Abstract
Mechanistic studies on gliotoxin biosynthesis and self-protection in Aspergillus fumigatus, both of which require the gliotoxin oxidoreductase GliT, have revealed a rich landscape of highly novel biochemistries, yet key aspects of this complex molecular architecture remain obscure. Here we show that an A. fumigatus ΔgliA strain is completely deficient in gliotoxin secretion but still retains the ability to efflux bisdethiobis(methylthio)gliotoxin (BmGT). This correlates with a significant increase in sensitivity to exogenous gliotoxin because gliotoxin trapped inside the cell leads to (i) activation of the gli cluster, as disabling gli cluster activation, via gliZ deletion, attenuates the sensitivity of an A. fumigatus ΔgliT strain to gliotoxin, thus implicating cluster activation as a factor in gliotoxin sensitivity, and (ii) increased methylation activity due to excess substrate (dithiol gliotoxin) for the gliotoxin bis-thiomethyltransferase GtmA. Intracellular dithiol gliotoxin is oxidized by GliT and subsequently effluxed by GliA. In the absence of GliA, gliotoxin persists in the cell and is converted to BmGT, with levels significantly higher than those in the wild type. Similarly, in the ΔgliT strain, gliotoxin oxidation is impeded, and methylation occurs unchecked, leading to significant S-adenosylmethionine (SAM) depletion and S-adenosylhomocysteine (SAH) overproduction. This in turn significantly contributes to the observed hypersensitivity of gliT-deficient A. fumigatus to gliotoxin. Our observations reveal a key role for GliT in preventing dysregulation of the methyl/methionine cycle to control intracellular SAM and SAH homeostasis during gliotoxin biosynthesis and exposure. Moreover, we reveal attenuated GliT abundance in the A. fumigatus ΔgliK strain, but not the ΔgliG strain, following exposure to gliotoxin, correlating with relative sensitivities. Overall, we illuminate new systems interactions that have evolved in gliotoxin-producing, compared to gliotoxin-naive, fungi to facilitate their cellular presence.
INTRODUCTION
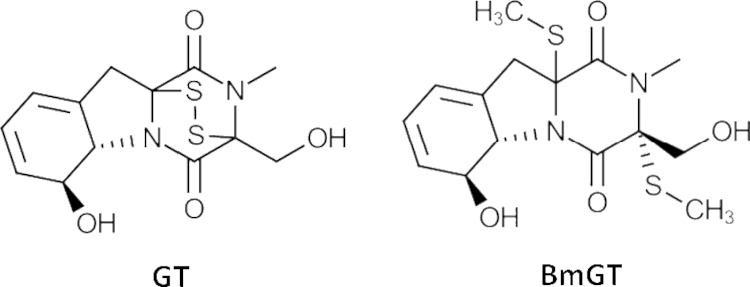
Biosynthesis, self-protection mechanisms, and functionality of gliotoxin and related epidithiodiketopiperazine (ETP) molecular species, such as chaetocin and acetylaranotin, are attracting ever-increasing attention as a consequence of findings from high-throughput genome sequencing projects, application of gene deletion technologies, and mass spectrometric analytical methodologies (1–5). Indeed, existing paradigms of gliotoxin (Fig. 1) as a toxin and the perspective of the disulfide bridge-containing (oxidized) form as the final, or only, product are undergoing significant reconsideration (6–11).
FIG 1.
Structures of gliotoxin (GT) and bisdethiobis(methylthio)gliotoxin (BmGT).
Self-protection against disulfide-containing metabolites appears to be essential in both fungi and bacteria. It has been demonstrated that the gliotoxin oxidoreductase GliT (12), encoded within the gli cluster, protects Aspergillus fumigatus against exogenous gliotoxin and is essential for gliotoxin biosynthesis (12, 13). A similar mechanism for self-protection against holomycin in Streptomyces clavuligerus has been described, where HlmI catalyzes disulfide bridge closure in holomycin (14). Deletion of hlmI impaired holomycin biosynthesis and sensitized S. clavuligerus to exogenous holomycin, as had been observed for gliotoxin in A. fumigatus. Additionally, Guo et al. revealed that in Aspergillus terreus, a fusion gene, ataTC, encodes both hydroxylation and disulfide bridge closure of biosynthetic intermediates during acetylaranotin formation (3). Wild-type A. terreus did not exhibit acetylaranotin sensitivity; however, no data were presented regarding the sensitivity of the ΔataTC strain to exogenous acetylaranotin. Deletion of a major facilitator superfamily (MFS) transporter, gliA, and gliK, a γ-glutamyl cyclotransferase, also sensitizes A. fumigatus to exogenous gliotoxin albeit to a lesser extent than in the absence of GliT (7, 15). While gliK deletion prevents gliotoxin biosynthesis (5, 7), interestingly, Wang et al. (15) noted only a reduction, not an abolition, of gliotoxin secretion by the A. fumigatus ΔgliA strain. Although gli cluster gene expression was shown previously to be activated by gliotoxin exposure (13, 16), no evidence of concomitant de novo gliotoxin biosynthesis had been detected. However, O'Keeffe et al. (17) demonstrated that de novo gliotoxin biosynthesis is induced by the addition of exogenous gliotoxin, which suggests that the significant inhibitory effect of exogenous gliotoxin on the A. fumigatus ΔgliT strain (12, 13) could, in part, also be due to the presence of newly synthesized gliotoxin or a gli pathway intermediate. However, surprisingly, the combined impact of the loss of gliotoxin biosynthesis, consequent to gliZ deletion, and GliT-mediated self-protection has not been explored to date.
In bacteria, thiomethylation has been posited to be an additional or backup strategy, for disulfide bridge closure, for self-protection during holomycin biosynthesis, and it has been proposed that S-methylation of biosynthetic intermediates, or possibly shunt metabolites, protects cellular components against these reactive species (14). Moreover, multiple S-methylated intermediates have been identified in wild-type A. fumigatus but not in an A. fumigatus strain deficient in gliotoxin biosynthesis (ΔgliZ) (9). Although bisdethiobis(methylthio)gliotoxin (BmGT) (Fig. 1) has not been reported in most studies relating to gliT, gliA, gliK, or gliG (a glutathione S-transferase responsible for biosynthetic intermediate sulfurization via bis-glutathionylation) gene cluster deletion (4, 5, 7, 13, 15, 18, 19), this metabolite is readily detectable in culture supernatants of A. fumigatus (8–10). Moreover, endogenous dithiol gliotoxin [GT-(SH)2] and exogenous gliotoxin can be converted to BmGT via a novel, S-adenosylmethionine (SAM)-dependent gliotoxin bis-thiomethyltransferase (GtmA) (10). Contrary to speculation in the literature (14) that deficiency of such an enzyme would induce gliotoxin sensitivity in A. fumigatus, as per deletion of gliT, no such phenotype has been observed. Indeed, gtmA deletion leads to the overproduction of gliotoxin, which positions BmGT formation as a negative regulatory mechanism of gliotoxin biosynthesis (10). However, the effects of gliotoxin bis-thiomethylation on the methionine cycle and cellular SAM and S-adenosylhomocysteine (SAH) levels in A. fumigatus remain obscure.
SAM is also involved in gliotoxin biosynthesis, where it provides a methyl group for N-methylation of the diketopiperazine scaffold during biosynthesis (4, 20, 21). Moreover, if S-methylation is involved in the modification of reactive biosynthetic intermediates, or shunt metabolites (9), SAM may also be the source of these methyl groups. SAM is derived from methionine, and while much work has been done on methionine biosynthesis in Aspergillus nidulans, until recently (22), few studies had specifically focused on sulfur metabolism in A. fumigatus. Moreover, there appears to be a dearth of literature pertaining to the functionality and detection of SAM in A. fumigatus. Enzyme-catalyzed S-methylation reactions using SAM yield SAH, which in A. nidulans and other organisms is subsequently hydrolyzed to homocysteine (Hcy) and adenosine via the action of S-adenosylhomocysteinase. The resultant Hcy is then reconverted to Met via the action of methionine synthase, with methylenetetrahydrofolate (from the methyl cycle) as the methyl source (23, 24). Notably, SAH is a competitive inhibitor of selected methyltransferases, and Hcy is cytotoxic, in A. nidulans (25, 26). These enzyme systems have received scant attention in A. fumigatus, which is of major significance because of the potential interplay between gliotoxin biosynthesis, SAM availability, and bis-thiomethylation, especially in the absence of gliT. Moreover, in the absence of gliotoxin biosynthesis in A. nidulans, the nature of the systems interactions between primary and so-called secondary metabolisms is refractory to investigation. The work presented here describes how gliotoxin biosynthesis, resistance, and secretion may be integrated into primary metabolism via bis-thiomethylation and SAM:SAH homeostasis. Furthermore, we reveal a key role for GliT in preventing dysregulation of SAM:SAH homeostasis.
MATERIALS AND METHODS
Gene deletion, complementation, and gene expression analyses of A. fumigatus.
A. fumigatus ΔgliA and ΔgliZ::ΔgliT strains were generated via the bipartite marker technique, using either the pyrithiamine resistance gene (ptrA) (ΔgliA and ΔgliZ::ΔgliT) or the hygromycin resistance gene (hph) (gliA complemented [gliAC] and ΔgliZ::ΔgliT::gliZ) for selection (27–29). All strains used are given in Table S1 in the supplemental material. Primers used for generating deletion constructs are given in Table S2 in the supplemental material. The ΔgliZ::ΔgliT double mutant and the ΔgliZ::ΔgliT::gliZ complemented strain were generated in the background of the A. fumigatus ΔgliZ strain, kindly provided by Nancy Keller (University of Wisconsin—Madison). The A. fumigatus ΔmetR strain was generously provided by Sven Krappmann (Erlangen, Germany). Fungal RNA isolation, DNase treatment, cDNA synthesis, and reverse transcription-quantitative PCR (qRT-PCR) were performed as described previously (30). Primers used for qRT-PCRs are listed in Table S2 in the supplemental material. qRT-PCR analysis was performed by using a Roche LightCycler 480 instrument.
LC-MS detection of gliotoxin and BmGT.
A. fumigatus wild-type, ΔgliA, and gliAC cultures were either grown for 72 to 96 h in Czapek Dox medium, to examine endogenous gliotoxin/BmGT production, or for 21 h in Czapek Dox medium followed by a 3-h challenge with gliotoxin (5-μg/ml final concentration), to examine the conversion of exogenous gliotoxin to BmGT. BmGT formation in the A. fumigatus ΔgliT strain was identically evaluated. Culture supernatants were subjected to organic extraction and liquid chromatography-mass spectrometry (LC-MS) analysis to detect gliotoxin and BmGT, as previously described (7, 10). GtmA activity in the A. fumigatus ΔgliK strain was determined as described previously for the A. fumigatus ΔgliT strain (10). Statistical analyses were carried out by using Student's t test.
Detection and quantification of S-adenosylmethionine and S-adenosylhomocysteine.
Czapek Dox medium was inoculated with 106 conidia/ml (from A. fumigatus wild-type, gene deletion, and complementation strains), in duplicate, and incubated at 37°C, with shaking 200 rpm, for 21 h. Gliotoxin (5-μg/ml final concentration) or the methanol control was added, and the cultures were incubated for a further 3 h before mycelia were harvested and snap-frozen in liquid N2. SAM and SAH were extracted according to a modified protocol (31, 32). Briefly, mycelia were ground under liquid N2 by using a pestle and mortar. A total of 0.1 M HCl (250 μl) was added to mycelia (100 mg), and the mixture was incubated on ice for 1 h with regular vortexing. Following centrifugation at 13,000 × g, protein was removed from the supernatant by trichloroacetic acid (TCA) precipitation. Samples were diluted in 0.1% (vol/vol) formic acid and analyzed by LC-tandem MS (MS/MS) using a porous graphitized carbon (PGC) chip on an Agilent 6340 Ion-Trap LC mass spectrometer (Agilent Technologies), using electrospray ionization. Quantification was enabled by using commercially available SAM and SAH obtained from Sigma-Aldrich. Intracellular gliotoxin and BmGT in wild-type and A. fumigatus ΔgliA strains were analyzed by using a C18 chip, and analysis of gliotoxin biosynthetic intermediates in the A. fumigatus ΔgliK strain, via PGC chip analysis, was also facilitated by using this extraction procedure. Statistical analyses were carried out by using Student's t test.
