Abstract
Very high ethanol tolerance is a distinctive trait of the yeast Saccharomyces cerevisiae with notable ecological and industrial importance. Although many genes have been shown to be required for moderate ethanol tolerance (i.e., 6 to 12%) in laboratory strains, little is known of the much higher ethanol tolerance (i.e., 16 to 20%) in natural and industrial strains. We have analyzed the genetic basis of very high ethanol tolerance in a Brazilian bioethanol production strain by genetic mapping with laboratory strains containing artificially inserted oligonucleotide markers. The first locus contained the ura3Δ0 mutation of the laboratory strain as the causative mutation. Analysis of other auxotrophies also revealed significant linkage for LYS2, LEU2, HIS3, and MET15. Tolerance to only very high ethanol concentrations was reduced by auxotrophies, while the effect was reversed at lower concentrations. Evaluation of other stress conditions showed that the link with auxotrophy is dependent on the type of stress and the type of auxotrophy. When the concentration of the auxotrophic nutrient is close to that limiting growth, more stress factors can inhibit growth of an auxotrophic strain. We show that very high ethanol concentrations inhibit the uptake of leucine more than that of uracil, but the 500-fold-lower uracil uptake activity may explain the strong linkage between uracil auxotrophy and ethanol sensitivity compared to leucine auxotrophy. Since very high concentrations of ethanol inhibit the uptake of auxotrophic nutrients, the active uptake of scarce nutrients may be a major limiting factor for growth under conditions of ethanol stress.
INTRODUCTION
Very high ethanol tolerance is an outstanding property of the yeast Saccharomyces cerevisiae. While laboratory strains of S. cerevisiae have low to moderate ethanol tolerance (i.e., 6 to 12% [vol/vol] ethanol), many natural and industrial strains are able to accumulate >16% ethanol (1, 2). This property is important in industrial processes such as sake brewing and bioethanol production, which rely on the accumulation of high concentrations of ethanol (3, 4). In bioethanol production, for example, very high ethanol tolerance of the yeast is a prerequisite to obtain a very high final ethanol titer, reducing distillation costs and contamination levels and decreasing liquid volumes in the factory. Very high ethanol tolerance is also important for maintaining a proper fermentation rate in the later stages of fermentation and for complete attenuation of the sugar. Adequate completion of fermentation is also of relevance for the production of beer, wine, and other alcoholic beverages by yeast fermentation.
Cellular membranes are believed to be the main targets of ethanol toxicity in S. cerevisiae, and this is related to the tendency of ethanol to preferentially accumulate in the hydrophobic part of the membranes (5, 6). The need for the cell to compensate for this effect is reflected in several experimental findings. For example, cells that are exposed to ethanol change the lipid composition of their membranes to counteract the detrimental effect of ethanol. In particular, changes in the compositions of fatty acids (7–9) and ergosterol (10) have been observed.
In addition to the crucial role of the plasma membrane, several amino acids have also been associated with ethanol tolerance in S. cerevisiae. In particular, it has been shown that increased accumulation of intracellular proline improved the cell viability of sake yeast in the presence of ethanol (11). Proline is known to enhance the stability of membranes and proteins (12) and to inhibit aggregation during protein refolding (13). Overexpression of either tryptophan biosynthesis genes or the tryptophan permease gene in a sake yeast strain as well as supplementation of tryptophan in the culture medium conferred increased tolerance to 5% ethanol (14). Nearly all studies on ethanol tolerance have been performed with laboratory yeast strains, which in general have lower fitness than do natural and industrial strains. Auxotrophic mutations have been indicated as a possible cause of reduced fitness (15, 16) but never in connection with ethanol tolerance.
Ethanol tolerance is a complex property that is determined by numerous genetic and environmental factors (1, 7, 14, 17–24). Several genome-wide analyses have been performed to decipher the genetic basis of ethanol tolerance in S. cerevisiae, as comprehensively reviewed by Stanley et al. (25). Virtually all of these studies focused on the tolerance of laboratory strains to moderate concentrations of ethanol (i.e., 6 to 12%) and were performed by determining the sensitivity of the yeast deletion strain collection to such ethanol concentrations under different conditions (17, 18, 20, 22, 23). Another study used genome-wide transcriptomics to study differences between the responses of natural S. cerevisiae strains to 5% ethanol (14). Although the above-mentioned studies identified hundreds of genes that are required for ethanol tolerance in S. cerevisiae, they showed little overlap between the genes identified under the different conditions used (14, 17, 18, 20, 22, 23). This discrepancy suggests that the mechanisms of ethanol sensitivity depend on the genetic background, the growth conditions, and the ethanol concentration used. Although the above-mentioned studies provided useful insights into the genetic determinants of tolerance to a range of ethanol concentrations, they did not give insight into those that are specific for very high ethanol concentrations (i.e., in the range of 16 to 18%) in natural and industrial strains.
We have focused on the very high ethanol tolerance of VR1, a yeast strain used in industrial bioethanol production, in comparison with the low tolerance of S288c-derived laboratory strains. The VR1 strain is a natural strain that occurred as a wild-yeast contaminant in a Brazilian bioethanol production plant (3). It was selected as a production strain because of its dominance in the yeast recycling system and other favorable characteristics for industrial bioethanol production, including very high ethanol tolerance. Analysis of genetic elements important for the very high ethanol tolerance of VR1 by linkage mapping with a series of S288c laboratory strains containing large numbers of artificially introduced genetic markers identified the ura3Δ0 mutation of the artificially marked strain as the causative mutation. Further exploration of this finding revealed that other auxotrophic mutations also reduce tolerance to very high ethanol concentrations (although to different extents) and that this effect is not observed with moderate concentrations. For uracil and leucine, the effect could be linked to the relative rate of uptake of the auxotrophic nutrient. Examination of other stress factors showed that the effect of auxotrophies also depends on the types of stress and auxotrophy and especially the concentration of the auxotrophic nutrient in the medium compared to the concentration that limits growth.
MATERIALS AND METHODS
Strains and cultivation conditions.
All S. cerevisiae strains were routinely maintained on solid yeast extract-peptone-dextrose (YPD) medium containing 1% yeast extract, 2% Bacto peptone, 2% glucose, and 1.5% agar. Ethanol tolerance assays were performed on solid yeast extract-peptone (YP) and YPD media containing ethanol at the indicated concentrations (vol/vol). Other stress tolerance assays were performed with solid YPD medium containing 0.3% or 0.4% (vol/vol) acetic acid at pH 5, YPD medium containing 4.0 mM or 4.5 mM H2O2, YPD medium containing 1.50 M or 1.75 M NaCl, and YP medium containing 25% or 30% glucose. Stress tolerance assays with limiting amounts of either uracil or leucine were performed on minimal medium containing a complete synthetic dropout amino acid-nucleotide mixture (CSM-AA; amounts according to the manufacturer's instructions), 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 2% glucose, 1.5% agar, and different amounts of uracil (0.005 to 0.400 mM) or leucine (0.05 to 2.00 mM), respectively. The pH of the medium was adjusted to 6.5 with 4 M KOH. After autoclaving, different concentrations of ethanol or NaCl were added to the medium as indicated. Transport assays were performed in minimal medium with 2% glucose or yeast nitrogen base (YNB) medium containing 0.17% yeast nitrogen base without amino acids and ammonium sulfate and 2% glucose. The pH of YNB medium was adjusted to 4.8 with HCl. All concentrations mentioned above are in weight per volume unless indicated differently.
Minimal medium for the selection of URA3 transformants and for linkage analysis of the auxotrophic alleles contained CSM-AA (amounts according to the manufacturer's instructions), 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 2% glucose, and 1.5% agar. The pH of the minimal medium was adjusted to 6.5 with 4 M KOH.
All strains used in this study are listed in Table 1. Strain pAMS(ura3Δ0::URA3) was constructed by transforming a partially artificially marked strain (pAMS) with a DNA fragment containing the URA3 open reading frame (ORF) flanked by a 496-bp upstream region and a 247-bp downstream region homologous to the borders of the ORF. The fragment was obtained by PCR amplification using primers URA3_upstream (5′-ATCATCTCATGGATCTGCAC) and URA3_downstream (5′-CGTCCATCTTTACAGTCCTG) on genomic DNA from VR1-5B. Transformants were selected on minimal medium without uracil. In the same way, URA3 was integrated into BY4741, BY4742, and seven segregants from the VR1-5B/BY4741 hybrid strain.
TABLE 1.
Strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 28 |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | 28 |
| BY | Hybrid strain obtained by crossing BY4741 with BY4742 | This study |
| S288c (a) | MATa prototroph | 28 |
| S288c (α) | MATα prototroph | 28 |
| S288c leu2Δ | MATa leu2Δ::attB-KanMX-attP | This study |
| S288c ura3-52 | MATa ura3-52 | 66 |
| pAMS | BY4741 or BY4742 with various numbers of artificial markers | This study |
| VR1 | Natural isolate and former production strain in Brazilian bioethanol production with sugar cane | Mario Lucio Lopes (Fermentec, Piracicaba) |
| VR1-5B | Haploid (MATα) segregant of VR1 with similarly high ethanol tolerance as its parent | 34 |
| pAMS(ura3Δ0::URA3) | pAMS in which the ura3Δ0 mutation has been reverted to URA3 | This study |
The S288c leu2Δ strain was constructed by transforming S288c (MATa) with a KanMX deletion cassette flanked by 50-bp regions homologous to the borders of the LEU2 ORF. The KanMX deletion cassette was obtained by PCR amplification using primers leu2-A1 (5′-TTTTCTTACCTTTTACATTTCAGCAATATATATATATATATTTCAAGGATATACCATTCTAGTGGTCGGCTGGAGATCGG) and leu2-A2 (5′-TTCTATTATGAATTTCATTTATAAAGTTTATGTACAAATATCATAAAAAAAGAGAATCTTTAGCCGTTATGGCGGGCATC) on pJET1,2-attB-KanMX-attP plasmid DNA (26). Transformants were selected on minimal medium without leucine. All transformations and DNA manipulations described above were performed according to standard methods (27) and verified by PCR and DNA sequence analysis.
