbelliferyl-β-d-N,N′-diacetylchitobiose (MUC2) and 4-methylumbelliferyl-β-d-N,N′,N″-triacetylchitotriose (MUC3) are substrates for chitinases such as Cts1, Cts2, and Cts3. It is expected that processive NAG activity contributes to degradation of MUC2 and MUC3, but higher enzyme concentrations are required (see Fig. S3 in the supplemental material). In U. maydis, all substrates were efficiently cleaved by intact yeast cells (Fig. 2B). In line with the prediction, strains lacking cts4 could not hydrolyze MUG, while the chitinase triple mutant cts1/2/3Δ retained full activity (Fig. 2B). Hence, Cts4 is the only N-acetyl-glucosaminidase active in yeast cells. Conversely, cts4Δ efficiently degraded the chitinase substrates MUC2 and MUC3, while the triple mutant lost chitinolytic activity toward these substrates (Fig. 2B; also see Fig. S3). Surprisingly, in this assay only Cts1 was active in degradation of the fluorescent chitinase substrates, while Cts2 did not show activity (Fig. 2C). This is consistent with previous studies (18) but inconsistent with the gel activity assays, where both Cts1 and Cts2 were active in extracts from yeast cells. To exclude that this difference in Cts2 activity is due to the measurement in intact cells, total protein extracts were tested for chitinase activity toward MUC2 and MUC3. As for intact cells, total protein extracts of mutants lacking cts1 completely lost chitinase activity (Fig. 2C), confirming that Cts1 is the only chitinase active towards these short-chain fluorescent substrates, whereas Cts2 seems to require longer chains or unlabeled chito-oligosaccharides.
To find out if Cts2 is active on unlabeled short (n ≤ 6) chito-oligosaccharides and to further characterize cleavage specificity of Cts1 and Cts2, total protein extracts were incubated with chitohexamer (GlcNAc)6 and chitotetramer (GlcNAc)4. The reaction products were analyzed by hydrophilic liquid chromatography-evaporative light scattering detection-electrospray ionization-mass spectrometry (HILIC-ELSD-ESI-MS) (26). Chitohexamer was indeed degraded to trimer (GlcNAc)3 and dimer (GlcNAc)2 in AB33, while the cts1/2Δ double mutant lacked chitinolytic activity (Table 2; also see Fig. S4 in the supplemental material), confirming that Cts3 does not contribute to the degradation of chito-oligomers. Intriguingly, both the cts1Δ and the cts2Δ single-deletion mutant still could degrade hexamer (GlcNAc)6 and tetramer (GlcNAc)4 into trimer (GlcNAc)3 and dimer (GlcNAc)2. The derived double and triple mutants degraded the chitohexamer in a corresponding pattern. Whenever cts1 or cts2 was present, degradation occurred, while the triple mutant lacks chitinase activity toward the hexamer (Table 2). Similar results were obtained using chitotetramer (GlcNAc)4 as a substrate (Table 2). Thus, Cts1 and Cts2 both are able to bind and process short chito-oligomers of n ≥ 4. In contrast, the fluorescent label from MUC3 is released only by Cts1, pointing to different enzymatic activities of the two enzymes. In support of this assumption, the (GlcNAc)2/(GlcNAc)3 ratio of strains lacking cts2Δ was significantly higher than that in strains lacking cts1Δ (Table 2). Thus, Cts1 and Cts2 seem to use different modes of chito-oligosaccharide processing, in that Cts1 preferentially releases chitodimers, while Cts2 seems to release more chitotrimers and is hindered by modification of the reducing end.
TABLE 2.
Cts1 and Cts2 degrade defined chito-oligosaccharides, but their processing mode differs
| Substrate | Detection ofa: |
Molar product ratiob |
||
|---|---|---|---|---|
| (GlcNAc)6 | (GlcNAc)4 | (GlcNAc)6 | (GlcNAc)4 | |
| AB33 | Y | Y | 8.3 ± 1.0 | 2.4 ± 0.1 |
| cts1Δ | Y | Y | 4.3 ± 0.8 | 1.3 ± 0.1 |
| cts2Δ | Y | Y | 8.2 ± 0.2 | 2.6 ± 0.2 |
| cts3Δ | Y | Y | ||
| cts1/2Δ | N | N | ||
| cts1/3Δ | Y | Y | 4.5 ± 1.4 | 1.3 ± 0.1 |
| cts2/3Δ | Y | Y | 8.3 ± 2.9 | 3.0 ± 0.2 |
| cts1/2/3Δ | N | N | ||
Incubation of chitohexamer (GlcNAc)6 or chitotetramer (GlcNAc)4 with U. maydis total protein extracts leads to production of chitodimer (GlcNAc)2 and chitotrimer (GlcNAc)3, which are detectable by HILIC-ELSD-ESI-MS (Y). This degradation is abolished in cts1Δ cts2Δ double mutants (N).
Since Cts1 and Cts2 are the only active chitinases, molar ratios between (GlcNAc)2 and (GlcNAc)3 were calculated for the deletion strains lacking either cts1 or cts2. They are reduced 2-fold in strains with Cts2 as the only active chitinase (cts1Δ and cts1/3Δ), suggesting different modes of processing for Cts1 and Cts2. Results are means and standard deviations from three independent experiments.
In summary, our activity data show that both Cts1 and Cts2 are active during yeast growth, whereas only Cts1 degrades chitin during filamentous growth. Both chitinases show activity against polymeric as well as short, oligomeric chitin derivatives, although the binding mode or cleavage site specificity might differ.
Cts1 and Cts2 act redundantly during cell separation.
To investigate the biological function of chitinases, we first looked at colony and cell morphology of AB33 cells during yeast growth and the corresponding chitinase-deficient derivatives. Wild-type AB33 and single-deletion mutants formed smooth colonies, whereas colonies of strains lacking both chitinase Cts1 and Cts2 (cts1/2Δ, cts1/2/3Δ, and cts1/2/3/4Δ) appeared rough and crinkly (see Fig. S5A in the supplemental material). The colony-morphology phenotype was accompanied by flocculation and faster sedimentation in liquid cultures (see Fig. S5A in the supplemental material). Such changes in colony morphology and sedimentation rate often are indicative of an altered cell wall composition or defects in cell separation during cytokinesis (2, 36, 37). Consistent with this, cts1/2Δ double-deletion mutants displayed a cell separation defect, while AB33 wild-type cells or cells lacking either cts1 or cts2 did not show altered cell morphology (Fig. 3). This phenotype correlates well with the observed redundancy of Cts1 and Cts2 in the in-gel activity assays. Interestingly, the growth rate in cts1/2Δ double mutants was comparable to that of the AB33 wild type (see Fig. S6), suggesting that incomplete cell division in the absence of chitinases does not feed back to the cell cycle.