A. fumigatus whole-protein extraction for 2D-PAGE, LC-MS, and protein identification.
Comparative two-dimensional PAGE (2D-PAGE) analysis of the A. fumigatus ΔgliK strain following exposure to gliotoxin was carried out to investigate gross proteome changes associated with gliotoxin sensitivity. The A. fumigatus ΔgliK strain was cultured in Sabouraud dextrose medium for 24 h before the addition of gliotoxin (10-μg/ml final concentration) or the equivalent volume of methanol as a control (n = 5 biological replicates). After 4 h, mycelia were harvested, and protein was extracted. Harvested mycelia were prepared for 2D-PAGE, protein spots were digested with trypsin, and LC-MS analysis (Agilent) was performed essentially as described previously (11, 29, 33).
Label-free quantitative proteomic analysis of A. fumigatus wild-type ΔgliK, ΔgliK, and ΔgliG strains.
A. fumigatus ATCC 26933, Af293, ATCC 26933 ΔgliK mutant (ΔgliKATCC 26933), and ΔgliGAf293 mutant strains were cultured in Sabouraud dextrose medium for 21 h, followed by the addition of gliotoxin (5-μg/ml final concentration) or methanol for 3 h (n = 3 to 4 biological replicates for all specimens). Comparative label-free quantitative (LFQ) proteomic analysis of A. fumigatus ATCC 26933 versus the A. fumigatus ΔgliAATCC 26933 strain was performed following culture in Czapek Dox medium for 72 h (n = 3 biological replicates). Mycelial lysates were prepared in lysis buffer (100 mM Tris-HCl, 50 mM NaCl, 20 mM EDTA, 10% [vol/vol] glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml pepstatin A [pH 7.5]) with grinding and sonication and were clarified by centrifugation. The resultant protein lysates were precipitated by using TCA-acetone and resuspended in 8 M urea. After dithiothreitol (DTT) reduction and iodoacetamide-mediated alkylation (11, 33), sequencing-grade trypsin combined with ProteaseMax surfactant was added. Digested samples were desalted prior to analysis by using either C18 spin columns (Thermo Scientific Pierce) or C18 ZipTips (Millipore). All peptide mixtures were analyzed via a Thermo Scientific Q-Exactive mass spectrometer coupled to a Dionex RSLCnano instrument. LC gradients from 4 to 35% or 10 to 35% solution B (solution A is 0.1% [vol/vol] formic acid, and solution B is 80% [vol/vol] acetonitrile plus 0.1% [vol/vol] formic acid) were run over 2 h, and data were collected by using the Top15 method for MS/MS scans. Comparative proteome abundance and data analyses were performed by using MaxQuant software (version 1.3.0.5) (34), with Andromeda being used for database searching and Perseus (version 1.4.1.3) being used to organize the data. Carbamidomethylation of cysteines was set as a fixed modification, while oxidation of methionine and acetylation of N termini were set as variable modifications. The maximum peptide/protein false discovery rates (FDRs) were set to 1% based on comparison to a reverse database. The LFQ algorithm was used to generate normalized spectral intensities and infer relative protein abundance. Proteins that matched to a contaminant database or the reverse database were removed, and proteins were retained for final analysis only if they were detected in at least three replicates from at least one sample. Quantitative analysis was performed by using a t test to compare pairs of samples, and proteins with significant changes in abundance (P value of <0.05; fold change of ≥2) were included in the quantitative results. Qualitative analysis was also performed to detect proteins that were found in at least 3 replicates for a particular sample but undetectable in the comparison sample. Gene Ontology (GO) term enrichment (P < 0.05) was investigated by using the FungiFun application (35). All subsets of proteins were compared to a background consisting of the total identified proteins from the respective studies.
Phenotypic assays.
Conidia (106), harvested aseptically from 1-week-old Aspergillus minimal medium (AMM) plates, were subject to a variety of phenotypic assays. Plates were incubated at 37°C. Colony diameters were measured periodically, and statistical analysis was carried out by using one-way analysis of variance (ANOVA).
RESULTS
Absence of GliA abolishes gliotoxin, but not BmGT, secretion.
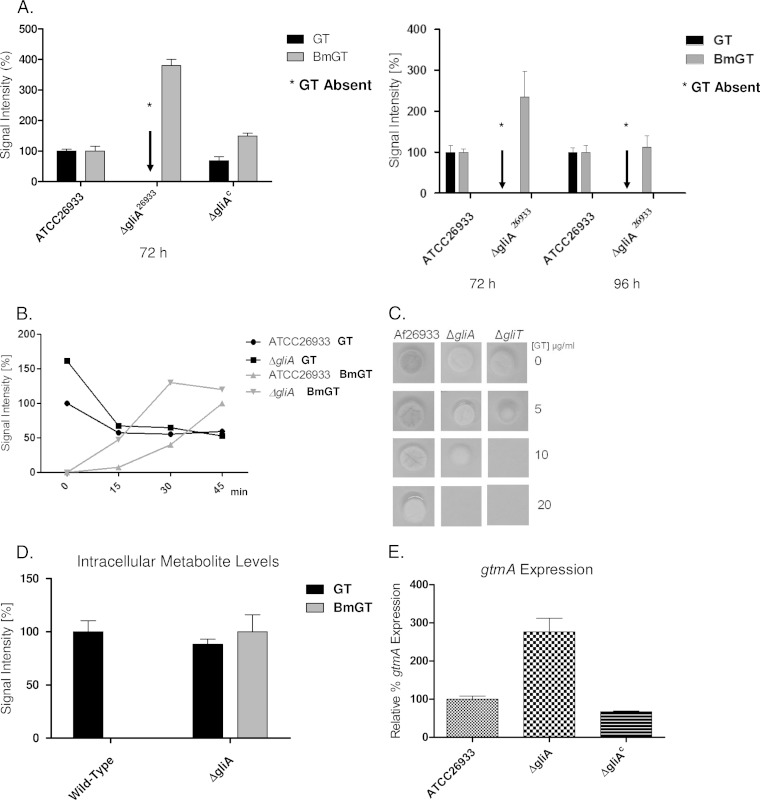
Deletion, complementation, and expression of A. fumigatus gliA, an MFS transporter proposed to be partly responsible for gliotoxin secretion (15), were confirmed by Southern blotting, PCR, and RT-PCR/qRT-PCR (see Fig. S1 in the supplemental material). Unexpectedly, the absence of gliA completely and specifically abolished the secretion of endogenous gliotoxin (Fig. 2A), yet BmGT was secreted by the A. fumigatus ΔgliA strain at significantly higher levels than those in the wild type (P < 0.0001) at 72 h (Fig. 2A, inset). Complementation of gliA in the A. fumigatus ΔgliA strain (see Fig. S1 in the supplemental material) restored gliotoxin secretion (Fig. 2A). Following exposure to exogenous gliotoxin in liquid culture, BmGT was produced by the A. fumigatus ΔgliA strain at significantly higher levels than those in the wild type at both 15 min (P = 0.005) and 30 min (P = 0.002), yet the ΔgliA strain was significantly sensitive (P < 0.0001) to exogenous gliotoxin (5 μg/ml) (Fig. 2B and C) albeit to a lesser extent than the A. fumigatus ΔgliT strain. This observation suggests that gliotoxin bis-thiomethylation is not primarily involved in, or sufficient for, protection against the growth-inhibitory effects of gliotoxin, in the absence of the ability to secrete gliotoxin.
FIG 2.
Characterization of the A. fumigatus ΔgliA strain. (A) Deletion of gliA completely abrogates gliotoxin, but not BmGT, secretion. gliA complementation restores gliotoxin secretion. The inset shows that gliA deletion also results in significantly increased BmGT secretion (P < 0.0001) at 72 h but not at 96 h of culture. (B) Exogenously added gliotoxin is converted to BmGT, and secreted, at a significantly higher rate in the A. fumigatus ΔgliA strain than in the wild type up to 30 min postaddition. (C) The A. fumigatus ΔgliA strain exhibits increased sensitivity to exogenous gliotoxin compared to the wild type. However, the A. fumigatus ΔgliA strain is not as sensitive to exogenous gliotoxin as the A. fumigatus ΔgliT strain (13). (D) Deletion of gliA results in accumulation of intracellular BmGT. (E) Comparative qRT-PCR analysis of gtmA expression in A. fumigatus wild-type, ΔgliA, and gliAC strains at 72 h. Significantly elevated gtmA expression levels are evident in the ΔgliA strain compared to those in the wild-type (P = 0.0392) and ΔgliAC (P = 0.0272) strains. Statistical analysis was performed by using Student's t test.
Intracellular gliotoxin and significantly elevated levels of intracellular BmGT were detectable in the A. fumigatus ΔgliA strain due to its inability to efflux gliotoxin, which results in significantly elevated expression levels of both the gli cluster (positive-feedback) (16) and gtmA (negative-feedback) (10) systems (Fig. 2D). Consequently, at between 48 and 96 h, this resulted in a significantly higher level of BmGT efflux in the A. fumigatus ΔgliA strain. Accordingly, although exogenous gliotoxin was not detected, Fig. 2E shows that the gtmA (10) expression level was significantly elevated (P = 0.0392) in the A. fumigatus ΔgliA strain at 72 h, as was the gliT expression level (P = 0.0060) (see Fig. S1 in the supplemental material), under conditions permissive for gliotoxin biosynthesis, which suggests cross talk between intracellular gliotoxin and gtmA expression. It is also notable that gtmA expression was restored to wild-type levels in the A. fumigatus gliAC strain (Fig. 2E).
gliA deletion alters the A. fumigatus proteome.
Label-free quantitative (LFQ) proteomic investigation of wild-type A. fumigatus versus the ΔgliA strain further underpinned the impact of the accrual of intracellular gliotoxin on gliA deletion. Compared to the wild type, the abundances of 92 proteins were significantly increased in the A. fumigatus ΔgliA strain, including GtmA (P < 0.00015), GliT (P < 0.01097), GliM (P < 0.00703), and GliN (P < 0.02596) (Table 1). To our knowledge, this is the first demonstration that disruption of the secretion of endogenous gliotoxin induces altered abundances of gliotoxin and BmGT biosynthetic enzymes in A. fumigatus. Moreover, this finding is entirely confluent with observations of increased gtmA (Fig. 2E) and gliT (see Fig. S1 in the supplemental material) (15) expression levels and significantly increased intracellular BmGT formation, as described above. Additionally, analysis of GO term enrichment revealed significant increases in the abundances of proteins involved in protein modification (3 proteins; P = 0.01312) and fatty acid biosynthetic processes (2 proteins; P = 0.0250) in the ΔgliA strain compared to the wild type. Proteins found in lower abundances in the ΔgliA strain than in the wild type were significantly enriched for translation (2 proteins; P = 0.0006), tRNA modification (2 proteins; P = 0.0087), and lipid metabolic processes (3 proteins; P = 0.0315). Methylation was significantly enriched among proteins with both increased (6 proteins; P = 0.00118) and decreased (7 proteins; P = 0.0145) abundances in the ΔgliA strain compared to the wild type.
TABLE 1.