Artificial genetic markers for linkage analysis.
We have integrated ∼550 artificial markers into the genomes of the commonly used laboratory strains BY4741 and BY4742 (28). The markers were integrated by using a ligase-free, PCR-based allele replacement method (GeneWeaver II) (29). The marker sequences are artificial, unique sequences of 20 bp that do not show sequence similarity to the S. cerevisiae genome. They are preceded by the 8-bp recognition sequence for the rare-cutting restriction enzyme FseI, which was originally added to allow specific cleavage of genomic fragments at the marker positions. The markers were integrated at a distance of ∼20 kb (∼7 centimorgans) at presumably neutral positions, i.e., outside open reading frames, promoter and terminator regions, and possible regulatory sequences.
The artificial markers were inserted in parallel in a series of BY4741/2 laboratory strains and subsequently concentrated by a combination of parallel crosses and serial backcrosses. In this way, we obtained a small number of strains with partially overlapping and different numbers of artificial markers that together cover the whole genome. These strains were called pAMSs. The artificial markers can be detected by PCR in which the direct primer is complementary to the marker sequence and the indirect primer is complementary to a sequence 200 to 400 bp downstream of the marker integration site.
Linkage mapping.
The highly ethanol-tolerant segregant VR1-5B was crossed with 28 pAMSs. The pAMSs were selected based on their total number of markers and the presence of unique markers. See Fig. S1 in the supplemental material for the genetic map combining all artificial markers present in the 28 pAMSs. The VR1-5B/pAMS hybrid strains were then allowed to sporulate, and segregants were isolated by using an MSM 300 dissection microscope from Singer Instrument Co. Ltd. (Watchet, United Kingdom). Mating, sporulation, and tetrad analyses were performed according to standard methods (30), and mating types were determined by diagnostic PCR for the MAT locus (31). The segregants were subsequently scored for ethanol tolerance, as described below in the section on stress tolerance assays. Next, the presence of the artificial markers in the highly ethanol-tolerant segregants was scored by PCR. For each marker, a deviation from 50% inheritance was weighed by using an exact binomial test with a confidence level of 95%. Correction for multiple testing was carried out by using a false discovery rate (FDR) control according to methods described previously by Benjamini and Yekutieli (32).
Stress tolerance assays.
Cells taken from solid YPD medium were transferred into 3 ml of liquid YPD or minimal medium in a glass tube and cultivated for 3 days in an orbital shaker at 200 rpm at 30°C. The cultures were then diluted in water to an optical density at 600 nm (OD600) of 0.5 and further serially diluted to obtain a 2-fold (100 until 8 × 10−3) or 10-fold (100 until 10−3) dilution range. Exactly 5 μl of each cell suspension of the dilution range was spotted onto solid YPD, YP, or minimal medium containing different stress factors, as indicated. Growth was scored after 1 day for control YPD plates and after 2 to 11 days, as indicated, for plates with different concentrations of ethanol or other stress factors. All plates were incubated at 30°C, except for the heat stress tolerance assay, in which case the plates were incubated at 39°C or 40°C. All stress tolerance assays were repeated at least three times, and representative results are shown.
Transport assays.
Cells of the S288c, S288c ura3-52, and S288c leu2Δ strains in 5 ml of minimal medium were cultivated overnight in an orbital shaker at 200 rpm at 30°C. Each preculture was then used to inoculate 200 ml of YNB medium containing 2% glucose (pH 4.8). The cells were grown to an OD600 of 1 to 2, after which they were harvested by centrifugation (3,000 rpm for 5 min), washed twice with morpholineethanesulfonic acid (MES)-KOH (25 mM; pH 6), and resuspended in fresh YNB medium to a final cell concentration of 160 mg ml−1. Fifty microliters of this cell suspension was transferred to a new centrifuge tube and incubated for 10 min at 30°C. In order to start transport measurements, a volume of 40 μl of a specific ethanol concentration was first added to the cell suspension. After 5 min of incubation, 10 μl of either 3H-labeled uracil to a final concentration of 0.05 mM (PerkinElmer) or 14C-labeled l-leucine to a final concentration of 2.5 mM (PerkinElmer) was added to the suspension. After 2 min of incubation, 5 ml of ice-cold water was added to stop the uptake of the radiolabeled nutrients. The cells were collected by using a glass microfiber filter (Whatman GF/C, with a retention particle size of 1.2 μm), saturated with unlabeled nutrient solution, and immediately washed twice with 5 ml of ice-cold water. For both transport assays (uracil and leucine), three samples with radiolabeled nutrient and two blanks (addition of 5 ml of ice-cold water followed by the addition of radiolabeled nutrient) were analyzed. The radioactivity on the filter was determined with a liquid scintillation counter (LS6500; Beckman Coulter). A 500-μl aliquot of the cell suspension was taken to determine protein content by using the Bradford method (33). Transport activity was expressed as nanomoles substrate transported per minute and milligram of protein and as a percentage of the transport rate in the absence of ethanol.
RESULTS
Identification of a segregant from VR1 with very high ethanol tolerance.
VR1 is a diploid strain, and thus, we first isolated a segregant with similarly high ethanol tolerance in order to be able to perform quantitative trait locus (QTL) mapping by crossing (34). We sporulated VR1 and isolated segregants using a dissection microscope. All segregants were haploid, indicating that VR1 is a heterothallic strain. To assay the ethanol tolerance of VR1 and its segregants, we scored their growth on solid YP medium containing different concentrations of ethanol. Ethanol tolerance can be scored in different ways; in previous work, ethanol tolerance has usually been determined by measuring growth on glucose in the presence of ethanol (1, 17, 18, 20, 22, 23). However, in the current study, high variability among biological replicates was observed when ethanol tolerance in the presence of glucose was scored. We therefore decided to determine ethanol tolerance by screening on a medium with only ethanol as a carbon source and no added sugar. Although it cannot be ruled out that segregants with high ethanol consumption rates are also selected under these conditions, these segregants must also contain genetic determinants of high ethanol tolerance. In fact, cells that grow well on high concentrations of ethanol are expected to also need high tolerance to such ethanol concentrations.
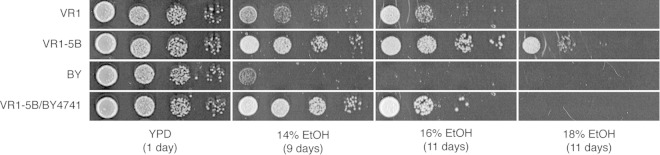
VR1 grew on medium containing up to 16% ethanol, which was significantly higher than the concentration tolerated by the diploid BY strain, which did not grow on more than 14% ethanol (Fig. 1). We identified a stable haploid segregant from VR1 with an even higher ethanol tolerance (referred to as VR1-5B) (Fig. 1) (34). The difference in ethanol tolerance between VR1 and VR1-5B may be due to the presence of recessive mutations in the diploid strain and/or to the difference in ploidy between the two strains. In fact, it was previously observed that haploid strains are slightly more ethanol tolerant than their corresponding homozygous diploids (35). The diploid VR1-5B/BY4741 hybrid displayed ethanol tolerance at least as high as that of VR1, which indicates that the very-high-ethanol-tolerance phenotype of VR1 is dominant (Fig. 1). The fact that VR1-5B/BY4741 has a slightly higher ethanol tolerance than VR1 supports the idea that the phenotype is determined (at least in part) by recessive mutations, of which phenotypic expression is not complemented in the hybrid background. Another explanation for the difference in tolerance between both strains might also be that alleles from BY4741 contribute to the ethanol tolerance phenotype of the hybrid.
FIG 1.
Growth on media containing different concentrations of ethanol (EtOH). A 10-fold dilution range was spotted onto YPD medium, as a control, and onto YP medium with 14%, 16%, and 18% ethanol. The diploid BY strain was obtained by crossing BY4741 with BY4742, and the diploid VR1-5B/BY4741 strain was obtained by crossing VR1-5B with BY4741.
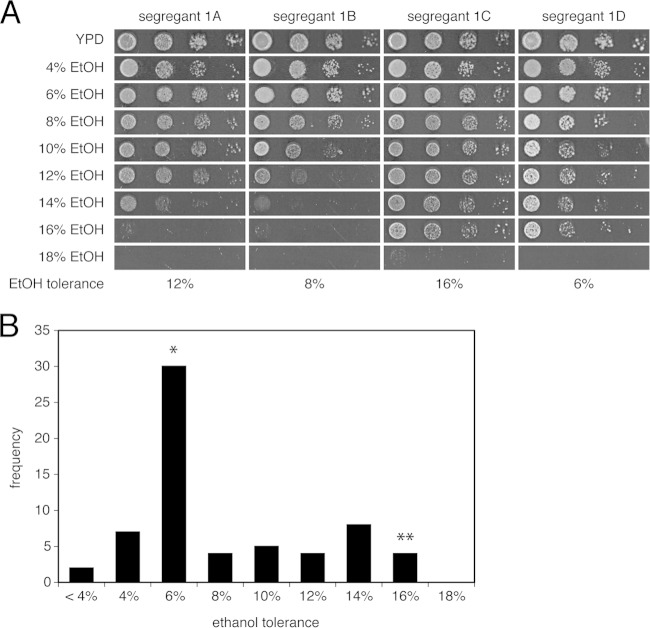
In order to determine the distribution of ethanol tolerance in segregants from the VR1-5B/BY4741 hybrid, we analyzed the growth of 16 complete tetrads on solid YP medium with ethanol concentrations ranging from 4% to 18%. The ethanol tolerance of each strain was scored as the maximum ethanol concentration at which its growth was still visually the same as that on the control plate without ethanol (Fig. 2A). The segregants exhibited a wide range of ethanol tolerance: the bulk of the segregants displayed tolerance to 6% ethanol, while the highest ethanol tolerance observed was to 16% ethanol, similar to that of VR1-5B (Fig. 2B). This indicates that ethanol tolerance is not a continuous trait, at least not in the segregants from this specific cross. Single or low numbers of causative mutant alleles may have only little effect in conferring high ethanol tolerance, while the presence of multiple mutant alleles might be required. Alternatively, a single mutant allele from the BY background may have a detrimental effect on high ethanol tolerance regardless of the other mutant alleles present. We decided to use growth in the presence of 16% ethanol for the selection of very highly ethanol-tolerant segregants for the genetic mapping experiment. Under these conditions, we assumed that the selected segregants must contain at least all major genetic elements from VR1-5B that are responsible for its very high ethanol tolerance.