FIG 3.
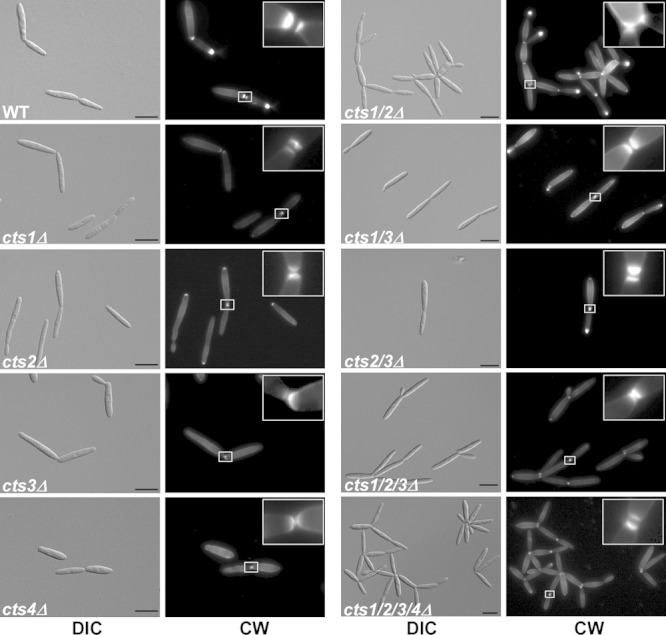
Cts1 and Cts2 act redundantly during cytokinesis. Cell morphology and septum formation of the chitinase deletion strains. cts1/2Δ strains exhibit a cytokinesis defect and form large aggregates, which is not due to lack of septum formation. Primary and secondary septa (see 5× enlargement in insets) were stained with CW prior to microscopy. Scale bars, 10 μm.
During cytokinesis, a primary septum between mother and daughter cells forms at the bud neck, followed by the secondary septum, which delimits the fragmentation zone and initiates cell separation (37). Several mutants have already been described in which cytokinesis is affected at the stage of secondary septum formation (37–39). To test for a role of the chitinases in septum formation, dividing cells were stained with Calcofluor White (CW). Both septa were inserted in all of the chitinase deletion strains (Fig. 3), indicating that formation of the fragmentation zone is not impaired; hence, cell division is affected at a later step, such as cell wall degradation.
Taken together, these observations suggest that during cell division, Cts1 and Cts2 act redundantly in cell wall degradation on the connecting chitin in the fragmentation zone.
Cts1 is translocated to the fragmentation zone by the daughter cell.
To investigate the localization of the chitinases, we generated translational C-terminal egfp fusions of cts1, cts2, and cts3 at their endogenous locus and analyzed expression, stability, and subcellular accumulation of the fusion proteins.
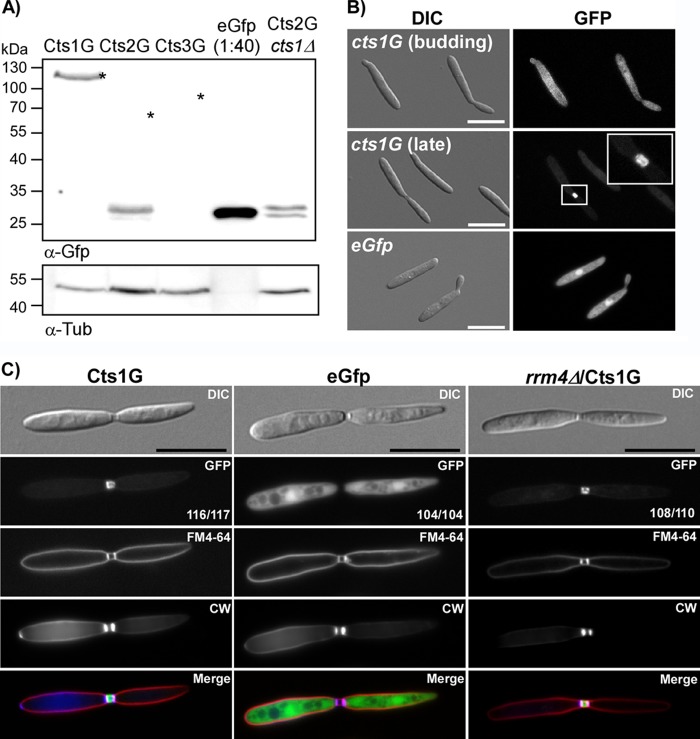
Western blot analysis confirmed that full-length Cts1G fusion protein was expressed (Fig. 4A). In contrast, for Cts2G the fusion protein appeared to be unstable, no full-length Cts2G was detected by Western blot analysis (Fig. 4A), and the GFP signal accumulated in endomembrane-like structures (data not shown). In this case, additional deletion of cts1 in the cts2-egfp strain should recapitulate the cell separation defect of the cts1/2Δ double mutant. Unexpectedly, such strains do not show a cell separation defect (data not shown), even though full-length Cts2G was not detected (Fig. 4A), suggesting that functional Cts2 is released from the fusion protein. We could not detect any expression for Cts3G. Consistent with this, semiquantitative reverse transcription-PCR (RT-PCR) showed the expression of cts1 and cts2, but not cts3, in yeast cells (see Fig. S2 in the supplemental material). Our semiquantitative data are in agreement with published microarray data (see Fig. S2) (40).
FIG 4.
Subcellular localization of chitinases. (A) Western blot analysis of chitinase fusion proteins. Cts1G can be detected as a stable fusion protein at 120 kDa. Cts2G is expressed, but stable fusion protein cannot be detected. Instead, a signal at the size of free EGFP can be observed, indicating the expression of the protein. Cts2G also is degraded in the cts1D background. The Cts3G fusion protein is not detectable as either the full length or as a degradation product. An asterisk indicates the expected size of fusion protein. (B) Cts1G is distributed evenly in the cytosol in budding cells and accumulates in the fragmentation zone between mother and daughter cells during late cell division stages. (C) Cells with a fully established fragmentation zone were identified based on the differential interference contrast (DIC) image. Subsequently, the localization of the GFP signal was scored in at least 100 cells for each strain (numbers are indicated in the GFP image). Cts1G localizes to the fragmentation zone between mother and daughter cells in AB33 and also in rrm4Δ mutants, while free cytosolic EGFP is never found in this fragmentation zone. To visualize the limits of the fragmentation zone, septa were stained with CW and the plasma membrane with FM4-64. The quantification was repeated three times with similar results. Scale bars, 10 μm.