LFQ proteomics analysis of proteins with increased abundance in, or that are unique to, the A. fumigatus ΔgliAATCC 26933 strain compared to wild-type Aspergillus fumigatus strain ATCC 26933a
| Protein description | Log2-fold increase | P value | No. of peptides | Sequence coverage (%) | CADRE identification no. |
|---|---|---|---|---|---|
| Gamma-glutamyltranspeptidase | Unique | NA | 4 | 13.3 | AFUA_4G13580 |
| Conserved hypothetical protein | Unique | NA | 3 | 15.3 | AFUA_5G02820 |
| Putative isopenicillin N-CoA epimerase; transcript upregulated in conidia exposed to neutrophils | Unique | NA | 2 | 8.9 | AFUA_5G03740 |
| Protein with ortholog(s) that has plasma membrane localization | Unique | NA | 4 | 28.3 | AFUA_5G06540 |
| Protein with ortholog(s) that has a role in cytoplasmic translation, and cell tip and cytoplasm localization | Unique | NA | 4 | 13 | AFUA_5G06770 |
| Protein with ortholog(s) that has mitochondrial ribosome localization | Unique | NA | 3 | 9.4 | AFUA_5G09500 |
| Protein with ortholog(s) that has an Elg1 RFC-like complex and cytosol and nucleus localization | Unique | NA | 3 | 15.2 | AFUA_6G05040 |
| Protein of unknown function; calcium downregulated | Unique | NA | 2 | 7.5 | AFUA_6G14280 |
| Putative 14-alpha demethylase with a predicted role in ergosterol biosynthesis; transcript upregulated in response to amphotericin B; SrbA regulated during hypoxia | Unique | NA | 2 | 5.3 | AFUA_7G03740 |
| Hypothetical protein | Unique | NA | 5 | 15 | AFUA_3G02430 |
| Putative pyridoxamine phosphate oxidase; transcript upregulated in conidia exposed to neutrophils | Unique | NA | 2 | 11.9 | AFUA_3G06670 |
| Putative aryl-alcohol dehydrogenase | Unique | NA | 9 | 31.4 | AFUA_4G00610 |
| Sterol 24-c-methyltransferase, putative | Unique | NA | 6 | 29.2 | AFUA_4G03630 |
| Protein with ortholog(s) that has DNA binding and tricarboxylate secondary active transmembrane transporter activity and roles in alpha-ketoglutarate transport, mitochondrial citrate transport, and mitochondrial genome maintenance | Unique | NA | 4 | 20.8 | AFUA_5G04220 |
| Conserved hypothetical protein | Unique | NA | 2 | 25.6 | AFUA_5G04336 |
| DUF453 domain protein | Unique | NA | 2 | 10.7 | AFUA_6G00360 |
| GliF | Unique | NA | 5 | 14.3 | AFUA_6G09730 |
| Putative cyclophilin; peptidyl-prolyl cis-trans-isomerase | Unique | NA | 3 | 21.6 | AFUA_6G10480 |
| Predicted aminopeptidase, metalloexopeptidase; encoded in the fma (fumagillin) secondary metabolite gene cluster | Unique | NA | 3 | 17.4 | AFUA_8G00460 |
| GtmA | 2.26265 | 0.00015 | 11 | 53.4 | AFUA_2G11120 |
| GliM | 2.08787 | 0.00703 | 17 | 65.4 | AFUA_6G09680 |
| GliT thioredoxin reductase | 1.52564 | 0.01097 | 15 | 80.2 | AFUA_6G09740 |
| GliN | 1.37247 | 0.02596 | 12 | 58.2 | AFUA_6G09720 |
Data are sorted by fold change, in descending order. Statistical analysis was performed by using Student's t test. NA, not applicable; CoA, coenzyme A; RFC, replication factor C.
Attenuated sensitivity to gliotoxin in the A. fumigatus ΔgliZ::ΔgliT strain and BmGT overproduction in the A. fumigatus ΔgliT strain.
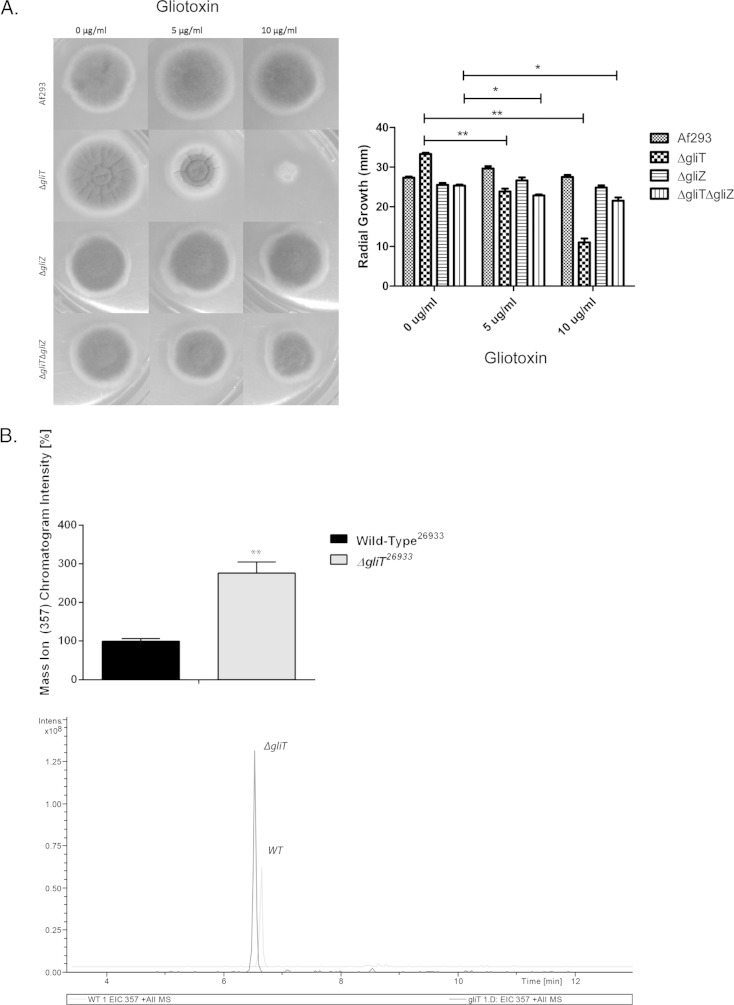
In order to evaluate the impact of a combined deficit in gliotoxin biosynthesis and self-protection against exogenous gliotoxin, an A. fumigatus ΔgliZ::ΔgliT double mutant was generated in a ΔgliZ background. As shown in Fig. S1 in the supplemental material, gliT loss in the A. fumigatus ΔgliZ strain was confirmed, whereby a 3.64-kb fragment was evident in both the double mutant and the A. fumigatus ΔgliTATCC 26933 strain (positive control) (13). While the A. fumigatus ΔgliZ::ΔgliT strain displayed mild sensitivity to exogenous gliotoxin (5 μg/ml), unexpectedly, this sensitivity was significantly lower (P = 0.0131) than that observed for the A. fumigatus ΔgliT strain (P = 0.0028) (Fig. 3A); thus, gli cluster expression and endogenous gliotoxin formation appear to be essential for the manifestation of gliotoxin sensitivity of the A. fumigatus ΔgliT strain. Exposure of the A. fumigatus ΔgliT strain to gliotoxin resulted in significantly increased production and secretion (P = 0.0039) of BmGT compared to wild-type exposure (Fig. 3B). Since SAM is required for BmGT biosynthesis, if this observation was accompanied by dysregulation of SAM:SAH homeostasis, then an important link between so-called primary and secondary metabolisms may be revealed.
FIG 3.
Evaluation of the effect of a combined deficit in biosynthesis and self-protection on A. fumigatus sensitivity to exogenous gliotoxin. (A) The A. fumigatus ΔgliZ::ΔgliT strain, deficient in both gliotoxin biosynthesis and self-protection systems, is significantly less sensitive (P = 0.0237) to exogenous gliotoxin than the A. fumigatus ΔgliT strain (P = 0.0011). (B) LC-MS analysis reveals significantly elevated BmGT secretion by the A. fumigatus ΔgliT compared to wild-type A. fumigatus when exposed to exogenous gliotoxin (5 μg/ml for 3 h; n = 3). The inset shows results for BmGT detection in the A. fumigatus ΔgliT strain following gliotoxin exposure by LC-MS. EIC, extracted ion chromatograph.
Exposure of A. fumigatus to gliotoxin depletes SAM and augments SAH levels in the absence of gliT.
Exogenous gliotoxin had no significant impact on the levels of SAM and SAH in A. fumigatus ATCC 46645 (low-gliotoxin producer), while in ATCC 26933 (high-gliotoxin producer), there was no alteration in SAM levels, but there was a significant increase (P = 0.0157) in SAH levels (Fig. 4A and B). No significant difference in cellular SAM and SAH levels was noted following exposure of A. fumigatus Af293 (36) to exogenous gliotoxin under conditions permissive for gliotoxin biosynthesis (Fig. 4C). The response of the A. fumigatus ΔgliG strain to gliotoxin exposure is equivalent to that of the background strain (Af293) (Fig. 4C), especially with respect to the maintenance of SAM levels. Under conditions that promote gliotoxin biosynthesis (7), gliotoxin exposure induced the depletion and production of SAM and SAH, respectively, in the A. fumigatus ΔgliT, ΔgliK, ΔgliZ, and ΔgliZ::ΔgliT strains (Fig. 4A to C). The altered SAM:SAH homeostasis in the ΔgliK strain is interesting, as this strain is sensitive to exogenous gliotoxin. Indeed, it is clear that cellular SAH levels are most significantly increased in the ΔgliTATCC 46645 and ΔgliTATCC 26933 strains (P = 0.0135 and P = 0.0002, respectively), with significant depletion of SAM levels in the ΔgliTATCC 46645 strain (P = 0.0359) and the ΔgliTATCC 26933 strain (P = 0.0028) (Fig. 4A and B), the gli single-deletion strain whose growth is most inhibited by exogenous gliotoxin, compared to all other strains tested. gliT complementation restores the wild-type scenario (Fig. 4A and B). Indeed, the deletion of gliT in the ΔgliZ strain resulted in further significant cellular SAM depletion compared to that in the ΔgliZ strain (P = 0.0377) upon exogenous gliotoxin exposure (Fig. 4C). We conclude that normal cellular SAM:SAH homeostasis requires GliT to preferentially oxidize GT-(SH)2 to gliotoxin, thereby preventing GtmA-mediated dysregulation of cellular SAM:SAH homeostasis. In the absence of GliT, it appears that increased SAM-dependent, GtmA-mediated bis-thiomethylation of GT-(SH)2 (10) rapidly depletes SAM and may result in dysregulation of the methyl/methionine cycle in A. fumigatus. Indeed, SAM depletion may also have further implications for global methylation reactions in A. fumigatus, and transcriptomic analysis of the ΔgliT strain exposed to exogenous gliotoxin revealed altered expression levels of a number of methyltransferases (17). Moreover, since GliG activity precedes GliK activity in the gliotoxin biosynthetic pathway, yet the respective deletion strains exhibit resistance or sensitivity to exogenous gliotoxin, respectively, we hypothesized that comparative proteomic analysis may allow dissection of these contrasting phenotypes.
FIG 4.
Quantitative determination of cellular SAM and SAH levels in A. fumigatus mycelial extracts by LC-MS analysis . (A) Effect of gliotoxin exposure (5 μg/ml for 3 h) on SAM and SAH levels in A. fumigatus ATCC 46645 and the ΔgliTATCC 46645, ΔgliTCATCC 46645, ΔgliKATCC 46645, and ΔgliKCATCC 46645 mutants. (B) Effect of gliotoxin exposure (5 μg/ml for 3 h) on SAM and SAH levels in A. fumigatus ATCC 26933 and the ΔgliTATCC 26933, gliTCATCC 26933, and ΔgliKATCC 26933 mutants. Compared to the wild-type and complemented strains, gliotoxin exposure results in significant or highly significant SAM depletion and SAH production in both the ΔgliK and ΔgliT strains, irrespective of the strain background. The level of SAH production is also significantly higher in the ΔgliTATCC 26933 strain than in the ΔgliKATCC 26933 strain. (C) Effect of gliotoxin exposure (5 μg/ml for 3 h) on SAM and SAH levels in A. fumigatus Af293 and the ΔgliZ, ΔgliZ::ΔgliT, and ΔgliGAf293 mutants. SAM depletion in the A. fumigatus ΔgliZ::ΔgliT strain is significantly enhanced compared to that in the A. fumigatus ΔgliZ strain. Although significantly depleted by the addition of gliotoxin, the absolute SAM level in the ΔgliG strain is higher than those in all other deletion mutants. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Statistical analysis was performed by using Student's t test. MeOH, methanol.
Gliotoxin exposure upregulates the expression of genes involved in sulfur metabolism in the A. fumigatus ΔgliT strain.