FIG 2.
Distribution of ethanol tolerance in segregants from VR1-5B/BY4741. (A) The segregants of 16 complete tetrads were grown on YP plates containing 4% to 18% ethanol (10-fold dilution range). The spot test for one complete tetrad is shown as an example. For each segregant, ethanol tolerance was defined as the maximum ethanol concentration at which growth was the same as that on the control YPD plate. (B) BY4741 showed tolerance to 6% ethanol (*), and VR1-5B showed tolerance to 16% ethanol (**). Most segregants showed ethanol tolerance similar to that of BY4741, very few showed the same high tolerance as that of VR1-5B, and the rest showed an intermediate tolerance or even a lower tolerance than that of BY4741. None of the segregants showed higher tolerance than that of VR1-5B.
Linkage analysis of very high ethanol tolerance using artificial markers.
In our previous study, we crossed the highly ethanol-tolerant segregant VR1-5B with a number of partial artificially marked strains (pAMSs) (34). These strains contain various numbers of unique 20-bp oligonucleotide markers that were inserted in presumably neutral genomic locations at intervals of ∼20 kb in the BY4741/2 genetic background (see Materials and Methods for a detailed description of the construction of the pAMSs). After sporulation, 5,974 segregants were isolated and subsequently scored for ethanol tolerance by assaying growth on 16% ethanol. The segregants displayed a wide range of ethanol tolerances, with ∼1 in 44 (i.e., 136 of 5,974 segregants phenotyped in total) displaying ethanol tolerance similar to that of the VR1-5B parent strain (16%). This ratio predicts 5 to 6 unlinked loci (1/44 = 1/25.5) that are responsible for the difference in ethanol tolerance between VR1-5B and BY4741/2. However, this is true only if all loci are truly unlinked and in case each locus has an indispensable contribution to the phenotype.
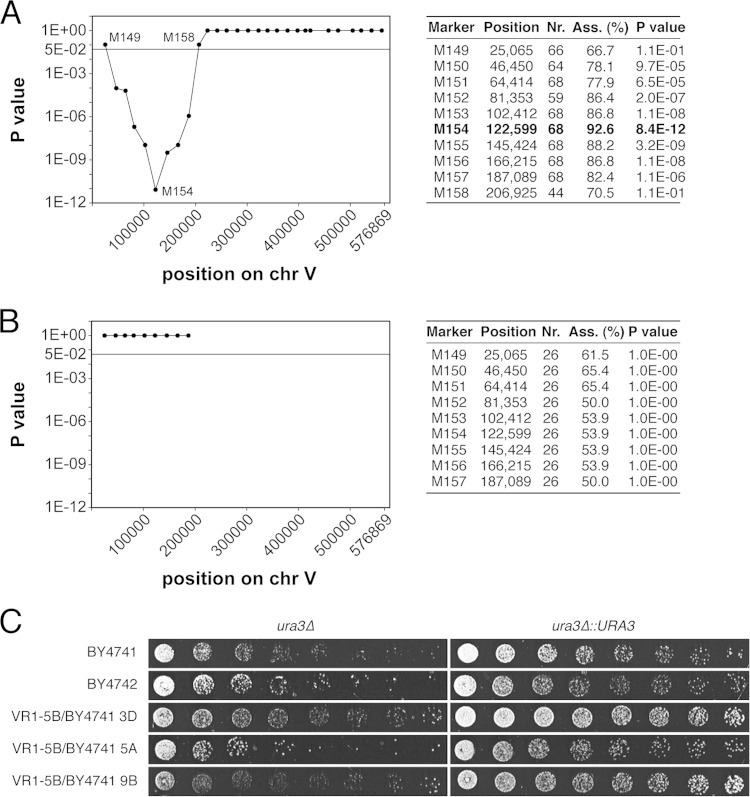
The highly ethanol-tolerant segregants were then applied for a QTL mapping experiment using pooled-segregant whole-genome sequence analysis, in which genetic differences that naturally occurred between the parent strains were used as markers (34). Our results identified three major QTL, and further downscaling of the QTL with the strongest linkage (on chromosome XIV) revealed three closely located genes affecting very high ethanol tolerance: MKT1, SWS2, and APJ1. In the current study, the same segregants were applied in a new QTL mapping experiment, with the difference that the artificially introduced sequences were used as markers. We first scored the presence of the artificial markers in each of the highly ethanol-tolerant segregants by PCR and then calculated the probability of random segregation for each marker (see Fig. S2 in the supplemental material). Significant linkage with the VR1-5B background, i.e., absence of the artificial markers in most of the segregants, was found for the region between markers 150 and 157 on chromosome V, with the lowest P value being found for marker 154 (8.4 × 10−12) (Fig. 3A). The auxotrophic gene URA3 is in close proximity to this marker and was an obvious candidate gene because of the presence of the ura3Δ0 mutation in the BY4741/2 genetic background of the pAMS.
FIG 3.
Linkage of the high-ethanol-tolerance phenotype of VR1-5B with a locus on chromosome V. (A) The probability of random segregation of 28 artificial markers on chromosome V in highly ethanol-tolerant segregants was calculated under two-sided binomial distribution with FDR adjustment according to the method of Benjamini and Yekutieli (32). The P values were considered to be significant if they were <0.05. The table shows the percent association (Ass.) (i.e., the percentage of segregants that contain the marker) and the corresponding P value for each marker in the mapped locus and for the two adjacent markers. The markers were checked in at least 44 highly ethanol-tolerant segregants (indicated as Marker nr.). (B) The probability of random segregation of markers 149 to 157 in 26 highly ethanol-tolerant segregants from the hybrid strain obtained by crossing VR1-5B with pAMS1(ura3Δ0::URA3) was determined as described above for panel A. The linkage with the locus is completely lost. (C) The VR1-5B allele of URA3 was integrated in its genomic position in BY4741, BY4742, and 7 segregants from the VR1-5B/BY4741 hybrid that were deleted of ura3 and showed different levels of ethanol tolerance. The ethanol tolerance of the untransformed (ura3Δ0) and transformed (ura3Δ0::URA3) strains was determined by growing them on different concentrations of ethanol (2-fold dilution range). The results for BY4741 (12% ethanol for 9 days), BY4742 (16% for 11 days), and 3 representative segregants (3D [14% for 9 days], 5A [16% for 11 days], and 9B [16% for 9 days]) are shown.
Contribution of URA3 to very high ethanol tolerance.
In order to evaluate whether URA3 was indeed the causative gene in the linked locus on chromosome V, we integrated the VR1-5B allele of URA3 at its original position in the BY4741 and BY4742 laboratory strains and in seven segregants from the VR1-5B/BY4741 hybrid strain that contained the ura3Δ0 mutation and showed different levels of ethanol tolerance. In all cases, an increase in ethanol tolerance was observed after integration of URA3, thereby confirming the relevance of this gene for the phenotype (Fig. 3C).
Next, we determined whether URA3 was the only causative gene in the locus by performing a new mapping experiment using a pAMS in which the ura3Δ0 mutation had been replaced by the VR1-5B allele of URA3. This strain contained artificial markers 149 to 157, which cover the entire locus. The strain was crossed with VR1-5B, and a total of 779 segregants were isolated and assayed for ethanol tolerance. Almost 1 in 30 segregants (i.e., 26 of 779 segregants phenotyped in total) displayed an ethanol tolerance phenotype similar to that of VR1-5B (16%), predicting 4.9 unlinked genetic loci (1/30 = 1/24.9). This number is lower than the previously calculated 5.5 unlinked genetic loci for the pAMS containing the ura3Δ0 mutation and would therefore be consistent with URA3 being the only causative gene in the identified locus. We scored the presence of artificial markers 149 to 157 in the 26 highly ethanol-tolerant segregants. Calculation of the P values resulted in a value of 1 for all markers (Fig. 3B), indicating that the linkage with the high-ethanol-tolerance phenotype was lost when the wild-type URA3 gene was present in this locus in both parent strains [i.e., VR1-5B and pAMS(ura3Δ0::URA3)]. This result indicated that URA3 was indeed the only genetic element in the linked locus on chromosome V that contributed to the difference in ethanol tolerance between VR1-5B and BY4741/2.
Contribution of other auxotrophic marker genes to very high ethanol tolerance.
After identifying the auxotrophic ura3Δ0 mutation as causing reduced tolerance to very high ethanol concentrations, we examined whether other auxotrophic mutations have a similar effect. The two strains that were used for the construction of the pAMSs were laboratory strains BY4741 and BY4742. These strains share three auxotrophic alleles (ura3Δ0, his3Δ1, and leu2Δ0) and differ in two auxotrophic alleles (met15Δ0 in BY4741 and lys2Δ0 in BY4742) (28).