Since unfortunately only Cts1G was stably expressed, we focused on its subcellular localization. As described before (18), Cts1G was distributed uniformly throughout the cytoplasm during early stages of cell division, when no fragmentation zone was visible. Importantly, during late stages of cell division, it accumulated at the mother-daughter cell boundary (Fig. 4B). This is consistent with the suggested activity on connecting chitin during cell separation. To analyze Cts1G localization in the fragmentation zone at the mother-daughter boundary in greater detail, we focused on cells in late division stages with a clearly established fragmentation zone between mother and daughter cells. Nearly all cells in this stage showed Cts1G signals in the fragmentation zone, whereas free cytosolic GFP never localized in this zone (Fig. 4C). Colocalization with the membrane dye FM4-64 and CW confirmed that the fragmentation zone indeed is an extracellular compartment delimited by septa and plasma membrane (Fig. 4C). Hence, Cts1G is actively secreted to this zone during late stages of cytokinesis. In filaments, but not in yeast cells, unconventional Cts1 secretion depends on Rrm4 (18). Nevertheless, we tested if Rrm4 is involved in this relocalization of Cts1 during cytokinesis. In the rrm4Δ deletion mutant, Cts1G accumulated in the fragmentation zone (Fig. 4C), showing that, as expected, Rrm4-dependent mRNA transport processes are dispensable for this Cts1 translocation.
During formation of the fragmentation zone in U. maydis, the mother cell first inserts the primary septum, followed by transfer of vesicles and proteins from the daughter cell to the newly establishing fragmentation zone, and finally formation of the secondary septum by the daughter cell (38, 41). Consistent with this, we detected asymmetric distribution of Cts1G into the fragmentation zone only from the daughter cell but never from the mother cell (see Fig. S7 in the supplemental material). Colocalization with FM4-64 and CW further confirmed that asymmetry of Cts1 occurred at early stages when only the primary septum was inserted (see Fig. S7D).
Taken together, Cts1 is translocated to the fragmentation zone during late cytokinesis prior to insertion of the secondary septum, where its activity supports the physical separation of mother and daughter cells.
Cts1 is the only functional chitinase during filamentous growth.
In order to infect its host plant, U. maydis switches from yeast to filamentous growth. Cts1 is the only active chitinase at this growth stage (Fig. 2A, lower), and in previous studies Cts1G localized to the subapical region of filaments (18), where it might contribute to cell elongation by loosening the chitin structure of the cell wall. To test if the other chitinases contribute to morphological changes in filament formation despite a lack of detectable chitinase activity, we investigated filamentous growth of the chitinase mutants in the AB33 background. Filaments elongated unipolarly and retraction septa were inserted, leaving empty sections at the basal pole even in the absence of all chitinases (Fig. 5A). This confirms that chitin degradation at the growth pole is not required for unipolar growth. Despite normal filament formation, in cts1Δ mutants the resulting filaments aggregate in liquid culture (18). In line with the activity data, this phenotype depends solely on Cts1 (see Fig. S5B in the supplemental material) and was not enhanced by the lack of other chitinases. This confirms the chitinase activity measurement and that the redundant function of cts1 and cts2 observed during yeast growth is not mirrored during filamentous growth.
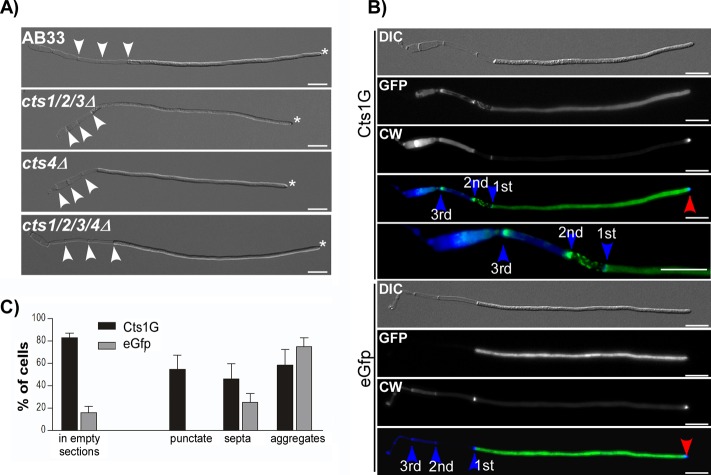
FIG 5.
Chitinases are dispensable for filament formation but still localize to empty sections. (A) Microscopic analysis of AB33 and corresponding chitinase-deficient strains. Even strains which lack all chitinolytic enzymes are able to establish unipolar filaments. All strains show a single growing pole (asterisks) and insert septa at the basal pole (arrowheads). Scale bars, 10 μm. (B) Fluorescence microscopic colocalization analysis of Cts1G in filaments. Cts1G localizes in the cytosol and in distinct patterns at the cell wall of empty sections, such as punctate localization (mainly the first empty section), accumulation at septa, and random aggregates (mainly in old empty sections beyond the second retraction septum). In the cytosol, Cts1G abundance gradually increases toward the hyphal tip, as described before (18). Free cytosolic EGFP is distributed equally throughout the cytoplasm. No accumulation is observable in empty sections. Septa are highlighted by blue and the growth poles by red arrowheads. Scale bars, 10 μm. (C) Quantification of localization patterns. Elongated filaments with at least three empty sections were identified based on the DIC image. Subsequently, the localization of the GFP signal in empty sections was scored in at least 100 images for each strain. A total of 83.1% of all cells expressed Cts1G (n = 181), but only 15.9% of EGFP cells (n = 139) showed EGFP signal in empty sections. Further scoring of cells with signal inside the empty sections (n = 150 for Cts1G, n = 22 for EGFP) into the different patterns clarified that free EGFP occasionally aggregates, whereas punctate localization, which is typical for Cts1G, was never observed in cytosolic EGFP-expressing cells. Error bars show standard deviations from three independent experiments. The quantification was repeated three times with similar results.
Filament aggregation could be caused by altered cell wall properties, which in turn might influence stress tolerance. To test for stress resistance in the chitinase mutants, we grew yeast and filamentous cells on different stress-inducing agents. CW and Congo red inhibit chitin microfibril assembly (42), SDS is a detergent inducing cell wall stress through perturbation of the membrane (43), H2O2 causes oxidative stress, and NaCl and sorbitol cause osmotic stress. No differences were observed in the stress responses of mutants in either the yeast or filamentous growth stages, suggesting that this major cell wall function is not dependent on chitinases (see Fig. S8 in the supplemental material).