Transcriptome sequencing (RNA-seq) analysis (17) revealed that exposure of the A. fumigatus ΔgliTATCC 46645 strain to gliotoxin resulted in significantly upregulated expression (P value of <5 × 10−5 to 0.005) of genes involved in sulfur assimilation and methionine and SAM biosynthesis (see Table S3 in the supplemental material). Specifically, the expression levels of phosphoadenylyl-sulfate reductase and sulfite reductase were upregulated 3.02 log2- and 2.55 log2-fold, respectively, while those of methylenetetrahydrofolate reductase (MTHFR) (mtrA), cobalamin-independent methionine synthase (metH), and S-adenosylmethionine synthetase (sasA) were also significantly increased (P value of <5 × 10−5 to 0.003) (see Table S3 in the supplemental material). Expressions of the bZIP transcription factor metR (22) and S-adenosylhomocysteinase (sahA) were also upregulated (P value of <0.00015 to 0.001), as were those of glycine dehydrogenase (GDH) (P < 0.0056) and serine hydroxymethyltransferase (SHMT) (P < 0.0001) (see Table S3 in the supplemental material). GDH and SHMT act in concert with the folate cycle to transfer methyl groups to Hcy for methionine biosynthesis (37). Interestingly, the A. fumigatus ΔmetR strain showed significantly increased sensitivity to gliotoxin (P < 0.05) compared to the wild type (ATCC 46645) (see Fig. S2 in the supplemental material), and qRT-PCR analysis revealed significantly attenuated gliT expression (P < 0.001) in response to gliotoxin exposure (see Fig. S2 in the supplemental material). Although the Hcy precursor cystathionine (0.5 to 1.5 mM) alone had no effect on the A. fumigatus ΔmetR strain (data not shown), coexposure with gliotoxin (10 μg/ml) resulted in a significant retardation of growth (1.5 mM cystathionine) (P < 0.01) at 72 h compared to that of ATCC 46645 (see Fig. S2 in the supplemental material), which suggests that impaired gliT expression may facilitate cystathionine and gliotoxin to interact to cause growth retardation. Overall, these observations reveal the impact of gliotoxin on, and the importance of GliT for, the control of sulfur metabolism. Moreover, these results underpin our observations regarding dysregulated SAM:SAH levels and strongly imply that gliotoxin exposure has an impact on the methyl/methionine cycle of A. fumigatus in the absence of gliT. This suggests enzyme functionality beyond gliotoxin biosynthesis. To explore this phenomenon further, proteomic approaches were adopted to ascertain the interplay between gliotoxin biosynthesis, resistance, and primary metabolism.
2D-PAGE and label-free quantitative proteomic investigations reveal that exogenous gliotoxin induces proteome remodeling in the A. fumigatus ΔgliK strain.
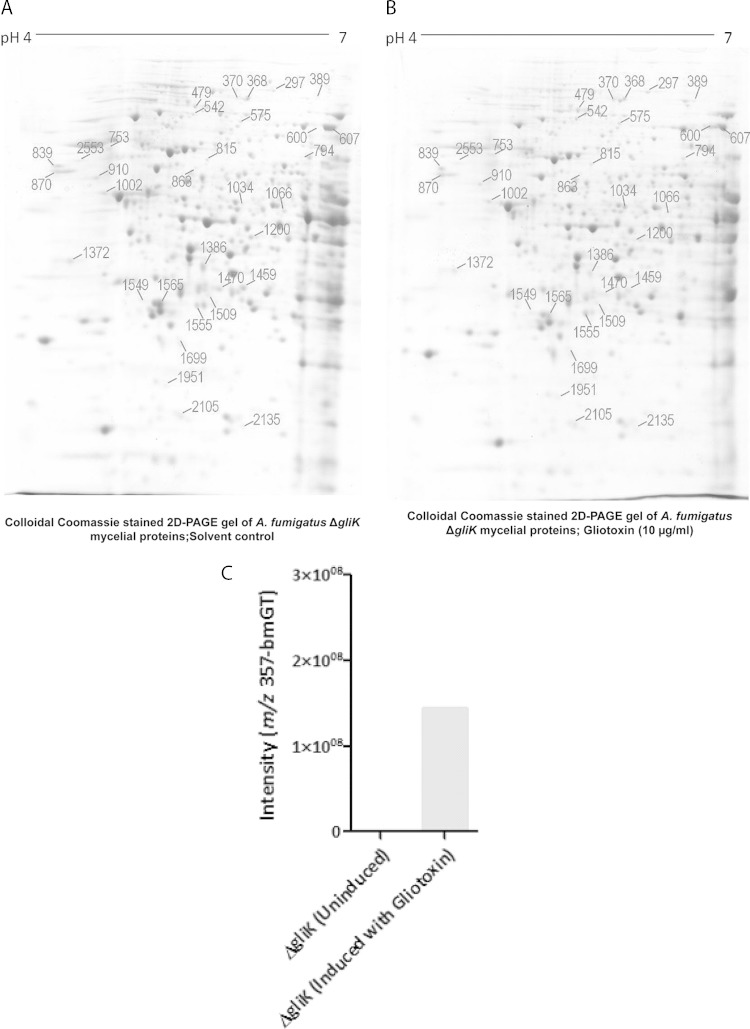
Following 2D-PAGE, 33 protein spots displayed significant changes in abundance in the A. fumigatus ΔgliKATCC 26933 strain (19 with ≥1.5-fold-increased and 14 with ≥1.5-fold-decreased abundances; P < 0.05) (7) upon exposure to gliotoxin (10 μg/ml) for 4 h (Fig. 5A and B and Table 2). LC-MS/MS analysis yielded identifications for 31 of these spots, corresponding to 30 distinct proteins. Overall, increased abundances of 18 protein spots, corresponding to 17 distinct A. fumigatus proteins, were observed for the ΔgliK strain in response to gliotoxin (Table 2). Proteins undergoing significant changes in abundance in the A. fumigatus ΔgliK strain following exposure to gliotoxin include those involved in translation and amino acid metabolism, those exhibiting regulatory roles, and endoplasmic reticulum (ER)-associated proteins (Table 2). However, of special significance, the abundance of the cobalamin-independent methionine synthase MetH/D (AFUA_4G07360) was significantly increased (1.9- to 2.2-fold), and that of MTHFR/MtrA was significantly increased (1.8-fold), in response to gliotoxin exposure. Both of these enzymes are essential for methionine biosynthesis (37, 38) and for the operation of the methyl/methionine cycle, which affects SAM biosynthesis.
FIG 5.
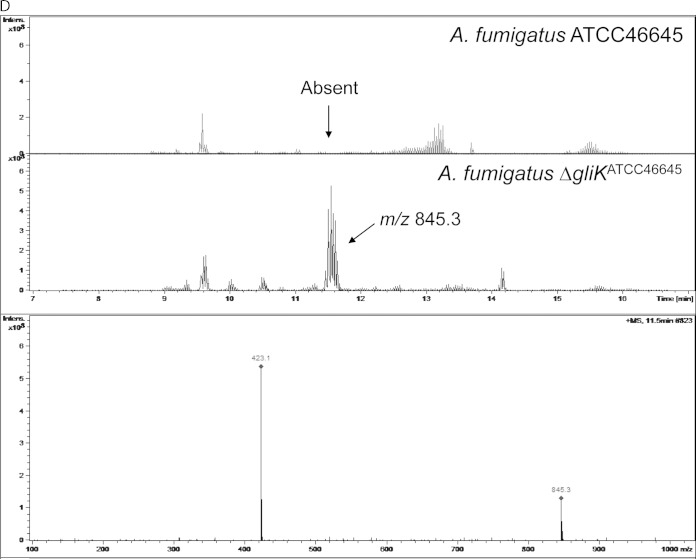
2D-PAGE analysis of the A. fumigatus ΔgliK strain. (A and B) Master 2D-PAGE of proteins with control treatment (A) or following exposure to gliotoxin (10 μg/ml) for 4 h (B). Proteins were first separated on pH 4 to 7 strips, followed by SDS-PAGE. Proteins found to be significantly differentially expressed (P < 0.05) after analysis by using Progenesis SameSpot software are numbered. (C) In vitro GtmA activity, as determined by BmGT formation in the presence of SAM and GT-(SH)2, is evident in mycelial lysates from A. fumigatus ΔgliK cells exposed to gliotoxin (5 μg/ml for 3 h). Assay conditions were described previously (10). (D) Detection of a bis-glutathionylated gliotoxin biosynthetic intermediate (m/z 845.3) in mycelial lysates of the A. fumigatus ΔgliK strain.
TABLE 2.
Proteins undergoing a significant change in abundance in the A. fumigatus ΔgliK strain following exposure to gliotoxin (10 μg/ml) relative to the solvent controla
| Protein | Fold change | Sequence coverage (%) | tMr (Da) | CADRE identification no. | Spot |
|---|---|---|---|---|---|
| Proteins with increased abundance following gliotoxin addition | |||||
| Translation elongation factor eEF-3 | ↑ 5.6 | 8 | 117,768.4 | AFUA_7G05660 | 297 |
| Alanyl-tRNA synthetase | ↑ 3.0 | 4 | 113,714.5 | AFUA_8G03880 | 368 |
| Eukaryotic translation initiation factor 3 subunit EifCb | ↑ 2.7 | 5 | 85,065.4 | AFUA_1G02030 | 542 |
| Translation elongation factor G1 | ↑ 2.5 | 19 | 87,877.7 | AFUA_4G08110 | 575 |
| Heat shock protein Hsp98/Hsp104/ClpA | ↑ 2.2 | 13 | 111,096.9 | AFUA_1G15270 | 370 |
| Glycine dehydrogenase | ↑ 2.2 | 18 | 115,203.5 | AFUA_4G03760 | 389 |
| Cobalamin-independent methionine synthase MetH/D | ↑ 2.2 | 5 | 87,736.8 | AFUA_4G07360 | 607 |
| Vesicular fusion protein Sec17 | ↑ 2.0 | 31 | 32,840.9 | AFUA_2G12870 | 1699 |
| CTP synthase | ↑ 2.0 | 27 | 64,869.3 | AFUA_7G05210 | 794 |
| Cobalamin-independent methionine synthase MetH/D | ↑ 1.9 | 14 | 87,736.8 | AFUA_4G07360 | 600 |
| Aminopeptidase | ↑ 1.9 | 35 | 106,227.3 | AFUA_4G09030 | 479 |
| Alanine aminotransferase | ↑ 1.9 | 31 | 55,135.5 | AFUA_6G07770 | 1034 |
| MTHFR | ↑ 1.8 | 39 | 69,278.9 | AFUA_2G11300 | 815 |
| Mitochondrial processing peptidase alpha subunit, putative | ↑ 1.7 | 8 | 63,997 | AFUA_1G11870 | 863 |
| Xanthine-guanine phosphoribosyl transferase Xpt1, putative | ↑ 1.7 | 29 | 19,505.5 | AFUA_4G04550 | 2105 |
| Isochorismatase family hydrolase | ↑ 1.7 | 26 | 20,904.6 | AFUA_6G12220 | 2135 |
| Pyruvate dehydrogenase complex component Pdx1 | ↑ 1.5 | 27 | 35,523.1 | AFUA_3G08270 | 1509 |
| GNAT family acetyltransferase | ↑ 1.5 | 39 | 29,061.1 | AFUA_5G00720 | 1951 |
| Proteins with decreased abundance following gliotoxin addition | |||||
| Pyruvate dehydrogenase E1 component alpha subunit, putative | ↓ 2.8 | 47 | 41,709.5 | AFUA_1G06960 | 1200 |
| Ran GTPase-activating protein 1 (RNA1 protein) | ↓ 2.4 | 8 | 46,228.2 | AFUA_3G07680 | 1002 |
| Protein phosphatase 2a 65-kDa regulatory subunit | ↓ 2.3 | 7 | 69,220 | AFUA_1G05610 | 753 |
| Diphthine synthase | ↓ 2.1 | 24 | 31,492.2 | AFUA_1G14020 | 1549 |
| Homogentisate 1,2-dioxygenase (HmgA) | ↓ 2.1 | 26 | 50,255.6 | AFUA_2G04220 | 1066 |
| Aspartic endopeptidase Pep2 | ↓ 2.1 | 20 | 43,355.1 | AFUA_3G11400 | 1372 |
| Zinc finger protein ZPR1 | ↓ 2.1 | 23 | 53,620 | AFUA_6G10470 | 2553 |
| CRAL/TRIO domain protein | ↓ 2.0 | 27 | 46,169.8 | AFUA_5G03690 | 910 |
| Translation elongation factor EF2 subunit | ↓ 1.9 | 11 | 93,428.8 | AFUA_2G13530 | 1459 |
| Thiamine biosynthesis protein (Nmt1) | ↓ 1.9 | 38 | 38,323 | AFUA_5G02470 | 1470 |
| Oxidoreductase, 2OG-Fe(II) oxygenase family | ↓ 1.8 | 13 | 43,013.2 | AFUA_1G01000 | 1386 |
| Nucleosome assembly protein Nap1 | ↓ 1.8 | 28 | 48,336.4 | AFUA_5G05540 | 839 |
| Protein disulfide isomerase Pdi1 | ↓ 1.6 | 31 | 56,187.2 | AFUA_2G06150 | 870 |
A change in abundance was considered significant at a P value of <0.05. Protein identification was achieved by 2D-PAGE and LC-MS/MS. Statistical analysis was performed by using Student's t test. ↑ and ↓ indicate fold increases and decreases, respectively, in protein abundance upon exposure to gliotoxin (10 μg/ml) relative to the solvent control. The CADRE gene identification number indicates A. fumigatus gene annotation nomenclature according to references 36 and 54. Spot indicates the number assigned to the protein in Fig. 5. tMr, theoretical molecular mass; 2OG, 2-oxoglutarate.