We investigated the linkage between the very-high-ethanol-tolerance phenotype and the additional auxotrophic mutations of the laboratory yeast strains. We therefore scored the presence of each mutation in the very highly ethanol-tolerant segregants from the VR1-5B/pAMS hybrid strains (in which the pAMS was deleted for the auxotrophic gene under study) by growth tests on minimal medium lacking the corresponding auxotrophic nutrient. In this way, significant linkage was found for all auxotrophic genes investigated (i.e., HIS3, LEU2, MET15, and LYS2) (Table 2); however, the linkage was weaker than that for URA3.
TABLE 2.
Linkage analysis of the auxotrophic alleles in BY4741 and BY4742 with the high-ethanol-tolerance phenotype of VR1-5Ba
| Auxotrophic allele | No. of segregants | Association (%) | P value |
|---|---|---|---|
| his3Δ1 | 136 | 36.8 | 2.6E−03 |
| leu2Δ0 | 136 | 36.8 | 2.6E−03 |
| met15Δ0 | 50 | 30.0 | 6.6E−03 |
| lys2Δ0 | 75 | 33.3 | 5.2E−03 |
For each auxotrophic allele, the probability of random segregation was determined for segregants from the diploid VR1-5B/pAMS hybrid strain in which the pAMS was deleted for the allele under study (two-sided binomial distribution). The P values were considered significant if they were <0.05 (P values for all four alleles were significant). The percent association is the percentage of segregants that contain the auxotrophic allele.
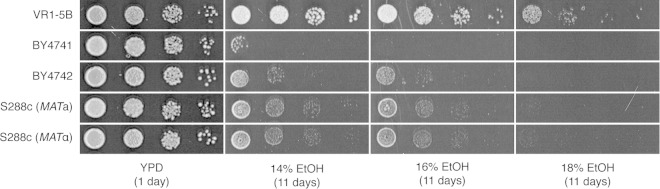
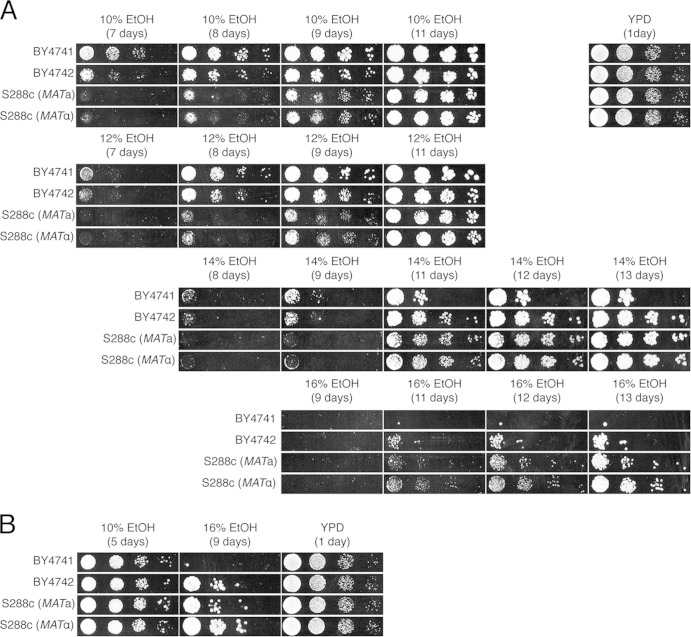
In addition, we grew the auxotrophic BY4741/2 strains and their prototrophic S288c counterparts (MATa and MATα) on YP medium containing very high concentrations of ethanol (Fig. 4). The prototrophic strains clearly showed a higher ethanol tolerance than did the auxotrophic strains, which supports our finding that auxotrophic mutations increase sensitivity to very high levels of ethanol stress.
FIG 4.
Growth of the laboratory BY4741/2 strains and their prototrophic counterparts (S288c MATa and S288c MATα) on media containing high concentrations of ethanol and comparison with VR1-5B. A 10-fold dilution range of each strain was spotted onto YP medium containing 14%, 16%, and 18% ethanol and subsequently grown for 11 days.
In summary, we have identified five auxotrophic mutations that are to some extent linked to the low ethanol tolerance of BY4741/2. However, the ethanol tolerance of VR1-5B was still much higher than that of the prototrophic S288c strains (Fig. 4), indicating that the auxotrophic mutations, although significant, are not the sole cause of the difference in ethanol tolerance between VR1-5B and BY4741/2. The identification of genetic elements other than the auxotrophic mutations that contribute to the very-high-ethanol-tolerance phenotype of VR1-5B has been reported previously (34).
Specificity of the auxotrophic mutations for tolerance to moderate as opposed to very high ethanol concentrations and to other stresses.
Virtually all previous studies on ethanol tolerance in S. cerevisiae have been performed with moderate concentrations of ethanol (i.e., 6% to 12%). As none of these studies associated auxotrophic mutations with ethanol tolerance, we examined whether this association is concentration dependent. This was studied in more detail by comparing the growth of the auxotrophic BY4741 and BY4742 strains with that of their prototrophic S288c counterparts on YP medium containing moderate to high concentrations of ethanol (10% to 16%) (Fig. 5). Interestingly, the auxotrophic strains grew better than the prototrophic strains on 10% and 12% ethanol, whereas the opposite was observed with 14% and 16% ethanol (Fig. 5A). When 2% glucose was added to the medium, no difference in growth in the presence of 10% ethanol was observed, whereas at higher concentrations, the prototrophic strains again grew better than the corresponding auxotrophic strains (Fig. 5B). The better growth of the prototrophic strains in the presence of high ethanol concentrations may be due to an inhibition of uptake of auxotrophic nutrients, but the better growth of the auxotrophic strains at low ethanol concentrations is more difficult to explain. Prototrophic strains must have a balance between uptake and biosynthesis of the nutrients that can be made available only by uptake in auxotrophic strains. Both systems, transport and biosynthesis, require energy, and the presence of ethanol may affect the balance toward a more unfavorable situation in the prototrophic strains. BY4741 and BY4742 are derivatives of S288c. These strains are in principle isogenic except for the auxotrophic markers. BY4741 and BY4742 have the same auxotrophies for his3Δ1, leu2Δ0, and ura3Δ0, while they vary for met15Δ0 and lys2Δ0, respectively. Hence, the small differences in ethanol tolerance between BY4741 and BY4742 may be due to this difference in auxotrophies or to background mutations spontaneously generated during cultivation of the strains.
FIG 5.
Growth of the laboratory BY4741/2 strains and their prototrophic counterparts (S288c MATa and S288c MATα) on media containing moderate to high concentrations of ethanol. (A) A 10-fold dilution range of each strain was spotted onto YP medium containing 10%, 12%, 14%, or 16% ethanol and subsequently grown for different periods of time, as indicated. (B) The 10-fold dilution range was also spotted onto YPD medium with 10% or 16% ethanol.
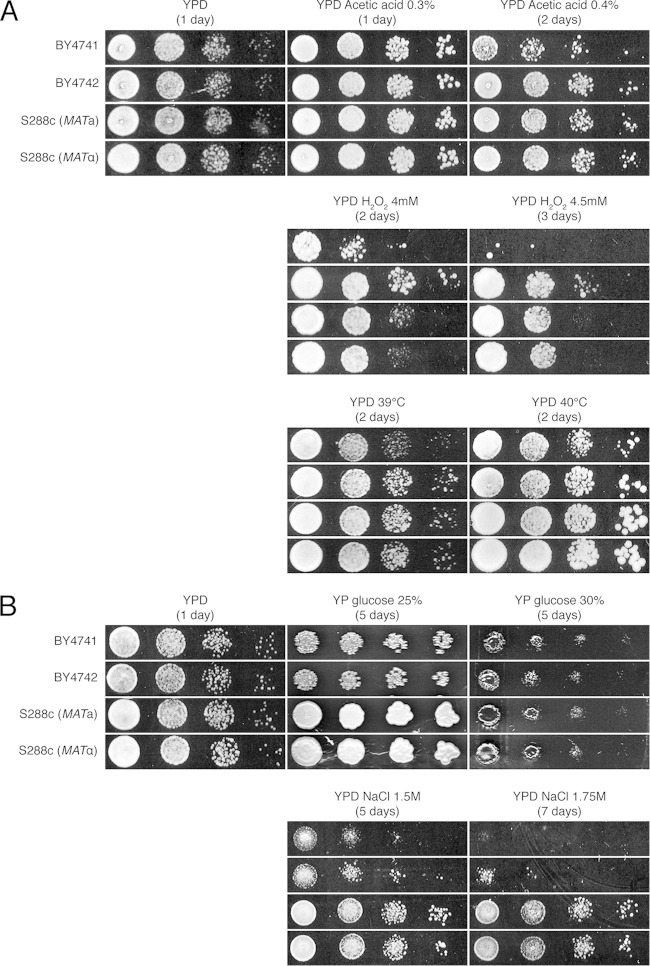
In addition, we determined tolerance to other stress factors on YPD medium: acetic acid at pH 5, hydrogen peroxide, high temperature, and osmotic stress. The doses that were applied are generally considered to cause high levels of stress, and under our conditions, they also caused a strong growth delay (Fig. 6). No significant difference between the auxotrophic and prototrophic strains was observed for acetic acid, hydrogen peroxide, and high-temperature stresses (under all conditions, BY4741 was somewhat more sensitive than BY4742 and both S288c strains) (Fig. 6A). On the other hand, under conditions of osmotic stress caused by either high glucose or NaCl concentrations, the prototrophic strains grew significantly better than the auxotrophic strains (Fig. 6B). Thus, under conditions of osmotic stress, a difference in growth between the auxotrophic and prototrophic strains similar to that under conditions of high levels of ethanol stress could be observed.