During filament elongation, retraction septa are inserted at the basal pole, leaving an empty section behind. Previously it was shown that Cts1G localizes to the hyphal tip, but aggregation of cts1Δ filaments occurs at the empty sections, also suggesting the action of Cts1 at this site (18). Therefore, we analyzed the localization of Cts1 in closer detail. The gradient of Cts1G toward the hyphal tip was confirmed, which is absent from free cytosolic EGFP (Fig. 5B). In addition, Cts1G localizes at the cell wall of empty compartments (Fig. 5B), which is consistent with the hypothesized role in aggregation. Interestingly, different patterns for Cts1G were observed in the empty sections. Punctate structures occurred mainly in the first empty section, accumulation at septa was most prominent at septa in the second empty section, and random accumulation was present mainly within older sections (Fig. 5C). Such patterns, except for random accumulations, were not observed in strains expressing cytosolic EGFP (Fig. 5C). Since Cts1 secretion depends on Rrm4 in filaments (18), we analyzed Cts1G localization in rrm4Δ. Surprisingly, its localization to the empty section is independent of Rrm4 (see Fig. S9 in the supplemental material), suggesting a different secretion pathway into the empty sections, which might resemble translocation into the fragmentation zone in yeast cells.
Collectively, our data showed that chitinases are unexpectedly dispensable for the establishment of unipolar growth. Instead, Cts1 is the single active chitinase that contributes to chitin degradation in empty sections, and it prevents aggregation.
Chitinases are dispensable during biotrophic development.
Mating-dependent filament formation is the first step in the infection process. During mating, chitinases could remodel the cells wall to allow growth of conjugation hyphae or fusion at the tip. We tested mating of AB33 and the chitinase-deficient strain with wild-type mating partners. Conjugation hyphae are induced in the chitinase-deficient strain by the compatible mating partner FB1, and they fuse to result in elongating filaments (see Fig. S10 in the supplemental material). The infectious filaments form appressorium-like structures for penetration of the plant surface and in planta proliferation. During these steps, the fungal cell wall establishes close contact with the host cell wall and plasma membrane (30). At these sites of intimate contact, fungal chitin can be recognized by the plant as a general elicitor of the immune system and trigger defense reactions (44). To test if chitinases contribute to pathogenic development and biotrophic growth, we generated chitinase deletion mutants in the solopathogenic strain SG200, which is able to infect the host plant Zea mays without prior mating (30). Virulence of the respective chitinase mutants was tested in seedling infection assays. All mutants were fully virulent. Hyphae proliferated and formed clamp cells in the maize leaves independently of chitinolytic activity (Fig. 6A). Tumors formed within 1 week after infection, even in mutants without any chitinase activity, as efficiently as in SG200, indicating that chitinases are dispensable during the infection process (Fig. 6B). During tumor formation, hyphal fragmentation is induced, leading to formation of black teliospores, which germinate and give rise to the promycelium from which haploid sporidia bud off. Both processes, hyphal fragmentation and teliospore germination, involve cell wall remodeling (45). To test if chitinases are required for teliospore formation or germination, we harvested tumors from the infected plants 4 weeks after infection and collected mature teliospores. The teliospores from both SG200 and the chitinase quadruple mutant germinated and subsequently gave rise to yeast-growing colonies (data not shown).
FIG 6.
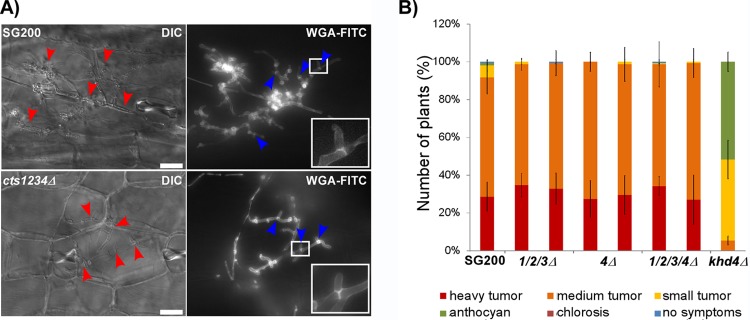
Chitinases are dispensable for pathogenesis. (A) Microscopic analysis of SG200 and the quadruple chitinase deletion mutant 3 days after infection. Even the strain lacking chitinolytic activity still is able to form clamp cells to proliferate inside the host plant. Fungal hyphae inside the plant are highlighted by red arrowheads. Clamp cells visualized by WGA-FITC straining are highlighted by blue arrowheads. Scale bars, 20 μm. (B) Disease rating of maize seedlings 9 days after infection with U. maydis. Two independent transformants were tested for each strain (n > 100). All chitinase-deficient mutants infected the host plant, leading to heavy tumor formation, as observed in wild-type infections. The solopathogenic strain SG200, which is fully pathogenic, was used as a reference. The khd4Δ mutant (74) was used as a reduced-virulence control strain. Error bars indicate standard deviations from three independent experiments.
Hence, by analyzing a quadruple mutant lacking all chitinolytic enzymes, we show unambiguously that against expectations, chitinolytic activity is not necessary for biotrophic development of U. maydis.
DISCUSSION
Although chitinases have been the subject of extensive research for decades, knowledge of their biological function and their role in fungal development still is scarce. Most information comes from studies in model ascomycetes (41, 46). To extend our understanding of fungal chitinases, we focused on the phytopathogenic basidiomycete U. maydis, whose complex life cycle comprises several morphological switches and represents a good model to study the role of chitinases in cell wall remodeling (47). Its cell wall contains 14 to 16% chitin (48), pointing to a need for chitinase activity during morphogenesis. Furthermore, the relatively small number of chitinase genes in this fungus allowed a comprehensive genetic approach and in vivo characterization of chitinase activity.
To biochemically characterize the enzymatic chitinase activity, we employed three different chitinase activity assays using chitinase deletion mutants. In-gel assays with polymeric chitin chains offer the most natural substrate. This led us to detect the activity of Cts1 and Cts2 during yeast growth and of Cts1 alone during filament formation. In addition to long-chain chitin, Cts1 and Cts2 both are able to cleave defined chito-oligomers of n ≥ 4. Differences in the resulting product lengths suggest different cleavage preferences of Cts1 and Cts2. Notably, the commercially available fluorescent substrate MUC3 is cleaved by Cts1 but not Cts2, making it an ideal substrate to distinguish activity of Cts1 and Cts2. Possibly, the fluorescent group hinders Cts2, but not Cts1, to cleave the pseudotetramer, further supporting the idea of different substrate preferences of the two chitinases.
By deleting all possible combinations of the four genes for chitinolytic enzymes, we could identify two roles for chitinases throughout the complete life cycle. First, Cts1 and Cts2 act redundantly during cell separation, and second, Cts1 degrades remnant chitin in empty sections during filamentous growth. Surprisingly, in contrast to our expectations, all chitinase mutants are able to finish the biotrophic part of the life cycle, i.e., they proceed through filament induction, plant infection, teliospore formation, and germination.
Role of chitinases in cell separation.