Interestingly, an increased abundance of the gliotoxin oxidoreductase GliT was not detectable by comparative 2D-PAGE analysis of the A. fumigatus ΔgliK strain in the presence of gliotoxin, although significantly elevated gliT expression levels under near-identical conditions were reported previously (7). This putative sensitivity limitation of 2D-PAGE led us to explore altered protein abundance in the A. fumigatus ΔgliK strain following gliotoxin exposure by a more sensitive approach, LFQ proteomics. These data reveal that gliotoxin (5 μg/ml for 3 h) significantly increases the abundance of GliT (log2 fold change = 3.837; P = 3.39 × 10−2), as well as effecting de novo expression of GtmA (10), in the A. fumigatus ΔgliK strain (Table 3). These findings are in complete accordance with the detection of GtmA activity in gliotoxin induced A. fumigatus ΔgliK mycelial lysates (Fig. 5C). Importantly, GliT abundance was increased significantly more (log2 fold change = 7.97; P = 1.35 × 10−5) following exposure of wild-type A. fumigatus to gliotoxin under identical conditions (see Table S4 in the supplemental material). In effect, the A. fumigatus ΔgliK strain is a “GliT-lite” mutant. Moreover, this observation supports the hypothesis of the requirement for de novo gliotoxin biosynthesis to effect maximum gliotoxin-mediated growth inhibition, as significantly less GliT is required for GT-(SH)2 oxidation to gliotoxin in the A. fumigatus ΔgliK strain, which is itself deficient in gliotoxin biosynthesis and accumulates a bis-glutathionylated gliotoxin biosynthetic intermediate at m/z 845.3 ([M + H]+) (Fig. 5D) (5). Finally, we show that AFUA_5G08600, a putative homoserine O-acetyltransferase that provides an alternative route for cystathionine (39) and, ultimately, Hcy biosynthesis, is uniquely expressed in the A. fumigatus ΔgliK strain following exposure to gliotoxin (Table 3). This may provide an alternative route for SAM formation and provides insight into the integrated nature of gliotoxin resistance/biosynthesis in primary cellular metabolism in A. fumigatus.
TABLE 3.
LFQ proteomics analysis of proteins with increased abundance in, or that are or unique to, the A. fumigatus ΔgliKATCC 26933 strain with gliotoxin compared to the ΔgliKATCC 26933 strain with methanola
| Protein description | Log2-fold increase | P value | No. of peptides | Sequence coverage (%) | CADRE identification no. |
|---|---|---|---|---|---|
| Ortholog of A. nidulans FGSC A4 protein AN9303, Aspergillus niger CBS 513.88 protein An07g06460, Aspergillus oryzae RIB40 protein AO090023000147, and Neosartorya fischeri NRRL 181 protein NFIA_064190 | Unique | NA | 9 | 40.4 | AFUA_3G13140 |
| Protein with domain(s) with predicted catalytic activity, coenzyme binding, nucleotide binding activity, and role in cellular metabolic processes | Unique | NA | 3 | 12.6 | AFUA_4G02810 |
| Putative aldehyde reductase with higher expression levels in biofilm grown for 48 h than in planktonic cells; repressed by gliotoxin exposure | Unique | NA | 15 | 65 | AFUA_5G02020 |
| Putative p-nitroreductase family protein; protein induced by heat shock; Yap1-dependent induction in response to hydrogen peroxide; induced by gliotoxin exposure | Unique | NA | 3 | 22.1 | AFUA_5G09910 |
| Protein with domain(s) with predicted hydrolase activity | Unique | NA | 5 | 28 | AFUA_5G12770 |
| Ortholog of A. nidulans FGSC A4 protein AN1460, A. niger CBS 513.88 protein An16g08480, and A. oryzae RIB40 protein AO090023000335 | Unique | NA | 7 | 45.4 | AFUA_8G04510 |
| Gliotoxin bis-thiomethyltransferase GtmAb | Unique | NA | 4 | 25.2 | AFUA_2G11120 |
| Protein with ortholog(s) that has homoserine O-acetyltransferase activity and role in cysteine metabolic processes | Unique | NA | 2 | 6.6 | AFUA_5G08600 |
| Protein with domain(s) with predicted nucleotide binding and oxidoreductase activities and role in metabolic processes | Unique | NA | 7 | 61.3 | AFUA_5G14000 |
| Protein with domain(s) with predicted hydrolase activity, acting on activity of ester bonds | Unique | NA | 5 | 20.7 | AFUA_7G04910 |
| Protein with domain(s) with predicted carbonate dehydratase activity, zinc ion binding activity, and role in carbon utilization | Unique | NA | 4 | 40.9 | AFUA_8G06554 |
| Gliotoxin oxidoreductase required for gliotoxin biosynthesis; encoded by the gliotoxin biosynthetic gene cluster; involved in self-protection against exogenous gliotoxin; induced in biofilm; immunoreactive (GliT) | 3.837 | 3.39E−02 | 13 | 71.3 | AFUA_6G09740 |
| Protein with domain(s) with predicted kynureninase activity, pyridoxal phosphate binding activity; role in NAD biosynthetic processes and tryptophan catabolic processes; cytoplasm localization | 2.221 | 1.43E−03 | 16 | 55.6 | AFUA_4G09840 |
| Glyceraldehyde-3-phosphate dehydrogenase, putative | 1.956 | 1.10E−02 | 24 | 74.5 | AFUB_049500 |
| Putative bifunctional catalase-peroxidase | 1.945 | 1.00E−04 | 26 | 54.4 | AFUA_8G01670 |
| Putative integral plasma membrane heat shock protein | 1.900 | 1.24E−02 | 6 | 58.5 | AFUA_6G06470 |
| Protein with domain(s) with predicted acid amino acid ligase activity and role in posttranslational protein modification | 1.391 | 2.93E−03 | 2 | 20.7 | AFUA_3G14430 |
| Protein with domain(s) with predicted catalytic activity and role in metabolic processes | 1.338 | 4.53E−02 | 7 | 21.9 | AFUA_3G09240 |
| Glyoxylase family protein, putative | 1.335 | 1.89E−02 | 3 | 16.9 | AFUB_090580 |
| Protein with ortholog(s) that has protein binding and bridging activity and roles in actin cortical patch assembly, axial cellular bud site selection, bipolar cellular bud site selection, and endocytosis | 1.286 | 1.37E−03 | 4 | 4.7 | AFUA_7G03870 |
| Putative 30-kDa heat shock protein; conidium-enriched protein | 1.278 | 1.23E−02 | 6 | 38.3 | AFUA_3G14540 |
| Protein with ortholog(s) that has cytoplasmic stress granule localization | 1.274 | 2.98E−03 | 2 | 17.7 | AFUA_6G02450 |
| Protein of unknown function; transcript upregulated in conidia exposed to neutrophils | 1.247 | 3.99E−02 | 2 | 9.1 | AFUA_1G11480 |
| Putative myoinositol-phosphate synthase; transcript upregulated in conidia exposed to neutrophils | 1.053 | 2.33E−04 | 21 | 60.3 | AFUA_2G01010 |
| Putative tripeptidyl-peptidase of the sedolisin family; predicted signal sequence for secretion | 1.051 | 9.37E−03 | 11 | 27.6 | AFUA_4G14000 |
Data are sorted by fold change, in descending order. Statistical analysis was performed by using Student's t test.
See reference 10.
The GliT-mediated self-protection system is operable in the A. fumigatus ΔgliG strain.
The A. fumigatus ΔgliG strain is incapable of gliotoxin biosynthesis, and growth is unaffected by the presence of gliotoxin (4). Consequently, this scenario represents an ideal model to further dissect the impact of gliotoxin on A. fumigatus metabolic systems. FunCat analysis of the proteins with increased abundances in the A. fumigatus ΔgliG strain following exogenous gliotoxin exposure revealed enrichment for proteins involved in the oxidative stress response. In addition to other FunCat categories, proteins involved in metabolism of peptide-derived compounds and RNA transport were also significantly represented among proteins with increased abundance upon exogenous gliotoxin exposure in the A. fumigatus ΔgliG strain. A number of proteins involved in RNA processing and RNA modification, along with some participating in DNA synthesis and replication, were significantly enriched for proteins with decreased abundance in the A. fumigatus ΔgliG strain exposed to exogenous gliotoxin. Other FunCat categories significantly enriched for proteins with decreased abundance included ATP binding, nucleotide/nucleoside/nucleobase binding, and respiration. Data in Table 4 confirm that GliT exhibits a highly significantly increased abundance (log2 fold change = 4.82; P < 8.77 × 10−5) in the A. fumigatus ΔgliG strain upon gliotoxin exposure. Interestingly, 10 additional proteins appear to be uniquely expressed in the A. fumigatus ΔgliG strain upon gliotoxin exposure, including a putative methyltransferase (AFUA_3G13140). Moreover, 8 proteins exhibit significantly increased abundances, including GliN, a catalase-peroxidase (AFUA_8G1670), and an oxidoreductase (AFUA_1G01000). Although not statistically significant, the abundance of GtmA was also increased by ∼5-fold upon gliotoxin exposure (data not shown). Exposure of the A. fumigatus ΔgliG strain to gliotoxin abolished the expression of 27 proteins, including 2 proteins (AFUA_8G00390 and AFUA_8G00440) encoded by the supercluster on chromosome 8 (40, 41), and resulted in decreased abundances of 5 additional proteins (Table 5).
TABLE 4.