FIG 6.
Growth of the laboratory BY4741/2 strains and their prototrophic counterparts (S288c MATa and S288c MATα) on YPD medium while being subjected to different stress factors. (A) A 10-fold dilution range was spotted onto YPD medium containing 0.3% or 0.4% (vol/vol) acetic acid at pH 5 and onto YPD medium containing 4.0 mM or 4.5 mM hydrogen peroxide. The cells were incubated for 2 days at 30°C. To investigate heat tolerance, the strains were spotted onto YPD medium and grown at 39°C and 40°C for 2 days. (B) The strains (10-fold dilution range) were also spotted onto YPD medium with 25% and 30% glucose and onto YPD medium containing 1.50 M or 1.75 M NaCl and incubated at 30°C for the indicated periods of time.
Exposure to very high ethanol concentrations inhibits nutrient uptake.
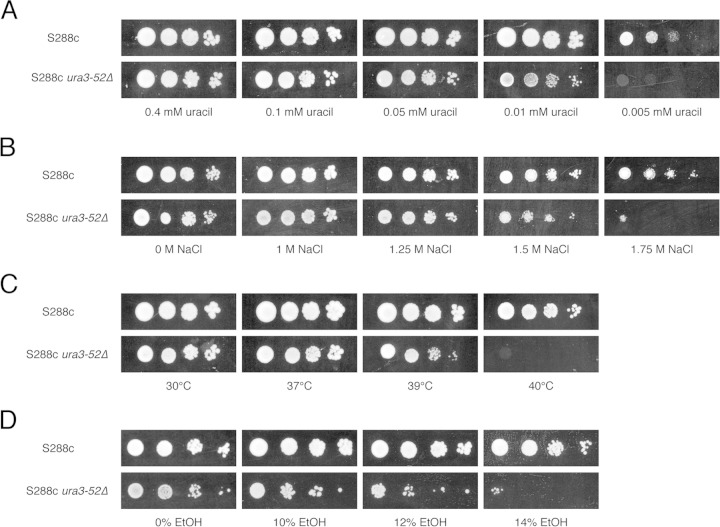
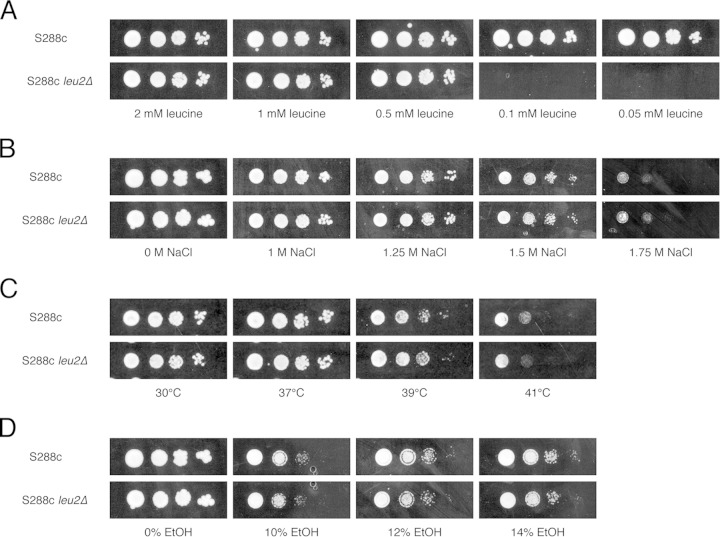
In order to determine whether the growth inhibition of auxotrophic strains under stress is due to an impairment of nutrient uptake, we compared the growth of the prototrophic S288c strain with that of the S288c ura3-52 and S288c leu2Δ strains on minimal medium with limiting amounts of uracil and leucine, respectively, combined with different stress factors (high ethanol and NaCl concentrations and also high temperature, as a control). First, we determined the minimal amounts of uracil and leucine that are required for growth of the respective auxotrophic strains by performing a spot assay on minimal medium with different concentrations of the auxotrophic nutrient. The S288c ura3-52 strain was not able to grow on minimal medium containing <0.05 mM uracil (Fig. 7A), while the S288c leu2Δ strain was not able to grow on minimal medium containing <0.5 mM leucine (Fig. 8A). In the case of the S288c ura3-52 strain, growth on minimal medium containing 0.05 mM uracil was inhibited by high osmotic stress (Fig. 7B), high temperature (Fig. 7C), and high ethanol concentrations (Fig. 7D). Hence, thermotolerance was also reduced, as opposed to what was previously observed for rich medium (Fig. 6A). On the other hand, no effect on growth of the S288c leu2Δ strain on minimal medium containing 0.5 mM leucine was observed under all the conditions mentioned above (Fig. 8B to D).
FIG 7.
Growth of prototrophic strain S288c and the auxotrophic S288c ura3-52 strain on media containing different stress factors. (A) The minimal concentration of uracil that is required to allow growth of the S288c ura3-52 strain on minimal medium was determined by growing the strain on CSM-URA with different concentrations of uracil. Strain S288c was included in the assay as a control. The cells were then incubated for 2 days at 30°C. (B to D) In order to investigate the effects of different stress factors on the growth of the strains under conditions of limited availability of uracil, a 10-fold dilution range of both strains was spotted onto minimal medium containing 0.05 mM uracil in combination with different concentrations of NaCl and incubated for 6 days at 30°C (B), grown with heat stress and incubated at the indicated temperatures for 3 days (C), and grown in the presence of different concentrations of ethanol and incubated for 7 days at 30°C (D).
FIG 8.
Growth of prototrophic strain S288c and the auxotrophic S288c leu2Δ strain on medium containing different stress factors. (A) In order to determine the minimal concentration of leucine that is required to allow growth of the S288c leu2Δ strain on minimal medium, a 10-fold dilution range of the strain was spotted onto CSM-LEU with different concentrations of leucine. The cells were incubated for 2 days at 30°C. (B to D) To investigate the effects of different stress factors on growth of the strains under conditions of limited availability of leucine, the dilutions were also spotted onto minimal medium containing 0.5 mM leucine in combination with different concentrations of NaCl and incubated for 5 days at 30°C (B), grown with heat stress and incubated at the indicated temperatures for 2 days (C), and grown in the presence of different concentrations of ethanol and incubated for 7 days at 30°C (D).
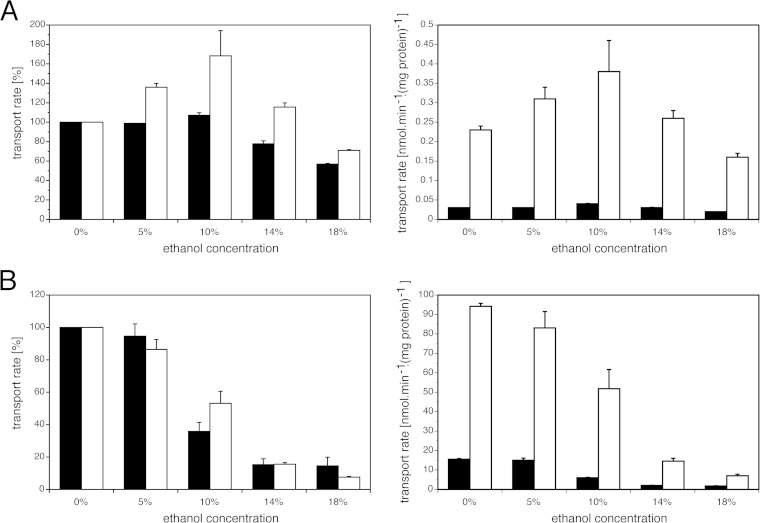
Furthermore, we investigated the effect of different ethanol concentrations on the rate of uptake of uracil and leucine. We found that the rate of transport of uracil is less sensitive to high ethanol concentrations than the rate of transport of leucine. The rate of uptake of radiolabeled uracil decreased ∼40% in the presence of 18% ethanol, while the rate of uptake of leucine decreased ∼80% under the same conditions (Fig. 9A and B). On the other hand, the absolute rate of transport of uracil was ∼500-fold lower than that of leucine, already under unstressed conditions, which can explain why strains with uracil auxotrophy are more sensitive to high ethanol stress than strains with leucine auxotrophy. The reason for the increase in the rate of uracil uptake with moderate ethanol concentrations is not clear. It could be due to a direct effect of ethanol on the protein structure of the uracil transporter or to an indirect effect of ethanol on the structure of the phospholipid membrane.
FIG 9.
Uptake of uracil and leucine in prototrophic and auxotrophic strains in the presence of different ethanol concentrations. Shown are rates of uptake of 0.05 mM uracil by the S288c (black bars) and S288c ura3-52 (white bars) strains (A) and rates of uptake of 2.5 mM leucine by the S288c (black bars) and S288c leu2Δ (white bars) strains (B) in the presence of different concentrations of ethanol. The transport activity measured in the presence of different concentrations of ethanol is expressed as a percentage of transport in the absence of ethanol (left) and in nanomoles per minute per milligram of protein (right). Error bars represent the standard deviations from three biological replicates.
DISCUSSION
Analyses of polygenic traits have been very cumbersome, even with a powerful genetic model organism such as S. cerevisiae. The interdependent and complex interactions and relationships between the genetic elements involved in such traits make it in practice almost impossible to identify and study these elements separately. Methodologies that allow the mapping of different genetic elements involved in polygenic traits simultaneously and with reasonable efficiency have only recently become available (36, 37). The most widely used molecular markers in yeast are natural variations detected as hybridization differences on high-density oligonucleotide arrays (38) or determined by whole-genome sequencing (39, 40). Several polygenic traits have been investigated by using these approaches, which led to the identification of loci, genes, and single nucleotide polymorphisms involved in high-temperature growth (39, 40), sporulation efficiency (41), mRNA expression profiles (42), acetic acid production (43), resistance to chemical agents (44), high ethanol tolerance (34), maximal ethanol accumulation (45), low glycerol production (46, 47), and thermotolerance (48).