Our study revealed a role of chitinases in cell separation of yeast cells in U. maydis. Such a function of chitinases has already been described in ascomycetes (reviewed in references 12 and 49), e.g., in the model yeast S. cerevisiae (11), the opportunistic pathogen Candida albicans (10), and the industrially used yeast Kluyveromyces lactis (50). In these species, single deletions of the corresponding chitinase lead to cell aggregation. In contrast, deletion of all chitinases in the basidiomycete Cryptococcus neoformans does not affect cytokinesis (51). Interestingly, we could show that in U. maydis, even two chitinases act redundantly in cell separation. Such redundant action of multiple cell wall-degrading enzymes for efficient cell separation to ensure rapid and complete degradation of residual cell wall material is already known for other cell wall-degrading enzymes. In the fission yeast S. pombe, two endoglucanases act redundantly during cytokinesis (52), and their expression is regulated by the transcription factor Ace2p (53, 54). In S. cerevisiae, localization of this transcription factor to the daughter nucleus additionally limits expression of the chitinase ScCts1p to the daughter cell (55, 56), thereby coordinating cell wall-degrading activities of different enzyme classes. In U. maydis the chitinases Cts1 and Cts2 are secreted by different pathways and have different catalytic activities, which indicate that they attack the chitin layer from different directions.
The fact that Cts1 and Cts2 are secreted by different mechanisms raises questions about the secretion pathways. How exactly secretion to the fragmentation zone is achieved during cell separation still remains unclear. The constant cytoplasmic pool of Cts1 and its specific translocation to the fragmentation zone from the daughter cell is reminiscent of protein translocation during formation of the secondary septum (38, 57), suggesting its deposition in the vesicle-rich region prior to insertion of the secondary septum. Clearly, Cts1 avoids the classical ER-Golgi apparatus-dependent secretory pathway (19). In contrast, Cts2 contains a secretion signal peptide. Interestingly, in contrast to Cts1G, the Cts2G fusion protein is unstable but the degradation product retains activity. Passage through the ER-Golgi apparatus pathway could lead to proteolytic processing of Cts2, similar to the zymogenic properties of yeast chitin synthase 3 (58) or activation of killer toxins by the protease Kex2 (59). Consistent with this, the S. cerevisiae chitinase ScCts1p, which mediates cytokinesis, is secreted in a signal peptide-dependent manner and requires glycosylation in the ER, whereas ScCts2p, like Cts1, lacks an N-terminal secretion signal (9, 19, 60). Hence, in general, chitinases seem to employ different modes of secretion, which might be required for their differential regulation or might be caused by their ability to bind sugar residues, which would interfere with glycosylation.
In filaments, remnant chitin is degraded at empty sections.
In filaments, Cts1 is the only active chitinase. This is in line with the expression data showing downregulation of cts2 expression in filaments and appressoria (61), raising questions about the biological role of chitinases during filamentous growth in this single chitinase. Previously it was reported that Cts1G localizes to the growth apex of filaments (18). However, the activity of chitinases at the growth cone does not seem to be essential, since filaments grow unipolarly even in the absence of all chitinases. In contrast, other fungi rely on chitinase activity for hyphal growth processes. In Neurospora crassa, Chit-1 localizes in the cell wall of vegetative hyphae (62) and deletion strains have a lower growth rate (63), suggesting that the chitinase remodels the chitin layer of the cell wall during hyphal elongation. Similarly, in A. nidulans, ChiA is required for colony growth (64) and localizes to hyphal tips (65). In addition to localization at the growth apex of U. maydis filaments, we observed localization of Cts1G in retraction septa and empty sections at the basal pole. These retraction septa are important for movement of the cytosol during filament elongation and for appressoria formation (14, 15). However, despite the accumulation of Cts1 at the septa, chitin remodeling by chitinases is not needed for filament elongation. Instead, Cts1 seems to act on the chitin remnant in empty sections. Modification of chitin in this compartment might alter the cell wall composition so that filaments do not aggregate and sediment in culture. Such chitin degradation in remnant cell wall material might be similar to autolytic processes observed for ChiB in A. nidulans and ChiB1 in A. fumigatus (65, 66). Taking these findings together, U. maydis maintains the expression of one nonconventionally secreted chitinase during its filamentous growth stage, even though its activity is dispensable for morphogenesis.
Chitinases are dispensable for biotrophic growth.
In phytopathogenic fungi, an additional role of chitinases during infection can be hypothesized. Chito-oligomers released from the fungal cell wall act as general elicitors of plant defense responses (44). Cladosporium fulvum secretes chitin binding proteins such as Avr4, which binds the fungal cell wall and prevents its degradation by plant chitinases, and Ecp6, which sequesters chito-oligomers (46, 67). Together they prevent detection of the fungus by the plant immune system. Chitinases might further contribute to successful immune evasion by degrading elicitor-active chito-oligomers.
Contradictory to this hypothesis, endogenous chitinases are not necessary for successful infection in U. maydis. This suggests that other virulence mechanisms are sufficient for immune suppression. Downstream of elicitor recognition, an early plant defense response is the production of reactive oxygen species. This production is efficiently inhibited by the Pep1 effector (68), indicating that U. maydis does not even need to mask individual elicitors such as chitin due to an efficient effector system but rather tackles signaling downstream of the convergence point.
Nevertheless, chitinases still could be a fitness factor that cannot be detected in laboratory infection experiments but may become relevant in nature. They could function in defeating endophytic fungi such as Fusarium verticillioides by attacking their cell walls inside the host plant, which otherwise would limit U. maydis growth (69). Similarly, antifungal chitinase activity could be directed against other pathogenic fungi, as described for the activity of ustilagic acid produced by U. maydis against Botrytis cinerea (70).
In conclusion, we extensively characterized the role of chitinases during the life cycle of the corn smut U. maydis. In contrast to an expected function of these enzymes during cell wall remodeling, U. maydis employs its chitinolytic machinery to degrade remnant chitin during saprophytic growth. This activity supports cell separation. Under natural conditions, complete separation of the yeast cells without mechanical force would provide benefits in access to nutrients or spreading of the yeast cells by wind and water (71). Despite extensive genetic characterization, the function of cts3 still remained elusive under the tested conditions. It might play a role in the interaction with other fungi, similar to the known role of chitinases that can act as biocontrol agents (72).
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Feldbrügge for supporting the work financially and scientifically, K. Schipper and all laboratory members for valuable discussion, and C. Haag for critical reading of the manuscript. Special thanks to K. Bösch, Z. Ye, S. Jankowski, S. Esch, and J. Stock for experimental support.
T.L. was supported by a doctoral fellowship of the DFG International Research Training Group 1525 iGRADplant. The laboratory of J.D.W. received funding from the Great Lakes Bioenergy Research Center (U.S. Department of Energy Office of Science BER DE-FC02-07ER64494) and from the MSU-Plant Research Laboratory (DOE Office of Basic Energy Sciences DE-FG02-91ER200021).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00022-15.