LFQ proteomics analysis of proteins with increased abundance in, or that are unique to, the A. fumigatus ΔgliGAf293 strain with gliotoxin compared to methanol-only exposurea
| Protein description | Log2-fold increase | P value | No. of peptides | Sequence coverage (%) | CADRE identification no. |
|---|---|---|---|---|---|
| GNAT family N-acetyltransferase, putative | Unique | 5 | 38.9 | AFUA_3G00870 | |
| Putative uncharacterized protein | Unique | 5 | 23.7 | AFUA_6G12780 | |
| Methyltransferase, putative | Unique | 10 | 42.4 | AFUA_3G13140 | |
| Putative uncharacterized protein | Unique | 2 | 10.7 | AFUA_6G02010 | |
| Guanyl-nucleotide exchange factor (Sec7) | Unique | 5 | 3 | AFUA_7G05700 | |
| NADH-dependent flavin oxidoreductase | Unique | 3 | 14.5 | AFUA_7G06420 | |
| Aspartyl-tRNA synthetase, cytoplasmic | Unique | 2 | 2.7 | AFUA_1G02570 | |
| Putative uncharacterized protein | Unique | 2 | 11.4 | AFUA_4G07680 | |
| Nucleoporin SONB, putative | Unique | 3 | 1.9 | AFUA_4G11070 | |
| Nuclear and cytoplasmic polyadenylated RNA binding protein Pub1 | Unique | 4 | 7.8 | AFUA_1G12000 | |
| Gliotoxin oxidoreductase (GliT) | 4.825 | 8.77E−05 | 19 | 66.5 | AFUA_6G09740 |
| Catalase-peroxidase | 3.725 | 0.000676 | 42 | 75.2 | AFUA_8G01670 |
| Oxidoreductase, 2OG-Fe(II) oxygenase family | 3.507 | 0.020389 | 9 | 34 | AFUA_1G01000 |
| N-Methyltransferase (GliN) | 2.561 | 0.00636 | 6 | 28.4 | AFUA_6G09720 |
| Short-chain dehydrogenase, putative | 1.781 | 0.014034 | 13 | 68.3 | AFUA_4G08710 |
| Heat shock protein Hsp30-like, putative | 1.558 | 0.026821 | 5 | 43 | AFUA_6G06470 |
| Putative uncharacterized protein | 1.549 | 0.008346 | 10 | 68.2 | AFUA_3G00960 |
| Lipase/esterase, putative | 1.549 | 0.00262 | 7 | 32.7 | AFUA_1G15430 |
| Class V chitinase, putative | 1.287 | 0.002555 | 7 | 27.8 | AFUA_3G11280 |
Data are sorted by fold change, in descending order. Statistical analysis was performed by using Student's t test.
TABLE 5.
LFQ proteomics analysis of proteins with decreased abundance in, or that are absent from, the A. fumigatus ΔgliGAf293 strain with gliotoxin compared to methanol-only exposurea
| Protein description | Log2-fold decrease | P value | No. of peptides | Sequence coverage (%) | CADRE identification no. |
|---|---|---|---|---|---|
| Putative uncharacterized protein | Absent | 7 | 8.4 | AFUA_2G05520 | |
| Alternative oxidase | Absent | 4 | 12.5 | AFUA_2G05060 | |
| Putative uncharacterized protein | Absent | 3 | 11.9 | AFUA_6G13500 | |
| ATP-dependent RNA helicase Mss116, mitochondrial | Absent | 3 | 6.6 | AFUA_1G15620 | |
| Ferrochelatase | Absent | 3 | 8.9 | AFUA_5G07750 | |
| tRNA [adenine(58)-N(1)]-methyltransferase catalytic subunit Trm61 | Absent | 2 | 5.8 | AFUA_5G09620 | |
| Mitochondrial phosphate carrier protein, putative | Absent | 4 | 9.2 | AFUA_3G08430 | |
| Catalase | Absent | 4 | 12.5 | AFUA_2G18030 | |
| U6 snRNA-associated Sm-like protein LSM4, putative | Absent | 3 | 31.1 | AFUA_2G12020 | |
| Regulator of nonsense transcripts, putative | Absent | 3 | 4 | AFUA_1G13060 | |
| Mitochondrial molecular chaperone (Atp12), putative | Absent | 5 | 16 | AFUA_7G02490 | |
| O-Methyltransferase, putative | Absent | 3 | 27 | AFUA_8G00390 | |
| Steroid monooxygenase, putative | Absent | 3 | 6.3 | AFUA_8G00440 | |
| tRNA-guanine transglycosylase family protein | Absent | 3 | 8.5 | AFUA_5G03470 | |
| Ubiquitin-conjugating enzyme (UbcH), putative | Absent | 3 | 42.8 | AFUA_5G04060 | |
| snRNP and snoRNP protein (Snu13), putative | Absent | 2 | 28.6 | AFUA_2G05950 | |
| Cellular morphogenesis regulator DopA | Absent | 3 | 1.4 | AFUA_2G05020 | |
| GTP binding protein, putative | Absent | 7 | 24.7 | AFUA_1G05560 | |
| Putative uncharacterized protein | Absent | 3 | 17.8 | AFUA_6G09950 | |
| mRNA splicing factor (Prp17), putative | Absent | 3 | 9.5 | AFUA_6G07300 | |
| Alpha,alpha-trehalase TreB/Nth1 | Absent | 4 | 5.9 | AFUA_4G13530 | |
| Hsp70 family protein | Absent | 2 | 3.3 | AFUA_1G15200 | |
| RNase III domain protein | Absent | 2 | 6.2 | AFUA_5G09670 | |
| Putative uncharacterized protein | Absent | 3 | 10.9 | AFUA_3G10710 | |
| DNA replication licensing factor Mcm2, putative | Absent | 4 | 5.8 | AFUA_3G14010 | |
| Nuclear cohesin complex subunit (Psc3), putative | Absent | 4 | 4 | AFUA_2G16080 | |
| Cyclic AMP-dependent protein kinase catalytic subunit PkaC1 | Absent | 3 | 10.6 | AFUA_2G12200 | |
| l-Amino acid oxidase LaoA | −1.198 | 0.026668 | 14 | 28 | AFUA_7G06810 |
| 40S ribosomal protein S25, putative | −1.300 | 0.048129 | 9 | 51.6 | AFUA_1G16523 |
| DUF636 domain protein | −1.348 | 0.049468 | 6 | 81.6 | AFUA_2G15290 |
| Hydrophobin RodA | −3.345 | 0.013418 | 3 | 31.4 | AFUA_5G09580 |
| Aldehyde dehydrogenase, putative | −3.410 | 0.04575 | 15 | 51.3 | AFUA_7G01000 |
Data are sorted by fold change, in descending order. Statistical analysis was performed by using Student's t test.
DISCUSSION
Here we reveal that deletion of A. fumigatus gliA completely disrupts the secretion of endogenous and exogenous gliotoxin, which results in the acquisition of a mild gliotoxin-sensitive phenotype but does not adversely affect BmGT formation or secretion. Moreover, it is revealed that elevated levels of endogenous gliotoxin induce transcriptome and proteomic changes in the A. fumigatus ΔgliA strain, including the induction of gtmA expression/GtmA abundance. We furthermore report that the A. fumigatus ΔgliZ::ΔgliT strain exhibits significantly greater resistance to gliotoxin than does the ΔgliT strain. This observation strongly suggests that an intact de novo gliotoxin biosynthesis pathway is necessary to effect maximum gliotoxin sensitivity, which engenders a highly deleterious situation in A. fumigatus. SAM depletion and SAH overproduction are most significant in the A. fumigatus ΔgliT strain, which suggests that GliT plays a critical role in preventing unrestricted GtmA-mediated GT-(SH)2 bis-thiomethylation, which would otherwise lead to a critical attenuation of the intracellular methylation capacity. Extensive transcriptomic and proteomic observations support the hypothesis that genes and proteins involved in the methyl/methionine cycle are overexpressed or have a significantly increased abundance, respectively, under conditions where either GliT is absent or its presence is significantly attenuated, as in the A. fumigatus ΔgliK strain upon gliotoxin exposure. Gliotoxin sensitivity has also been observed for an A. fumigatus ΔmetR strain, which secretes BmGT (10) but exhibits impaired gliT induction upon exposure to gliotoxin. Thus, we now propose that in vivo, gliotoxin biosynthesis, resistance, and primary metabolism require regulation, via GliT and GliA functionality, to control gliotoxin oxidation and efflux, respectively, which in turn prevent SAM depletion and the concomitant overproduction of potentially inhibitory levels of SAH (Fig. 4A to C). We also conclude that bis-thiomethylation insufficiency is not primarily responsible for the gliotoxin sensitivity phenotype and that BmGT secretion occurs via an unknown, but GliA-independent, mechanism.
Wang et al. (15) convincingly reported that gliA deletion from A. fumigatus Afs35 led to significantly reduced secretion of gliotoxin, the acquisition of a gliotoxin-sensitive phenotype, significantly increased gliZ expression levels, and a reduction in intracellular gliotoxin levels. Wang et al. also noted elevated gliT expression in the A. fumigatus ΔgliA strain and overall concluded that gliA- and gliT-mediated self-protection were distinct mechanisms. Our data in part agree with those reported by Wang et al.; however, our distinct findings regarding elevated intracellular BmGT levels, that BmGT secretion remains intact, along with significant upregulation of gtmA expression by elevated intracellular gliotoxin levels, in the A. fumigatus ΔgliA strain lead us to alternative conclusions. In our model, we propose that gliA deletion results in elevated intracellular gliotoxin levels, which in turn induce gliT (as also observed previously [15]) along with gtmA, thereby increasing the formation, intracellular accumulation, and secretion of BmGT. Our hypothesis is in complete accordance with both of our data sets; however, it is based on the conclusion that GliT and GliA work in concert, as opposed to by distinct mechanisms, to effect gliotoxin resistance in A. fumigatus. A novel C2H2 transcription factor that also regulates gliA expression, with gliZ, has been identified (42), which may also function to protect A. fumigatus against exogenous gliotoxin.
The absence of both gliZ and gliT from A. fumigatus results in a codeficiency in gliotoxin biosynthesis (43) and self-protection (12, 13) and leads to significantly improved resistance to exogenous gliotoxin compared to that observed for the A. fumigatus ΔgliT strain. This apparently puzzling result means that the growth-inhibitory phenotype observed upon gliotoxin exposure requires both a functional biosynthetic system and an inactive resistance system. Otherwise, if gliZ expression is intact, exogenous gliotoxin induces gli cluster expression (13, 16, 17) and leads to the de novo production of GT-(SH)2 and thus an exacerbation of the growth-inhibitory effect of exogenous gliotoxin. This observation also explains why the A. fumigatus ΔgliT strain grows normally in the absence of exogenous gliotoxin (12, 13). Thus, growth of the A. fumigatus ΔgliT strain is inhibited only by exogenous gliotoxin, which induces gli cluster activity (13, 17), because in the absence of GliT-mediated oxidation, gliotoxin cannot be secreted via GliA and so persists in the cell. We propose that the additional presence of endogenous GT-(SH)2, combined with that formed intracellularly upon the reduction of exogenous gliotoxin, results in SAM depletion and SAH overproduction as a consequence of GtmA activity, which in turn contributes to growth inhibition of the A. fumigatus ΔgliT strain. This further underpins our hypothesis that bis-thiomethylation of gliotoxin is not a backup strategy mediated by GliM or GliN, as has been proposed for A. fumigatus (14), but serves to negatively regulate gliotoxin biosynthesis via the non-gli cluster gene gtmA (10). In the absence of gliT and in the presence of exogenous gliotoxin, this negative regulatory system causes a dysregulation of SAM:SAH homeostasis (Fig. 6).
FIG 6.
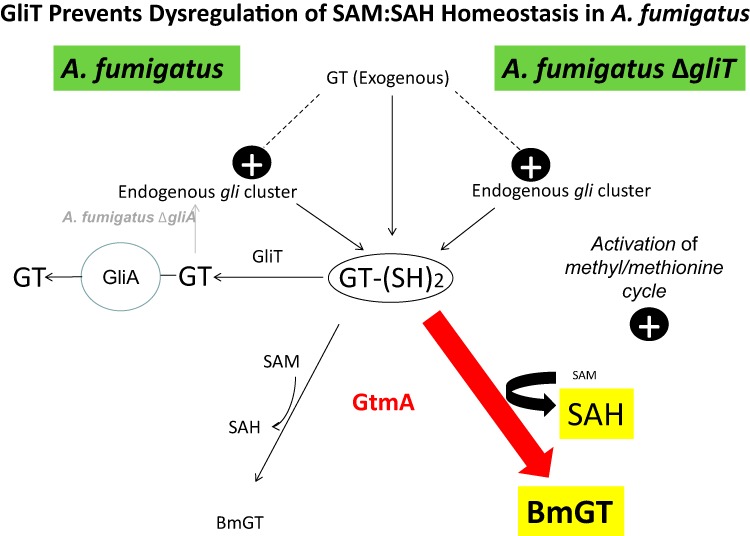
GliT prevents dysregulation of SAM:SAH homeostasis in A. fumigatus. This model is in accordance with all experimental observations whereby exogenous and/or endogenous gliotoxin results in GT-(SH)2 formation followed by conversion to gliotoxin and secretion via GliA. Alternatively, GT-(SH)2 can be converted to BmGT via GtmA under controlled conditions (10, 20, 53). In the absence of GliT (ΔgliT), GtmA-mediated BmGT formation at nonphysiological levels of occurs, leading to SAM depletion and SAH overproduction. In the A. fumigatus ΔgliA strain, increases in gli cluster activity and GtmA abundance also occur, leading to elevated intra- and extracellular BmGT levels. Note that because the absence of GliT is so deleterious (53), the attenuated levels of GliT present in the A. fumigatus ΔgliK strain provided an alternative experimental system to reveal the link to the methyl/methionine cycle (Tables 2 and 3; see also Fig. S3 in the supplemental material).