We have constructed laboratory strains of S. cerevisiae with artificial genetic markers inserted in the genome at predetermined, presumably neutral positions at a relative distance of ∼20 kb. These equidistant markers provide a constant and universal genetic map and can be scored in an easy and reliable manner. We have applied this technology to a crucial polygenic trait of yeast, i.e., very high ethanol tolerance, which is of both fundamental and applied interest. Although the presence of the artificial markers in a laboratory strain may at first seem to be a disadvantage, for the analysis of very high ethanol tolerance, it provided us with two parent strains that are clearly different regarding the phenotype of interest. Ethanol tolerance in practice can be quite variable, and for genetic mapping purposes, it is essential that phenotypic differences between segregants are due to genetic differences and not to experimental variability. In this regard, we noticed that the variability between the measurements of ethanol tolerance was smaller when only ethanol was provided as a carbon source than when ethanol was added together with glucose. All segregants selected for very high ethanol tolerance were phenotyped multiple times in order to ensure that the phenotype was truly inherent to the strain and thus had a genetic basis.
Our results show that the artificial markers provide a convenient approach for linkage analysis. In one of the first mapping experiments, a significant linkage with markers 150 to 157 on chromosome V was identified. Calculation of the P values revealed that at least 11 segregants with very high ethanol tolerance are required to obtain a statistically significant conclusion about the presence of linkage between the trait and specific markers. This is the case when a locus is indispensable for the phenotype and therefore the markers in this locus are absent or present in all segregants. The lower the contribution of the locus to the trait, the more segregants are required to demonstrate significance of the linkage. Our results also revealed that with a relative distance of 20 kb, several consecutive markers showed linkage with very high ethanol tolerance. Therefore, any genetic element with an important contribution to the trait of interest will be revealed with high reliability by this technology.
The observation that the URA3 gene is linked to the very-high-ethanol-tolerance phenotype of the VR1 bioethanol production strain raised several interesting questions. First, how could an auxotrophic mutation affect ethanol tolerance? Since the plasma membrane is generally considered an important target of ethanol toxicity (1), the very high ethanol concentrations that have been used in our assays may have inhibited nutrient uptake across the plasma membrane. Therefore, an auxotrophic strain would be more compromised for growth in the presence of very high ethanol concentrations than a prototrophic strain. Inhibition of nutrient uptake in the presence of ethanol has been described previously, for instance, in the case of amino acid uptake by the proton symporter Gap1 (49). Since ethanol increases the passive influx of protons across the plasma membrane (50), it can be expected that symport is more sensitive to ethanol inhibition than facilitated diffusion. This indeed seems to be reflected in several experimental findings. In the case of acetic acid, it has been shown that uptake by symport is inhibited by ethanol, whereas uptake of the compound by passive influx is actually enhanced by ethanol (51). In addition, in S. cerevisiae, where glucose transport occurs by facilitated diffusion, transport is not inhibited by ethanol concentrations of up to 13% (52), while in mammalian systems, where glucose uptake occurs by Na+ symport, inhibition by ethanol has been shown (53, 54). These results point to the electrochemical ion gradient over the plasma membrane as a major target for high ethanol stress. On the other hand, we cannot exclude that very high ethanol concentrations also have a direct destabilizing effect on the structure and/or functionality of the transporters in the plasma membrane.
A second question raised by our results was whether other auxotrophic mutations also reduce very high ethanol tolerance. If this were the case, it would support the hypothesis that compromised nutrient uptake is responsible for the impaired growth of auxotrophic strains in the presence of very high concentrations of ethanol. Our results showed that other auxotrophic genes are also linked to the ethanol tolerance phenotype, and this was further supported by our observation that prototrophic S288c strains are clearly more ethanol tolerant than their auxotrophic counterparts. The linkage with very high ethanol tolerance was much stronger for the pyrimidine biosynthesis gene URA3 than for the amino acid biosynthesis genes HIS3, LEU2, MET15, and LYS2. A possible explanation might have been that uracil uptake in yeast is more sensitive to very high ethanol levels than the uptake of amino acids. We confirmed that the rates of uptake of uracil and leucine are reduced with increasing ethanol concentrations. However, the rate of uptake of uracil was actually less affected by high ethanol concentrations than that of leucine, but the absolute activity of uracil uptake was 500-fold lower than that of leucine (in both the ura3Δ0 mutant and the wild-type strain and in the absence of ethanol). This may explain why ura3 mutants are more sensitive to ethanol than leu2 mutants; their uracil uptake activity may easily drop to levels that limit growth. Uracil uptake is carried out by only one carrier, Fur4, and since natural yeast strains are prototrophic for uracil, uptake of uracil by Fur4 might be an auxiliary, dispensable function (55). This would explain the very low uptake rate. Amino acid uptake, on the other hand, seems to be much more important for yeast. It is actually essential in media without ammonium and also limits growth in media with low levels of ammonium. All the protein amino acids can be taken up by multiple amino acid permeases (56). The difference in absolute uptake rates and the negative effect of high levels of ethanol on uracil and leucine uptake may not be the only factors responsible for the higher ethanol sensitivity of ura3 mutants than of leu2 mutants. One or more enzymes involved in uracil incorporation into metabolism may be more sensitive to high ethanol concentrations than the enzymes involved in leucine utilization, which would further compromise the limiting availability of uracil for cell multiplication. We propose that very high ethanol concentrations generally compromise the uptake of nutrients required to fulfill auxotrophic requirements. The ethanol tolerance of VR1-5B, however, was still appreciably higher than that of the prototrophic laboratory strains, indicating that other genetic elements in the VR1-5B background contribute to its very high ethanol tolerance (34).
In previous research on the molecular basis of ethanol tolerance, a linkage with auxotrophic mutations has never been noticed. However, this research was performed solely with laboratory yeast strains displaying moderate ethanol tolerance, and therefore, only moderate ethanol concentrations were added to the medium (17, 18, 20, 22–24). We have now confirmed that auxotrophies are not disadvantageous for growth in the presence of low to moderate ethanol concentrations and that with specific ethanol concentrations, they even improve growth. It is likely that low to moderate ethanol concentrations have little or no effect on nutrient uptake across the plasma membrane and that, therefore, auxotrophic requirements, which strongly depend on nutrient uptake, have always remained unnoticed as an important factor in ethanol tolerance. Another QTL mapping experiment in a cross between the sake strain CBS 1585 and the laboratory BY strain identified URA3 as a causative gene for maximal ethanol accumulation capacity, but it had only a very weak contribution to ethanol tolerance for cell proliferation in the presence of 18 to 20% ethanol (45). This result may at first seem surprising, but it is not contradictory to our findings because a different genetic background was used in this study.
Several previous reports have called for caution when using auxotrophic parent strains in genetic experiments (15, 16, 57–59). We have now shown that in rich medium, the auxotrophic mutations do not seem to interfere with tolerance to any of the other stress factors tested, i.e., acetic acid at pH 5, hydrogen peroxide, and high temperature, except for osmotic stress caused by either high glucose or NaCl concentrations. Hence, osmotic stress may have a similar inhibiting effect on the uptake of auxotrophic nutrients as that of high ethanol stress in rich medium. However, when the ura3-52Δ strain was grown in minimal medium with a uracil concentration close to that limiting growth, not only high ethanol and high salt concentrations but also high temperature inhibited growth. This suggests that stress factors inhibit the growth of auxotrophic strains depending on the growth conditions and that they do this by reducing the uptake of the auxotrophic nutrient to a level below what is required for a regular growth rate under the specific growth conditions used.
Cohen and Engelberg (60) reported previously that some commonly used S. cerevisiae laboratory strains, including BY4741, do not grow on synthetic complete medium. By using a 2μ library, they showed that this growth defect could be complemented by enhancing leucine transport (BAP2 and TAT1) or by restoring the ability to synthesize leucine (LEU2). In another study, a mutated dominant allele of the transcription factor Spt15 that conferred increased ethanol/glucose tolerance to BY4741 was identified (61). In an attempt to use this mutated allele to improve the ethanol tolerance of various other yeast strains, it was discovered that the improved growth was manifested only in medium with limiting amounts of leucine, irrespective of the presence of ethanol (62). Thus, the supposedly improved ethanol tolerance of BY4741 with a mutated SPT15 allele could be attributed to an improved uptake of leucine in medium with a limiting concentration of this nutrient. In these two cases, the growth defect of the leucine-auxotrophic BY4741 strain was observed only in media with limiting amounts of leucine, suggesting that sufficient leucine in the medium can overcome poor leucine transport in BY4741. In our study, on the other hand, we found a significant linkage between LEU2 and growth on rich complex medium (YP) containing 16% ethanol. We therefore assume that very high ethanol concentrations have such a detrimental effect on plasma membrane transport that they prevent sufficient leucine uptake to support a high growth rate, irrespective of the amount of leucine present in the medium. Probably, the same conclusion can be drawn for the other auxotrophies in the BY4741/2 background.