REFERENCES
- 1.Bowman SM, Free SJ. 2006. The structure and synthesis of the fungal cell wall. Bioessays 28:799–808. doi: 10.1002/bies.20441. [DOI] [PubMed] [Google Scholar]
- 2.Klis FM, Boorsma A, De Groot PWJ. 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast 23:185–202. doi: 10.1002/yea.1349. [DOI] [PubMed] [Google Scholar]
- 3.Seidl V. 2008. Chitinases of filamentous fungi: a large group of diverse proteins with multiple physiological functions. Fungal Biol Rev 22:36–42. doi: 10.1016/j.fbr.2008.03.002. [DOI] [Google Scholar]
- 4.Karlsson M, Stenlid J. 2008. Comparative evolutionary histories of the fungal chitinase gene family reveal non-random size expansions and contractions due to adaptive natural selection. Evol Bioinformatics 4:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aam BB, Heggset EB, Norberg AL, Sorlie M, Varum KM, Eijsink VGH. 2010. Production of chitooligosaccharides and their potential applications in medicine. Mar Drugs 8:1482–1517. doi: 10.3390/md8051482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermosa R, Viterbo A, Chet I, Monte E. 2012. Plant-beneficial effects of Trichoderma and of its genes. Microbiology 158:17–25. doi: 10.1099/mic.0.052274-0. [DOI] [PubMed] [Google Scholar]
- 7.Neeraja C, Anil K, Purushotham P, Suma K, Sarma P, Moerschbacher BM, Podile AR. 2010. Biotechnological approaches to develop bacterial chitinases as a bioshield against fungal diseases of plants. Crit Rev Biotechnol 30:231–241. doi: 10.3109/07388551.2010.487258. [DOI] [PubMed] [Google Scholar]
- 8.Alcazar-Fuoli L, Clavaud C, Lamarre C, Aimanianda V, Seidl-Seiboth V, Mellado E, Latge JP. 2011. Functional analysis of the fungal/plant class chitinase family in Aspergillus fumigatus. Fungal Genet Biol 48:418–429. doi: 10.1016/j.fgb.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Dünkler A, Jorde S, Wendland J. 2008. An Ashbya gossypii cts2 mutant deficient in a sporulation-specific chitinase can be complemented by Candida albicans CHT4. Microbiol Res 163:701–710. doi: 10.1016/j.micres.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Dünkler A, Walther A, Specht CA, Wendland J. 2005. Candida albicans CHT3 encodes the functional homolog of the Cts1 chitinase of Saccharomyces cerevisiae. Fungal Genet Biol 42:935–947. doi: 10.1016/j.fgb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Kuranda MJ, Robbins PW. 1991. Chitinase is required for cell-separation during growth of Saccharomyces cerevisiae. J Biol Chem 266:19758–19767. [PubMed] [Google Scholar]
- 12.Hartl L, Zach S, Seidl-Seiboth V. 2012. Fungal chitinases: diversity, mechanistic properties and biotechnological potential. Appl Microbiol Biotechnol 93:533–543. doi: 10.1007/s00253-011-3723-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vollmeister E, Schipper K, Baumann S, Haag C, Pohlmann T, Stock J, Feldbrügge M. 2012. Fungal development of the plant pathogen Ustilago maydis. FEMS Microbiol Rev 36:59–77. doi: 10.1111/j.1574-6976.2011.00296.x. [DOI] [PubMed] [Google Scholar]
- 14.Freitag J, Lanver D, Bohmer C, Schink KO, Bolker M, Sandrock B. 2011. Septation of infectious hyphae is critical for appressoria formation and virulence in the smut fungus Ustilago maydis. PLoS Pathog 7:e1002044. doi: 10.1371/journal.ppat.1002044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinberg G, Schliwa M, Lehmler C, Bolker M, Kahmann R, McIntosh JR. 1998. Kinesin from the plant pathogenic fungus Ustilago maydis is involved in vacuole formation and cytoplasmic migration. J Cell Sci 111:2235–2246. [DOI] [PubMed] [Google Scholar]
- 16.Weber I, Assmann D, Thines E, Steinberg G. 2006. Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell 18:225–242. doi: 10.1105/tpc.105.037341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumann S, Pohlmann T, Jungbluth M, Brachmann A, Feldbrugge M. 2012. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J Cell Sci 125:2740–2752. doi: 10.1242/jcs.101212. [DOI] [PubMed] [Google Scholar]
- 18.Koepke J, Kaffarnik F, Haag C, Zarnack K, Luscombe NM, König J, Ule J, Kellner R, Begerow D, Feldbrügge M. 2011. The RNA-binding protein Rrm4 is essential for efficient secretion of endochitinase Cts1. Mol Cell Proteomics 10:M111.011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stock J, Sarkari P, Kreibich S, Brefort T, Feldbrügge M, Schipper K. 2012. Applying unconventional secretion of the endochitinase Cts1 to export heterologous proteins in Ustilago maydis. J Biotechnol 161:80–91. doi: 10.1016/j.jbiotec.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 21.Terfrüchte M, Joehnk B, Fajardo-Somera R, Braus GH, Riquelme M, Schipper K, Feldbrügge M. 2014. Establishing a versatile Golden Gate cloning system for genetic engineering in fungi. Fungal Genet Biol 62:1–10. doi: 10.1016/j.fgb.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Holliday R. 1974. Ustilago maydis, p 575–595. In King RC. (ed), Handbook of genetics, vol 1 Plenum Press, New York, NY. [Google Scholar]
- 23.Brachmann A, König J, Julius C, Feldbrügge M. 2004. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol Genet Genomics 272:216–226. [DOI] [PubMed] [Google Scholar]
- 24.Kämper J. 2004. A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol Genet Genomics 271:103–110. doi: 10.1007/s00438-003-0962-8. [DOI] [PubMed] [Google Scholar]
- 25.Trudel J, Asselin A. 1989. Detection of chitinase activity after polyacrylamide gel electrophoresis. Anal Biochem 178:362–366. doi: 10.1016/0003-2697(89)90653-2. [DOI] [PubMed] [Google Scholar]
- 26.Hamer SN, Cord-Landwehr S, Biarnés X, Planas A, Waegeman H, Moerschbacher BM, Kolkenbrock S. 2015. Enzymatic production of defined chitosan oligomers with a specific pattern of acetylation using a combination of chitin oligosaccharide deacetylases. Sci Rep 5:8716. doi: 10.1038/srep08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mengerink Y, Deman HCJ, Vanderwal S. 1991. Use of an evaporative light-scattering detector in reversed-phase high-performance liquid-chromatography of oligomeric surfactants. J Chromatogr 552:593–604. doi: 10.1016/S0021-9673(01)95975-8. [DOI] [Google Scholar]