Cellular SAM and SAH levels were also significantly decreased and increased, respectively (P = 0.0106 and P = 0.0390, respectively) in the A. fumigatus ΔgliG strain (Fig. 4C), which was previously shown to be resistant to the growth-inhibitory effects of exogenous gliotoxin (4); however, cellular SAM levels were comparable to those in Af293 exposed to exogenous gliotoxin. This finding suggests that the ΔgliG strain retains the ability to maintain cellular SAM levels consistent with those in the wild type upon exogenous gliotoxin exposure, unlike other gli mutants, and consequently is resistant to the growth-inhibitory effects. GliG mediates the conjugation of glutathione (GSH) to a gliotoxin biosynthetic intermediate during synthesis (4, 18, 44, 45). This reaction is effectively inhibited in the ΔgliG strain; thus, it is possible that in the ΔgliG strain, surplus GSH can be converted to Hcy and, consequently, SAM through the transsulfuration process, accounting for the maintenance of cellular SAM levels upon the addition of exogenous gliotoxin. It has been shown that a number of methylated gliotoxin-related intermediates/shunt metabolites are present in A. fumigatus, which are absent in the A. fumigatus ΔgliZ strain (9). We hypothesized that if gliotoxin could induce gli cluster expression in the A. fumigatus ΔgliZ strain (i.e., independently of gliZ), the production of these intermediates could lead to SAM depletion and SAH production. However, the addition of gliotoxin to the A. fumigatus ΔgliZ strain did not induce gliotoxin biosynthesis and secretion, as 13C-labeled gliotoxin was undetectable in A. fumigatus ΔgliZ culture supernatants following feeding experiments with [13C]phenylalanine (data not shown). However, SAM depletion and SAH production were evident in the A. fumigatus ΔgliZ strain upon exposure to exogenous gliotoxin (Fig. 4C).
The addition of gliotoxin significantly upregulates the expression of genes involved in the methyl/methionine cycle in the A. fumigatus ΔgliT strain (17) (see Table S3 in the supplemental material). As shown here, significantly elevated SAH levels are also uniquely observed in this strain in response to gliotoxin exposure. Methionine produced from Hcy can either be used in protein synthesis or, alternatively, enter the methionine cycle, whereby it is S-adenylated to form SAM (37). SAM is a ubiquitous cellular methyl donor and is utilized by methyltransferases for DNA, protein, and metabolite methylation reactions (46). Elevated SAH levels prevent the production of toxic Hcy (26) and may lead to the inhibition of cellular methyltransferases via competitive inhibition of SAM binding. Our data are consistent with a scenario whereby exogenous and de novo-produced gliotoxin undergoes bis-thiomethylation in an unregulated manner in the absence of GliT, leading to elevated SAH and attenuated SAM levels, respectively, which activate the methyl/methionine cycle to increase the SAM supply (Fig. 6). Interestingly, in the A. fumigatus ΔmetR strain, the addition of cystathionine potentiates gliotoxin sensitivity, even though the mutant is not significantly sensitive to the addition of cystathionine alone, compared to ATCC 46645. Cystathionine feeds into the methyl/methionine cycle, which suggests that increased levels of cycle components, in conjunction with upregulated methyl/methionine cycle genes due to the addition of gliotoxin, exacerbate gliotoxin sensitivity. Indeed, it was reported recently that SAM depletion in mammalian cells can induce cell cycle arrest in the G1 phase (47).
Comparative 2D-PAGE-LC-MS/MS analysis of mycelial protein extracts of the A. fumigatus ΔgliK strain, which is partially sensitive to exogenous gliotoxin (7), revealed a significantly increased abundance of cobalamin-independent methionine synthase (MetH) (present in two protein spots, with 1.9- and 2.2-fold-increased abundances, respectively). MetH couples the methyl cycle and the methionine cycle through the methylation of Hcy and the subsequent regeneration of tetrahydrofolate (THF) (see Fig. S3 in the supplemental material). An additional step in the methyl cycle, involving the conversion of THF to 5,10-methylene-THF, is usually catalyzed by SHMT, with the simultaneous metabolism of serine and formation of glycine (48). However, in the presence of excess glycine or a limited pool of 5,10-methylene-THF, this step can be catalyzed by the mitochondrial glycine decarboxylase complex (GDC) (49). The abundance of a subunit of this complex, glycine dehydrogenase (see Fig. S3 in the supplemental material), was also increased in the A. fumigatus ΔgliK strain following incubation with gliotoxin (2.2-fold; P = 0.025), suggesting an overall upregulation of the methyl cycle in the A. fumigatus ΔgliK strain upon gliotoxin exposure. MetH has also been shown to be a target of the transcriptional activator yap1 in A. fumigatus, and expression of this protein is induced in the presence of oxidative stress (50). In fungi, the production of methionine from Hcy is catalyzed by MetH/D, and in Candida albicans, this enzyme has been shown to be essential for cell viability (37). Indeed, Hcy induces the upregulation of A. nidulans methionine synthase (51), in addition to the positive regulation of MTHFR/MtrA expression (38). Interestingly, a significantly increased abundance of MTHFR/MtrA was observed in gliotoxin-exposed ΔgliK cells (1.8-fold; P < 0.05). This scenario is in contrast to that observed for wild-type A. fumigatus, whereby downregulation of MTHFR/MtrA was noted in response to gliotoxin and H2O2 combined, relative to the solvent control (11). Although an altered GliT abundance was not detectable in the A. fumigatus ΔgliK strain during the above-mentioned 2-DE-LC-MS/MS analysis, it has been reported that elevated gliT expression occurs in response to gliotoxin exposure in this mutant (7). Here, parallel LFQ proteomic investigations of the A. fumigatus ΔgliK strain, in the presence and absence of gliotoxin, revealed a 14- to 15-fold (log2-fold change = 3.837)-increased abundance of GliT, compared to the 250-fold increase in wild-type A. fumigatus following gliotoxin exposure. Thus, we hypothesize that the apparent gliotoxin sensitivity of the A. fumigatus ΔgliK strain (7) can in part be explained by the attenuated expression of GliT, compared to that of the wild type, and a consequent deficit in gliotoxin secretion, as noted by the authors of that study.
Figure 6 presents a model for how GliT prevents dysregulation of SAM:SAH homeostasis in A. fumigatus. GT-(SH)2 formation (10) is mediated by gli cluster activity, which in turn is induced by gliotoxin. GT-(SH)2 is either oxidized to gliotoxin by GliT, followed by GliA-mediated secretion, or converted to BmGT by the SAM-dependent activity of GtmA. Notably, GT-(SH)2 dismutation to BmGT consumes 2 SAM molecules. BmGT is secreted from A. fumigatus; concomitantly, its biosynthesis results in SAM depletion and SAH formation. Under normal circumstances, SAH is reconverted to SAM via the methionine cycle, which in turn necessitates the activity of the methyl cycle (Fig. 6; see also Fig. S3 in the supplemental material). Thus, GtmA appears to link the regulation of gliotoxin biosynthesis to what are conventionally considered to be primary metabolic processes. In doing so, a highly precarious situation materializes in A. fumigatus, whereby any scenario which increases GT-(SH)2 levels (e.g., gliT deletion) could potentially lead to SAM depletion and SAH overproduction and the subsequent occurrence of unwanted downstream consequences. Regulation of gliotoxin biosynthesis is indeed a high-risk strategy for A. fumigatus and has necessitated the evolution, and integration, of multiple facilitatory mechanisms to prevent deleterious eventualities.
Finally, it has been proposed that epigenetic modifications, including methylation, may affect cellular metabolism by limiting SAM availability for essential reactions (52). Conversely, it has not escaped our attention that SAM limitation as a consequence of excessive BmGT formation could impact the epigenetic regulation of chromatin structure and secondary metabolite formation, especially since altered fumagillin, pseurotin A, and brevianamide F levels have been observed in the A. fumigatus ΔgliT strain (17).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by a Science Foundation Ireland Principal Investigator award to S.D. (PI/11/1188). R.A.O. was funded by a 3U Partnership award (DCU/NUIM/RCSI). S.H. and S.K.D. are recipients of Irish Research Council Embark Ph.D. fellowships. LC-MS facilities were funded by competitive awards from Science Foundation Ireland (12/RI/2346 [3]) and the Irish Higher Education Authority. qRT-PCR instrumentation was funded by Science Foundation Ireland (SFI/07/RFP/GEN/F571/ECO7). The image station for the visualization of Southern blots was funded by Science Foundation Ireland career development award 13/CDA/2142 to Ozgur Bayram.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00055-15.