Very high ethanol tolerance of yeast is a property of great industrial relevance. A general issue in industrial yeast fermentations, whether for the production of alcoholic beverages or for the production of bioethanol, is the conspicuous drop in the fermentation rate once the ethanol concentration surpasses 8 to 12%. A lower fermentation rate decreases the productivity of the ethanol factory, increases the chance of contamination, and especially affects the conversion of the residual sugars. In starch-based fermentations (e.g., with beer wort or with corn starch hydrolysate), the last sugar taken up is maltotriose, which is transported by the low-capacity symporter Agt1 (63, 64). Also, in wine fermentations, incomplete usage of residual fructose is a common cause of sluggish and stuck fermentations (65). Our results suggest that compromised plasma membrane transport may be a common cause of reduced or failing sugar consumption in the second phase of industrial yeast fermentations. Although the auxotrophic mutations clearly point to inadequate nutrient uptake as a possible cause of ethanol sensitivity, they are not directly useful for further improvement of ethanol tolerance in industrial yeast strains. On the other hand, our results suggest that enhanced expression of transporters for micronutrients that limit growth or for residual fermentation substrates may improve substrate-to-ethanol conversion and thus increase the yield of the fermentations. In addition, causative genes in the other mapped loci have to be identified. This has recently been accomplished, which resulted in the identification of several previously unidentified genes that are involved in tolerance to very high ethanol concentrations (34).
Supplementary Material
ACKNOWLEDGMENTS
We thank Henrique Amorim and Mario Lucio Lopes (Fermentec, Piracicaba, Brazil) for the gift of the VR1 strain and for fruitful discussions. We thank Catherina Coun, Kjell Lenaers, Ilse Palmans, Paul Vandecruys, Evy Vanderheyden, and Willy Verheyden for technical help with the experiments and Nico Vangoethem for preparation of the figures.
This work has been supported by a predoctoral fellowship to S.S. and a postdoctoral fellowship to K.S. from the Agency for Innovation by Science and Technology (IWT-Flanders) and by SBO grants (IWT 50148 and IWT 90043) from IWT-Flanders, the EC 7th Framework program (NEMO project), the IOF-Knowledge platform (IKP/10/002 ZKC 1836), and BOF-Program financing (project NATAR) to J.M.T.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00053-15.
REFERENCES
- 1.Casey GP, Ingledew WM. 1986. Ethanol tolerance in yeasts. Crit Rev Microbiol 13:219–280. doi: 10.3109/10408418609108739. [DOI] [PubMed] [Google Scholar]
- 2.Garay-Arroyo A, Covarrubias AA, Clark I, Nino I, Gosset G, Martinez A. 2004. Response to different environmental stress conditions of industrial and laboratory Saccharomyces cerevisiae strains. Appl Microbiol Biotechnol 63:734–741. doi: 10.1007/s00253-003-1414-4. [DOI] [PubMed] [Google Scholar]
- 3.Basso LC, de Amorim HV, de Oliveira AJ, Lopes ML. 2008. Yeast selection for fuel ethanol production in Brazil. FEMS Yeast Res 8:1155–1163. doi: 10.1111/j.1567-1364.2008.00428.x. [DOI] [PubMed] [Google Scholar]
- 4.Kodama K. 1993. Sake-brewing yeast, p 129–168. In Rose AH, Harrison JS (ed), The yeasts, vol 3 Academic Press, London, United Kingdom. [Google Scholar]
- 5.Beney L, Gervais P. 2001. Influence of the fluidity of the membrane on the response of microorganisms to environmental stresses. Appl Microbiol Biotechnol 57:34–42. doi: 10.1007/s002530100754. [DOI] [PubMed] [Google Scholar]
- 6.Weber FJ, de Bont JA. 1996. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim Biophys Acta 1286:225–245. doi: 10.1016/S0304-4157(96)00010-X. [DOI] [PubMed] [Google Scholar]
- 7.Alexandre H, Rousseaux I, Charpentier C. 1994. Relationship between ethanol tolerance, lipid composition and plasma membrane fluidity in Saccharomyces cerevisiae and Kloeckera apiculata. FEMS Microbiol Lett 124:17–22. doi: 10.1111/j.1574-6968.1994.tb07255.x. [DOI] [PubMed] [Google Scholar]
- 8.Thomas DS, Hossack JA, Rose AH. 1978. Plasma-membrane lipid composition and ethanol tolerance in Saccharomyces cerevisiae. Arch Microbiol 117:239–245. doi: 10.1007/BF00738541. [DOI] [PubMed] [Google Scholar]
- 9.You KM, Rosenfield CL, Knipple DC. 2003. Ethanol tolerance in the yeast Saccharomyces cerevisiae is dependent on cellular oleic acid content. Appl Environ Microbiol 69:1499–1503. doi: 10.1128/AEM.69.3.1499-1503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swan TM, Watson K. 1998. Stress tolerance in a yeast sterol auxotroph: role of ergosterol, heat shock proteins and trehalose. FEMS Microbiol Lett 169:191–197. doi: 10.1111/j.1574-6968.1998.tb13317.x. [DOI] [PubMed] [Google Scholar]
- 11.Takagi H, Takaoka M, Kawaguchi A, Kubo Y. 2005. Effect of l-proline on sake brewing and ethanol stress in Saccharomyces cerevisiae. Appl Environ Microbiol 71:8656–8662. doi: 10.1128/AEM.71.12.8656-8662.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudolph AS, Crowe JH. 1985. Membrane stabilization during freezing: the role of two natural cryoprotectants, trehalose and proline. Cryobiology 22:367–377. doi: 10.1016/0011-2240(85)90184-1. [DOI] [PubMed] [Google Scholar]
- 13.Samuel D, Kumar TK, Ganesh G, Jayaraman G, Yang PW, Chang MM, Trivedi VD, Wang SL, Hwang KC, Chang DK, Yu C. 2000. Proline inhibits aggregation during protein refolding. Protein Sci 9:344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirasawa T, Yoshikawa K, Nakakura Y, Nagahisa K, Furusawa C, Katakura Y, Shimizu H, Shioya S. 2007. Identification of target genes conferring ethanol stress tolerance to Saccharomyces cerevisiae based on DNA microarray data analysis. J Biotechnol 131:34–44. doi: 10.1016/j.jbiotec.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Pronk JT. 2002. Auxotrophic yeast strains in fundamental and applied research. Appl Environ Microbiol 68:2095–2100. doi: 10.1128/AEM.68.5.2095-2100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cakar ZP, Sauer U, Bailey JE. 1999. Metabolic engineering of yeast: the perils of auxotrophic hosts. Biotechnol Lett 21:611–616. doi: 10.1023/A:1005576004215. [DOI] [Google Scholar]
- 17.Auesukaree C, Damnernsawad A, Kruatrachue M, Pokethitiyook P, Boonchird C, Kaneko Y, Harashima S. 2009. Genome-wide identification of genes involved in tolerance to various environmental stresses in Saccharomyces cerevisiae. J Appl Genet 50:301–310. doi: 10.1007/BF03195688. [DOI] [PubMed] [Google Scholar]
- 18.Fujita K, Matsuyama A, Kobayashi Y, Iwahashi H. 2006. The genome-wide screening of yeast deletion mutants to identify the genes required for tolerance to ethanol and other alcohols. FEMS Yeast Res 6:744–750. doi: 10.1111/j.1567-1364.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- 19.Ingram LO, Buttke TM. 1984. Effects of alcohols on micro-organisms. Adv Microb Physiol 25:253–300. [DOI] [PubMed] [Google Scholar]
- 20.Kubota S, Takeo I, Kume K, Kanai M, Shitamukai A, Mizunuma M, Miyakawa T, Shimoi H, Iefuji H, Hirata D. 2004. Effect of ethanol on cell growth of budding yeast: genes that are important for cell growth in the presence of ethanol. Biosci Biotechnol Biochem 68:968–972. doi: 10.1271/bbb.68.968. [DOI] [PubMed] [Google Scholar]
- 21.Mishra P. 1993. Tolerance of fungi to ethanol, p 189–208. In Jennings DH. (ed), Stress tolerance of fungi. Marcel Dekker, New York, NY. [Google Scholar]
- 22.Teixeira MC, Raposo LR, Mira NP, Lourenco AB, Sa-Correia I. 2009. Genome-wide identification of Saccharomyces cerevisiae genes required for maximal tolerance to ethanol. Appl Environ Microbiol 75:5761–5772. doi: 10.1128/AEM.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Voorst F, Houghton-Larsen J, Jonson L, Kielland-Brandt MC, Brandt A. 2006. Genome-wide identification of genes required for growth of Saccharomyces cerevisiae under ethanol stress. Yeast 23:351–359. doi: 10.1002/yea.1359. [DOI] [PubMed] [Google Scholar]
- 24.Yoshikawa K, Tanaka T, Furusawa C, Nagahisa K, Hirasawa T, Shimizu H. 2009. Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res 9:32–44. doi: 10.1111/j.1567-1364.2008.00456.x. [DOI] [PubMed] [Google Scholar]
- 25.Stanley D, Bandara A, Fraser S, Chambers PJ, Stanley GA. 2010. The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J Appl Microbiol 109:13–24. doi: 10.1111/j.1365-2672.2009.04657.x. [DOI] [PubMed] [Google Scholar]
- 26.Wallace-Salinas V, Signori L, Li YY, Ask M, Bettiga M, Porro D, Thevelein JM, Branduardi P, Foulquie-Moreno MR, Gorwa-Grauslund M. 2014. Re-assessment of YAP1 and MCR1 contributions to inhibitor tolerance in robust engineered Saccharomyces cerevisiae fermenting undetoxified lignocellulosic hydrolysate. AMB Express 4:56. doi: 10.1186/s13568-014-0056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 28.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132. doi:. [DOI] [PubMed] [Google Scholar]