- 28.Decroos K, Vincken JP, Heng L, Bakker R, Gruppen H, Verstraete W. 2005. Simultaneous quantification of differently glycosylated, acetylated, and 2,3-dihydro-2,5-dihydroxy-6-methyl-4H-pyran-4-one-conjugated soyasaponins using reversed-phase high-performance liquid chromatography with evaporative light scattering detection. J Chromatogr A 1072:185–193. doi: 10.1016/j.chroma.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 29.Remoroza C, Cord-Landwehr S, Leijdekkers AGM, Moerschbacher BM, Schols HA, Gruppen H. 2012. Combined HILIC-ELSD/ESI-MSn enables the separation, identification and quantification of sugar beet pectin derived oligomers. Carbohydr Polym 90:41–48. doi: 10.1016/j.carbpol.2012.04.058. [DOI] [PubMed] [Google Scholar]
- 30.Kämper J, Kahmann R, Bölker M, Ma LJ, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Muller O, Perlin MH, Wosten HAB, de Vries R, Ruiz-Herrera J, Reynaga-Pena CG, Snetselaar K, McCann M, Perez-Martin J, Feldbrügge M, Basse CW, Steinberg G, Ibeas JI, Holloman W, Guzman P, Farman M, Stajich JE, Sentandreu R, Gonzalez-Prieto JM, Kennell JC, Molina L, Schirawski J, Mendoza-Mendoza A, Greilinger D, Munch K, Rossel N, Scherer M, Vranes M, Ladendorf O, Vincon V, Fuchs U, Sandrock B, Meng S, Ho ECH, Cahill MJ, Boyce KJ, Klose J, Klosterman SJ, Deelstra HJ, Ortiz-Castellanos L, Li W, Sanchez-Alonso P, Schreier PH, Häuser-Hahn I, Vaupel M, Koopmann E, Friedrich G, Voss H, Schlüter T, Margolis J, Platt D, Swimmer C, Gnirke A, Chen F, Vysotskaia V, Mannhaupt G, Güldener U, Münsterkötter M, Haase D, Oesterheld M, Mewes HW, Mauceli EW, DeCaprio D, Wade CM, Butler J, Young S, Jaffe DB, Calvo S, Nusbaum C, Galagan J, Birren BW. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- 31.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Brachmann A, Weinzierl G, Kamper J, Kahmann R. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol Microbiol 42:1047–1063. doi: 10.1046/j.1365-2958.2001.02699.x. [DOI] [PubMed] [Google Scholar]
- 35.Stöveken J, Singh R, Kolkenbrock S, Zakrzewski M, Wibberg D, Eikmeyer FG, Pühler A, Schlüter A, Moerschbacher BM. 2015. Successful heterologous expression of a novel chitinase identified by sequence analyses of the metagenome from a chitin-enriched soil sample. J Biotechnol 201:60–68. doi: 10.1016/j.jbiotec.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 36.Voth WP, Olsen AE, Sbia M, Freedman KH, Stillman DJ. 2005. ACE2, CBK1, and BUD4 in budding and cell separation. Eukaryot Cell 4:1018–1028. doi: 10.1128/EC.4.6.1018-1028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinzierl G, Leveleki L, Hassel A, Kost G, Wanner G, Bolker M. 2002. Regulation of cell separation in the dimorphic fungus Ustilago maydis. Mol Microbiol 45:219–231. doi: 10.1046/j.1365-2958.2002.03010.x. [DOI] [PubMed] [Google Scholar]
- 38.Böhmer C, Böhmer M, Bölker M, Sandrock B. 2008. Cdc42 and the Ste20-like kinase Don3 act independently in triggering cytokinesis in Ustilago maydis. J Cell Sci 121:143–148. doi: 10.1242/jcs.014449. [DOI] [PubMed] [Google Scholar]
- 39.Mahlert M, Leveleki L, Hlubek A, Sandrock B, Bolker M. 2006. Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol Microbiol 59:567–578. doi: 10.1111/j.1365-2958.2005.04952.x. [DOI] [PubMed] [Google Scholar]
- 40.Heimel K, Scherer M, Vranes M, Wahl R, Pothiratana C, Schuler D, Vincon V, Finkernagel F, Flor-Parra I, Kamper J. 2010. The transcription factor Rbf1 is the master regulator for b-mating type controlled pathogenic development in Ustilago maydis. PLoS Pathog 6:e1001035. doi: 10.1371/journal.ppat.1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schink KO, Bolker M. 2009. Coordination of cytokinesis and cell separation by endosomal targeting of a Cdc42-specific guanine nucleotide exchange factor in Ustilago maydis. Mol Biol Cell 20:1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roncero C, Duran A. 1985. Effect of Calcofluor White and Congo Red on fungal cell-wall morphogenesis–in vivo activation of chitin polymerization. J Bacteriol 163:1180–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ram AF, Arentshorst M, Damveld RA, van Kuyk PA, Klis FM, van den Hondel CA. 2004. The cell wall stress response in Aspergillus niger involves increased expression of the glutamine: fructose-6-phosphate amidotransferase-encoding gene (gfaA) and increased deposition of chitin in the cell wall. Microbiology 150:3315–3326. doi: 10.1099/mic.0.27249-0. [DOI] [PubMed] [Google Scholar]
- 44.Wan J, Zhang XC, Stacey G. 2008. Chitin signaling and plant disease resistance. Plant Signal Behav 3:831–833. doi: 10.4161/psb.3.10.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banuett F, Herskowitz I. 1996. Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development 122:2965–2976. [DOI] [PubMed] [Google Scholar]
- 46.van den Burg HA, Harrison SJ, Joosten MHAJ, Vervoort J, de Wit PJGM. 2006. Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe Interact 19:1420–1430. doi: 10.1094/MPMI-19-1420. [DOI] [PubMed] [Google Scholar]
- 47.Feldbrügge M, Kamper J, Steinberg G, Kahmann R. 2004. Regulation of mating and pathogenic development in Ustilago maydis. Curr Opin Microbiol 7:666–672. doi: 10.1016/j.mib.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz-Herrera J, Leon CG, Carabez-Trejo A, Reyes-Salinas E. 1996. Structure and chemical composition of the cell walls from the haploid yeast and mycelial forms of Ustilago maydis. Fungal Genet Biol 20:133–142. doi: 10.1006/fgbi.1996.0028. [DOI] [PubMed] [Google Scholar]
- 49.Adams DJ. 2004. Fungal cell wall chitinases and glucanases. Microbiology 150:2029–2035. doi: 10.1099/mic.0.26980-0. [DOI] [PubMed] [Google Scholar]