REFERENCES
- 1.Gerken T, Walsh CT. 2013. Cloning and sequencing of the chaetocin biosynthetic gene cluster. Chembiochem 14:2256–2258. doi: 10.1002/cbic.201300513. [DOI] [PubMed] [Google Scholar]
- 2.Chang S-L, Chiang Y-M, Yeh H-H, Wu T-K, Wang CCC. 2013. Reconstitution of the early steps of gliotoxin biosynthesis in Aspergillus nidulans reveals the role of the monooxygenase GliC. Bioorg Med Chem Lett 23:2155–2157. doi: 10.1016/j.bmcl.2013.01.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo CJ, Yeh HH, Chiang YM, Sanchez JF, Chang SL, Bruno KS, Wang CC. 2013. Biosynthetic pathway for the epipolythiodioxopiperazine acetylaranotin in Aspergillus terreus revealed by genome-based deletion analysis. J Am Chem Soc 135:7205–7213. doi: 10.1021/ja3123653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis C, Carberry S, Schrettl M, Singh I, Stephens JC, Barry SM, Kavanagh K, Challis GL, Brougham D, Doyle S. 2011. The role of glutathione S-transferase GliG in gliotoxin biosynthesis in Aspergillus fumigatus. Chem Biol 18:542–552. doi: 10.1016/j.chembiol.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 5.Scharf DH, Chankhamjon P, Scherlach K, Heinekamp T, Willing K, Brakhage AA, Hertweck C. 2013. Epidithiodiketopiperazine biosynthesis: a four-enzyme cascade converts glutathione conjugates into transannular disulfide bridges. Angew Chem Int Ed Engl 52:11092–11095. doi: 10.1002/anie.201305059. [DOI] [PubMed] [Google Scholar]
- 6.Choi HS, Shim JS, Kim J-A, Kang SW, Kwon HJ. 2007. Discovery of gliotoxin as a new small molecule targeting thioredoxin redox system. Biochem Biophys Res Commun 359:523–528. doi: 10.1016/j.bbrc.2007.05.139. [DOI] [PubMed] [Google Scholar]
- 7.Gallagher L, Owens RA, O'Keeffe G, Dolan SK, Schrettl M, Kavanagh K, Jones G, Doyle S. 2012. The Aspergillus fumigatus protein GliK protects against oxidative stress and is essential for gliotoxin biosynthesis. Eukaryot Cell 11:1226–1238. doi: 10.1128/EC.00113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domingo MP, Colmenarejo C, Martínez-Lostao L, Müllbacher A, Jarne C, Revillo MJ, Delgado P, Roc L, Meis JF, Rezusta A, Pardo J, Gálvez EM. 2012. Bis(methyl)gliotoxin proves to be a more stable and reliable marker for invasive aspergillosis than gliotoxin and suitable for use in diagnosis. Diagn Microbiol Infect Dis 73:57–64. doi: 10.1016/j.diagmicrobio.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Forseth RR, Fox EM, Chung D, Howlett BJ, Keller NP, Schroeder FC. 2011. Identification of cryptic products of the gliotoxin gene cluster using NMR-based comparative metabolomics and a model for gliotoxin biosynthesis. J Am Chem Soc 133:9678–9681. doi: 10.1021/ja2029987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolan SK, Owens RA, O'Keeffe G, Hammel S, Fitzpatrick DA, Jones GW, Doyle S. 2014. Regulation of non-ribosomal peptide synthesis: bis-thiomethylation attenuates gliotoxin biosynthesis in Aspergillus fumigatus. Chem Biol 21:999–1012. doi: 10.1016/j.chembiol.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Owens RA, Hammel S, Sheridan KJ, Jones GW, Doyle SA. 2014. Proteomic approach to investigating gene cluster expression and secondary metabolite functionality in Aspergillus fumigatus. PLoS One 9:e106942. doi: 10.1371/journal.pone.0106942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scharf DH, Remme N, Heinekamp T, Hortschansky P, Brakhage AA, Hertweck C. 2010. Transannular disulfide formation in gliotoxin biosynthesis and its role in self-resistance of the human pathogen Aspergillus fumigatus. J Am Chem Soc 132:10136–10141. doi: 10.1021/ja103262m. [DOI] [PubMed] [Google Scholar]
- 13.Schrettl M, Carberry S, Kavanagh K, Haas H, Jones GW, O'Brien J, Nolan A, Stephens J, Fenelon O, Doyle S. 2010. Self-protection against gliotoxin—a component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog 6:e1000952. doi: 10.1371/journal.ppat.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Forseth RR, Bowers AA, Schroeder FC, Walsh CT. 2012. A backup plan for self-protection: S-methylation of holomycin biosynthetic intermediates in Streptomyces clavuligerus. Chembiochem 13:2521–2526. doi: 10.1002/cbic.201200536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Toyotome T, Muraosa Y, Watanabe A, Wuren T, Bunsupa S, Aoyagi K, Yamazaki M, Takino M, Kamei K. 2014. GliA in Aspergillus fumigatus is required for its tolerance to gliotoxin and affects the amount of extracellular and intracellular gliotoxin. Med Mycol 52:506–518. doi: 10.1093/mmy/myu007. [DOI] [PubMed] [Google Scholar]
- 16.Cramer RA, Gamcsik MP, Brooking RM, Najvar LK, Kirkpatrick WR, Patterson TF, Balibar CJ, Graybill JR, Perfect JR, Abraham SN, Steinbach WJ. 2006. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot Cell 5:972–980. doi: 10.1128/EC.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Keeffe G, Hammel S, Owens RA, Keane TM, Fitzpatrick DA, Jones GW, Doyle S. 2014. RNA-seq reveals the pan-transcriptomic impact of attenuating the gliotoxin self-protection mechanism in Aspergillus fumigatus. BMC Genomics 15:894. doi: 10.1186/1471-2164-15-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scharf DH, Remme N, Habel A, Chankhamjon P, Scherlach K, Heinekamp T, Hortschansky P, Brakhage AA, Hertweck C. 2011. A dedicated glutathione S-transferase mediates carbon-sulfur bond formation in gliotoxin biosynthesis. J Am Chem Soc 133:12322–12325. doi: 10.1021/ja201311d. [DOI] [PubMed] [Google Scholar]
- 19.Scharf DH, Chankhamjon P, Scherlach K, Heinekamp T, Roth M, Brakhage AA, Hertweck C. 2012. Epidithiol formation by an unprecedented twin carbon-sulfur lyase in the gliotoxin pathway. Angew Chemie Int Ed Engl 51:10064–10068. doi: 10.1002/anie.201205041. [DOI] [PubMed] [Google Scholar]
- 20.Scharf DH, Habel A, Heinekamp T, Brakhage AA, Hertweck C. 2014. Opposed effects of enzymatic gliotoxin N- and S-methylations. J Am Chem Soc 136:11674–11679. doi: 10.1021/ja5033106. [DOI] [PubMed] [Google Scholar]
- 21.Owens RA, O'Keeffe G, O'Hanlon KA, Gallagher L, Doyle S. 2014. Virulence characteristics of Aspergillus fumigatus, p 163–194. In Sullivan DJ, Moran GP (ed), Human pathogenic fungi: molecular biology and pathogenic mechanisms. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 22.Amich J, Schafferer L, Haas H, Krappmann S. 2013. Regulation of sulphur assimilation is essential for virulence and affects iron homeostasis of the human-pathogenic mould Aspergillus fumigatus. PLoS Pathog 9:e1003573. doi: 10.1371/journal.ppat.1003573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brzywczy J, Kacprzak MM, Paszewski A. 2011. Novel mutations reveal two important regions in Aspergillus nidulans transcriptional activator MetR. Fungal Genet Biol 48:104–112. doi: 10.1016/j.fgb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Sauter M, Moffatt B, Saechao MC, Hell R, Wirtz M. 2013. Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem J 451:145–154. doi: 10.1042/BJ20121744. [DOI] [PubMed] [Google Scholar]
- 25.Dorgan KM, Wooderchak WL, Wynn DP, Karschner EL, Alfaro JF, Cui Y, Zhou ZS, Hevel JM. 2006. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem 350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Sieńko M, Natorff R, Owczarek S, Olewiecki I, Paszewski A. 2009. Aspergillus nidulans genes encoding reverse transsulfuration enzymes belong to homocysteine regulon. Curr Genet 55:561–570. doi: 10.1007/s00294-009-0269-3. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen ML, Albertsen L, Lettier G, Nielsen JB, Mortensen UH. 2006. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet Biol 43:54–64. doi: 10.1016/j.fgb.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Kubodera T, Yamashita N, Nishimura A. 2000. Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: cloning, characterization and application as a dominant selectable marker for transformation. Biosci Biotechnol Biochem 64:1416. doi: 10.1271/bbb.64.1416. [DOI] [PubMed] [Google Scholar]
- 29.O'Keeffe G, Jöchl C, Kavanagh K, Doyle S. 2013. Extensive proteomic remodeling is induced by eukaryotic translation elongation factor 1Bγ deletion in Aspergillus fumigatus. Protein Sci 22:1612–1622. doi: 10.1002/pro.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Hanlon KA, Cairns T, Stack D, Schrettl M, Bignell EM, Kavanagh K, Miggin SM, O'Keeffe G, Larsen TO, Doyle S. 2011. Targeted disruption of nonribosomal peptide synthetase pes3 augments the virulence of Aspergillus fumigatus. Infect Immun 79:3978–3992. doi: 10.1128/IAI.00192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roeder S, Dreschler K, Wirtz M, Cristescu SM, van Harren FJM, Hell R, Piechulla B. 2009. SAM levels, gene expression of SAM synthetase, methionine synthase and ACC oxidase, and ethylene emission from N. suaveolens flowers. Plant Mol Biol 70:535–546. doi: 10.1007/s11103-009-9490-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, Waszkuc T, Garbis S, Mohammed F. 2002. Liquid chromatographic determination of S-adenosyl-L-methionine in dietary supplement tablets. J AOAC Int 85:901–905. [PubMed] [Google Scholar]
- 33.Collins C, Keane TM, Turner DJ, O'Keeffe G, Fitzpatrick DA, Doyle S. 2013. Genomic and proteomic dissection of the ubiquitous plant pathogen, Armillaria mellea: toward a new infection model system. J Proteome Res 12:2552–2570. doi: 10.1021/pr301131t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 35.Priebe S, Linde J, Albrecht D, Guthke R, Brakhage AA. 2011. FungiFun: a Web-based application for functional categorization of fungal genes and proteins. Fungal Genet Biol 48:353–358. doi: 10.1016/j.fgb.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, García JL, García MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jiménez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Lafton A, Latgé J-P, Li W, Lord A, et al. . 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 37.Suliman HS, Appling DR, Robertus JD. 2007. The gene for cobalamin-independent methionine synthase is essential in Candida albicans: a potential antifungal target. Arch Biochem Biophys 467:218–226. doi: 10.1016/j.abb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sieńko M, Natorff R, Zieliński Z, Hejduk A, Paszewski A. 2007. Two Aspergillus nidulans genes encoding methylenetetrahydrofolate reductases are up-regulated by homocysteine. Fungal Genet Biol 44:691–700. doi: 10.1016/j.fgb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Grynberg M, Topczewski J, Godzik A. 2000. The Aspergillus nidulans cysA gene encodes a novel type of serine O-acetyltransferase which is homologous to homoserine O-acetyltransferases. Microbiology 146:2695–2703. [DOI] [PubMed] [Google Scholar]
- 40.Perrin RM, Fedorova ND, Bok JW, Cramer RA, Wortman JR, Kim HS, Nierman WC, Keller NP. 2007. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog 3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiemann P, Guo C, Palmer JM, Sekonyela R, Wang CCC. 2013. Prototype of an intertwined secondary-metabolite supercluster. Proc Natl Acad Sci U S A 110:17065–17070. doi: 10.1073/pnas.1313258110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schoberle TJ, Nguyen-Coleman CK, Herold J, Yang A, Weirauch M, Hughes TR, McMurray JS, May GS. 2014. A novel C2H2 transcription factor that regulates gliA expression interdependently with GliZ in Aspergillus fumigatus. PLoS Genet 10:e1004336. doi: 10.1371/journal.pgen.1004336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bok JW, Chung D, Balajee SA, Marr KA, Andes D, Nielsen KF, Frisvad JC, Kirby KA, Keller NP. 2006. GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect Immun 74:6761–6768. doi: 10.1128/IAI.00780-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welch TR, Williams RM. 2014. Epidithiodioxopiperazines. Occurrence, synthesis and biogenesis. Nat Prod Rep 31:1376–1404. doi: 10.1039/C3NP70097F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amatov T, Jahn U. 2014. Gliotoxin: nature's way of making the epidithio bridge. Angew Chem Int Ed Engl 53:3312–3314. doi: 10.1002/anie.201310982. [DOI] [PubMed] [Google Scholar]
- 46.Grillo MA, Colombatto S. 2008. S-Adenosylmethionine and its products. Amino Acids 34:187–193. doi: 10.1007/s00726-007-0500-9. [DOI] [PubMed] [Google Scholar]
- 47.Lin D-W, Chung BP, Kaiser P. 2014. S-Adenosylmethionine limitation induces p38 mitogen-activated protein kinase and triggers cell cycle arrest in G1. J Cell Sci 127:50–59. doi: 10.1242/jcs.127811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacFarlane AJ, Liu X, Perry CA, Flodby P, Allen RH, Stabler SP, Stover PJ. 2008. Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J Biol Chem 283:25846–25853. doi: 10.1074/jbc.M802671200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piper MD, Hong SP, Ball GE, Dawes IW. 2000. Regulation of the balance of one-carbon metabolism in Saccharomyces cerevisiae. J Biol Chem 275:30987–30995. doi: 10.1074/jbc.M004248200. [DOI] [PubMed] [Google Scholar]
- 50.Lessing F, Kniemeyer O, Wozniok I, Loeffler J, Kurzai O, Haertl A, Brakhage AA. 2007. The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot Cell 6:2290–2302. doi: 10.1128/EC.00267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kacprzak MM, Lewandowska I, Matthews RG, Paszewski A. 2003. Transcriptional regulation of methionine synthase by homocysteine and choline in Aspergillus nidulans. Biochem J 376:517–524. doi: 10.1042/BJ20030747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez-Pastor B, Cosentino C, Mostoslavsky R. 2013. A tale of metabolites: the cross-talk between chromatin and energy metabolism. Cancer Discov 3:497–501. doi: 10.1158/2159-8290.CD-13-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dolan SK, O'Keeffe G, Jones GW, Doyle S. 2015. Resistance is not futile: gliotoxin biosynthesis, functionality and utility. Trends Microbiol 23:419–428. doi: 10.1016/j.tim.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 54.Mabey Gilsenan J, Cooley J, Bowyer P. 2012. CADRE: the Central Aspergillus Data REpository 2012. Nucleic Acids Res 40:D660–D666. doi: 10.1093/nar/gkr971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.