- 29.Erdeniz N, Mortensen UH, Rothstein R. 1997. Cloning-free PCR-based allele replacement methods. Genome Res 7:1174–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherman F, Hicks J. 1991. Micromanipulation and dissection of asci. Methods Enzymol 194:21–37. doi: 10.1016/0076-6879(91)94005-W. [DOI] [PubMed] [Google Scholar]
- 31.Huxley C, Green ED, Dunham I. 1990. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet 6:236. doi: 10.1016/0168-9525(90)90190-H. [DOI] [PubMed] [Google Scholar]
- 32.Benjamini Y, Yekutieli D. 2005. Quantitative trait loci analysis using the false discovery rate. Genetics 171:783–790. doi: 10.1534/genetics.104.036699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 34.Swinnen S, Schaerlaekens K, Pais T, Claesen J, Hubmann G, Yang Y, Demeke M, Foulquie-Moreno MR, Goovaerts A, Souvereyns K, Clement L, Dumortier F, Thevelein JM. 2012. Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res 22:975–984. doi: 10.1101/gr.131698.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu XH, Wang MH, Tan T, Li JR, Yang H, Leach L, Zhang RM, Luo ZW. 2007. Genetic dissection of ethanol tolerance in the budding yeast Saccharomyces cerevisiae. Genetics 175:1479–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swinnen S, Thevelein JM, Nevoigt E. 2012. Genetic mapping of quantitative phenotypic traits in Saccharomyces cerevisiae. FEMS Yeast Res 12:215–227. doi: 10.1111/j.1567-1364.2011.00777.x. [DOI] [PubMed] [Google Scholar]
- 37.Liti G, Louis EJ. 2012. Advances in quantitative trait analysis in yeast. PLoS Genet 8:e1002912. doi: 10.1371/journal.pgen.1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winzeler EA, Richards DR, Conway AR, Goldstein AL, Kalman S, McCullough MJ, McCusker JH, Stevens DA, Wodicka L, Lockhart DJ, Davis RW. 1998. Direct allelic variation scanning of the yeast genome. Science 281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- 39.Parts L, Cubillos FA, Warringer J, Jain K, Salinas F, Bumpstead SJ, Molin M, Zia A, Simpson JT, Quail MA, Moses A, Louis EJ, Durbin R, Liti G. 2011. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res 21:1131–1138. doi: 10.1101/gr.116731.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH, Davis RW. 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 41.Deutschbauer AM, Davis RW. 2005. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat Genet 37:1333–1340. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- 42.Brem RB, Yvert G, Clinton R, Kruglyak L. 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 43.Marullo P, Aigle M, Bely M, Masneuf-Pomarede I, Durrens P, Dubourdieu D, Yvert G. 2007. Single QTL mapping and nucleotide-level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res 7:941–952. doi: 10.1111/j.1567-1364.2007.00252.x. [DOI] [PubMed] [Google Scholar]
- 44.Ehrenreich IM, Torabi N, Jia Y, Kent J, Martis S, Shapiro JA, Gresham D, Caudy AA, Kruglyak L. 2010. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464:1039–1042. doi: 10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pais TM, Foulquie-Moreno MR, Hubmann G, Duitama J, Swinnen S, Goovaerts A, Yang Y, Dumortier F, Thevelein JM. 2013. Comparative polygenic analysis of maximal ethanol accumulation capacity and tolerance to high ethanol levels of cell proliferation in yeast. PLoS Genet 9:e1003548. doi: 10.1371/journal.pgen.1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hubmann G, Foulquie-Moreno MR, Nevoigt E, Duitama J, Meurens N, Pais TM, Mathe L, Saerens S, Nguyen HT, Swinnen S, Verstrepen KJ, Concilio L, de Troostembergh JC, Thevelein JM. 2013. Quantitative trait analysis of yeast biodiversity yields novel gene tools for metabolic engineering. Metab Eng 17:68–81. doi: 10.1016/j.ymben.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Hubmann G, Mathe L, Foulquie-Moreno MR, Duitama J, Nevoigt E, Thevelein JM. 2013. Identification of multiple interacting alleles conferring low glycerol and high ethanol yield in Saccharomyces cerevisiae ethanolic fermentation. Biotechnol Biofuels 6:87. doi: 10.1186/1754-6834-6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Foulquie-Moreno MR, Clement L, Erdei E, Tanghe A, Schaerlaekens K, Dumortier F, Thevelein JM. 2013. QTL analysis of high thermotolerance with superior and downgraded parental yeast strains reveals new minor QTLs and converges on novel causative alleles involved in RNA processing. PLoS Genet 9:e1003693. doi: 10.1371/journal.pgen.1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferreras JM, Iglesias R, Girbes T. 1989. Effect of the chronic ethanol action on the activity of the general amino-acid permease from Saccharomyces cerevisiae var. ellipsoideus. Biochim Biophys Acta 979:375–377. doi: 10.1016/0005-2736(89)90260-5. [DOI] [PubMed] [Google Scholar]
- 50.Leao C, Van Uden N. 1984. Effects of ethanol and other alkanols on passive proton influx in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 774:43–48. doi: 10.1016/0005-2736(84)90272-4. [DOI] [PubMed] [Google Scholar]
- 51.Casal M, Cardoso H, Leao C. 1998. Effects of ethanol and other alkanols on transport of acetic acid in Saccharomyces cerevisiae. Appl Environ Microbiol 64:665–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sousa MJ, Mota M, Leao C. 1995. Effects of ethanol and acetic acid on the transport of malic acid and glucose in the yeast Schizosaccharomyces pombe: implications in wine deacidification. FEMS Microbiol Lett 126:197–202. doi: 10.1111/j.1574-6968.1995.tb07416.x. [DOI] [PubMed] [Google Scholar]
- 53.Dinda PK, Beck IT. 1981. Ethanol-induced inhibition of glucose transport across the isolated brush-border membrane of hamster jejunum. Dig Dis Sci 26:23–32. doi: 10.1007/BF01307972. [DOI] [PubMed] [Google Scholar]
- 54.Tillotson LG, Carter EA, Inui KI, Isselbacher KJ. 1981. Inhibition of Na+-stimulated glucose transport function and perturbation of intestinal microvillus membrane vesicles by ethanol and acetaldehyde. Arch Biochem Biophys 207:360–370. doi: 10.1016/0003-9861(81)90043-6. [DOI] [PubMed] [Google Scholar]
- 55.Jund R, Weber E, Chevallier MR. 1988. Primary structure of the uracil transport protein of Saccharomyces cerevisiae. Eur J Biochem 171:417–424. doi: 10.1111/j.1432-1033.1988.tb13806.x. [DOI] [PubMed] [Google Scholar]
- 56.Regenberg B, During-Olsen L, Kielland-Brandt MC, Holmberg S. 1999. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr Genet 36:317–328. doi: 10.1007/s002940050506. [DOI] [PubMed] [Google Scholar]
- 57.Basso TO, Dario MG, Tonso A, Stambuk BU, Gombert AK. 2010. Insufficient uracil supply in fully aerobic chemostat cultures of Saccharomyces cerevisiae leads to respiro-fermentative metabolism and double nutrient-limitation. Biotechnol Lett 32:973–977. doi: 10.1007/s10529-010-0248-2. [DOI] [PubMed] [Google Scholar]
- 58.Gonzalez A, Larroy C, Biosca JA, Arino J. 2008. Use of the TRP1 auxotrophic marker for gene disruption and phenotypic analysis in yeast: a note of warning. FEMS Yeast Res 8:2–5. doi: 10.1111/j.1567-1364.2007.00315.x. [DOI] [PubMed] [Google Scholar]
- 59.Ding J, Bierma J, Smith MR, Poliner E, Wolfe C, Hadduck AN, Zara S, Jirikovic M, van Zee K, Penner MH, Patton-Vogt J, Bakalinsky AT. 2013. Acetic acid inhibits nutrient uptake in Saccharomyces cerevisiae: auxotrophy confounds the use of yeast deletion libraries for strain improvement. Appl Microbiol Biotechnol 97:7405–7416. doi: 10.1007/s00253-013-5071-y. [DOI] [PubMed] [Google Scholar]
- 60.Cohen R, Engelberg D. 2007. Commonly used Saccharomyces cerevisiae strains (e.g. BY4741, W303) are growth sensitive on synthetic complete medium due to poor leucine uptake. FEMS Microbiol Lett 273:239–243. doi: 10.1111/j.1574-6968.2007.00798.x. [DOI] [PubMed] [Google Scholar]
- 61.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. 2006. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 62.Baerends RJ, Qiu JL, Rasmussen S, Nielsen HB, Brandt A. 2009. Impaired uptake and/or utilization of leucine by Saccharomyces cerevisiae is suppressed by the SPT15-300 allele of the TATA-binding protein gene. Appl Environ Microbiol 75:6055–6061. doi: 10.1128/AEM.00989-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alves SL Jr, Herberts RA, Hollatz C, Trichez D, Miletti LC, de Araujo PS, Stambuk BU. 2008. Molecular analysis of maltotriose active transport and fermentation by Saccharomyces cerevisiae reveals a determinant role for the AGT1 permease. Appl Environ Microbiol 74:1494–1501. doi: 10.1128/AEM.02570-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stambuk BU, da Silva MA, Panek AD, de Araujo PS. 1999. Active alpha-glucoside transport in Saccharomyces cerevisiae. FEMS Microbiol Lett 170:105–110. doi: 10.1111/j.1574-6968.1999.tb13361.x. [DOI] [PubMed] [Google Scholar]
- 65.Gafner J, Schütz M. 1996. Impact of glucose-fructose-ratio on stuck fermentations: practical experiences to restart stuck fermentations. Vitic Enol Sci 51:214–218. [Google Scholar]
- 66.Grundmann O, Mosch HU, Braus GH. 2001. Repression of GCN4 mRNA translation by nitrogen starvation in Saccharomyces cerevisiae. J Biol Chem 276:25661–25671. doi: 10.1074/jbc.M101068200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.