- 50.Colussi PA, Specht CA, Taron CH. 2005. Characterization of a nucleus-encoded chitinase from the yeast Kluyveromyces lactis. Appl Environ Microbiol 71:2862–2869. doi: 10.1128/AEM.71.6.2862-2869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker LG, Specht CA, Lodge JK. 2009. Chitinases are essential for sexual development but not vegetative growth in Cryptococcus neoformans. Eukaryot Cell 8:1692–1705. doi: 10.1128/EC.00227-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dekker N, Speijer D, Grun CH, van den Berg M, de Haan A, Hochstenbach F. 2004. Role of the alpha-glucanase Agn1p in fission-yeast cell separation. Mol Biol Cell 15:3903–3914. doi: 10.1091/mbc.E04-04-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alonso-Nunez ML, An HB, Martin-Cuadrado AB, Mehta S, Petit C, Sipiczki M, del Rey F, Gould KL, de Aldana CRV. 2005. Ace2p controls the expression of genes required for cell separation in Schizosaccharomyces pombe. Mol Biol Cell 16:2003–2017. doi: 10.1091/mbc.E04-06-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dekker N, de Haan A, Hochstenbach F. 2006. Transcription regulation of the alpha-glucanase gene agn1 by cell separation transcription factor Ace2p in fission yeast. FEBS Lett 580:3099–3106. doi: 10.1016/j.febslet.2006.04.061. [DOI] [PubMed] [Google Scholar]
- 55.Colman-Lerner A, Chin TE, Brent R. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107:739–750. doi: 10.1016/S0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- 56.Doolin MT, Johnson AL, Johnston LH, Butler G. 2001. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol Microbiol 40:422–432. doi: 10.1046/j.1365-2958.2001.02388.x. [DOI] [PubMed] [Google Scholar]
- 57.Sandrock B, Böhmer C, Bölker M. 2006. Dual function of the germinal centre kinase Don3 during mitosis and cytokinesis in Ustilago maydis. Mol Microbiol 62:655–666. doi: 10.1111/j.1365-2958.2006.05405.x. [DOI] [PubMed] [Google Scholar]
- 58.Choi WJ, Sburlati A, Cabib E. 1994. Chitin synthase 3 from yeast has zymogenic properties that depend on both the CAL1 and the CAL3 genes. Proc Natl Acad Sci U S A 91:4727–4730. doi: 10.1073/pnas.91.11.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tao J, Ginsberg I, Banerjee N, Held W, Koltin Y, Bruenn JA. 1990. Ustilago maydis KP6 killer toxin: structure, expression in Saccharomyces cerevisiae, and relationship to other cellular toxins. Mol Cell Biol 10:1373–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarkari P, Reindl M, Stock J, Muller O, Kahmann R, Feldbrügge M, Schipper K. 2014. Improved expression of single-chain antibodies in Ustilago maydis. J Biotechnol 191:165–175. doi: 10.1016/j.jbiotec.2014.06.028. [DOI] [PubMed] [Google Scholar]
- 61.Lanver D, Berndt P, Tollot M, Naik V, Vranes M, Warmann T, Munch K, Rossel N, Kahmann R. 2014. Plant surface cues prime Ustilago maydis for biotrophic development. PLoS Pathog 10:e1004272. doi: 10.1371/journal.ppat.1004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maddi A, Bowman SM, Free SJ. 2009. Trifluoromethanesulfonic acid-based proteomic analysis of cell wall and secreted proteins of the ascomycetous fungi Neurospora crassa and Candida albicans. Fungal Genet Biol 46:768–781. doi: 10.1016/j.fgb.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tzelepis GD, Melin P, Jensen DF, Stenlid J, Karlsson M. 2012. Functional analysis of glycoside hydrolase family 18 and 20 genes in Neurospora crassa. Fungal Genet Biol 49:717–730. doi: 10.1016/j.fgb.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 64.Takaya N, Yamazaki D, Horiuchi H, Ohta A, Takagi M. 1998. Cloning and characterization of a chitinase-encoding gene chiA from Aspergillus nidulans, disruption of which decreases germination frequency and hyphal growth. Biosci Biotechnol Biochem 62:60–65. doi: 10.1271/bbb.62.60. [DOI] [PubMed] [Google Scholar]
- 65.Yamazaki H, Tanaka A, Kaneko J, Ohta A, Horiuchi H. 2008. Aspergillus nidulans ChiA is a glycosylphosphatidylinositol (GPI)-anchored chitinase specifically localized at polarized growth sites. Fungal Genet Biol 45:963–972. doi: 10.1016/j.fgb.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 66.Jaques AK, Fukamizo T, Hall D, Barton RC, Escott GM, Parkinson T, Hitchcock CA, Adams DJ. 2003. Disruption of the gene encoding the ChiB1 chitinase of Aspergillus fumigatus and characterization of a recombinant gene product. Microbiology 149:2931–2939. doi: 10.1099/mic.0.26476-0. [DOI] [PubMed] [Google Scholar]
- 67.de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R, van der Krol S, Shibuya N, Joosten MHAJ, Thomma BPHJ. 2010. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329:953–955. doi: 10.1126/science.1190859. [DOI] [PubMed] [Google Scholar]
- 68.Hemetsberger C, Herrberger C, Zechmann B, Hillmer M, Doehlemann G. 2012. The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog 8:e1002684. doi: 10.1371/journal.ppat.1002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodriguez Estrada AE, Jonkers W, Corby Kistler H, May G. 2012. Interactions between Fusarium verticillioides, Ustilago maydis, and Zea mays: an endophyte, a pathogen, and their shared plant host. Fungal Genet Biol 49:578–587. doi: 10.1016/j.fgb.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 70.Teichmann B, Linne U, Hewald S, Marahiel MA, Bölker M. 2007. A biosynthetic gene cluster for a secreted cellobiose lipid with antifungal activity from Ustilago maydis. Mol Microbiol 66:525–533. doi: 10.1111/j.1365-2958.2007.05941.x. [DOI] [PubMed] [Google Scholar]
- 71.Banuett F. 1995. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu Rev Genet 29:179–208. doi: 10.1146/annurev.ge.29.120195.001143. [DOI] [PubMed] [Google Scholar]
- 72.Gruber S, Seidl-Seiboth V. 2012. Self versus non-self: fungal cell wall degradation in Trichoderma. Microbiology 158:26–34. doi: 10.1099/mic.0.052613-0. [DOI] [PubMed] [Google Scholar]
- 73.Spellig T, Bottin A, Kahmann R. 1996. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol Gen Genet 252:503–509. [DOI] [PubMed] [Google Scholar]
- 74.Vollmeister E, Haag C, Zarnack K, Baumann S, König J, Mannhaupt G, Feldbrügge M. 2009. Tandem KH domains of Khd4 recognize AUACCC and are essential for regulation of morphology as well as pathogenicity in Ustilago maydis. RNA 15:2206–2218. doi: 10.1261/rna.1817609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.